Abstract
Genome-wide association studies have been an important approach used to localize trait loci, with primary focus on common variants. The multiple rare variant–common disease hypothesis may explain the missing heritability remaining after accounting for identified common variants. Advances of sequencing technologies with their decreasing costs, coupled with methodological advances in the context of association studies in large samples, now make the study of rare variants at a genome-wide scale feasible. The resurgence of family-based association designs because of their advantage in studying rare variants has also stimulated more methods development, mainly based on linear mixed models (LMMs). Other tests such as score tests can have advantages over the LMMs, but to date have mainly been proposed for single-marker association tests. In this article, we extend several score tests to the multiple variant association framework. We evaluate and compare their statistical performances relative with the LMM. Moreover, we show that three tests can be cast as the difference between marker allele frequencies (AFs) estimated in each of the group of affected and unaffected subjects. We show that these tests are flexible, as they can be based on related, unrelated or both related and unrelated subjects. They also make feasible an increasingly common design that only sequences a subset of affected subjects (related or unrelated) and uses for comparison publicly available AFs estimated in a group of healthy subjects. Finally, we show the great impact of linkage disequilibrium on the performance of all these tests.
Keywords: linear mixed models, score test, pedigree data, next-generation sequencing, association test, shared controls
Introduction
The identification of DNA variants involved in complex trait etiologies is a major focus of current genetic studies. For the past decade, genome-wide association studies (GWAS) have been the predominant approach used to localize trait loci [1, 2]. The majority of variants discovered by GWAS are common, and explain only a small fraction of heritability of traits studied so far, with rare variants likely to explain part of the remaining heritability [3–5]. Advances of sequencing techniques and decreasing costs have recently made the study of rare variants affordable. Ideally, one wants to have a large sample of sequenced affected and unaffected subjects. Owing to budget constraints, one of the designs implemented is a design that sequences affected subjects only, selected from pedigrees with GWAS data [6, 7]. The aim of such a design, which does not sequence healthy controls, is to increase the chance of capturing rare causal mutations present in affected subjects. The joint impact of the underlying effects of rare variants is, of course, a key factor in determining appropriate designs and analytical approaches that may lead to identification of genes relevant to complex traits.
Association analyses in GWAS can be carried out on unrelated subjects, related subjects or both related and unrelated subjects [8–11]. However, association analysis is known to be underpowered to detect rare variants with low-penetrant alleles [4]. In contrast, family-based linkage analysis has led to discovery of many rare variants with highly penetrant genotypes for many traits [12, 13]. These variants with high effects may be identified by association approaches, with much smaller sample sizes required in the case of regional follow-up of a strong linkage analysis result than when a GWAS is used for localization. Currently, population-based association designs that use data of unrelated subjects are more common than family-based association designs because of the difficulty and expensive need to sample family members [14]. Nevertheless, family-based association designs may sometimes be more useful for detecting rare variants because of the extra information provided by segregation of alleles within pedigrees, yielding enrichment of copies of specific rare variants, and also possibly the presence of rare variants with higher penetrant forms [14].
In association studies based on data from unrelated subjects, statistical testing approaches are generally based on linear regression for continuous traits and logistic regression for binary traits (case-control designs). For family data in which subjects are related, with therefore correlated observations, these regression models in their simplest forms are not valid because they are based on the assumption that observations are independent (or uncorrelated) [15–18]. In this case, mixed models, which account for correlation (relatedness) among subjects, can be used. The relatedness between subjects can be measured through the kinship matrix, which can be obtained from the defined pedigree structure, or the analogous genetic relationship matrix or GRM, which is estimated from genetic marker data [e.g. single-nucleotide polymorphism (SNP) data] [10, 19, 20]. In this article, except where noted, we do not differentiate between the kinship matrix and the GRM. Linear mixed models (LMMs) can be easily used for continuous traits and are widely implemented in multiple software packages [9, 10, 21–23]. However for binary traits, the most appropriate models, called generalized linear mixed models (GLMMs), may be computationally infeasible when the pedigree structure is too large for the use of exact-likelihood calculation [24]. In this case, performing LMM analysis on binary traits is still straightforward, as it provides similar P-values with the GLMM model (e.g. as implemented in the GMMAT package [23]) when the effect sizes are small/moderate and when there is no population stratification and no substantial ascertainment bias [10, 25–27]. Note that in a design that sequences cases only, both the LMM and GMMAT models do not work to test association, as this requires the genotype data for both cases and controls.
Other testing approaches that are computationally feasible have been proposed for binary traits (e.g. ) [20, 24, 28, 29] and are attractive for two reasons. First, they do not require the kinship matrix to be positive semidefinite as required in LMMs. This is important because the kinship matrix estimated from genotype data (the GRM) may not be always positive semidefinite, depending on the estimation method used [10, 20]. Second, we show later that these tests make feasible a design that only sequences a subset of affected subjects (related or unrelated) and uses for comparison publicly available allele frequencies (AFs) estimated in a group of healthy subjects, such as the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (http://evs.gs.washington.edu/EVS/). This is important because many study designs focus on only sequencing cases [6, 7]. Therefore, the proposed tests allow the comparison between the sequenced cases and any set of already sequenced public controls that provide at least the AFs.
These tests, which we call here ‘Allele Frequency Comparison (AFC) tests’, are based on the difference of SNP AFs in the cases (affected) versus controls (unaffected). These tests also use the kinship matrix to account for family relationships between subjects. AFC tests were originally proposed for single SNP tests (e.g. ). Currently, the focus is more on multiple rare variants that are assumed to contribute to complex trait etiologies [4, 30, 31]. These rare variants are accessible through the high-throughput sequencing that is now available. As such, it is important to extend the AFC tests to the multiple rare variant testing framework. This can be done through a burden test by using the difference of sums of AFs between cases and controls for the SNPs in a region of interest and also by accounting for correlation between SNPs in the region, which reflects linkage disequilibrium (LD). A recent study [32] proposed an extension of the test to the multiple rare variant testing framework, and also proposed a sequence kernel association test (SKAT) for binary trait in family data.
In this article, we show that the and SKAT tests can be written as a function of AFs in affected and unaffected subjects. In doing so, we open the door to testing for association using AFs estimated in controls instead of directly having their genotypes for comparison. This also makes association testing more flexible because in additional scenarios, we do not even need the genotypes in cases, as the AFs in both groups of cases and controls suffice to test the association.
We also propose an extension of the statistical test for multiple rare variant testing for binary traits. In the same context (association between multiple rare variants and binary traits), we evaluate the statistical properties of and SKAT in several simulation scenarios in both population- and family-based designs. We compare the statistical performance of these tests along with the LMM used as a baseline. We implemented these three tests ( and SKAT) in R. The source code and examples are available at http://faculty.washington.edu/wijsman/progdists/AFC/.
Materials and methods
Notation
First, we introduce the following notation:
are the frequencies of the minor allele in affected, unaffected and all subjects, respectively, at marker j (j = 1,…,m). The minor allele is defined based on the frequency estimated in the set of all subjects.
are the number of affected, unaffected and all subjects, respectively.
is the matrix of twice the kinship coefficients between pairs of subjects, where is the kinship between subject i and j. is a phenotype vector of length N with ith entry () 1 if subject i is affected and 0 if subject i is unaffected, and is a vector of length N with all entries equal to 1.
is the genotype matrix of m SNPs in the region of interest, where is the genotype of the ith subject at the jth SNP coded as 0, 1 or 2 copies of the minor allele. Similarly, we define the weighted genotype matrix as: , where , and is a prespecified weight for SNP j. If all m weights are equal to one, then is equal to. Several weighting approaches have been proposed in the literature to up-weight/down-weight rare and common variants. For example, proposed by [33] and proposed by [34]. To simplify without loss of generality, subjects are grouped by affectation status where affected subjects are stored before unaffected subjects in all matrices and vectors ().
Test statistic
We write a general form of the test statistic as follows:
| (1) |
where
Note that the variance and covariance estimates become more accurate and more representative of the population variance and population-covariance (estimated in the whole population) as sample size increases.
We considered two forms of the matrix:
, and
.
The use of and leads to the and tests, respectively, which are described below.
Wcorrected test
This test is equivalent to the test proposed initially for the single-marker test framework. It is constructed using the first form of V (i.e. V1) and that leads to the following test statistic:
After some algebra, it is straightforward to show that can be written as a function of the minor allele frequencies (MAFs) estimated in affected subjects () and all subjects ():
| (2) |
This test is the same as that proposed by [32] and equivalent to the test proposed for the single SNP test in [24]. To perform this test, we need (1) the AFs in both affected and unaffected subjects, (2) the number of affected and unaffected subjects (Na and Nu), (3) the kinship matrix and (4) the correlation between SNPs. The correlation between SNPs can be estimated in the whole data (affected and unaffected), or in the absence of unaffected subject genotypes, in the group of affected subjects or from an external database with genetic data from the same population under study. To choose an appropriate external database, one can use multidimensional reduction techniques such as principal component analysis (PCA). This can be done by applying PCA on the combined data of the study affected subjects and the external database subjects, and further determine if both types of subjects cluster together and therefore belong to the same population. If only AFs are available, one can determine whether two populations are genetically similar by computing a metric based on the distance between the two populations’ AFs (Fpc) as proposed by [35]. If a variant is present in the affected subject group and absent in the external database, one might assume, especially if the database sample size (e.g. Nu) is relatively large, that this variant is monomorphic in this database, and thus, the MAF would be set to zero in the unaffected subjects. If all subjects are unrelated, the kinship matrix will be the identity matrix, and therefore, the test statistic can be simply written as follows:
| (3) |
WQLS test
The second form of V (i.e. V2) leads to the test statistic proposed in [24] and discussed in [36]:
If all subjects are unrelated, it is easy to see that is equal to , and therefore, is equivalent to . In this case, AFs, correlations between SNPs and the number of affected and unaffected subjects are enough to perform the test (there is no need to have direct access to genotypes). If one group of subjects (affected or unaffected) does not have accessible genotypes, then for the test to be performed, this group of subjects can only contain unrelated subjects. However, if this group contains unknown cryptic relatedness between subjects, which is impossible to determine, as genotypes are not available, the allele frequency estimates may be inaccurate, with the rare variants being the most affected. In addition, the correlation between SNPs and the matrix (through the misspecification of kinship coefficients) may be affected as well. In this case, will most certainly lead to an increase of type 1 error. In a common design in human genetics where only one has access to the affected genotypes (case-only design) whether the affected subjects are related, we can use unaffected subjects that are unrelated and have available AFs to conduct . We show in Supplementary Note 1 that we can replace the unaffected unrelated subjects’ genotypes by the AFs estimated in the unaffected subject group (). The final form of this equation can be written as follows:
The denominator of depends on the relationship between subjects, which is represented by the kinship matrix, and the correlation between SNPs. This test is similar to the Wald test for a GLMM.
SKAT test
The SKAT test was extended to binary traits in family data by [32]. The score test statistic is written as:
As shown in Supplementary Note 2, we demonstrate that the score statistic Q can be written as a function of (1) the AFs in the group of affected and the group of all subjects, (2) SNP weights and (3) the number of affected and unaffected subjects (the number of unaffected subjects should be known to calculate ). The score statistic is: As shown in [32], the distribution of Q can be approximated by a Satterwaite approximation [33, 37] for the distribution of quadratic forms by estimating the and , where , with being a vector of m entries: , is the correlation matrix between SNPs, the symbols * and denote a scalar multiplication and element-wise matrix multiplication, respectively, and finally, with the term ‘sum’ being the sum of all matrix elements. To calculate the matrix , we do not need to have access to the subjects’ genotypes if we have the correlation matrix estimated from an external source such as the pedigree structure. Nonetheless, the correlation matrix can be estimated via a GRM using genotypes of affected subjects, if this is the only available group of subjects. We estimate the distribution of Q by a scaled distribution as follows: where and . Alternatively, the Q statistic is asymptotically distributed as a mixture of independent distributions:, where is the ith eigenvalue of (Davies approximation [33, 38]). If all subjects are unrelated, it is easy to show that this SKAT model is equivalent to the original proposed SKAT model for data with unrelated subjects [33] (Supplementary Note 3).
Simulation
To evaluate the type 1 error and power of these tests, we simulated sequence data on a collection of extended pedigrees (1000 subjects from 35 pedigrees, with mean size of 30 subjects, and size ranging from 20 to 53 subjects) extracted from an Alzheimer’s disease cohort under study at the University of Washington. We simulated three sets of sequence data on these pedigrees to obtain 3000 familial subjects (in 105 pedigrees). Also, we simulated sequence data on 3000 unrelated subjects.
We used the same simulation strategy used in a previous study [39] to obtain 100 semi-realistic sequence data sets (each contains 3000 familial and 3000 unrelated subjects) that mimic the LD structure and the allele frequency spectrum in European descent subjects of the 1000 Genomes Project sequence data [40]. Briefly, we simulated 20 000 haplotypes for a region of 10 Mb. In the center of this region, we considered a gene of 30 kilo base (kb) length (313 SNPs or sequence variants). We considered two LD pattern scenarios: LowLD and HighLD. From the pool of 20 000 haplotypes, we started by randomly selecting haplotypes, without replacement, for the unrelated founders and unrelated subjects. Then in pedigrees, we dropped the haplotypes from founders down through the —three to five generations in pedigrees using a recombination rate of 1% [per centiMorgan (cM)] per meiosis under the assumption that 1 cM is equal to 1000 kb. We repeated this last step 100 times for each LD pattern to obtain 100 sequence data sets, each consisting of 6000 subjects. Finally, for each simulated sequence data set, we simulated 100 quantitative traits under the null hypothesis of no association and under the alternative hypothesis of association for both pedigree and unrelated data.
Type 1 error and power simulation
We fitted the model , where follows a multivariate normal distribution, with for pedigree data and for unrelated data, where is the identity matrix, and is the heritability. We fixed the genetic variance because of polygenic effects as and the residual variance as, so the heritability is. This model has genetic variation defined by the pedigree structure but not by the gene. We assigned affected and unaffected subjects based on their quantitative trait values: the highest and lowest trait values were used to assign affected and unaffected subjects, respectively. We fixed the number of unaffected subjects to 1000 unrelated subjects, and we varied the number of affected subjects between 100, 200 and 500, related or unrelated subjects, so we had the scenarios described in the following table:
| #Affected subjects | Related? | |
|---|---|---|
| Rel100 | 100 | Yes |
| Rel200 | 200 | Yes |
| Rel500 | 500 | Yes |
| Unrel100 | 100 | No |
| Unrel200 | 200 | No |
| Unrel500 | 500 | No |
Finally, we varied the number of non-associated SNPs (U) included in the association model as 10, 20 and 30.
Under the alternative hypothesis of association, we simulated 100 quantitative traits for each simulated sequence data set by fitting the model: , where is the vector of effect sizes of the A associated SNPs, is the matrix of their genotypes and is defined above. The effect sizes of the associated SNPs were determined by the function , where is the MAF of the associated SNP j estimated in the generated sequence data, and is the total additive variance of all associated SNPs combined. In our simulation, we set the total additive variance to 1%, so 99% of the genetic variance is not explained by SNPs in the gene. We tuned the value of the total additive variance in such a way that the power of the association test is neither very low nor very high to maximize potential power differences among the methods. We randomly selected associated SNPs from the list of rare SNPs (MAF < 0.05) in the gene, and we varied their numbers (A) as 10, 20 and 30. We also added nonassociated SNPs into the model. We set the number of nonassociated SNPs (U) as equal to the number of associated SNPs (U = A). Finally, we considered two scenarios of SNP effect directions: mix0, in which all associated SNPs have the same direction of effect, and mix30, in which 30% of associated SNPs have a positive direction of effect and the remaining 70% have a negative direction of effect. To assign affected and unaffected subjects, we followed the same strategy explained in the type 1 error section above.
We estimated both type 1 error and power at threshold as the proportion of replicates for which the P-value of the association test is lower than .
Results
For all scenarios, we considered (Rel100, Rel200, Rel500, Unrel100, Unrel200 and Unrel500), we performed five association tests: Wcorrected, WQLS, LMM, generalized linear model (GLM) and SKAT. For these tests, we repeated the analysis three times, estimating the correlation between SNPs in three data sets: (1) the data set of combined affected and unaffected subjects (Cor1), (2) the data set of affected subjects plus 300 randomly selected unaffected subjects (Cor2; 300 unrelated subjects, similar to the 1000 Genomes Project Data) and (3) the data set of affected subjects only (Cor3). For the SKAT analysis, we compared both the Satterwaite and Davies approximations in the Cor1 scenario. Then, we used the Davies approximation to compare SKAT with all other tests. All type 1 error and power results were estimated for an.
Type 1 error results
Type1 error results are shown in Table 1 and Supplementary Table S1. For ‘Rel’ scenarios (Table 1) and both LowLD and HighLD patterns, we observed an inflation of the type 1 error for the logistic regression (GLM), which is expected because the GLM does not account for relatedness between subjects. The LMM, which accounts for relatedness between subjects, had controlled type 1 errors. For Wcorrected and WQLS, the type 1 errors were well controlled in Cor1 and Cor2. However, in Cor3, in which the correlation among SNPs was estimated in small data sets (affected subject data set), the type 1 errors tend to be slightly inflated. This is explained by the underestimation of correlations between SNPs in small sample sizes (Supplementary Figure S1; the term cs in the equations is smaller in Cor3 compared with Cor1, and thus, both statistics will be bigger in Cor3). In the same correlation scenario (Cor3), the type 1 error rates were closer to what we expect (0.01) for bigger sample sizes (e.g. Na = 500). For the SKAT analysis in the Cor1 scenario, our results showed that the Satterwaite approximation led to a greater type 1 error rate than did the Davies approximation. Therefore, we decided to focus on results from the Davies approximation. For all correlation scenarios under the LowLD pattern, the type 1 error rates of SKAT tend to be slightly higher than expected for smaller sample size (Na = 100) than for large sample size. Interestingly, the type 1 errors are generally well controlled under the HighLD pattern. This likely is because of the more accurate estimation of correlations between SNPs under the HighLD pattern, even in small data sets. Considering the 99% confidence interval of the type 1 error rate on 10 000 simulations [i.e. (0.008, 0.013)], we observed that the inflated type 1 error rates that are not within this interval are in the scenario of small sample sizes and LowLD. This suggests that SKAT would achieve appropriate type 1 error rates for relatively large data sets. Nonetheless, for Wcorrected and WQLS, the type 1 errors were similar in both LD patterns. This means that SKAT is more affected by LD compared with the other tests used. For the ‘Unrel’ scenarios (Supplementary Table S1), the type 1 errors were more controlled than those in the ‘Rel’ scenarios. All tests had type 1 error rates close to what we expect, except for Wcorrected and WQLS in the Cor3 scenario and SKAT with the Satterwaite approximation.
Table 1.
Related affected, 1000 unrelated unaffected, SNPs included in the test have MAF < 0.05
| LD | Na | nSNP |
Wcorrected |
WQLS |
LMM | GLM | SKAT (Satterwaite) | SKAT (Davies) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cor1 | Cor2 | Cor3 | Cor1 | Cor2 | Cor3 | – | – | Cor1 | Cor1 | Cor2 | Cor3 | |||
| Low | 100 | 20 | 0.009 | 0.010 | 0.016 | 0.009 | 0.009 | 0.016 | 0.009 | 0.016 | 0.022 | 0.017 | 0.013 | 0.016 |
| 40 | 0.010 | 0.010 | 0.020 | 0.010 | 0.011 | 0.020 | 0.010 | 0.017 | 0.026 | 0.019 | 0.014 | 0.018 | ||
| 60 | 0.010 | 0.009 | 0.021 | 0.009 | 0.009 | 0.021 | 0.010 | 0.015 | 0.027 | 0.020 | 0.012 | 0.018 | ||
| 200 | 20 | 0.011 | 0.011 | 0.014 | 0.010 | 0.010 | 0.014 | 0.011 | 0.025 | 0.018 | 0.014 | 0.013 | 0.013 | |
| 40 | 0.011 | 0.010 | 0.016 | 0.009 | 0.010 | 0.014 | 0.012 | 0.023 | 0.020 | 0.014 | 0.012 | 0.014 | ||
| 60 | 0.010 | 0.010 | 0.017 | 0.009 | 0.010 | 0.016 | 0.010 | 0.024 | 0.023 | 0.015 | 0.011 | 0.012 | ||
| 500 | 20 | 0.010 | 0.010 | 0.013 | 0.011 | 0.011 | 0.013 | 0.011 | 0.040 | 0.016 | 0.012 | 0.011 | 0.012 | |
| 40 | 0.010 | 0.011 | 0.013 | 0.008 | 0.009 | 0.011 | 0.010 | 0.040 | 0.019 | 0.013 | 0.011 | 0.012 | ||
| 60 | 0.012 | 0.011 | 0.013 | 0.009 | 0.010 | 0.012 | 0.011 | 0.041 | 0.019 | 0.011 | 0.010 | 0.010 | ||
| High | 100 | 20 | 0.010 | 0.010 | 0.018 | 0.011 | 0.010 | 0.018 | 0.010 | 0.017 | 0.019 | 0.015 | 0.013 | 0.015 |
| 40 | 0.008 | 0.007 | 0.021 | 0.008 | 0.008 | 0.021 | 0.008 | 0.014 | 0.019 | 0.013 | 0.011 | 0.013 | ||
| 60 | 0.008 | 0.009 | 0.025 | 0.009 | 0.009 | 0.025 | 0.009 | 0.015 | 0.021 | 0.015 | 0.011 | 0.014 | ||
| 200 | 20 | 0.009 | 0.009 | 0.014 | 0.009 | 0.009 | 0.013 | 0.009 | 0.023 | 0.015 | 0.011 | 0.010 | 0.011 | |
| 40 | 0.009 | 0.009 | 0.015 | 0.009 | 0.009 | 0.015 | 0.009 | 0.022 | 0.019 | 0.012 | 0.010 | 0.011 | ||
| 60 | 0.008 | 0.009 | 0.018 | 0.008 | 0.010 | 0.018 | 0.008 | 0.023 | 0.019 | 0.012 | 0.011 | 0.011 | ||
| 500 | 20 | 0.011 | 0.011 | 0.014 | 0.010 | 0.010 | 0.012 | 0.011 | 0.043 | 0.015 | 0.012 | 0.013 | 0.013 | |
| 40 | 0.009 | 0.010 | 0.012 | 0.008 | 0.008 | 0.011 | 0.010 | 0.043 | 0.016 | 0.011 | 0.011 | 0.011 | ||
| 60 | 0.010 | 0.011 | 0.013 | 0.009 | 0.009 | 0.013 | 0.010 | 0.039 | 0.016 | 0.010 | 0.010 | 0.010 | ||
Note: nSNP = number of SNPs included in the model; Na = number of affected subjects; Cor1: correlation in affected and unaffected subjects combined; Cor2: correlation in affected subjects plus 300 randomly selected unaffected subjects; Cor3: correlation in affected subjects only.
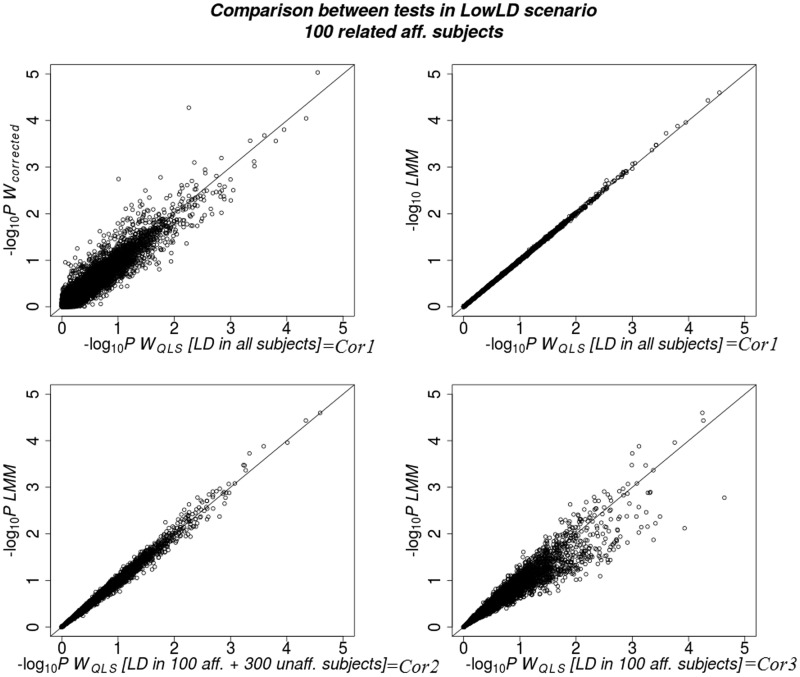
For the ‘Rel’ scenario, the WQLS and LMM tests have similar P-values under Cor1, where the correlations between SNPs were estimated in the same group of data by the two tests (Figure 1, top right). For Cor2, the P-values are still similar but less correlated than in Cor1 scenario (Figure 1, bottom left). For Cor3, WQLS and LMM become substantially different with WQLS yielding smaller P-values and thus more false positives. In addition, WQLS shows high similarity with Wcorrected (Figure 1, top left) but less than with LMM. As expected and shown in the equations in the ‘Materials and methods’ section, the P-values of WQLS are identical to those of Wcorrected in the ‘Unrel’ scenarios (results not shown). Moreover, WQLSP-values are more similar to LMM P-values than to the logistic regression (GLM) P-values. These results are shown in Supplementary Figure S2. In addition, we show the corresponding QQ plots, which represent a different way to evaluate the statistical distributions in Supplementary Figures S3 and S4. All QQ plots show that the distribution of test statistics is well behaved except for SKAT. These figures are shown for the 100 affected subjects, 20 SNPs included in the test and LowLD scenario. The other scenarios show the same general results (results not shown).
Figure 1.
Comparison between tests’ P-values (represented as −log10 P) in the 100 affected related subjects, LowLD scenario and nonassociated SNPs (20 SNPs) included in the test. Top left: WQLS versus Wcorrected in Cor1 scenario; top right: WQLS versus LMM in Cor1 scenario; bottom left: WQLS versus LMM in Cor2; bottom right: WQLS versus LMM in Cor3.
Power results
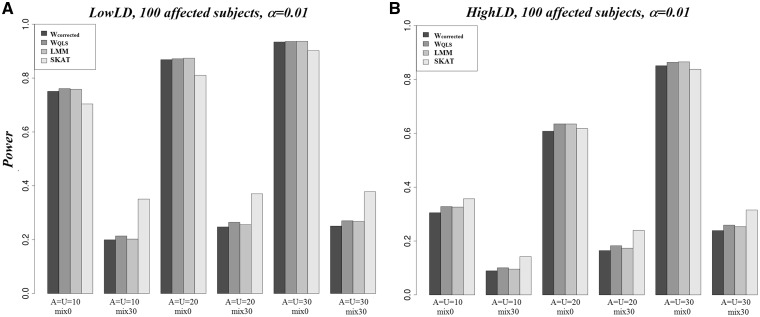
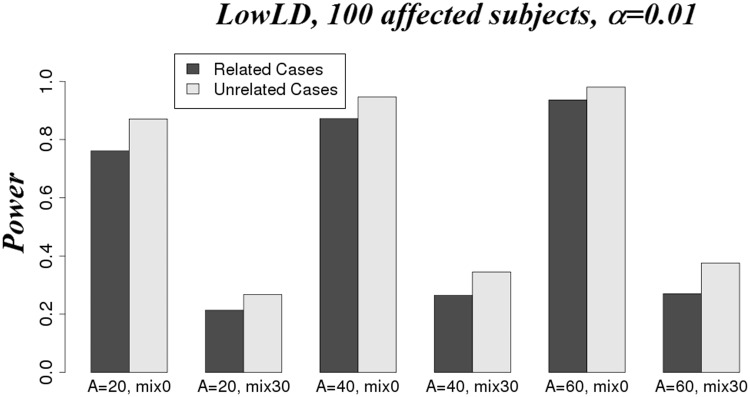
We compared the power of Wcorrected, WQLS, LMM and SKAT (Davies approximation) for related and unrelated affected scenarios. For the ‘Rel’ scenarios, the first three tests have similar power rates for all scenarios we considered (Figure 2). Nonetheless, Wcorrected tends to be slightly less powerful, and it becomes more apparent in the HighLD scenario. These three tests are more powerful than SKAT for the scenarios with the same effect direction of associated SNPs. Not surprisingly, SKAT performs better when 30% of associated SNPs are protective and the remaining 70% of associated SNPs are risk variants. Under the LowLD pattern, the power of the four tests is greater than that under the HighLD pattern (Figure 2A versus B). This result is congruent with what was observed in [32], which reported that the power of these tests increases when the LD decreases. This observation might be explained by the fact that with high LD between SNPs, the term cs in the denominator of the Equations (2) and (3) is greater than that estimated when the LD is low between SNPs (Supplementary Figure S1). Therefore, the statistics tend to be smaller and so does the power. In other words, the accumulation of evidence for association across variants that are correlated tells us less than across variants that are uncorrelated. The same trends were observed for ‘Unrel’ scenarios (results not shown). Our results show greater power for unrelated affected subjects compared with related affected subjects (Figure 3). This is because of our simulation strategy in which we chose the cases based on their quantitative trait values, and we did not simulate the trait in a way that favors the selection of cases from same pedigrees. Therefore, the enrichment of rare variants in familial cases is minimal. In this case, case-control studies in unrelated data are likely to be more powerful than in pedigree data for the same sample size.
Figure 2.
Power of Wcorrected, WQLS, LMM and SKAT in the scenario of 100 related affected subjects under: (A) LowLD, (B) HighLD patterns. Power was estimated using α = 0.01. Mix0: all SNPs have positive direction of effects. Mix30: 30% of the SNPs have positive direction of effects. A: number of associated SNPs. U: number of nonassociated SNPs.
Figure 3.
Power of WQLS in the scenario of 100 affected (related: dark grey; unrelated: light grey) subjects under LowLD patterns. Power was estimated using α = 0.01.
Discussion
With the advances of sequencing technologies and decreasing associated costs, searching for genome-wide association between rare variants and complex traits is now feasible. These new molecular advances also require advances in statistical methodology to allow efficient analysis of the resulting data. Here, we have provided three flexible tests that allow flexible use of related and unrelated subjects, either observed genotypes or availability of AFs. This will accelerate and facilitate the data analysis and will omit several steps, while obtaining the same results.
The approach we have taken for association analysis of rare variants in family-data is based on a score testing framework. An alternative framework, based on the GLMM, would also be suitable, and has been implemented in many packages such as EMMAX, GMMAT and GMMAX [9, 10, 23]. In our article, we extended the three models behind the Wcorrected, WQLS and SKAT tests to the multiple rare variant context, especially the WQLS test. The two tests Wcorrected and WQLS are theoretically similar and provided similar statistical performance as shown in the ‘Results’ section. The main motivation of developing these tests was to extend them to the rare variant testing framework and also to cast them as the difference between AFs of SNPs estimated in each of the group of affected and unaffected subjects. This will make performing association tests easier/feasible in population- and family-based designs that depend on use of shared controls. We also show that three tests can be cast as the difference between AFs of SNPs estimated in each of the group of affected and unaffected subjects. We therefore refer to them as AFC tests. Note that SKAT appeared to be more sensitive to LD and small sample sizes than Wcorrected, WQLS and should be used with caution. For relatively large data sets, the type 1 error rates of SKAT will most likely be well controlled, regardless of the LD level. These AFC tests should facilitate sharing genetic data between research groups, as only summary statistics are needed to carry out the test: AFs of the SNPs or SNVs to be included, the data set sample size and the kinship matrix (if related subjects exist). Note that in the case of Wcorrected, the kinship matrix can be used to calculate one value [denominator of Equation (2) without cs], where this value will be used for each test. That means that if one does not want to share the kinship matrix, Wcorrected would still be feasible by only providing the cs values. Like all family-based association tests, AFC tests require the use of an accurate kinship matrix to avoid an increase of false-positive rates or a decrease of statistical power. In both related and unrelated subject data, the use of a GRM (plus the theoretical kinship matrix if available) is advisable to account for any possible cryptic relatedness between subjects. However, the method used to estimate empirical kinship coefficients should be the same in both affected and unaffected groups to avoid creating a mismatch between the two groups.
It is important to note that these AFC tests have some limitations. First, they may be not valid if genotypes depart from Hardy–Weinberg equilibrium (HWE) [41]. However in GWAS studies, SNPs that deviate strongly from HWE are typically removed during the quality control steps and therefore not considered for association. Second, AFC tests may give incorrect results if correlation between SNPs is not appropriately taken into account. For instance, the type 1 error rates of AFC tests were higher than expected when the correlation between SNPs was computed using small sample sizes. These rates were even similar to the GLM (which ignores the family relationships between subjects) type 1 error rates, especially for the smallest sample sizes. This highlights the importance of using adequate sample sizes for both cases and controls to achieve appropriate type 1 error rates. Ideally, estimation of the correlation should be done in a large sample that contains both cases and controls. Publicly available genotypes such as the 1000 Genomes Project data [40] can be used to estimate the correlation for samples that have ancestries similar to those in the database. The selection of matching ancestries can be done by checking how the affected subjects cluster with the 1000 Genomes Project’s different ancestries by performing PCA on the combined genotype data. If the correlation information does not exist or is inaccurate, these tests may be compromised. Finally, high-throughput sequencing results can be affected by batch effects present both in laboratory and analytical procedures. Results obtained by using these methods with reference to allele frequency information from shared controls will need to be verified by other means.
The AFC tests can obviously be applied when genotype data of cases and controls are available. In our study, we reformulated the equations of these tests to handle the design that only sequences cases and uses public allele frequency estimates. The general forms of the equations allow the use of data that contain unrelated subjects, related subjects or even both. We evaluated the statistical performance (power and type 1 error) of these tests along with an LMM for comparison, and showed that the results obtained by the WQLS test, in particular, are similar to those obtained by the LMM model, across a number of simulation scenarios. We did not yet deeply evaluate the AFC tests in real data. This awaits future work that requires extensive pedigree sequence data with known truths, extensive comparison between the AFC results using several available public databases, testing the different scenarios of estimating the correlation between SNPs and also the availability of AFs only in both affected and unaffected subjects. Such application also requires use of data generated by identical experimental protocols to avoid issues such as those described elsewhere [42].
Key Points
The multiple rare variant–common disease hypothesis may explain the missing heritability remaining after accounting for identified common variants.
Next-generation sequencing technologies allow the genotyping of rare variants.
Family-based association designs are important to study rare variants because of their enrichment in pedigrees, which has stimulated methods development, mainly based on LMMs.
We extend several score tests , we call AFC tests that can have advantages over the LMMs to the multiple variant association framework.
These tests make feasible an increasingly common design that only sequences a subset of affected subjects and uses for comparison publicly available AFs estimated in a group of healthy subjects.
Funding
Funded in part by US National Institutes of Health grants P50 AG005136, U01 AG049507 and R01 MH094293.
Biographies
Mohamad Saad, PhD, is a Scientist at QCRI. His area of expertise is Biostatistics and Statistical Genetics. His research work focuses on GWAS and imputation of missing genotypes in population- and family-based designs. This work started when he was at the University of Washington.
Ellen M Wijsman, PhD, is a Professor in Medicine and Biostatistics at the University of Washington. Her research interest includes statistical genetics, computational genetics, genetic epidemiology, population genetics and Alzheimer’s disease.
References
- 1. Hirschhorn JN, Lohmueller K, Byrne E. A comprehensive review of genetic association studies. Genet Med 2002;4:45–61. [DOI] [PubMed] [Google Scholar]
- 2. Evangelou E, Ioannidis JP.. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet 2013;14:379–89. [DOI] [PubMed] [Google Scholar]
- 3. Manolio TA, Collins FS, Cox NJ. Finding the missing heritability of complex diseases. Nature 2009;461:747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bansal V, Libiger O, Torkamani A. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet 2010;11:773–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet 2012;13:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saad M, Brkanac Z, Wijsman EM.. Family-based genome scan for age at onset of late-onset Alzheimer's disease in whole exome sequencing data. Genes Brain Behav 2015;14:607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu L, Schaid DJ, Sicotte H. Case-only exome sequencing and complex disease susceptibility gene discovery: study design considerations. J Med Genet 2015;52:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ott J, Kamatani Y, Lathrop M.. Family-based designs for genome-wide association studies. Nat Rev Genet 2011;12:465–74. [DOI] [PubMed] [Google Scholar]
- 9. Zhou X, Stephens M.. Genome-wide efficient mixed-model analysis for association studies. Nat Genet 2012;44:821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kang HM, Sul JH, Service SK. Variance component model to account for sample structure in genome-wide association studies. Nat Genet 2010;42:348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCarthy MI, Abecasis GR, Cardon LR. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–69. [DOI] [PubMed] [Google Scholar]
- 12. Jorde LB. Linkage disequilibrium and the search for complex disease genes. Genome Res 2000;10:1435–44. [DOI] [PubMed] [Google Scholar]
- 13. Botstein D, Risch N.. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet 2003;33:228–37. [DOI] [PubMed] [Google Scholar]
- 14. Wijsman EM. The role of large pedigrees in an era of high-throughput sequencing. Hum Genet 2012;131:1555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laird NM, Ware JH.. Random-effects models for longitudinal data. Biometrics 1982;38:963–74. [PubMed] [Google Scholar]
- 16. Boerwinkle E, Chakraborty R, Sing CF.. The use of measured genotype information in the analysis of quantitative phenotypes in man. Ann Hum Genet 1986;50:181–94. [DOI] [PubMed] [Google Scholar]
- 17. Abney M, Ober C, McPeek MS.. Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites. Am J Hum Genet 2002;70:920–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen WM, Abecasis GR.. Family-based association tests for genomewide association scans. Am J Hum Genet 2007;81:913–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amin N, van Duijn CM, Aulchenko YS.. A genomic background based method for association analysis in related individuals. PLoS One 2007;2:e1274.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi Y, Wijsman EM, Weir BS.. Case-control association testing in the presence of unknown relationships. Genet Epidemiol 2009;33:668–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aulchenko YS, de Koning DJ, Haley C.. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 2007;177:577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lippert C, Listgarten J, Liu Y. FAST linear mixed models for genome-wide association studies. Nat Methods 2011;8:833–5. [DOI] [PubMed] [Google Scholar]
- 23. Chen H, Wang C, Conomos MP. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet 2016;98:653–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bourgain C, Hoffjan S, Nicolae R. Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am J Hum Genet 2003;73:612–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sawcer S, Hellenthal G, Pirinen M. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011;476:214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Widmer C, Lippert C, Weissbrod O. Further improvements to linear mixed models for genome-wide association studies. Sci Rep 2014;4:6874.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Agresti A. Categorical Data Analysis. 2002. John Wiley and Sons, New York.
- 28. Thornton T, McPeek MS.. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am J Hum Genet 2010;86:172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jakobsdottir J, McPeek MS.. MASTOR: mixed-model association mapping of quantitative traits in samples with related individuals. Am J Hum Genet 2013;92:652–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee S, Abecasis GR, Boehnke M. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet 2014;95:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li B, Leal SM.. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet 2008;83:311–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schaid DJ, McDonnell SK, Sinnwell JP. Multiple genetic variant association testing by collapsing and kernel methods with pedigree or population structured data. Genet Epidemiol 2013;37:409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu MC, Lee S, Cai T. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 2011;89:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Madsen BE, Browning SR.. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet 2009;5:e1000384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen GB, Lee SH, Robinson MR. Across-cohort QC analyses of GWAS summary statistics from complex traits. Eur J Hum Genet 2016;25:137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thornton T, McPeek MS.. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet 2007;81:321–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwee LC, Liu D, Lin X. A powerful and flexible multilocus association test for quantitative traits. Am J Hum Genet 2008;82:386–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davies RB. Algorithm as 155: the distribution of a linear combination of χ2 random variables. Appl Stat 1980;29:323–33. [Google Scholar]
- 39. Saad M, Wijsman EM.. Combining family- and population-based imputation data for association analysis of rare and common variants in large pedigrees. Genet Epidemiol 2014;38:579–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abecasis GR, Altshuler D, Auton A. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sasieni PD. From genotypes to genes: doubling the sample size. Biometrics 1997;53:1253–61. [PubMed] [Google Scholar]
- 42. Derkach A, Chiang T, Gong J. Association analysis using next-generation sequence data from publicly available control groups: the robust variance score statistic. Bioinformatics 2014;30:2179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]