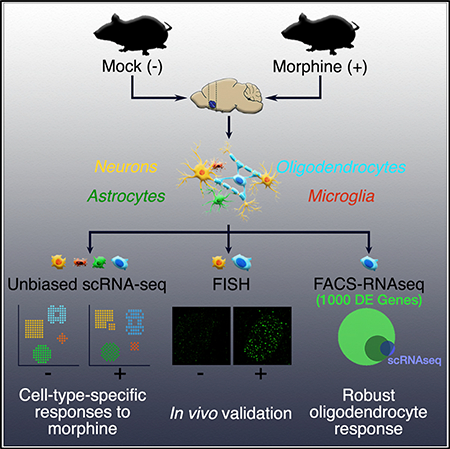
SUMMARY
Molecular and behavioral responses to opioids are thought to be primarily mediated by neurons, although there is accumulating evidence that other cell types play a prominent role in drug addiction. To investigate cell-type-specific opioid responses, we performed single-cell RNA sequencing (scRNA-seq) of the nucleus accumbens of mice following acute morphine treatment. Differential expression analysis uncovered unique morphine-dependent transcriptional responses by oligodendrocytes and astrocytes. We examined the expression of selected genes, including Cdkn1a and Sgk1, by FISH, confirming their induction by morphine in oligodendrocytes. Further analysis using RNA-seq of FACS-purified oligodendrocytes revealed a large cohort of morphine-regulated genes. The affected genes are enriched for roles in cellular pathways intimately linked to oligodendrocyte maturation and myelination, including the unfolded protein response. Altogether, our data illuminate the morphine-dependent transcriptional response by oligodendrocytes and offer mechanistic insights into myelination defects associated with opioid abuse.
In Brief
Avey et al. use single-cell RNA-seq to uncover cell-type-specific responses to morphine in the brains of mice, revealing a unique and robust response by oligodendrocytes, which they further validate by multiple approaches. These analyses offer insights into the molecular mechanisms underlying oligodendrocyte dysfunction in the context of opioid abuse
Graphical Abstract
INTRODUCTION
Drug addiction is a global health concern associated with considerable adverse impacts. Affected individuals experience severe physical and emotional hardship, which also takes a toll on their families and society (Bauer et al., 2015; Dart et al., 2015). The fiscal consequences in the United States alone have been estimated to exceed $600 billion a year (https://www.drugabuse.gov/publications/principles-drug-addiction-treatment-researchbased-guide-third-edition/preface). The current epidemic of prescription opioid abuse has contributed to an alarming rise in overdose death rates (Dart etal., 2015).Opioid-dependent individuals also exhibit increased comorbidity with other psychiatric disorders, including depression, as well as impaired white matter integrity and consequent cognitive deficits (Bora et al., 2012; Liu et al., 2008). Although environmental risk factors contribute to drug addiction, it is well-established that acute drug exposure induces gene expression changes in the CNS and that these changes affect susceptibility to addiction (Lüscher and Malenka, 2011).
All addictive drugs modulate the mesolimbic pathway of the brain by elevating dopamine levels in the nucleus accumbens (NAc), a region in the ventral striatum with a central role in processing motivation, reward, aversion, and reinforcement learning (Volkow and Morales, 2015). Because of this, researchers have aimed to elucidate the effects of various addictive drugs, including methamphetamine (Piechota et al., 2010; Zhu et al., 2015), cocaine (Albertson et al., 2006; Piechota et al., 2010), and opioids (Albertson et al., 2006; Korostynski et al., 2007; Piechota et al., 2010), on gene expression in the NAc. However, because the brain is an exceptionally heterogeneous tissue, previous experiments using bulk tissue preparations could not confidently ascribe gene expression changes to a certain cell type, thereby precluding the elucidation of cellular and molecular mechanisms of drug abuse. Advances in microfluidics and sequencing technology make it feasible to analyze thousands of single-cell transcriptomes in a single experiment. Various single-cell RNA sequencing (scRNA-seq) platforms have been developed (Cao et al., 2017; Islam et al., 2014; Klein et al., 2015; Zheng et al., 2017), enabling the characterization of dozens of molecularly distinct CNS cell types from multiple regions.
We profiled the gene expression of 23,276 individual cells from the NAc of mice using Drop-seq (Macosko et al., 2015). Differential expression analysis uncovered numerous cell-type-specific changes in gene expression due to acute morphine treatment. We observed a dramatic morphine-dependent transcriptional response in oligodendrocytes, which was an unorthodox result given that no study has described opioid-dependent gene expression changes unique to oligodendrocytes. We validated the morphine-dependent and cell-type-specific expression of a subset of genes by using fluorescence in situ hybridization (FISH). We also performed bulk RNA sequencing (RNA-seq) of oligodendrocytes isolated from mock- or morphine-treated mice, which confirmed a robust transcriptional response to morphine in these cells. We identified key molecular pathways modulated by morphine in oligodendrocytes that are likely to underlie the myelination defects associated with heroin abuse in human patients.
RESULTS
Unbiased scRNA-Seq Analysis Identified 18Distinct Cell Types in the NAc of Mice
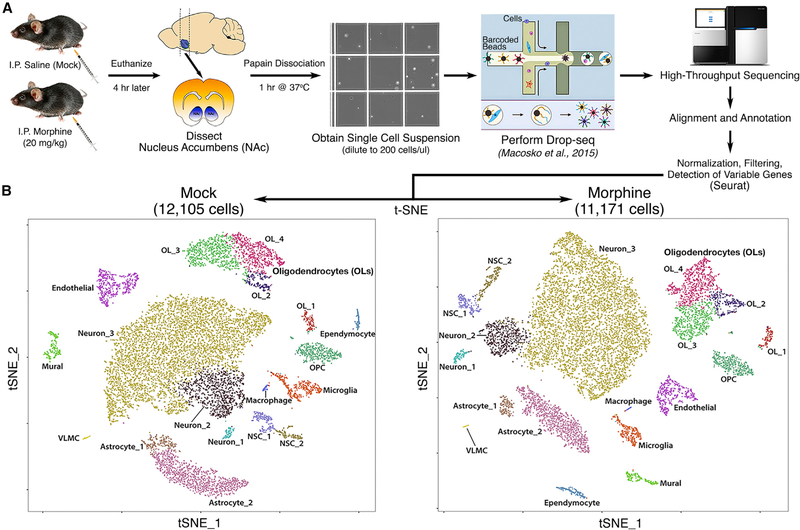
The molecular responses to morphine and other drugs of abuse by the neurons of the NAc have been well studied. However, the responses by specific neuronal or glial subpopulations have not been investigated in an unbiased and high-throughput manner. To address this, we treated mice with morphine or saline as a mock control and 4 hr later dissected the NAc. We chose 4 hr because it is the time at which the maximal number and magnitude of gene expression changes were observed in previous studies of the striatal transcriptional response to morphine (Korostynski et al., 2007; Piechota et al., 2010). A single-cell suspension was prepared from the dissociated tissue, and Drop-seq (Macosko et al., 2015) was used to obtain transcriptomes from 23,276 single cells (Figure 1A). We sequenced these cells to an average depth of ~67,000 reads/cell and detected 1,612 mean genes and 3,228 mean transcripts (Figure S1). Using principalcomponent analysis (PCA), graph-based clustering, and dimensionality reduction for 2D visualization through t-distributed stochastic neighbor embedding (t-SNE) (van der Maaten, 2008), we identified 18 distinct clusters (Figure 1B), all of which were present at nearly identical ratios in mock- and morphine-treated mice and at comparable ratios among biological replicates (Table S1). Next, we assigned a major CNS cell-type identity to each cluster (Figure 1B) by cross-referencing genes that were enriched in specific clusters with published gene expression datasets (Gokce et al., 2016; Zhang et al., 2014). Many clusterenriched genes have been previously identified as markers for specific cell types (Table S1), and the overall gene expression profiles of our samples correlate well with bulk RNA-seq (Hodes et al., 2015) or scRNA-seq (Gokce et al., 2016) analyses of the striatum (Figure S1). Clusters from mock- and morphine-treated mice exhibited striking similarities in cell number, number of genes detected per cell, and expression levels of clusterenriched genes (Figure S2), indicating that the overall cell-type composition of the NAc is unchanged by acute morphine treatment.
Figure 1. Unbiased Single-Cell RNA-Seq Analysis Identified 18 Distinct Cell Types in the Nucleus Accumbens of Mice.
(A) Flowchart showing an overview of the single-cell RNA-seq experimental design.
(B) Spectral t-SNE plots of all cells analyzed from mock-treated mice (12,105) or morphine-treated mice (11,171), colored by density clustering and annotated by cell-type identity. OPC, oligodendrocyte progenitor cell; NSC, neural stem cell; VLMC, vascular and leptomeningeal cell
Subclustering of Neurons Enables the Identification of a Subpopulation of Morphine-Activated MSNs
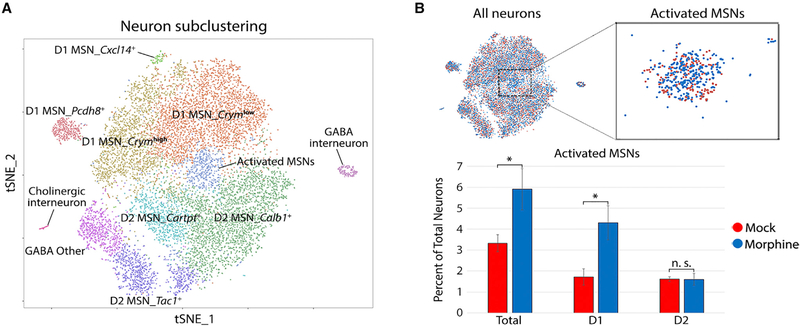
The NAc is composed of many neuronal subtypes, yet our initial t-SNE analysis resolved only three neuronal clusters. To increase resolution, we re-clustered all cells initially identified as neurons, revealing 11 clusters (Figure 2A; Table S1). These clusters directly correspond to the previously identified neuronal subtypes of the NAc. For example, γ-aminobutyric acid-ergic (GABAergic) D1 and D2 medium spiny neurons (MSNs) are known to comprise ~95% of the neurons in the NAc (Lobo et al., 2006); accordingly, we found two major populations of neurons that were enriched for expression of either Drd1a, Tac1, and Pdyn (D1 MSNs) or Drd2, Penk, and Adora2a (D2 MSNs) (Figure S3; Table S1). Another ~2% of the neurons we identified were GABAergic (Sst+/Npy+) and cholinergic (Chat+/Slc5a7+) interneurons. The ubiquitous expression of Gad2 (GABA neuron marker) and Ppp1r1b (striatal neuron marker) and sparse detection of the glutamate neuron-specific gene Slc17a7 indicate that our dissections of the NAc were highly accurate, containing minimal contamination with adjacent tissues such as the cortex (Figure S3). Overall, the neuronal subtype diversity that we uncovered is highly concordant with previous studies of cellular diversity in the NAc, including a previously described single-cell analysis of the striatum (Gokce et al., 2016).
Figure 2. Iterative Clustering of Neurons Enables the Identification of a Subpopulation of Morphine-Activated MSNs.
(A) Spectral t-SNE plot of 13,033 neurons, annotated by cell-type identity. MSN, medium spiny neuron.
(B) Zoomed view of all-neuron t-SNE with cells colored by treatment. Bar graph displays the percentage of total neurons in the activated MSN cluster, stratified bytreatment and expression of the dopamine receptor (D1 versus D2). Error bars show SD among four biological replicates (*p = 0.028, paired t test, two-sided).
We were intrigued by a cluster of neurons in the center of the t-SNE plot characterized by high expression of several immediate early genes (IEGs), such as Fos (c-fos), Junb, Arc, and Nr4a1 (Figure 2; Figure S3). Morphine treatment resulted in a statistically significant two-fold increase in the proportion of these activated MSNs (Figure 2B). It has been reported that c-fos protein expression is upregulated specifically in D1 MSNs upon acute morphine treatment (Enoksson et al., 2012). Consistent with this, Drd1a-expressing cells account for the morphine-dependent increase in the proportion of activated MSNs (Figure 2B). Furthermore, gene set enrichment analysis indicated that genes enriched in this cluster are associated with several terms related to opioid addiction, including morphine dependence and opioidrelated disorders (Table S3). Thus, we identified a subpopulation of neurons displaying a canonical transcriptional response to acute morphine treatment.
Cell-Type-Specific Differential Expression Analysis Uncovered a Robust Morphine-Dependent Transcriptional Response in Oligodendrocytes
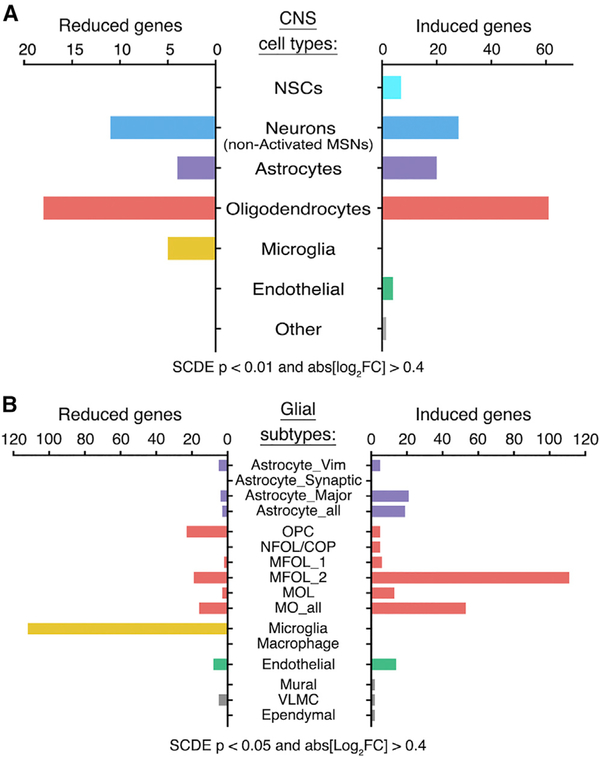
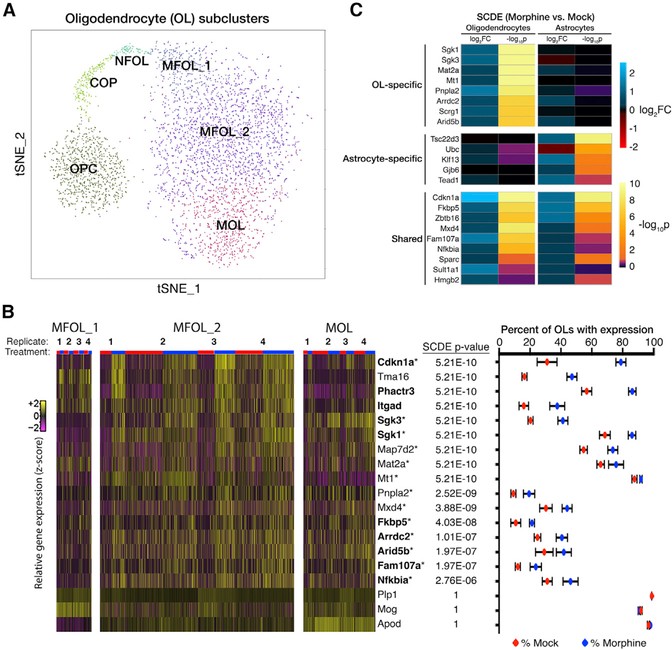
The primary goal of this study was to identify cell-type-specific alterations in gene expression induced by morphine. To accomplish this, we used single-cell differential expression (SCDE) software that uses a Bayesian approach to detect differentially expressed (DE) genes from single-cell data (Kharchenko et al., 2014). We presumed that MSNs would exhibit the strongest transcriptional response, because these cells are known to express opioid receptors, display an electrophysiological response to morphine, and mediate morphine-induced behavioral phenotypes. We found that morphine induces the expression of several hundred genes, including the classical markers Fos, Junb, and Nr4a1, in D1 neurons of the activated MSN cluster compared to other D1 neurons (Table S2). We also detected morphine-dependent changes in gene expression in nearly every glial cell type, with the most numerous changes found in oligodendrocytes (OLs) (Figure 3). Morphine elicited significant alterations in gene expression in each of six clusters of OLs (Figure 3B; Figure S4), which correspond to developmental states in the continuum from oligodendrocyte progenitor cells (OPCs) to myelin-forming OLs (MFOLs) and mature OLs (MOLs) (Figure 4A) (Marques et al., 2016). Many of the morphine-induced genes identified in MFOLs and MOLs, including Cdkn1a, Phactr3, and serum/ glucocorticoid-regulated kinase 1 (Sgk1), have been previously reported to be upregulated by morphine in the striatum (Korostynski et al., 2007; Piechota et al., 2010; Skupio et al., 2017), but these changes could not be confidently ascribed to a specific cell type (Figure 4B, bolded genes). 13 of the top 30 morphineinduced genes in OLs (SCDE p value < 0.0001) are reported targets of the glucocorticoid receptor (GR) (Figure 4B, marked by asterisks). GR signaling is known to play a key role in morphinedependent gene expression and behaviors in mice (Marinelli et al., 1998) and was shown to be critical for a large subset of morphine-induced transcriptional responses in cultured astrocytes (Slezak et al., 2013). We found a significant enrichment of GR targets among morphine-induced genes in astrocytes but also uncovered a distinct subset of GR target genes in OLs (Figure 4C; Table S3). Altogether, these data suggest a robust and previously underappreciated transcriptional response to morphine in OLs. Because multiple studies suggest myelin pathology in human opioid addicts (Bora et al., 2012; Li et al., 2016; Liu et al., 2008; Upadhyay et al., 2010; Wang et al., 2011), we pursued this observation, with the goal of illuminating the molecular mechanisms by which morphine influences OL function.
Figure 3. SCDE Analysis Uncovered Cell-Type-Specific Responses to Morphine.
(A and B) We performed single-cell differential expression (SCDE) (morphine versus mock) on each t-SNE cluster. The total number of unique genes significantly induced or reduced by morphine is shown, grouped by (A) major CNS cell type or (B) glial subpopulation. Astrocyte_Vim, Astrocyte_Synaptic, and Astrocyte_Major refer to three subclusters of astrocytes, and Astrocyte_all is the combined analysis of all astrocytes. MO_all denotes the combined analysis of all MFOLs and/or mature oligodendrocytes (e.g., MFOL_1, MFOL_2, and MOL). See STAR Methods and Table S2 for additional details. Full terms for abbreviations of oligodendrocyte clusters (red) are given in the legend of Figure 4
Figure 4. Iterative Clustering and SCDE Revealed a Robust Morphine-Dependent Transcriptional Response by Oligodendro-cytes.
(A) Iterative clustering of all 3,817 oligodendrocytes (OLs) reveals subpopulations corresponding to OL maturation state. OPC, oligodendrocyte progenitor cell; COP, differentiation-committed OL progenitor; NFOL, newly formed OL; MFOL, myelin-forming OL; MOL, mature OL.
(B) Morphine robustly modulates the expression of several genes in OLs. (Left) Heatmap of relative gene expression for a subset of genes in three clusters of MFOLs and/or mature OLs. Columns representing individual cells are ordered horizontally by biological replicate and treatment (red, mock; blue, morphine). Bolded genes have been previously reported to be induced by morphine in the mouse striatum, and those marked with an asterisk are reported targets of the glucocorticoid receptor (GR). (Right) SCDE p value and expression percentages for combined analysis of all MFOLs and/or mature OLs (MFOL_1, MFOL_2, and MOL). Expression is defined as detection of ≥1 transcript. Dots and error bars are, respectively, the mean and SD among four biological replicates.
(C) Morphine regulates the expression of distinct subsets of GR target genes in OLs and astrocytes
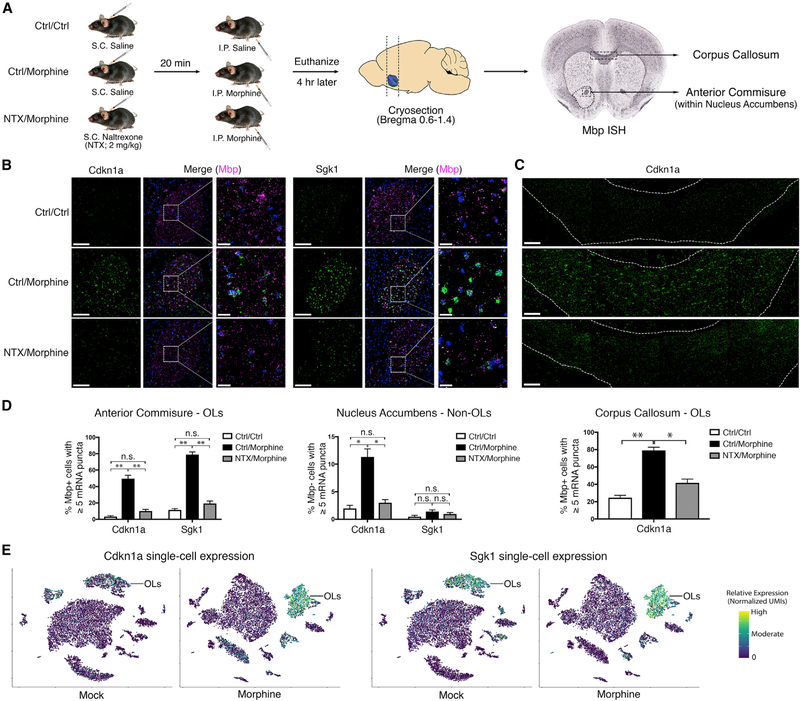
In Vivo Validation of Cell-Type-Specific MorphineDependent Changes in Gene Expression by FISH
Although single-cell mRNA profiling is widely used for the identification of new cellular subtypes, calling DE genes from these data can be challenging due to technical issues such as dropout and dynamic range compression (Dal Molin et al., 2017). Therefore, we sought to independently validate some of the most dramatic morphine-dependent changes in gene expression. We used FISH to costain Cdkn1a or Sgk1 and Mbp (myelin basic protein), a highly expressed marker of OLs (Figure 5A). The Cdkn1a and Sgk1 signal was sparsely detected in mock-treated mice, while morphine treatment elicited a dramatic increase in puncta number selectively in OLs (Figures 5B–5D). The morphine-dependent increase was observed throughout the NAc and dorsal striatum but was especially apparent in oligodendrocyte-dense white matter tracts, such as the anterior commissure (Figure 5B) and corpus callosum (Figure 5C). This suggests that the OL response to morphine is likely not restricted to the NAc. The anterior commissure was included within our initial NAc dissections; thus, OL lineage cells from this region comprise a significant fraction of the total OLs in our scRNA-seq dataset (roughly 25% based on estimates from the Mbp FISH signal). When mice were pre-treated with naltrexone (NTX), an opioid receptor antagonist, the morphine-dependent induction of Cdkn1a and Sgk1 was dramatically reduced (Figures 5A–5D), indicating that their induction occurs in an opioid receptor-dependent manner. As expected, morphine-dependent induction of the immediate-early genes Fos, Junb, and Nr4a1 was observed in the NAc and dorsal striatum, predominantly in Drd1-expressing cells (D1 MSNs), and was similarly reduced by NTX pre-treatment (Figure S5). Altogether, these results validate the cell-type-specific transcriptional responses we identified by scRNA-seq and suggest that both neuron- and OL-specific changes are mediated by opioid receptor activation.
Figure 5. In Vivo Validation of Oligodendrocyte-Specific Morphine-Dependent Gene Expression by FISH.
(A) Schematic of experimental design. Pre-treatment with naltrexone (NTX) should block morphine responses mediated by opioid receptor activation. Mbp in situ hybridization (ISH) (from the Allen Brain Atlas database) is shown to illustrate regions with high densities of OLs.
(B and C) Expression of Cdkn1a and Sgk1 is induced by morphine in OLs in an opioid receptor-dependent manner. Representative images of the anterior commissure (B) and corpus callosum (C) are shown. Signals are pseudocolored by expression of Cdkn1a (green), Sgk1 (green), Mbp (magenta), and/or DAPI (blue). Scale bars, 100 μm (left panels) and 20 mm (right panels and zoomed view).
(D) Quantification of the percentage of OLs (left panel: n = 781 cells; right panel: n = 1,374 cells) or of non-OLs (middle panel: n = 1,000 cells) with ≥5 puncta of Cdkn1a or Sgk1 (3 mice per condition; paired t test, two-sided; NS, not significant; **p < 1e4, *p < 0.05).
t-SNE plots of all cells from mock- or morphine-treated mice overlaid with expression of Cdkn1a or Sgk1 indicates that the morphine-dependent induction of these genes occurs predominantly in OLs. In the case of Cdkn1a, a modest induction was also detected in astrocytes and endothelial cells.
Bulk RNA-Seq of FACS-Isolated OLs Revealed a Remarkable Extent of Transcriptional Regulation by Morphine
Because our Drop-seq and FISH data indicated a robust oligodendrocyte transcriptional response to morphine in vivo, we sought to validate this finding in vitro. We treated purified cultures of mouse OLs with morphine and assessed expression of Cdkn1a, Sgk1, and Phactr3 by qRT-PCR. Expression of these genes was not appreciably affected by morphine (Figure S6), suggesting that the response we observed in vivo might be mediated by an indirect or non-cell-autonomous mechanism. To further characterize this response, we adapted an immunostaining and fluorescence-activated cell sorting (FACS)-based approach (Robinson et al., 2014) to isolate OLs from the entire brains of six mice, three each mock or morphine treated. Saline or morphine injections and tissue dissociation were performed exactly as in our scRNA-seq experiments to enable cross-validation between the two datasets. We gated out CD45+ and PDGFRa+ populations to remove immune cells and OPCs, respectively, and then isolated MFOLs and/or MOLs based on their dual expression of GALC and MOG (Figure S7A). RNA was purified from each of the six samples and examined by qRT-PCR. We found 10- to 20-fold enrichment of the OL markers Cnp and Mog, which was comparable to that observed following affinity purification of ribosome-associated mRNAs from CnpCre x HA-Rpl22 (Ribotag) mice (Sanz et al., 2009). A significant reduction of the neuronal marker Rbfox3/NeuN and the astrocytic marker Gfap in these FACS samples indicated enhanced specificity of this approach compared to the Ribotag method (Figures S7B and S7C). Assured that this was an enriched OL population, we tested the morphine-dependent upregulation of Cdkn1a and Sgk1 by qRT-PCR and found that they were induced by more than 10-fold (Figure S7C).
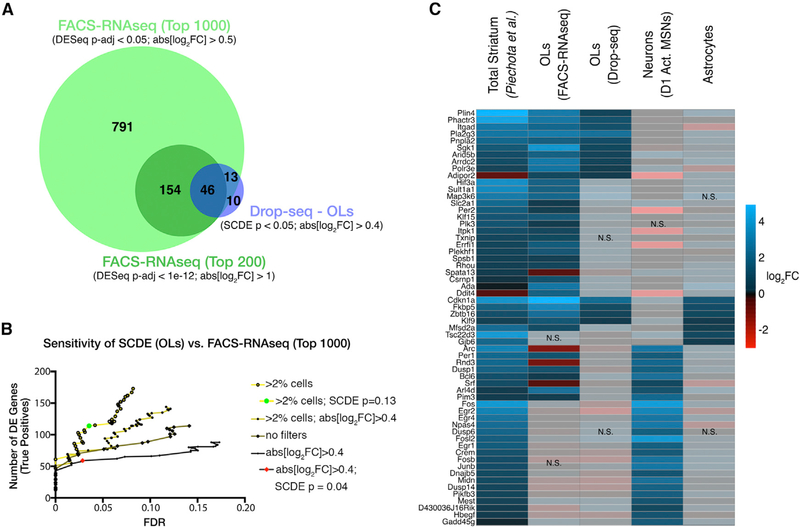
To ascertain the full extent of morphine-dependent transcriptional regulation in OLs, we next performed RNA-seq on these sorted samples. We reasoned that the FACS population should be most comparable to the three MFOLs and/or MOL clusters observed in our single-cell transcriptional profiling (MFOL_1, MFOL_2, and MOL), because these clusters specifically and ubiquitously express Mog. Differential expression analysis of the bulk FACS-RNA-seq data confirmed 85.5% (59/69) of the changes detected by single-cell analysis (SCDE) of OLs. In addition, we uncovered an additional 935 genes significantly affected by acute morphine treatment (DESeq adjusted p value [p-adj] < 0.05 and abs[log2FC] > 0.5) (Figure 6A; Table S5). These results demonstrate that most morphine-dependent expression changes detected by single-cell RNA-seq were bona fide; in addition, our bulk RNA-seq experiments identified a large set of expression changes that were not observed in our single-cell experiments, illustrating the increased sensitivity of the the FACS-RNA-seq dataset as the gold standard upon which to benchmark single-cell DE analysis, we optimized the sensitivity of single-cell DE calling by filtering out lowly expressed genes (those detected in less than 2% of cells), removing a fold-change cutoff, and relaxing the SCDE p value cutoff. These criteria effectively doubled the number of DE genes detected in MFOLs and/or MOLs without compromising specificity (Figure 6B, compare green dot and red diamond). Of the most significant morphine-regulated genes identified by a previous microarray analysis of the striatum (Piechota et al., 2010), nearly half of them can be attributed to OLs (Figure 6C). In addition, our single-cell analysis allowed us to ascribe other reported morphine-dependent gene expression changes to neurons or astrocytes (Figure 6C). A comparison of our data to changes observed at various time points following acute morphine treatment (1, 2, 4, or 8 hr) indicates cell-type-specific differences in the temporal response to morphine (Figure S8). These analyses underscore the utility of cell-type-specific profiling strategies and suggest that oligodendrocyte-specific responses to opioids have been underappreciated.
Figure 6. Bulk RNA-Seq of FACS-Isolated OLs Revealed a Remarkable Extent of Transcriptional Regulation by Morphine.
(A) Venn diagram illustrating significant overlap between DE genes identified from Drop-seq (OLs) or FACS-RNA-seq.
(B) We improved the sensitivity of single-cell DE analysis with minimal loss in specificity. The number of DE genes (true positives: present in the FACS-RNA-seq top 1,000 list) detected by SCDE of OLs was plotted versus the empirical false discovery rate (FDR; STAR Methods). We optimized SCDE by filtering out genes expressed in less than 2% of cells, removing a fold-change cutoff, and relaxing the p value cutoff to 0.13 (green circle).
(C) Left column of the heatmap shows the relative morphine-dependent induction (log2FC morphine versus saline: 4 hr) of a subset of genes previously reported to be induced by morphine, as assessed by microarray of total striatum (Piechota et al., 2010). We filtered the top 100 morphine-regulated genes from that dataset for those significantly affected in OLs (FACS-RNA-seq and/or Drop-seq), Drd1a-expressing activated MSNs (D1 Act. MSNs), or astrocytes to obtain the 60 genes shown here. Opaque blocks (labeled NS) indicate changes that were not statistically significant (p < 0.05)
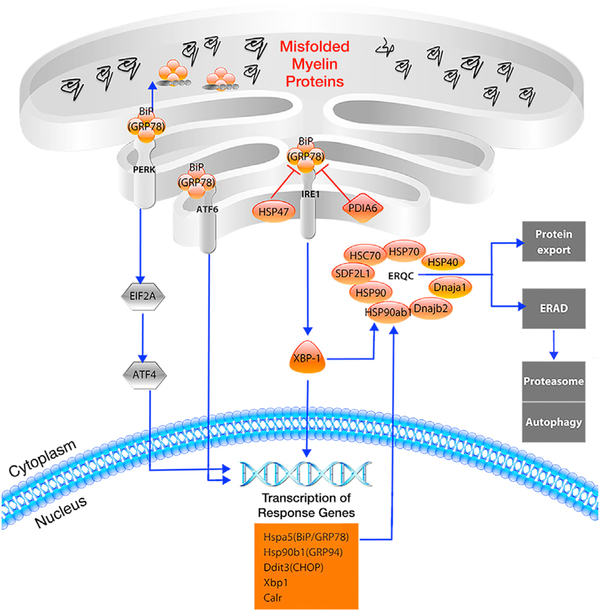
To obtain biological insights into the response to morphine by OLs, we performed gene set enrichment and pathway analyses of our FACS-RNA-seq data. This confirmed the enrichment of GR targets, including all GR-related genes we identified by Drop-seq, among the morphine-induced genes in OLs (Figure S9; Table S3). We also found that several morphinerepressed genes encode endoplasmic reticulum (ER) chaperone proteins with critical functions in ER quality control (ERQC) and the unfolded protein response (UPR) (Figure 7; Figure S9). MFOLs are thought to be particularly susceptible to ER stress because of their high rates of protein and lipid synthesis (Lin and Popko, 2009). Accordingly, components of the UPR machinery are dysregulated in various demyelinating diseases of the peripheral and CNS (Lin and Popko, 2009). The morphinedependent downregulation of ERQC and UPR could lead to an increase in the intracellular accumulation and/or export of misfolded proteins, as well as downstream consequences on oligodendrocyte survival, maturation, and/or myelination. This has considerable clinical implications in light of several studies describing impaired white matter integrity in the brains of human opioid users (Bora et al., 2012; Li et al., 2016; Liu et al., 2008; Upadhyay et al., 2010; Wang et al., 2011).
Figure 7. Several Components of the UPR Are Downregulated by Morphine in OLs.
Genes downregulated by morphine in OLs (p-adj < 1e-8, log2FC < −0.5) are shown in orange, and unaffected factors are shown in gray. The expression of Xbp1 (XBP-1), as well as numerous XBP-1-induced genes (shown in the orange box), are reduced by morphine, as are several endoplasmic reticulum (ER) chaperone proteins involved in ER quality control (ERQC). These chaperones assist in the proper folding and export of proteins or target misfolded proteins to the proteasome by a process known as ER-associated degradation (ERAD). The downregulation of these factors might lead to the increased accumulation and/or export of misfolded proteins, including myelin constituents
DISCUSSION
Molecular and behavioral responses to opioids have long been presumed to be mediated by neurons. The finding that OLs and astrocytes express opioid receptors in vitro and in vivo suggested that they too might respond to opioids (Knapp et al., 1998; StieneMartin et al., 2001). Astrocyte proliferation is reduced by stimulation of mu opioid receptor (MOR), kappa opioid receptor (KOR), or delta opioid receptor (DOR) (Stiene-Martin and Hauser, 1991; Hauser et al., 1996) and morphine-induced transcriptional responses of primary astrocytes have been described (Slezak et al., 2013). MOR agonists enhance proliferation of immature OLs, but not MOLs (Knapp and Hauser, 1996), while KOR agonists appear to promote OL differentiation and myelination (Knapp et al., 1998; Mei et al., 2016; Du et al., 2016). Altogether, previous reports indicate that opioids differentially influence the survival, proliferation, and/or differentiation of OLs and astrocytes, but the molecular mechanisms remain poorly understood. Brain imaging of heroin users has revealed impaired white matter integrity, implicating OLs in the pathophysiology of opioid dependence (Bora et al., 2012; Li et al., 2016; Liu et al., 2008; Upadhyay et al., 2010; Wang et al., 2011). Heroin abuse has also been linked to leukoencephalopathies that are likely mediated by OL dysfunction (Bach et al., 2012). We performed unbiased profiling of cell-type-specific transcriptional responses to morphine and uncovered a remarkable number of alterations in gene expression that are unique to OLs. Many of the gene expression changes we observed in OLs are also altered following chronic opioid morphine exposure (Skupio et al., 2017). Our results are consistent with previous reports suggesting a role for opioid signaling in OL maturation and myelination. Moreover, our detailed characterization of the OL-specific response to morphine offers insights into the putative mechanisms of OL dysfunction in opioid-induced myelin pathology.
The most significantly DE gene we detected in OLs is Cdkn1a, which encodes p21. The major function of p21 is the inhibition of cyclin-dependent kinases that leads to cell-cycle arrest. In OLs, its expression is required for differentiation and myelination (Zezula et al., 2001). It has been demonstrated that autonomous transforming growth factor β (TGF-β) signaling drives oligodendrocyte differentiation and CNS myelination during postnatal development via SMAD3/4-dependent transcriptional activation of Cdkn1a and repression of Myc (c-myc) (Palazuelos et al., 2014). We observed morphine-dependent reduction in the OL-specific expression of Myc, as well as Sox2 and Sox10, key drivers of OPC identity. Sox10 expression is downregulated in the brains of rats following heroin self-administration (Martin et al., 2018). Our single-cell analyses revealed that a significant number of GR-regulated genes are induced by morphine in OLs. Morphine rapidly increases serum levels of corticosterone (Simon et al., 1975), which in turn induces expression of Sgk1 (Miyata et al., 2011). GR signaling is critical for morphinedependent dopamine release and locomotor activation in mice (Marinelli etal., 1998), but the contributions of OLs to these phenotypes have not been examined. Presumably, the morphinedependent induction of Sgk1 and a subset of the other genes we identified occurs via GR-dependent transcriptional activation. TherobustinductionofSgk1weobservedisparticularlyintriguing, because this gene facilitates morphological changes in OLs that are associated with depressive-like behaviors (Miyata et al., 2015). Alterations in OL morphology have also been observed in morphine-treated mice (Hauser et al., 2009). Thus, the morphine-dependent induction of Sgk1 specifically in OLs constitutes a putative molecular mechanism underlying depressive-like behaviors in opioid-dependent individuals. It was previously suggested that morphine- and GR-dependent gene expression changes (including Sgk1 induction) are largely confined to astrocytes (Slezak et al., 2013). However, our single-cell analyses indicate that many GR-dependent genes are induced uniquely in either OLs (i.e., Sgk1 and Sgk3) or astrocytes (i.e., Tsc22d3 and Gjb6), whereas others are induced in both of these cell types (i.e., Fkbp5 and Nfkbia). Cell-type-specific expression patterns of opioid receptor subtypes and GR may explain the shared and unique gene expression changes we observed in neurons, astrocytes, and OLs. Unfortunately, because opioid receptor genes are relatively lowly expressed, they were only sparsely detected in our single-cell data, obfuscating our attempts to correlate patterns of opioid receptor expression with the observed transcriptional response to morphine. Regarding the mechanism of the response by OLs, our FISH data suggest that opioid receptor activation is critical, but the relative contributions of OL-,neuron-,and/or astrocyte-derived opioid receptors remain unknown. Our in vitro data indicate that a non-cell-autonomous mechanism is likely, but they do not exclude the possibility of OL-specific, GR-mediated effects. We also cannot rule out the putative contributions of other indirect mechanisms, including neuronal activity-dependent responses and paracrine regulation by neurons, astrocytes, and/or microglia.
Our pathway analysis revealed highly significant morphinedependent downregulation of genes encoding heat shock proteins, ER chaperones, and other factors critically involved in the UPR and ER quality control (ERQC). The expression of heat shock proteins, including Hsp70 (Hspa1a) and Hsc70 (Hspa8), has previously been shown to be modulated by morphine treatment of rats (Ammon-Treiber et al., 2004; Salas et al., 2011). Proper regulation of the UPR is essential for efficient myelination, as evidenced by the apparent dysregulation of UPR in multiple myelin disorders, including Charcot-Marie-Tooth disease, vanishing white matter disease, and multiple sclerosis (Lin and Popko, 2009; Volpi et al., 2017). Under normal conditions, GRP78/BiP (encoded by Hspa5) is thought to initiate the UPR by sensing misfolded proteins in the ER (Volpi et al., 2017), and its conditional ablation in OLs or Schwann cells results in cell death and severe hypomyelination (Hussien et al., 2015). Coincidentally, Hspa5 is also the most significantly morphinerepressed gene that we detected in OLs. The reduced expression of downstream response genes, including the anti-survival gene CHOP (Ddit3) and several ER chaperone proteins, suggests that morphine potently suppresses the canonical UPR, which normally entails attenuation of global protein synthesis, degradation of misfolded proteins, and under severe ER stress, autophagy or apoptosis. Previous studies suggested that perinatal exposure to other opioid drugs, specifically buprenorphine and methadone, appears to promote myelination (Eschenroeder et al., 2012; Sanchez et al., 2008; Vestal-Laborde et al., 2014). Perhaps in the context of chronic opioid exposure, persistent upregulation of myelin synthesis and concomitant downregulation of ERQC leads to increased accumulation and/or export of misfolded myelin constituents and therefore altered myelin composition. The integrity and composition of myelin have not been investigated in models of long-term morphine exposure.
In summary, we used scRNA-seq to enable the elucidation of cell-type-specific responses to morphine in the NAc of mice. We identified a dramatic transcriptional response by OLs and shed light on the underlying molecular mechanisms of this response, specifically implicating GR signaling and the UPR as potential links between gene expression changes we observed and opioid-mediated myelin pathology. Our single-cell and bulk RNA-seq datasets should serve as valuable resources for research aiming to assess transcriptional responses to opioids or other drugs of abuse. Future studies are necessary to investigate the mechanisms, functional consequences, and physiological significance of oligodendrocyte-opioid interactions.
STAR⋆METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mog | Millipore | Cat# MAB5680; RRID:AB_1587278 |
| Anti-GalC-FITC | Millipore | Cat# FCMAB312F; RRID:AB_10806360 |
| Anti-PDGFRα-PE | BD Biosciences | Cat # 562776; RRID:AB_657615 |
| Anti-CD45-Pacific Blue | BioLegend | Cat# 103126; RRID:AB_493535 |
| CD140a (PDGFRα) MicroBead Kit | Miltenyi Biotech | 130–101-547 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trehalose | Sigma-Aldrich | T5251; CAS: 6138–23-4 |
| Neural Tissue Dissociation Kit | Miltenyi Biotech | 103–092-628 |
| Critical Commercial Assays | ||
| Lightning Link Rapid Cy5 | Novus Biosciences | 342–0010 |
| ViewRNA ISH Cell Assay | Thermo Fisher | QVC-0001 |
| MiniMACS Separator | Miltenyi Biotech | 130–042-102 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE118918 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 mice (male; 6–12 weeks) | Jackson Laboratory | N/A |
| Cnp-Cre mice | N/A | N/A |
| Rpl22-HA (Ribotag) mice | Sanz et al., 2009 | N/A |
| Cnp-Cre x Ribotag mice | This paper | N/A |
| Oligonucleotides | ||
| *See Table S4 for list of primers used for qRT-PCR | N/A | N/A |
| Software and Algorithms | ||
| Seurat | Satija et al., 2015 | http://satijalab.org/seurat/ |
| Monocle | Qiu et al., 2017 | http://cole-trapnell-lab.github.io/monocle-release/ |
| SCDE | Kharchenko et al., 2014 | http://hms-dbmi.github.io/scde/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Robi Mitra (rmitra@wustl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal care and experimental procedures were approved in advance by the National Institute of Health and Washington University School of Medicine Institutional Animal Care and Use Committee (Protocol #20170030). Male C57BL/6 mice (6–12 weeks old) were used for all experiments, and all animals were socially housed on a 12 h light dark cycle with water and food ad libitum. Mice were injected with saline or morphine sulfate (20 mg/kg) intraperitoneally, and/or naltrexone hydrochloride (2 mg/kg) subcutaneously. Cnp-Cre mice were crossed with Rpl22-HA (“RiboTag”) (Sanz et al., 2009) mice (both in the C57BL/6 background) to generate Cnp-Cre x RiboTag mice, facilitating the immunoprecipitation of oligodendrocyte-enriched mRNAs for qRT-PCR analysis.
METHOD DETAILS
Dissection and tissue dissociation
Four h after saline (mock) or morphine injection, mice were anesthetized by isoflurane and euthanized by decapitation. Brains were rapidly extracted, cooled in ice-cold Hibernate A (-Ca) media for 5 min, then placed ventral surface up onto a chilled Petri dish on ice.
The rostral and caudal boundaries of the NAc were visually approximated and a ~1 mm-thick coronal section was obtained using a chilled razor blade. The NAc was microdissected at its dorsolateral borders and pooled by experimental condition (two mice per replicate). Pooled tissue samples were digested in papain/DNase I solution for 1 h at 37°C, according to a previously published protocol (Saxena et al., 2012). The final volume was 1 mL per mouse NAc. Digestion was stopped by dilution with EBSS #2 (Saxena et al., 2012), which contains ovomucoid protease inhibitor. Tissue was centrifuged at 300g for 5 min, resuspended in EBSS #2, then gently triturated with a P200 pipette. The resulting cell suspension was divided between two microfuge tubes and diluted with Drop-seq buffer to a volume of 1 mL per tube (per 2 pooled samples). Drop-seq buffer consisted of 5% (wt/vol) trehalose, Hank’s buffered salt solution (HBSS; magnesium- and calcium-free), 2.13 mM MgCl2, 2 mM MgSO4, 1.26 mM CaCl2, 1 mM glucose and 0.02% bovine serum albumin. Cells were washed twice by centrifugation at 300g for 5 min, followed by resuspension in 1 mL Drop-seq buffer. Cells were then passed through a 40-mm mesh filter, diluted to 210 cells/μL using cell concentration estimates from a hemocytometer, and kept on ice until use (< 30 min).
Drop-seq library generation and QC
Drop-seq was performed as previously described (Campbell et al., 2017). Mouse NAc suspensions were processed through Drop-Seq in eight separate groups over four separate batches (one batch per day). Libraries were sequenced on the Illumina HiSeq3000 or HiSeq2500. Read 1 was 25 bp (bases 1–12 cell barcode, bases 13–20 UMI – unique molecular identifier), read 2 (paired end) was 75 bp, and the index primer was 7 bp. Raw sequence data were first filtered to remove all read pairs with a barcode base quality of less than 10. The second read (75 bp) was then trimmed at the 5ʹ end to remove any TSO adaptor sequence and at the 3ʹ end to remove poly(A) tails of length 6 or greater, then aligned to the mouse (mm10) genome using STAR v2.4.2a with default settings. Uniquely mapped reads were grouped by cell barcode. A list of UMIs in each gene, within each cell, was assembled, and UMIs within ED = 1 were merged together. The total number of unique UMI sequences was counted, and this number was reported as the number of transcripts of that gene for a given cell.
All cell barcodes in which 500 or more genes and 1000 or more transcripts (UMIs) were detected were used in downstream analysis. Raw digital expression matrices were generated separately, then the replicates from each experimental group were merged together into a single matrix. Cells with high expression levels of one or more marker gene(s) of more than one cell type (i.e., neuron, oligodendrocyte, astrocyte, endothelial cell, etc.), representing likely cell doublets/multiplets, were removed (~4% of all STAMPS), resulting in a 12,105-cell barcode dataset for mock treatment and a 11,171-cell barcode dataset for morphine treatment. Gene expression was normalized in Scater (McCarthy et al., 2017) using the method described by Lun et al. (Lun et al., 2016), and genes with expression in ≥ 50 cells were retained for clustering. Before clustering, we removed unwanted variation due to number of UMIs, ratio of reads mapping to mitochondrial genes, and batch effects using Seurat software (Satija et al., 2015).
Unsupervised clustering and dimensionality reduction
We used Seurat to perform clustering (Butler et al., 2018; Satija et al., 2015). We identified genes that were most variable across the entire dataset, controlling for the known relationship between mean expression and variance. We calculated the mean and dispersion (variance/mean) for each gene across all cells, then identified outlier genes. To distinguish principal components (PCs) for further analysis, we used the PCElbowPlot() function, and found that PCs beyond 15 accounted for negligible variance. We identified molecularly distinct clusters using the FindClusters() function with the Louvain algorithm with multilevel refinement, using a high resolution (2.0) to detect weakly distinguishable clusters. We then built a classification hierarchy, merged clusters with higher than 0.05 Out-of-bag error (OOBE) from a random forest classifier, and performed t-distributed stochastic neighbor embedding (t-SNE) (van der Maaten, 2008) in Seurat with default settings. This allowed us to assign cells into a total of 18 cell type clusters each for mock and morphine. To further resolve cell subtypes, cells corresponding to neuronal, oligodendrocyte, or other clusters were combined, then iteratively clustered as above using CCA (Butler et al., 2018). Altogether, we identified 29 molecularly distinct cell types, all of which were shared between mock- and morphine-treated mice.
scRNA-seq analysis
To analyze differential gene expression between morphine and mock datasets, we performed pairwise differential expression analysis on specific cell subpopulations using the SCDE package, which fits individual error models to account for stochastic detection of lowly expressed genes (Kharchenko et al., 2014). We included all Seurat-generated clusters containing > 10 cells from each of 8 biological replicates (Mock1–4, Morphine1–4), and that had relatively uniform distribution across all replicates. Only 3 neuronal subtypes (cholinergic interneurons, D1 MSN_Cxcl14+, and D1 MSN_Pcdh8+) did not meet these criteria. We used default settings with the exception that we varied the min.detected parameter of the clean.counts function to filter out genes detected in less than 2% of cells. We included the results of every cluster for all gene with p value < 0.5 in Table S2.
Morphine-activated score
Score was generated for each cell of the OL lineage using quadratic programming, as previously described (Treutlein et al., 2016) with the following modifications. Briefly, we log-transformed transcript counts from FACS-RNaseq data of OLs (average of 3 mock or 3 morphine samples). We used this as mock and morphine reference profiles after filtering out genes not appreciably detected by Drop-seq of OLs (genes detected in < 5% of cells). The R package QuadProg was used for quadratic programming to assign a mock score and a morphine score to each single cell from the OL lineage. The values shown in Figure S4 were calculated by dividing the morphine score by the sum of the mock score and the morphine score to obtain a scaled morphine-activated score (from 0 to 1).
In situ hybridization
Morphine and mock treatments were performed as described above, except that mock treatment was preceded by a subcutaneous (s.c.) saline injection (Ctrl/Ctrl), and morphine treatment was preceded by injection with s.c. saline (Ctrl/Morphine) or naltrexone (NTX/Morphine). After 4 h, brains were harvested and immediately flash-frozen in OCT (optimal cutting temperature compound) in liquid nitrogen. We stored frozen tissue blocks at –80°C prior to sectioning. Twelve-micron thick coronal sections were cut at –20°C and adhered to positively charged Leica slides (one of each of the three treatment conditions per slide), then stored at –80°C. Frozen tissue sections were washed briefly with 1X PBS and fixed in 3.7% para-formaldehyde for 1 h. Following fixation, slides were washed twice with 1X PBS and then submerged in 70% ethanol overnight at 4°C for permeabilization. RNA FISH was performed according to the Affymetrix ViewRNA protocol.
Imaging and image analysis
We imaged FISH AND IHC slides on a Zeiss LSM 880 II confocal microscope at the Washington University Center for Cellular Imaging (WUCCI). Image files were processed and analyzed in Fiji/ImageJ (Schindelin et al., 2012) software. RGB channels corresponding to the two Affymetrix probes and DAPI were split. RNA particles in each channel were then identified in a semi-automated manner by selecting an intensity threshold above which a spot is considered an RNA particle. Numbers of puncta per cell were quantified from the left and right anterior commissures, the corpus callosum, and the NAc (n = 3 mice per treatment).
In vitro OL culture
We isolated, expanded, and differentiated OPCs into OLs as previously described (Dincman et al., 2012). Briefly, we dissociated cells from postnatal day 6 (P6) mouse cortices using the papain-based Neural Tissue Dissociation Kit (Miltenyi) and mechanical trituration, then enriched for OPCs using magnetic cell sorting (MACS) with anti-PDGFRα-conjugated microbeads and a miniMACS separator. OPCs were expanded for 3 days, passaged to new plates, then differentiated for 6 days (or maintained in proliferation media and harvested as OPCs). On the sixth day (D6), cells were mock-treated with media, or treated with a low (0.5 μg/ml) or high (5 μg/ml) dose of morphine sulfate, then harvested for qRT-PCR at 3 h post-treatment. Values shown in Figure S6 are means from three biological replicates.
Cnp-Cre/Ribotag RNA isolation
Mock/morphine treatment and dissection was performed as described above, except that the entire striatum, corpus callosum, or 40 mg of cortex were harvested and transferred to microcentrifuge tubes containing 500 μL polysome lysis buffer. Lysis and immunoprecipitation were performed as previously described (Sanz et al., 2009). Lysates were applied to Machery Nagel Nucleospin RNA columns and RNA was purified per the manufacturer’s protocol.
Quantitative real-time PCR
RNA isolated from Cnp-Cre x Ribotag mice or FACS-isolated OLs (see below) was reverse transcribed using qScript cDNA Supermix from QuantaBio according to the manufacturer’s protocol. cDNA was mixed with Fast SYBR Green master mix from ThermoFisher and primers to the indicated gene (see Table S4 for primer sequences) in a 96-well plate and quantified using the Quant Studio 3 from Applied Biosciences (in technical triplicates). Differences in relative gene expression were calculated using the delta delta cT method after normalization to beta-actin.
Immunostaining and FACS of OLs
WT mice were injected with saline (mock) or morphine as described above. Four h later, mice were euthanized and entire brains were removed, minced with a razor blade, and dissociated with papain solution (400 units of papain in 8 mL total volume). After 50 min dissociation, suspensions were diluted with EBSS #2 (Saxena et al., 2012), serially passed through 100 μm and 40 μm filters, and centrifuged at 300g for 5 min. The pellet was resuspended in 35% Percoll solution prepared in EBSS containing 0.1% BSA and 5% trehalose, then centrifuged at 800g for 20 min at 18°C. The supernatant containing myelin and cell debris was carefully removed and the pellet was resuspended in flow cytometry buffer (1% normal mouse serum, 1% normal rat serum, 5% trehalose, 0.1% BSA, 1 mM EDTA in DPBS). Cells were counted using a hemocytometer and ~3e6 cells were transferred to a fresh tube, pelleted by brief centrifugation at 300g, and resuspended in 200 μL flow cytometry buffer containing the following fluorophore-conjugated antibodies (or isotype controls): GalC-FITC (4 μg; Millipore), PDGFRa-PE (2 μg; BD Biosciences), Mog-Cy5 (4 μg; Millipore; conjugated with Lightning Link kit from Novus Bio), and CD45-PacBlue (2 μg; BioLegend). Tubes were incubated at 4C with gentle rotation for 30 min, protected from light. Cells were washed twice with and resuspended in flow cytometry buffer (500 μL) and stored on ice protected from light until sorting (< 30 min). Samples were sorted into flow cytometry buffer using the Sony SY3200 “Synergy” with 100 μm nozzle size at the Siteman Cancer Center Flow Cytometry Core.
RNA purification, library prep, and sequencing
Immediately following FACS, cells (~100K per sample) were centrifuged at 300g for 5 min at 4C, and the supernatant was carefully removed. The pellet was resuspended in 300 μL Trizol reagent and RNA was purified according to the manufacturer’s protocol. Sequencing libraries were generated using the SeqPlex RNA Amplification Kit (Sigma) according to the manufacturer’s instructions and sequenced to a depth of ~30 million raw reads per sample (single-end, 50 bp) on the Illumina HiSeq2500.
RNaseq alignment and DE analysis
Reads were mapped to the mouse genome mm10 using STAR. We used htseq to generate gene-count tables for all samples, then performed differential expression between the 3 replicates each from mock- or morphine-treated mice using DESeq. We first removed genes whose basemean expression was < 30, leaving ~12,000 genes. For Figure 4A, we filtered the genes by adjusted p value (p-adj) and log2(fold-change) to generate three lists with increasing stringency: “Top 1000” – p-adj < 0.05 and abs[log2FC] > 0.5; “Top 500” – p-adj < 1e-7 and abs[log2FC] > 0.5; “Top 200” – p-adj < 1e-12 and abs[log2FC] > 1. SCDE false discovery rate (FDR) was determined by dividing the number of true positives (which were defined as those present in the Top 1000 list) by the total number of genes at a given SCDE p value cutoff. FDR calculations were iterated over the full range of SCDE p value thresholds. An empirical FDR of ~3.5% corresponded to an SCDE p value cutoff of 0.13.
Pathway analysis of RNaseq data
Pathway analysis was performed using Genomatix GeneRanker software. For single-cell data, we used as input lists: (i) genes enriched in D1 MSNs of the activated MSN cluster compared to all other D1 MSNs (SCDE p < 0.001; 256 genes), (ii) morphine-regulated genes in OLs (SCDE p < 0.05, abs[log2FC] > 0.4; 69 genes), or (iii) morphine-regulated genes in astrocytes (SCDE p < 0.3; 81 genes). For bulk RNaseq analysis, we used the top 200 morphine-induced (p-adj < 1e-12, log2FC > 1) or the top 148 morphinerepressed (p-adj < 1e-8, log2FC < 0.5) genes.
QUANTIFICATION AND STATISTICAL ANALYSIS
Single-cell analysis
Error bars in Figures 2B, 4B, and S2 represent the standard deviation among four biological replicates per treatment.
FISH
Quantification in Figures 5 and S5 was performed using Fiji/ImageJ software (Schindelin et al., 2012). RGB channels corresponding to the two Affymetrix probes and DAPI were split. RNA particles in each channel were then identified in a semi-automated manner by selecting an intensity threshold above which a spot is considered an RNA particle. The number of puncta, their average size, and total area of signal was calculated for each probe. Cell number was quantified by DAPI signal using the NucleusCounter plugin in ImageJ. For Figure S5, positive cells were defined as having at least 5 puncta for the designated gene within 1 micron of the nucleus. For Figure 5, OLs were defined as having at least 2 Mbp puncta within 1 micron of the nucleus. By these criteria, ~95% of the cells in the anterior commissure and corpus callosum and ~10% of cells in the NAc were classified as OLs. Error bars represent the standard deviation among three biological replicates (3 mice per condition) and statistical significance was assessed by paired t test, two-sided.
qRT-PCR
Differences in relative gene expression were calculated using the delta delta cT method after normalization to beta-actin. Values shown are the mean plus SEM of three biological replicates (Figure S6) or three replicate plates (Figure S7), with each sample run in technical triplicate.
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw and processed sequencing data reported in this paper is GEO: GSE118918
Supplementary Material
Highlights.
Single-cell RNA-seq uncovers cell-type-specific responses to morphine in mice
Among glia, the most robust transcriptional response are in oligodendrocytes
Oligodendrocyte- and neuron-specific responses to morphine are validated by FISH
Bulk RNA-seq of purified oligodendrocytes reveals ~1,000 morphine-regulated genes
ACKNOWLEDGMENTS
This work was supported by NIH grants CA009547–33 (to D.A.), AGO13730 and NS099314 (to J.M.), and 5U01MH109133, RF1MH117070, and R01GM129126 (to R.D.M.) and the Intellectual and Developmental Disabilities Research Center at Washington University (U54 HD087011). We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis for the use of the Siteman Flow Cytometry core. The Siteman Cancer Center is supported in part by an NCI Cancer Center support grant (P30 CA091842). We thank Peter Wang and Gwendalyn Randolph for sharing the CD45-PacBlue antibody
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes nine figures and five tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.08.080
REFERENCES
- Albertson DN, Schmidt CJ, Kapatos G, and Bannon MJ (2006). Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology 31, 2304–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammon-Treiber S, Grecksch G, Stumm R, Riechert U, Tischmeyer H, Reichenauer A, and Höllt V (2004). Rapid, transient, and dose-dependent expression of hsp70 messenger RNA in the rat brain after morphine treatment.Cell Stress Chaperones 9, 182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach AG, Jordan B, Wegener NA, Rusner C, Kornhuber M, Abbas J, and Surov A (2012). Heroin spongiform leukoencephalopathy (HSLE). Clin. Neuroradiol. 22, 345–349. [DOI] [PubMed] [Google Scholar]
- Bauer IE, Soares JC, and Nielsen DA (2015). The role of opioidergic genes in the treatment outcome of drug addiction pharmacotherapy: a systematic review. Am. J. Addict 24, 15–23. [DOI] [PubMed] [Google Scholar]
- Bora E, Yücel M, Fornito A, Pantelis C, Harrison BJ, Cocchi L, Pell G, and Lubman DI (2012). White matter microstructure in opiate addiction. Addict. Biol 17, 141–148. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AM, Resch JM, McCarroll SA, et al. (2017). A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci 20, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. (2017). Comprehensive singlecell transcriptional profiling of a multicellular organism. Science 357, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Molin A, Baruzzo G, and Di Camillo B (2017). Single-cell RNAsequencing: assessment of differential expression analysis methods. Front. Genet 8, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart RC, Surratt HL, Cicero TJ, Parrino MW, Severtson SG, BucherBartelson B, and Green JL (2015). Trends in opioid analgesic abuse and mortality in the United States. N. Engl. J. Med 372, 241–248. [DOI] [PubMed] [Google Scholar]
- Dincman TA, Beare JE, Ohri SS, and Whittemore SR (2012). Isolation of cortical mouse oligodendrocyte precursor cells. J. Neurosci. Methods 209, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Duan Y, Wei W, Cai Y, Chai H, Lv J, Du X, Zhu J, and Xie X (2016). Kappa opioid receptor activation alleviates experimental autoimmune encephalomyelitis and promotes oligodendrocyte-mediated remyelination. Nat. Commun 7, 11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoksson T, Bertran-Gonzalez J, and Christie MJ (2012). Nucleus accumbens D2- and D1-receptor expressing medium spiny neurons are selectively activated by morphine withdrawal and acute morphine, respectively. Neuropharmacology 62, 2463–2471. [DOI] [PubMed] [Google Scholar]
- Eschenroeder AC, Vestal-Laborde AA, Sanchez ES, Robinson SE, and Sato-Bigbee C (2012). Oligodendrocyte responses to buprenorphine uncover novel and opposing roles of m-opioid- and nociceptin/orphanin FQ receptors in cell development: implications for drug addiction treatment during pregnancy. Glia 60, 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Su€dhof TC, and Quake SR (2016). Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep. 16, 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser KF, Stiene-Martin A, Mattson MP, Elde RP, Ryan SE, and Godleske CC (1996). mu-Opioid receptor-induced Ca2+ mobilization and astroglial development: morphine inhibits DNA synthesis and stimulates cellular hypertrophy through a Ca(2+)-dependent mechanism. Brain Res. 720, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser KF, Hahn YK, Adjan VV, Zou S, Buch SK, Nath A, BruceKeller AJ, and Knapp PE (2009). HIV-1 Tat and morphine have interactive effects on oligodendrocyte survival and morphology. Glia 57, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, Magida J, Brancato A, Takahashi A, Flanigan ME, et al. (2015). Sex differences in nucleus accumbens transcriptome profiles associated with susceptibility versus resilience to subchronic variable stress. J. Neurosci 35, 16362–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussien Y, Podojil JR, Robinson AP, Lee AS, Miller SD, and Popko B (2015). ER chaperone BiP/GRP78 is required for myelinating cell survival and provides protection during experimental autoimmune encephalomyelitis. J. Neurosci 35, 15921–15933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, and Linnarsson S (2014). Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, and Scadden DT (2014). Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, and Kirschner MW (2015). Droplet barcoding for singlecell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp PE, and Hauser KF (1996). mu-Opioid receptor activation enhances DNA synthesis in immature oligodendrocytes. Brain Res. 743, 341–345. [DOI] [PubMed] [Google Scholar]
- Knapp PE, Maderspach K, and Hauser KF (1998). Endogenous opioid system in developing normal and jimpy oligodendrocytes: mu and kappa opioid receptors mediate differential mitogenic and growth responses. Glia 22, 189–201. [DOI] [PubMed] [Google Scholar]
- Korostynski M, Piechota M, Kaminska D, Solecki W, and Przewlocki R (2007). Morphine effects on striatal transcriptome in mice. Genome Biol. 8, R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhu J, Li Q, Ye J, Chen J, Liu J, Li Z, Li Y, Yan X, Wang Y, and Wang W (2016). Brain white matter integrity in heroin addicts during methadone maintenance treatment is related to relapse propensity. Brain Behav. 6, e00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, and Popko B (2009). Endoplasmic reticulum stress in disorders of myelinating cells. Nat. Neurosci 12, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li L, Hao Y, Cao D, Xu L, Rohrbaugh R, Xue Z, Hao W, Shan B, and Liu Z (2008). Disrupted white matter integrity in heroin dependence: a controlled study utilizing diffusion tensor imaging. Am. J. Drug Alcohol Abuse 34, 562–575. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, and Yang XW (2006). FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci. 9, 443–452. [DOI] [PubMed] [Google Scholar]
- Lun AT, Bach K, and Marioni JC (2016). Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C, and Malenka RC (2011). Drug-evoked synaptic plasticity in addiction: from molecular changes to circuit remodeling. Neuron 69, 650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli M, Aouizerate B, Barrot M, Le Moal M, and Piazza PV (1998). Dopamine-dependent responses to morphine depend on glucocorticoid receptors. Proc. Natl. Acad. Sci. USA 95, 7742–7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, Caccamise A, Werner CT, Viswanathan R, Polanco JJ, Stewart AF, Thomas SA, Sim FJ, and Dietz DM (2018). A novel role for oligodendrocyte precursor cells (OPCs) and Sox10 in mediating cellular and behavioral responses to heroin. Neuropsychopharmacology 43, 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun AT, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Mayoral SR, Nobuta H, Wang F, Desponts C, Lorrain DS, Xiao L, Green AJ, Rowitch D, Whistler J, and Chan JR (2016). Identification of the kappa-opioid receptor as a therapeutic target for oligodendrocyte remyelination. J. Neurosci 36, 7925–7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Koyama Y, Takemoto K, Yoshikawa K, Ishikawa T, Taniguchi M, Inoue K, Aoki M, Hori O, Katayama T, and Tohyama M (2011). Plasma corticosterone activates SGK1 and induces morphological changes in oligodendrocytes in corpus callosum. PLoS ONE 6, e19859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Hattori T, Shimizu S, Ito A, and Tohyama M (2015). Disturbance of oligodendrocyte function plays a key role in the pathogenesis of schizophrenia and major depressive disorder. BioMed Res. Int 2015, 492367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazuelos J, Klingener M, and Aguirre A (2014). TGFβ signaling regulates the timing of CNS myelination by modulating oligodendrocyte progenitor cell cycle exit through SMAD3/4/FoxO1/Sp1. J. Neurosci 34, 7917–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechota M, Korostynski M, Solecki W, Gieryk A, Slezak M, Bilecki W, Ziolkowska B, Kostrzewa E, Cymerman I, Swiech L, et al. (2010). The dissection of transcriptional modules regulated by various drugs of abuse in the mouse striatum. Genome Biol. 11, R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AP, Rodgers JM, Goings GE, and Miller SD (2014). Characterization of oligodendroglial populations in mouse demyelinating disease using flow cytometry: clues for MS pathogenesis. PLoS ONE 9, e107649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas E, Bocos C, Del Castillo C, Pérez-García C, Morales L, and Alguacil LF (2011). Gene expression analysis of heat shock proteins in the nucleus accumbens of rats with different morphine seeking behaviours. Behav. Brain Res 225, 71–76. [DOI] [PubMed] [Google Scholar]
- Sanchez ES, Bigbee JW, Fobbs W, Robinson SE, and Sato-Bigbee C (2008). Opioid addiction and pregnancy: perinatal exposure to buprenorphine affects myelination in the developing brain. Glia 56, 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, and Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA 106, 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Wagatsuma A, Noro Y, Kuji T, Asaka-Oba A, Watahiki A, Gurnot C, Fagiolini M, Hensch TK, and Carninci P (2012). Trehaloseenhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques 52, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, George R, and Garcia J (1975). Acute morphine effects on regional brain amines, growth hormone and corticosterone. Eur. J. Pharmacol 34, 21–26. [DOI] [PubMed] [Google Scholar]
- Skupio U, Sikora M, Korostynski M, Wawrzczak-Bargiela A, Piechota M, Ficek J, and Przewlocki R (2017). Behavioral and transcriptional patterns of protracted opioid self-administration in mice. Addict. Biol 22, 1802–1816. [DOI] [PubMed] [Google Scholar]
- Slezak M, Korostynski M, Gieryk A, Golda S, Dzbek J, Piechota M, Wlazlo E, Bilecki W, and Przewlocki R (2013). Astrocytes are a neural target of morphine action via glucocorticoid receptor-dependent signaling. Glia 61, 623–635. [DOI] [PubMed] [Google Scholar]
- Stiene-Martin A, and Hauser KF (1991). Glial growth is regulated by agonists selective for multiple opioid receptor types in vitro. J. Neurosci. Res 29, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiene-Martin A, Knapp PE, Martin K, Gurwell JA, Ryan S, Thornton SR, Smith FL, and Hauser KF (2001). Opioid system diversity in developing neurons, astroglia, and oligodendroglia in the subventricular zone and striatum: impact on gliogenesis in vivo. Glia 36, 78–88. [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SA, Sim S, Neff NF, Skotheim JM, Wernig M, and Quake SR (2016). Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay J, Maleki N, Potter J, Elman I, Rudrauf D, Knudsen J, Wallin D, Pendse G, McDonald L, Griffin M, et al. (2010). Alterations in brain structure and functional connectivity in prescription opioid-dependent patients. Brain 133, 2098–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten LHG (2008). Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Vestal-Laborde AA, Eschenroeder AC, Bigbee JW, Robinson SE, and Sato-Bigbee C (2014). The opioid system and brain development: effects of methadone on the oligodendrocyte lineage and the early stages of myelination. Dev. Neurosci 36, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, and Morales M (2015). The brain on drugs: from reward to addiction. Cell 162, 712–725. [DOI] [PubMed] [Google Scholar]
- Volpi VG, Touvier T, and D’Antonio M (2017). Endoplasmic reticulum protein quality control failure in myelin disorders. Front. Mol. Neurosci 9, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li W, Li Q, Yang W, Zhu J, and Wang W (2011). White matter impairment in heroin addicts undergoing methadone maintenance treatment and prolonged abstinence: a preliminary DTI study. Neurosci. Lett 494, 49–53. [DOI] [PubMed] [Google Scholar]
- Zezula J, Casaccia-Bonnefil P, Ezhevsky SA, Osterhout DJ, Levine JM, Dowdy SF, Chao MV, and Koff A (2001). p21cip1 is required for the differentiation of oligodendrocytes independently of cell cycle withdrawal. EMBO Rep. 2, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhu J, Liu Y, Chen Y, Li Y, Chen S, Li T, Dang Y, and Chen T (2015). Chronic methamphetamine regulates the expression of MicroRNAs and putative target genes in the nucleus accumbens of mice. J. Neurosci. Res 93, 1600–1610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw and processed sequencing data reported in this paper is GEO: GSE118918