Abstract
Background & Aims
Hepatocellular carcinoma (HCC) typically arises in fibrotic or cirrhotic livers, which are characterized by pathogenic angiogenesis. Myeloid immune cells, specifically tumor-associated macrophages (TAMs), may represent potential novel therapeutic targets in HCC, complementing current ablative or immune therapies. However, the detailed functions of TAM subsets in hepatocarcinogenesis have remained obscure.
Methods
TAM subsets were analyzed in-depth in human HCC samples and a combined fibrosis–HCC mouse model, established by i.p. injection with diethylnitrosamine after birth and repetitive carbon tetrachloride (CCl4) treatment for 16 weeks. Based on comprehensively phenotyping TAM subsets (fluorescence-activated cell sorter, transcriptomics) in mice, the function of CCR2+ TAM was assessed by a pharmacologic chemokine inhibitor. Angiogenesis was evaluated by contrast-enhanced micro–computed tomography and histology.
Results
We show that human CCR2+ TAM accumulate at the highly vascularized HCC border and express the inflammatory marker S100A9, whereas CD163+ immune-suppressive TAM accrue in the HCC center. In the fibrosis–cancer mouse model, we identified 3 major hepatic myeloid cell populations with distinct messenger RNA profiles, of which CCR2+ TAM particularly showed activated inflammatory and angiogenic pathways. Inhibiting CCR2+ TAM infiltration using a pharmacologic chemokine CCL2 antagonist in the fibrosis–HCC model significantly reduced pathogenic vascularization and hepatic blood volume, alongside attenuated tumor volume.
Conclusions
The HCC microenvironment in human patients and mice is characterized by functionally distinct macrophage populations, of which the CCR2+ inflammatory TAM subset has pro-angiogenic properties. Understanding the functional differentiation of myeloid cell subsets in chronically inflamed liver may provide novel opportunities for modulating hepatic macrophages to inhibit tumor-promoting pathogenic angiogenesis.
Keywords: Tumor-Associated Macrophages, Fibrosis, HCC, Chemokine, Therapy, Angiogenesis
Abbreviations used in this paper: CCL2, C-C motif chemokine ligand 2; CCL2i, inhibitor for C-C motif chemokine ligand 2; CCR2, C-C motif chemokine receptor 2; DEN, diethylnitrosamine; HCC, hepatocellular carcinoma; KC, Kupffer cell; MHCII, major histocompatibility complex class II; MI, myeloid infiltrate; MoMF, monocyte-derived macrophage; mRNA, messenger RNA; μ-CT, micro–computed tomography; TAM, tumor-associated macrophage; 3D, 3-dimensional
Graphical abstract
See editorial on page 293.
Summary.
Hepatocellular carcinoma typically arises in livers affected by long-standing chronic injury, persistent inflammation, and tissue scarring (fibrosis). We herein define in human samples and in a fibrosis–cancer mouse model a specialized population of infiltrating inflammatory cells, termed macrophages, that constitute the tumor environment and provide molecular signals for blood vessel formation to support tumor growth, termed angiogenesis. Inhibiting the macrophage infiltration of fibrotic livers by blocking chemotactic signals may provide a new therapeutic strategy to suppress pathogenic angiogenesis.
Liver cancer is the second leading cause of cancer-related death, with a dramatically increasing incidence in industrialized countries.1 Hepatocellular carcinoma (HCC) usually occur in patients with cirrhosis, in which persistent inflammation, hepatocyte cell death, and compensatory regeneration, as well as excessive fibrosis contribute to a tumor-promoting microenvironment.2 Enhanced angiogenesis with the formation of a pathogenic vasculature is a common feature of liver fibrosis3 and is considered a key component of a tumor-promoting or tumor-sustaining stroma.4 We previously showed that pathogenic angiogenesis in hepatic fibrosis is driven by liver-infiltrating, monocyte-derived macrophages that are being recruited to chronically injured livers via the C-C motif chemokine receptor 2 (CCR2).5 It therefore is conceivable that modulating hepatic macrophages could effectively remodel the tumor microenvironment,6, 7 which may prevent HCC development or may enhance the efficacy of chemotherapy, immune checkpoint inhibition, or anti-angiogenic therapies.8, 9
The tumor microenvironment contains multiple myeloid cell types with overall immune-suppressive functions, including neutrophils, dendritic cells, and tumor-associated macrophages (TAMs).10 Analogous to the classification of Th1 and Th2 T-helper cell subsets, TAMs often are characterized as ‘alternatively activated’ M2 type macrophages because of their expression of CD163, T-cell suppression, and release of anti-inflammatory cytokines.11 However, experimental models of different types of solid tumors identified heterogeneous subpopulations of TAMs, some of which also express a ‘classically activated’ inflammatory M1 signature.12 Nonetheless, a high density of TAMs have been related consistently to poor patient prognosis in many types of cancer,13 including observations in human HCC.14, 15 In line with this, high expression of the C-C motif chemokine ligand 2 (CCL2), the ligand for CCR2, in the tumor and its surrounding tissue has been linked to poor survival in a large cohort of patients with HCC,16 indicating that CCR2+ TAM may promote liver cancer. In experimental animal models of HCC, however, macrophages can exert protumoral as well as antitumoral functions during hepatocarcinogenesis.17 For instance, CCR2+ monocyte-derived macrophages can suppress early tumor formation by clearing senescent hepatocytes, although they may promote hepatocarcinogenesis in established tumors.18 Similarly, inhibiting CCR2+ macrophage recruitment reduced the tumor burden in models of subcutaneous implantation of hepatoma cells into mice.16, 19 In agreement, inhibition of CCL2 by a neutralizing antibody also reduced the spontaneous development of liver tumors in microRNA122-deficient mice.20
In this study, we set out to dissect the TAM subtypes involved in HCC progression with a particular focus on characterizing the role of CCR2+ monocyte-derived TAMs in the tumor-prone environment of fibrotic livers. We show that CCR2+ TAMs co-localize with inflammatory markers and pathogenic vasculature at the tumor border in human HCC, whereas suppressive CD163+ CCR2- TAMs are found primarily within tumor nodes. We used a combined mouse model of endogenous liver cancer, which is initiated by diethylnitrosamine (DEN) exposure after birth and accelerated by repetitive injections of carbon tetrachloride (CCl4) causing liver fibrosis. We show that pharmacologic inhibition of the chemokine CCL2 reduced CCR2+ TAMs, alongside significantly reduced pathogenic vascularization, hepatic blood volume, and, subsequently, tumor volume. Detailed characterization of TAM subsets from the fibrosis–cancer model identified a functional heterogeneity among 3 major subsets, with angiogenesis-promoting features in the CCR2+ TAMs, thereby indicating the potential of targeting macrophage subsets to prevent or reduce hepatocarcinogenesis in patients suffering from liver fibrosis.
Results
CCR2+ TAMs Accumulate in the Stroma of Human HCC and Co-localize With Pathogenic Vascularization
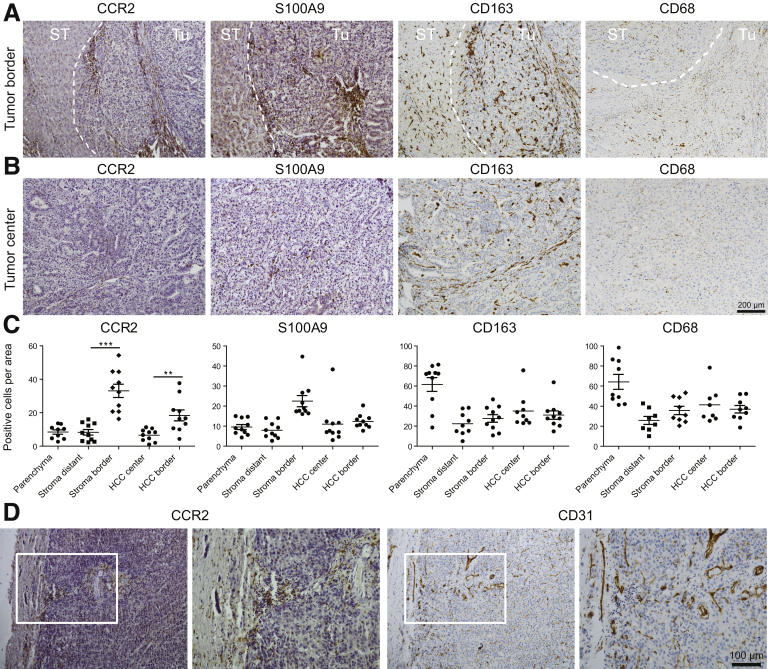
The relevance of TAM for the progression of human HCC has been proposed from studies linking the accumulation of stromal macrophages,14 activated monocytes,15 and the expression of the chemoattractant CCL216 to a decreased survival rate in patients with HCC. Given the heterogeneity of TAMs in experimental cancer models,21 we first analyzed TAM subsets in sections of resected human HCC by staining for the chemokine receptor CCR2, the inflammatory marker S100 calcium binding protein A9 (S100A9), the anti-inflammatory marker CD163, and the pan-macrophage marker CD68 (see Table 1 for patient characteristics). CCR2+ TAMs significantly accumulated at the interface between tumor and surrounding tissue, and were found to infiltrate the epithelial tumor tissue (Figure 1A). The CCR2+ cells at the tumor border showed a round monocyte-like morphology, and a substantial fraction of them expressed S100A9, indicating an inflammatory polarization.22 Fewer CCR2 or S100A9 expressing TAMs were found at the tumor center compared with the border, whereas CD163+ and CD68+ cells were numerous in the center as well and showed a stretched morphology (Figure 1B), in agreement with a mature, suppressive macrophage subset.23 Statistical evaluation confirmed that the CCR2+ TAMs were significantly enriched at the stroma and HCC borders, similar to the S100A9+ TAMs. In contrast, the CD163+ as well as the CD68+ TAMs were more numerous in parenchyma and distributed more evenly throughout tumor and stroma areas (Figure 1C). Because these data indicated a specific accumulation of CCR2+ TAMs at the stroma/tumor-interface, we also stained for the endothelial marker CD31, which largely marks pathogenic vessel formation in fibrotic liver.5 CCR2+ TAMs clearly co-localized with CD31+ endothelial cells in areas of dense vascularization (Figure 1D).
Table 1.
Characterization of the Study Cohort Comprising Human HCC Patients
| Parameters | HCC patients |
|---|---|
| Number, n | 10 |
| Sex, male/female, n | 8/2 |
| Age, median (range), y | 63 (57–79) |
| Body mass index, median (range), kg/m2 | 27.8 (20–35) |
| Type 2 diabetes, n (%) | 3 (30) |
| Cirrhosis, n (%) | 9 (90) |
| Child score, median (range), points | 5 (5–10) |
| Disease etiology | |
| Alcoholic liver disease, n (%) | 3 (30) |
| Nonalcoholic steatohepatitis, n (%) | 1 (10) |
| Viral hepatitis (HBV/HCV), n (%) | 5 (50) |
| Genetic or cryptogenic, n (%) | 2 (20) |
| BCLC stage A/B/C, n (%) | A 7 (70)/B 0 (0)/C 3 (30) |
HBV, hepatitis B virus; HCV, hepatitis C virus; BCLC, Barcelona Clinic Liver Cancer.
Figure 1.
Hepatic tumor-associated macrophages and colocalization with endothelial cells in human liver cancer. Human HCC resection specimens were stained for CCR2, S100A9, CD163 (subsets of macrophages), and CD31 (endothelial cells). (A) Representative sections of human HCC with surrounding noncancerous tissue showing the different localizations of TAMs. Dashed line indicates border between tumor (Tu) and surrounding tissue (ST). (B) Representative example of TAMs in tumor center regions. (C) Statistical evaluation of TAM locations in stromal and cancerous regions. (D) Colocalization of CCR2+ and CD31+ cells at the tumor border. Data represent means ± SD of N = 10. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, calculated for intraregion analysis (HCC or stroma) (1-way analysis of variance). ST, surrounding tissue; Tu, tumor.
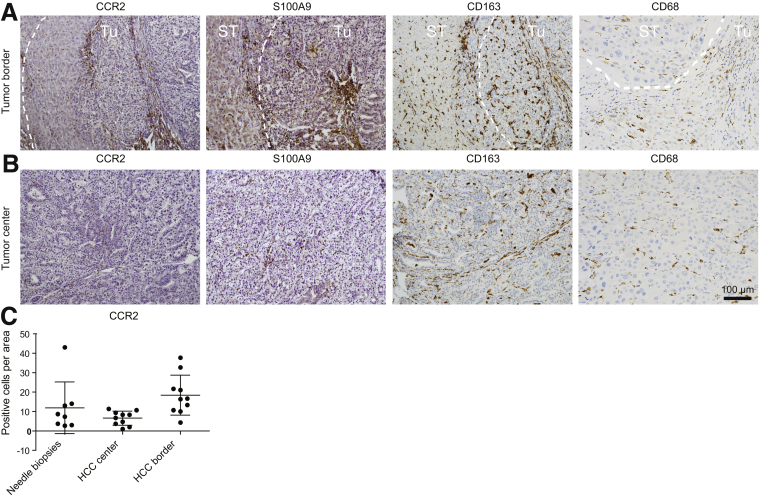
Enlarged views on the different tissue sections clarify the distinct differences in cell morphology between macrophages in the tumor border (mostly round) (Figure 2A) and tumor center (stretched) (Figure 2B). Importantly, we excluded that the infiltration of immune cells was triggered by the surgical resection because similar numbers of CCR2+ cells were found in needle biopsy specimens (Figure 2C).
Figure 2.
Subpopulations of tumor-associated macrophages in human liver cancer. Human HCC resection specimens (n = 10) and needle biopsy specimens (n = 8) were stained for CCR2, S100A9, and CD163. (A) Representative sections of human HCC with surrounding noncancerous tissue showing the different localizations of TAMs. (B) Enlarged view (representative example) of TAMs in tumor center regions. (C) Quantifications of CCR2 in needle biopsy specimens (mostly reflecting tumor center) and resections (separated for HCC center and border, as separated by white dashed lines).
CCL2 Inhibition Reduces Hepatic Blood Volume and Tumor-Supporting Pathogenic Angiogenesis
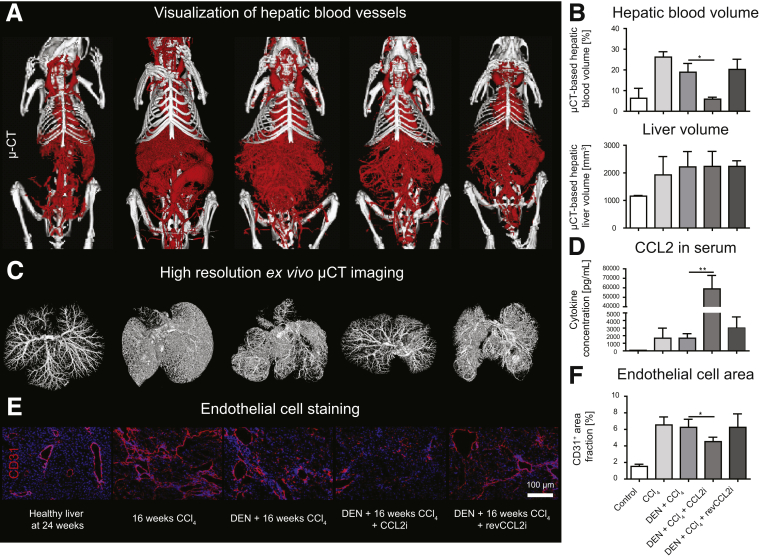
To study the role of CCR2+ TAMs in tumor progression, we used a mouse model for autochthonous liver cancer in fibrotic liver, in which multifocal tumor formation was initiated by DEN and accelerated by fibrosis induction as a result of repetitive injections of CCl4.24 Inhibition of the CCR2-dependent TAMs was achieved by the RNA aptamer-based inhibitor for CCL2 (CCL2i), mNOX-E36, over 16 weeks, which we have used before in models of chronic liver injury.5 The vascularization of the liver (and other organs) was assessed by contrast-enhanced micro–computed tomography (μ-CT)–based methodology. CCL2 inhibition led to a remarkable reduction in the vascular bed of the liver and, consequently, the hepatic blood volume, as clearly visible in 3-dimensional (3D) reconstructions of the in vivo imaging (Figure 3A, Supplementary Videos 1–5 provide 3D reconstructed livers). Quantification of the hepatic blood volume showed that the liver volume remained similar, whereas the hepatic blood volume was reduced significantly by pharmacologic CCL2 inhibition (Figure 3B). These observations were confirmed by high-resolution ex vivo μ-CT scans of tumor-bearing fibrotic livers, by which livers from DEN-CCl4–treated mice were able to preserve the vascular structure upon CCL2 inhibition (Figure 3C). Measurement of CCL2 serum levels confirmed strong target engagement of the inhibitor by significantly increased CCL2 in serum (Figure 3D), which consists of inactive CCL2/mNOX-E36 complexes.25 In agreement with reduced hepatic blood volume, the staining pattern for CD31+ endothelial cells was partially abrogated in CCL2i-treated mice (Figure 3E), corresponding to significantly reduced numbers of hepatic endothelial cells (Figure 3F). Importantly, CCL2 inhibition does not affect proliferation of primary hepatic endothelial cells directly or angiogenic sprouting of blood vessels in vitro,5 indicating that CCR2+ TAMs promote pathogenic angiogenesis.
Figure 3.
CCL2 inhibition in a combined fibrosis–HCC model results in reduced hepatic angiogenesis in vivo. Mice received vehicle (corn oil) and were left either untreated for 24 weeks; received 16 weeks of twice-weekly injections of CCl4; received DEN at 14 days of age and were challenged with CCl4 for 16 weeks; were treated with DEN and CCl4 and received CCL2i mNOXE36 3 times/wk subcutaneously; or received DEN, CCl4, and revCCL2i. Mice were killed 48 hours after the last injection of oil or CCl4. (A) 3D volume renderings of hepatic blood vessels using an iodine-based blood pool contrast agent (eXIA160XL), resulting in a spatial resolution of 35-μm voxel side length. (B) μ-CT–based quantifications of relative hepatic blood volume and liver volume. (C) High-resolution ex vivo μ-CT imaging (after perfusion with Microfil, a lead-containing radiopaque contrast agent) enables a detailed 3D examination of the vascular microarchitecture. (D) CCL2 levels were analyzed from serum by a cytokine multiplex assay. (E) Immunofluorescence staining of liver cryosections for the endothelial cell marker CD31 and (F) quantification of the CD31+ cells based on the area fraction. Data represent means ± SD of N = 6. *P < .05 (1-way ANOVA).
Inhibition of CCL2 Affects Tissue Remodeling and Tumor Volume in the Combined Fibrosis–Cancer Model
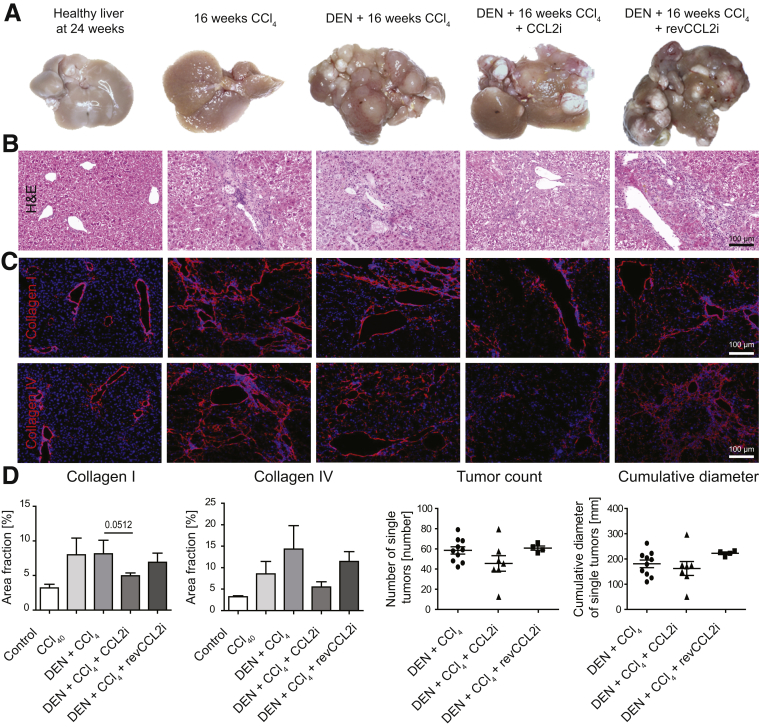
Macroscopic analysis of livers from mice subjected to the combined fibrosis–HCC model showed that CCL2 inhibition led to a slight reduction in the number of hepatic tumors (Figure 4A). Treatment with CCl4 alone did not induce tumors, and the inactive control substance (reverse sequence, revmNOX-E36, revCCL2i) did not affect tumor number (Figure 4A). Histologic analysis based on H&E staining showed a less disturbed tissue architecture in the CCL2i-exposed animals compared with DEN and CCl4 or revCCL2i (Figure 4B). Staining of collagen I (indicative for fibrosis) and collagen IV (strongly expressed in liver tumors) showed reduced extracellular matrix fibers by CCL2 inhibition (Figure 4C), although these differences did not reach statistical significance (Figure 4D). Macroscopic evaluation further showed a tendency toward reduced tumor burden (Figure 4D).
Figure 4.
Effects of CCL2 inhibition on the progression of liver cancer. Mice received vehicle (corn oil) and were left either untreated for 24 weeks, or received 16 weeks of twice-weekly injections of CCl4; received DEN at 14 days of age and were challenged with CCl4 for 16 weeks; were treated with DEN and CCl4 and received CCL2i mNOXE36 3 times/wk subcutaneously; or received DEN, CCl4, and revCCL2i. Mice were killed 48 hours after the last injection of oil or CCl4. (A) Representative macroscopic analysis of the different conditions (valid for all subfigures). (B) H&E staining. (C) Immunofluorescence staining of collagen I and IV. (D) Quantification of the collagen I and IV area fraction and of the cumulative tumor diameter from macroscopy. Data represent means ± SD of N = 6.
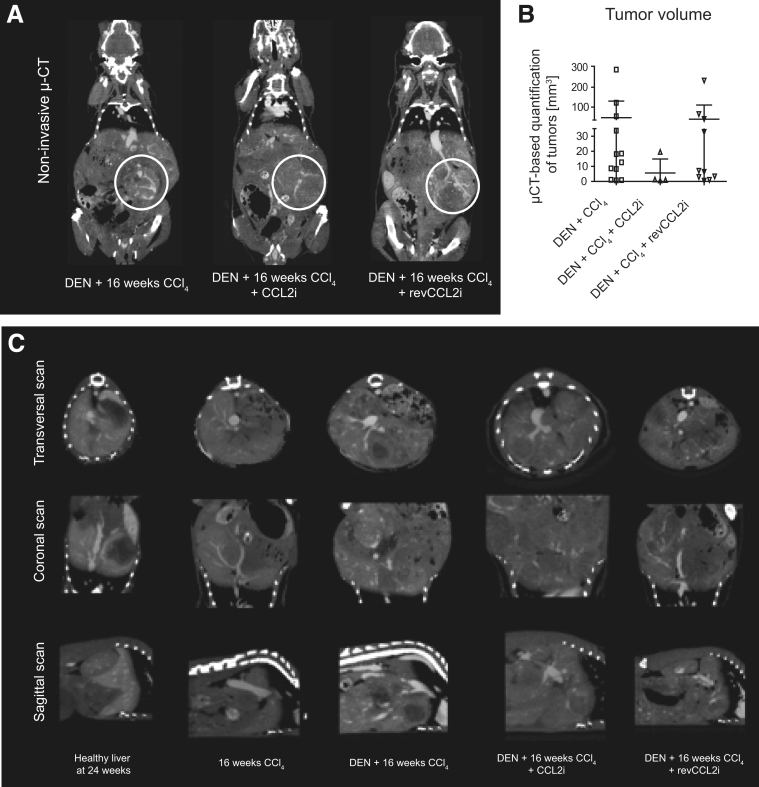
Most interestingly, contrast-enhanced μ-CT–based evaluation of intrahepatic tumors of 2 representative animals showed that CCL2 inhibition led to tumor nodules with central necrosis (Figure 5A, Supplementary Videos 6–10 show representative CT scans through the livers), alongside reduced central tumor perfusion compared with DEN-CCl4– or revCCL2i-treated mice. This corresponded to an overall reduced volume of radiologically measured intrahepatic tumors (Figure 5B), which was based on 3D reconstructions of conventional 2-dimensional CT scans (transversal, coronal, and sagittal) (representative pictures are shown in Figure 5C). Collectively, these data indicate that CCR2+ TAMs remodel liver tissue and their inhibition leads to liver tissue preservation and reduced tumor progression.
Figure 5.
Effects of CCL2 inhibition on the tumor volume in the fibrosis–cancer model. Mice received DEN at 14 days of age and were challenged with CCl4 for 16 weeks; treated with DEN and CCl4 and received CCL2i mNOXE36 3 times/wk subcutaneously; or received DEN, CCl4, and revCCL2i. Mice were killed 48 hours after the last injection of oil or CCl4. (A) Two-dimensional coronal scans of 2 representative animals for intrahepatic tumor volume quantification, white circles label liver tumor nodules. (B) Quantifications of the tumor volumes calculated from μ-CT data. (C) Conventional transversal, coronal, and sagittal CT scans of the liver tumors (representative pictures).
Impact of CCL2 Inhibition on TAM Composition in the Fibrosis–Cancer Model
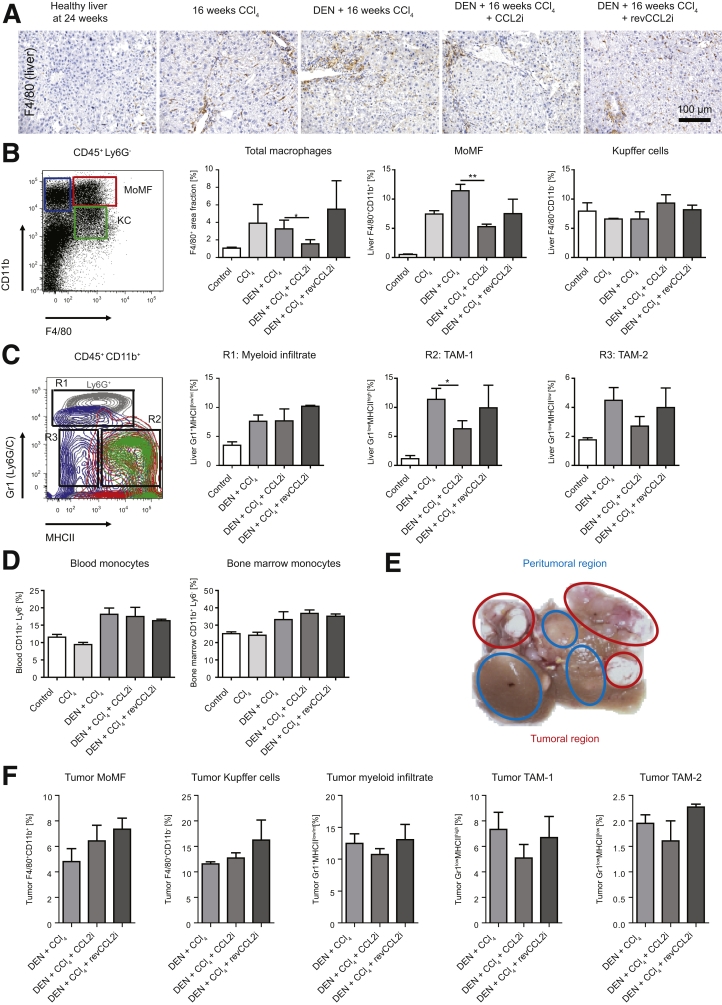
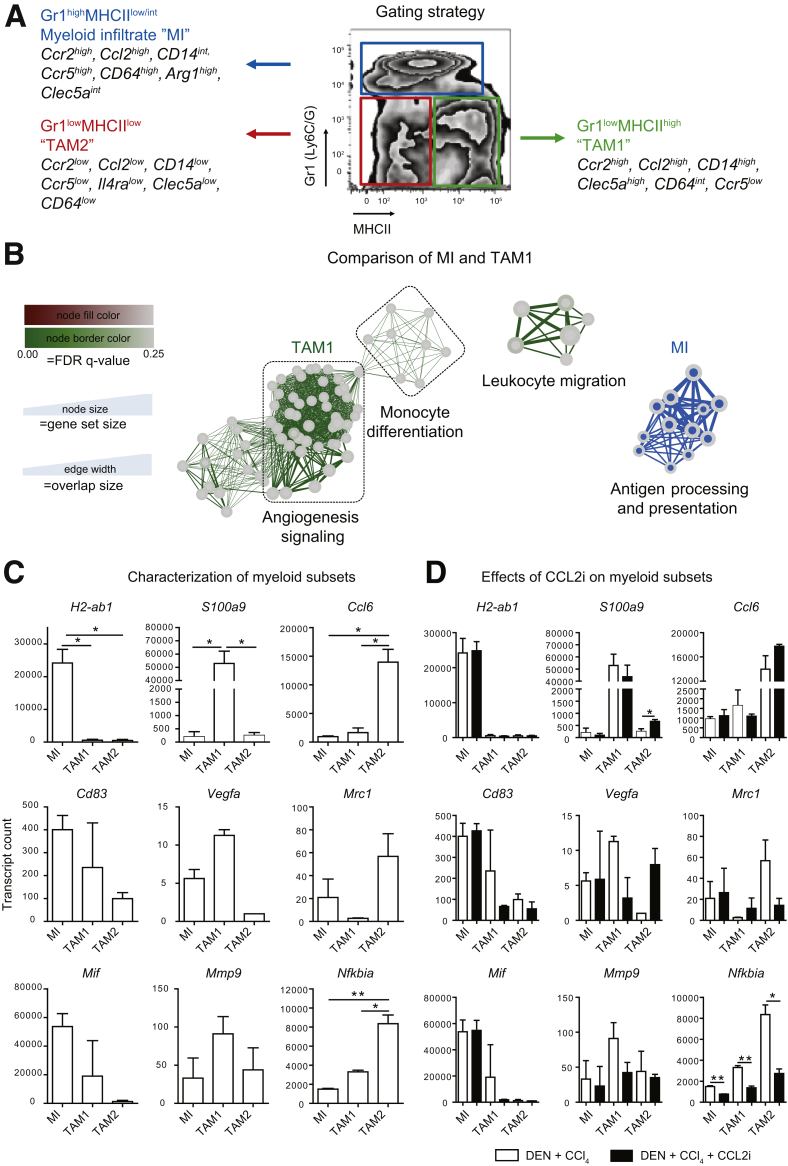
To link the effects of CCL2 inhibition on tissue remodeling, tumor volume, and angiogenesis to specific TAM populations, we analyzed the hepatic immune cell composition in the fibrosis–HCC model in peritumoral tissue. Immunohistochemical staining of the pan-macrophage marker F4/80 showed a clear increase in hepatic macrophages induced by CCl4, DEN-CCl4, and the respective revCCL2i mice compared with healthy livers. In contrast, F4/80+ cells were strongly reduced by CCL2i treatment (Figure 6A and B). When hepatic macrophages were classified as CD45+ Ly6G- cells according to their expression of F4/80 and CD11b by flow cytometry,26 CD11b+F4/80+ monocyte-derived macrophages (MoMFs) were reduced significantly by CCL2i, whereas CD11bintF4/80+ Kupffer cells (KCs) remained unaffected by CCL2 inhibition (Figure 6B). Because different subsets of myeloid cells, including neutrophils, are functionally important in the tumor microenvironment, we applied a different flow-cytometric gating strategy that differentiated CD45+ CD11b+ myeloid cells further by their expression of Gr1 (Ly6C/G) and major histocompatibility complex class II (MHCII).27, 28 This analysis identified a myeloid infiltrate containing Ly6C+ monocytes and Ly6G+ neutrophils (R1) as well as 2 populations of TAMs (R2 and R3) (Figure 6C). The Gr1lowMHCIIhigh TAM population, herein termed TAM1, contained MoMF and KCs as per a traditional gating strategy and was reduced upon CCL2 inhibition (R2) (Figure 6C). The Gr1lowMHCIIlow TAM population, herein termed TAM2, differed from conventional macrophages in non–tumor-containing livers and was only slightly affected by CCL2 inhibition (R3) (Figure 6C). The fibrosis–tumor model leads to a general induction of monocytes in blood and bone marrow. In contrast, the CCL2i specifically reduced myeloid populations in the liver, but did not affect blood and bone marrow monocytes (Figure 6D).
Figure 6.
Analysis of hepatic tumor–associated myeloid cells. Mice received vehicle (corn oil) and were left either untreated for 24 weeks; received 16 weeks of twice-weekly injections of CCl4; received DEN at 14 days of age and were challenged with CCl4 for 16 weeks; were treated with DEN and CCl4 and received CCL2i mNOXE36 3 times/wk subcutaneously; or received DEN, CCl4, and revCCL2i. Mice were killed 48 hours after the last injection of oil or CCl4. (A) Immunohistochemical staining of F4/80+ hepatic macrophages in peritumoral regions. (B) Gating of the hepatic peritumoral macrophage subsets (pregated on CD45+ and subsequently Ly6G-), MoMFs identified as CD11b+F4/80+ and KCs identified as CD11bintF4/80+, and quantifications of the subsets in 3 different experiments. (C) Allocation of peritumoral MoMF (red circle) and KC (green circle) to subsets of TAMs (pregated as CD45+CD11b+) by back-gating analysis. Myeloid cells include a myeloid infiltrate (composed of monocytes and neutrophils) scoring Gr1highMHCIIlow/int, proinflammatory TAM1 expressing Gr1lowMHCIIhigh, and anti-inflammatory TAM2, which are Gr1lowMHCIIlow. (D) Effects on CD45+CD11b+Ly6G- monocytes in blood and bone marrow. (E) Selection of peritumoral (blue outline) and tumoral tissue (red outline) for flow cytometry, and (F) quantifications of the myeloid subsets in the tumor tissue. Data represent means ± SD of N = 6. ∗P < .05, ∗∗P < .01 (1-way analysis of variance).
We further analyzed the myeloid cells directly from the tumor tissue (Figure 6E). Quantifications of the cellular subsets showed that, compared with the peritumoral tissue, the MoMF, TAM1, and TAM2 were reduced, whereas the KCs and myeloid infiltrate (MI) were increased in the tumor regions. However, the changes induced in the cell populations triggered by CCL2i and revCCL2i were similar to the peritumoral tissue (Figure 6F).
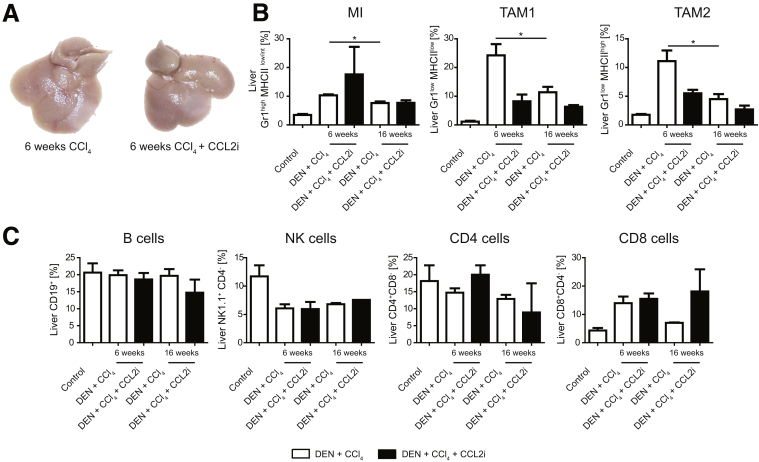
Interestingly, already after 6 weeks of treatment with CCl4, before tumors became visible (Figure 7A), the myeloid cell populations (Figure 7B), but not the lymphoid cells, were increased significantly compared with the 16-week time point (Figure 7C), corroborating the central role of myeloid cells during fibrogenesis and early stages of tumor development.
Figure 7.
Myeloid and lymphoid cells at early stages of cancer development. Mice at 14 days of age received DEN and twice-weekly injections of CCl4 for 6 weeks. In addition, mice were treated with CCL2i mNOXE36 or revCCL2i 3 times/wk subcutaneously. Mice were killed 48 hours after the last injection of CCl4, and immune cells were isolated. (A) Macroscopic analysis of livers, showing no tumors at this time point. (B) Flow cytometric analysis of myeloid and (C) lymphoid liver leukocytes. Data represent means ± SD of N = 6. *P < .05 (1-way analysis of variance).
CCR2+ TAMs Show an Inflammatory and Angiogenic Phenotype
To classify the functionality of the TAM subsets in more detail, we subjected highly purified populations of MI, TAM1, and TAM2 after fluorescence-activated cell sorting from livers of the fibrosis–cancer mouse model to Nanostring (Seattle, WA) arrays including 561 immunology-related messenger RNA (mRNA) transcripts. The different myeloid cell subsets significantly differed in their mRNA expression of typical macrophage markers (summarized in Figure 8A). Using unbiased gene set enrichment analysis, the pathways most strongly regulated in the TAM1 subset included mRNAs involved in angiogenesis signaling and monocyte differentiation, whereas the MI (which also express Ccr2) prominently up-regulated mRNAs involved in antigen processing and presentation (Figure 8B). In detail, the MI significantly up-regulated mRNA associated with the immunologic synapse such as H2-ab1 and Cd83, and highly up-regulated the pleiotropic marker macrophage migration inhibitory factor. Key mRNA up-regulated by TAM1 were inflammatory mediators such as S100a9 and Il1β, but also Vegfa and matrix metalloproteinase 9 (Mmp9), which are associated with initiating angiogenesis in carcinogenesis. TAM2 expressed the Ccl6 chemokine, which is involved in immune cell recruitment, and nuclear factor-κB inhibitor a (NfκBia), which negatively regulates the important nuclear factor-κB pathway that is associated with many inflammatory processes (Figure 8C). These data suggest that each myeloid subset in liver cancer is involved in specific biological processes. Interestingly, the CCL2i had a stronger effect on the TAM1 and TAM2 subsets than on the MI, despite the fact that the MI also express high levels of CCR2, the ligand of CCL2. In particular, it was noted that the CCL2i-induced down-regulation of specific markers including S100a9 and Vegfa were down-regulated by the CCL2i in TAM1, but up-regulated these in the TAM2 subset by the CCL2i (Figure 8D).
Figure 8.
Analysis of subpopulations of hepatic tumor–associated myeloid cell populations using cell sorting and mRNA profiling. Mice received vehicle (corn oil) and were left either untreated for 24 weeks; received 16 weeks of twice-weekly injections of CCl4; received DEN at 14 days of age and were challenged with CCl4 for 16 weeks; were treated with DEN and CCl4 and received CCL2i mNOXE36 3 times/wk subcutaneously; or received DEN, CCl4, and revCCL2i. Mice were killed 48 hours after the last injection of oil or CCl4. (A) Gating strategy for subsets of peritumoral myeloid cells: a MI composed of monocytes and neutrophils scoring Gr1highMHCIIlow/int, proinflammatory TAM1 being Gr1lowMHCIIhigh, or anti-inflammatory TAM2, which are Gr1lowMHCIIlow. (B) Unbiased quantitative analysis of inflammatory mRNA expression of TAMs. (C) Characteristic mRNA expressed by TAM1. (D) Changes in the mRNA profiles of TAM subsets induced by CCL2i. Data represent means ± SD of 2 independent experiments. *P < .05 (1-way analysis of variance). FDR, false discovery rate.
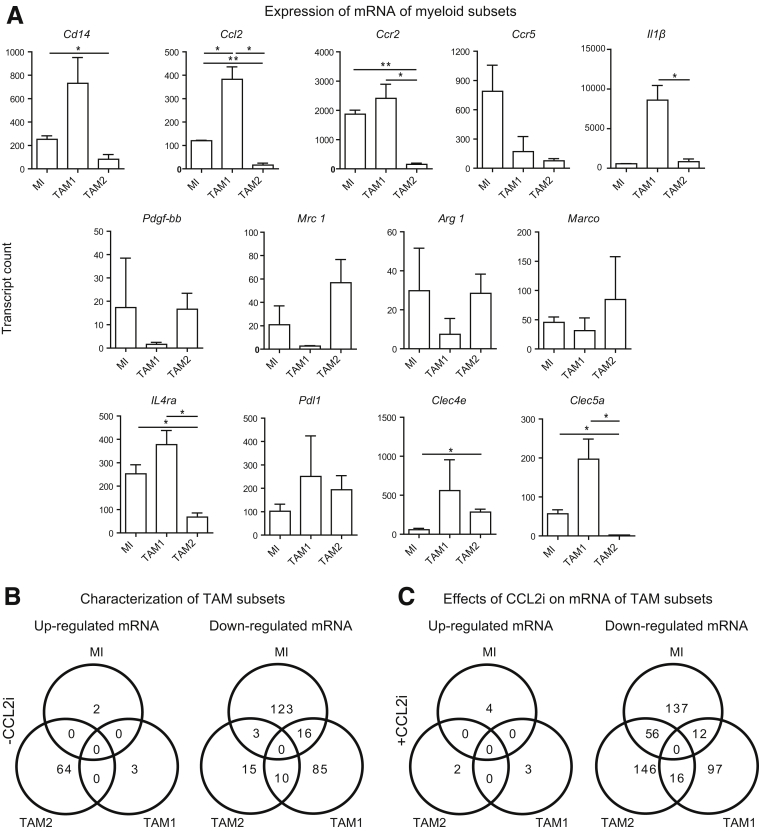
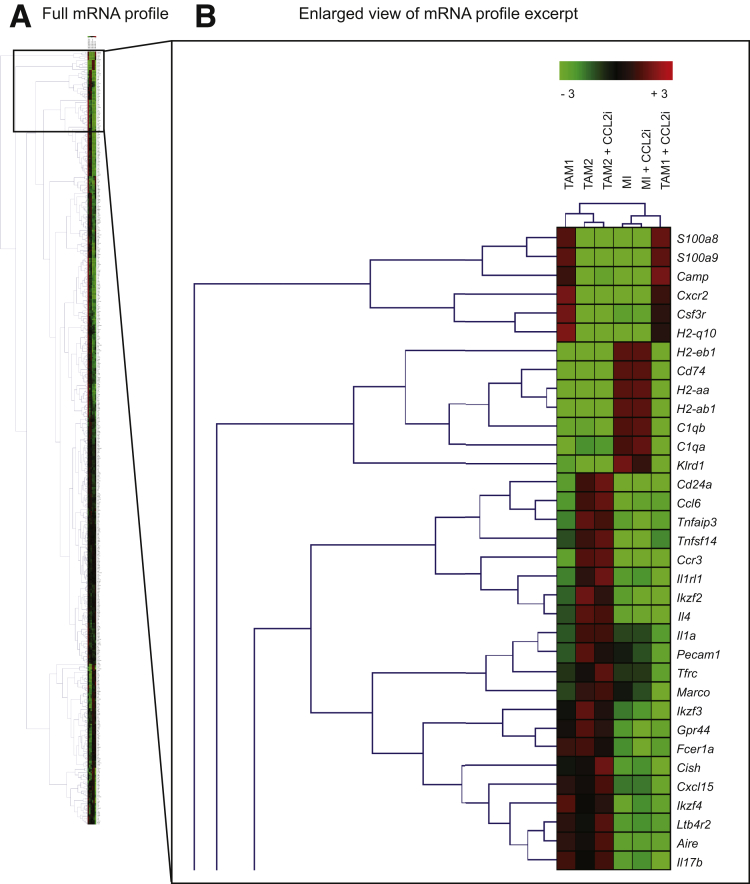
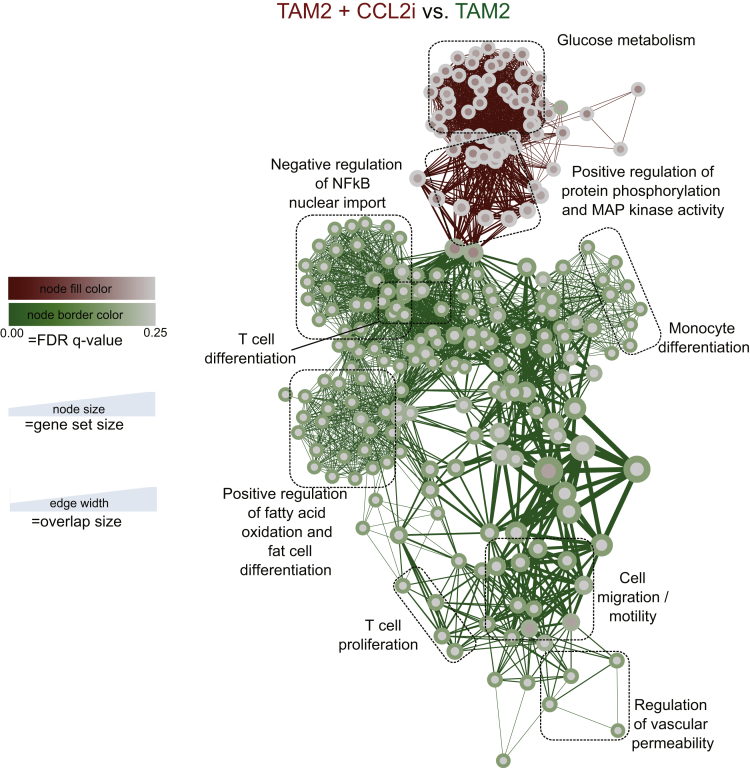
Detailed analyses illustrate the strong differences in the expression of mRNA by the different myeloid cells: although the MI and TAM1 highly expressed Ccr2/Ccl2, the TAM2 expressed very low levels of Ccr2/Ccl2. In addition to strong Ccr2 and Cd14 expression, another hallmark of the TAM1 subset was high Clec5a expression. Notably, the TAM2 subset expressed many myeloid markers (Ccr2, Ccl2, Il4r, Cd64) at a very low level (Figure 9A), but showed markers of a mature, rather immune-suppressive phenotype such as Mannose receptor-1 (Mrc1) or Arginase-1 (Arg1) (Figure 9A). Venn diagrams reflect summaries on the mRNA regulation in the different myeloid subsets and the effects of the CCL2i (Figure 9B and C). Only very few changes occurred in the MI by the CCL2i, but unexpectedly, some mRNAs in TAM2 also were affected (Tables 2 and 3). However, hierarchical clustering analysis (based on the similarity of mRNA expression) identified the most striking changes induced by CCL2i to the TAM1 populations (Figure 10). Key pathways of TAM2 cells that were affected by CCL2i related to inflammatory pathways (Figure 11). These data indicate that CCL2 inhibition promotes characteristic changes in the TAM2 population, too, which could be a compensatory response of TAM2 caused by reduced TAM1 cells or a direct result of CCL2 inhibition (eg, by reducing the cellular precursors of TAM2 from the TAM1 population).
Figure 9.
Expression of mRNA by TAM subsets and Venn diagram. Mice at 14 days of age received DEN and twice-weekly injections of CCl4 for 16 weeks. Peritumoral hepatic myeloid cells were isolated and subjected to quantitative analysis of mRNA expression (Nanostring immunology kit). (A) Changes are shown in selected mRNA of the 3 subsets of myeloid infiltrators, defined as Gr1+MHCIIlow/int, TAM1 as Gr1lowMHCIIhigh, and TAM2, which were classified as Gr1lowMHCIIlow. The MI was defined as Gr1+MHCIIlow/int, TAM1 as Gr1lowMHCIIhigh, and TAM2 as Gr1lowMHCIIlow. Three-fold changes in mean gene expression were used to characterize the subsets. (B) Summary of mRNA expression by the different subsets and (C) of the effects of the CCL2i. Data in panel A represent means ± SD. *P < .05, **P < .01, calculated against DEN–CCl4 disease control (1-way analysis of variance).
Table 2.
List of mRNA Up-regulated by the TAM2 Subset (Myeloid Cells Scoring Gr1lowMHCIIlow), but No Longer Up-regulated Upon Treatment With the CCL2i Subjected for Biocarta-Based Gene Group Analysis
| Gene symbol | Full name |
|---|---|
| Abcb1a | ATP-binding cassette, subfamily B (MDR/TAP), member 1A |
| Btnl1 | Butyrophilin-like 1 |
| C2 | Complement component 2 (within H-2S) |
| C4bp | Complement component 4 binding protein |
| C7 | Complement component 7 |
| C8b | Complement component 8, β polypeptide |
| Ccl26 | Chemokine (C-C motif) ligand 26 |
| Ccr6 | Chemokine (C-C motif) receptor 6 |
| Cd209g | CD209g antigen |
| Cd27 | CD27 antigen |
| Cd5 | CD5 antigen |
| Cd53 | CD53 antigen |
| Cd6 | CD6 antigen |
| Cd96 | CD96 antigen |
| Csf3r | Colony stimulating factor 3 receptor (granulocyte) |
| Cxcl11 | Chemokine (C-X-C motif) ligand 11 |
| Cxcr1 | Chemokine (C-X-C motif) receptor 1 |
| Cxcr2 | Chemokine (C-X-C motif) receptor 2 |
| Eomes | Eomesodermin |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Folr4 | IZUMO1 receptor, JUNO |
| Foxp3 | Forkhead box P3 |
| Frmpd4 | FERM and PDZ domain containing 4 |
| Gp1bb | Glycoprotein Ib, β polypeptide |
| Gzma | Granzyme A |
| H2-Q10 | Histocompatibility 2, Q region locus 10 |
| Icam4 | Intercellular adhesion molecule 4, Landsteiner–Wiener blood group |
| Icam5 | Intercellular adhesion molecule 5, telencephalin |
| Ifitm1 | Interferon-induced transmembrane protein 1 |
| Ifna1 | Interferon α1 |
| Il17a | Interleukin 17A |
| Il18r1 | Interleukin 18 receptor 1 |
| Il18rap | Interleukin 18 receptor accessory protein |
| Il1b | Interleukin 1β |
| Il1r1 | Interleukin 1 receptor, type I |
| Il1r2 | Interleukin 1 receptor, type II |
| Il1rap | Interleukin 1 receptor accessory protein |
| Il21 | Interleukin 21 |
| Il22 | Interleukin 22 |
| Il22ra2 | Interleukin 22 receptor, α 2 |
| Il23a | Interleukin 23, α subunit p19 |
| Il2ra | Interleukin 2 receptor, α chain |
| Il3 | Interleukin 3 |
| Il33 | Interleukin 3 |
| Il9 | Interleukin 9 |
| Itln1 | Intelectin 1 (galactofuranose binding) |
| Kir3dl1 | Killer cell immunoglobulin-like receptor, 3 domains, long cytoplasmic tail, 1 |
| Klra21 | Killer cell lectin-like receptor subfamily A, member 21 |
| Klra5 | Killer cell lectin-like receptor, subfamily A, member 5 |
| Klra6 | Killer cell lectin-like receptor, subfamily A, member 6 |
| Klra8 | Killer cell lectin-like receptor, subfamily A, member 8 |
| Lef1 | Lymphoid enhancer binding factor 1 |
| Masp1 | Mannan-binding lectin serine peptidase 1 |
| Masp2 | Mannan-binding lectin serine peptidase 2 |
| Pla2g2e | Phospholipase A2, group IIE |
| Psmb11 | Proteasome (prosome, macropain) subunit, β type, 11 |
| Rag2 | Recombination activating gene 2 |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) |
| S100a9 | S100 calcium binding protein A9 (calgranulin B) |
| Sele | Selectin, endothelial cell |
| Tnfrsf9 | Tumor necrosis factor receptor superfamily, member 9 |
| Tnfsf15 | Tumor necrosis factor (ligand) superfamily, member 15 |
| Trem1 | Triggering receptor expressed on myeloid cells 1 |
| Xcl1 | Chemokine (C motif) ligand 1 |
NOTE. Mice at 14 days of age received DEN and twice-weekly injections of CCl4 for 16 weeks. In addition, mice were treated with CCL2i mNOXE36 or control inhibitor (revCCL2i) 3 times/wk subcutaneously. Peritumoral hepatic myeloid cells were isolated and subjected to quantitative analysis of mRNA expression (Nanostring immunology kit).
ATP, adenosine triphosphate; IZUMO1, Izumo sperm-egg fusion 1; JUNO, folate receptor 4, folate receptor delta or IZUMO1R; MDR, multidrug resistance; TNF, tumor necrosis factor.
Table 3.
List of mRNA Down-regulated by the TAM2 Subset (Myeloid Cells Scoring Gr1lowMHCIIlow) After Treatment With the CCL2 Inhibitor Subjected for Biocarta-Based Gene Group Analysis
| Gene symbol | Full name |
|---|---|
| Aire | Autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
| Atm | Ataxia telangiectasia mutated |
| Blnk | B-cell linker |
| C1ra | Complement component 1, r subcomponent A |
| C1s | Complement component 1s |
| C2 | Complement component 2 (within H-2S) |
| C4bp | Complement component 4 binding protein |
| C6 | Complement component 6 |
| C7 | Complement component 7 |
| C9 | Complement component 9 |
| Ccl22 | Chemokine (C-C motif) ligand 22 |
| Ccl26 | Chemokine (C-C motif) ligand 26 |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Ccr6 | Chemokine (C-C motif) receptor 6 |
| Ccr7 | Chemokine (C-C motif) receptor 7 |
| Ccr9 | Chemokine (C-C motif) receptor 9 |
| Ccrl1 | C-C chemokine receptor type 11 |
| Cd163 | CD163 antigen |
| Cd19 | CD19 antigen |
| Cd1d1 | CD1d1 antigen |
| Cd209g | CD209g antigen |
| Cd22 | CD22 antigen |
| Cd247 | CD247 antigen |
| Cd28 | CD28 antigen |
| Cd34 | CD34 antigen |
| Cd3d | CD3 antigen, δ polypeptide |
| Cd4 | CD4 antigen |
| Cd40lg | CD40 ligand |
| Cd7 | CD7 antigen |
| Cd83 | CD83 antigen |
| Cd8a | CD8 antigen, α chain |
| Cd96 | CD96 antigen |
| Cdh5 | Cadherin 5 |
| Cfd | Complement factor D (adipsin) |
| Cish | Cytokine inducible SH2-containing protein |
| Ctla4 | Cytotoxic T-lymphocyte–associated protein 4 |
| Cxcl1 | Chemokine (C-X-C motif) ligand 1 |
| Cxcl12 | Chemokine (C-X-C motif) ligand 12 |
| Cxcl15 | Chemokine (C-X-C motif) ligand 15 |
| Defb1 | Defensin β 1 |
| Eomes | Eomesodermin |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Fcer1a | Fc receptor, IgE, high-affinity I, α polypeptide |
| Foxp3 | Forkhead box P3 |
| Gata3 | GATA binding protein 3 |
| Gp1bb | Glycoprotein Ib, β polypeptide |
| Gpr44 | Prostaglandin D2 receptor 2 |
| H2-Ob | Histocompatibility 2, O region β locus |
| H60a | Histocompatibility 60a |
| Hamp | Hepcidin antimicrobial peptide |
| Hcst | Hematopoietic cell signal transducer |
| Icam5 | Intercellular adhesion molecule 5, telencephalin |
| Icos | Inducible T-cell co-stimulator |
| Ifna1 | Interferon α 1 |
| Ikzf2 | IKAROS family zinc finger 2 |
| Ikzf3 | IKAROS family zinc finger 3 |
| Ikzf4 | IKAROS family zinc finger 4 |
| Il10 | Interleukin 10 |
| Il12a | Interleukin 12a |
| Il12rb1 | Interleukin 12 receptor, β 1 |
| Il12rb2 | Interleukin 12 receptor, β 2 |
| Il13 | Interleukin 13 |
| Il17a | Interleukin 17A |
| Il17b | Interleukin 17B |
| Il17f | Interleukin 17F |
| Il1a | Interleukin 1 α |
| Il1r1 | Interleukin 1 receptor, type I |
| Il1rl1 | Interleukin 1 receptor-like 1 |
| Il21 | Interleukin 21 |
| Il23a | Interleukin 23, α subunit p19 |
| Il28a | Interferon λ 2 |
| Il2ra | Interleukin 2 receptor, α chain |
| Il4 | Interleukin 4 |
| Il7r | Interleukin 7 receptor |
| Itga2b | Integrin α 2b |
| Itga6 | Integrin α 6 |
| Itgax | Integrin α X |
| Itln1 | Intelectin 1 (galactofuranose binding) |
| Kir3dl1 | Killer cell immunoglobulin-like receptor, 3 domains, long cytoplasmic tail, 1 |
| Kit | Kit oncogene |
| Klra21 | Killer cell lectin-like receptor subfamily A, member 21 |
| Klra5 | Killer cell lectin-like receptor, subfamily A, member 5 |
| Klrc1 | Killer cell lectin-like receptor subfamily C, member 1 |
| Klrc2 | Killer cell lectin-like receptor subfamily C, member 2 |
| Klrk1 | Killer cell lectin-like receptor subfamily K, member 1 |
| Lilra5 | Leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| Ltb4r2 | Leukotriene B4 receptor 2 |
| Marco | Macrophage receptor with collagenous structure |
| Masp1 | Mannan-binding lectin serine peptidase 1 |
| Masp2 | Mannan-binding lectin serine peptidase 2 |
| Ms4a1 | Membrane-spanning 4-domains, subfamily A, member 1 |
| Nos2 | Nitric oxide synthase 2, inducible |
| Nox1 | NADPH oxidase 1 |
| Nox4 | NADPH oxidase 4 |
| Pdcd1 | Programmed cell death 1 |
| Pdgfb | Platelet-derived growth factor, B polypeptide |
| Pdgfrb | Platelet-derived growth factor receptor, β polypeptide |
| Pecam1 | Platelet/endothelial cell adhesion molecule 1 |
| Pigr | Polymeric immunoglobulin receptor |
| Pla2g2a | Phospholipase A2, group IIA (platelets, synovial fluid) |
| Pparg | Peroxisome proliferator activated receptor γ |
| Prf1 | Perforin 1 (pore-forming protein) |
| Prim1 | DNA primase, p49 subunit |
| Psmb11 | Proteasome (prosome, macropain) subunit, β type, 11 |
| Ptk2 | PTK2 protein tyrosine kinase 2 |
| Rag1 | Recombination activating gene 1 |
| Rorc | RAR-related orphan receptor γ |
| Runx3 | Runt-related transcription factor 3 |
| Sele | Selectin, endothelial cell |
| Sh2d1a | SH2 domain containing 1A |
| Slamf7 | SLAM family member 7 |
| Src | Rous sarcoma oncogene |
| Tbx21 | T-box 21 |
| Tcf7 | Transcription factor 7, T-cell specific |
| Tfrc | Transferrin receptor |
| Tgfb2 | Transforming growth factor, β |
| Thy1 | Thymus cell antigen 1, θ |
| Tigit | T-cell immunoreceptor with Ig and ITIM domains |
| Tlr3 | Toll-like receptor 3 |
| Tnfaip3 | Tumor necrosis factor, α-induced protein 3 |
| Tnfrsf13c | Tumor necrosis factor receptor superfamily, member 13c |
| Tnfrsf17 | Tumor necrosis factor receptor superfamily, member 17 |
| Tnfrsf9 | Tumor necrosis factor receptor superfamily, member 9 |
| Tnfsf10 | Tumor necrosis factor (ligand) superfamily, member 10 |
| Tnfsf11 | Tumor necrosis factor (ligand) superfamily, member 11 |
| Tnfsf14 | Tumor necrosis factor (ligand) superfamily, member 14 |
| Tnfsf15 | Tumor necrosis factor (ligand) superfamily, member 15 |
| Tnfsf8 | Tumor necrosis factor (ligand) superfamily, member 8 |
| Xcl1 | Chemokine (C motif) ligand 1 |
| Xcr1 | Chemokine (C motif) receptor 1 |
| Zap70 | Ζ-chain (TCR)–associated protein kinase |
NOTE. Mice at 14 days of age received DEN and twice-weekly injections of CCl4 for 16 weeks. In addition, mice were treated with CCL2i mNOXE36 or control inhibitor (revCCL2i) 3 times/wk subcutaneously. Peritumoral hepatic myeloid cells were isolated and subjected to quantitative analysis of mRNA expression (Nanostring immunology kit).
GATA, glutaminyl-tRNA synthase; ITIM, immunoreceptor tyrosine-based inhibition motif; NADPH, Nicotinamide adenine dinucleotide phosphate; PTK2, protein tyrosine kinase 2; RAR, retinoic acid receptor; SH2, Src Homology 2; SLAM, signaling lymphocytic activation molecule; TCR, T-cell receptor; TM, transmembrane; TNF, tumor necrosis factor.
Figure 10.
Hierarchical clustering analysis of gene expression of tumor-associated macrophages. Mice at 14 days of age received DEN and twice-weekly injections of CCl4 for 16 weeks. In addition, mice were treated with CCL2i mNOXE36 or control inhibitor revCCL2i 3 times/wk subcutaneously. Peritumoral hepatic myeloid cells were isolated and subjected to quantitative analysis of mRNA expression (Nanostring immunology kit). (A) Changes are shown in selected mRNA of the 3 MI subsets, defined as Gr1+MHCIIlow/int, TAM1 as Gr1lowMHCIIhigh, and TAM2, which were analyzed as Gr1lowMHCIIlow. (B) Enlarged view of the most differentially expressed mRNA.
Figure 11.
Analysis of the effects of CCL2 inhibition on the mRNA expression of anti-inflammatory hepatic tumor–associated macrophages. Mice at 14 days of age received DEN and twice-weekly injections of CCl4 for 16 weeks. In addition, mice were treated with CCL2i mNOXE36 or control inhibitor (revCCL2i) 3 times/wk subcutaneously. Peritumoral hepatic myeloid cells were isolated and subjected to quantitative analysis of mRNA expression (Nanostring immunology kit). (A) Unbiased clustered gene enrichment maps for analysis of the effects of CCL2 inhibition on the Gr1lowMHCIIlow TAM2. (B) Changes in selected mRNA in the TAM2 subset.
Discussion
Tumor-associated macrophages increasingly are recognized as emerging targets to improve anticancer therapies.29 For instance, the reprogramming of TAM toward inflammatory cells via CD40 stimulation and colony-stimulating factor 1 receptor inhibition boosted antitumor immunity in mice.30, 31 Such findings support the notion that myeloid targeting interventions have the potential to augment current cytoreductive approaches such as chemotherapy, radiation, or local ablation, as well as to complement immune checkpoint-inhibitor therapies in many solid tumors.29 HCCs arise almost exclusively in chronically inflamed and fibrotic tissue, in which different macrophage subsets exert key roles in sustaining inflammation, promoting disease progression, and provoking tissue remodeling.17 Although myeloid cells in livers with established tumors typically are immune-suppressive and associated with a poor prognosis,21 the characteristic sequence of long-standing chronic liver disease, fibrosis, and, ultimately, HCC development, offers the potential for chemopreventive approaches by modulating hepatic macrophages.2, 7 Hepatic macrophages, however, show a remarkable heterogeneity. The classic KCs derive from local precursors, whereas a large fraction of macrophages in inflamed liver are descendants of tissue-infiltrating monocytes of bone marrow origin.17 In the current work, we hypothesized that targeting the monocyte-derived macrophage fraction by inhibiting the chemokine CCL2 in a combined liver fibrosis–cancer model in mice would modify progression of primary liver tumors.
In line with previous reports from patient cohorts with HCC,14, 15, 16 we observed CCR2+ macrophages at the interface of tumors and surrounding hepatic tissue in HCC resections. However, these macrophages expressed the inflammatory marker S100A9 rather than CD163, indicating that these TAMs do not belong to the typical suppressive M2-like myeloid population described in human HCC. Strikingly, these CCR2+ TAMs co-localized with newly formed blood vessels, as identified by (pathogenic) CD31 expression. In a combined liver fibrosis–cancer model in mice, pharmacologic inhibition of monocyte recruitment via CCL2 strongly suppressed pathogenic angiogenesis. These data are well in agreement with earlier work showing a critical role of CCR2+ monocyte-derived macrophages for angiogenesis in liver fibrosis.5
Angiogenesis is considered a hallmark of cancer,32 but also is linked to fibrosis progression in the liver.4 Tumor-associated macrophages may promote pathogenic angiogenesis in many types of cancer,33, 34 but conflicting data exist on the exact phenotype of angiogenesis-promoting macrophages. In vitro as well as after implantation into mice, M2-polarized macrophages had a higher angiogenic potential than their M1-polarized counterparts.35 In a similar direction, CD163-expressing macrophages were associated with plaque-related angiogenesis in human and murine atherosclerosis,36 and repolarization of TAMs toward an M1-phenotype normalized aberrant blood vessels in a murine melanoma model.37 Our data corroborate that the traditional M1/M2 paradigm is not suitable to apprehend the heterogeneity and functional diversity of macrophages in the environment of a chronically injured liver. By comparing 3 major myeloid cell compartments from tumor-bearing fibrotic livers, based on surface characteristics proposed for TAMs,28 it became apparent that functional pathways related to inflammation, angiogenesis, and immunity were regulated differentially between the 3 subsets. Although the CCR2-dependent TAM1 population activated pathways of angiogenesis as well as specific inflammatory markers (eg, S100a9 or Il1β), their antigen processing and presentation abilities appeared reduced, supporting the notion that TAM1 do not bear strong immunogenic and T-cell regulating properties. On the other hand, the CCR2- TAM2 population showed many characteristics of immune-suppressive macrophages. Consequently, pharmacologic inhibition of CCL2, which mainly suppressed the numbers of CCR2+ TAM1 in the fibrosis–cancer model, inhibited pathogenic tumor vascularization in vivo.
Interestingly, the pharmacologic inhibition of CCL2 not only affected CCR2+ TAM1, but also induced changes in the CCR2- TAM2 population. For instance, TAM2 up-regulated S100a9, an inflammatory molecule related to NK cell activation in tumors,38 in tumor-bearing mice treated with the CCL2 inhibitor. These data imply a remarkable plasticity of the hepatic myeloid cells, including compensatory activation when suppressing distinct subsets. In studies based on 4T1 tumor cell line implantation in mice, TAM2 were shown to originate from TAM1, which were derived from infiltrating monocytes.28 The close relationship between myeloid cell subsets in the liver and their partial functional redundancy may have major implications for chemopreventive or therapeutic strategies.7 As shown in our study, inhibiting CCL2-dependent macrophage accumulation affected fibrosis and angiogenesis in the DEN-CCl4 model, and lead to reduced tumors sizes. The mouse model of multifocal autochthonous HCC development in a fibrotic liver investigated in our study substantially differed from the simple injection of hepatoma cells, in which CCR2-targeting approaches were reported to convey beneficial effects on tumor burden.16, 19 It is noteworthy that the CCR2/CCR5 inhibitor cenicriviroc is currently under phase 3 clinical investigation in patients with nonalcoholic steatohepatitis and advanced fibrosis,39 who represent a high-risk group for liver cancer. Based on the observation from our study, the immunologic changes induced by CCL2 inhibition already are present early in the fibrosis–cancer model (after 6 weeks of CCl4, before tumors are apparent), beneficial effects of CCR2/CCR5 or CCL2 inhibition on tissue remodeling and angiogenesis could be anticipated in these patients. The clear link between CCR2+ macrophages and pathogenic tumor vascularization supports the exploration of combination therapies (eg, combining CCR2 or CCL2 inhibition with conventional HCC treatment modalities), and with novel programmed cell death protein 1-directed immunotherapies.
Material and Methods
Human Liver Cancer Specimens
Formalin-fixed, paraffin-embedded liver samples from HCC patients (resection specimens, n = 10; diagnostic needle biopsy specimens, n = 8) were analyzed as approved by the Newcastle and North Tyneside Regional Ethics Committee, the Newcastle Academic Health Partners Bioresource, and the Newcastle upon Tyne National Health Service Foundation Trust Research and Development department (10/H0906/41; Newcastle Academic Health Partners Bioresource project 48; REC 12/NE/0395; R&D 6579; and Human Tissue Act license 12534). Informed consent was obtained from all patients. Detailed information on patient characteristics is shown in Table 1. Sections were stained using antibodies directed against human CCR2 (ab176390; Abcam, Cambridge, UK), S100A9 (ab63818; Abcam), CD163 (760-4437, MRQ-26; Ventana Medical Systems, Inc, Tucson, AZ), CD68 (790-2931; Ventana), and CD31 (760-4378, JC70; Ventana). CCR2 and S100A9 stainings were performed manually using Envision Flex+ reagent (Dako, Glostrup Denmark); CD163, CD68, and CD31 stainings were performed on the Benchmark Ultra (Ventana). The tumor border was defined as HCC cells adjacent to the tumor capsule and/or the non-neoplastic surrounding tissue, visible within a 200× magnification field. On a tissue slide, an area was considered as the center of the HCC when no tumor capsule or surrounding tissue could be observed using 50× magnification. Stroma was defined as fibrous tissue either within the tumor or at the border. Immunopositive cells were quantified in 3 high-power fields at 400× magnification.
Mice
C57BL6/J wild-type mice were housed in a specific pathogen-free environment. All experiments were approved by the appropriate authorities according to German legal requirements (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, Recklinghausen, Germany). Reporting of animal studies is based on the Animal Research: Reporting of In Vivo Experiments guidelines. All mice were analyzed in a blinded and anonymized fashion.
Pharmacologic CCL2 Inhibition
The RNA-aptamer (Spiegelmer) mNOX-E36 (abbreviated as CCL2i in this study), which specifically binds to murine CCL2, was kindly provided by NOXXON Pharma AG (Berlin, Germany). This 50-nucleotide L-RNA oligonucleotide (5’-GGCGACAUUGGUUGGGCAUGAGGCGAGGCCCUUUGAUGAAUCCG CGGCCA-3’) inhibits the infiltration of CCR2-dependent monocytes and macrophages, as shown earlier.5 A control molecule with reverse nucleotide sequence was used as inactive control Spiegelmer, termed rev-mNOX-E36 (revCCL2i). The oligonucleotide part of both, the active drug and the control molecule, is linked covalently with 40-kilodalton polyethylene glycol at the 3'-end to prevent rapid renal elimination. The compounds were injected subcutaneously at a dose of 20 mg/kg body weight (diluted in 5% glucose solution) 3 times/wk. All mNOX-E36 (CCL2i) and revmNOX-E36 (revCCL2i) doses and concentrations refer to the oligonucleotide part of the molecules.
Liver Fibrosis–Cancer Model
Multifocal liver cancer development was induced by the carcinogen DEN, which was administered intraperitoneally into mice 14 days postpartum at 25 mg/kg body weight.40 Starting at the age of 8 weeks (6 weeks after DEN exposure), chronic liver injury was induced using repetitive intraperitoneal CCl4 (Merck, Darmstadt, Germany) applications twice weekly for 16 weeks. CCl4 (Merck) was mixed with corn oil at a dose of 0.6 mL/kg body weight. Mice were killed 48 hours after the last injection of CCl4 or corn oil (vehicle). CCL2 was inhibited using the CCL2i (or revCCL2i) over the whole 16-week period.
μ-CT
Mice were anesthetized with 1.5% isoflurane in oxygen-enriched air and received 100 μL eXIA160XL intravenously (Binitio Biomedical, Ottawa, Ontario, Canada). Animals were scanned via a dual-energy scan at 41 and 65 kV (at 0.5 and 1 mA), obtaining 2880 projections sized 1032 × 1024 over 6 minutes. A Feldkamp-type reconstruction algorithm was applied with a voxel size of 35 × 35 × 35 μm3, including ring artefact correction. Visualization of reconstructed data was performed with Imalytics (Aachen, NRW, Germany) Preclinical software.41 The relative blood volume was determined as published earlier,5 and liver and tumor volumes were quantified on cross-sectional images in transversal, coronal, and sagittal planes.
Ex Vivo μ-CT
Mice were perfused intracardially with Microfil (Flow Tech, Carver, MA), a lead-containing silicone rubber CT contrast agent for high-resolution 3D investigation of the microarchitecture of blood vessels in the liver. Microfil replaces the blood volume and polymerizes 20 minutes after application, resulting in vascular casting. The liver was excised, formalin-fixed, and scanned using a high-resolution SkyScan 1272 μ-CT system (SkyScan, Kontich, Belgium) as published earlier.5 After 3D volume rendering of reconstructed high-resolution μ-CT data sets, 3D micromorphology of vessels were analyzed semiautomatically, using Imalytics Preclinical software.41
Cell Isolation and Sorting
Single-cell suspensions of livers were generated by mincing the organ into small pieces less than 1 mm and 30 minutes of digestion with collagenase type IV (Worthington, Lakewood, NJ). Right ventricle blood was subjected to red blood cell lysis using Pharm Lyse (Becton Dickinson, Franklin Lakes, NJ), filtered using a 70-μm mesh, and stained for flow cytometry using CD45, CD11b, Ly6G, Gr1, F4/80, and MHCII (all BD Biosciences). Count beads (BD) were added to single-cell suspensions to determine absolute cell numbers in different organs.
Gene Expression Profiling of Tumor-Associated Macrophage Subsets
Leukocytes were isolated from tumors as described earlier, and CD11b+ leukocytes were purified using magnetic-assisted cell sorting (Miltenyi Biotec, Bergisch, Gladbach, Germany). To further increase purity for sorting using a BD Aria-II, lineage staining was performed using a collection of common lymphoid cells (CD4, CD8, B220, and NK1.1; all BD Biosciences). In each NanoString assay, 20,000 sorted cells were analyzed. Housekeeping mRNA was used to normalize data and to generate transcript counts. Differential gene expression was calculated using the R package DESeq2. A log2 fold change threshold ≥2 and an adjusted P value of ≤.01 for comparison of TAM populations or a P value ≤.05 for comparison of CCL2i-treated against untreated TAM was used. Gene set enrichment analysis was performed using the Cytoscape plug-in BinGO,42 and the plugin Enrichment Map was used (P < .05, false discovery rate Q-value < 0.25, and a similarity with a Jaccard coefficient cut-off value of 0.5. Functional annotations of were performed using Gene Ontology and Biocarta from the Database for Annotation, Visualization and Integrated Discovery database.43 GO and Biocarta pathway terms then were analyzed for their frequency in the 2 gene sets. Three-fold expression was used for the Venn diagrams and hierarchical clustering analysis was based on the mean value of all experiments.
Statistical Analysis
All data are presented as means ± SD. Differences between groups were assessed using the appropriate statistical tests (GraphPad Prism 5, LaJolla, CA).
Acknowledgments
The authors thank Aline Roggenkamp and the SFB/TRR57 Q3 cell isolation facility (Carmen Tag, Sibille Sauer-Lehnen) for excellent technical assistance; and NOXXON (Berlin, Germany) for providing the CCL2 inhibitor mNOX-E36.
Footnotes
Author contributions Matthias Bartneck, Peter Schrammen, and Frank Tacke were responsible for the study concept and design; Matthias Bartneck, Peter Schrammen, Diana Möckel, Olivier Govaere, Anke Liepelt, Oliver Krenkel, Can Ergen, Misti Vanette McCain, and Dirk Eulberg aquired data; Matthias Bartneck, Peter Schrammen, Helen Reeves, Twan Lammers, and Frank Tacke wrote the article; and Matthias Bartneck, Peter Schrammen, Diana Möckel, Olivier Govaere, and Frank Tacke analyzed and interpreted data.
Conflicts of interest These authors disclose the following: Dirk Eulberg is an employee of NOXXON Pharma AG, and work in the laboratory of Frank Tacke has received financial support from Allergan/Tobira, Bristol-Myers Squibb, Inventiva, and Galapagos. The remaining authors disclose no conflicts.
Funding This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft Ta434/3-1 and SFB/TRR57 to F.T.); the Wilhelm Sander Foundation (2015.124.1 to M.B.), the Cooperation in Science and Technology Action BM1404 Mye-EUNITER (http://www.mye-euniter.eu), which is part of the European Union Framework Program Horizon 2020 (M.B.); the Interdisciplinary Center for Clinical Research Aachen (F.T. and T.La.); and by the Europäischer Fonds für regionale Entwicklung. NRW initiative (I3-STM to F.T. and T.La.).
Supplementary Material
3D rendering of a mouse liver at 26 weeks of age.
3D rendering of a mouse liver treated with CCl4.
3D rendering of a mouse liver treated with DEN and CCl4.
3D rendering of a mouse liver treated with DEN, CCl4, and CCL2i.
3D rendering of a mouse liver treated with DEN, CCl4, and revCCL2i.
CT scan of a liver of a control mouse at 26 weeks of age.
CT scan of a mouse liver treated with CCl4.
CT scan of a mouse liver treated with DEN and CCl4.
CT scan of a mouse liver treated with DEN, CCl4, and CCL2i.
CT scan of a mouse liver treated with DEN, CCl4, and revCCL2i.
References
- 1.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 2.Ringelhan M., Pfister D., O'Connor T., Pikarsky E., Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19:222–232. doi: 10.1038/s41590-018-0044-z. [DOI] [PubMed] [Google Scholar]
- 3.Coulon S., Heindryckx F., Geerts A., Van Steenkiste C., Colle I., Van Vlierberghe H. Angiogenesis in chronic liver disease and its complications. Liver Int. 2011;31:146–162. doi: 10.1111/j.1478-3231.2010.02369.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang D.Y., Friedman S.L. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology. 2012;56:769–775. doi: 10.1002/hep.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehling J., Bartneck M., Wei X., Gremse F., Fech V., Mockel D., Baeck C., Hittatiya K., Eulberg D., Luedde T., Kiessling F., Trautwein C., Lammers T., Tacke F. CCL2-dependent infiltrating macrophages promote angiogenesis in progressive liver fibrosis. Gut. 2014;63:1960–1971. doi: 10.1136/gutjnl-2013-306294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afik R., Zigmond E., Vugman M., Klepfish M., Shimshoni E., Pasmanik-Chor M., Shenoy A., Bassat E., Halpern Z., Geiger T., Sagi I., Varol C. Tumor macrophages are pivotal constructors of tumor collagenous matrix. J Exp Med. 2016;213:2315–2331. doi: 10.1084/jem.20151193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol. 2017;66:1300–1312. doi: 10.1016/j.jhep.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 8.Gerbes A., Zoulim F., Tilg H., Dufour J.F., Bruix J., Paradis V., Salem R., Peck-Radosavljevic M., Galle P.R., Greten T.F., Nault J.C., Avila M.A. Gut roundtable meeting paper: selected recent advances in hepatocellular carcinoma. Gut. 2018;67:380–388. doi: 10.1136/gutjnl-2017-315068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murdoch C., Muthana M., Coffelt S.B., Lewis C.E. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8:618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 10.Schupp J., Krebs F.K., Zimmer N., Trzeciak E., Schuppan D., Tuettenberg A. Targeting myeloid cells in the tumor sustaining microenvironment. Cell Immunol. 2017 doi: 10.1016/j.cellimm.2017.10.013. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez-Gea V., Toffanin S., Friedman S.L., Llovet J.M. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512–527. doi: 10.1053/j.gastro.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolli E., Movahedi K., Laoui D., Van Ginderachter J.A. Novel insights in the regulation and function of macrophages in the tumor microenvironment. Curr Opin Oncol. 2017;29:55–61. doi: 10.1097/CCO.0000000000000344. [DOI] [PubMed] [Google Scholar]
- 13.Takeya M., Komohara Y. Role of tumor-associated macrophages in human malignancies: friend or foe? Pathol Int. 2016;66:491–505. doi: 10.1111/pin.12440. [DOI] [PubMed] [Google Scholar]
- 14.Ding T., Xu J., Wang F., Shi M., Zhang Y., Li S.P., Zheng L. High tumor-infiltrating macrophage density predicts poor prognosis in patients with primary hepatocellular carcinoma after resection. Hum Pathol. 2009;40:381–389. doi: 10.1016/j.humpath.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Kuang D.M., Zhao Q., Peng C., Xu J., Zhang J.P., Wu C., Zheng L. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med. 2009;206:1327–1337. doi: 10.1084/jem.20082173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Yao W., Yuan Y., Chen P., Li B., Li J., Chu R., Song H., Xie D., Jiang X., Wang H. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–167. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 17.Krenkel O., Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17:306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 18.Eggert T., Wolter K., Ji J., Ma C., Yevsa T., Klotz S., Medina-Echeverz J., Longerich T., Forgues M., Reisinger F., Heikenwalder M., Wang X.W., Zender L., Greten T.F. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell. 2016;30:533–547. doi: 10.1016/j.ccell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao W., Ba Q., Li X., Li H., Zhang S., Yuan Y., Wang F., Duan X., Li J., Zhang W., Wang H. A natural CCR2 antagonist relieves tumor-associated macrophage-mediated immunosuppression to produce a therapeutic effect for liver cancer. EBioMedicine. 2017;22:58–67. doi: 10.1016/j.ebiom.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teng K.Y., Han J., Zhang X., Hsu S.H., He S., Wani N.A., Barajas J.M., Snyder L.A., Frankel W.L., Caligiuri M.A., Jacob S.T., Yu J., Ghoshal K. Blocking the CCL2-CCR2 axis using CCL2-neutralizing antibody is an effective therapy for hepatocellular cancer in a mouse model. Mol Cancer Ther. 2017;16:312–322. doi: 10.1158/1535-7163.MCT-16-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan S., Kuo N., Kryczek I., Zou W., Welling T.H. Myeloid cells in hepatocellular carcinoma. Hepatology. 2015;62:1304–1312. doi: 10.1002/hep.27867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mossanen J.C., Krenkel O., Ergen C., Govaere O., Liepelt A., Puengel T., Heymann F., Kalthoff S., Lefebvre E., Eulberg D., Luedde T., Marx G., Strassburg C.P., Roskams T., Trautwein C., Tacke F. Chemokine (C-C motif) receptor 2-positive monocytes aggravate the early phase of acetaminophen-induced acute liver injury. Hepatology. 2016;64:1667–1682. doi: 10.1002/hep.28682. [DOI] [PubMed] [Google Scholar]
- 23.Yeung O.W., Lo C.M., Ling C.C., Qi X., Geng W., Li C.X., Ng K.T., Forbes S.J., Guan X.Y., Poon R.T., Fan S.T., Man K. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. 2015;62:607–616. doi: 10.1016/j.jhep.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 24.Dapito D.H., Mencin A., Gwak G.Y., Pradere J.P., Jang M.K., Mederacke I., Caviglia J.M., Khiabanian H., Adeyemi A., Bataller R., Lefkowitch J.H., Bower M., Friedman R., Sartor R.B., Rabadan R., Schwabe R.F. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulkarni O., Eulberg D., Selve N., Zollner S., Allam R., Pawar R.D., Pfeiffer S., Segerer S., Klussmann S., Anders H.J. Anti-Ccl2 Spiegelmer permits 75% dose reduction of cyclophosphamide to control diffuse proliferative lupus nephritis and pneumonitis in MRL-Fas(lpr) mice. J Pharmacol Exp Ther. 2009;328:371–377. doi: 10.1124/jpet.108.142711. [DOI] [PubMed] [Google Scholar]
- 26.Bartneck M., Fech V., Ehling J., Govaere O., Warzecha K.T., Hittatiya K., Vucur M., Gautheron J., Luedde T., Trautwein C., Lammers T., Roskams T., Jahnen-Dechent W., Tacke F. Histidine-rich glycoprotein promotes macrophage activation and inflammation in chronic liver disease. Hepatology. 2016;63:1310–1324. doi: 10.1002/hep.28418. [DOI] [PubMed] [Google Scholar]
- 27.Georgoudaki A.M., Prokopec K.E., Boura V.F., Hellqvist E., Sohn S., Ostling J., Dahan R., Harris R.A., Rantalainen M., Klevebring D., Sund M., Brage S.E., Fuxe J., Rolny C., Li F., Ravetch J.V., Karlsson M.C. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 2016;15:2000–2011. doi: 10.1016/j.celrep.2016.04.084. [DOI] [PubMed] [Google Scholar]
- 28.Movahedi K., Laoui D., Gysemans C., Baeten M., Stange G., Van den Bossche J., Mack M., Pipeleers D., In't Veld P., De Baetselier P., Van Ginderachter J.A. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70:5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 29.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoves S., Ooi C.H., Wolter C., Sade H., Bissinger S., Schmittnaegel M., Ast O., Giusti A.M., Wartha K., Runza V., Xu W., Kienast Y., Cannarile M.A., Levitsky H., Romagnoli S., De Palma M., Ruttinger D., Ries C.H. Rapid activation of tumor-associated macrophages boosts preexisting tumor immunity. J Exp Med. 2018;215:859–876. doi: 10.1084/jem.20171440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry C.J., Munoz-Rojas A.R., Meeth K.M., Kellman L.N., Amezquita R.A., Thakral D., Du V.Y., Wang J.X., Damsky W., Kuhlmann A.L., Sher J.W., Bosenberg M., Miller-Jensen K., Kaech S.M. Myeloid-targeted immunotherapies act in synergy to induce inflammation and antitumor immunity. J Exp Med. 2018;215:877–893. doi: 10.1084/jem.20171435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Abou-Elkacem L., Arns S., Brix G., Gremse F., Zopf D., Kiessling F., Lederle W. Regorafenib inhibits growth, angiogenesis, and metastasis in a highly aggressive, orthotopic colon cancer model. Mol Cancer Ther. 2013;12:1322–1331. doi: 10.1158/1535-7163.MCT-12-1162. [DOI] [PubMed] [Google Scholar]
- 34.Noy R., Pollard J.W. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jetten N., Verbruggen S., Gijbels M.J., Post M.J., De Winther M.P., Donners M.M. Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis. 2014;17:109–118. doi: 10.1007/s10456-013-9381-6. [DOI] [PubMed] [Google Scholar]
- 36.Guo L., Akahori H., Harari E., Smith S.L., Polavarapu R., Karmali V., Otsuka F., Gannon R.L., Braumann R.E., Dickinson M.H., Gupta A., Jenkins A.L., Lipinski M.J., Kim J., Chhour P., de Vries P.S., Jinnouchi H., Kutys R., Mori H., Kutyna M.D., Torii S., Sakamoto A., Choi C.U., Cheng Q., Grove M.L., Sawan M.A., Zhang Y., Cao Y., Kolodgie F.D., Cormode D.P., Arking D.E., Boerwinkle E., Morrison A.C., Erdmann J., Sotoodehnia N., Virmani R., Finn A.V. CD163+ macrophages promote angiogenesis and vascular permeability accompanied by inflammation in atherosclerosis. J Clin Invest. 2018;128:1106–1124. doi: 10.1172/JCI93025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarosz-Biej M., Kaminska N., Matuszczak S., Cichon T., Pamula-Pilat J., Czapla J., Smolarczyk R., Skwarzynska D., Kulik K., Szala S. M1-like macrophages change tumor blood vessels and microenvironment in murine melanoma. PLoS One. 2018;13:e0191012. doi: 10.1371/journal.pone.0191012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narumi K., Miyakawa R., Ueda R., Hashimoto H., Yamamoto Y., Yoshida T., Aoki K. Proinflammatory proteins S100A8/S100A9 activate NK cells via interaction with RAGE. J Immunol. 2015;194:5539–5548. doi: 10.4049/jimmunol.1402301. [DOI] [PubMed] [Google Scholar]
- 39.Tacke F. Cenicriviroc for the treatment of non-alcoholic steatohepatitis and liver fibrosis. Expert Opin Investig Drugs. 2018;27:301–311. doi: 10.1080/13543784.2018.1442436. [DOI] [PubMed] [Google Scholar]
- 40.Schneider C., Teufel A., Yevsa T., Staib F., Hohmeyer A., Walenda G., Zimmermann H.W., Vucur M., Huss S., Gassler N., Wasmuth H.E., Lira S.A., Zender L., Luedde T., Trautwein C., Tacke F. Adaptive immunity suppresses formation and progression of diethylnitrosamine-induced liver cancer. Gut. 2012;61:1733–1743. doi: 10.1136/gutjnl-2011-301116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gremse F., Stark M., Ehling J., Menzel J.R., Lammers T., Kiessling F. Imalytics preclinical: interactive analysis of biomedical volume data. Theranostics. 2016;6:328–341. doi: 10.7150/thno.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 43.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D rendering of a mouse liver at 26 weeks of age.
3D rendering of a mouse liver treated with CCl4.
3D rendering of a mouse liver treated with DEN and CCl4.
3D rendering of a mouse liver treated with DEN, CCl4, and CCL2i.
3D rendering of a mouse liver treated with DEN, CCl4, and revCCL2i.
CT scan of a liver of a control mouse at 26 weeks of age.
CT scan of a mouse liver treated with CCl4.
CT scan of a mouse liver treated with DEN and CCl4.
CT scan of a mouse liver treated with DEN, CCl4, and CCL2i.
CT scan of a mouse liver treated with DEN, CCl4, and revCCL2i.