Abstract
We hypothesized that expert epileptologists can detect seizures directly by visually analyzing EEG plot images, unlike automated methods that analyze spectro-temporal features or complex, non-stationary features of EEG signals. If so, seizure detection could benefit from convolutional neural networks because their visual recognition ability is comparable to that of humans. We explored image-based seizure detection by applying convolutional neural networks to long-term EEG that included epileptic seizures. After filtering, EEG data were divided into short segments based on a given time window and converted into plot EEG images, each of which was classified by convolutional neural networks as ‘seizure’ or ‘non-seizure’. These resultant labels were then used to design a clinically practical index for seizure detection. The best true positive rate was obtained using a 1-s time window. The median true positive rate of convolutional neural networks labelling by seconds was 74%, which was higher than that of commercially available seizure detection software (20% by BESA and 31% by Persyst). For practical use, the median of detected seizure rate by minutes was 100% by convolutional neural networks, which was higher than the 73.3% by BESA and 81.7% by Persyst. The false alarm of convolutional neural networks' seizure detection was issued at 0.2 per hour, which appears acceptable for clinical practice. Moreover, we demonstrated that seizure detection improved when training was performed using EEG patterns similar to those of testing data, suggesting that adding a variety of seizure patterns to the training dataset will improve our method. Thus, artificial visual recognition by convolutional neural networks allows for seizure detection, which otherwise currently relies on skillful visual inspection by expert epileptologists during clinical diagnosis.
Keywords: Convolutional neural networks, Seizure detection, Deep learning, Scalp electroencephalogram, Epileptic seizure
Abbreviation: CNN, convolutional neural networks; PWE, patients with epilepsy; ROC, receiver operating characteristic; RSA, rhythmic slow activity; PFA, paroxysmal fast activity; RED, repetitive epileptiform discharge; Sup, suppression; OLE, occipital lobe epilepsy; TLE, temporal lobe epilepsy; PLE, parietal lobe epilepsy; MF, multifocal; DMF, diffuse multifocal; FLE, frontal lobe epilepsy; FBTCS, focal to bilateral tonic-clonic seizure; FIAS, focal impaired awareness seizure; FAS, focal aware seizure; TS, tonic seizure; Sb, subject; Sz, seizure; rec, recording; STD, standard deviation
Graphical abstract
Highlights
-
•
Artificial visual recognition of scalp EEG plot images successfully detects seizures.
-
•
CNN-based automatic detection performed better than commercial software.
-
•
Customized CNN learning using large datasets improves detection.
1. Introduction
Scalp electroencephalogram (EEG) has played a major role in the diagnosis of patients with epilepsy (PWE) since it was first attempted by Gibbs in 1935 (Gibbs et al., 1935; Smith, 2005). More recently, the use of digital EEG monitoring systems with extensive memory has allowed the establishment of long-term video-EEG monitoring as routine clinical care for PWE in classifying epilepsy and determining the appropriate therapeutic strategy (Aminoff, 2012; Cascino, 2002). The average amount of time needed for successful clinically relevant long-term monitoring ranges between 4.8 and 7.6 days (Velis et al., 2007). Seizures are identified by epileptologists who read extensive EEGs, which is a time-consuming task requiring experience (Benbadis, 2010). Automatic seizure detection is therefore a key technology to save time and effort associated with EEG reading. Furthermore, automatic seizure detection may open a new therapeutic avenue for future treatments of epilepsy such as on-demand neurostimulation (Theodore and Fisher, 2004) and drug delivery (Stein et al., 2000).
A number of EEG signal features have been considered to represent seizures (Ahmedt-Aristizabal et al., 2017; Baldassano et al., 2017; Orosco et al., 2013; Venkataraman et al., 2014); e.g., time-frequency analysis (Anusha et al., 2012; Gao et al., 2017; Li et al., 2018), wavelet transform (Adeli et al., 2003, Adeli et al., 2007; Adeli and Ghosh-Dastidar, 2010; Ayoubian et al., 2013; Faust et al., 2015; Sharma et al., 2014; Yuan et al., 2018), and nonlinear analysis (Ghosh-Dastidar et al., 2007; Takahashi et al., 2012). The detection accuracy has also improved with advances in machine learning algorithms such as the support vector machine (Satapathy et al., 2016), logistic regression (Lam et al., 2016), and neural networks (Adeli and Ghosh-Dastidar, 2010; Ghosh-Dastidar et al., 2008; Ghosh-Dastidar and Adeli, 2007, Ghosh-Dastidar and Adeli, 2009; Juárez-Guerra et al., 2015). However, because the actual patterns of epileptic EEG differ from patient to patient, the efficacy of most of these traditional methods is patient-specific. Furthermore, because the amount of data in seizure states is very limited (i.e., seizures are typically observed only for a few min in 24 h during EEG monitoring) and because EEG contains noise and artifacts, seizure detection by EEG remains challenging. For example, Varsavsky et al. compared four commercially available seizure detection algorithms–Monitor (Gotman, 1990), CNet (Gabor, 1998), Reveal (Wilson et al., 2004), and Saab (Saab and Gotman, 2005)–using the same dataset and showed that the true positive rate ranged from 71 to 76% with a false positive rate of 9.65 to 2.24 alarms per hour, which is not acceptable for clinical applications (Varsavsky et al., 2016). Thus, no existing hand-crafted features appear universally applicable so far.
To overcome these difficulties, deep learning technology has emerged, by which relevant features are automatically learnt in a supervised learning framework (Lecun et al., 2015). A number of recent studies demonstrated the efficacy of deep learning in the classification of EEG signals (AliMardani et al., 2016; Dvey-Aharon et al., 2017; Jirayucharoensak et al., 2014; Ma et al., 2015; Schirrmeister et al., 2017). Yet, the performance of seizure detection by deep learning still requires improvement (Acharya et al., 2018; Ahmedt-Aristizabal et al., 2017, 2018; Thodoroff et al., 2016) compared to the level of human performance in certain visual recognition tasks (Dodge and Karam, 2017; Esteva et al., 2017).
Here, we hypothesized that expert epileptologists detect seizure states directly by visually analyzing EEG plot images, rather than employing automatic seizure detection based on spectro-temporal features or complex, non-stationary features in EEG signals. If so, seizure detection could benefit from convolutional neural networks (CNNs) that show performance comparable to that of human experts in visual recognition (Litjens et al., 2017). In the present study, we demonstrated the efficacy of image-based seizure detection for scalp EEG, in which EEG data were converted into a series of plot images, analyzed by epileptologists, then the CNN was used to classify each image as seizure or non-seizure.
2. Methods
We attempted image-based seizure detection by applying CNN to long-term EEG that included epileptic seizure states as shown in Fig. 1A. After filtering, EEG data were divided into short segments using a given time window and converted into plot images of EEG, each of which was classified by CNN as ‘seizure’ or ‘non-seizure’. These resultant labels were then used to make a clinically practical index for seizure detection.
Fig. 1.
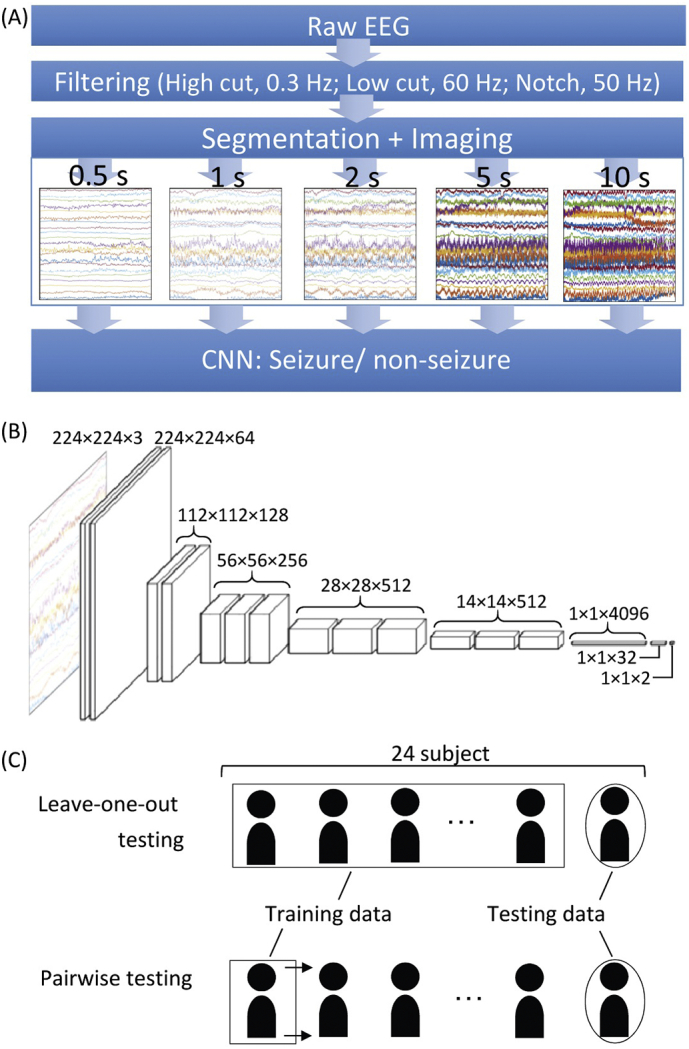
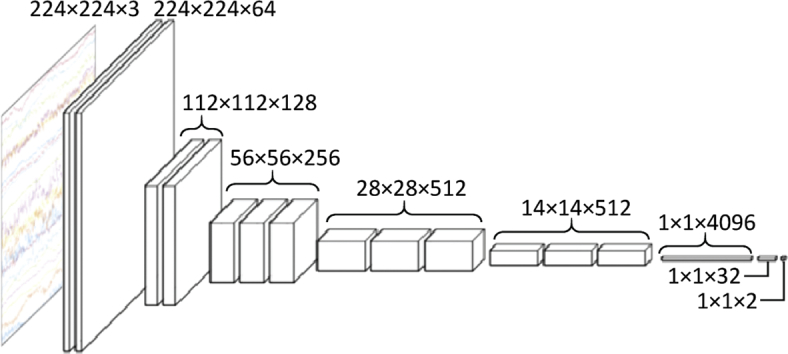
Seizure detection by image-based CNN of scalp EEG. (A) The flow of seizure detection. The raw EEG was pre-processed with 0.3-Hz high-cut, 60-Hz low-cut, and 50-Hz notch filters. EEG signals were segmented using a given time window (i.e., 0.5 s, 1 s, 2 s, 5 s, and 10 s), and converted to a time series of images. CNN then classified each image into ‘seizure’ or ‘non-seizure’. (B) CNN architecture. VGG-16 model was modified in this study. (C) Testing methods. In leave-one-out testing, a CNN was constructed with EEG data from 23 out of 24 subjects and tested with EEG data from the last remaining subject. In pairwise testing, a CNN was constructed with a single subject's data, and tested with one of other subjects.
2.1. Subjects
Long-term video-EEG monitoring was carried out as part of a phase one evaluation of PWE in the NTT Medical Center Tokyo (eight subjects) and the University of Tokyo Hospital (16 subjects). Written informed consent was obtained from all patients for research use of the EEG data. Research use of EEG data for this study was approved by the local ethics committee.
EEG was obtained with 19 channels based on the 10–20 system with two additional zygomatic electrodes (Rzyg and Lzyg) (Manzano et al., 1986) (except for subject #14) and one channel for electrocardiography (ECG). The sampling rate was 1000 Hz for the patients from the NTT Medical Center Tokyo and 500 Hz for the University of Tokyo Hospital. The EEG data were filtered with 0.3-Hz high-pass, 60-Hz low-pass and 50-Hz notch filters.
The information relevant to the subjects enrolled in this study is summarized in Table 1. The patients diagnosed as having focal seizures based on long-term video-EEG with acceptable recording quality were included in this study. For six patients, long-term video-EEG failed to localize seizure focus due to multiregional or widespread abnormalities on EEG scans. The total recording time of EEG analyzed in this study was of 1124.3 h during which 97 seizures were recorded. The total seizure state duration was 6950 s. For each subject, seizures were identified based on the agreement among at least two expert epileptologists, and the patterns of seizure onset were categorized into one of the following five categories (Tanaka et al., 2017): (i) rhythmic slow activity (RSA), i.e., sinusoidal activity at <13 Hz; (ii) paroxysmal fast activity (PFA), i.e., sinusoidal activity at ≥13 Hz; (iii) repetitive epileptiform discharge (RED), i.e., spike-and-wave or sharp-and-wave activity or repetitive spikes at <13 Hz without any visible sinusoidal activity; (iv) suppression (Sup), i.e., suppression of background activity to ≤10 μV; and (v) artifact: no visible EEG pattern because of artifacts at seizure onset.
Table 1.
Subject information. (A) Individual subjects. (B) Summary of patient demographics.
| (A) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sb# | Age | Sex | Sz time (mean), s | # of Sz | Total Sz time, s | Total rec time, h | Sz onset pattern | Epileptic focus | Sz type |
| 1 | 8 | F | 93 | 1 | 94 | 88.2 | RSA | OLE | FBTCS |
| 2 | 18 | F | 103 | 6 | 624 | 47.7 | PFA | TLE | FIAS |
| 3 | 34 | F | 376 | 5 | 1889 | 39.9 | RSA | TLE | FIAS |
| 4 | 39 | M | 12 | 11 | 153 | 20.9 | PFA | PLE | FIAS |
| 5 | 19 | M | 53 | 3 | 163 | 113.6 | RSA | OLE | FIAS |
| 6 | 62 | F | 172 | 5 | 866 | 66.8 | RED | MF | FAS |
| 7 | 34 | M | 94 | 3 | 287 | 90.0 | PFA | MF | FIAS |
| 8 | 37 | M | 12 | 4 | 52 | 71.3 | RSA | TLE | FIAS |
| 9 | 20 | M | 120 | 1 | 121 | 94.8 | RSA | TLE | FIAS |
| 10 | 30 | M | 50 | 1 | 51 | 19.6 | RSA | TLE | FIAS |
| 11 | 24 | F | 42 | 15 | 656 | 60.1 | Sup | MF | FIAS |
| 12 | 44 | F | 51 | 2 | 105 | 59.4 | PFA | TLE | FIAS |
| 13 | 20 | M | 29 | 12 | 371 | 38.2 | Sup | MF | FIAS |
| 14 | 43 | F | 188 | 2 | 378 | 27.8 | RSA | TLE | FBTCS |
| 15 | 17 | M | 40 | 3 | 125 | 16.8 | Sup | OLE | FIAS |
| 16 | 32 | F | 41 | 4 | 168 | 20.5 | RSA | TLE | FIAS |
| 17 | 11 | M | 27 | 2 | 57 | 21.4 | PFA | TLE | FIAS |
| 18 | 19 | F | 48 | 1 | 49 | 41.9 | RSA | TLE | FIAS |
| 19 | 39 | M | 69 | 1 | 70 | 5.0 | Sup | TLE | FBTCS |
| 20 | 30 | M | 120 | 2 | 243 | 43.4 | RSA | TLE | FIAS |
| 21 | 18 | M | 14 | 3 | 47 | 20.9 | Sup | DMF | TS |
| 22 | 37 | M | 16 | 6 | 106 | 22.0 | PFA | FLE | FAS |
| 23 | 49 | F | 115 | 1 | 116 | 24.5 | RSA | PLE | FIAS |
| 24 | 34 | M | 52 | 3 | 159 | 69.8 | PFA | MF | FIAS |
| (B) | ||
|---|---|---|
| Age (mean ± STD) | 30 ± 13 | |
| Sex (n) | M | 14 |
| F | 10 | |
| Sz Time (s) | 81 ± 79 | |
| # of Sz | 4 ± 3.7 | |
| Total Sz time (s) | 290 ± 403 | |
| Total rec time (h) | 46.8 ± 29.4 | |
| Sz onset pattern | RSA | 11 |
| PFA | 7 | |
| RED | 1 | |
| Sup | 5 | |
| Artifact | – | |
| Epileptic focus | OLE | 3 |
| TLE | 12 | |
| PLE | 2 | |
| MF | 5 | |
| DMF | 1 | |
| FLE | 1 | |
| Sz type | FAS | 2 |
| FIAS | 18 | |
| FBTCS | 3 | |
| TS | 1 | |
Abbreviations: RSA, Rhythmic slow activity; PFA, Paroxysmal fast activity; RED, Repetitive epileptiform discharge; Sup, Suppression; OLE, Occipital lobe epilepsy; TLE, Temporal lobe epilepsy; PLE, Parietal lobe epilepsy; MF, Multifocal; DMF, Diffuse multifocal; FLE, Frontal lobe epilepsy; FBTCS, Focal to bilateral tonic-clonic seizure; FIAS, Focal impaired awareness seizure; FAS, Focal aware seizure; TS, Tonic seizure; Sb, Subject; Sz, Seizure; rec, recording; STD, Standard deviation.
2.2. CNN for seizure detection
EEG data were segmented using a given time window and converted into a series of plot EEG images with 224 × 224 pixels. The time window served as a parameter, ranging from 0.5 s to 10 s (0.5, 1, 2, 5, and 10 s). Assuming normal printing resolution with 300 dots per inch (DPI), 0.5, 1, 2, 5, and 10 s time windows corresponded to the time bases of 38, 19, 9.5, 3.8 and 1.9 mm/s, respectively. When the size of the time window was ≤1 s, EEG data were segmented without overlaps; otherwise, data were segmented every second with overlaps with previous segments, e.g., a 9-s overlap for a 10-s segment.
A chainer v.1.24, that provides a flexible Python-based deep learning framework, was used for programming. VGG-16 was used as a CNN architecture (Simonyan and Zisserman, 2014) because very small (3 × 3) convolution filters efficiently detect small EEG waves. The original model of VGG-16 was pre-trained using an ImageNet database to differentiate 1000 classes, and classification errors were 7.4% in ILSVRC-2014 (Russakovsky et al., 2015). Because the original VGG-16 was designed to differentiate among 1000 classes, the last two layers had 4096 and 1000 dimensional vectors. We modified these last two layers as having 32 and two dimensional vectors, respectively, because we had only two target classes, i.e., seizure and non-seizure (Fig. 1B). The initial weights of the first 14 layers were the same as the pre-trained VGG-16, and those of the last two layers were independent and identically distributed random variables with a mean of zero. For the training optimizer, the Adam algorithm (Kingma and Ba, 2014) was used with an alpha (step size) of 10−5, a beta1 (exponential decay rate of the first order moment) of 0.9, and a beta2 (exponential decay rate of the second order moment) of 0.999 because this algorithm was computationally efficient and handled non-stationary objectives. The model was trained using a mini-batch of 40 images for 50 epochs, and the model of the best epoch with minimal loss was used for testing. One epoch served as a complete presentation of the training data set to be learned by the CNN model. The training time depended on the number of training data images; for reference example, 58,418 images from 23 subjects were used for training to construct a model for testing subject #2, resulting in 1461 mini-batches on each epoch. The total training time for 50 epochs was approximately 11 h with GPU of GeForce GTX 1080 Ti (Nvidia Corp., Santa Clara, CA). We also confirmed that the validation accuracy increased with reduced training loss, when the validation data had a seizure/non-seizure ratio of one to one (Fig. 3).
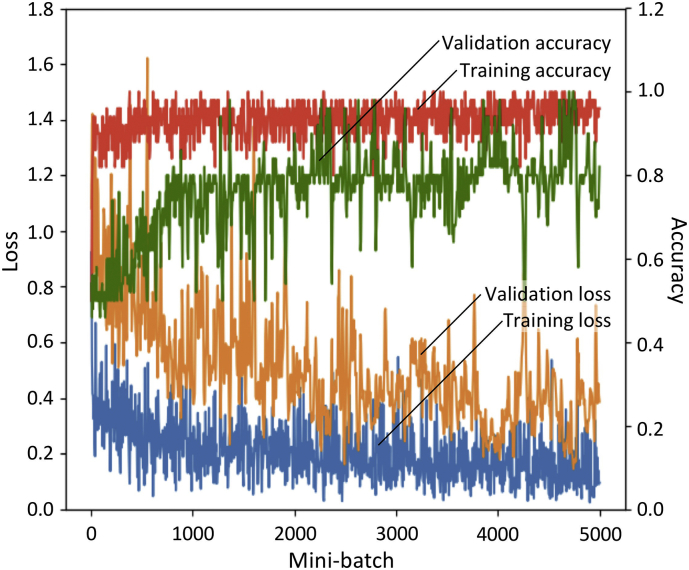
Fig. 3.
Accuracy of validation (green), accuracy of training (red), loss of validation (orange), and loss of training (blue) during the first 5000 mini-batches for subject #2. The seizure/non-seizure ratio in training data and validation data were one to eight and one to one, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To train CNN models, all the segments fully within seizure states were used to model seizure class, while the segments 8 times longer than the seizure period were randomly selected from inter-ictal states, defined based on the separation from the seizure state by >1 h, to model non-seizure class during CNN learning.
The trained CNN classified every segment of EEG, i.e., every 0.5 s for the 0.5-s segment and every second for the remaining segments, as either seizure or non-seizure. To quantify the performance of the CNN classification, the seizure/non-seizure labels by CNN were compared with those assigned by epileptologists. When deriving the true positive and true negative rates, segments including a seizure period were considered as seizure states, and those not including seizure periods as non-seizure states. We evaluated the classification performance either in leave-one-out testing or pairwise testing (Fig. 1C). In the leave-one-out testing, a model was trained with EEG data from 23 out of 24 subjects and tested with EEG data from the last remaining subject. In the pairwise testing, a model was constructed from a single subject's data and tested with EEG data from each subject individually.
For practical use, we defined a seizure index as the number of seizure labels within 10 s and issued a seizure alarm when the index exceeded a given discrimination threshold (0−10). Based on a receiver operating characteristic (ROC) curve, defined as a cumulative distribution function of the true positive rate vs. true negative rate at a given discrimination threshold, the best discrimination threshold for each subject was identified as a threshold with the minimum false alarm rate under the best detected seizure rate. The accuracy of the seizure alarm was then evaluated by minutes: when a given time window of 1 min contained a seizure period with alarms, this window was labelled as having a true positive alarm; the window with neither seizure states nor alarms was labelled as true negative; the windows including either seizure states only or alarms only, were labelled false negative or false positive, respectively. For comparison, we also used commercially-available software(BESA (Hopfengärtner et al., 2014) and Persyst (Wilson, 2004)) and evaluated the accuracy of seizure alarms as described for our seizure alarm.
3. Results
3.1. Leave-one-out testing
We first evaluated the performance of image-based seizure detection by CNN in a leave-one-out cross-validation. A CNN model was trained with EEG data from 23 out of 24 subjects and used to test EEG data from the last remaining subject, which was completely new data for the model. Therefore, a total of 24 CNN models were constructed and tested. The output of CNN was a label of ‘seizure’ or ‘non-seizure’ for a given test EEG image. The true-positive rates of this classification depended on the length of the time window ranging 0.5–10 s (Fig. 2A; Kruskal-Wallis test, p < 10−5). The best true positive rate was obtained using a 1-s time window; 0.5-s time window was too short to identify EEG features of seizure state (0.5 s vs. 1 s: post-hoc Dunn's test, p < 10−6), while using long segments of 10 s led to a failure to detect the onset of seizure state, resulting in tendency of deterioration of the true positive rate (1 s vs. 10 s: post-hoc Dunn's test, p = .083). Based on these results, the time window was fixed at 1 s hereafter.
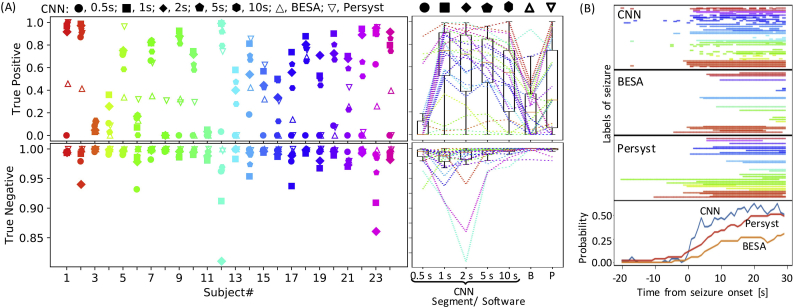
Fig. 2.
Leave-one-out testing. (A) Classification accuracy of CNN in leave-one-out testing. True positive and true negative rates are given. For CNN, the length of the time window served as a parameter (0.5–10 s). For comparison, classification accuracies of existing software, i.e., BESA (B) and Persyst (P), are also indicated. Insets on the left show classification accuracy in individual subjects. The length of the time window is indicated by symbols. Data from each subject are depicted in a different color here and hereafter. The right insets depict boxplots of classification accuracy as a function of the length of the time window and software. Each broken line refers to data from an identical subject. A boxplot depicts the median as the central mark of the box, and the 25th and 75th percentiles as the edges of the box with the whiskers extending to the most extreme data points. (B) Seizure label around the onset of seizure state. In the upper 3 insets, each line indicates a period with seizure label from an indicated method. Each row corresponds to a single seizure, and 53 seizures with a duration of 30 s or longer were examined. Different subjects are indicated by colors. The bottom inset shows the probability of seizure label around the onset of seizure state for each method.
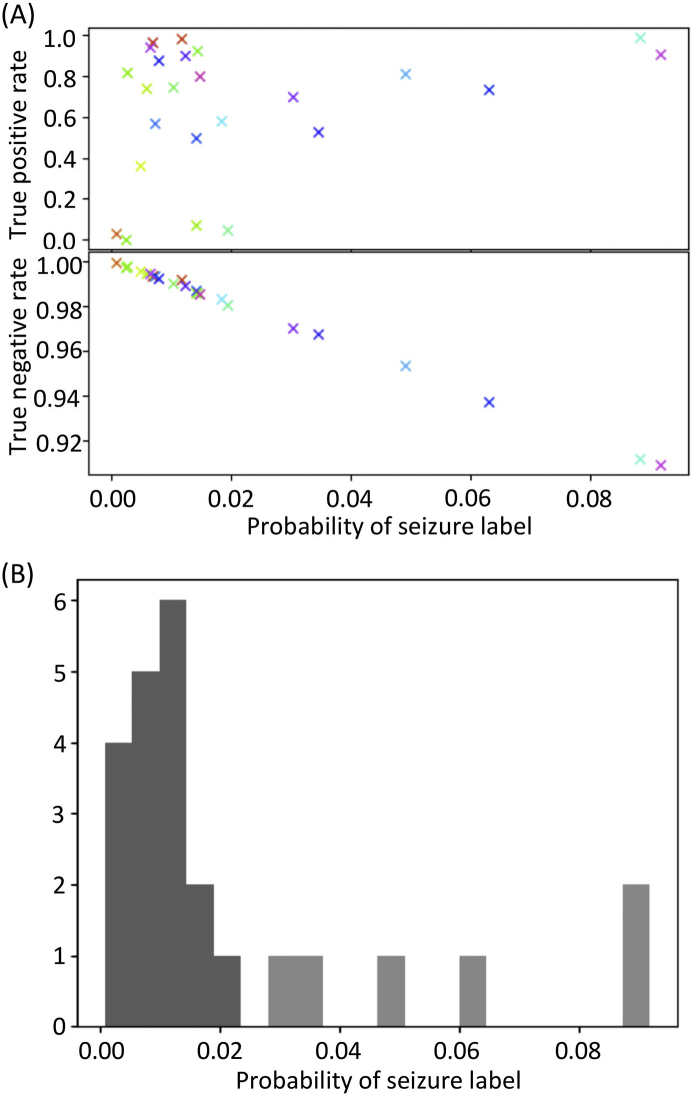
For comparison, the performances of BESA and Persyst were also quantified by seconds (Fig. 2A): As compared to those commercially available algorithms, our CNN performed better in terms of true positive rates (Kruskal-Wallis test, p = .0003; post-hoc Dunn's test: CNN vs. BESA, p = .00006; CNN vs. Persyst, p = .019; BESA vs. Persyst, p = .096), but worse in terms of true negative rates (Kruskal-Wallis test, p < 10−6; post-hoc Dunn's test: CNN vs. BESA, p < 10−6; CNN vs. Persyst, p = .00023; BESA vs. Persyst, p = .006). A poor performance in true negative rates was highly correlated with the probability of seizure label throughout all monitoring, which did not correlate with the true positive rate (Fig. 4A), suggesting that the inter-ictal EEG contained seizure-like characteristics and was hard to discriminate from a seizure. Specifically, 6 subjects with the lowest true negative rates were estimated as outliers in terms of seizure label probability (Fig. 4B; Smirnov-Grubbs one-sided (max) test, p < .05), which is why CNN might have issued too many false alarms in these subjects.
Fig. 4.
Classification accuracy vs. probability of seizure label. (A) True negative rate, but not true positive rate, correlates with the probability of seizure label. (B) Histogram showing the probability of seizure label. Six subjects were statistically considered as outliers (light gray), for which the accuracy of classification by CNN was low.
The CNN attributed the seizure label successively following the onset of seizure state (Fig. 2B), but also accidentally in inter-ictal states. Because the output of CNN was obtained every second for a given 1-s time window, the true negative rate of 0.99 (Fig. 2A), i.e., the false positive rate of 0.01, indicated that false alarms were raised every 100 s. To reduce such frequent false alarms, we defined an index for seizure detection as the number of seizure labels within 10 s. During long-term monitoring, the seizure index exhibited a rapid increase selectively at the onset of seizure state in the presence of confusing artifacts (Figs. 5A and 6A) or during the seizure period (Fig. 5B) in certain subjects, but was not accurate for others (Figs. 5C and 6B). When the detection performance was poor, the seizure index often fluctuated widely during inter-ictal states (e.g., up to 4 in Fig. 5C; Fig. 4).
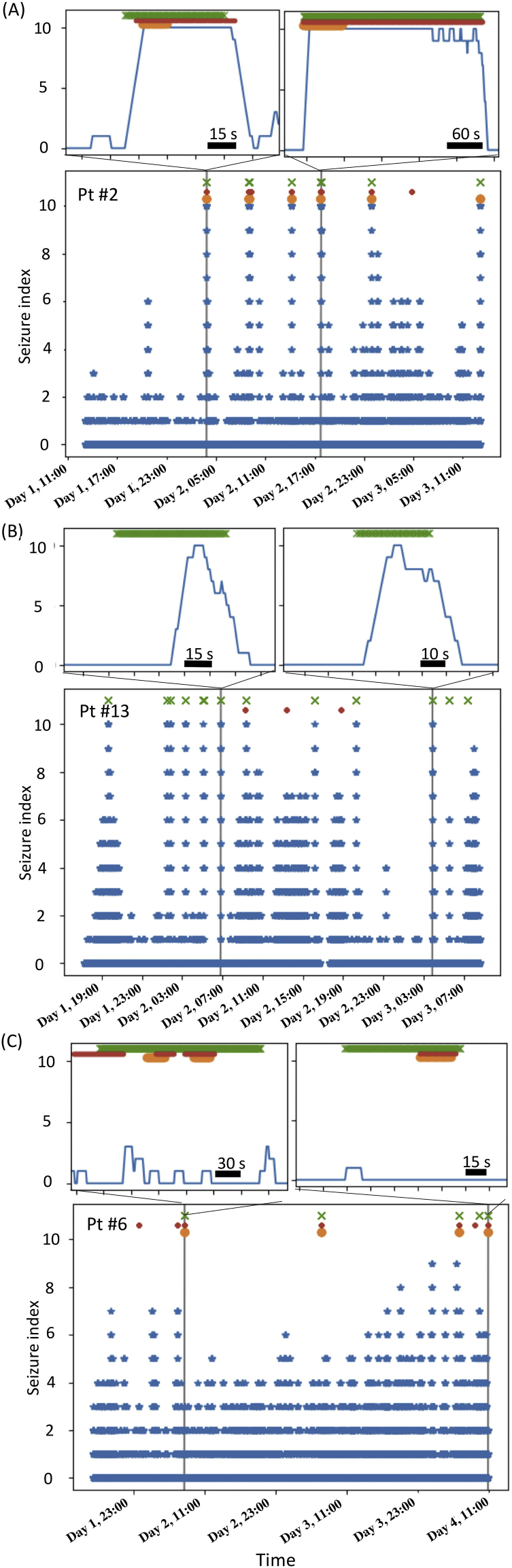
Fig. 5.
Representative data of long-term monitoring of seizure index. Cross marks show seizures diagnosed by epileptologists. Circles show seizure labels given by the available seizure detection algorithms, BESA (orange circles) and Persyst (red circles). Upper insets show magnification around several seizure onsets. (A) Seizures for subject #2 were detected without delays by both our seizure index and the available algorithms. (B) Seizures for subject #13 were detected by our seizure index with some delays but overlooked by other algorithms. (C) Reliable seizure detection was difficult by both our seizure index and available algorithms for subject #6.
Fig. 6.
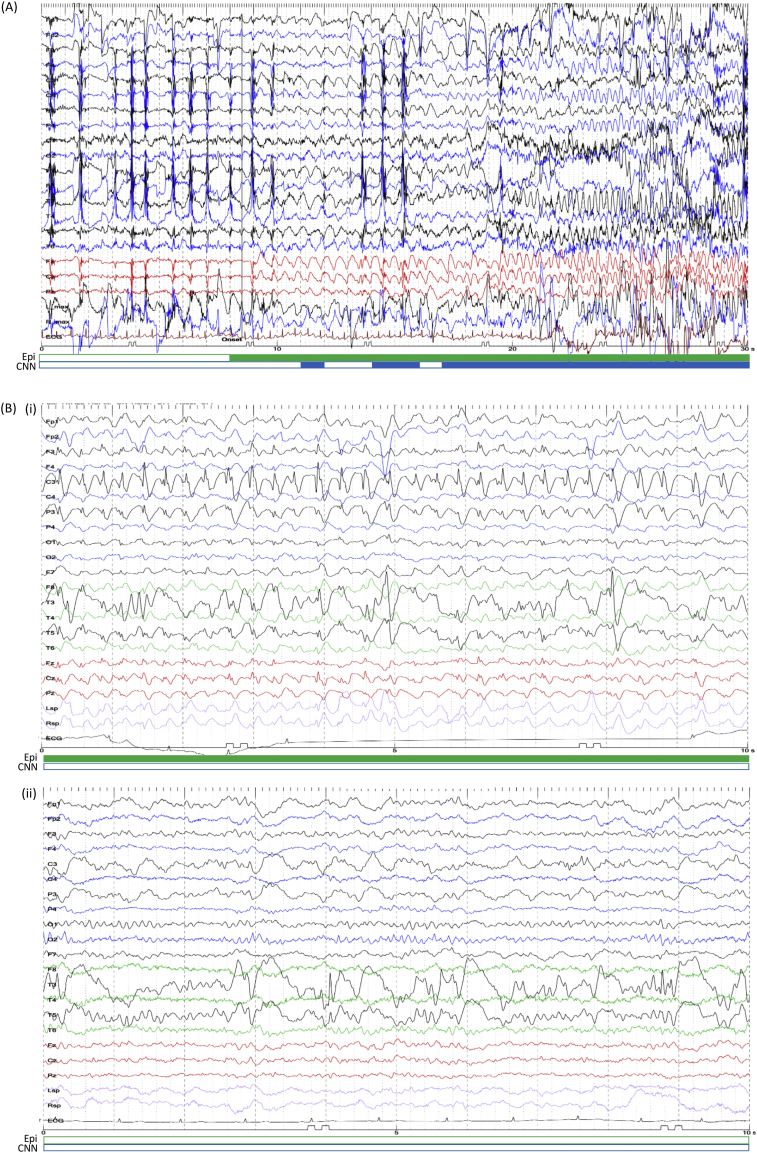
Representative EEG. (A) A seizure in subject #18 occurred during eating. Artifacts due to chewing were observed in the beginning, followed by focal epileptic discharges at the seizure onset and focal to bilateral tonic-clonic seizure in the end. (B) A seizure in subject #6 (Fig. 5C) was overlooked by CNN. An example of inter-ictal EEG from the same subject is shown in the bottom inset. The filled period in the green bar below EEG traces was labelled as a seizure by epileptologists, and the blue by CNN. The unfilled period was labelled as a non-seizure state. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
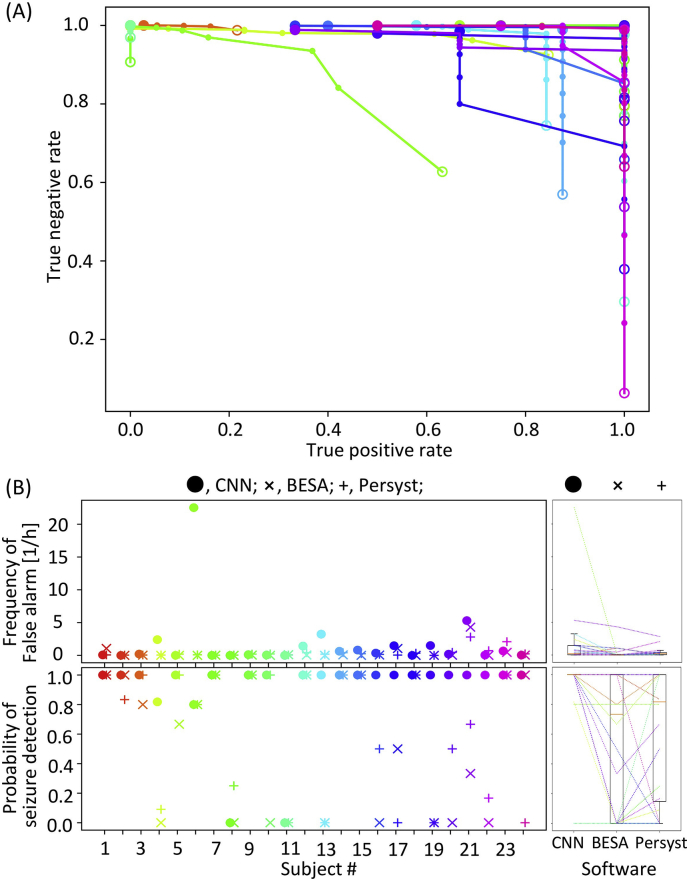
We then evaluated the quality of seizure alarms, issued when the seizure index exceeded a given discrimination threshold (0–10). To identify the best threshold, we obtained a ROC curve of seizure alarm (Fig. 7A), as a cumulative plot with a true positive rate on the abscissa vs. a true negative rate on the ordinate at a given discrimination threshold. To draw this ROC curve, the true positive and true negative rates were defined by minutes: when a given time window of 1 min contained seizure period with alarms, this window was labelled as having a true positive alarm; the window with neither seizure states nor alarms was labelled as true negative; the windows with seizure states only were labelled as false negative, and those with alarms only as false positive. Because this seizure alarm led to detection delays (e.g., Fig. 5B), the performance was not always consistent with the segment-based CNN accuracy in Fig. 2A. Consequently, using an optimal discrimination threshold, all seizures were detected without any false alarms in half of test subjects (Fig. 7B). As compared with BESA and Persyst, our method performed best in terms of the seizure detection rate (Kruskal-Wallis test, p = .0054; post-hoc Dunn's test: CNN vs. BESA, p = .0023; CNN vs. Persyst, p = .01416; BESA vs. Persyst, p = .55), but in turn, showed higher rates of false alarm than BESA (Kruskal-Wallis test, p = .0038; post-hoc Dunn's test: CNN vs. BESA, p = .0035; CNN vs. Persyst, p = .95; BESA vs. Persyst, p = .0043).
Fig. 7.
The performance of seizure detection. (A) ROC curve plotting true positive vs. true negative rates at a given threshold of seizure index ranging from 0 (an empty circle) to 10 (a filled circle). (B) The frequency of false alarms and the probability of seizure detection at the best threshold of seizure index. The conventions comply with Fig. 2B.
3.2. Pairwise testing
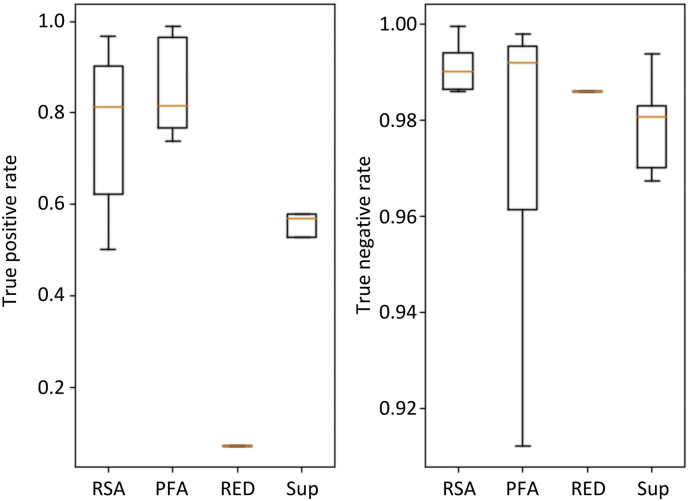
The performance of seizure detection by CNN was poor in certain subjects. Several possibilities underlying these negative results can be considered. First, the inter-ictal EEG was too aberrant to be differentiated from the seizure state (e.g., Fig. 5B). In such cases, the seizure index should fluctuate widely during the inter-ictal state, resulting in a relatively high false-positive rate (Fig. 5C). Second, the seizure pattern in the test data might be different from those in the training data, resulting in low true-positive rates. In fact, RSA (11 subjects) and PFA (seven subjects) were more common than Sup (five subjects) and RED (one subject) in our dataset, and CNN tended to perform better for RSA and PFA than for Sup and RED (Fig. 8; Kruskal-Wallis test for true positive rates, p = .104). To overcome the second possibility, a CNN model should be trained with a wide variety of seizure patterns. If this was the case, a CNN model based on single subject data should successfully detect seizure states in subjects with a similar seizure pattern. To verify this hypothesis, we attempted a pairwise testing, wherein a CNN model constructed from single subject data was used to test the models based on data from other subjects.
Fig. 8.
The performance of CNN as a function of clinically diagnosed seizure pattern (Table 1).
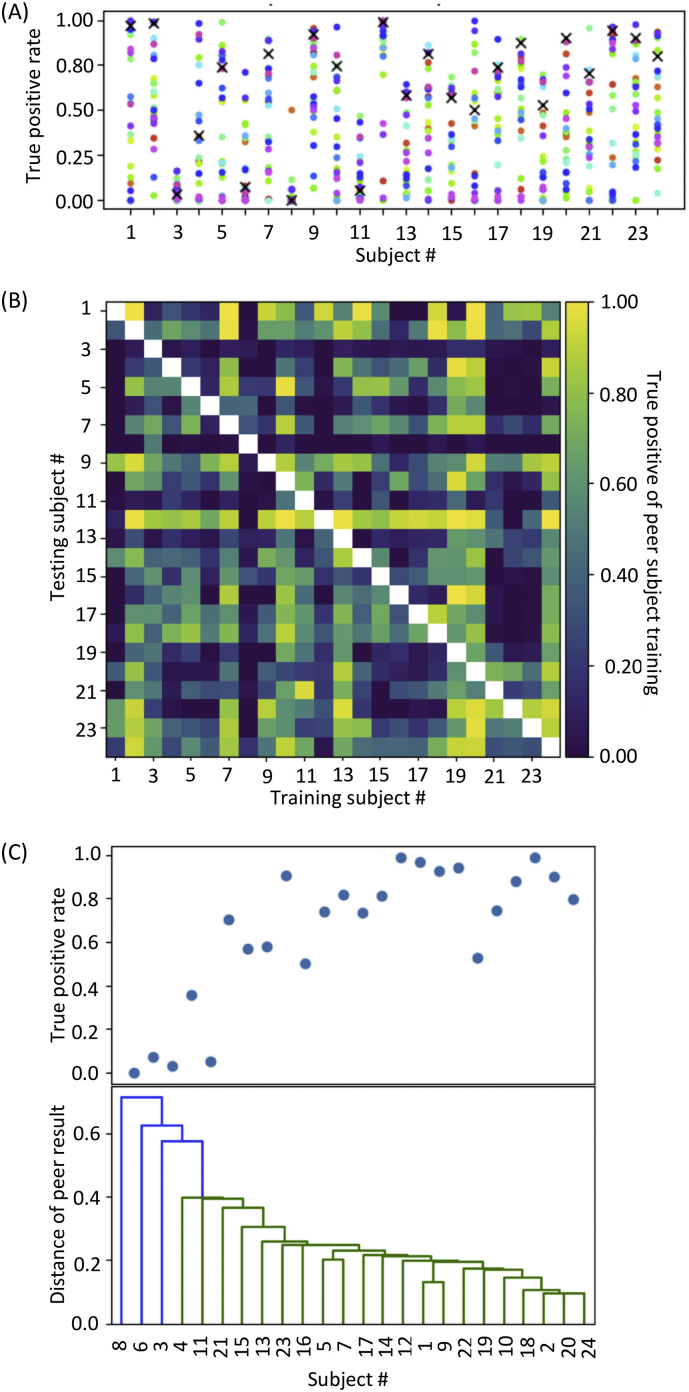
In pairwise testing, the performance of seizure detection varied among the subjects as expected (Fig. 9A and Fig. 9B). However, the best performance in the pairwise testing was comparable or superior to the performance in leave-one-out testing for all subjects. For each test pair of subjects, the better true positive rate was defined as a distance among each pair. The distance of each subject data was then visualized as a dendrogram in Fig. 9C, which demonstrated that the true positive rate in leave-one-out testing was high when a seizure was successfully detected by a pairwise testing. On the contrary, when given subject data were distant from other subject data, i.e., low performance in all sets of pairwise testing, leave-one-out testing resulted in poor performance. These results support our hypothesis that the performance of seizure detection depends on the similarity of seizure patterns between the training dataset of the CNN model and the test data. Thus, training a CNN model with a large amount of data containing a variety of seizure patterns should improve image-based seizure detection and other EEG-based diagnosis systems.
Fig. 9.
Pairwise testing. (A) True positive rates of CNN trained using data from an individual subject. A CNN model was trained using data from each subject on the abscissa and tested using data from other subjects. Of note, the best classification accuracy in pairwise testing outperformed that in leave-one-out testing (cross marks). (B) Classification accuracy matrix. CNN models were trained using data on the abscissa and tested using data on the ordinate. (C) A similarity of seizure patterns among the subjects as visualized by dendrogram based on the classification accuracy in pairwise testing. The performance of the leave-one-out testing (the upper inset) was poor when the distance from any other subject, based on the classification accuracy in pairwise testing, was large.
4. Discussion
To our knowledge, the present study is the first comprehensive attempt to automatically evaluate EEG as plot images. We demonstrated that artificial visual recognition can achieve a satisfactory level of seizure detection, which currently relies on skillful visual inspection by expert epileptologists in clinical diagnosis. The median of the true positive rate of CNN labelling by seconds was 74%, which was higher than that of commercially available seizure detection software, i.e., 20% for BESA and 31% for Persyst. For practical use, the median of detected seizure rate by minutes was 100% for CNN, which was higher than the 73.3% for BESA and 81.7% for Persyst. We also demonstrated that the best performance in pairwise testing was comparable or superior to the performance in leave-one-out testing in all subjects, suggesting that adding new data with a variety of seizure patterns should improve the performance of our method.
Our dataset included subjects with different backgrounds, which made automatic seizure detection challenging; for example, the ages of the subjects ranged from 8 to 62 years; all subjects had focal epilepsy, which makes it more difficult to detect seizure states than generalized epilepsy; and each seizure had a different epileptic focus and EEG patterns. For such a wide range of data, the performance of hand-made numerical features may not always be satisfactory (Ahmedt-Aristizabal et al., 2018). To address this problem, some pioneering works have already reported the first attempt of deep learning with CNN for automatic seizure detection (Acharya et al., 2018; Thodoroff et al., 2016). For example, Thodoroff et al. (2016) reported that CNN classifiers were successful in patient-specific but not in cross-patient seizure detection; Acharya et al. (2018) achieved 95% accuracy in seizure detection by CNN, but the data was limited to five subjects, each of which had 300 segments of single-channel EEG signals with a duration of 23.6 s (7080 s in total). The large amount of data (1124.3-h continuous multichannel EEG data from 24 subjects) differentiated our work from these previous studies, and was a key success factor in verifying our hypothesis that EEG morphology in plot images contains universal features of seizure state used by epileptologists.
Additionally, visual seizure detection requires experience (Benbadis, 2010). Inexperienced trainees and traditional seizure detection algorithms sometimes recognize artifacts, e.g., chewing artifacts with rhythmicity, as pathological discharges (Henry and Sha, 2012; Saab and Gotman, 2005). CNN could differentiate physiological artifacts from pathological discharges (Fig. 6A), suggesting that the empirical criterion for distinguishing physiological and pathological features has been achieved through training with large amounts of data.
The false alarm of CNN seizure detection was issued at 0.2 per hour, i.e., 1 false alarm per 5 h, which was comparable to that of Persyst, but was 10 times higher than that of BESA. This level of false alarm resulted in unnecessary review of 1-min EEG in 5 h of recording, which appears acceptable for practical use. Nevertheless, false alarms in certain subjects were issued too frequently for practical use. In these subjects, inter-ictal discharges may have appeared similar to ictal EEG of others, and therefore, seizure labels of CNN output were observed more frequently. In our dataset, as true negative rates deteriorated linearly with the probability of seizure label, the results by CNN were not reliable when the probability of seizure label was higher than 0.02 (Fig. 4).
For classification of plot images in our study, the optimal time window was 1 s, while epileptologists tend to use 10 s/page (30 mm/s) (Foldvary et al., 2000). This discrepancy was due to several reasons. First, in the CNN model used in this study, the image size was fixed to 224 × 224 pixels, which was too small to plot a 10-s EEG with visible epileptic discharges. A high-resolution supported CNN could therefore improve the performance in this condition. Second, although all the segments fully within the seizure states were used as the training model for the seizures label, the segments including seizure states were also partially considered as segments with seizure labels in testing. To recognize seizure portions in those segments, more sophisticated algorithms such as region-based CNN could offer a solution.
The main advantage of CNN is that retraining with new data improves the performance. For example, in subject #6, after left frontotemporal craniotomy, irregular slow wave or high amplitude spikes continuously appeared in the inter-ictal state in the left temporal lobe (Fig. 6B), and during ictal state, central repetitive slow wave appeared in the inter-ictal discharges (Fig. 6C). CNN correctly recognized irregular inter-ictal discharges as non-seizures, but could not detect the ictal state, possibly because our dataset did not contain this type of repetitive slow wave pattern in other subjects. Adding EEG data with similar patterns may improve the performance in this subject. Conversely, false-positive EEG images could be used to retrain the CNN model to have them recognized as non-seizures. Furthermore, identical datasets could be used to train another CNN with a different montage, by which some types of seizures would become easier to detect.
EEG images in our analysis were not evaluated as time series, but temporal information such as subtle changes in pre-ictal period is critical for epileptologists to detect a seizure onset. For example, in Fig. 6A, epileptologists first detected a continuing theta oscillation and checked back to the previous EEG to identify the slower oscillation as the seizure onset. Such epileptologist-like strategy could be implemented by recurrent neural network combined with our CNN model.
Acknowledgements
This work was partially supported by KAKENHI grants (26242040, 17H04305), AMED grant (JP18dm0307009) and Tateisi Science and Technology Foundation.
Contributor Information
Kensuke Kawai, Email: kenkawai-tky@umin.net.
Hirokazu Takahashi, Email: takahashi@i.u-tokyo.ac.jp.
References
- Acharya U.R., Oh S.L., Hagiwara Y., Tan J.H., Adeli H. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput. Biol. Med. 2018;100:270–278. doi: 10.1016/j.compbiomed.2017.09.017. [DOI] [PubMed] [Google Scholar]
- Adeli H., Ghosh-Dastidar S. CRC Press; 2010. Automated EEG-Based Diagnosis of Neurological Disorders. [Google Scholar]
- Adeli H., Zhou Z., Dadmehr N. Analysis of EEG records in an epileptic patient using wavelet transform. J. Neurosci. Methods. 2003;123:69–87. doi: 10.1016/s0165-0270(02)00340-0. [DOI] [PubMed] [Google Scholar]
- Adeli H., Ghosh-Dastidar S., Dadmehr N. A wavelet-chaos methodology for analysis of EEGs and EEG subbands to detect seizure and epilepsy. IEEE Trans. Biomed. Eng. 2007;54:205–211. doi: 10.1109/TBME.2006.886855. [DOI] [PubMed] [Google Scholar]
- Ahmedt-Aristizabal D., Fookes C., Dionisio S., Nguyen K., Cunha J.P.S., Sridharan S. Automated analysis of seizure semiology and brain electrical activity in presurgery evaluation of epilepsy: a focused survey. Epilepsia. 2017;58:1817–1831. doi: 10.1111/epi.13907. [DOI] [PubMed] [Google Scholar]
- Ahmedt-Aristizabal D., Fookes C., Nguyen K., Sridharan S. Deep classification of epileptic signals. arXiv Prepr. 2018 doi: 10.1109/EMBC.2018.8512249. (arXiv1801.03610) [DOI] [PubMed] [Google Scholar]
- AliMardani F., Boostani R., Blankertz B. Presenting a spatial-geometric EEG feature to classify BMD and schizophrenic patients. Int. J. Adv. Telecommun. Electrotech. Signals Syst. 2016;5:79–85. [Google Scholar]
- Aminoff M.J. Elsevier; 2012. Aminoff’s Electrodiagnosis in Clinical Neurology. [Google Scholar]
- Anusha K.S., Mathews M.T., Puthankattil D.S. 2012 International Conference on advances in Computing and Communications (ICACC) IEEE; 2012. Classification of normal and epileptic EEG signal using time & frequency domain features through artificial neural network; pp. 98–101. [Google Scholar]
- Ayoubian L., Lacoma H., Gotman J. Automatic seizure detection in SEEG using high frequency activities in wavelet domain. Med. Eng. Phys. 2013;35:319–328. doi: 10.1016/j.medengphy.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassano S.N., Brinkmann B.H., Ung H., Blevins T., Conrad E.C., Leyde K., Cook M.J., Khambhati A.N., Wagenaar J.B., Worrell G.A., Litt B. Crowdsourcing seizure detection: algorithm development and validation on human implanted device recordings. Brain. 2017;140:1680–1691. doi: 10.1093/brain/awx098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbadis S.R. The tragedy of over-read EEGs and wrong diagnoses of epilepsy. Expert. Rev. Neurother. 2010;10:343–346. doi: 10.1586/ern.09.157. [DOI] [PubMed] [Google Scholar]
- Cascino G.D. Mayo Clinic Proceedings. Elsevier; 2002. Clinical indications and diagnostic yield of video-electroencephalographic monitoring in patients with seizures and spells; pp. 1111–1120. [DOI] [PubMed] [Google Scholar]
- Dodge S., Karam L. 2017 26th International Conference on Computer Communication and Networks (ICCCN) IEEE; 2017. A study and comparison of human and deep learning recognition performance under visual distortions; pp. 1–7. [Google Scholar]
- Dvey-Aharon Z., Fogelson N., Peled A., Intrator N. Connectivity maps based analysis of EEG for the advanced diagnosis of schizophrenia attributes. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;546:115. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust O., Acharya U.R., Adeli H., Adeli A. Wavelet-based EEG processing for computer-aided seizure detection and epilepsy diagnosis. Seizure. 2015;26:56–64. doi: 10.1016/j.seizure.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Foldvary N., Caruso A.C., Mascha E., Perry M., Klem G., McCarthy V., Qureshi F., Dinner D. Identifying montages that best detect electrographic seizure activity during polysomnography. Sleep. 2000;23:1–9. [PubMed] [Google Scholar]
- Gabor A. Seizures detection using a self organizing neural network: validation and comparison with other detection strategies. Electroencephalogr. Clin. Neurophysiol. 1998;107:27–32. doi: 10.1016/s0013-4694(98)00043-1. [DOI] [PubMed] [Google Scholar]
- Gao Z.-K., Cai Q., Yang Y.-X., Dong N., Zhang S.-S. Visibility graph from adaptive optimal kernel time-frequency representation for classification of epileptiform EEG. Int. J. Neural Syst. 2017;27:1750005. doi: 10.1142/S0129065717500058. [DOI] [PubMed] [Google Scholar]
- Ghosh-Dastidar S., Adeli H. Improved spiking neural networks for EEG classification and epilepsy and seizure detection. Integr. Comput. Aided. Eng. 2007;14:187–212. [Google Scholar]
- Ghosh-Dastidar S., Adeli H. A new supervised learning algorithm for multiple spiking neural networks with application in epilepsy and seizure detection. Neural Netw. 2009;22:1419–1431. doi: 10.1016/j.neunet.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Ghosh-Dastidar S., Adeli H., Dadmehr N. Mixed-band wavelet-chaos-neural network methodology for epilepsy and epileptic seizure detection. IEEE Trans. Biomed. Eng. 2007;54:1545–1551. doi: 10.1109/TBME.2007.891945. [DOI] [PubMed] [Google Scholar]
- Ghosh-Dastidar S., Adeli H., Dadmehr N. Principal component analysis-enhanced cosine radial basis function neural network for robust epilepsy and seizure detection. IEEE Trans. Biomed. Eng. 2008;55:512–518. doi: 10.1109/TBME.2007.905490. [DOI] [PubMed] [Google Scholar]
- Gibbs F.A., Davis H., Lennox W.G. The electro-encephalogram in epilepsy and in conditions of impaired consciousness. Arch. Neurol. Psychiatr. 1935;34:1133–1148. [Google Scholar]
- Gotman J. Automatic seizure detection: improvements and evaluation. Electroencephalogr. Clin. Neurophysiol. 1990;76:317–324. doi: 10.1016/0013-4694(90)90032-f. [DOI] [PubMed] [Google Scholar]
- Henry Thomas R., Sha Z. Seizures and epilepsy: electrophysiological diagnosis. Epilepsy Board Rev. Man. 2012;1:1–45. [Google Scholar]
- Hopfengärtner R., Kasper B., Graf W., Gollwitzer S., Kreiselmeyer G., Stefan H., Hamer H. Automatic seizure detection in long-term scalp EEG using an adaptive thresholding technique: a validation study for clinical routine. Clin. Neurophysiol. 2014;125:1346–1352. doi: 10.1016/j.clinph.2013.12.104. [DOI] [PubMed] [Google Scholar]
- Jirayucharoensak S., Pan-Ngum S., Israsena P. EEG-based emotion recognition using deep learning network with principal component based covariate shift adaptation. Sci. World J. 2014;2014 doi: 10.1155/2014/627892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Guerra E., Alarcon-Aquino V., Gómez-Gil P. Epilepsy seizure detection in EEG signals using wavelet transforms and neural networks. Lect. Notes Electr. Eng. 2015;312:261–269. [Google Scholar]
- Kingma Diederik P., Ba J. Adam: a method for stochastic optimization. arXiv Prepr. 2014 (arXiv1412.6980) [Google Scholar]
- Lam A.D., Zepeda R., Cole A.J., Cash S.S. Widespread changes in network activity allow non-invasive detection of mesial temporal lobe seizures. Brain. 2016;139:2679–2693. doi: 10.1093/brain/aww198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015 doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Li Y., Cui W., Luo M., Li K., Wang L. Epileptic seizure detection based on time-frequency images of EEG signals using Gaussian mixture model and gray level co-occurrence matrix features. Int. J. Neural Syst. 2018;28:1850003. doi: 10.1142/S012906571850003X. [DOI] [PubMed] [Google Scholar]
- Litjens G., Kooi T., Bejnordi B.E., Arindra A., Setio A., Ciompi F., Ghafoorian M., Van Der Laak J.A.W.M., Van Ginneken B., Sánchez C.I., Van Der Laak J.A.W.M., Van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Ma Lan, Minett J.W., Blu T., Wang W.S.-Y. 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE; 2015. Resting state EEG-based biometrics for individual identification using convolutional neural networks; pp. 2848–2851. [DOI] [PubMed] [Google Scholar]
- Manzano G.M., Ragazzo P.C., Tavares S.M., Marino R. Anterior zygomatic electrodes: a special electrode for the study of temporal lobe epilepsy. Stereotact. Funct. Neurosurg. 1986;49:213–217. doi: 10.1159/000100148. [DOI] [PubMed] [Google Scholar]
- Orosco L., Correa A.G., Laciar E. Review: a survey of performance and techniques for automatic epilepsy detection. J. Med. Biol. Eng. 2013 [Google Scholar]
- Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A.C., Fei-Fei L. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. [Google Scholar]
- Saab M.E., Gotman J. A system to detect the onset of epileptic seizures in scalp EEG. Clin. Neurophysiol. 2005;116:427–442. doi: 10.1016/j.clinph.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Satapathy S.K., Dehuri S., Jagadev A.K. An empirical analysis of different machine learning techniques for classification of EEG signal to detect epileptic seizure. Int. J. Appl. Eng. Res. 2016;11:973–4562. [Google Scholar]
- Schirrmeister R.T., Springenberg J.T., Fiederer L.D.J., Glasstetter M., Eggensperger K., Tangermann M., Hutter F., Burgard W., Ball T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017;38:5391–5420. doi: 10.1002/hbm.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Khan Y.U., Farooq O., Tripathi M., Adeli H. A wavelet-statistical features approach for nonconvulsive seizure detection. Clin. EEG Neurosci. 2014;45:274–284. doi: 10.1177/1550059414535465. [DOI] [PubMed] [Google Scholar]
- Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. 2014 1409.1556. [Google Scholar]
- Smith S.J.M. EEG in the diagnosis, classification, and management of patients with epilepsy. J. Neurol. Neurosurg. Psychiatry. 2005;76:ii2–ii7. doi: 10.1136/jnnp.2005.069245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A.A.G., Eder H.H.G., Blum D.D.E., Drachev A., Research R.F.-E. An automated drug delivery system for focal epilepsy. Epilepsy Res. 2000;39:103–114. doi: 10.1016/s0920-1211(99)00107-2. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Takahashi S., Kanzaki R., Kawai K. State-dependent precursors of seizures in correlation-based functional networks of electrocorticograms of patients with temporal lobe epilepsy. Neurol. Sci. 2012;33:1355–1364. doi: 10.1007/s10072-012-0949-5. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Khoo H.M., Dubeau F., Gotman J. Association between scalp and intracerebral electroencephalographic seizure-onset patterns: a study in different lesional pathological substrates. Epilepsia. 2017;59:420–430. doi: 10.1111/epi.13979. [DOI] [PubMed] [Google Scholar]
- Theodore W.H., Fisher R.S. Brain stimulation for epilepsy. Lancet Neurol. 2004;3:111–118. doi: 10.1016/s1474-4422(03)00664-1. [DOI] [PubMed] [Google Scholar]
- Thodoroff P., Pineau J., Lim A. Learning Robust Features using Deep Learning for Automatic Seizure Detection. In: Doshi-Velez F., Fackler J., Kale D., Wallace B., Wiens J., editors. Proceedings of the 1st Machine Learning for Healthcare Conference, Proceedings of Machine Learning Research. PMLR, Children’s Hospital LA; Los Angeles, CA, USA: 2016. pp. 178–190. [Google Scholar]
- Varsavsky A., Mareels I., Cook M. CRC Press; 2016. Epileptic Seizures and the EEG : Measurement, Models, Detection and Prediction. [Google Scholar]
- Velis D., Plouin P., Gotman J., Da Silva F.L. Recommendations regarding the requirements and applications for long-term recordings in epilepsy. Epilepsia. 2007;48:379–384. doi: 10.1111/j.1528-1167.2007.00920.x. [DOI] [PubMed] [Google Scholar]
- Venkataraman V., Vlachos I., Faith A., Krishnan B., Tsakalis K., Treiman D., Iasemidis L. 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2014. Brain dynamics based automated epileptic seizure detection; pp. 946–949. [DOI] [PubMed] [Google Scholar]
- Wilson S. 2004. Method and System for Detecting Seizures Using Electroencephalograms. (US Pat) [Google Scholar]
- Wilson S.B., Scheuer M.L., Emerson R.G., Gabor A.J. Seizure detection: evaluation of the Reveal algorithm. Clin. Neurophysiol. 2004;115:2280–2291. doi: 10.1016/j.clinph.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Yuan Q., Zhou W., Xu F., Leng Y., Wei D. Epileptic EEG Identification via LBP Operators on Wavelet Coefficients. Int. J. Neural Syst. 2018;28:1850010. doi: 10.1142/S0129065718500107. [DOI] [PubMed] [Google Scholar]