Abstract
BATF functions in T cells and B cells to control the host response to antigen and promote the production of class switched immunoglobulins. In this study, we demonstrate that BATF expression increases rapidly, and transiently, following B cell stimulation and use an inducible murine model of BATF deletion to show that this induction is necessary, and sufficient, for immunoglobulin (Ig) class switch recombination (CSR). We examine two genes (Nfil3 and miR155gh) that are positively regulated, and one gene (Wnt10a) that is negatively regulated by BATF during CSR. These genes play essential roles in CSR and each impacts the expression and/or function of the other. Our observations allow these targets of BATF regulation to be positioned in a network upstream of the activation of germline transcripts (GLT) from the IgH locus and of transcriptional activation of Aicda – the gene encoding the enzyme directing Ig gene rearrangements. This work extends the knowledge of the molecular control of CSR and, importantly, positions the induction and function of BATF as an early event in this process.
Keywords: BATF, B cells, class switch recombination, transcription regulation, signaling pathways
Introduction
BATF is the founding member of the BATF family of basic leucine zipper (bZIP) transcription factors (reviewed in [1,2]). BATF proteins function to regulate gene transcription as heterodimers with other bZIP proteins, notably the JUN proteins [3–5]. BATF:JUN heterodimers preferentially bind consensus AP-1 DNA [3] or extended AP-1 elements, designated AICE, where members of the interferon regulatory factor (IRF) family of helix-turn-helix transcription factors are recruited through interaction with BATF [6]. Current models suggest that negative regulation of gene expression occurs when BATF:JUN dimers bind AP-1 sites, while positive gene regulation is associated with IRF:BATF:JUN complexes bound to AICE [1]. However, as the genes impacted by BATF in vivo continue to be identified, this model will undergo modification. In this regard, recent studies show that sequences proximal to AICE control how genes respond to variable levels, or to an alternative composition, of the IRF:BATF:JUN complex [7].
BATF expression is restricted to immune cells and is modulated by developmental transitions and environmental cues [1,2]. Data from BATF transgenic [8,9] and Batf knock-out mice [10–12] show that BATF is essential for both innate and adaptive immune responses. BATF plays a prominent role in the CD4+ T helper (Th)-17 and T follicular helper (Tfh) cell lineages [10,11], but is also critical for the antibody response in B cells [11,13], the control of CD8+ T cell “exhaustion” [14], the production of interleukin (IL)-9 by Th9 cells [15] and for the development of IL-3 dependent mast cells [16] and type1 regulatory T cells [17]. BATF is expressed in hematopoietic stem cells where it limits cell renewal and promotes lymphoid differentiation in response to DNA damage [18]. Experimentally blocking the activities of BATF protects against inflammatory, autoimmune and allergic diseases [10,15,19,20], disrupts anti-viral responses [14,21–23] and extends allograft survival [24]. Although BATF deletion can be compensated for by other members of the BATF protein family [25], co-expression of BATF proteins is rarely observed in vivo, and where it is, redundancy appears to be directed toward a restricted set of genes [7].
Understanding the role of BATF in specific cell types hinges on identifying BATF target genes. This approach proved valuable for understanding the regulatory network specifying Th17 cells [26] and for discovering that it is through an interaction with IRFs that BATF:JUN heterodimers activate gene expression [6]. In this study, we apply this approach to B cells. We observed biphasic regulation of BATF mRNA and protein following B cell stimulation and used a conditional Batf knock-out model to show that the early, transient induction of BATF is sufficient to initiate events that culminate in Ig CSR. Here we show that Nfil3, Wnt10a and microRNA 155 (miR155hg) are targets regulated by BATF early in CSR and that this regulation is essential for CSR. Our studies extend the knowledge of the molecular network controlling CSR in B cells and position BATF induction and function as critical, early events in the process.
Results
BATF displays a biphasic pattern of expression in stimulated B cells
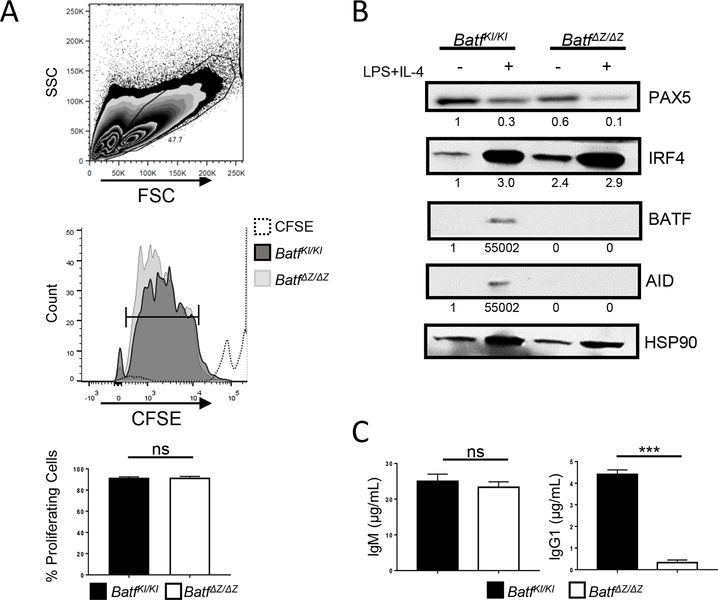
Previous studies with BATF deficient mice revealed a profound defect in antigen triggered germinal center formation and in the generation of class switched antibodies [11,13]. B cells isolated from BATF deficient mice (BatfΔZ/ΔZ) proliferate appropriately when cultured in the presence of 20 μg/mL lipopolysaccharide (LPS) and 20 ng/mL IL-4 [11](Figure 1A). These cells properly regulate the expression of many B cell genes, but do not express activation induced cytidine deaminase (AID), the enzyme required for Ig CSR (Figure 1B). Media harvested from stimulated BatfΔZ/ΔZ B cells contains IgM but, as expected, no detectable IgG1 (Figure 1C), indicating that the BATF null cells do not undergo CSR.
Figure 1. BATF is required for B cells to produce IgG.
Splenic B cells, isolated from BatfKI/KI and BatfΔZ/ΔZ mice were labeled with CFSE and cultured with LPS and IL-4. CFSE dilution was analyzed by flow cytometry. The gating strategies to identify lymphocytes using (upper panel) and the percent of those cells that are proliferating (bar, middle panel) are shown. Results in the histogram (lower panel) are the mean±SEM of data collected from four independent CFSE experiments, each using cells from one mouse per genotype. SSC (side scatter); FSC (forward scatter) (B) B cells isolated from BatfKI/KI and BatfΔZ/ΔZ spleens were cultured in the presence of absence of LPS and IL-4 and protein extracts immunoblotted to visualize the indicated proteins. Signals were normalized to HSP90 and expressed relative to unstimulated BatfKI/KI B cells (set to 1.0). Shown is a representative blot from two independent experiments using protein from two mice per genotype. (C) Media from cultured BatfKI/KI and BatfΔZ/ΔZ B cells were screened by ELISA for the indicated Ig. Results shown are the mean±SEM pooled from two independent experiments, each using cells from two mice per genotype. (***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test)
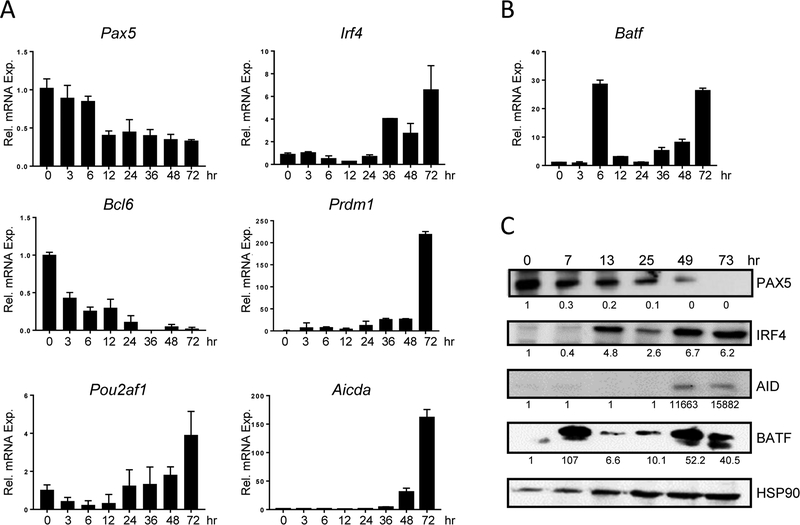
The gene expression program required for CSR is well known (reviewed in [27]), yet the role of BATF in that program is poorly characterized. A time-course study was performed to track BATF expression in B cells following stimulation. Transcripts downregulated (Pax5, Bcl6, Pou2af1) or elevated (Irf4, Prdm1, Aicda - the gene encoding AID) during CSR were used as controls (Figure 2A). Batf mRNA was expressed at low levels in resting cells, but accumulated to a high level 6 hrs after stimulation (Figure 2B). Almost as rapidly as Batf mRNA was induced, levels were down-regulated, before gradually increasing again as Aicda mRNA accumulates (Figure 2A and B). The pattern of BATF protein expression in B cells (Figure 2C) mirrors the biphasic pattern of Batf mRNA expression. This demonstrates, as was previously shown in T cells, that BATF protein is rapidly turned over in B cells and that Batf mRNA accurately reflects levels of BATF protein [9].
Figure 2. BATF has a biphasic expression pattern.
(A, B) RNA was prepared from WT splenic B cells at the indicated times following stimulation. RT-qPCR, performed in duplicate, was used to detect the indicated transcripts. Results were normalized to β-actin and expressed relative to time 0 (set to 1.0). Results shown are the mean±SEM of data obtained from four independent experiments, each using RNA from one mouse. (C) Protein was isolated from WT B cells at the indicated times following stimulation. Immunoblotting was used to detect the expression of proteins. Signals were normalized to HSP90 and expressed relative to time 0 (set to 1.0). Shown is a representative blot from four independent experiments, each using protein pooled from two mice.
Early induction of BATF in stimulated B cells has functional significance
The BATF expression data from stimulated B cells are consistent with observations in other immune cell types and reflects the current notion that BATF is a “pioneering” transcription factor that reshapes the genetic landscape of differentiating immune cells for subsequent gene expression events [17,26,28–30]. To test if the early spike in BATF expression has functional significance with regard to CSR in B cells, we generated IzT mice (BatfKI/KIROSA26mT/mGUBCCre-ERT2). Treatment of IzT mice with tamoxifen (TAM) daily over a five day period in vivo (Suppl. Figure 1A) leads to the CRE recombinase-mediated expression of green fluorescent protein (GFP) in all tissues tested and to the deletion of the floxed Batf alleles (Suppl. Figure 1B and data not shown). GFP+ splenocytes isolated from TAM-treated IzT mice express Irf4 and Pax5 but, as expected, do not express Batf or Aicda (Suppl. Figure 1C).
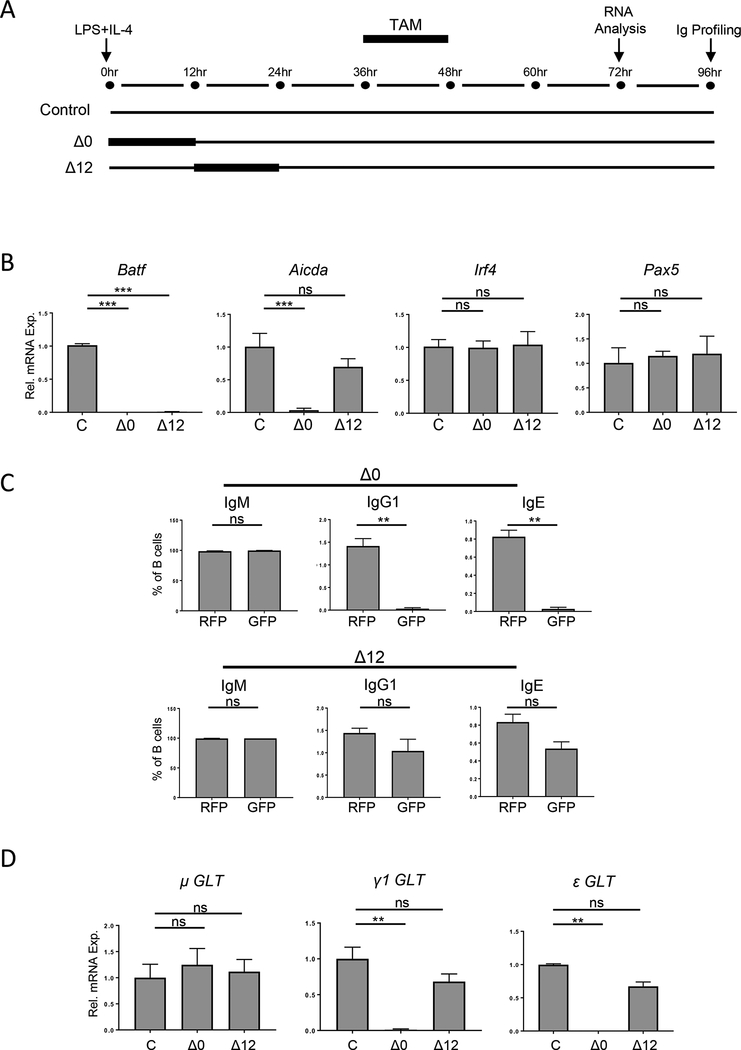
To delete Batf in naïve B cells at different time points following stimulation, the onset of exposure to TAM was varied (Figure 3A). Stimulated, non-TAM treated cells served as the control. Δ0 cells deplete Batf at the time of stimulation and should behave as BatfΔZ/ΔZ B cells. Δ12 cells preserve the induction of Batf expression for 6 hrs post-stimulation, but cannot induce the second wave of Batf expression that begins after 24 hrs. At 72 hrs, GFP+ B cells from the Δ0 and Δ12 groups were purified by flow activated cell sorting (FACS) (gating as in Suppl. Figure 2A) and RNA harvested from all three groups. As expected, Batf mRNA was undetectable in both the Δ0 and Δ12 groups, while Irf4 and Pax5 mRNA levels were expressed normally (Figure 3B). Interestingly, Aicda mRNA was not induced in the Δ0 cells, but was induced in Δ12 cells (Figure 3B). To confirm that the level of Aicda expression in Δ12 cells was driving CSR, control, Δ0 and Δ12 cells were cultured for 96 hrs, after which the levels of IgG1 and IgE on GFP+ and RFP+ cells from the same groups were measured (Figure 3C, gating shown in Suppl. Figure 2A and 2B). Indeed, while class switched Ig were barely detectable on GFP+ Δ0 cells, GFP+ Δ12 cells displayed levels comparable to their RFP+ counterparts. Additionally, RNA from the three groups was analyzed for the GLT induced from I-region promoters of the IgH locus following stimulation with LPS and IL-4 [31] (Figure 3D). All groups transcribe the μ promoter region for IgM, as expected. However, while γ1 (IgG1) and ε (IgE) were detected in both the control and Δ12 cells, these GLTs were not present in Δ0 cells. These results indicate that the early peak of BATF expression following B cell stimulation is sufficient to set in motion a program of gene expression that leads to GLT, AID expression and the production of class switched Ig.
Figure 3. Early expression of BATF is necessary and sufficient for CSR.
(A) Schematic of TAM treatment to generate the timed deletion of Batf in the Δ0 and Δ12 IzT B cell cultures. (B and D) Control (C), Δ0 and Δ12 cells were sorted and RNA analyzed by RT-qPCR for the indicated transcripts, in duplicate. Results are normalized to β-actin (B) or Gapdh (D) and expressed relative to the control (set to 1.0). Results shown are the mean±SEM of data from four independent experiments, using RNA from one mouse per treatment. (C) Δ0 and Δ12 B cells were surface stained for IgM, IgG1, and IgE and profiled by flow cytometry. Gating strategy is in Suppl. Figure 2. Results are presented as the mean±SEM of data from six independent experiments, each using cells from one mouse per treatment. (**p<0.01; ***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test)
BATF regulates gene targets critical for CSR early after B cell activation
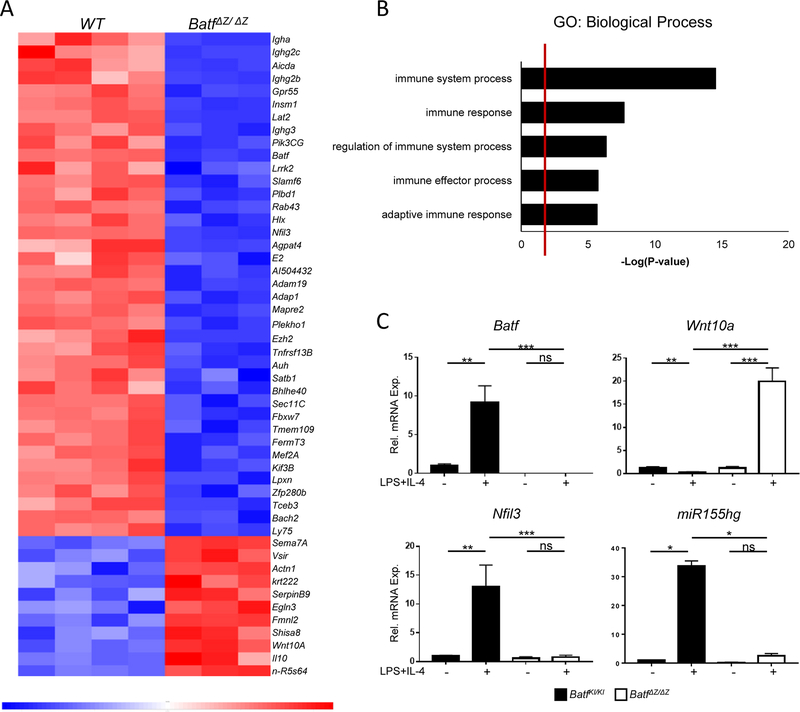
Given that the early induction of BATF expression is sufficient to drive CSR, RNA-Seq was performed using WT and BatfΔZ/ΔZ B cells stimulated for only 6 hrs (see Materials and Methods for details). EdgeR analysis identified 353 differentially expressed genes, with 133 upregulated and 220 downregulated in the BatfΔZ/ΔZ cells compared to WT (GSE106427). The top 50 significantly altered genes are presented in a heat map (Figure 4A). 44% of the Gene Ontology (GO) categories containing these altered genes are immune system-related, including the five listed in Figure 4B.
Figure 4. Analysis of Batf ΔZ/ΔZ B cell transcripts by RNA-Seq.
(A) Heatmap of the top 50 significantly altered genes in BatfΔZ/ΔZ B cells at 6hrs post-stimulation. (B) Shown are the top 5 GO:Biological Processes pathways into which genes identified by the RNA-Seq cluster. (C) RNA isolated from BatfKI/KI and BatfΔZ/ΔZ splenic B cells cultured in the presence or absence of LPS and IL-4 was analyzed by RT-qPCR, in duplicate, to confirm mis-expression of three putative BATF targets genes. Results were normalized to β-actin and expressed relative to unstimulated BatfKI/KI B cells (set to 1.0). Results shown are the mean±SEM of data collected from two independent experiments, each using RNA from two mice per genotype. (*p<0.05; **p<0.01; ***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test)
Among the differentially expressed genes in BatfΔZ/ΔZ B cells, two were chosen for further study. Nfil3 encodes a bZIP transcription factor which, like BATF, is required for Th17 differentiation [32] and for B cell CSR [33]. Nfil3 shows a 1.7 log fold decrease in BatfΔZ/ΔZ B cells and contains several AICE motifs (Supplemental Figure 5A). Wnt10a encodes a secreted glycoprotein that is expressed at low levels in normal lymphocytes, but shows increased expression in human leukemia cell lines [34,35]. Hyperactivation of WNT signaling is associated with impaired antibody production by B cells [36]. Wnt10a contains AP-1 sites and shows a 1.5 log fold increase in BatfΔZ/ΔZ B cells. The third molecule selected is miR155 which is transcribed from the miR155gh (Bic) gene (reviewed in [37]). MicroRNAs were not captured in the starting material prepared for the RNA-Seq, yet a previous, unpublished study from our laboratory noted that miR155 is barely detected in stimulated BatfΔZ/ΔZ B cells. Since there is disagreement in the literature as to whether miR155 functions as a positive, or negative, regulator of CSR [38–40] (reviewed in [37]), additional investigation of the AICE-containing mir155gh gene in BatfΔZ/ΔZ B cells was warranted. Standard RT-qPCR with RNA isolated from BatfKI/KI and BatfΔZ/ΔZ B cells stimulated for 48hrs confirmed that the loss of BATF is correlated with dramatically reduced levels of Nfil3 and miR155gh transcripts, along with increased levels of Wnt10a mRNA (Figure 4C).
Expressing BATF in BatfΔZ/ΔZ B cells restores the proper expression of Nfil3, miR155hg, and Wnt10a
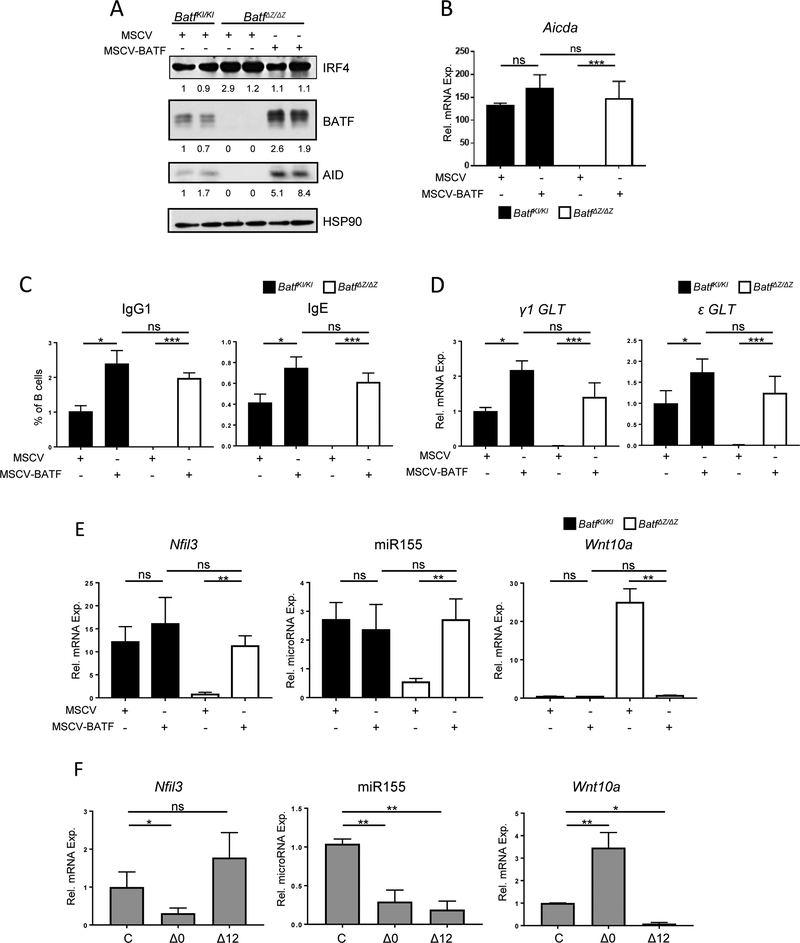
Resting B cells from BatfKI/KI and BatfΔZ/ΔZ mice were cultured in vitro and transduced with a murine stem cell virus (MSCV) expressing the Thy1.1 selectable marker only, or Thy1.1 and HA-tagged BATF (MSCV-BATF). Thy1.1+ cells were isolated by FACS and returned to culture with LPS and IL-4. MSCV-BATF restored robust levels of BATF protein in BatfΔZ/ΔZ B cells and resulted in the induction of AID protein (Figure 5A) and Aicda transcripts (Figure 5B). Expression of other CSR associated genes remained intact (Suppl. Figure 3). Restoring BATF expression in BatfΔZ/ΔZ B cells also generated cultures capable of producing class switched IgG1 (Figure 5C) and expressing γ1 and ε GLT (Figure 5D).
Figure 5: BATF re-expression restores gene expression and CSR in BatfΔZ/ΔZ B cells.
Stimulated B cells isolated from spleens of BatfKI/KI and BatfΔZ/ΔZ mice were transduced with either MSCV or MSCV-BATF and Thy1.1 + cells isolated by FACS. (A) Protein extracts were immunoblotted, in duplicate, to detect the indicated proteins. Signals were normalized to HSP90 and expressed relative to MSCV-transduced, stimulated BatfKI/KI B cells (set to 1.0). Shown is a representative blot from three individual experiments in which protein from two BatfKI/KI or BatfΔZ/ΔΖ mice were blotted in parallel for each treatment. (B, D and E) RNA and microRNA from each group was analyzed by RT-qPCR to detect the indicated transcripts, in duplicate. Aicda, Nfil3 and Wnt10a transcripts are normalized to β-actin (B and E), miR155 to RNU6 (E) and GLT transcripts to Gapdh (D) and are expressed relative to levels in unstimulated BatfKI/KI B cells (B and E) (set to 1.0, but not shown) or MSCV-transduced BatfKI/KI B cells (set to 1.0) (D). Results shown are the mean±SEM of data collected from three independent experiments, each using RNA or microRNA from two mice per genotype and treatment. (C) Transduced cells from each group were profiled for Thy1.1 and surface IgG1 and IgE expression by flow cytometry. See Suppl. Figure 7 for gating strategy. Results shown are the mean±SEM of data collected from three independent experiments, each using cells from two mice per genotype and treatment. (F) RNA and microRNA were prepared from control (C) and TAM-treated Δ0 and Δ12 GFP+ IzT B cells (see Figure 3). RT-qPCR was used to detect the indicated RNAs, in duplicate. Results are normalized to β-actin (mRNA) or RNU6 (miRNA) and expressed relative to levels in Izt control cells (set to 1.0). Results are presented as the mean±SEM of data from four independent experiments, using RNA or microRNA isolated from one mouse per treatment. (*p<0.05; **p<0.01; ***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test)
To investigate the BATF-dependent regulation of Nfil3, Wnt10a and miR155hg, two experiments were performed. In the first, MSCV-BATF was used to re-establish BATF expression in stimulated BatfΔZ/ΔZ B cells. This resulted in the accumulation of Nfil3 transcripts and miR155, as well as in the dramatic reduction of Wnt10a mRNA (Figure 5E). The second approach utilized TAM treatment of IzT B cells (as in Figure 2A) to test if the WT expression pattern of each of these molecules is sustained exclusively by the early spike in BATF expression. The results were dramatic (Figure 5F). Both the induction of Nfil3 mRNA and the repression of Wnt10a transcripts required BATF expression at 6 hrs, but did not rely on sustained BATF expression after that time. In contrast, miR155 levels were repressed in both treatment groups, suggesting that BATF activity is required continuously to maintain miR155hg transcription. This result with miR155 is consistent with the delayed accumulation of miR155hg mRNA in stimulated B cells (Suppl. Figure 4).
BATF binds as predicted to target gene loci
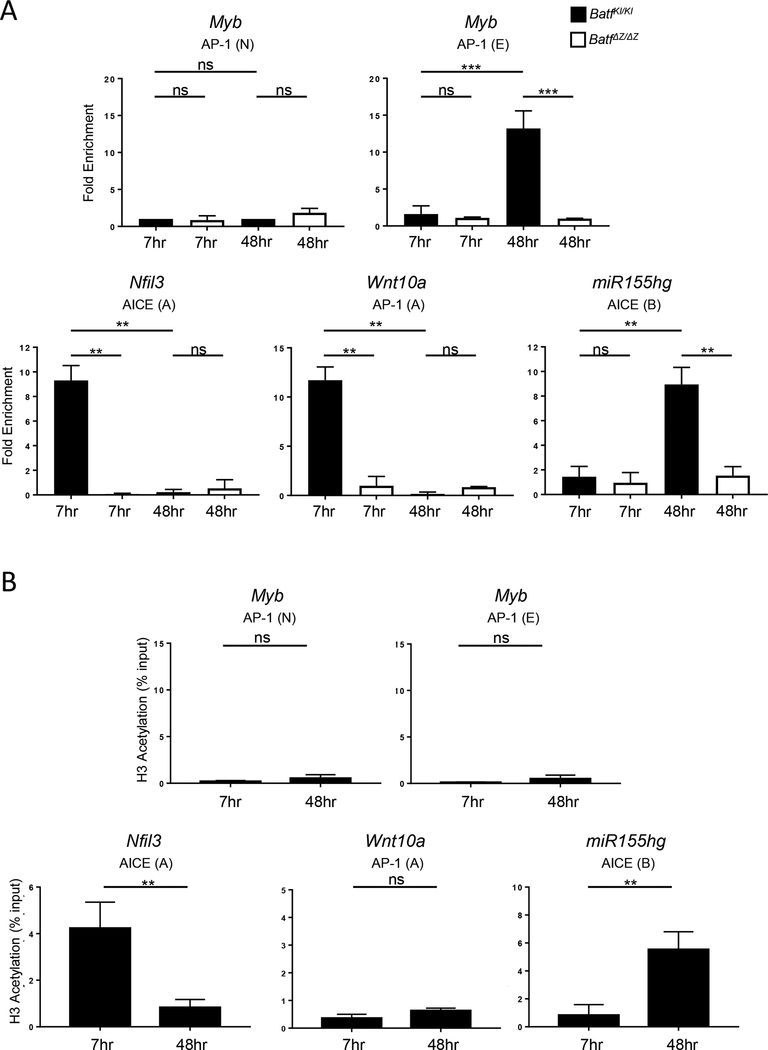
Nfil3, Wnt10a and mir155gh contain consensus sequences for binding BATF containing transcription complexes associated with the activation (AICE) or repression (AP-1) of gene transcription (Suppl. Figure 5A). To test if BATF directly binds to any of these sites, chromatin immunoprecipitation (ChIP) was performed using WT and BatfΔZ/ΔZ B cells and a BATF monoclonal antibody (see Materials and Methods for details). Purified DNA was analyzed by PCR for the presence of fragments spanning key AICE or AP-1 motifs. A previous study identified an AP-1 site in the Myb promoter as a direct target for repression by BATF:JUN in mouse myeloid leukemia cells [29]. Since Myb is expressed in B cells [41], detection of AP-1(E) DNA served as a positive control, while AP-1(N), a second AP-1 site from Myb that is not bound by BATF:JUN, served as the negative control [29]. Figure 6A shows that Myb AP-1(E) is detected in DNA precipitated from WT B cells, 48 hrs following stimulation. The absence of AP-1(E) amplification in DNA prepared from BatfΔZ/ΔZ cells demonstrates the specificity of the BATF antibody.
Figure 6: BATF binding accompanies regulation of Nfil3, miR155hg and Wnt10a expression.
Chromatin was prepared from B cells isolated from spleens of WT and BatfΔZ/ΔZ mice and stimulated with LPS and IL-4. ChIP was performed to detect binding of BATF (A) or acetylation of H3K27 (B) at the indicated AP-1 or AICE motifs identified within Nfil3, mir155gh and Wnt10a (see Suppl. Figure 5A). BATF binding to AP-1(E) of Myb was used as a positive control while non-BATF bound AP-1 (N) of Myb was used as a normalization control for background [29]. (B) DNA associated with H3K27 acetylation is expressed as a % of total input. Results shown are the mean±SEM of data obtained from two independent ChIP experiments, each performed with DNA isolated from two mice per genotype and time point. (**p<0.01; ***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test)
ChIP-qPCR revealed that BATF binds to AICE(A) of Nfil3 and to AP-1(A) of Wnt10a at 7 hrs following B cell stimulation, but not at 48 hrs post-activation (Figure 6A). Other consensus sites identified within these genes showed no significant amplification (Suppl. Figure 5B). Conversely, BATF binds strongly to the AICE(B) motif of miR155hg at 48hr, but not at 7 hrs post activation. Again, binding was not observed at the upstream AICE(A) of miR155hg at either time point (Suppl. Figure 5B). ChIP of material from stimulated WT B cells using an antibody to detect acetylation of histone H3 at lysine 27 confirmed that the BATF binding at AICE(A) of Nfil3 and AICE(B) of miR155hg is associated with an active chromatin configuration (Figure 6B). As expected, H3 acetylation was not a feature of the AP-1(A) of Wnt10a or the AP-1(E) of Myb (Figure 6C) since BATF binding here correlates with the repression of gene transcription. H3 acetylation also was not associated with any of the other sites tested from the Nfil3, miR155gh or Wnt10a genes (Suppl. Figure 5C).
NFIL3, miR155, and WNT10A influence AID expression and GLT
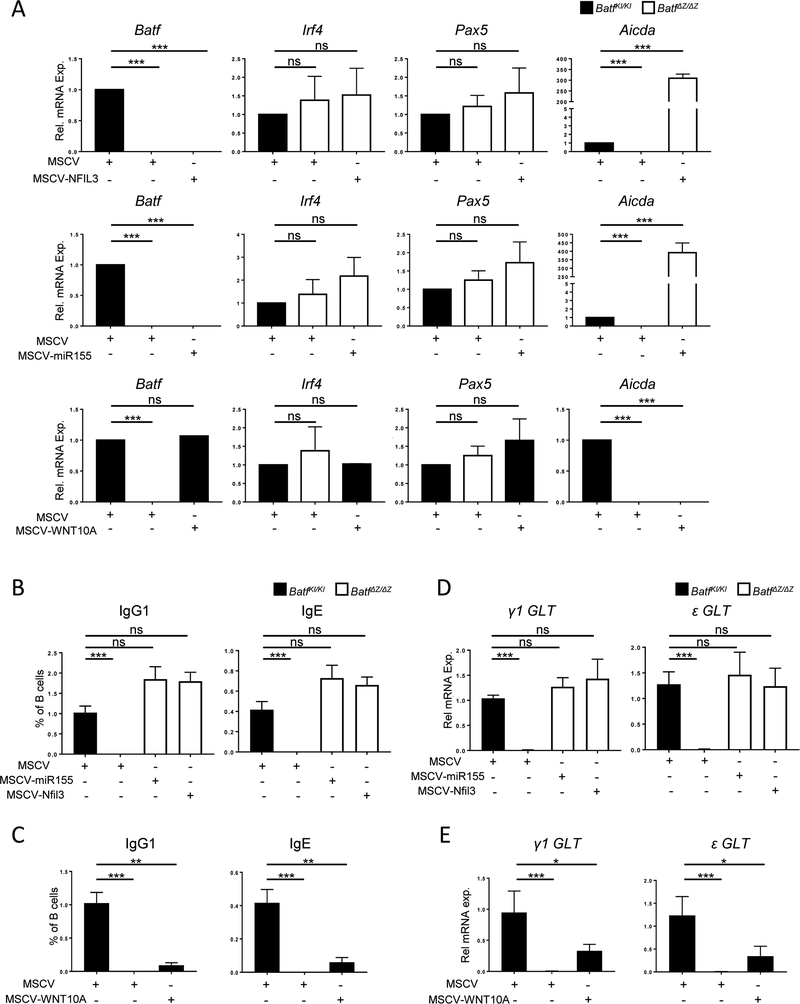
To further understand how the CSR-deficient phenotype of BatfΔZ/ΔZ B cells relies on the function of BATF target molecules, we employed MSCV-Thy1.1 vectors expressing NFIL3, miR155, and WNT10A (Suppl. Figure 6A and B) to restore NFIL3 and miR155 in BatfΔZ/ΔZ B cells or constitutively express WNT10A in WT (BatfKI/KI) B cells. Transduction with these viruses had no deleterious effect on B cell proliferation in response to LPS and IL-4 (Suppl. Figure 6C). Exogenous expression of NFIL3 or miR155 in stimulated BatfΔZ/ΔZ B cells rescued Aicda mRNA expression (Figure 7A) within 48 hrs and allowed for the production of both IgG1 and IgE by 96 hrs (Figure 7B and Suppl. Figure 7). Similarly, expression of WNT10A in BatfKI/KI cells blocked Aicda expression and CSR (Figure 7A and 7C), despite the cells retaining normal levels of BATF. Expression of γ1 and ε GLT was assayed and transcripts were detected in the cells transduced with MSCV-NFIL3 and MSCV-miR155 (Figure 7D). Interestingly, for the WNT10A expressing cells, where AID is fully silenced, low levels of GLT expression were retained (Figure 7E).
Figure 7: NFIL3, miR155 and WNT10A impact Aicda expression and CSR.
Stimulated B cells isolated from spleens of BatfKI/KI and BatfΔZ/ΔZ mice were transduced with the indicated retroviruses. Transduced, Thy1.1 cells isolated by FACS. (A, D and E) RNA from each group was analyzed by RT-qPCR to detect the indicated transcripts, in duplicate. Results are normalized to β-actin (A) or Gapdh (D and E) and are expressed relative to levels in MSCV-transduced, BatfKI/KI B cells (set to 1.0). Results are presented as the mean±SEM of data collected from three independent experiments, each using RNA from two mice per genotype. (B and C) Transduced cells from each group were profiled for Thy1.1 and surface IgG1 and IgE expression by flow cytometry. Gating strategy is in Suppl. Figure 7. Results shown are the mean±SEM of data obtained from six independent experiments, each using cells from one mouse per treatment. (*p<0.05; **p<0.01; ***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test)
Positioning BATF and its targets within the CSR network
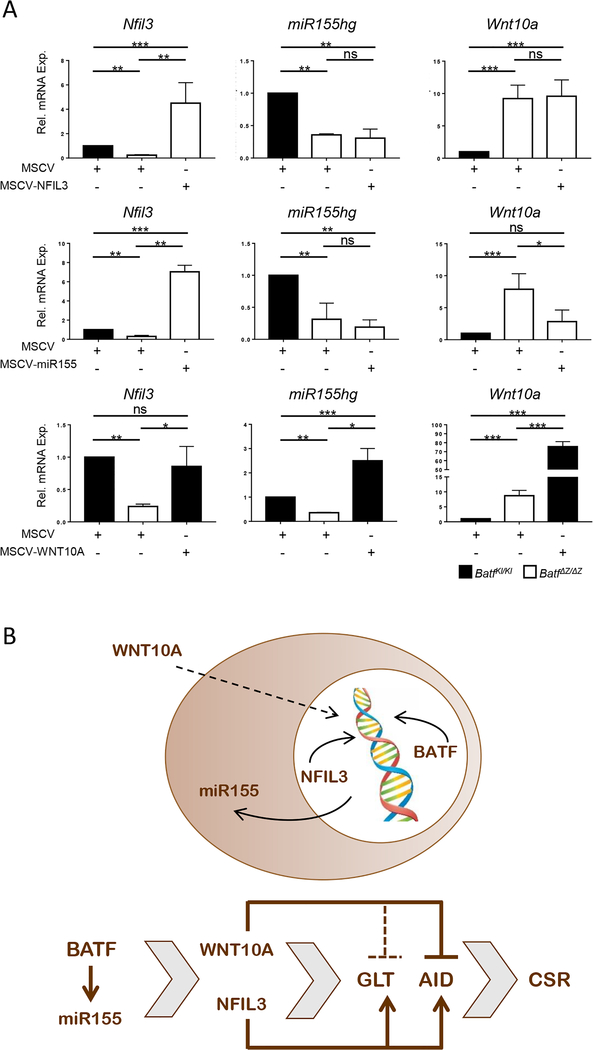
These studies suggest that BATF functions during the early stages of CSR to regulate the expression of molecules that will subsequently coordinate transcription (NFIL3), modulate cellular signaling pathways (WNT10A) and influence the efficiency of mRNA accumulation and/or translation (miR155). To position these BATF targets in the CSR network downstream of BATF, RNA isolated from BatfΔZ/ΔZ B cells transduced with MSCV-NFIL3 or MSCV-miR155 and from BatfKI/KI cells transduced with MSCV-WNT10A were examined for the expression of all three molecules. As shown in Figure 8A, restoring NFIL3 expression does not alter Wnt10a or miR155hg mRNA levels in these cells, allowing for the positioning of NFIL3 as a proximal inducer of GLT, AID and CSR (Figure 8B). On the other hand, MSCV-miR155 expression (which is not reflected as endogenous miR155gh mRNA) rescues induction of Nfil3 and leads to a significant repression of Wnt10a in BatfΔZ/ΔZ B cells (Figure 8A), suggesting that miR155 can substitute for BATF as an initiator of all of these events (Figure 8B). Interestingly, while expression of WNT10A in BatfKI/KI B cells efficiently blocks AID induction and CSR (Figure 7A and C), it has no impact on Batf (Figure 7A) and Nfil3, and causes elevated expression of miR155hg (Figure 8A). The ability of WNT10A to suppress GLT in BatfKI/KI cells is not complete, however, implying there are differences in the pathways used by NFIL3 and WNT10A to influence GLT. On the other hand, the effects of WNT10A and NFIL3 on AID point to opposing roles for these two molecules in events most proximal to the induction of CSR (Figure 8B), suggesting that the relative levels of each protein in cells may dictate the dominance of one pathway over the other.
Figure 8: Hierarchy of BATF, NFIL3, miR155 and WNT10A in regulating CSR.
(A) Stimulated B cells from BatfKI/KI and BatfΔZ/ΔZ spleens were transduced with the indicated retroviruses and Thy1.1 cells isolated by FACS. RNA from each group was analyzed by RT-qPCR to detect the indicated transcripts, in duplicate. Results are normalized to β-actin and expressed relative to the level in MSCV-transduced BatfKI/KI B cells (set to 1.0). Results shown are the mean±SEM of data collected from three independent experiments, each with RNA from two mice per genotype. (*p<0.05; **p<0.01; ***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test) (B) Working model depicting the primary site of action of the molecules examined in this study (upper) and the hierarchy of their actions in inducing IgH GLT, AID expression and CSR in B cells (lower).
Discussion
Dozens of molecular events are required for CSR and these events are coordinated in a network initiated by a stimulus that induces B cell proliferation, the production of GLT from the IgH locus, AID expression and the recombination of CH genes to produce class switched Ig (reviewed in [27]). There are several ways to induce CSR in cultured B cells, including treatment with LPS and IL-4, which was used here and in previous work to show that Batf deficient B cells proliferate, yet are unable to execute subsequent steps in CSR [11,13]. In this study, we show that B cells stimulated with LPS and IL-4 express BATF in a biphasic pattern, with an early spike in expression followed by a rapid decline and a more steady accumulation as IgM producing B cells undergo GLT, express AID and acquire the ability to produce IgG and IgE. A similar pattern of Batf mRNA and protein induction was first noted in a myeloid cell model of macrophage differentiation, and later in Th17, CD8+ T and type I regulatory T cells, where BATF was categorized as a pioneering transcription factor responsible for reshaping the genetic landscape of differentiating T cells [17,26,28–30]. Our current work supports the role of BATF as a pioneering transcription factor in B cells. Whereas the induced deletion of Batf at the time of stimulation completely blocks GLT, AID expression and CSR, silencing BATF after 6 hrs of stimulation supports all functions leading to CSR. Unfortunately, the IzT mice used this study do not allow for additional fine-tuning of BATF expression during this process. Efforts to design an in vivo tetracycline-regulated system of BATF expression are underway.
To shed light on the function of BATF in the early events leading to CSR, we investigated two positive targets and one negative target of BATF regulation. These targets were selected for their diverse functions, plus their history with regard to B cell CSR. The NFIL3 transcription factor was shown to be essential for the induction of B cell CSR by LPS and IL-4 [33], but a direct link to regulation by BATF had not been described. Signaling through the canonical WNT pathway inhibits IgG production in B cells [36], but there was no documented role for an endogenously expressed WNT protein in activating that pathway. The role of miR155 in CSR is controversial (review in [37]). Results from miR155 deficient mice provide evidence that miR155 targets PU.1 transcripts to induce CSR [39], while separate studies have shown that miR155 directly regulates Aicda transcripts to inhibit CSR [38,40]. Restoring BATF expression in BatfΔZ/ΔZ B cells resulted in the appropriate induction, or repression, of all three BATF targets and BATF binds directly to sequences within the regulatory regions of all three genes. When each target molecule was expressed individually in BatfΔZ/ΔZ B cells (NFIL3 and miR155) or in BatfKI/KI B cells (WNT10A), CSR was induced or repressed as expected. The ability of miR155 to dramatically induce Aicda mRNA and protein in BatfΔZ/ΔZ B cells (Figure 7A and data not shown) demonstrates that in our experimental model, miR155 is not a negative regulator of AID and CSR. Clearly, more studies are needed to sort out the pleiotropic effects of miR155 in B cells and explain why different experimental models of CSR are generating such disparate results.
Perhaps the most interesting data are the observed responses of these targets to each other. For miR155, expression in BatfΔZ/ΔZ B cells resulted in a significant repression of Wnt10a and induction of Nfil3, apparently through a BATF-independent mechanism. Expression of NFIL3 exerted no influence on the high level of Wnt10a mRNA or the low level of miR155hg in BatfΔZ/ΔZ B cells, yet robustly restored GLT, AID and CSR. Similarly, when WNT10A was expressed constitutively in BatfKI/KI cells, AID expression and CSR did not occur, despite the induction of cell proliferation and sustained expression of Batf, Nfil3 and miR155gh. Interestingly, WNT10A overexpression in BatfKI/KI B cells did not fully silence GLT, suggesting that at least a part of the GLT induction pathway mediated by BATF (through up-regulation of NFIL3) cannot be targeted for negative regulation by WNT10A. Taken together, we propose a working model to describe a hierarchy within the CSR network regulated by BATF, with the more widespread positive impact of a microRNA (miR155) functioning at a level equivalent to BATF, while NFIL3 and WNT10A function downstream, in bifurcating, antagonistic pathways to impact GLT, AID expression and CSR. Experiments in which the levels of NFIL3 and WNT10A are varied relative to each other will be necessary to test this model. We also are intrigued by the ability of miR155 to restore the pattern of Nfil3 and Wnt10a gene expression in the absence of BATF, as defining the mechanism underlying this observation might also explain how the expression profile of these two genes (and not miR155gh) is maintained in CSR when BATF is experimentally silenced (Figure 5E).
Although the process of CSR in B cells is complex, we have clarified the role of BATF in the process. The positioning of BATF as an early, or pioneering, regulator of a diverse gene set impacting transcription events (Nfil3), post-transcriptional events (miR155gh), and cellular signaling (Wnt10a) is very exciting. While the negative impact of canonical WNT signaling on CSR was noted previously [36], a role for WNT10A expression was not, and the availability of recombinant WNT10A and WNT pathway inhibitors should facilitate future investigations of this pathway in CSR. Finally, as BATF, NFIL3 and miR155 are known regulators Th2, Th17 and Tfh cell differentiation and function [10,11,13,42,43], it is not surprising that the BATF targets studied here are also misregulated in BatfΔZ/ΔZ CD4+ T cells (Morman and Taparowsky, in preparation). It will be important to define the composition of the pioneering complexes containing BATF as a first step towards understanding how these complexes function to reshape the landscape of the genome in ways that allow additional signals to drive cells toward different cellular fates.
Materials and Methods
Mouse lines and in vivo studies
C57BL/6J mice were provided by the Transgenic Mouse Core Facility (TMCF) of the Purdue University Center for Cancer Research. C57BL/6J BatfKI/KI mice, homozygous for a floxed Batf exon 3, and C57BL/6J BatfΔZ/ΔZ mice, derived by in vivo CRE-mediated deletion of the floxed BatfKI alleles, were described previously [11]. C57BL/6 ROSAmT/mG [Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo; JAX Stock No: 007576] and UBC-Cre-ERT2 [Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb; JAX Stock No: 007001] mice were used in crosses with BatfKI/KI mice to obtain the IzT (BatfKI/KI ROSA26mT/mG UBCCre-ERT2) mice. Experiments used 9–10 week old animals, sex-matched when possible. Oral gavage was used to treat Izt mice with tamoxifen (Sigma-Aldrich), 0.5uM/g body weight, daily for 5 days. Corn oil was administered as the control. On day 6, spleen, thymus and pancreas were fixed and processed for immunofluorescent staining as described [11]. Antibodies are listed in Suppl. Table 1. Nuclei were visualized using VECTASHIELD mounting medium containing DAPI (Vector Laboratories). All experiments were conducted in compliance with NIH and Purdue University IACUC guidelines. Protocols used [1110000057 (EJT) and 1110000037 (SFK)] were approved by the IACUC of Purdue University.
Cell culture
Primary splenic B cells were isolated with CD43(Ly48) microbeads (Miltenyi Biotec) and cultured at 5×105 cells/mL in RPMI 1640 (Gibco) supplemented with 100U/mL penicillin-streptomycin (Gibco), 10% heat inactivated fetal bovine serum (Gibco) and 50μM 2-mercaptoethanol (Sigma-Aldrich). Media with 20μg/mL LPS (Sigma) and 20ng/mL IL-4 (BD Biosciences) was used to stimulate cells and induce CSR. HeLa 229 cells (CCL-2.1; ATCC) were maintained in DMEM (HyClone), 100U/mL penicillin-streptomycin and 10% fetal bovine serum. B cells at a density of 10×106 cells/mL were labeled with 1μM CFSE (Cell Division Tracker Kit, BD Biosciences) and stimulation-induced proliferation was profiled after 72hrs using flow cytometry and analyzed using FlowJo software (TreeStar).
Immunoblot
Protein was isolated from B cells cultured as indicated for 48hrs using standard RIPA lysis buffer. Immunoblots were performed as described [9]. Antibodies are listed in Suppl. Table 1.
ELISA
Media collected from B cells stimulated for 72 hrs was diluted in Assay Diluent (eBioscience) and ELISA performed as previously described [11]. Absorbance was measured at 405nm with a Multiskan FC (Thermo Fisher).
Analysis of mRNA and microRNA
B cells were stimulated for 48hrs and RNA or microRNA prepared using the RNeasy mini kit or miRNeasy mini kit (Qiagen). Reverse transcription (RT) with the iSCRIPT Kit (Bio-Rad) or miSCRIPT II RT kit (Qiagen) was used to prepare cDNA. Specific sequences were quantified using quantitative polymerase chain reaction (qPCR) with FastStart Universal SYBR Green Master (Roche) and a Light Cycler 96 System (Roche). Primer sequences are listed in Suppl. Table 2. ΔΔCT values were used to calculate relative expression of each mRNA or microRNA.
Flow cytometry
Profiling for Ig expression in GFP+ or Thy1.1+ B cells was performed after 96 hrs of stimulation using a LSRFortessa (BD Biosciences). Sorting Thy1.1, GFP or RFP B cells for RNA or protein analyses was performed using a FACSAria III (BD Biosciences). Fc block (BD Biosciences) at a 1:100 dilution prevented non-specific interaction. Antibodies used are listed in Suppl. Table 1. Data were analyzed with FlowJo software (TreeStar).
RNA-Seq
B cells from 4 WT and 3 BatfΔZ/ΔZ mice were stimulated for 6 hrs and RNA isolated using E.Z.N.A. Total RNA Mini Kit (Omega Bio-Tek). The Purdue Genomics Core Facility generated a paired-end library for each RNA sample and the sequences determined using an Illumina HiSeq System to a depth of >30 million reads per sample. Reads were filtered using the FASTX toolkit (v 0.0.13) to remove bases with a Phred score below 30 and then aligned with Tophat (v 2.0.6) to the mouse genome (mm10). Read counts for each gene were generated using HTSeq (v 0.5.3p7). Differential expression was determined using edgeR (v 3.03) with a false discovery rate of < 0.05 and a fold-change cut-off of 2. The heatmap was created with the edgeR heatmap.2 function from the gplots package. Gene Ontology analysis was performed using the PANTHER Classification System webtool with p-values adjusted for multiple testing using the Bonferroni correction.
Retroviral infection
Derivatives of the MSCV-Thy1.1-IRES vector (Addgene #17442) containing hemagglutinin-tagged mouse BATF (MSCV-BATF), MYC-tagged mouse NFIL3 (MSCV-NFIL3) and MYC-tagged mouse WNT10A (MSCV-WNT10A) were generated by PCR cloning. MSCV-Thy1.1-IRES-miR155 was generated by PCR cloning using the MSCV-miR155-eGPF vector as a template (gift of Ricardo Aguiar [44]). Virus supernatants were produced by the Sanford Burnham Prebys Medical Discovery Institute. B cells stimulated for 4 hrs were transduced using a one-hour spinfection protocol. For RNA and protein isolation, transduced cells were cultured with stimulation for 24 hrs, sorted and returned to culture for another 24 hours. For Ig expression analyses, transduced cells were cultured with stimulation for 96 hours and profiled by flow cytometry.
ChIP assay
B cells were stimulated for 7hrs or 48hrs and chromatin prepared from the cells using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling). Chromatin was fragmented by sonicating 3 times for 30 seconds each on ice. Antibodies for immunoprecipitation are listed in Suppl. Table 1. ChIP-qPCR was used to quantify specific sequences within the isolated DNA. Primer sequences are listed in Suppl. Table 3.
Statistical Analysis
Graphpad Prisim 5 was used to perform all statistical analyses.
Supplementary Material
Supplemental Figure 1. In vivo treatment of IzT mice with TAM deletes Batf. (A) Schematic showing TAM-treatment schedule of IzT mice by oral gavage. (B) Spleen, thymus and pancreas from four control treated and four TAM treated IzT mice were fixed, sectioned and stained with anti-RFP or anti-GFP antibodies. DAPI was used to visualize nuclei. Representative images are shown. Bars represent 50μM. (C) B220+ GFP+ cells were sorted from the spleens of control TAM-treated IzT mice and stimulated in culture for 48hrs. Column purified B cells from the spleens of non-TAM IzT mice and BatfΔZ/ΔZ mice served as controls. RNA was prepared RT-qPCR used to detect transcripts from the indicated genes. Results are normalized to β-actin and expressed relative to the levels in the non-TAM IzT control (set to 1.0). Results shown are the mean±SEM of data collected from two independent experiments, each with RNA from two mice per genotype. (***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test)
Supplemental Figure 2. Gating strategy to quantify surface Ig on stimulated B cells. (A) Non-TAM (Control) and TAM-treated Δ0 and Δ12 B cell cultures derived from IzT mice (see Figure 3A) were profiled by flow cytometry at to detect cells expressing RFP or the CRE-activated, GFP reporter gene. SSC (side scatter); FSC (forward scatter). (B) Cells gated as GFP+ (Δ0 and Δ12; control not shown) or RFP+ (control; Δ0 and Δ12 not shown) were profiled to detect surface IgM and IgG1 (top) or IgM and IgE (bottom). Antibodies used for staining are listed in Suppl. Table 1.
Supplemental Figure 3. B cell gene expression remains normal following expression of MSCV-BATF. B cells from BatfKI/KI and BatfΔZ/ΔZ spleens were transduced with the indicated retroviruses and Thy1.1 cells isolated by FACS. RNA was isolated and RT-qPCR used to detect levels of the indicated transcripts, in duplicate. Results are normalized to β-actin and expressed relative to levels in unstimulated BatfKI/KI B cells (set to 1.0, but not shown). Results shown are the mean±SEM of data collected from three independent experiments, each with RNA from two mice per genotype and treatment. (ns=not significant; two-tailed, unpaired Student’s t-test)
Supplemental Figure 4. Time-course of Nfil3, miR155hg and Wnt10a expression in stimulated B cells. WT B cells were stimulated and RNA prepared at the indicated time points. RT-qPCR was used to assay for the indicated transcripts, in duplicate. Results are normalized using β-actin and expressed relative to time 0 (set to 1.0). Results shown are the mean±SEM of data collected from two independent experiments, each with RNA from two mice.
Supplemental Figure 5. BATF does not bind all AP-1 and AICE motifs in Nfil3, miR155hg, and Wnt10a. (A) Location of key AP-1 and AICE containing DNA fragments within the indicated genes. Exons are represented by numbered black boxes. The nucleotide sequence of each motif is presented, along with the positions of the motif relative to the transcription start site. Genes are not drawn to scale and not all are complete. (B and C) ChIP-qPCR was performed on DNA fragments isolated from the chromatin of stimulated WT B cells that had been precipitated using an antibody to BATF (B) or H3K27 (C) (Suppl. Table 1). For BATF, DNA fragment amplification was quantified as the fold enrichment over binding to the unoccupied AP-1(N) of Myb (See Figure 6). H3K27specific fragment amplification was quantified as a % of input. Results shown are the mean±SEM of data collected from two independent experiments, each using DNA from two mice per genotype. (ns=not significant; two-tailed, unpaired Student’s t-test)
Supplemental Figure 6. Retroviruses drive expression of NFIL3, WNT10A and miR155. (A) microRNA was isolated from BatfKI/KI and BatfΔZ/ΔZ B cells that were transduced with MSCV or MSCV-miR155 and sorted as Thy1.1+ (gating strategy as in Suppl. Figure 7A). RT-qPCR was used to detect miR155. Results were normalized to RNU6 and expressed relative to levels detected in MSCV-transduced BatfKI/KI B cells (set to 1.0). Results shown are the mean±SEM of data collected from two independent experiments, each with miRNA from two mice per genotype. (*p<0.05; ns=not significant; two-tailed, unpaired Student’s t-test) (B) HeLa cells were transduced with MSCV-NFIL3 or MSCV-WNT10A. Protein extracts were prepared and immunoblotting performed to detect expression of mouse NFIL3 and WNT10A (antibodies listed in Suppl. Table 1). Samples were run in duplicate and signals expressed relative to each other following normalization to HSP90. Shown is a representative blot from two independent experiments using protein from two mice per genotype. (C) BatfKI/KI and BatfΔZ/ΔZ splenic B cells were CFSE labeled and transduced with the indicated retroviruses as detailed in Materials and Methods. Lymphocyte gating for CSFE analysis is shown in the upper panel of Figure 1A. CFSE dilution was analyzed by flow cytometry. Gating (indicated by the bar) was quantified and represents the percent proliferating cells. Results in the histogram are the mean±SEM of data collected from two independent experiments, each using two groups of cells transduced with each of the indicated viruses.
Supplemental Figure 7. Gating strategy to quantify surface Ig on MSCV infected B cells. (A) BatfΔZ/ΔZ B cells transduced with MSCV or MSCV-BATF and BatfKI/KI B cells transduced with MSCV (not shown) were profiled for Thy1.1 expression by flow cytometry after 96 hrs. SSC (side scatter); FSC (forward scatter) (B) Cells gated as Thy1.1+ were profiled to detect surface IgM and IgG1 (top) or IgM and IgE (bottom) by flow cytometry. Antibodies used for profiling are listed in Suppl. Table 1.
Acknowledgements
This work was supported by NIH A1 105620 (EJT) and a by grant awarded to EJT by the Indiana Clinical and Translational Sciences Institute (NIH UL1 TR001108). Core facilities supported by the Purdue University Center for Cancer Research (NIH P30 CA023168) assisted with the care and generation of mice, the collection of flow cytometry data, RNA-Seq, and DNA sequencing. The authors thank M. Olson, S. Poston and B. Damsz for assistance with technical aspects of the project. All experiments were performed by REM. RNA-Seq data analysis was performed by PGS and REM with assistance from J. Shreve, K. Bhide and J. Thimmapuram of the Purdue Bioinformatics Core. EJT and SFK provided financial support for the project. Experimental design and the preparation of the manuscript was a joint effort between REM, SFK and EJT.
Abbreviations:
- AID
activation induced cytidine deaminase
- AICE
(AP-1)-IRF composite element
- AP-1
activator protein-1
- bZIP
basic leucine zipper
- CFSE
carboxyfluorescein succinimidyl ester
- ChIP
chromatin immunoprecipitation
- CSR
class switch recombination
- FACS
flow activated cell sorting
- GFP
green fluorescent protein
- GLT
germ line transcript
- Ig
immunoglobulin
- IL
interleukin
- IRF
interferon regulatory factor
- LPS
lipopolysaccharide
- MSCV
murine stem cell virus
- RFP
red fluorescent protein
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- TAM
tamoxifen
- Th
T helper cell
- Tfh
T follicular helper cell
- WT
wild type
Footnotes
Conflict of Interest
The authors declare no commercial or financial conflict of interest.
References
- 1.Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 2013; 13:499–509. [DOI] [PubMed] [Google Scholar]
- 2.Sopel N, Graser A, Mousset S, Finotto S. The transcription factor BATF modulates cytokine-mediated responses in T cells. Cytokine Growth Factor Rev. 2016; 30:39–45. [DOI] [PubMed] [Google Scholar]
- 3.Dorsey M, Tae H, Sollenberger K, Mascarenhas N, Johansen L, Taparowsky E. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene 1995; 11:2255–2265. [PubMed] [Google Scholar]
- 4.Wang H, Xie Z, Scott RE. Differentiation modulates the balance of positive and negative Jun AP-1 DNA binding activities to regulate cellular proliferative potential: different effects in nontransformed and transformed cells. J. Cell Biol. 1996; 135:1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuwahara M, Ise W, Ochi M, Suzuki J, Kometani K, Maruyama S, et al. Bach2-BATF interactions control Th2-type immune response by regulating the IL-4 amplification loop. Nat. Commun. 2016; 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glasmacher E, Agrawal S, Chang AB, Theresa L, Spooner C, Rutz S, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science (80-. ). 2012; 338:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwata A, Durai V, Tussiwand R, Briseño CG, Wu X, Grajales-reyes GE, et al. Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF – IRF4 transcription factor complex. Nat. Immunol. 2017; 18:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams KL, Nanda I, Lyons GE, Kuo CT, Schmid M, Leiden JM, et al. Characterization of murine BATF : a negative regulator of activator protein-1 activity in the thymus. Eur. J. Immunol. 2001; 31:1620–1627. [DOI] [PubMed] [Google Scholar]
- 9.Thornton TM, Zullo AJ, Williams KL, Taparowsky EJ. Direct manipulation of activator protein-1 controls thymocyte proliferation in vitro. Eur. J. Immunol. 2006; 36:160–169. [DOI] [PubMed] [Google Scholar]
- 10.Schraml BU, Hildner K, Ise W, Lee W-L, Smith W a-E, Solomon B, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 2009; 460:405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 2010; 207:933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuroda S, Yamazaki M, Abe M, Sakimura K, Takayanagi H, Iwai Y. Basic leucine zipper transcription factor, ATF-like (BATF) regulates epigenetically and energetically effector CD8 T-cell differentiation via Sirt1 expression. Proc. Natl. Acad. Sci. U. S. A. 2011; 108:14885–14889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 2011; 12:536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, et al. Transcriptional analysis of HIV-specific CD8 + T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 2010; 16:1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jabeen R, Goswami R, Awe O, Kulkarni A, Nguyen ET, Attenasio A, et al. Th9 cell development requires a BATF-regulated transcriptional network. J. Clin. Invest. 2013; 123:4641–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ubel C, Sopel N, Graser A, Hildner K, Reinhardt C, Zimmermann T, et al. The activating protein 1 transcription factor basic leucine zipper transcription factor, ATF-like (BATF), regulates lymphocyte-and mast cell–driven immune responses in the setting of allergic asthma. J Allergy Clin Immunol 2014; 133:198–206. [DOI] [PubMed] [Google Scholar]
- 17.Karwacz K, Miraldi ER, Pokrovskii M, Madi A, Yosef N, Wortman I, et al. Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat. Immunol. 2017; 18:412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Sun Q, Morita Y, Jiang H, Groß A, Lechel A, et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 2012; 148:1001–1014. [DOI] [PubMed] [Google Scholar]
- 19.Rhee J, Park S-H, Kim S-K, Kim J-H, Ha C-W, Chun C-H, et al. Inhibition of BATF/JUN transcriptional activity protects against osteoarthritic cartilage destruction. Ann Rheum Dis 2017; 76:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Thangamani S, Kim M, Gu B-H, Lee JH, Taparowsky EJ, et al. BATF is required for normal expression of gut-homing receptors by T helper cells in response to retinoic acid. J. Exp. Med. 2013; 210:475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, et al. CRISPR/Cas9 screens reveal Epstein-Barr Virus-transformed B cell host dependency factors. Cell Host Microbe 2017; 21:580–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grusdat M, McIlwain DR, Xu HC, Pozdeev VI, Knievel J, Crome SQ, et al. IRF4 and BATF are critical for CD8(+) T-cell function following infection with LCMV. Cell Death Differ. 2014; 21:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xin G, Schauder D, Lainez B, Weinstein J, Dai Z, Chen Y, et al. A Critical Role of IL-21 Induced BATF in Sustaining CD8 T cell Mediated Chronic Viral Control. Cell Rep 2015; 13:1118–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B, He F, Dai C, Tan R, Ma D, Wang Z, et al. BATF inhibition prevent acute allograft rejection after cardiac transplantation. Am. J. Transl. Res. 2016; 8:3603–3613. [PMC free article] [PubMed] [Google Scholar]
- 25.Tussiwand R, Lee W-L, Murphy TL, Mashayekhi M, Kc W, Albring JC, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 2012; 490:502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciofani M, Madar A, Galan C, Sellars M, MacE K, Pauli F, et al. A validated regulatory network for Th17 cell specification. Cell 2012; 151:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008; 26:261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 2014; 15:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao J, Humphrey SE, Poston S, Taparowsky EJ. Batf promotes growth arrest and terminal differentiation of mouse myeloid leukemia cells. Mol. Cancer Res. 2011; 9:350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godec J, Cowley GS, Barnitz RA, Root DE, Sharpe AH, Haining WN. Inducible RNAi in vivo reveals that the transcription factor BATF is required to initiate but not maintain CD8+ T-cell effector differentiation. Proc. Natl. Acad. Sci. U. S. A. 2015; 112:512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manis JP, Tian M, Alt FW. Mechanism and control of class-switch recombination. Trends Immunol. 2002; 23:31–39. [DOI] [PubMed] [Google Scholar]
- 32.Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, et al. Th17 cell differentiation is regulated by the circadian clock. Science (80-. ). 2013; 342:727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kashiwada M, Levy DM, McKeag L, Murray K, Schröder AJ, Canfield SM, et al. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc. Natl. Acad. Sci. U. S. A. 2010; 107:821–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sercan Z, Pehlivan M, Sercan HO. Expression profile of WNT, FZD and sFRP genes in human hematopoietic cells. Leuk. Res. 2010; 34:946–949. [DOI] [PubMed] [Google Scholar]
- 35.Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, et al. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2004; 101:3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sertorio M, Amarachintha S, Wilson A, Pang Q. Loss of Fancc impairs antibody secreting cell differentiation in mice through deregulating Wnt signaling pathway. J. Immunol. Res. 2014; 4:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta - Mol. Basis Dis. 2009; 1792:497–505. [DOI] [PubMed] [Google Scholar]
- 38.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 2008; 28:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007; 27:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity 2008; 28:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. C-Myb Is critical for B cell development and maintenance of follicular B cells. Immunity 2005; 23:275–286. [DOI] [PubMed] [Google Scholar]
- 42.Male V, Nisoli I, Gascoyne DM, Brady HJM. E4BP4: An unexpected player in the immune response. Trends Immunol. 2012; 33:98–102. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science (80-. ). 2007; 316:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouamar H, Jiang D, Wang L, Lin AP, Ortega M, Aguiar RC. MicroRNA 155 control of p53 activity is context dependent and mediated by Aicda and Socs1. Mol Cell Biol 2015; 35:1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. In vivo treatment of IzT mice with TAM deletes Batf. (A) Schematic showing TAM-treatment schedule of IzT mice by oral gavage. (B) Spleen, thymus and pancreas from four control treated and four TAM treated IzT mice were fixed, sectioned and stained with anti-RFP or anti-GFP antibodies. DAPI was used to visualize nuclei. Representative images are shown. Bars represent 50μM. (C) B220+ GFP+ cells were sorted from the spleens of control TAM-treated IzT mice and stimulated in culture for 48hrs. Column purified B cells from the spleens of non-TAM IzT mice and BatfΔZ/ΔZ mice served as controls. RNA was prepared RT-qPCR used to detect transcripts from the indicated genes. Results are normalized to β-actin and expressed relative to the levels in the non-TAM IzT control (set to 1.0). Results shown are the mean±SEM of data collected from two independent experiments, each with RNA from two mice per genotype. (***p<0.0001; ns=not significant; two-tailed, unpaired Student’s t-test)
Supplemental Figure 2. Gating strategy to quantify surface Ig on stimulated B cells. (A) Non-TAM (Control) and TAM-treated Δ0 and Δ12 B cell cultures derived from IzT mice (see Figure 3A) were profiled by flow cytometry at to detect cells expressing RFP or the CRE-activated, GFP reporter gene. SSC (side scatter); FSC (forward scatter). (B) Cells gated as GFP+ (Δ0 and Δ12; control not shown) or RFP+ (control; Δ0 and Δ12 not shown) were profiled to detect surface IgM and IgG1 (top) or IgM and IgE (bottom). Antibodies used for staining are listed in Suppl. Table 1.
Supplemental Figure 3. B cell gene expression remains normal following expression of MSCV-BATF. B cells from BatfKI/KI and BatfΔZ/ΔZ spleens were transduced with the indicated retroviruses and Thy1.1 cells isolated by FACS. RNA was isolated and RT-qPCR used to detect levels of the indicated transcripts, in duplicate. Results are normalized to β-actin and expressed relative to levels in unstimulated BatfKI/KI B cells (set to 1.0, but not shown). Results shown are the mean±SEM of data collected from three independent experiments, each with RNA from two mice per genotype and treatment. (ns=not significant; two-tailed, unpaired Student’s t-test)
Supplemental Figure 4. Time-course of Nfil3, miR155hg and Wnt10a expression in stimulated B cells. WT B cells were stimulated and RNA prepared at the indicated time points. RT-qPCR was used to assay for the indicated transcripts, in duplicate. Results are normalized using β-actin and expressed relative to time 0 (set to 1.0). Results shown are the mean±SEM of data collected from two independent experiments, each with RNA from two mice.
Supplemental Figure 5. BATF does not bind all AP-1 and AICE motifs in Nfil3, miR155hg, and Wnt10a. (A) Location of key AP-1 and AICE containing DNA fragments within the indicated genes. Exons are represented by numbered black boxes. The nucleotide sequence of each motif is presented, along with the positions of the motif relative to the transcription start site. Genes are not drawn to scale and not all are complete. (B and C) ChIP-qPCR was performed on DNA fragments isolated from the chromatin of stimulated WT B cells that had been precipitated using an antibody to BATF (B) or H3K27 (C) (Suppl. Table 1). For BATF, DNA fragment amplification was quantified as the fold enrichment over binding to the unoccupied AP-1(N) of Myb (See Figure 6). H3K27specific fragment amplification was quantified as a % of input. Results shown are the mean±SEM of data collected from two independent experiments, each using DNA from two mice per genotype. (ns=not significant; two-tailed, unpaired Student’s t-test)
Supplemental Figure 6. Retroviruses drive expression of NFIL3, WNT10A and miR155. (A) microRNA was isolated from BatfKI/KI and BatfΔZ/ΔZ B cells that were transduced with MSCV or MSCV-miR155 and sorted as Thy1.1+ (gating strategy as in Suppl. Figure 7A). RT-qPCR was used to detect miR155. Results were normalized to RNU6 and expressed relative to levels detected in MSCV-transduced BatfKI/KI B cells (set to 1.0). Results shown are the mean±SEM of data collected from two independent experiments, each with miRNA from two mice per genotype. (*p<0.05; ns=not significant; two-tailed, unpaired Student’s t-test) (B) HeLa cells were transduced with MSCV-NFIL3 or MSCV-WNT10A. Protein extracts were prepared and immunoblotting performed to detect expression of mouse NFIL3 and WNT10A (antibodies listed in Suppl. Table 1). Samples were run in duplicate and signals expressed relative to each other following normalization to HSP90. Shown is a representative blot from two independent experiments using protein from two mice per genotype. (C) BatfKI/KI and BatfΔZ/ΔZ splenic B cells were CFSE labeled and transduced with the indicated retroviruses as detailed in Materials and Methods. Lymphocyte gating for CSFE analysis is shown in the upper panel of Figure 1A. CFSE dilution was analyzed by flow cytometry. Gating (indicated by the bar) was quantified and represents the percent proliferating cells. Results in the histogram are the mean±SEM of data collected from two independent experiments, each using two groups of cells transduced with each of the indicated viruses.
Supplemental Figure 7. Gating strategy to quantify surface Ig on MSCV infected B cells. (A) BatfΔZ/ΔZ B cells transduced with MSCV or MSCV-BATF and BatfKI/KI B cells transduced with MSCV (not shown) were profiled for Thy1.1 expression by flow cytometry after 96 hrs. SSC (side scatter); FSC (forward scatter) (B) Cells gated as Thy1.1+ were profiled to detect surface IgM and IgG1 (top) or IgM and IgE (bottom) by flow cytometry. Antibodies used for profiling are listed in Suppl. Table 1.