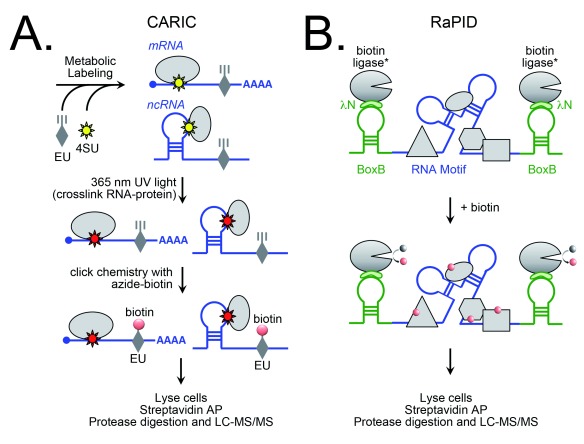
Figure 2. Biotin-based approaches to map ribonucleoprotein complexes.
( A) CARIC (click chemistry-assisted RNA interactome capture) is a global screening method for RNA-binding proteins that is based on the incorporation of the uridine analogs 5-ethynyl-uridine (EU) and 4-thiouridine (4SU) into RNA (both messenger RNA and non-coding RNA) during transcription. Exposure to 365 nm ultraviolet (UV) light induces RNA–protein crosslinking via the photoactivatable 4SU. The RNA is then biotinylated by chemosensitive reaction of the alkyne-containing EU with azide biotin via copper(I)-catalyzed azide-alkyne cycloaddition (click chemistry) for efficient capture on a streptavidin affinity matrix. RNA-associated proteins are identified by protease digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. ( B) RaPID (RNA–protein interaction detection) is a targeted BioID approach in which an RNA motif of interest is flanked by two bacteriophage lambda BoxB stem loops. Expression of a promiscuous biotin ligase fused to the 22-amino-acid λN peptide, which binds the stem loops with high affinity, results in the recruitment of the fusion protein to the RNA motif and subsequent biotinylation of proteins in close proximity. These proteins then can be captured by streptavidin-based affinity purification (AP) for identification by LC-MS/MS.