Abstract
Purpose
The Radiological Society of North America (RSNA) Pediatric Bone Age Machine Learning Challenge was created to show an application of machine learning (ML) and artificial intelligence (AI) in medical imaging, promote collaboration to catalyze AI model creation, and identify innovators in medical imaging.
Materials and Methods
The goal of this challenge was to solicit individuals and teams to create an algorithm or model using ML techniques that would accurately determine skeletal age in a curated data set of pediatric hand radiographs. The primary evaluation measure was the mean absolute distance (MAD) in months, which was calculated as the mean of the absolute values of the difference between the model estimates and those of the reference standard, bone age.
Results
A data set consisting of 14 236 hand radiographs (12 611 training set, 1425 validation set, 200 test set) was made available to registered challenge participants. A total of 260 individuals or teams registered on the Challenge website. A total of 105 submissions were uploaded from 48 unique users during the training, validation, and test phases. Almost all methods used deep neural network techniques based on one or more convolutional neural networks (CNNs). The best five results based on MAD were 4.2, 4.4, 4.4, 4.5, and 4.5 months, respectively.
Conclusion
The RSNA Pediatric Bone Age Machine Learning Challenge showed how a coordinated approach to solving a medical imaging problem can be successfully conducted. Future ML challenges will catalyze collaboration and development of ML tools and methods that can potentially improve diagnostic accuracy and patient care.
© RSNA, 2018
Online supplemental material is available for this article.
See also the editorial by Siegel in this issue.
Summary
The RSNA Pediatric Bone Age Machine Learning Challenge showed the application of machine learning in medical imaging, promoted ways in which these emerging tools and methods can improve diagnostic care, and identified innovators in machine learning applications in medical imaging.
Implication for Patient Care
■ Machine learning challenges will stimulate collaboration and development of machine learning tools and methods that can improve diagnostic care.
Introduction
Artificial intelligence (AI) algorithms have existed for decades and have recently been propelled to the forefront of medical imaging research. To a large extent, this is related to improvements in computing power, availability of a large amount of training data, and innovative and improved neural network architectures, with the recognition that certain types of algorithms are well suited to image analysis. The latter discovery was accelerated by the ImageNet competition and represents a fundamental transformation in research mechanics and methods in computer vision.
Currently, in most studies, researchers collect data, perform analysis, and publish results. The same researchers may continue to augment and expand the data set and perform subsequent analysis with resulting publications. The data for each study are held quite closely and are rarely shared among institutions outside of multicenter trials. Competitions represent a different model of research: Research data are made available to the public, usually with a baseline performance metric. Groups around the world are invited to analyze the data and create algorithms to beat the performance of the prior generation. For example, the baseline performance metric for this challenge was set by the previous skeletal age model developed by Larson et al (1).
The Radiological Society of North America (RSNA) Pediatric Bone Age Machine Learning Challenge was created to evaluate the performance of computer algorithms in executing a common image analysis activity that is familiar to many pediatric radiologists: estimating the bone age of pediatric patients based on radiographs of their hand (1–5). This challenge used a data set of pediatric hand radiographs with associated bone age assessments provided by multiple expert reviewers.
The RSNA promotes excellence in patient care and health care delivery through education, research, and technologic innovation. Machine learning (ML) competitions align with the research and technologic innovation aspects of the RSNA mission.
The aim of this article is to describe how and why the RSNA Informatics Committee and volunteers created, organized, implemented, and evaluated this challenge. The challenge was created to (a) show the application of ML and AI in medical imaging, (b) promote ways in which these emerging tools and methods might improve diagnostic care, and (c) identify innovators in ML and AI applications in medical imaging. The organizers and sponsors of the challenge are shown in Appendix E1 [online], and the timeline of the competition is shown in Appendix E2 [online].
Most ML challenges consist of three phases—(a) the training or learning phase, (b) the validation phase, and (c) the test phase—that correspond to similarly named data sets. In the training phase, a model (eg, a neural net) is trained on a known data set by using a supervised learning method (eg, gradient descent or stochastic gradient descent). Subsequently, the fitted model is used to predict responses for the observations in a second smaller data set, which is termed the validation data set. Performance of the model is assessed through validation to determine if the training phase was effective. Finally, a test data set is used to perform an unbiased evaluation of the performance of the trained model. Participants in the challenge had the option to display their results on a public leaderboard during the validation and test phases of the competition. The final results were based on the test phase submissions. Challenge terms, conditions, and rules are shown in Appendix E3 (online).
Materials and Methods
The institutional review boards at Stanford University and the University of Colorado approved the curation and use of pediatric hand radiographs for the purposes of this ML competition. Patient consent was waived after approval by the institutional review board.
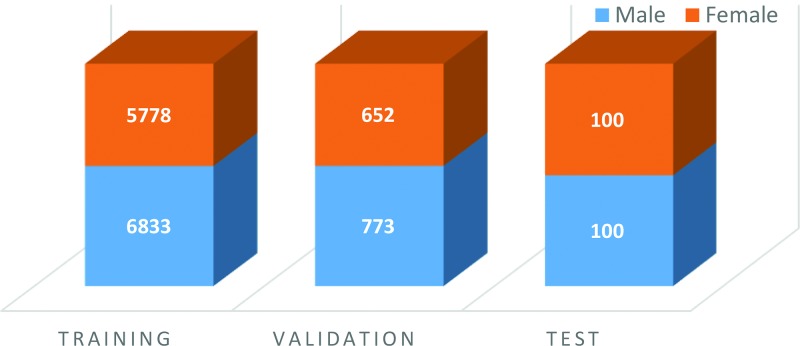
The data sets (Fig 1) made available to participants were composed of an initial training set that contained 12 611 (mean patient age, 127 months) deidentified hand radiographs and a validation set that contained 1425 images (mean patient age, 127 months). Images for the training and validation sets were obtained from Children’s Hospital Colorado (Aurora, Colo), and Lucile Packard Children’s Hospital at Stanford. The images were labeled, with skeletal age estimates and sex from the accompanying clinical radiology report provided at the time of imaging.
Figure 1:
Sex distribution and number of images in the training, validation, and test bone age data sets.
A separate test set containing 200 images (mean age, 132 months) was used to evaluate the performance of the submitted algorithms. Images for the test set were obtained from Lucile Packard Children’s Hospital.
The ground truth skeletal age estimates for all image sets were based on six separate estimates for each image that consisted of (a) the clinical radiology report from their respective institution, (b) four pediatric radiologists who reviewed the cases independently (two pediatric radiologists from each institution), and (c) a second review by one of the pediatric radiologists who reviewed the cases approximately 1 year after the first review. The Greulich and Pyle standard (2) was used by reviewers to determine the ground truth bone age.
The ground truth estimate for each case in the test data set was determined in two steps: First, a preliminary ground truth estimate was obtained as the simple mean of the six reviewers’ estimates. The performance of each reviewer was evaluated by determining the mean difference and the mean absolute difference (MAD) between the reviewer’s performance and the mean of all reviewers’ estimates, which ranged from −0.75 to 1.16 months and from 4.8 to 7.0 months, respectively. Next, each reviewer’s estimate was corrected for bias, and a reviewer weight was determined as the inverse of the MAD (1/MAD) and ranged from 0.14 for the reviewer with the highest MAD to 0.21 for the reviewer with the lowest MAD. The final ground truth estimate was determined by calculating the weighted mean of the corrected reviewer estimates. These data were the foundation for the work by Larson et al (1).
The details of this challenge’s web-based platform are described in Appendix E4 (online).
Results
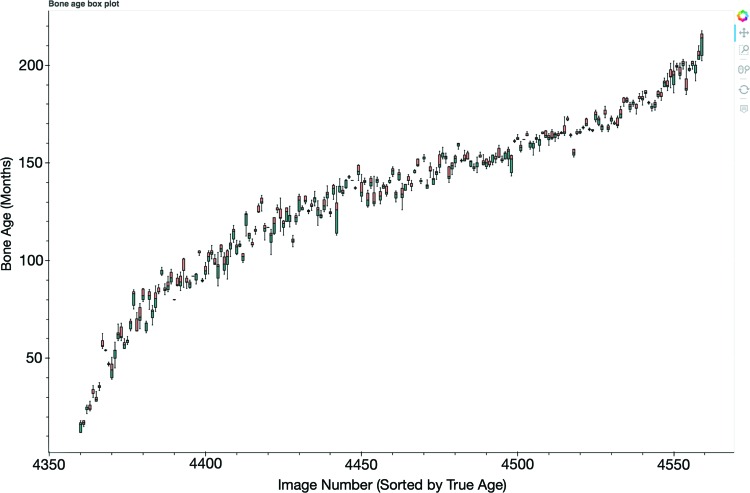
A total of 260 individuals or teams from around the world registered on the challenge website. A total of 105 submissions were uploaded from 48 unique users during the training, validation, and test phases. The 10 best submissions and MADs are listed in Appendix E5 (online). The box plot of the five best submissions comparing predicted bone age with ground truth is shown in Figure 2.
Figure 2:
Box plot shows the five best submissions comparing predicted bone age with ground truth. The x-axis represents the image number from the test data set, and the y-axis represents the bone age predicted by the competitors’ models. A full interactive summary of challenge data and analytics, including this box plot, can be accessed at https://rsnachallenges.cloudapp.net:5006/rsna_interactive.
First Place: Alexander Bilbily, MD, BHSc, FRCPC; Mark Cicero, MD, BESC, FRCPC (Canada)
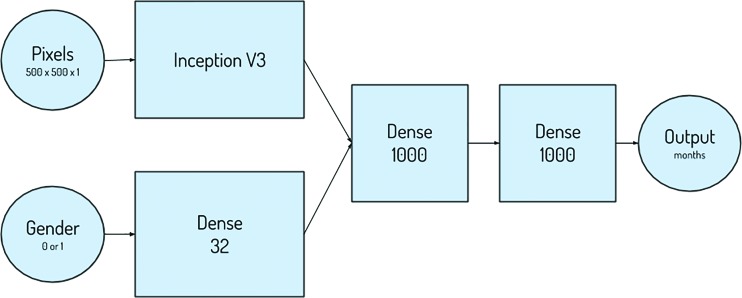
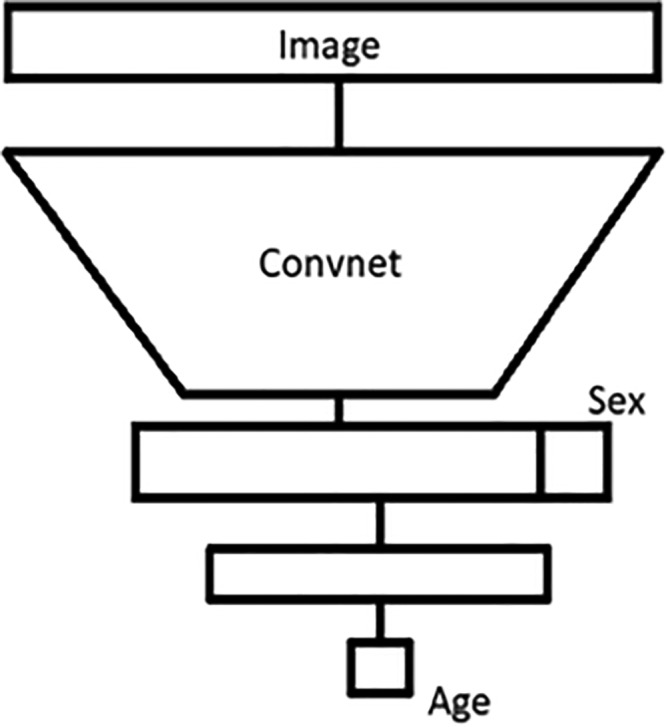
The winning approach used both the pixel and sex information in the same network at an image size of 500 × 500 pixels (Fig 3). The Inception V3 architecture was used for the pixel information and was concatenated with the sex information, with additional dense layers after concatenation, to enable the network to learn the relationship between pixel and sex information (6,7). Data augmentation proved to be a necessary step for success in this challenge, as it was used in many of the submissions. Finally, the combination of multiple high-performing models in an ensemble approach at test time also improved overall performance.
Figure 3:
Depiction of the inputs, outputs, and layers of the first-place network design.
Second Place: Ian Pan, MA (United States)
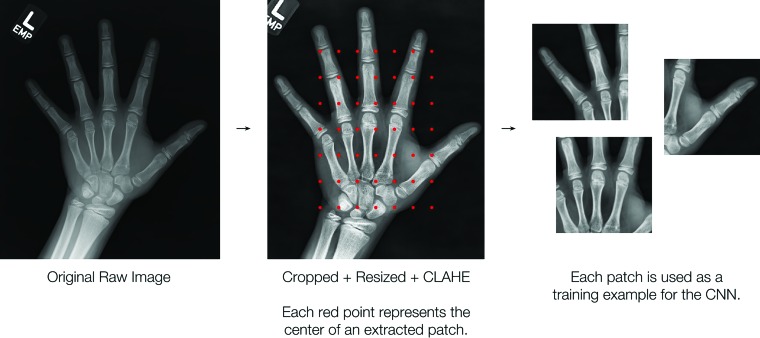
The second-place approach trained sex-specific models by using contrast-enhanced image patches of 224 × 224 pixels instead of the entire image. Each image was divided into 49 overlapping patches (Fig 4, 5). The final prediction was calculated by taking the Xth percentile of the patch predictions, where X was typically around the 50th percentile (ie, the median). This approach used transfer learning and fine-tuned ResNet-50 architectures pretrained on the ImageNet data set. As in other approaches, data augmentation and ensembling (nine models) were leveraged to avoid overfitting and to improve performance.
Figure 4:
Preprocessing pipeline for the second-place method used to construct inputs to the neural network. The image is manually cropped and resized to a length of 560 pixels, and the contrast is enhanced; this is followed by extraction of 49 patches of 224 × 224 pixels. CLAHE = contrast limited adaptive histogram equalization, CNN = convolutional neural network.
Figure 5:
Network architecture of the third-place team shows a convolutional neural network followed by two dense layers. The convolutional layers extract imaging features. The dense layers use imaging features and sex input to output the predicted age.
Third Place: Felipe Campos Kitamura, MD, MSc; Lucas Araújo Pereira, BSc; Rafael Teixeira Sousa, MSc; Larissa Vasconcellos De Moraes, BSc; Anderson Da Silva Soares, PhD; Nitamar Abdala, MD, PhD; Gabriel Alencar De Oliveira; Igor Rafael Martins Dos Santos, MD (Brazil)
The third-place model did not use any known deep learning model (ie, Inception, ResNet). This group developed a new variant of a convolutional neural network by creating the Ice Module (Fig 6). This model is considerably smaller than Inception V4 (approximately 1% of the number of parameters). They split the data set into five parts and trained a model on each part. The best four parts were used for prediction on the test set, and their average score comprised the final output (simple ensemble).
Figure 6:
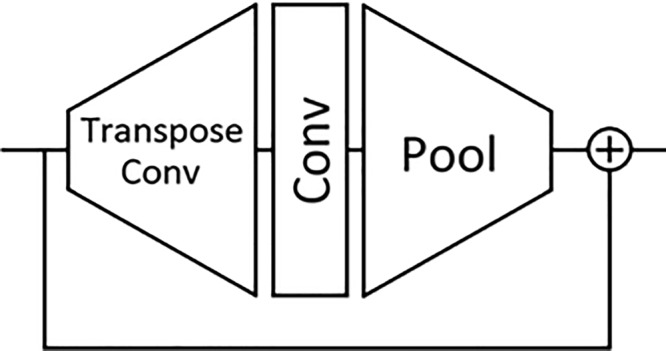
The Ice Module architecture is composed of a transpose convolution followed by a convolutional and pooling layer bypassed by a residual connection. This Ice Module is used in the convolutional part of the network architecture of the third-place team.
Fourth Place: Hans Henrik Thodberg, PhD (Denmark)
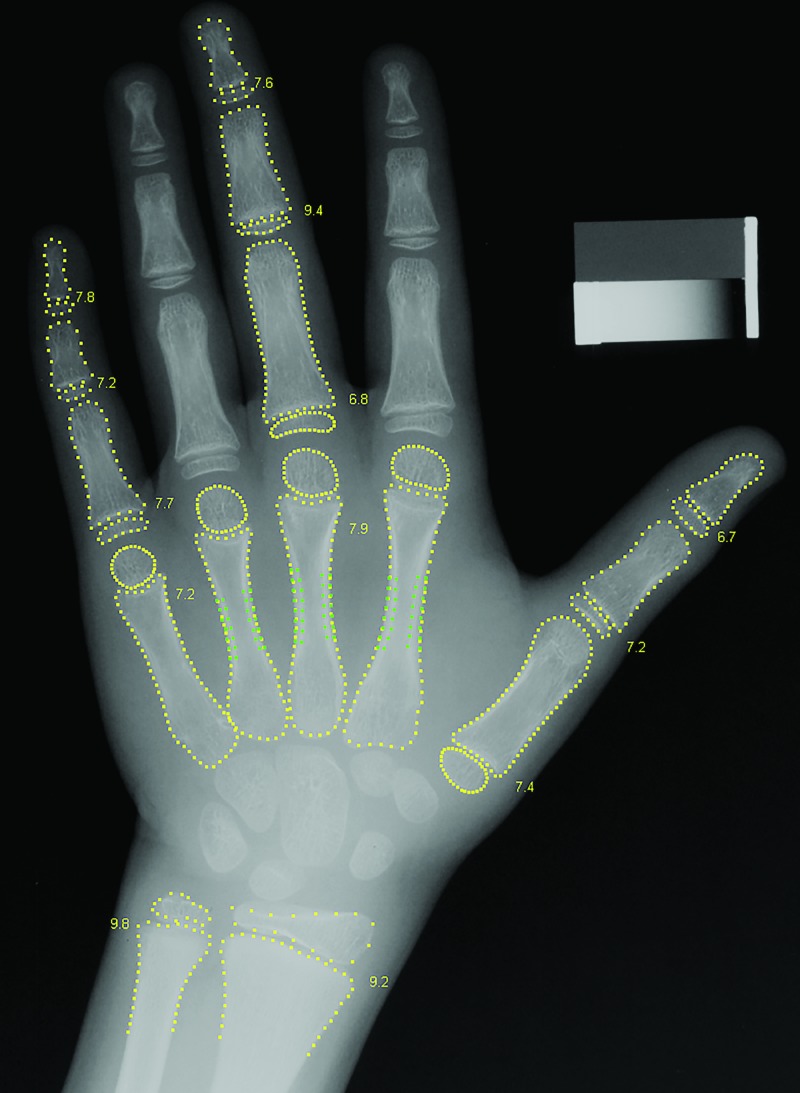
The fourth-place approach used conventional (nondeep) ML. The image preprocessing segmented the hand image into 15 bones. Bone age was estimated in each of the 13 bones by using hand-crafted features as opposed to features learned by deep learning (Fig 7). Three kinds of features were used: the shape of the bone, the intensity pattern across the growth zone, and the pattern of Gabor texture energies across the growth zone.
Figure 7:
Radiograph of a boy’s left hand shows the preprocessing method used by the fourth-place participant. A bone age value is estimated for each of the 13 bones.
Fifth Place: Leon Chen, MD; George Shih, MD (United States)
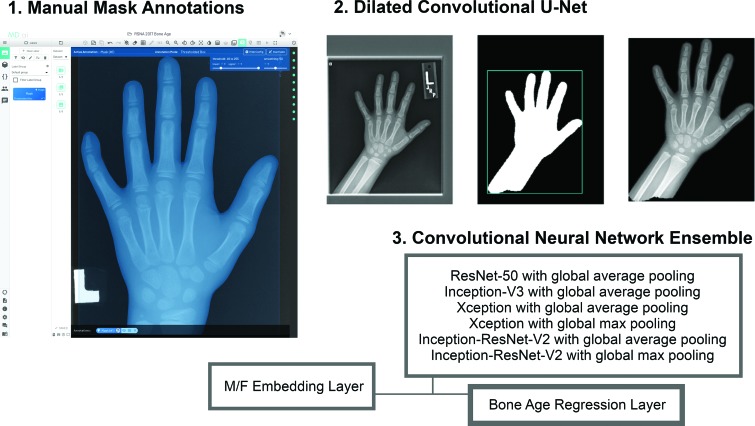
The fifth-place approach was unique due to the creation of a segmentation mask module. A total of 400 manual segmentation masks for the hand, wrist, and distal forearm were created to train a dilated convolutional u-net to predict segmentation masks for the entire data set (Fig 8). A bone age prediction network was then trained on masked images, which consisted of an ensemble of convolution neural networks with a final regression layer and a sex-embedding layer.
Figure 8:
Approximately 400 manual mask annotations were used by the fifth-place team to train a dilated convolutional u-net to generate masks for the remaining (approximately 12 000) hand radiographs. This was then used to train the convolutional neural networks.
The technical details of the five best methods are described in Appendixes E6–E10 (online).
Discussion
The field of computer vision has been evolving at a dramatic pace in recent years. While the majority of advancements and discoveries in this area were based on standard color photographic images containing common objects (eg, cars, airplanes, fruits), the recent successes have fostered tremendous interest in applying these principles to diagnostic imaging. Although many image types share important similarities, medical images impose several distinct challenges to ML applications. These challenges vary from technical difficulties related to processing Digital Imaging and Communications in Medicine files to a variety of clinical considerations, such as normal variations in human anatomy and different clinical presentations of the same disease, image quality degradation secondary to artifact, the unwarranted variability in image interpretation, and patient privacy concerns. Additionally, use of crowdsourcing to create annotations for medical images is particularly challenging because of the specific domain knowledge required to interpret these images.
The RSNA Pediatric Bone Age Challenge was made possible by the availability of a large data set curated by radiologists with subspecialty training. The data set was initially created for the group’s own research but, more importantly, it was made freely available to the public so that other investigators could take advantage of this valuable resource. In recent years, greater emphasis has been placed on collaborative science and related mandates to make all research data open access to encourage reproducibility and to provide greater legitimacy to research results. ML and related data science initiatives for medical imaging will succeed with greater access to accurately curated and publicly available data sets. The broad availability of the data set allows individuals with different backgrounds to explore nontraditional solutions, accelerating discoveries at a pace that is more rapid than that of the traditional scientific method. This is even more apparent in image challenge competitions, where individuals collaborate to solve a specific clinical problem and often share discoveries and methods with other participants in forums or blog posts. Although some data science challenges offer monetary rewards for the best results, the worldwide enthusiasm and spirit of collaboration and competition remains novel in the scientific community and is the driving force behind participation.
The winning methods featured in this challenge represented a substantial performance improvement compared with the best performing algorithm published only a year earlier and produced a smaller rating error when compared with that of one radiologist. The performance of the five best algorithms in this competition was not statistically different. However, each of the solutions used distinct approaches, including deep learning and traditional ML methods. This suggests that specific ML algorithms and data processing techniques should not be overemphasized at the expense of others, as no clearly superior method has been identified yet for medical images. ML techniques are often complementary to each other depending on the task. This is further demonstrated by the common use of algorithm ensembles in the challenge, a technique that uses a weighted vote or an average of multiple algorithms to better generalize predictions.
All five winning algorithms used a preprocessing step, in which images were normalized or important anatomic areas were selected prior to algorithm training. Preprocessing seems to be an important component for generalizability of the algorithm. This is shown with some of the rotated images available in the original data set. Algorithms that used a preprocessing step that segmented the appropriate anatomy or randomly rotated the images seemed to be less prone to errors when images were rotated in the test data set.
Although ensembles were popular in this challenge and have likely contributed to improvements in algorithm performance, it is important to carefully determine whether the added benefit in performance outweighs the additional effort required for clinical implementation when portability and speed of the application are important considerations. For example, a 1% increase in performance may not be as advantageous in the clinical setting if it significantly affects execution speed. On the other hand, a 10% improvement in performance may be clinically advantageous, even if it doubles computational time. Further details of using ensembles to improve bone age prediction are described in Appendix E11 (online).
Despite the similar results between the models, there are numerous potential effects of such models on clinical care. These models could systematically standardize image interpretation tasks, such as bone age assessment, to yield more reliable and reproducible results (8). In nonlinear image tasks, the models may help alert the interpreter that an image deviates from a certain standard or norm. Ultimately, ML models may identify and diagnose diseases based on imaging findings with great precision.
As ML algorithms become more embedded in clinical practice, radiologists will need to expand their understanding of these methods and what functions the models were created to perform. More importantly, it will be imperative to understand the limitations of these tools and models in the patient care continuum. Education in this field may need to start early in residency or even medical school and should cover traditional and newer ML methods as they become more common.
On the basis of our initial experience, the RSNA image challenge competition has a unique opportunity to foster scientific advancements through a new form of independent scientific collaboration and validation in the field of medical imaging ML but may generate enough thoughtful discussions to ensure that future clinically endorsed algorithms would be scientifically validated and will provide meaningful clinical improvements to patient care.
In conclusion, the first RSNA Pediatric Bone Age Machine Learning Challenge successfully achieved the following objectives set forth by the organizers: (a) to show the application of ML and AI in medical imaging, (b) to promote ways in which these emerging tools and methods may improve diagnostic care, and (c) to identify innovators in ML and AI applications in medical imaging. This challenge showed the power of sharing data publicly to solve a problem and create a tool that can provide more accurate, efficient, and timely diagnosis. This will hopefully be one of many challenges sponsored by the RSNA and other medical societies.
APPENDIX
Acknowledgments
Acknowledgments
We thank Matthew C. Chen, MS, for his assistance curating and organizing the data sets (hand radiographs and annotations) for the RSNA Pediatric Bone Age Machine Learning Challenge. This challenge was hosted on the MedICI platform (built CodaLab) provided by Jayashree Kalpathy-Cramer and Massachusetts General Hospital and funded through a contract with Leidos
Supported by the National Institute for Health Research (U24CA180927).
Disclosures of Conflicts of Interest: S.S.H. disclosed no relevant relationships. L.M.P. disclosed no relevant relationships. J.K.C. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is a consultant for Infotech Software. Other relationships: disclosed no relevant relationships. A.B.M. disclosed no relevant relationships. A.B. Activities related to the present article: is the CEO and cofounder of 16 Bit. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. M.C. Activities related to the present article: is a shareholder and board member of 16 Bit. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. I.P. disclosed no relevant relationships. L.A.P. disclosed no relevant relationships. R.T.S. disclosed no relevant relationships. N.A. disclosed no relevant relationships. F.C.K. disclosed no relevant relationships. H.H.T. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: disclosed no relevant relationships. Other relationships: is the owner of Visiana, which develops and markets the BoneXpert method for automated bone age assessment. L.C. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is on the board and has stock options for MD.ai. Other relationships: disclosed no relevant relationships. G.S. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is a board member of and shareholder in MD.ai. Other relationships: disclosed no relevant relationships. K.A. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: disclosed no relevant relationships. Other relationships: is the Director of Research Strategy and Operations at the Massachusetts General Hospital & Brigham and Women’s Hospital Center for Clinical Data Science, which is funded in part by monies and resources from Nvidia, General Electric, and Nuance. M.D.K. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: Spoke at a Gilead Sciences event. Other relationships: disclosed no relevant relationships. B.J.E. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: received an NVidia Global Impact award. Other relationships: disclosed no relevant relationships. A.E.F. disclosed no relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- MAD
- mean absolute difference
- ML
- machine learning
References
- 1.Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology 2018;287(1):313–322. [DOI] [PubMed] [Google Scholar]
- 2.Greulich WW, Pyle SI. Radiographic atlas of skeletal development of the hand and wrist. Stanford, Calif: Stanford University Press, 1999. [Google Scholar]
- 3.Lee H, Tajmir S, Lee J, et al. Fully automated deep learning system for bone age assessment. J Digit Imaging 2017;30(4):427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JR, Shim WH, Yoon HM, et al. Computerized bone age estimation using deep learning based program: evaluation of the accuracy and efficiency. AJR Am J Roentgenol 2017;209(6):1374–1380. [DOI] [PubMed] [Google Scholar]
- 5.Mutasa S, Chang PD, Ruzal-Shapiro C, Ayyala R. MABAL: a novel deep-learning architecture for machine-assisted bone age labeling. J Digit Imaging 2018;31(4):513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Arxiv: 1512.03385 [preprint]. https://arxiv.org/abs/1512.03385. Posted December 15, 2015. Accessed November 3, 2017.
- 7.Krishevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 2012;25:1106–1114. [Google Scholar]
- 8.Tajmir SH, Lee H, Shailam R, et al. Artificial intelligence-assisted interpretation of bone age radiographs improves accuracy and decreases variability. Skeletal Radiol 2018 Aug 1 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.