Abstract
Purpose
To develop and validate a deep learning algorithm that predicts the final diagnosis of Alzheimer disease (AD), mild cognitive impairment, or neither at fluorine 18 (18F) fluorodeoxyglucose (FDG) PET of the brain and compare its performance to that of radiologic readers.
Materials and Methods
Prospective 18F-FDG PET brain images from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (2109 imaging studies from 2005 to 2017, 1002 patients) and retrospective independent test set (40 imaging studies from 2006 to 2016, 40 patients) were collected. Final clinical diagnosis at follow-up was recorded. Convolutional neural network of InceptionV3 architecture was trained on 90% of ADNI data set and tested on the remaining 10%, as well as the independent test set, with performance compared to radiologic readers. Model was analyzed with sensitivity, specificity, receiver operating characteristic (ROC), saliency map, and t-distributed stochastic neighbor embedding.
Results
The algorithm achieved area under the ROC curve of 0.98 (95% confidence interval: 0.94, 1.00) when evaluated on predicting the final clinical diagnosis of AD in the independent test set (82% specificity at 100% sensitivity), an average of 75.8 months prior to the final diagnosis, which in ROC space outperformed reader performance (57% [four of seven] sensitivity, 91% [30 of 33] specificity; P < .05). Saliency map demonstrated attention to known areas of interest but with focus on the entire brain.
Conclusion
By using fluorine 18 fluorodeoxyglucose PET of the brain, a deep learning algorithm developed for early prediction of Alzheimer disease achieved 82% specificity at 100% sensitivity, an average of 75.8 months prior to the final diagnosis.
© RSNA, 2018
Online supplemental material is available for this article.
See also the editorial by Larvie in this issue.
Summary
By using fluorine 18 fluorodeoxyglucose PET of the brain, a deep learning algorithm developed for early prediction of Alzheimer disease achieved 82% specificity at 100% sensitivity, an average of 75.8 months prior to the final diagnosis.
Implications for Patient Care
■ A deep learning algorithm can be used to improve the accuracy of predicting the diagnosis of Alzheimer disease from fluorine 18 fluorodeoxyglucose PET of the brain.
■ A deep learning algorithm can be used as an early prediction tool for Alzheimer disease, especially in conjunction with other biochemical and imaging tests, thereby providing an opportunity for early therapeutic intervention.
Introduction
Although Alzheimer disease (AD) remains a diagnosis based on clinical grounds (1,2), advancements in diagnostic technology such as PET with fluorine 18 (18F) fluorodeoxyglucose (FDG) allow earlier diagnosis and treatments, when they may be most effective (3). There is a continuous spectrum from normal cognition to AD, including mild cognitive impairment (MCI) as a prodromal stage of AD (4,5). Classically, patients with AD tend to show hypometabolism on 18F-FDG PET scans in regions of the posterior cingulate, parietotemporal cortices, and frontal lobes, while patients with MCI often show posterior cingulate and parietotemporal hypometabolism with variable frontal lobe involvement. However, there is substantial overlap of findings as both entities lie along a continuum (5). In current practice, 18F-FDG PET requires interpretation by specialists in nuclear medicine and neuroimaging to make pattern recognition decisions mostly using qualitative readings. This is particularly challenging in the setting of a disease that involves a wide continuous spectrum, from normal cognition to MCI to AD, with only a fraction of patients with MCI eventually advancing to AD. At present, there is no definite marker to determine this eventual progress.
There is wide recognition that deep learning may assist in addressing the increasing complexity and volume of imaging data, as well as the varying expertise of trained imaging physicians (6). There has been substantial effort to apply deep learning in many diseases and imaging types such as breast cancer detection with mammography, pulmonary nodule detection with CT, and hip osteoarthritis classification with radiography, though integration into clinical flow is yet to be developed and validated (7–10). The application of machine learning technology to complex patterns of findings, such as those found at functional PET imaging of the brain, is only beginning to be explored.
In this study, we aimed to evaluate whether a deep learning algorithm could be trained to predict the final clinical diagnoses in patients who underwent 18F-FDG PET of the brain and, once trained, how the deep learning algorithm compares with the current standard clinical reading methods in differentiation of patients with final diagnoses of AD, MCI, or no evidence of dementia. We hypothesized that the deep learning algorithm could detect features or patterns that are not evident on standard clinical review of images and thereby improve the final diagnostic classification of individuals.
Materials and Methods
Data Acquisition
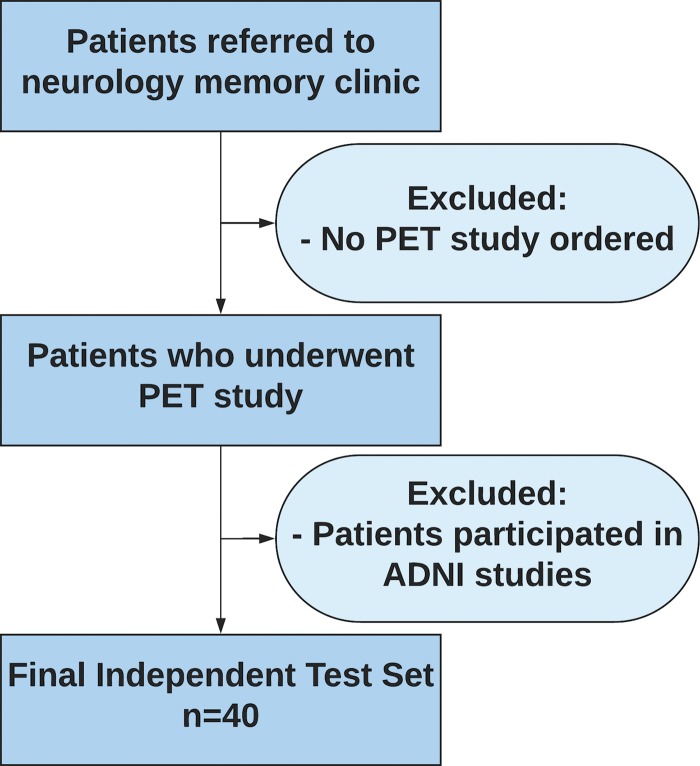
This institutional review board approved, written informed consent waived, and Health Insurance Portability and Accountability Act compliant study involved retrospective analysis of prospectively collected 2109 18F-FDG PET imaging studies from 1002 patients, most patients with multiple scans, with dates ranging from May 2005 to January 2017, across Alzheimer’s Disease Neuroimaging Initiative (ADNI)-1, ADNI-2, and ADNI-GO (Grand Opportunities) studies (Appendix E1 [online]). Data regarding the patient’s final diagnoses were downloaded from the ADNI web portal (adni.loni.ucla.edu) (11). Detailed 18F-FDG PET imaging protocols can be found at http://adni.loni.usc.edu/methods/documents/ (12–14). Ninety percent (1921 imaging studies, 899 patients) of this data set was used for model training and internal validation. The remaining 10% (188 imaging studies, 103 patients) was used for model testing, which we call 10% ADNI hold-out test set, serving as the internal test set from the perspective of the algorithm. An additional test set was obtained from the author’s own institution, which we call independent test set, serving as the external test set from the perspective of the algorithm. The independent test set (Fig 1) comprised 40 18F-FDG PET imaging studies from 40 patients who were not enrolled in the ADNI, with imaging study dates ranging from 2006 to 2016. Approximately 45 minutes after intravenous administration of 8–10 mCi 18F-FDG, following standard clinical guidelines, images were acquired as dedicated PET emission-only images (ECAT HR+; Siemens, Knoxville, Tenn) or as part of PET-CT (Discovery VCT, General Electric, Wakesha, Wis; or Biograph 16, Siemens). Only PET emission images were utilized in the test set to remain consistent with the training set. Necropsy data were used as the final diagnosis in one patient for which they were available. None of the patients had a diagnosis of a dementia of the non-Alzheimer type. For both data sets, final clinical diagnosis after all follow-up examinations was used as the ground truth label.
Figure 1:
Inclusion and exclusion criteria for the independent test set. Patient must have had at least one follow-up with a neurologist at our local institution. ADNI = Alzheimer’s Disease Neuroimaging Initiative.
Data Preprocessing
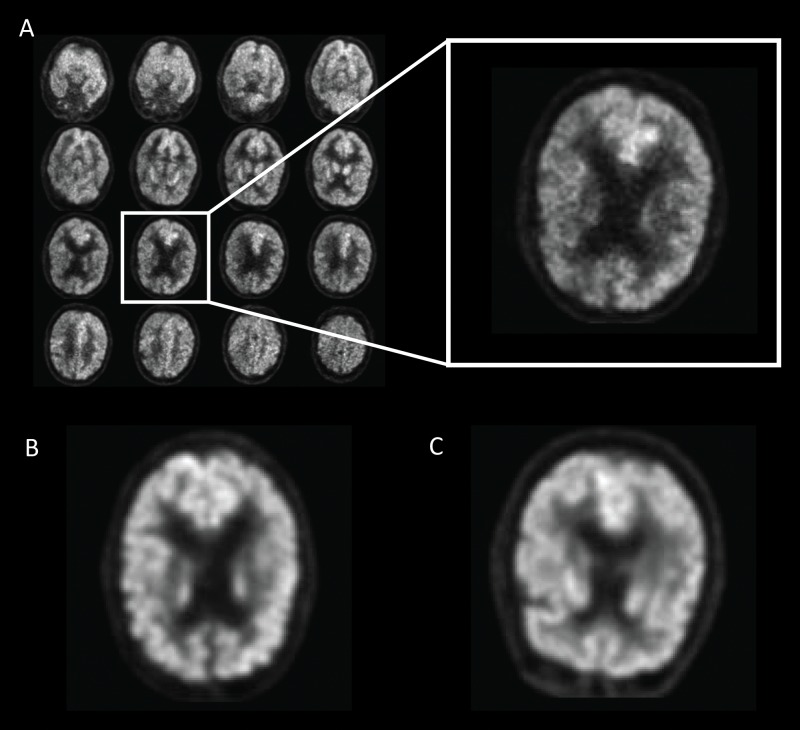
The imaging data were preprocessed by using a grid method (Fig 2). Images were resampled to 2-mm isotropic voxels and cropped to a 100 × 100 × 90-pixel grid resulting in a 200 × 200 × 180-mm3 volume. An Otsu threshold was utilized to select brain voxels. Connected component analysis was used to derive the relevant imaging volume by selecting the cranial-most and caudal-most sections representing more than 100 × 100 mm2 of brain parenchyma. The total volume was then divided into 16 evenly spaced sections, rounded to the nearest axial location, and distributed into a 4 × 4 grid with the cranial-most section in the top left and caudal-most section in the bottom right. All preprocessing steps were conducted in Python (Python 2.7; Python Software Foundation, Wilmington, Del; 2009) using package SciPy (http://www.scipy.org).
Figure 2:
Example of fluorine 18 fluorodeoxyglucose PET images from Alzheimer’s Disease Neuroimaging Initiative set preprocessed with the grid method for patients with Alzheimer disease (AD). One representative zoomed-in section was provided for each of three example patients: A, 76-year-old man with AD, B, 83-year-old woman with mild cognitive impairment (MCI), and, C, 80-year-old man with non-AD/MCI. In this example, the patient with AD presented slightly less gray matter than did the patient with non-AD/MCI. The difference between the patient with MCI and the patient with non-AD/MCI appeared minimal to the naked eye.
Model Training
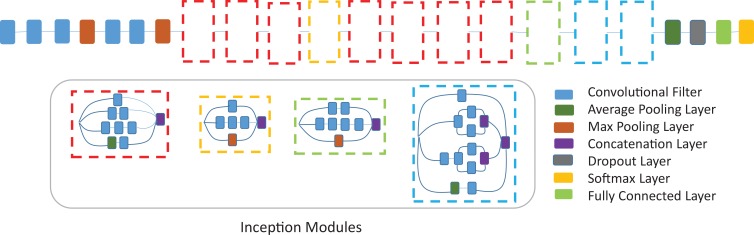
After preprocessing, the images were 512 × 512 matrix size and were loaded onto a machine with Linux operating system (Ubuntu 14.04; Canonical, London, England). The machine has six-core Intel i7 5930k 3.5-gHz processor (Intel, Santa Clara, Calif), 64 GB of DDR4 SDRAM, and a NVIDIA Pascal Titan X graphical processing unit (Nvidia Corporation, Santa Clara, Calif) with CUDA 8.0 and CuDNN 6.0 (Nvidia). Convolutional neural network architecture Inception V3 was used in the study (15). The network was pretrained on ImageNet, an everyday image data set containing 14 million images of 1000 classes, before being fine-tuned using 90% of the ADNI data set (1921 imaging studies). Data augmentation, including random width and height shift (range, 0%–10%) and zooming (range, 0%–8%), was performed on the training set. Dropout layer with a rate of 0.6 was added before the fully connected layers at the end of the network as means of regulation. The neural network architecture is shown in Figure 3 and Appendix E1 (online).
Figure 3:
Convolutional neural network architecture, Inception v3, used in this study. Inception v3 network stacks 11 inception modules where each module consists of pooling layers and convolutional filters with rectified linear units as activation function. The input of the model is two-dimensional images of 16 horizontal sections of the brain placed on 4 × 4 grids as produced by the preprocessing step. Three fully connected layers of size 1024, 512, and 3 are added to the final concatenation layer. A dropout with rate of 0.6 is applied before the fully connected layers as means of regularization. The model is pretrained on ImageNet dataset and further fine-tuned with a batch size of 8 and learning rate of 0.0001.
Adam, a first-order gradient-based stochastic optimization algorithm, with a learning rate of 0.0001, categorical cross entropy loss function, and batch size of 8 was used for model training (16). The trained algorithm was tested by the accuracy on the held-out ADNI data set (n = 188) and the independent test set (n = 40). Keras 2.0 (2017; Google, Mountain View, Calif) with Tensorflow 1.3 (2015; Google) backend was used for designing neural networks and loading pretrained weights. All programs were run in Python 2.7.
Model Interpretation and Data Visualization
To gain further intuition into how the network derived its decisions, one average saliency map taken across 10% ADNI test set and one across independent test set were shown. Saliency map plots the gradient of AD class score regarding each input pixel and thereby visualizes areas on the images that were deemed important for the classification result (17). To illustrate the connection between the saliency map and anatomy, an additional example individual saliency map was presented with anatomy overlay. All saliency maps were produced by using Keras 2.0.
t-Distributed stochastic neighbor embedding (t-SNE) (18), a dimension reduction method that preserves relative closeness of data points, was then performed on features extracted by the deep learning network on training data. By using package scikit-learn (19), the 1024 features were first reduced to dimension 30 with principal component analysis before t-SNE was applied with learning rate 200 and 1000 iterations to reduce the dimension further to 2.
Clinical Interpretation of 18F-FDG PET
To obtain reader performance on the independent test set, three board-certified nuclear medicine physicians (R.A.H., nuclear medicine; B.L.F., nuclear medicine; S.C.B., abdominal imaging and nuclear medicine) with 36, 14, and 5 years of experience, respectively, performed independent interpretations of the 40 18F-FDG PET imaging studies from the independent test set. Interpretations consisted of two components: qualitative interpretation of the PET emission images in axial, sagittal, and coronal planes, followed by a semiquantitative regional metabolic analysis using a commercially available clinical neuro-analysis software package (MIM Software, Cleveland, Ohio). Only 18F-FDG PET imaging data, name, age, and date of scan were visible to the readers. Qualitative and quantitative interpretations for one patient were performed consecutively before moving on to the next patient. If any of the three qualitative interpretations disagreed, the imaging study was interpreted by two additional radiology readers (L.N, nuclear medicine; C.M.A., nuclear medicine) with 1 year and 13 years of experience, respectively. The diagnosis of the majority of the five radiology readers was taken as the final clinical imaging diagnosis.
Model Testing and Statistical Analysis
The trained deep learning model was tested on two test data sets: 10% ADNI set as internal hold-out test set and independent test set from local institution as external test set. Probability that an image belongs to class AD, MCI, and non-AD/MCI was output by the model, and the class with the highest probability was chosen as the classification result.
Receiver operating characteristic (ROC) curves of the model on 10% ADNI set were plotted and the area under the ROC curve (AUC) was calculated. To compare the performance of deep learning model to reader performance, the ROC curves of deep learning model on independent test set were plotted with 95% confidence interval (CI), calculated by using package pROC 1.12.1 in R 3.5.1 with 200 iterations of bootstrapping (20,21). The sensitivity and specificity of reader performance were plotted in the same ROC space. If clinical reader’s sensitivity and specificity point lies outside of the CI space of the ROC curves, then the result was deemed as statistically significant. Sensitivity, specificity, precision, and F1 score were reported for both deep learning model and radiology readers. Model training, model testing, and model visualization were performed by Y.D., J.H.S., and M.K. Statistical analysis was performed by Y.D. and J.H.S.
Results
Demographics
As shown in Table 1, the ADNI set was composed of 2109 imaging studies from 1002 patients. The average age of the patients was 76 years (range, 55–93 years) for men and 75 years (range, 55–96 years) for women (P < .001), with an average age of 77 years (range, 56–92 years) for men and 75 years (range, 55–93 years) for women in the AD group (P = .04), 76 years (range, 55–93 years) for men and 74 years (range, 57–91 years) for women in the MCI group (P = .01), and 76 years (range, 60–90 years) for men and 75 years (range, 60–96 years) for women in the non-AD/MCI group (P = .14). The overall percentage of men was 54% (547 of 1002) by patient and 58% (1225 of 2109) by imaging study. The average follow-up period of the patients was 54 months by patient and 62 months by imaging study.
Table 1:
Demographics of Datasets

Note.–Unless otherwise indicated, data are averages ± standard deviation. ADNI = Alzheimer’s Disease Neuroimaging Initiative, AD = Alzheimer disease, MCI = mild cognitive impairment, Non-AD/MCI = neither Alzheimer disease nor mild cognitive impairment. NA = not applicable.
*Data in parentheses are the range.
†Data in parentheses are the percentage of male patients.
The independent test set was composed of 40 patients, with seven clinically diagnosed as having AD, seven as having MCI, and 26 as having non-AD/MCI at the end of the follow-up period. The average age of the 40 test patients was 66 years (range, 48–84 years) for men and 71 years (range, 41–84 years) for women, with an average age of 69 years (range, 56–79 years) for men and 73 years (range, 73–73 years) for women in the AD group, 63 years (range, 48–83 years) for men and 68 years (range, 68–68 years) for women in the MCI group, and 66 years (range, 55–84 years) for men and 71 years (range, 41–84 years) for women in the non-AD/MCI group. The overall percentage of men was 58% (23 of 40), while the percentage in the AD, MCI, and non-AD/MCI group was 86% (six of seven), 86% (six of seven), and 42% (11 of 26), respectively. The average follow-up period of the patients was 76 months, with an average of 82 months in the AD group, 75 months in the MCI group, and 74 months in the non-AD/MCI group.
Results of Model Training
The ROC curves of the inception V3 network trained on 90% of ADNI data and tested on the remaining 10% are shown in Figure 4a. The AUC for prediction of AD, MCI, and non-AD/MCI was 0.92, 0.63, and 0.73 respectively. The above AUCs indicate that the deep learning network had reasonable ability to distinguish patients who finally progressed to AD at the time of imaging from those who stayed to have MCI or non-AD/MCI, but was weaker at discriminating patients with MCI from the others. As shown in Table 2, in the prediction of AD, MCI, and non-AD/MCI, the respective sensitivity was 81% (29 of 36), 54% (43 of 79), and 59% (43 of 73), specificity was 94% (143 of 152), 68% (74 of 109), and 75% (86 of 115), and precision was 76% (29 of 38), 55% (43 of 78), and 60% (43 of 72).
Figure 4a:
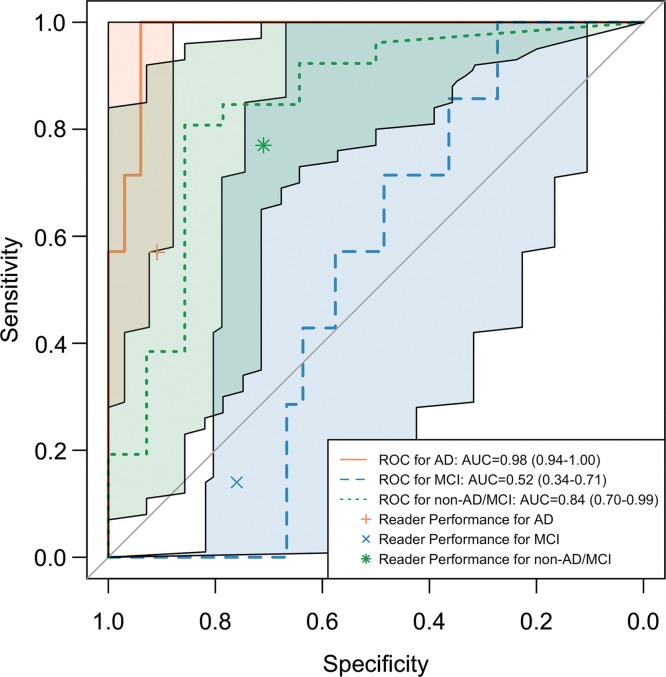
Receiver operating characteristic (ROC) curves of deep learning model Inception V3 trained on 90% of Alzheimer’s Disease Neuroimaging Initiative (ADNI) data and tested on the remaining 10% of ADNI set and independent test set. (a) ROC curves of trained deep learning model tested on the remaining 10% of ADNI set. ROC curve labeled AD (Alzheimer disease) represents the core model performance for distinguishing AD versus all other cases. ROC curves for mild cognitive impairment (MCI) and non-AD/MCI are also reported for technical completeness. (b) ROC curves including the 95% confidence interval of trained deep learning model tested on the independent test set together with reader performance plotted on ROC space. The deep learning algorithm performs statistically significantly better at recognizing patients with AD on the independent test set. The algorithm is also better at recognizing patient with non-AD/MCI and worse at recognizing patients with MCI, but did not reach statistical significance.
Table 2:
Performance Comparison of Deep Learning Algorithm and Radiology Readers

Note.–Unless otherwise indicated, data are averages ± standard deviation. ADNI = Alzheimer’s Disease Neuroimaging Initiative, AD = Alzheimer disease, MCI = mild cognitive impairment, Non-AD/MCI = neither Alzheimer disease nor mild cognitive impairment.
*Numbers in parentheses are raw data used to calculate the percentage.
†Numbers indicate higher performance from deep learning algorithm compared with reader performance on independent test set.
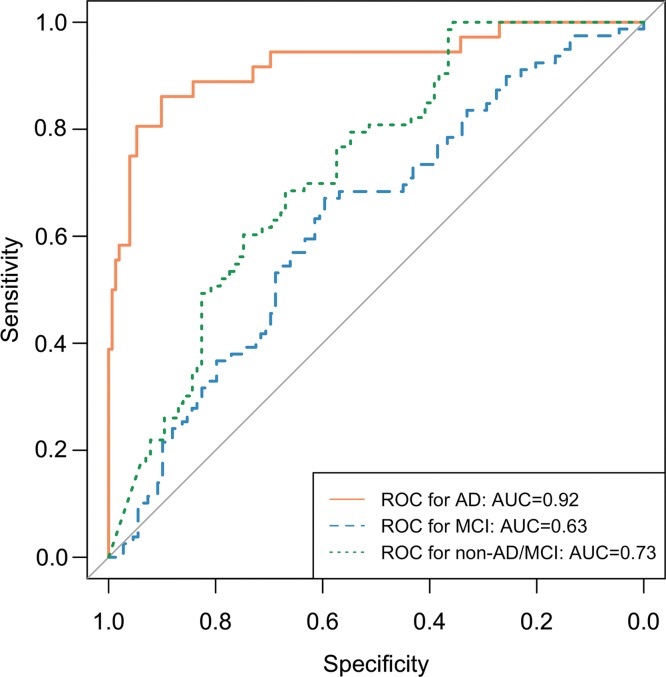
The ROC curves of the inception V3 network trained on 90% ADNI data and tested on independent test set with 95% CI are shown in Figure 4b. The AUC for the prediction of AD, MCI, and non-AD/MCI was 0.98 (95% CI: 0.94, 1.00), 0.52 (95% CI: 0.34, 0.71), and 0.84 (95 CI: 0.70, 0.99), respectively. Choosing the class with the highest probability as the classification result, in the prediction of AD, MCI, and non-AD/MCI, respectively, the sensitivity was 100% (seven of seven), 43% (three of seven), and 35% (nine of 26), the specificity was 82% (27 of 33), 58% (19 of 33), and 93% (13 of 14), and the precision was 54% (seven of 13), 18% (three of 17), and 90% (nine of 10). With a perfect sensitivity rate and reasonable specificity on AD, the model preserves a strong ability to predict the final diagnoses prior to the full follow-up period that, on average, concluded 76 months later.
Figure 4b:
Receiver operating characteristic (ROC) curves of deep learning model Inception V3 trained on 90% of Alzheimer’s Disease Neuroimaging Initiative (ADNI) data and tested on the remaining 10% of ADNI set and independent test set. (a) ROC curves of trained deep learning model tested on the remaining 10% of ADNI set. ROC curve labeled AD (Alzheimer disease) represents the core model performance for distinguishing AD versus all other cases. ROC curves for mild cognitive impairment (MCI) and non-AD/MCI are also reported for technical completeness. (b) ROC curves including the 95% confidence interval of trained deep learning model tested on the independent test set together with reader performance plotted on ROC space. The deep learning algorithm performs statistically significantly better at recognizing patients with AD on the independent test set. The algorithm is also better at recognizing patient with non-AD/MCI and worse at recognizing patients with MCI, but did not reach statistical significance.
Model Interpretation: Saliency Map and t-SNE Plot
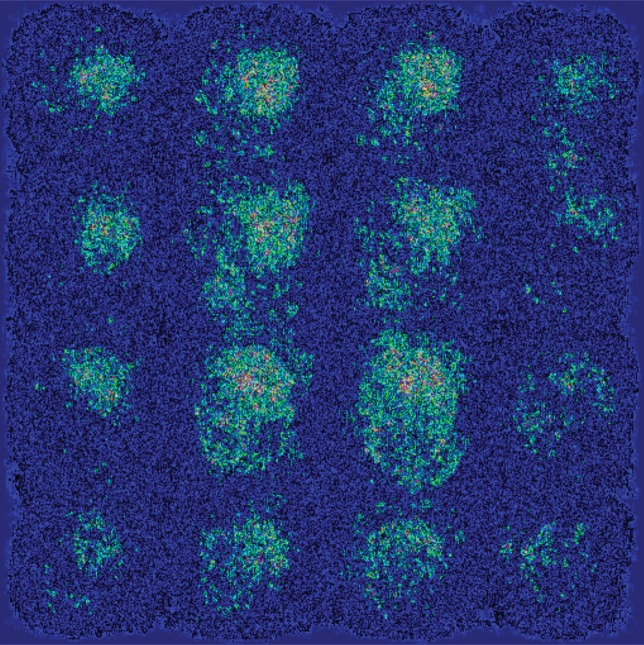
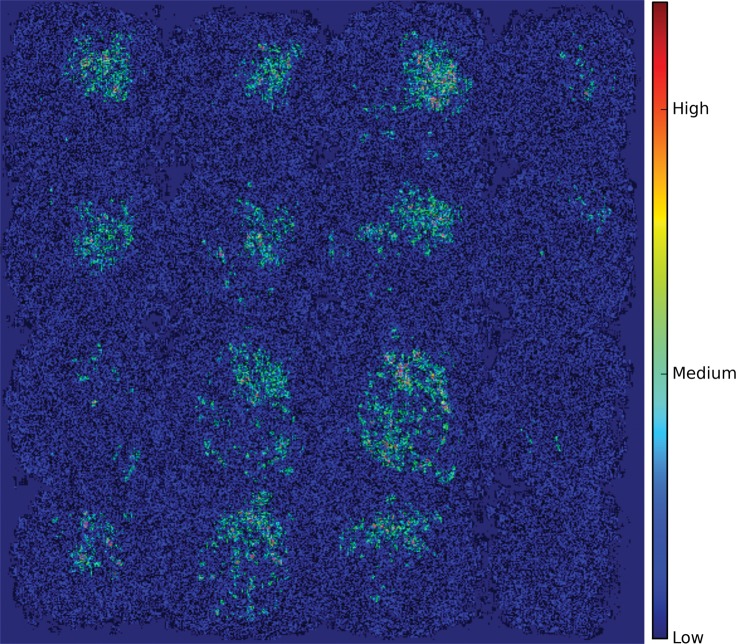
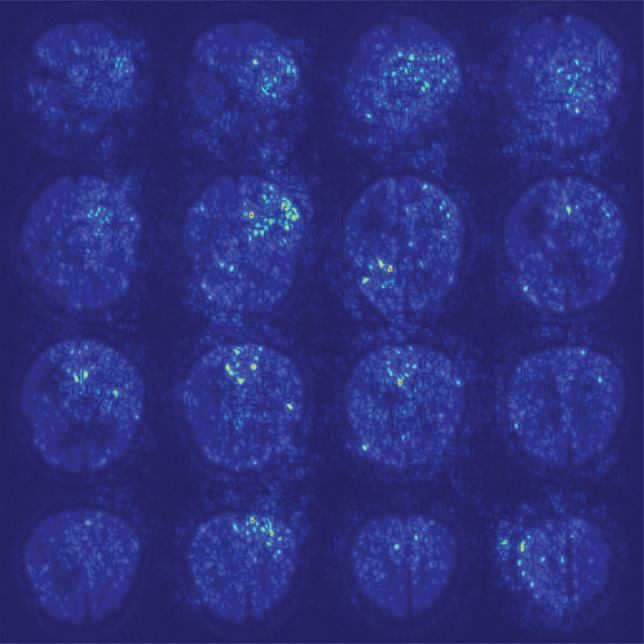
As shown in the saliency map in Figure 5b and 5c, the second and third sections in the third row demonstrate the most intense signals among the scattered areas of signal. The result indicates their importance in the decision of classifying a patient with AD, which is in line with the clinical implication that more caudal sections in the parietotemporal regions are informative of AD. However, the patterns are not specific enough to extract a unified human-interpretable imaging biomarker, and overall, the saliency map suggests that the deep learning model considered the whole brain when making the prediction, as presented in Figure 5a.
Figure 5b:
Saliency map of deep learning model Inception V3 on the classification of Alzheimer disease. (a) A representative saliency map with anatomic overlay in 77-year-old man. (b) Average saliency map over 10% of Alzheimer’s Disease Neuroimaging Initiative set. (c) Average saliency map over independent test set. The closer a pixel color is to the "High" end of the color bar in the image, the more influence it has on the prediction of Alzheimer disease class.
Figure 5c:
Saliency map of deep learning model Inception V3 on the classification of Alzheimer disease. (a) A representative saliency map with anatomic overlay in 77-year-old man. (b) Average saliency map over 10% of Alzheimer’s Disease Neuroimaging Initiative set. (c) Average saliency map over independent test set. The closer a pixel color is to the "High" end of the color bar in the image, the more influence it has on the prediction of Alzheimer disease class.
Figure 5a:
Saliency map of deep learning model Inception V3 on the classification of Alzheimer disease. (a) A representative saliency map with anatomic overlay in 77-year-old man. (b) Average saliency map over 10% of Alzheimer’s Disease Neuroimaging Initiative set. (c) Average saliency map over independent test set. The closer a pixel color is to the "High" end of the color bar in the image, the more influence it has on the prediction of Alzheimer disease class.
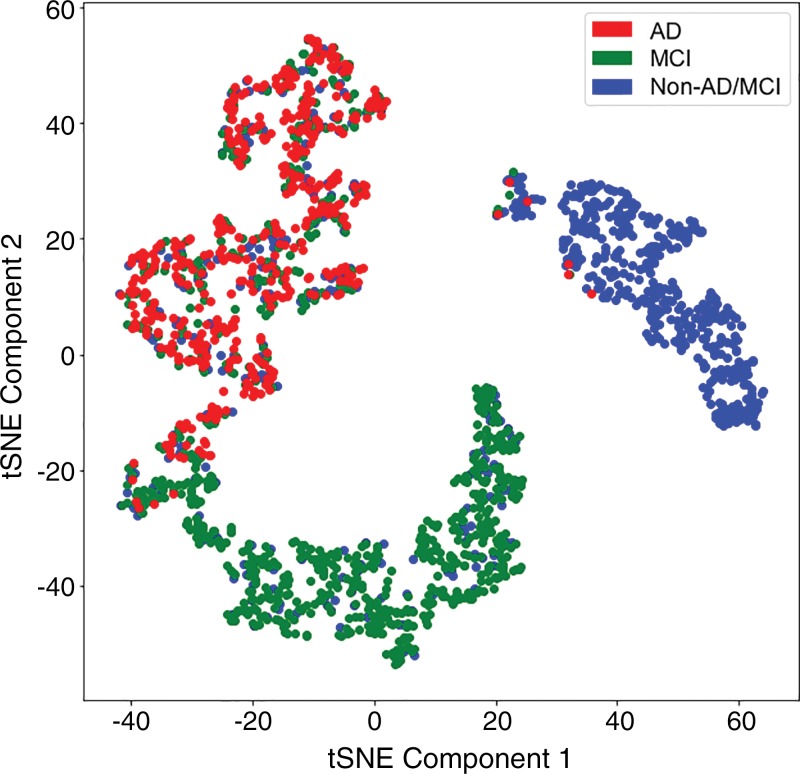
As shown in Figure 6, after dimension reduction with t-SNE, the features extracted by Inception V3 network separated the three classes into approximately three clusters. While the cluster of non-AD/MCI was almost pure, the cluster of MCI was mixed with patients with non-AD/MCI and patients with AD, and the cluster of AD was mixed with the other two classes. This gives insight to the behavior of the model at test time: We obtained a high sensitivity rate on AD class because nearly all patients with AD were located in the AD cluster; we obtained a relatively high precision rate on non-AD/MCI class because the non-AD/MCI cluster was almost pure.
Figure 6:
Visualization of training set after dimension reduction with t-distributed stochastic neighbor embedding (t-SNE). Each dot represents the 1024 features output by the final fully connected layer of the Inception V3 network. Red dots represent samples from Alzheimer disease (AD), green dots represent samples from mild cognitive impairment (MCI), and blue dots represent samples from neither classes (non-AD/MCI).
Comparison to Clinical Interpretations
As reported in Table 2, the sensitivity, specificity, and precision for reader performance were 57% (four of seven), 91% (30 of 33), and 57% (four of seven) for class AD; 14% (one of seven), 76% (25 of 33), and 11% (one of nine) for class MCI; and 77% (20 of 26), 71% (10 of 14), and 83% (20 of 24) for class non-AD/MCI. By plotting reader performance and ROC curves for model performance in the same ROC space as in Figure 4b for class AD in independent test set, reader performance lies below the model ROC curve and outside its 95% CI. While for class MCI and non-AD/MCI, reader performance lies above and below the model ROC curves, respectively, but both within the 95% CI of the ROC curve. Therefore, compared with radiology readers, the deep learning model performed better, with statistical significance, at recognizing patients who would go on to have a clinical diagnosis of AD. On the independent test set, it also performs better at recognizing patient with neither AD nor MCI, while worse at recognizing patients who would develop MCI but would not advance to AD, though without statistical significance.
Discussion
There is a growing number of patients living with AD, and it has been forecasted that more than 2% of the U.S. population and 1% of the world’s population will have AD by 2050 (22,23). Unfortunately, early identification of those patients who will have a final diagnosis of AD is challenging. The deep learning algorithm developed and tested in our study was shown to be robust across ADNI hold-out test set and independent test set, with 100% sensitivity (95% CI: 65%, 100%) for AD prediction on the independent test set. Furthermore, in predicting the final diagnosis of AD on the independent test set, it outperformed three radiology readers in ROC space, with statistical significance. With further validation with larger and more diverse datasets, this algorithm may be able to augment radiologist reader performance and improve the prediction of AD diagnosis, providing an opportunity for early intervention.
Multiple previous studies have shown that the distinctive distribution of areas of cortical hypometabolism on 18F-FDG PET images has implications in differentiating AD or MCI from a normal brain; however, 18F-FDG itself is not a definitive imaging biomarker for AD or MCI. The past decade has produced several tools for the early diagnosis of AD, including increasingly specific biomarkers of the disease (24,25). For example, β-amyloid (Aβ), a marker of AD, can be detected in the cerebral spinal fluid or at imaging with PET by using radiolabeled Aβ ligands, such as 18F-florbetapir, flutemetamol, and florbetaben (3,26,27). However, these innovations are associated with a high cost that may not be reimbursed by a patient’s health insurance or may not be universally available; hence, enhancement of the diagnostic and predictive ability of a long-established imaging technique, such as 18F-FDG PET, using a deep learning algorithm offers the opportunity to provide clinically relevant molecular imaging data across a multitude of populations and settings worldwide.
Substantial work in the area of computer-aided diagnosis and risk classification has been performed by using structural imaging of the brain (28,29). But less work has been devoted to applying deep learning methods to functional imaging alone to classify patients with symptoms of dementia. To our knowledge, the method in our present study has not previously been emphasized in the literature. After training the deep learning model on 90% of the ADNI dataset, validation of the model using the remaining 10% of the ADNI 10% hold-out dataset yielded discrimination of AD of more than 90% as shown by the AUC. Notably, the pooled sensitivity and specificity of 18F-FDG PET imaging in identifying mild AD as the cause of a patient’s symptoms across several studies are reported as 90% and 89%, respectively (30–32).
Application of the model to standard clinical 18F-FDG PET imaging studies performed on a cohort of patients for the indication of memory loss (referred to as independent test set) yielded high predictive ability for those patients who were ultimately diagnosed with AD (92% in ADNI test set and 98% in the independent test set) and those who were non-AD/MCI (73% in ADNI test set and 84% in the independent test set). Arguably, these two groups are the most important to classify correctly. However, the model’s predictive ability for those patients who were ultimately diagnosed with MCI was lower (63% in ADNI test set and 52% in the independent test set). This is not unexpected given the high degree of variability in the diagnosis of MCI and its existence on a continuum with AD. The lower diagnostic power can also be caused by the fact that patients who carried final diagnosis of MCI may have been at a state too early to show clinical signs of AD or may be those who will not progress to AD.
It is noteworthy that model visualization with saliency map did not reveal a distinctly human interpretable imaging biomarker that appears influential for AD prediction. Instead, the deep learning algorithm apparently utilized the whole brain with varying degrees of influence from various anatomic areas to make its final decision. This highlights the strength of the deep learning algorithm that considers the brain as a pixel-by-pixel volume in its classification, implying that the deep learning algorithm arrives at the diagnosis distinct from how humans interpret the imaging studies.
Our study had several limitations. First, our independent test data were relatively small (n = 40) and were not collected as part of a clinical trial. Most notably, this was a highly selected cohort in that all patients must have been referred to the memory clinic and neurologist must have decided that a PET study of the brain would be useful in clinical management. This effectively excluded most non-AD neurodegenerative cases and other neurologic disorders such as stroke that could affect memory function. Arguably, such cohort of patients would be the most relevant group to test the deep learning algorithm, but the algorithm’s performance on a more general patient population remains untested and unproven, hence the pilot nature of this study.
Second, the deep learning algorithm’s robustness is inherently limited by the clinical distribution of the training set from ADNI. The algorithm achieved strong performance on a small independent test set, where the population substantially differed from the ADNI test set; however, its performance and robustness cannot yet be guaranteed on prospective, unselected, and real-life scenario patient cohorts. Further validation with larger and prospective external test set must be performed before actual clinical use. Furthermore, this training set from ADNI did not include non-AD neurodegenerative cases, limiting the utility of the algorithm in such patient population. Third, the deep learning algorithm did not yield a human interpretable imaging biomarker despite visualization with saliency map, which highlights the inherent black-box limitation of deep learning algorithms. The algorithm instead made predictions based on holistic features of the imaging study, distinct from the human expert approaches. Fourth, MCI and non-AD/MCI were inherently unstable diagnoses in that their accuracy is dependent on the length of follow-up. For example, some of the MCI patients, if followed up for long enough time, may have eventually progressed to AD.
Overall, our study demonstrates that a deep learning algorithm can predict the final diagnosis of AD from 18F-FDG PET imaging studies of the brain with high accuracy and robustness across external test data. Furthermore, this study proposes a working deep learning approaches and a set of convolutional neural network hyperparameters, validated on a public dataset, that can be the groundwork for further model improvement. With further large-scale external validation on multi-institutional data and model calibration, the algorithm may be integrated into clinical workflow and serve as an important decision support tool to aid radiology readers and clinicians with early prediction of AD from 18F-FDG PET imaging studies.
Up-to-date source code can be found on the Big Data in Radiology (BDRad) Git Repository: http://bit.ly/2o0JSRb.
As of 8/1/2018, there are 1,005 publications that references the ADNI dataset used in this study. The full list can be found at http://adni.loni.usc.edu/news-publications/publications/.
H.T. supported by Foundation for the National Institutes of Health fellowship (5T32EB001631-10). Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI), National Institutes of Health (U01 AG024904) and U.S. Department of Defense (W81XWH-12-2-0012). J.H.S. supported by University of California, San Francisco (CTSI Resident Research Grant 2017, Radiology & Biomedical Imaging Seed Grant #17-11). ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Data used in the preparation of this article were obtained from the ADNI database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report.
From the 2017 RSNA Annual Meeting.
Disclosures of Conflicts of Interest: Y.D. disclosed no relevant relationships. J.H.S. Activities related to the present article: received grants from UCSF. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. M.G.K. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employed by Genentech and has stock options from Roche. Other relationships: disclosed no relevant relationships. H.T. disclosed no relevant relationships. R.H. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employed by UCSF Radiology department as a programmer, analyst, algorithm developer. Other relationships: disclosed no relevant relationships. N.W.J. disclosed no relevant relationships. D.L. Activities related to the present article: received payment from NVIDIA for travel to meetings. Activities not related to the present article: received payment from HUST Science Forum UC Berkeley for lectures including service on speakers bureaus. Other relationships: disclosed no relevant relationships. T.P.C. disclosed no relevant relationships. M.S.A. disclosed no relevant relationships. C.M.A. disclosed no relevant relationships. S.C.B. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: institution received research grant from GE Healthcare. Other relationships: disclosed no relevant relationships. R.R.F. disclosed no relevant relationships. S.Y. H. disclosed no relevant relationships. K.A.Z. disclosed no relevant relationships. L.N. disclosed no relevant relationships. Y.S. disclosed no relevant relationships. R.A.H. disclosed no relevant relationships. M.H.P. disclosed no relevant relationships. D.H. disclosed no relevant relationships. B.L.F. disclosed no relevant relationships.
Abbreviations:
- AD
- Alzheimer disease
- ADNI
- Alzheimer’s Disease Neuroimaging Initiative
- AUC
- area under the ROC curve
- CI
- confidence interval
- FDG
- fluorodeoxyglucose
- MCI
- mild cognitive impairment
- ROC
- receiver operating characteristic
- t-SNE
- t-distributed stochastic neighbor embedding
References
- 1.Jack CR, Jr, Albert MS, Knopman DS, et al. Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011;7(3):257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011;7(3):263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheltens P, Blennow K, Breteler MMB, et al. Alzheimer’s disease. Lancet 2016;388(10043):505–517. [DOI] [PubMed] [Google Scholar]
- 4.Matsuda H. The role of neuroimaging in mild cognitive impairment. Neuropathology 2007;27(6):570–577. [DOI] [PubMed] [Google Scholar]
- 5.Mosconi L, Tsui WH, Herholz K, et al. Multicenter standardized 18F-FDG PET diagnosis of mild cognitive impairment, Alzheimer’s disease, and other dementias. J Nucl Med 2008;49(3):390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Summers RM. Machine learning and radiology. Med Image Anal 2012;16(5):933–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhungel N, Carneiro G, Bradley AP. A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med Image Anal 2017;37:114–128. [DOI] [PubMed] [Google Scholar]
- 8.Setio AAA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 2016;35(5):1160–1169. [DOI] [PubMed] [Google Scholar]
- 9.Xu L, Wu X, Chen K, Yao L. Multi-modality sparse representation-based classification for Alzheimer’s disease and mild cognitive impairment. Comput Methods Programs Biomed 2015;122(2):182–190. [DOI] [PubMed] [Google Scholar]
- 10.Xue Y, Zhang R, Deng Y, Chen K, Jiang T. A preliminary examination of the diagnostic value of deep learning in hip osteoarthritis. PLoS One 2017;12(6):e0178992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller SG, Weiner MW, Thal LJ, et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am 2005;15(4):869–877, xi–xii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alzheimer’s Disease Neuroimaging Initiative PET technical procedures manual . http://adni.loni.usc.edu/wp-content/uploads/2010/09/PET-Tech_Procedures_Manual_v9.5.pdf. Published 2006. Accessed July 18, 2018.
- 13.ADNI 2 PET technical procedures manual for FDG and AV-45 ADNI 2 PET technical procedures manual AV-45 (Florbetapir F 18) & FDG. http://adni.loni.usc.edu/wp-content/uploads/2010/05/ADNI2_PET_Tech_Manual_0142011.pdf. Published 2011. Accessed July 18, 2018.
- 14.ADNI-GO PET technical procedures manual for FDG and AV-45 ADNI-GO PET Technical Procedures Manual . http://adni.loni.usc.edu/wp-content/uploads/2010/05/ADNIGO_PET_Tech_Manual_01142011.pdf. Published 2011. Accessed July 18, 2018.
- 15.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. IEEE Conference on Computer Vision and Pattern Recognition, 2015; 2818–2826. [Google Scholar]
- 16.Kingma DP, Ba J. Adam: A method for stochastic optimization. https://arxiv.org/abs/1412.6980. Published December 22, 2014. Accessed February 20, 2018.
- 17.Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps. https://arxiv.org/abs/1312.6034. Published December 20, 2013. Accessed March 18, 2018.
- 18.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res 2008;9(Nov):2579–2605. [Google Scholar]
- 19.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res 2011;12(Oct):2825–2830. [Google Scholar]
- 20.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The R Core Team. R : A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2018. [Google Scholar]
- 22.Brookmeyer R, Gray S, Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health 1998;88(9):1337–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement 2007;3(3):186–191. [DOI] [PubMed] [Google Scholar]
- 24.Nordberg A. Dementia in 2014. Towards early diagnosis in Alzheimer disease. Nat Rev Neurol 2015;11(2):69–70. [DOI] [PubMed] [Google Scholar]
- 25.Westman E, Muehlboeck J-S, Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. Neuroimage 2012;62(1):229–238. [DOI] [PubMed] [Google Scholar]
- 26.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984;34(7):939–944. [DOI] [PubMed] [Google Scholar]
- 27.Morris E, Chalkidou A, Hammers A, Peacock J, Summers J, Keevil S. Diagnostic accuracy of (18)F amyloid PET tracers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. Eur J Nucl Med Mol Imaging 2016;43(2):374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie D, Zhang H, Adeli E, Liu L, Shen D. 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients. Med Image Comput Comput Assist Interv 2016;9901:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen W, Zhou M, Yang F, Yang C, Tian J. Multi-scale convolutional neural networks for lung nodule classification. Inf Process Med Imaging 2015;24:588–599. [DOI] [PubMed] [Google Scholar]
- 30.Bloudek LM, Spackman DE, Blankenburg M, Sullivan SD. Review and meta-analysis of biomarkers and diagnostic imaging in Alzheimer’s disease. J Alzheimers Dis 2011;26(4):627–645. [DOI] [PubMed] [Google Scholar]
- 31.Marcus C, Mena E, Subramaniam RM. Brain PET in the diagnosis of Alzheimer’s disease. Clin Nucl Med 2014;39(10):e413–e422; quiz e423–e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shivamurthy VK, Tahari AK, Marcus C, Subramaniam RM. Brain FDG PET and the diagnosis of dementia. AJR Am J Roentgenol 2015;204(1):W76–W85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Up-to-date source code can be found on the Big Data in Radiology (BDRad) Git Repository: http://bit.ly/2o0JSRb.
As of 8/1/2018, there are 1,005 publications that references the ADNI dataset used in this study. The full list can be found at http://adni.loni.usc.edu/news-publications/publications/.