Abstract
A polydimethylsiloxane-based microfluidic device has been developed for the multiplex detection of viral envelope proteins such as Zika and chikungunya on a single platform using aptamer–analyte interactions. The channel is integrated with microsized pillars that increase the surface area allowing more aptamers to attach to the incoming envelope protein molecules, thus increasing the overall sensitivity of the system. The working of the device depends on the formation of protein-mediated sandwich morphology that is obtained using an aptamer and aptamer-functionalized gold nanoparticle (AuNP) pair. The colorimetric signal is obtained upon introduction of silver reagents into the channel, which are selectively deposited on the AuNP surface, providing a gray contrast in the testing zone. The microfluidic channel approach successfully detected clinically relevant concentrations of Zika and chikungunya envelope proteins in phosphine-buffered saline (1 pM) and calf blood (100 pM) with high specificity using gold-decorated aptamers integrated in a microfluidic channel.
1. Introduction
Zika fever is a mosquito-borne illness caused by Zika virus (ZIKV) that is vectored by Aedes (Ae.) genus mosquitoes such as Ae. aegypti.1,2 ZIKV is known to pose a serious threat to the population worldwide; for example, nearly 440000 cases of Zika fever associated with microcephaly and central nervous system abnormalities were reported during the Zika outbreak in Brazil in 2015.3,4 Besides Brazil, cases associated with ZIKV infection have also been reported in 66 other countries to date.5 The World Health Organization (WHO) declared a Public Health Emergency of International Concern (PHEIC) in 2016 because of the increase in microcephaly cases and neurological disorders that were associated with ZIKV infection.6 More than 5000 cases since 2016 have been confirmed in United States alone as reported by the Pan American Health Organization.3 The transmission of ZIKV occurs through the bite of a ZIKV-infected mosquito, in utero from mother to fetus and via sexual contact with an infected person.2,7−9 ZIKV infection is usually asymptomatic; however, nonspecific symptoms such as rash, fever, headache, vomiting, and joint pains of varying degrees are sometimes observed.10,11 These symptoms overlap significantly with other arboviral infections such as chikungunya (CHIKV). Because of a shared mosquito vector as well as nonspecific symptoms, adequate diagnosis of ZIKV infection at an early stage poses a serious challenge, especially in resource-constrained communities where both the viral infections are endemic.
The current diagnostic tools for the detection of ZIKV are mainly reverse transcription polymerase chain reaction (RT-PCR),10 reverse transcription loop-mediated isothermal amplification (RT-LAMP),12−16 and reverse transcription recombinase polymerase amplification (RT-RPA)17 methods, coupled with external power source and a display unit such as smart phone for an easy readout signal.18−24 These techniques often require expensive instrumentation and skilled labor and sometimes face major disadvantages of false negative/positive results arising from new strains or contamination.25 Other methods for diagnosis of ZIKV infection such as IgM-capture ELISA (enzyme-linked immunosorbent assay) and plaque reduction neutralization test face a major challenge of cross-reactivity with other flaviviruses25 leading to ambiguous results. Therefore, there is a dire need for the development of novel, compact, and passive point-of-care (POC) testing tools which can provide early diagnosis of a disease leading to a more consumer-driven health care.
To this end, we have fabricated a flexible, portable, easy-to-use aptamer-based device for the multiplex detection of recombinant ZIKV and CHIKV envelope proteins. Aptamers are short single-stranded oligonucleotides that are designed using in vitro SELEX (systematic evolution of ligands by exponential enrichment) process to bind to a specific target with high specificity and affinity.26,27 Aptamers offer several advantages over antibodies such as stability in a wider range of environments, are more readily synthesized and functionalized, have higher batch-to-batch consistency, and can be generated against a wider variety of targets.28−32 Herein, these aptamers are integrated in a microfluidic platform, which allows for the manipulation of fluids at the micron scale and is frequently used to distill/translate/convert operations that originally required large, expensive equipment onto a micron-sized device.33,34 For the present study, the compatibility of polydimethylsiloxane (PDMS) with microfluidic fabrication offers flexibility and optical transparency, which facilitates observation of the colorimetric signals produced in the presence of analytes during the test.
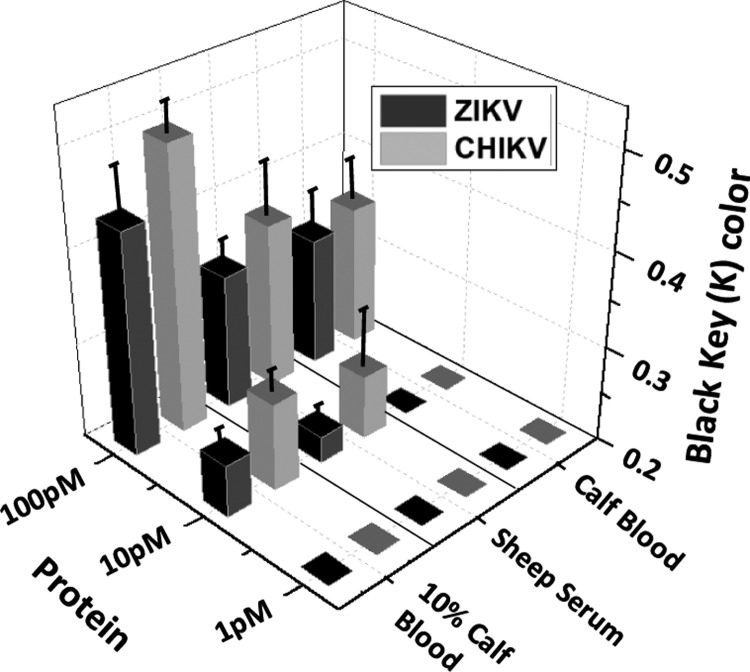
The current sensor depends on the engineered sandwich morphology consisting of aptamer1–antigen–aptamer2 for the detection of ZIKA and CHIKV antigens (Figure 1). Aptamer1 is immobilized on the microfluidic channel, which captures the incoming protein present in the samples. Aptamer2 conjugated with gold nanoparticles (Apt-AuNPs) is used to report the presence of antigen by generating an output signal in the form of color change. A silver staining technique has been used to further amplify the color change facilitating colorimetric detection by eye.35,36 In close proximity of silver ions, AuNPs serve as an electron-transferring agent from the reducing agent (e.g., hydroquinone) to silver ions.36,37 This leads to the deposition of metal silver over the surface of AuNPs, which catalyzes the subsequent reactions themselves. This process leads to continuous deposition of silver over gold, which produces gray color in the testing zone. The intensity of developed color depends on the concentration of analyte-bound Apt-AuNPs over the channel surface. Thus, the present technique provides a quantitative and qualitative strategy toward detecting arboviral proteins (antigens). The major advantage of using an antigen capture sandwich assay is high sensitivity and colorimetric detection capabilities with crude sample preparation.38 The approach is similar to the ELISA technique but avoids the use of expensive plates and plate readers and is very easy to handle. Also, the replacement of antibodies with aptamers makes this technique more robust while maintaining the same level of accuracy and reliability of the approach.
Figure 1.
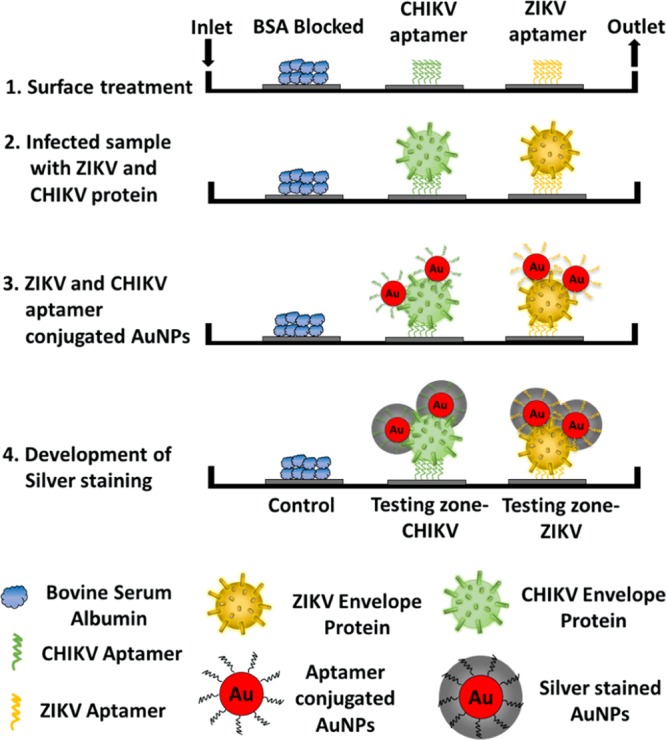
Schematic representation of the working of current approach based on sandwich morphology for the detection of multiple viral envelope proteins on a single platform. The surface is first conjugated with a linker molecule, PMPI, that attaches to the thiolated end of the aptamers (1); the control site is blocked with bovine serum albumin (BSA). Sample containing viral envelope protein(s) is introduced in the channel where the aptamers specific for this protein bind the target (2). AuNPs conjugated with protein-specific aptamers are then flowed in and bind to other free epitopes on the previously captured envelope protein(s) to form a sandwich morphology (3); if no arboviral envelope protein is present, AuNPs will be washed away. Finally, silver reagents are introduced into the channel, which get deposited onto the surface of bound AuNPs, thereby generating a colorimetric signal indicating the presence of arboviral envelope protein (4).
2. Materials and Methods
2.1. Chemical Reagents
Recombinant Zika virus (ZIKV) envelope protein (ZKV-005) and chikungunya virus (CHIKV) type E1 envelope protein (CHI-001) were obtained from ProSpec (Rehovot, Israel). The protein samples were provided to Base Pair Biotechnologies (BPB, Pearland, TX) for synthesizing custom-made aptamers with thiol functionalization. The aptamers showing highest binding affinity were used for experimentation. Aptamer folding buffer, reducing buffer, and resuspension buffer were also provided by BPB to prepare the aptamers for immobilization. Other chemicals such as gold(III) chloride trihydrate, trisodium citrate, phosphine-buffered saline (PBS) tablets, sodium hydroxide, hexamethyldisilazane (HMDS), acetonitrile, and magnesium chloride (MgCl2) were purchased from Sigma-Aldrich (St. Louis, MO). PMPI (p-maleimidophenyl isocyanate) used for the functionalization of PDMS was obtained from Thermo Fisher (Waltham, MA). Dow Corning Sylgard 184 Silicone Encapsulant Clear Kit for the fabrication of microfluidic channels was purchased from Ellsworth (Germantown, WI). SU8 and SU8 developer used during photolithography were purchased from MicroChem (Westborough, MA). The masks were printed by CAD/Art Services (Bandon, OR). The silver enhancement kit was purchased from Cytodiagnostics (Burlington, ON). Silicon wafers were obtained from Waferworld Inc. (West Palm Beach, FL). Sterile, mechanically defibrinated bovine/calf blood was obtained from Rockland Immunochemicals Inc. (Limerick, PA) and sheep serum was purchased from Sigma-Aldrich. The entire study was done using DNAse–RNAse-free water, vials and pipette tips, obtained from Thermo Fisher.
2.2. Fabrication of Microfluidic Channel
2.2.1. Fabrication of Master Mold
PDMS-based microfluidic channels were fabricated using a soft lithography technique (Figure S1).39 The silicon wafers were cleaned thoroughly by rinsing with acetone, isopropanol, and water, followed by nitrogen blow. These were baked at 80 °C for 15 min to evaporate any residual water. After cleaning, the wafers were pretreated with HMDS overnight to improve the adhesion between the photoresist layer and wafer surface. The HMDS-treated wafers were deposited with SU8 photoresist for the development of master mold. The wafer was centered on the rotating stage of the spin coater and a dollop of SU8-2050 was poured in the center. SU-8 was spin-coated over the wafer at 2500 rpm for 50 s, which gave an average thickness of 40–60 μm of the layer. After spinning, the wafer was soft-baked at 65 °C for 3 min, followed by 95 °C for 8 min. The wafer was allowed to cool at room temperature for 5 min. The next step in the lithography is to transfer the pattern of the mask onto the SU8-deposited Si wafer. The mask was placed over the Si wafer in the mask aligner and exposed to UV (275 nm) for 50 s. After UV exposure, the silicon wafers were baked for 1 min at 65 °C, followed by a 6 min bake at 95 °C. Upon cooling, the wafers were developed in copious amount of SU-8 developer for 5 min, followed by rinsing with isopropanol. If white streaks were seen, the wafers were again kept in a fresh developer solution for few seconds and then rinsed with isopropyl alcohol. The developed wafers were dried using nitrogen blow and baked at 95 °C for 3 min. The fabricated master molds were silanized using tricholorsilane for 2 h under vacuum to make the peeling step easier.
2.2.2. Development of Microfluidic Channels
The PDMS mixture was formed by adding the base and cross-linker (provided in the Dow Corning kit) in 10:1 weight ratio in a disposable plastic weigh boat. The mixture was thoroughly mixed with a plastic-stir rod until it became cloudy and suspended with air bubbles. The air bubbles were removed by placing the boat under vacuum in a desiccator and venting the desiccator periodically. Once all the air bubbles were removed, the PDMS mixture was poured slowly onto the master mold in a Petri dish. The Petri dish was then placed in a preheated oven at 65 °C for 1 h to cure. After curing, the PDMS hardened over the mold and took the features of the mold as shown in Figure S2. It was then peeled off carefully and individual channels were cut using a sharp blade. The entire channel length was 4.5 cm and the depth was measured to be around 45 μm using a Dektak instrument (as shown in Figure S3). The diameter of the testing zone was maintained at 0.5 cm. Figure S4 shows a digital image of a single microfluidic channel. The red color solution represents the AuNP solution in the channel.
2.2.3. Surface Treatment of Microfluidic Channels
The microfluidic channels were treated with 1 mM NaOH solution at room temperature for 16 h under gentle shaking. The samples were then washed thoroughly with DNase–RNase-free water, followed by drying with nitrogen blow. After this, PMPI dissolved in acetonitrile (10 mg/mL) was applied onto the testing regions and kept for 3 h at room temperature. The surface treatment of PDMS with PMPI offers a free maleimide group, which shows high affinity toward the thiol bond present at the end of aptamers. The samples were washed with acetonitrile and working buffer (1 mM MgCl2, 1× PBS, pH 7.5) thrice before conjugating with aptamers.
2.3. Preparation of Aptamers and Immobilization
The obtained thiolated aptamers (CHIKV E1 and ZIKV envelope) were dissolved in resuspension buffer to achieve a 100 μM stock concentration. The aptamers were diluted to 50 μM using a folding buffer and heated at 95 °C for 5 min. Then, these were cooled to room temperature for 15 min. This allows the aptamers to fold properly and attain their native conformation required for binding to the target molecule. After 15 min, the aptamer solution was mixed with the reducing buffer (tris(2-carboxyethyl)phosphine, TCEP) in 1:1 ratio (v/v) to obtain a final concentration of 25 μM and kept at room temperature for another 20 min. TCEP is used to cleave the disulfide bonds (−S–S−), which allows for free −SH group. The −SH group plays a major role in binding of the aptamers to the AuNPs as well as the PDMS surface.
2.3.1. Immobilization on AuNPs
AuNPs were synthesized using a standard Turkevich method in which a gold chloride precursor salt is reduced using trisodium citrate at high temperature.40−42 The aptamers were immobilized on the AuNP surface by incubating 1 mL of AuNPs (0.2 mg/mL) with 2 μM (10 μL) of reduced aptamers at room temperature with gentle stirring overnight. The AuNP solution was then centrifuged down (11 000 rcf, 11 min) to remove unbound aptamers and resuspended in working buffer. The conjugation of aptamers was confirmed by performing gel electrophoresis on bare and aptamer-conjugated AuNPs.
2.3.2. Immobilization on PDMS
The prepared aptamers were added directly to the functionalized testing zone on the microfluidic channel and allowed to sit for 3 h with gentle stirring. The final concentration of the aptamers used for each testing zone was 25 μM, 20 μL. After 3 h of incubation, the channels were washed gently with working buffer to remove unbound aptamers. To avoid nonspecific binding of the protein, the channels were then passivated using 1% BSA for 1 h.
2.4. Silver Staining Procedure
The silver staining kit containing two different solutions—solution A (silver precursor) and solution B (reducing agent, e.g., hydroquinone)—was obtained commercially. These solutions were diluted in half and mixed in equal ratios (1:1) right before adding in the microfluidic channel. This mixture was allowed to flow through the channel for 10 min by maintaining a constant flow rate of 80 μL/min. At the end, the channel was washed with deionized water for 15 min to stop the reaction and remove excess silver from the channel. The presence of AuNPs allowed the deposition of silver in the testing zone, which provided gray color in the channel.
3. Results and Discussions
The fabricated microfluidic channels were embedded with micro-sized pillars to increase the surface area in the testing region. The increased surface area ensured large numbers of aptamers in the testing zone, which increased the binding sites for the protein molecules and thus enhanced the sensitivity of the approach. Scanning electron microscopy (SEM) analysis shown in Figure 2A confirms the presence of pillars in the magnified view of the testing zone. The analysis of the vertical and lateral view (inset) indicates that the pillars have an average diameter and height of 50 and 45 μm, respectively.
Figure 2.
Schematic representation of the microfluidic channel and detailed characterization of different components of the present sensor. (A) Image of a single microfluidic channel. The SEM image representing a magnified view of the testing zone showing the presence of pillars with an average diameter of 50 μm. (B) FTIR spectroscopy of the microfluidic channel after PMPI and aptamer treatments. Peaks at 1698 and 1531 cm–1 represent amide I and amide II bands in PMPI, which indicate the presence of urethane linkages. (C) Schematic representation of the reaction for functionalization of the channel using PMPI as the linker molecule. (D,E) UV–visible spectroscopy and TEM analysis of the Turkevich method-based AuNPs. Inset shows narrow size distribution of the particles in the range of 15–20 nm as measured by dynamic light-scattering measurement. (F) Gel electrophoresis of bare and aptamer-conjugated AuNPs. Migration is observed in the aptamer-conjugated samples because of the negative charge on the particle surface.
The microfluidic channels were treated with aqueous solution of NaOH to make the surface hydrophilic. Treatment with NaOH creates a hydroxyl group (−OH) on the surface, which not only improves the flow but also facilitates the functionalization of the surface with other chemicals.43−45 The major advantage of using NaOH over traditional ozone treatment is the enhanced stability of the samples irrespective of the storage condition.43 The −OH functionalized surface was then reacted with PMPI (p-maleimidophenyl isocyanate), that was used as a linker molecule between aptamers and the PDMS surface. The isocyanate end (−N=C=O) of PMPI reacts with −OH and forms urethane linkage. This renders the maleimide end available for coupling with the thiol group present at the end of aptamers.46−48 Fourier transform infrared spectroscopy (FTIR) is shown in Figure 2B along with a schematic illustration of the reaction in Figure 2C. The FTIR spectrum of PDMS–NaOH–PMPI showed peaks at 1698 and 1531 cm–1, which corresponds to amide I and amide II bands (C=O stretching and −N–H bending), respectively.47,49,50 A distinct peak at 1585 cm–1 is also observed, which indicates the presence of C=C in the maleimide group. After the immobilization of the aptamers, a significant peak at ∼3410 cm–1 is observed, which is due to large number of hydroxyl groups present on the aptamers. A peak at 1648 cm–1 is also seen, which indicates the presence of purine and pyrimidine rings in the aptamers.51 The FTIR data confirmed the immobilization of aptamers over PDMS using PMPI as the linker molecule.
The characterization of the synthesized AuNPs is shown in Figure 2D,E. The UV–vis spectrum (Figure 2D) shows a distinctive absorbance peak at 520 nm, which represents the surface plasmon band of the AuNPs. The transmission electron microscopy (TEM) results indicate that the particles were uniformly distributed and spherical in nature with an average diameter of 13 ± 2.1 nm. These particles were conjugated with ZIKV and CHIKV envelope protein-specific aptamers using a standard protocol at a concentration of 20 nM.40 A high aptamer density results in repulsion among the AuNPs, whereas lower aptamer density provides fewer binding sites.28 The successful conjugation of the aptamers onto the AuNPs was confirmed by electrophoresis of the particles through a 0.5% agarose gel (Figure 2F). It was observed that Apt-AuNPs migrated through the gel (red arrow), whereas bare (unconjugated) AuNPs remained in the well (black arrow). The migration of functionalized particles is due to the overall negative particle charge imparted by the conjugated aptamers, which enables these Apt-AuNPs to be driven through the gel in response to the applied voltage. Because of insufficient negative charges on bare AuNPs,52 immediate aggregation and color change was seen in the AuNPs in the presence of running buffer (Tris), which inhibited the migration of the bare AuNPs through the gel.
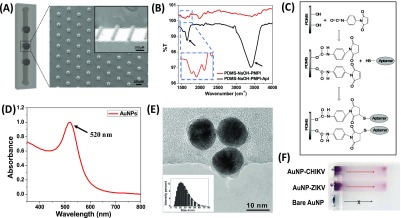
The present approach was first tested on a PDMS substrate to examine the sensitivity and selectivity of aptamers for the target arboviral envelope proteins. A distinct color development was observed from 500 pM of CHIKV envelope protein, which intensified with increasing protein concentrations (Figure S6). The approach was then extended to the microfluidic channels with a controlled flow rate (80 μL/min). The aptamer-conjugated microfluidic channels were tested against different concentrations (10 fM to 1 nM) of both CHIKV and ZIKV envelope proteins. Figure 3A shows distinct development of silver in the testing zones represented by circles. The “+” and “–” signs indicate the presence or absence of the specific arboviral envelope protein in the testing solution. For 0 nM, PBS was spiked with BSA, which was used as the negative control to test nonspecific interactions of AuNPs in the channel. No color development was observed in the negative control regions in any of the channels, which indicates high specificity of Apt-AuNPs (Figure S7). Also, for the last two channels, only one viral envelope protein (either ZIKV or CHIKV) was injected as per the (+) or (−) symbol. It can be seen that the ZIKV envelope protein at a very high concentration of 10 nM did not interfere with the CHIKV aptamers and vice versa. This clearly suggests that the current approach is not only highly sensitive with a detection limit of 1 pM, but also exhibits high level of selectivity among different arboviruses such as ZIKV and CHIKV.
Figure 3.
(A) Digital image of microfluidic channels representing color change in the testing zones in the presence of different concentrations of protein (1, 10, 100 pM, and 1 nM). The last two channels represent the specificity of the current sensor. The sample containing only Zika protein (10 nM) was introduced in the channel and no color change in the CHIKV region was observed and vice versa. This shows that the aptamers are highly specific in nature. Negative control did not produce any color. (B) Graph represents the change in key value (CMYK color model) calculated using ImageJ as a function of concentration of protein. Black and white are represented as 100 and 0, respectively. It can be seen that the key value moves toward black attributing to the increased deposition of silver in the testing zone.
The color change in the channels was also measured using an image processing software to calculate the CMYK (cyan, magenta, yellow, and key) values. The key value represents the black color, which was used as the measure of silver development in microfluidic channels. The more number of AuNPs results in increased silver deposition in the testing zone, leading to an intense color change. Figure 3B shows the key value with respect to concentration of protein. It can be seen that with increasing protein concentration, the K value increases. The measure of K value can be used for a quantitative estimation of an unknown viral protein concentration sample by plotting a standard calibration curve using a known protein concentration and the respective key value, given similar experimental conditions.
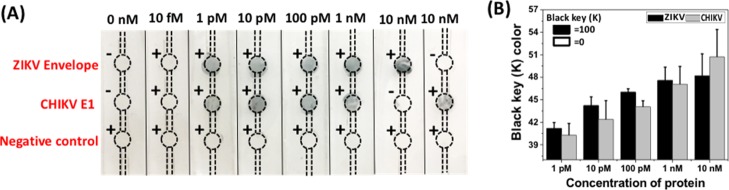
The microfluidic device was also tested against sterile, mechanically defibrinated calf blood and complete sheep serum spiked with the given ZIKV and CHIKV recombinant envelope proteins as surrogates for infected human blood/blood products. The results are represented as black key color (K) as a function of protein concentration (Figure 4). The testing showed a response to as low as 1 pM of the recombinant protein in PBS and 10 pM when tested against spiked 10% calf blood (diluted with working buffer) or sheep serum. In the case of undiluted (i.e., 100%) calf blood, a slight increase in K value was observed in the presence of 100 pM (∼5 ng/mL) of the protein. Diluting the blood with the binding buffer therefore increased the sensitivity of the system by ∼10-fold. A comparative analysis of different detection techniques is shown in Table S1. The present sensor gives a minimum detection limit of 1 pM (50 pg/mL) for ZIKV envelope protein and CHIKV envelope protein E1 in PBS, which is the lowest obtained so far. A similar detection limit (in buffer) was also obtained by Zeng et al. using an antibody–aptamer-mediated sandwich assay on a 96-well ELISA plate.53 The current approach abrogates the use of expensive plates or a sophisticated plate reader, making the device more adoptable in resource-constrained areas.
Figure 4.
Development of silver in microfluidic channels as a function of different concentrations of ZIKV and CHIKV envelop proteins (1, 10, and 100 pM) has been represented in terms of key value (CMYK color code). No change in K values was observed in the presence of 1 pM of protein for both calf blood and sheep serum samples. Serum and 10% calf blood exhibited a slight color development at 10 pM of recombinant protein. In the case of 100 pM, a distinct color development was observed in all the three cases.
4. Conclusions
A multiplex detection system for ZIKV and CHIKV arboviral envelope proteins has been developed using an aptamer-conjugated microfluidic channel approach. This novel method implements the immobilizing aptamers to the PDMS substrate using PMPI as the linker molecule. The channels are patterned with microsized pillars that facilitate an enhanced sensitivity of the approach down to detections of 1 pM (50 pg/mL) of viral envelope protein targets in PBS and 10 pM in dilute (10%) mechanically defibrinated calf blood. The detection is based on a silver enhancement technique that deposits a thick silver shell over the surface of target-bound Apt-AuNPs. This enhancement enables significant color changes on the PDMS substrate, indicating the presence of target protein. The device also exhibited high specificity toward the target protein with no interference from other, off-target arboviral proteins. The use of aptamers as the biorecognition molecule provides flexibility to incorporate additional or alternative targets by designing the specific aptamers and immobilizing them in the microfluidic channel. The present approach can be extended to detect other clinically important biomarkers for which there are few POC options available.
Acknowledgments
The authors are grateful to REU, NSF, for financial support. The authors would also like to acknowledge Abraham Vazquez-Guardado, Nanoscience Technology Center, University of Central Florida, for his help during testing and sample preparation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b03277.
Detailed fabrication protocol for the microfluidic channel, synthesis of nanoparticles, and conjugation techniques (PDF)
National Science Foundation, REU EEC 1560007, Florida Department of Agriculture and Consumer Services (FDACS #023593, #24236) and Bill & Melinda Gates Foundation Grand Challenges Explorations Grant, Investment ID OPP1138590.
The authors declare no competing financial interest.
Supplementary Material
References
- Campos G. S.; Bandeira A. C.; Sardi S. I. Zika virus outbreak, Bahia, Brazil. Emerging Infect. Dis. 2015, 21, 1885. 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J.; Korva M.; Tul N.; Popović M.; Poljšak-Prijatelj M.; Mraz J.; Kolenc M.; Resman Rus K.; Vesnaver Vipotnik T.; Fabjan Vodušek V.; Vizjak A.; Pižem J.; Petrovec M.; Avšič Županc T. Zika virus associated with microcephaly. N. Engl. J. Med. 2016, 374, 951–958. 10.1056/nejmoa1600651. [DOI] [PubMed] [Google Scholar]
- Pan American Health Organization/World Health Organization . Zika—Epidemiological Report. United States of America June 2017; PAHO/WHO: Washington, D.C., 2017.
- European Centre for Disease Prevention and Control . Rapid risk Assessment: Zika Virus Epidemic in the Americas: Potential Association with Microcephaly and Guillain-Barré Syndrome—10 December 2015; ECDC: Stockholm, 2015.
- Ramos da Silva S.; Gao S.-J. Zika virus: an update on epidemiology, pathology, molecular biology, and animal model. J. Med. Virol. 2016, 88, 1291–1296. 10.1002/jmv.24563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . WHO statement on the First Meeting of the International Health Regulations (2005) (IHR 2005) Emergency Committee on Zika Virus and Observed Increase in Neurological Disorders and Neonatal Malformations, 2016.
- Musso D.; Roche C.; Robin E.; Nhan T.; Teissier A.; Cao-Lormeau V.-M. Potential sexual transmission of Zika virus. Emerging Infect. Dis. 2015, 21, 359. 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blohm G. M.; Lednicky J. A.; Márquez M.; White S. K.; Loeb J. C.; Pacheco C. A.; Nolan D. J.; Paisie T.; Salemi M.; Rodríguez-Morales A. J.; Glenn Morris J.; Pulliam J. R. C.; Paniz-Mondolfi A. E. Evidence for mother-to-child transmission of zika virus through breast milk. Clin. Infect. Dis. 2017, 66, 1120–1121. 10.1093/cid/cix968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory C. J.; Oduyebo T.; Brault A. C.; Brooks J. T.; Chung K.-W.; Hills S.; Kuehnert M. J.; Mead P.; Meaney-Delman D.; Rabe I.; Staples E.; Petersen L. R. Modes of transmission of Zika virus. J. Infect. Dis. 2017, 216, S875–S883. 10.1093/infdis/jix396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessler J.; Chaisson L. H.; Kucirka L. M.; Bi Q.; Grantz K.; Salje H.; Carcelen A. C.; Ott C. T.; Sheffield J. S.; Ferguson N. M.; Cummings D. A. T.; Metcalf C. J. E.; Rodriguez-Barraquer I. Assessing the global threat from Zika virus. Science 2016, 353, aaf8160. 10.1126/science.aaf8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden V. Description of 13 infants born during October 2015–January 2016 with congenital Zika virus infection without microcephaly at birth—Brazil. Morb. Mortal. Wkly. Rep. 2016, 65, 1343–1348. 10.15585/mmwr.mm6547e2. [DOI] [PubMed] [Google Scholar]
- Tian B.; Qiu Z.; Ma J.; Zardán Gómez de la Torre T.; Johansson C.; Svedlindh P.; Strömberg M. Attomolar Zika virus oligonucleotide detection based on loop-mediated isothermal amplification and AC susceptometry. Biosens. Bioelectron. 2016, 86, 420–425. 10.1016/j.bios.2016.06.085. [DOI] [PubMed] [Google Scholar]
- Song J.; Mauk M. G.; Hackett B. A.; Cherry S.; Bau H. H.; Liu C. Instrument-free point-of-care molecular detection of Zika virus. Anal. Chem. 2016, 88, 7289–7294. 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Yin F.; Bi Y.; Cheng G.; Li J.; Hou L.; Li Y.; Yang B.; Liu W.; Yang L. Rapid and sensitive detection of Zika virus by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods 2016, 238, 86–93. 10.1016/j.jviromet.2016.10.010. [DOI] [PubMed] [Google Scholar]
- Lee D.; Shin Y.; Chung S.; Hwang K. S.; Yoon D. S.; Lee J. H. Simple and highly sensitive molecular diagnosis of Zika virus by lateral flow assays. Anal. Chem. 2016, 88, 12272–12278. 10.1021/acs.analchem.6b03460. [DOI] [PubMed] [Google Scholar]
- Priye A.; Bird S. W.; Light Y. K.; Ball C. S.; Negrete O. A.; Meagher R. J. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci. Rep. 2017, 7, 44778. 10.1038/srep44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.; Weaver S. C.; Wong P.-Y.; Lie S.; Wang E.; Guerbois M.; Vayugundla S. P.; Wong S. Rapid, affordable and portable medium-throughput molecular device for Zika virus. Sci. Rep. 2016, 6, 38223. 10.1038/srep38223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priye A.; Bird S. W.; Light Y. K.; Ball C. S.; Negrete O. A.; Meaghar R. J. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci. Rep. 2017, 7, 44778. 10.1038/srep44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.; Weaver S. C.; Wong P.-Y.; Lie S.; Wang E.; Guerbois M.; Vayugundla S. P.; Wong S. Rapid, Affordable and Portable Medium-Throughput Molecular Device for Zika Virus. Sci. Rep. 2016, 6, 38223. 10.1038/srep38223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K.; Green A. A.; Takahashi M. K.; Braff D.; Lambert G.; Lee J. W.; Ferrante T.; Ma D.; Donghia N.; Fan M.; Daringer N. M.; Bosch I.; Dudley D. M.; O’Connor D. H.; Gehrke L.; Collins J. J. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Song J.; Mauk M. G.; Hackett B. A.; Cherry S.; Bau H. H.; Liu C. Instrument-Free Point-of-Care Molecular Detection of Zika Virus. Anal. Chem. 2016, 88, 7289–7294. 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli A.; Ornob A.; Yu H.; Damhorst G. L.; Chen W.; Sun F.; Bhuiya A.; Cunningham B. T.; Bashir R. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point-of-care. Biomed. Microdevices 2017, 19, 73. 10.1007/s10544-017-0209-9. [DOI] [PubMed] [Google Scholar]
- Bedin F.; Boulet L.; Voilin E.; Theillet G.; Rubens A.; Rozand C. Paper-based point-of-care testing for cost-effective diagnosis of acute flavivirus infections. Med. Virol. 2017, 89, 1520. 10.1002/jmv.24806. [DOI] [PubMed] [Google Scholar]
- Lee K. H.; Zeng H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal Chem 2017, 89, 12743–12748. 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
- Nicolini A. M.; McCracken K. E.; Yoon J.-Y. Future developments in biosensors for field-ready Zika virus diagnostics. J. Biol. Eng. 2017, 11, 7. 10.1186/s13036-016-0046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo Z.; Yu Y.; Wang M.; Li J.; Zhang Z.; Liu J.; Wu X.; Lu A.; Zhang G.; Zhang B. Recent advances in SELEX technology and aptamer applications in biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. 10.3390/ijms18102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. X.; Kwon Y. J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. 10.1016/j.ymeth.2016.04.020. [DOI] [PubMed] [Google Scholar]
- Saraf N.; Woods E. R.; Peppler M.; Seal S. Highly selective aptamer based organic electrochemical biosensor with pico-level detection. Biosens. Bioelectron. 2018, 117, 40–46. 10.1016/j.bios.2018.05.031. [DOI] [PubMed] [Google Scholar]
- Toh S. Y.; Citartan M.; Gopinath S. C. B.; Tang T.-H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403. 10.1016/j.bios.2014.09.026. [DOI] [PubMed] [Google Scholar]
- Baker M. Reproducibility crisis: Blame it on the antibodies. Nature 2015, 521, 274. 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- Famulok M.; Mayer G. Aptamers and SELEX in Chemistry & Biology. Chem. Biol. 2014, 21, 1055–1058. 10.1016/j.chembiol.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Song K.-M.; Lee S.; Ban C. Aptamers and Their Biological Applications. Sensors 2012, 12, 612–631. 10.3390/s120100612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark D.; Haeberle S.; Roth G.; Von Stetten F.; Zengerle R.. Microfluidic lab-on-a-chip platforms: requirements, characteristics and applications. In Microfluidics Based Microsystems; Springer, 2010; pp 305–376. [DOI] [PubMed] [Google Scholar]
- Ohno K.-i.; Tachikawa K.; Manz A. Microfluidics: applications for analytical purposes in chemistry and biochemistry. Electrophoresis 2008, 29, 4443–4453. 10.1002/elps.200800121. [DOI] [PubMed] [Google Scholar]
- Chin C. D.; Laksanasopin T.; Cheung Y. K.; Steinmiller D.; Linder V.; Parsa H.; Wang J.; Moore H.; Rouse R.; Umviligihozo G.; Karita E.; Mwambarangwe L.; Braunstein S. L.; van de Wijgert J.; Sahabo R.; Justman J. E.; El-Sadr W.; Sia S. K. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 2011, 17, 1015. 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- Liu R.; Zhang Y.; Zhang S.; Qiu W.; Gao Y. Silver enhancement of gold nanoparticles for biosensing: from qualitative to quantitative. Appl. Spectrosc. Rev. 2014, 49, 121–138. 10.1080/05704928.2013.807817. [DOI] [Google Scholar]
- Yu Q.; Li H.; Li C.; Zhang S.; Shen J.; Wang Z. Gold nanoparticles-based lateral flow immunoassay with silver staining for simultaneous detection of fumonisin B1 and deoxynivalenol. Food Control 2015, 54, 347–352. 10.1016/j.foodcont.2015.02.019. [DOI] [Google Scholar]
- Souf S.Recent advances in diagnostic testing for viral infections. Biosci. Horiz., 2016, 9. 10.1093/biohorizons/hzw010 [DOI] [Google Scholar]
- Lake M.; Narciso C.; Cowdrick K.; Storey T.; Zhang S.; Zartman J.; Hoelzle D.. Microfluidic device design, fabrication, and testing protocols. Protoc. Exch., 2015, 10. 10.1038/protex.2015.069 [DOI] [Google Scholar]
- Saraf N.; Bosak A.; Willenberg A.; Das S.; Willenberg B. J.; Seal S. Colorimetric detection of epinephrine using an optimized paper-based aptasensor. RSC Adv. 2017, 7, 49133–49143. 10.1039/c7ra10272k. [DOI] [Google Scholar]
- Turkevich J.; Stevenson P. C.; Hillier J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951, 11, 55–75. 10.1039/df9511100055. [DOI] [Google Scholar]
- Niu S.; Lv Z.; Liu J.; Bai W.; Yang S.; Chen A. Colorimetric aptasensor using unmodified gold nanoparticles for homogeneous multiplex detection. PLoS One 2014, 9, e109263. 10.1371/journal.pone.0109263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek I.; Tho F.; Arnold W. M. Sodium hydroxide treatment of PDMS based microfluidic devices. Lab Chip 2010, 10, 2283–2285. 10.1039/c004769d. [DOI] [PubMed] [Google Scholar]
- Ren X.; Bachman M.; Sims C.; Li G. P.; Allbritton N. Electroosmotic properties of microfluidic channels composed of poly(dimethylsiloxane). J. Chromatogr. B: Biomed. Sci. Appl. 2001, 762, 117–125. 10.1016/s0378-4347(01)00327-9. [DOI] [PubMed] [Google Scholar]
- Thorslund S.; Lindberg P.; Andrén P. E.; Nikolajeff F.; Bergquist J. Electrokinetic-driven microfluidic system in poly(dimethylsiloxane) for mass spectrometry detection integrating sample injection, capillary electrophoresis, and electrospray emitter on-chip. Electrophoresis 2005, 26, 4674–4683. 10.1002/elps.200500338. [DOI] [PubMed] [Google Scholar]
- Weinert U.; Günther T.; Raff J.; Pollmann K. Self assembling proteins as matrix for the construction of optical devices. WIT Trans. Modell. Simul. 2011, 51, 569–575. 10.2495/cmem110501. [DOI] [Google Scholar]
- Thatiparti T. R.; Averell N.; Overstreet D.; von Recum H. A. Multiplexing interactions to control antibiotic release from cyclodextrin hydrogels. Macromol Biosci. 2011, 11, 1544–1552. 10.1002/mabi.201100159. [DOI] [PubMed] [Google Scholar]
- Li D.; Teoh W. Y.; Gooding J. J.; Selomulya C.; Amal R. Functionalization strategies for protease immobilization on magnetic nanoparticles. Adv. Funct. Mater. 2010, 20, 1767–1777. 10.1002/adfm.201000188. [DOI] [Google Scholar]
- Soares J. W.; Steeves D. M.; Singh J.; Im J.; Whitten J. E. In Effect of Surface Modification on the Optical Properties of Nanocrystalline Zinc Oxide Materials, Oxide-Based Materials and Devices; International Society for Optics and Photonics, 2010; p 76031L. [Google Scholar]
- Shen G.; Anand M. F. G.; Levicky R. X-ray photoelectron spectroscopy and infrared spectroscopy study of maleimide-activated supports for immobilization of oligodeoxyribonucleotides. Nucleic Acids Res. 2004, 32, 5973–5980. 10.1093/nar/gkh932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Han L.; Yao Y.; Li Y.; Liu X. Key factors in FTIR spectroscopic analysis of DNA: the sampling technique, pretreatment temperature and sample concentration. Anal. Methods 2018, 10, 2436–2443. 10.1039/c8ay00386f. [DOI] [Google Scholar]
- Crew E.; Yan H.; Lin L.; Yin J.; Skeete Z.; Kotlyar T.; Tchah N.; Lee J.; Bellavia M.; Goodshaw I.; Joseph P.; Luo J.; Gal S.; Zhong C.-J. DNA assembly and enzymatic cutting in solutions: A gold nanoparticle based SERS detection strategy. Analyst 2013, 138, 4941–4949. 10.1039/c3an00683b. [DOI] [PubMed] [Google Scholar]
- Lee K. H.; Zeng H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal. Chem. 2017, 89, 12743–12748. 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.