Abstract
Genomic imprinting is important for normal brain development and aberrant imprinting has been associated with impaired cognition. We studied the imprinting status in selected imprints (H19, IGF2, SNRPN, PEG3, MEST1, NESPAS, KvDMR, IG-DMR and ZAC1) by pyrosequencing in blood samples from longitudinal cohorts born in 1936 (n = 485) and 1921 (n = 223), and anterior hippocampus, posterior hippocampus, periventricular white matter, and thalamus from brains donated to the Aberdeen Brain Bank (n = 4). MEST1 imprint methylation was related to childhood cognitive ability score (-0.416 95% CI -0.792,-0.041; p = 0.030), with the strongest effect evident in males (-0.929 95% CI -1.531,-0.326; p = 0.003). SNRPN imprint methylation was also related to childhood cognitive ability (+0.335 95%CI 0.008,0.663; p = 0.045). A significant association was also observed for SNRPN methylation and adult crystallised cognitive ability (+0.262 95%CI 0.007,0.517; p = 0.044). Further testing of significant findings in a second cohort from the same region, but born in 1921, resulted in similar effect sizes and greater significance when the cohorts were combined (MEST1; -0.371 95% CI -0.677,-0.065; p = 0.017; SNRPN; +0.361 95% CI 0.079,0.643; p = 0.012). For SNRPN and MEST1 and four other imprints the methylation levels in blood and in the five brain regions were similar. Methylation of the paternally expressed, maternally methylated genes SNRPN and MEST1 in adult blood was associated with cognitive ability in childhood. This is consistent with the known importance of the SNRPN containing 15q11-q13 and the MEST1 containing 7q31-34 regions in cognitive function. These findings, and their sex specific nature in MEST1, point to new mechanisms through which complex phenotypes such as cognitive ability may be inherited. These mechanisms are potentially relevant to both the heritable and non-heritable components of cognitive ability. The process of epigenetic imprinting—within SNRPN and MEST1 in particular—and the factors that influence it, are worthy of further study in relation to the determinants of cognitive ability.
Introduction
Human cognitive ability is an important determinant of educational and occupational success, social mobility, health, and longevity [1] though it is not clear whether higher cognitive ability leads to better health and longevity through improved lifestyle choices and life opportunities or whether there is a common biological basis to a well-functioning brain and body. In addition to uncertainty over the direction of causality, the transgenerational nature of these links suggests that causal pathways may span more than one generation. Cognitive ability is influenced both by genetics and the environment [1]. Epigenetic states are generally erased between generations. However, examples of heritability from parent to offspring with possible relevance to transgenerational transmission of phenotypes have been reported in mouse and human [2,3].
One class of epigenetics—imprinting—is particularly relevant to transgenerational and early life effects since imprints are established in the germline, maintained during the preimplantation reprogramming phase, and then passed on through the somatic cell lineages, where they can influence genome function and gene expression [4]. The sensitivity of some imprints to environmental effects prior to birth [5–8] suggests the potential for epigenetic states and their related biological functions to also be influenced by the early environment in a way that may persist in later life. Imprints as a class are also generally stable over time, and for some imprints the original signal persists in a wide range of cell types many divisions and decades later [9,10]. These characteristics make imprints particularly amenable to study in longitudinal cohort designs where only blood samples may be available and it may not be possible to sample the tissues, such as the brain, which is the primary interest in the case of cognitive phenotypes. Imprints are particularly promising candidates in the study of cognition as they are known to be important for neurogenesis, brain function and behaviour [11–15], and they have even been proposed as possible mediators of reported generational increases in intelligence test scores [16].
This study investigates the link between epigenetic imprinting and cognitive ability at age 11 and in adulthood using data from a well-characterised cohort born in 1936 and recruited at 64 years of age. A second cohort born in the same geographical region in 1921 and recruited at 78 years of age was used to further assess significant findings in the 1936 cohort. We focused on nine selected imprints (H19, IGF2, SNRPN, PEG3, MEST1, NESPAS, KvDMR, IG-DMR and ZAC1). All except IGF2 are germline imprints [17]. These genes, or the chromosomal regions containing them, have been implicated in cognitive development, or early life outcomes such as birth weight which have been related to cognitive outcomes, in humans and animal models [8,18–24]. We also compared the methylation level in the blood and selected regions of human brain sampled post-mortem for each imprint. The blood methylation levels were related to cognitive ability in childhood in the cohort born in 1936 and significant results were further evaluated using the cohort born in 1921.
Materials and methods
Participants were members of the Aberdeen Birth Cohorts of 1936 (ABC36) and 1921 (ABC21) from whom DNA was collected (n = 485 and n = 223, respectively) [25]. Ethical approval for the study was obtained from the Multi-Centre Research Ethics Committee for Scotland (MREC/01/0/56) and Grampian Research Ethics Committee (LREC/01/0299). The research was conducted in compliance with the Helsinki Declaration and all participants gave written, informed consent.
All children born in 1921 and 1936 and attending school in Scotland on 1st June 1932 or 4th June 1947 respectively were tested at age 11 (±0.5) years for general cognitive ability [26]. The test administered was a version of the Moray House Test No. 12, which was concurrently validated against the Terman-Merrill revision of the Binet Scales with a coefficient of approximately 0.8. At age 64(±1) years (ABC36) and age 78(±1) years (ABC21), crystallised cognitive ability was assessed by the National Adult Reading Test (NART) [27] which tests word knowledge; and fluid, non-verbal reasoning ability was assessed by Raven’s Progressive Matrices [28]. Participants also gave blood samples at this age, and weight and height were measured by trained research nurses. The Scottish Index of Multiple Deprivation (SIMD) decile was used as the measure of socioeconomic circumstances at the time of blood sampling.
Tissues were sampled from 4 brains donated to the Aberdeen Brain Bank post-mortem. These were not from ABC participants, however the donors were from the same region and were approximately contemporary with the ABC participants. Brain 1 was from a male with typical Alzheimer’s disease aged 65 years, brain 2 from a mixed Alzheimer’s and cerebral small vessel disease (CSVD) male aged 75 years, and brains 3 and 4 were from two control samples, both male, aged 67 and 70, with no evidence of neurodegeneration beyond the normal expectation for that age. Samples were taken in duplicate from the anterior hippocampus (AH), basal ganglia (BG), posterior hippocampus (PH), periventricular white matter (PWM) and thalamus (TH) of each brain with sterile scalpel and forceps. Genomic DNA was extracted using EZ1 DNA Tissue kits (Qiagen, Crawley, UK) and bisulphite conversion using Zymo EZ DNA Methylation-Gold Bisulfite kits (Zymo Research, California, USA).
Genomic DNA was extracted from lymphocytes using QIAamp DNA Mini Blood kits (Qiagen, Crawley, UK) and DNA methylation was measured by pyrosequencing using a PyroMark MD system (Qiagen, Crawley, UK) following bisulphite conversion of DNA using Zymo EZ DNA Methylation-Gold Bisulfite kits (Zymo Research, California, USA). Bisulphite conversion efficiency was evaluated by including a non-CpG cytosine base as a conversion control in each assay.
DNA methylation was measured in nine imprints–IGF2, H19ICR, SNRPN, PEG3, MEST1, NESPAS, KvDMR, IG-DMR and ZAC1. Assays were carried out using previously published primer sets when these were available; IGF2 [29], PEG3 [30], SNRPN [31], KvDMR, IG-DMR and ZAC1 [10]. Avoidance of SNPs is one of the criteria for assay designs. In the previously developed ZAC1 assay, methylation site 8 of 11 corresponded to a SNP (rs77574073) and this site was excluded from the analyses. FASTA sequences were used to design the H19ICR assay in house using PyroMark Assay Design Software (version 2.0, Qiagen, Crawley, UK). MEST1 and NESPAS primer sets were designed by Dr Mitsutero Ito at Cambridge University. The following primers were used; H19ICR assay: forward 5’-TGGGGATTTTGATGGGGTTA-3’, reverse 5’-biotin-CCTACTCCAAACATTATAAAAAAAACTAAC-3’ and sequencing primer 5’-GATGGTTAGGGTGTGTT-3’; MEST1 assay: forward 5’-AAGGGGGTTTTGTTTTTTTAATTGTG-3’ [10], reverse 5’-biotin-AAACTCTATTAAACCCACCACCAAACTAAT-3’ and sequencing primer 5’-TTGTTGTAAAGGAAATTT-3’; NESPAS assay: forward 5’-TGTGTATATATTAAGGTTATTAGGTG-3’, reverse 5’-biotin-TAATCAATCAACTCCTTTAACCCC-3’ and sequencing primer 5’-GGTTAGTTTTGAGTTTAT-3’. The genomic locations of the assays are shown in S1 Table.
All the assays met our criteria for conversion efficiency and there was no correlation between this and any of the imprint methylation results. The technical replication in our assays is such that routine repeat analyses have a minimal effect on study power therefore methylation in samples is determined singly. Outliers and individual data points with significant leverage on the results are assessed and, where appropriate, reanalysed but this is rarely required. All of the assays included adjustment for plate run to avoid any batch effects during the methylation analysis. Plate was included as a factor in all the regression analyses. The methylation level at adjacent CpG sites within each imprint was significantly correlated (p<0.001) in all the genes studied and an average methylation level for each imprint was used in the analysis. The number of CpG sites averaged was 4 for SNRPN and IGF2, 5 for IG-DMR, 6 for H19ICR and MEST1, 7 for PEG3, 8 for NESPAS, 10 for ZAC1 and 12 for KvDMR.
Statistical analysis was carried out using STATA/MP version 15 (Stata Corp, College Station, Texas, USA). Multivariate linear regression was carried out independently for each imprint with adjustment for sex and adult socioeconomic circumstance. Methylation was the explanatory variable in all regressions. Outliers with potentially high leverage on regression outcomes were re-analysed. Where the 5% statistical significance cut-off was observed in the cohort as a whole, the regressions were also stratified by sex and tested for an interaction term between methylation and sex by ANOVA in the combined cohorts. Cohort was included as an additional factor in the combined cohort regressions. The proportion of variance accounted for in models (adjusted R2) is reported for significant results.
Results
Imprint methylation data obtained for ABC36 participants are described along with the participant cognitive test scores in Table 1. There were no significant sex differences in MHT, NART or Raven’s test scores. The ABC36 cohort consisted of 50.5% females. Mean weights were 67.5 (SD 12.7) kg in females and 79.3 (SD 11.4) kg in males, (p<0.001). Mean heights were 158.9 (SD 6.1) cm in females and 171.9 (SD 6.0) cm in males, (p<0.001). Mean BMI was 26.8 (SD 4.9) kgm-2 in females and 26.8 (SD 3.5) kgm-2 in males and the difference was not statistically significant.
Table 1. ABC36 cognitive test scores and imprint methylation levels in blood DNA.
| Mean | Standard Deviation | n | |
|---|---|---|---|
| 1Measures of cognitive ability | |||
| Moray House Test | 42.0 | 13.5 | 480 |
| National Adult Reading Test | 108.9 | 10.6 | 474 |
| Raven’s Progressive Matrices | 35.6 | 8.8 | 462 |
| 2Average methylation | |||
| H19ICR | 58.0 | 4.3 | 462 |
| IGF2 | 49.3 | 5.8 | 467 |
| SNRPN | 46.6 | 3.7 | 464 |
| PEG3 | 51.2 | 2.8 | 474 |
| MEST1 | 48.2 | 3.1 | 469 |
| NESPAS | 47.8 | 2.9 | 472 |
| KvDMR | 47.8 | 2.7 | 461 |
| IG-DMR | 63.1 | 1.8 | 478 |
| ZAC1 | 43.8 | 3.6 | 477 |
1Cognitive ability was measured in children using the Moray House Test (a test of general intelligence), and in adults using the National Adult Reading Test (a measure of crystallised intelligence) and Raven’s Matrices (a measure of fluid intelligence).
2Mean methylation across measured CpG sites within each imprint.
Imprint methylation data obtained for ABC21 participants are described along with the participant cognitive test scores in Table 2. There were no significant sex differences in MHT, NART or Raven’s test scores. The ABC21 cohort consisted of 47.5% females. Mean weights were 64.0 (SD 13.0) kg in females and 72.4 (SD 10.0) kg in males, (p<0.001). Mean heights were 156.3 (SD 5.8) cm in females and 167.5 (SD 6.6) cm in males, (p<0.001). Mean BMI was 26.4 (SD 4.8) kgm-2 in females and 25.8 (SD 3.2) kgm-2 in males and the difference was not statistically significant.
Table 2. ABC21 cognitive test scores and imprint methylation levels in blood DNA.
| Mean | Standard Deviation | n | |
|---|---|---|---|
| 1Measures of cognitive ability | |||
| Moray House Test | 37.9 | 13.1 | 204 |
| National Adult Reading Test | 110.1 | 9.4 | 167 |
| Raven’s Progressive Matrices | 26.5 | 8.6 | 167 |
| 2Average methylation | |||
| SNRPN | 42.9 | 3.3 | 220 |
| MEST1 | 43.2 | 3.9 | 201 |
1Cognitive ability was measured in children using the Moray House Test (a test of general intelligence), and in adults using the National Adult Reading Test (a measure of crystallised intelligence) and Raven’s Matrices (a measure of fluid intelligence).
2Mean methylation across measured CpG sites within each imprint.
No significant associations were identified between childhood or adult cognitive test scores and weight, BMI or analysis plate with test scores as the dependent variable. Raven’s test score was associated with height in ABC21 (+0.177 95% CI 0.019,0.336; p = 0.029; R2 = 0.026) and in ABC36 (+0.127 95% CI 0.039,0.215; p = 0.005; R2 = 0.015). All cognitive test scores were positively associated with SIMD in ABC21 (MHT: +0.734 95% CI 0.042,1.426; p = 0.038; R2 = 0.018; NART: +1.230 95% CI 0.711,1.749; p<0.001; R2 = 0.122; Raven’s: +0.538 95% CI 0.042,1.034; p = 0.034; R2 = 0.023) and ABC36 (MHT: +1.385 95% CI 0.995,1.776; p<0.001; R2 = 0.093; NART: +1.227 95% CI 0.923,1.530; p<0.001; R2 = 0.120; Raven’s +0.854 95% CI 0.597,1.112; p<0.001; R2 = 0.085). All of the associations between methylation and cognition were adjusted for SIMD, and adjustment for height did not change the significance of Raven’s associations with methylation.
The methylation levels within the imprinted genes were all close to the 50% level characteristic of imprints and within the range of 33–70% typically reported for imprints [32] (Tables 1 and 2). There was no evidence of a difference in the level of methylation for the imprints between sexes with the exception of H19ICR methylation in ABC36 which was significantly higher in females (-1.086 95% CI -1.881,-0.290; p = 0.008; R2 = 0.003). There was no evidence of an association between the level of methylation and SIMD with the exception of H19ICR methylation in ABC36 (-0.175 95% CI -0.310,-0.040; p = 0.011; R2 = 0.012). KvDMR (+0.093 95% CI 0.036,0.149; p = 0.001; R2 = 0.020) and MEST1 (-0.078 95% CI -0.144,-0.012; p = 0.020; R2 = 0.009) methylation were associated with BMI in ABC36. Adjustment of the MEST1 associations with cognition for BMI did not change the level of significance. No further associations were observed between methylation and weight, BMI, height or SIMD.
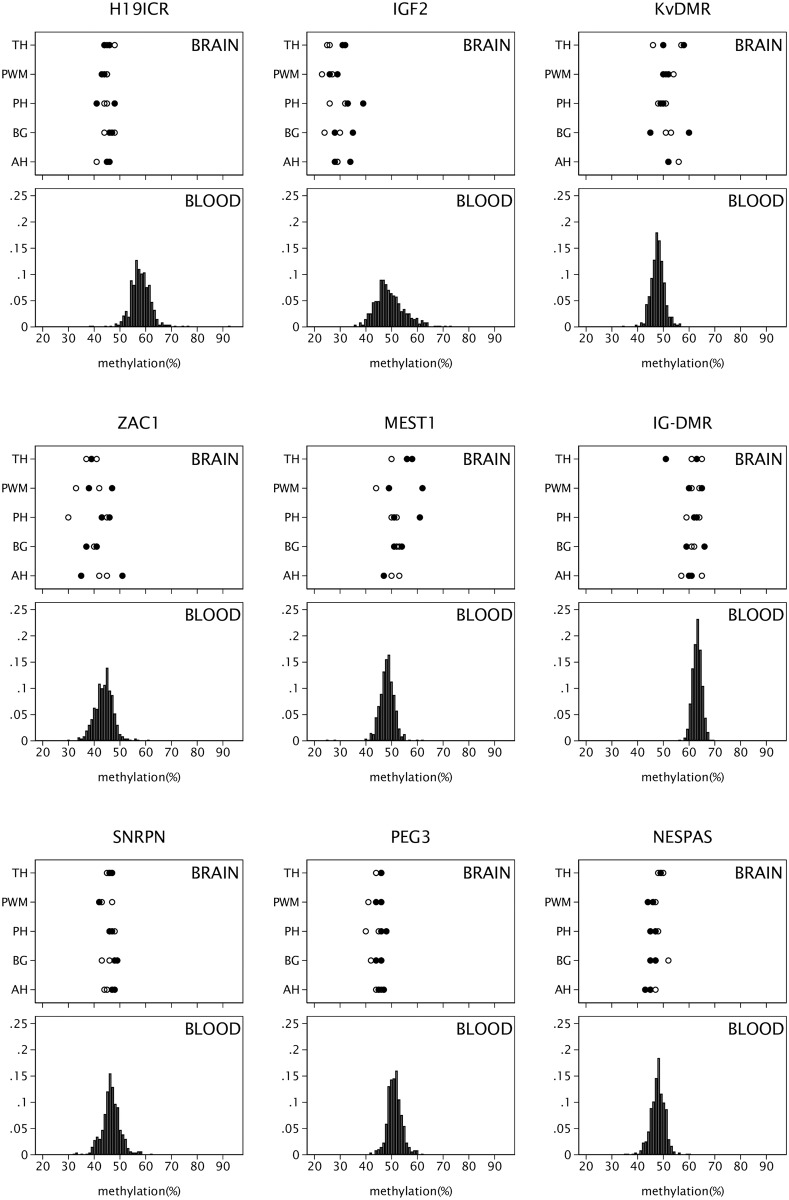
Methylation status for the nine imprints in the cohort blood samples were compared with values in a range of anatomical sites (anterior hippocampus, posterior hippocampus, periventricular white matter, and thalamus) in post-mortem samples from the brains of four adults from the same geographical location (Fig 1). The number of brains studied was small but the results demonstrate that the methylation levels for each imprint were similar for all of the brain anatomical sites studied and there was no separation by Alzheimer’s Disease status. This is consistent with the fact that none of the imprinted loci studied here have been reported to differ by Alzheimer’s Disease status [33]. In addition, the overall level was similar to that in the blood, with the exception of H19ICR and IGF2. In the majority of the imprints the methylation in the brain was similar to the 50% level expected for imprinted genes but for IGF2 in particular the brain methylation level was lower at around 30%. Some methylation signatures differ between blood cell types but the imprinted loci studied here were not identified as differentially methylated by cell type in studies using the 450k array [34]. No comparison was possible for the IG-DMR locus as this was not present on the 450K array but it was notable there was agreement between blood and brain for this imprint, though the level of methylation was high at around 60%, and none of the results reported here for IG-DMR and the outcomes of interest were significant.
Fig 1. Brain and blood methylation comparison.
DNA methylation in the imprints H19ICR, IGF2, IG-DMR, KvDMR, MEST1, NESPAS, PEG3, SNRPN and ZAC1 in blood from ABC36 (n = 485) and five selected regions from four brains sampled post-mortem (AH: Anterior Hippocampus. BG: Basal ganglia. PH: Posterior Hippocampus. PWM: Periventricular white matter. TH: Thalamus). The individual methylation values for each brain sample are shown with cases (individuals with evidence of Alzheimer’s Disease) represented by open circles and controls (no apparent neurodegeneration beyond that expected for age) by closed circles. The population distribution of methylation levels in blood are shown on the same methylation scale.
The relationships between imprinting methylation and measures of cognitive ability in childhood and adulthood are shown in Table 3. The results are expressed as score point change per methylation %. In the ABC36 cohort, SNRPN methylation was positively related to childhood MHT score (+0.335 95%CI 0.008,0.663; p = 0.045; R2 = 0.097) and NART score in adults (+0.262 95% CI 0.007,0.517; p = 0.044; R2 = 0.127). The MEST1 methylation was negatively related to childhood MHT score only (-0.416 95% CI -0.792,-0.041; p = 0.030; R2 = 0.106). The association between SNRPN and NART was no longer significant when adjusted for childhood MHT, though this might be expected as the childhood MHT score was highly correlated with the adult scores for NART (+0.575 95% CI 0.526,0.623; p = 0.000; R2 = 0.534) and Ravens (+0.403 95% CI 0.355,0.452; p = 0.000; R2 = 0.367). If a False Discovery Rate of 25% is applied to the full 9 MHT imprints tested the significant p values are retained.
Table 3. DNA methylation in imprints and measures of childhood and adult cognitive ability in ABC36.
| Moray House Test | National Adult Reading Test | Raven’s Progressive Matrices | ||||
|---|---|---|---|---|---|---|
| Imprint | Coefficient [95% CI] |
p value | Coefficient [95% CI] |
p value | Coefficient [95% CI] |
p value |
| H19ICR | -0.104 [-0.383,0.176] |
0.466 | -0.103 [-0.321,0.116] |
0.355 | 0.045 [-0.141,0.230] |
0.635 |
| IGF2 | 0.026 [-0.182,0.233] |
0.809 | 0.151 [-0.009,0.312] |
0.065 | 0.042 [-0.095,0.179] |
0.544 |
| SNRPN | 0.335* [0.008,0.663] |
0.045 | 0.262* [0.007,0.517] |
0.044 | 0.195 [-0.025,0.416] |
0.083 |
| PEG3 | -0.008 [-0.430,0.415] |
0.971 | -0.040 [-0.369,0.289] |
0.810 | -0.182 [-0.465,0.102] |
0.209 |
| MEST1 | -0.416* [-0.792,-0.041] |
0.030 | -0.184 [-0.480,0.111] |
0.220 | -0.071 [-0.329,0.186] |
0.587 |
| NESPAS | 0.008 [-0.396,0.411] |
0.970 | 0.065 [-0.251,0.381] |
0.687 | 0.112 [-0.161,0.385] |
0.419 |
| KvDMR | -0.123 [-0.568,0.323] |
0.589 | -0.217 [-0.563,0.128] |
0.217 | 0.033 [-0.262,0.327] |
0.827 |
| ZAC1 | -0.117 [-0.449,0.215] |
0.489 | -0.116 [-0.371,0.139] |
0.373 | 0.031 [-0.185,0.247] |
0.779 |
| IG-DMR | -0.036 [-0.684,0.613] |
0.914 | 0.110 [-0.398,0.617] |
0.671 | 0.207 [-0.222,0.636] |
0.344 |
Multivariate linear regression was carried out with imprint methylation as the explanatory variable and cognitive ability at age 11 and in adulthood as the dependent variable for each imprint in ABC36. Cognitive ability was measured at age 11 using the Moray House Test (a test of general intelligence), and in adults using the National Adult Reading Test (a measure of crystallised intelligence) and Raven’s Progressive Matrices (a measure of fluid intelligence). All regressions were adjusted for sex, adult socioeconomic status (index of multiple deprivation decile) and plate. The results are expressed as score point change per methylation %. P values are not adjusted for multiple testing correction.
* p<0.05.
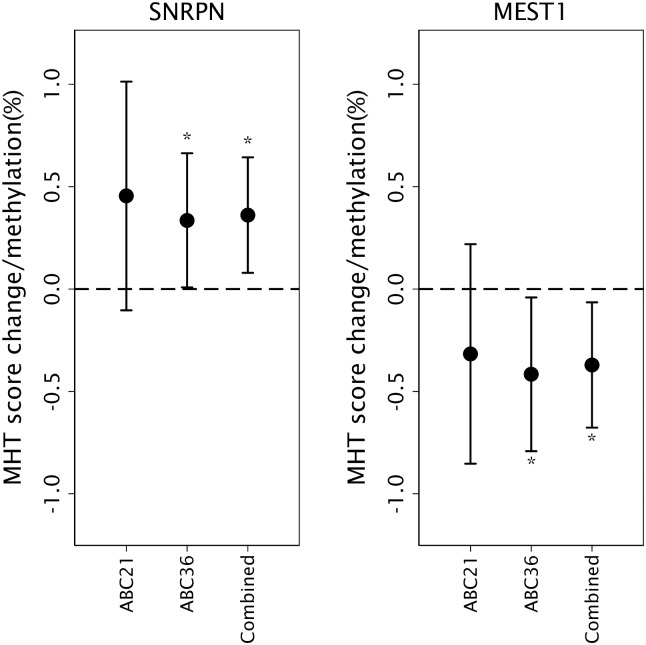
Further analysis of the significant ABC36 findings in the ABC21 cohort alone indicated that the effect size and direction were similar for SNRPN (MHT +0.455 95% CI -0.104,1.013; NART +0.182 95% CI -0.231,0.594) and MEST1 (MHT -0.317 95% CI -0.853,0.219) (Fig 2). The ABC21 cohort was around half the size of ABC36 and the results in ABC21 alone were not significant but in the combined cohorts the MHT results were more significant for both SNRPN (+0.361 95%CI 0.079,0.643; p = 0.012; R2 = 0.090) and MEST1 (-0.371 95%CI -0.677,-0.065; p = 0.017 R2 = 0.096); regressions additionally adjusted for cohort (Fig 2).The SNRPN association with NART was also more significant in the combined analysis (+0.241 95% CI 0.025,0.457; p = 0.029; R2 = 0.137) but, as with ABC36 alone, the association was not significant with adjustment for childhood MHT.
Fig 2. SNRPN and MEST1 methylation in ABC cohorts.
Regression coefficients with 95% CIs from regressions between SNRPN and MEST1 methylation in ABC36, ABC21 and the combined cohort and childhood cognitive ability measured at age 11 using the Moray House Test (MHT). All regressions were adjusted for sex, adult socioeconomic circumstance (index of multiple deprivation decile), plate and the combined cohort was additionally adjusted for cohort.
There was no evidence of an interaction with sex for the SNRPN effect on MHT in ABC36 alone or the combined cohort. There was a significant interaction with sex for MEST1 and MHT in ABC36 alone (p = 0.023), though the effect was attenuated in the combined cohort analysis (p = 0.063). Further analysis of the sex interaction in MEST1 indicated a highly significant result in males for MHT in ABC36 alone (-0.929 95% CI -1.531,-0.326; p = 0.003; R2 = 0.152) and in the combined cohort analysis (-0.807 95% CI -1.268,-0.346; p = 0.001; R2 = 0.138).
Discussion
Methylation of the paternally expressed, maternally methylated genes SNRPN and MEST1 in adulthood was associated with cognitive ability in childhood in a longitudinal cohort. The childhood cognitive associations with SNRPN were positive in both sexes, while the associations with MEST1 were negative and strongest in males. This is the first report of an association between SNRPN and MEST1 imprinting methylation and cognitive ability in a normal population. Some strengths and limitations of the current study should be noted. This longitudinal cohort study provides invaluable cognitive data from single individuals across the life course. The numbers in the analyses are effectively halved when considering the sexes separately but this is more likely to result in false negatives, due to reduced power, than in false positives. We did not adjust for multiple testing, but we checked the consistency of significant results in a second older cohort and carried out a combined cohort analysis with adjustment for cohort membership. Participants from both cohorts were recruited from a small geographical area and were born in the same year, 1921 or 1936. They undertook the same childhood cognitive test at the same age, and had the same information collected following recruitment. In terms of wider significance, imprinting is an established biological process that is essential in normal development and therefore the factors that influence imprinting are likely to operate via conserved mechanisms across populations.
Although the post-mortem brain sample size is small, the inclusion of the same imprint methylation analysis, using the same assays, in brain samples is a strength of the study as it demonstrates similar methylation profiles in different cell types and brain regions and the assays in which it is valid to assume similar levels in blood and brain. In all but two of the assays (H19ICR and IGF2) methylation levels in blood and in five separate anatomical regions of brain were similar to each other and similar to methylation levels in the same genes in cord blood at birth in a contemporary cohort [6,8]. Tissue-specific methylation patterns are sometimes observed in secondary, or somatic DMRs, of which the IGF2 imprint is an example, indicating that differential methylation was established post-fertilization and therefore may not be inherited by all tissues [4]. Establishment of secondary DMRs requires the presence of a ubiquitous germline DMR [35] and in the case of IGF2, this is the H19 imprint control region (ICR).
Evidence from imprinting syndromes supports a role for SNRPN in cognitive ability. Prader-Willi syndrome (PWS; OMIM 176270) and Angelman syndrome (AS; OMIM 105830) are clinically distinct imprinting disorders characterised primarily by neurological and behavioural features [21]. These conditions can result from microdeletion, uniparental disomy, and epigenetic changes in the 15q11-q13 chromosomal region [21,22,36,37]. Imprinting of 15q11-q13 is regulated by an imprinting control centre which contains a differentially methylated region at the 5’ end of SNRPN that controls expression throughout 15q11-q13 [38] and which was assayed here. SNRPN has been proposed as the most likely candidate gene [21,22] for PWS, which results in an approximately normal distribution of IQ but with a mean 40 points below that of the general population, with the IQ of unaffected siblings being correlated with that of the PWS cases [22,37,39]. Autism spectrum disorders with varying degrees of learning disability have also been linked to duplication of chromosome region 15q11q-13 [40]. Epigenetic inheritance from fathers may also be relevant as the risk of autism in human offspring is predicted by epigenetic differences in genes within the SNRPN containing gene cluster in sperm [41].
A pathway for SNRPN involvement in postnatal brain development has recently been identified, where SNRPN expression directly regulates the expression level of the nuclear receptor Nr4a1 which is critical for the proper development of cortical neurons [42]. Overexpression of SNRPN results in increased dendritic spine density, and morphological characteristics of dendritic spines contribute to cognitive ability [43]. This presents a potential pathway by which imprint methylation may impact brain function but more research is needed on the mechanism.
A third imprinting syndrome, Silver–Russell Syndrome (SRS; OMIM 180860) has been associated with changes in several chromosomal regions, including 11p15 [44]—a region which contains an imprinting cluster regulated by two ICRs, H19ICR and KvDMR1—and the 7q31 [45,46] which contains MEST1. Pathological cognitive impairment is not generally associated with SRS but a sibling controlled study investigating cognitive development in SRS associated with chromosome 7 imprinting regions, including MEST1, found a significant difference in IQ in direct analysis of paired differences between the subsample of children with SRS and a non-affected sibling [47]. In animal studies, MEST1-deficient female mice demonstrated impaired maternal care for their offspring, suggesting a link between MEST1 imprinting and brain function [19]. A role for MEST1 imprinting in human cognition is further supported by genome-wide association, which suggests an autism susceptibility locus within the MEST1 containing chromosomal region 7q31-q32 in males on the autism spectrum [24]. MEST1 methylation has also been associated with maternal stress exposure in utero [48], raising the possibility that the association between MEST1 methylation and cognitive ability may reflect a more general response to in utero exposure to stress.
The stronger effect of MEST1 in males was notable. Maternal stress has been reported to influence MEST1 methylation in opposite directions in male and female offspring [48]. Furthermore, the effect of the genomic changes in the SNRPN containing PWS region on intelligence was different in magnitude in males and females. The correlation between the intelligence of PWS offspring and that of their mothers in males was found to be over four times that observed in female offspring, and two fold higher in a selected sub-group of typically developing children [37]. Furthermore, a sex-specific association between maternal depression and PEG3 expression has been reported in the placenta of male, but not female, offspring [49]. The mechanisms underlying these sex effects are not understood. The difference in the direction of significant association for SNRPN and MEST1 is also noteworthy. This observed difference in direction, and an opposite direction of response in imprinted gene methylation to folic acid [6] suggest that the regulation of methylation is not necessarily uniform across all imprints.
The associations were strongest between SNRPN and MEST1 and a childhood measure of cognition (MHT). Of the tests taken in later life, the imprint-cognition associations were stronger for measures of crystallised (NART) rather than fluid (Raven’s) intelligence. These results were no longer significant when adjusted for MHT but the MHT score is itself highly correlated with NART and Raven’s. Crystallized abilities (such as word knowledge) depend more on experience and knowledge and increase throughout young adulthood and middle-age, then plateau, showing little or no further gain into old age [50]. Fluid abilities show a pattern of increase throughout young adulthood, followed by slow decline beginning in middle-adulthood which continues throughout old age [50].
The methylation changes observed were subtle but comparable in magnitude to those observed in similar studies, for example, in response to prenatal obesity [51,52], prenatal exposure to famine [53], and periconceptual folic acid intake [6]. It is worth noting that large change in methylation at imprint loci typically results in pathology and that was not the focus of this study. Whilst these changes are modest they may have significant effects. Previous analysis in cord blood has suggested that a 1% change in methylation at the IGF2 DMR in response to prenatal exposure to cigarette smoke, corresponded to an approximately 2-fold change in IGF2 transcription [54]. There is also the potential for small changes in imprinting methylation to influence the wider genome through long range interactions over thousands of bases.
The magnitude of the cognitive effects can be illustrated by translating the key childhood cognitive ability score into the commonly used IQ type scale, although it should be noted that we are not reporting IQ values. A 1% change in SNRPN and MEST1 methylation translates into a 0.4 point change in IQ type scale whilst two standard deviations (approximately full range) in the population distribution of SNRPN and MEST1 translates into around 3 points on the IQ type scale. The SNRPN methylation assay used here was developed as a diagnostic test for PWS and AS [31], which manifest predominantly as cognitive phenotypes, and the methylation level of SNRPN in blood from the cohort studied here was similar to that in five different brain anatomical locations (including two hippocampal regions) measured in post-mortem samples. The levels of SNRPN methylation reported here in later life are also similar to those measured in cord blood in a large contemporary birth cohort from the same city [6]. Cross sectional studies have demonstrated stability in children over the first 7 years of life [7] and in adult females between age 25 years and 85 years [55]. This evidence supports the conclusion that, for the regions reported here, the imprints are stable once set.
Imprints can be influenced by both genetics and the environment. For genetic effects, epigenetic states may be influenced by variation in the sequence underlying the epigenetic regions being measured [56], in proximal elements [57], or in the epigenetic machinery involved in setting and maintaining the methylation pattern. The SNRPN and MEST1 assays used here specifically excluded genetic variants within the region measured and we have no evidence for a genetic effect underlying the epigenetic associations with cognitive ability but this remains a possibility that cannot yet be ruled out. There is evidence that both SNRPN and MEST methylation may be influenced by the early environment. Animal studies indicate that paternal alcohol intake influences SNRPN and PEG3 methylation in the brains of their offspring [20], while human studies have shown that SNRPN methylation is altered in children conceived by intracytoplasmic sperm injection to overcome male infertility [7] and is influenced by the IVF culture media used [58]. Maternal stress during pregnancy has been associated with higher infant DNA methylation at the MEST DMR with some evidence for a sex bias in the response [48] Folate intake during pregnancy is linked to IGF2 and PEG3 but not SNRPN methylation [6] indicating that sensitivity to the early environment depends on the timing and nature of the environmental exposure.
The findings reported here are consistent with the known importance of the SNRPN containing 15q11-q13 and the MEST1 containing 7q31-34 regions in cognitive function. These findings, and their sex specific nature in MEST1, point to new mechanisms through which complex phenotypes such as cognitive ability may be inherited and the potential role of the environment in modifying that heritability. The process of epigenetic imprinting—within SNRPN and MEST1 in particular—and the factors that influence it, are worthy of further study in relation to the determinants of cognitive ability.
Supporting information
Chromosome, start and end coordinates for each differentially methylated region (DMR) are tabulated.
(DOCX)
Methylation levels at H19ICR, IGF2, SNRPN, PEG3, MEST1, NESPAS, KvDMR, ZAC1 and IGDMR1 imprints determined by pyrosequencing in blood samples from the ABC21 and ABC36 cohorts alongside cohort (ABCgroup), sex and analysis plate.
(XLSX)
Acknowledgments
The authors would like to thank the participants of the Aberdeen 1936 and 1921 Birth Cohorts, without whom this research would not have been possible.
Data Availability
Anonymized data are available as Supporting Information files. Access to the full dataset may be requested by contacting the Aberdeen Birth Cohort steering committee (c.mcneil@abdn.ac.uk).
Funding Statement
This work was supported by the Economic and Social Research Council/Biotechnology and Biological Sciences Research Council BioSocial initiative (grant number ES/N00048X/1; PH, ADM, LHP, RS, AF-S, MR). PH and GH acknowledge the support of the Scottish Government’s Rural and Environment Science and Analytical Services Division (RESAS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Deary IJ. Intelligence. Annu Rev Psychol. 2012;63: 453–482. 10.1146/annurev-psych-120710-100353 [DOI] [PubMed] [Google Scholar]
- 2.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. The Dynamics of Genome-wide DNA Methylation Reprogramming in Mouse Primordial Germ Cells. Mol Cell. 2012;48: 849–862. 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang WC, Dietmann S, Irie N, Leitch H, Floros V, Bradshaw C, et al. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell. 2015;161: 1453–1467. 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson-Smith A. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12: 565–575. 10.1038/nrg3032 [DOI] [PubMed] [Google Scholar]
- 5.Haggarty P, Ferguson-Smith A. Life course epigenetics and healthy ageing In: Kuh D, Cooper R, Hardy R, Richards M, Ben-Shlomo Y, editors. A Life Course Approach to Healthy Ageing. Oxford: Oxford University Press; 2013. [Google Scholar]
- 6.Haggarty P, Hoad G, Campbell DM, Horgan GW, Piyathilake C, McNeill G. Folate in pregnancy and imprinted gene and repeat element methylation in the offspring. Am J Clin Nutr. 2013;97: 94–99. 10.3945/ajcn.112.042572 [DOI] [PubMed] [Google Scholar]
- 7.Whitelaw N, Bhattacharya S, Hoad G, Horgan GW, Hamilton M, Haggarty P. Epigenetic status in the offspring of spontaneous and assisted conception. Hum Reprod. 2014;29: 1452–1458. 10.1093/humrep/deu094 [DOI] [PubMed] [Google Scholar]
- 8.Haggarty P, Hoad G, Horgan GW, Campbell DM. DNA Methyltransferase Candidate Polymorphisms, Imprinting Methylation, and Birth Outcome. PLoS One. 2013;8: e68896 10.1371/journal.pone.0068896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coolen MW, Statham AL, Qu W, Campbell MJ, Henders AK, Montgomery GW, et al. Impact of the Genome on the Epigenome Is Manifested in DNA Methylation Patterns of Imprinted Regions in Monozygotic and Dizygotic Twins. PLoS One. 2011;6: e25590 10.1371/journal.pone.0025590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodfine K, Huddleston JE, Murrell A. Quantitative analysis of DNA methylation at all human imprinted regions reveals preservation of epigenetic stability in adult somatic tissue. Epigenetics Chromatin. 2011;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6: 108–118. 10.1038/nrn1604 [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8: 832–843. 10.1038/nrn2235 [DOI] [PubMed] [Google Scholar]
- 13.Badcock C. The imprinted brain: how genes set the balance between autism and psychosis. Epigenomics. 2011;3: 345–359. 10.2217/epi.11.19 [DOI] [PubMed] [Google Scholar]
- 14.Ferrón SR, Charalambous M, Radford E, McEwen K, Wildner H, Hind E, et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 2011;475: 381–385. 10.1038/nature10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopsida E, Mikaelsson MA, Davies W. The role of imprinted genes in mediating susceptibility to neuropsychiatric disorders. Horm Behav. 2011;59: 375–382. 10.1016/j.yhbeh.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Pietschnig J, Voracek M. One Century of Global IQ Gains: A Formal Meta-Analysis of the Flynn Effect (1909–2013). Perspect Psychol Sci. 2015;10: 282–306. 10.1177/1745691615577701 [DOI] [PubMed] [Google Scholar]
- 17.Hanna CW, Peñaherrera MS., Saadeh H, Andrews S, McFadden DE, Kelsey G, et al. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 2016;26: 756–767. 10.1101/gr.196139.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rijlaarsdam J, Cecil CAM, Walton E, Mesirow MSC, Relton CL, Gaunt TR, et al. Prenatal unhealthy diet, insulin-like growth factor 2 gene (IGF2) methylation, and attention deficit hyperactivity disorder symptoms in youth with early-onset conduct problems. Journal of Child Psychology and Psychiatry. 2017;58: 19–27. 10.1111/jcpp.12589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20: 163–169. 10.1038/2464 [DOI] [PubMed] [Google Scholar]
- 20.Liang F, Diao L, Liu J, Jiang N, Zhang J, Wang H, et al. Paternal ethanol exposure and behavioral abnormities in offspring: Associated alterations in imprinted gene methylation. Neuropharmacology. 2014;81: 126–133. 10.1016/j.neuropharm.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Cassidy SB, Dykens E, Williams CA. Prader-Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet. 2000;97: 136–146. [DOI] [PubMed] [Google Scholar]
- 22.Whittington J, Holland A, Webb T. Relationship between the IQ of people with Prader–Willi syndrome and that of their siblings: evidence for imprinted gene effects. J Intellect Disabil Res. 2009;53: 411–418. 10.1111/j.1365-2788.2009.01157.x [DOI] [PubMed] [Google Scholar]
- 23.Ubeda F, Gardner A. A model for genomic imprinting in the social brain: juveniles. Evolution. 2010;64: 2587–2600. 10.1111/j.1558-5646.2010.01015.x [DOI] [PubMed] [Google Scholar]
- 24.Lamb JA, Barnby G, Bonora E, Sykes N, Bacchelli E, Blasi F, et al. Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J Med Genet. 2005;42: 132–137. 10.1136/jmg.2004.025668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whalley LJ, Murray AD, Staff RT, Starr JM, Deary IJ, Fox HC, et al. How the 1932 and 1947 mental surveys of Aberdeen schoolchildren provide a framework to explore the childhood origins of late onset disease and disability. Maturitas. 2011;69: 365–372. 10.1016/j.maturitas.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Penrose LS. The Trend of Scottish Intelligence: A Comparison of the 1947 and 1932 Surveys of the Intelligence of Eleven-year-old Pupils. Scottish Council for Research in Education. Univ. of London Press Ltd. 1949. Pp. 151 + xxviii. Price 7s. 6d. Annals of Eugenics. 1949;15: 186–187. [Google Scholar]
- 27.Nelson HE. The National Adult Reading Test (NART): test manual. Windsor: NFER-Nelson; 1982.
- 28.Raven JC, Court JH, Raven J. Manual for Raven’s Progressive Matrices and Vocabulary Scales. London: Lewis; 1977. [Google Scholar]
- 29.Dupont J, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem. 2004;333: 119–127. 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JJ, et al. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112: 1489–1502. 10.1002/cncr.23323 [DOI] [PubMed] [Google Scholar]
- 31.White HE, Durston VJ, Harvey JF, Cross NCP. Quantitative Analysis of SRNPN Gene Methylation by Pyrosequencing as a Diagnostic Test for Prader-Willi Syndrome and Angelman Syndrome. Clin Chem. 2006;52: 1005–1013. 10.1373/clinchem.2005.065086 [DOI] [PubMed] [Google Scholar]
- 32.Das R, Lee YK, Strogantsev R, Jin S, Lim YC, Ng PY, et al. DNMT1 and AIM1 Imprinting in human placenta revealed through a genome-wide screen for allele-specific DNA methylation. BMC Genomics. 2013;14: 685 10.1186/1471-2164-14-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Jager P,L., Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17: 1156–1163. 10.1038/nn.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13: 86 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Current Opinion in Cell Biology. 2007;19: 281–289. 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Margolis SS, Sell GL, Zbinden MA, Bird LM. Angelman Syndrome. Neurotherapeutics. 2015;12: 641–650. 10.1007/s13311-015-0361-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittington J, Holland A. Cognition in people with Prader-Willi syndrome: Insights into genetic influences on cognitive and social development. Neurosci Biobehav Rev. 2017;72: 153–167. 10.1016/j.neubiorev.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Sutcliffe JS, Nakao M, Christian S, Örstavik KH, Tommerup N, Ledbetter DH, et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8: 52–58. 10.1038/ng0994-52 [DOI] [PubMed] [Google Scholar]
- 39.Whittington J, Holland A, Webb T, Butler J, Clarke D, Boer H. Cognitive abilities and genotype in a population-based sample of people with Prader–Willi syndrome. Journal of Intellectual Disability Research. 2004;48: 172–187. [DOI] [PubMed] [Google Scholar]
- 40.Piard J, Philippe C, Marvier M, Beneteau C, Roth V, Valduga M, et al. Clinical and molecular characterization of a large family with an interstitial 15q11q13 duplication. Am J Med Genet A. 2010;152A: 1933–1941. 10.1002/ajmg.a.33521 [DOI] [PubMed] [Google Scholar]
- 41.Feinberg JI, Bakulski KM, Jaffe AE, Tryggvadottir R, Brown SC, Goldman LR, et al. Paternal sperm DNA methylation associated with early signs of autism risk in an autism-enriched cohort. Int J Epidemiol. 2015;44: 1199–1210. 10.1093/ije/dyv028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Zhao P, Xu Q, Shan S, Hu C, Qiu Z, et al. The autism-related gene SNRPN regulates cortical and spine development via controlling nuclear receptor Nr4a1. Scientific Reports. 2016;6: 29878 10.1038/srep29878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends in Neurosciences. 2010;33: 121–129. 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Eggermann T, Begemann M, Binder G, Spengler S. Silver-Russell syndrome: genetic basis and molecular genetic testing. Orphanet J Rare Dis. 2010;5: 19 10.1186/1750-1172-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannula K, Kere J, Pirinen S, Holmberg C, Lipsanen-Nyman M. Do patients with maternal uniparental disomy for chromosome 7 have a distinct mild Silver-Russell phenotype? J Med Genet. 2001;38: 273–278. 10.1136/jmg.38.4.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eggermann T, Schönherr N, Eggermann K, Buiting K, Ranke M, Wollmann H, et al. Use of multiplex ligation-dependent probe amplification increases the detection rate for 11p15 epigenetic alterations in Silver–Russell syndrome. Clin Genet. 2008;73: 79–84. 10.1111/j.1399-0004.2007.00930.x [DOI] [PubMed] [Google Scholar]
- 47.Noeker M, Wollmann HA. Cognitive development in Silver-Russell syndrome: a sibling-controlled study. Dev Med Child Neurol. 2004;46: 340–346. [PubMed] [Google Scholar]
- 48.Vidal AC, Neelon SEB, Liu Y, Tuli AM, Fuemmeler BF, Hoyo C, et al. Maternal Stress, Preterm Birth, and DNA Methylation at Imprint Regulatory Sequences in Humans. Genet Epigenet. 2014;6: GEG.S18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssen AB, Capron LE, O’Donnell K, Tunster SJ, Ramchandani PG, Heazell AEP, et al. Maternal prenatal depression is associated with decreased placental expression of the imprinted gene PEG3. Psychol Med. 2016;46: 2999–3011. 10.1017/S0033291716001598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S, Lindenberger U, Hommel B, Aschersleben G, Prinz W, Baltes PB. Transformations in the Couplings Among Intellectual Abilities and Constituent Cognitive Processes Across the Life Span. Psychol Sci. 2004;15: 155–163. 10.1111/j.0956-7976.2004.01503003.x [DOI] [PubMed] [Google Scholar]
- 51.Soubry A, Murphy SK, Wang F, Huang Z, Vidal AC, Fuemmeler BF, et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes. International Journal of Obesity (2005). 2013;39: 650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soubry A, Guo L, Huang Z, Hoyo C, Romanus S, Price T, et al. Obesity-related DNA methylation at imprinted genes in human sperm: Results from the TIEGER study. Clinical Epigenetics. 2016;8: 51 10.1186/s13148-016-0217-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105: 17046 10.1073/pnas.0806560105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murphy SK, Adigun A, Huang Z, Overcash F, Wang F, Jirtle RL, et al. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene. 2012;494: 36–43. 10.1016/j.gene.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrison K, Hoad G, Scott P, Simpson L, Horgan GW, Smyth E, et al. Breast cancer risk and imprinting methylation in blood. Clin Epigenetics. 2015;7: 92 10.1186/s13148-015-0125-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517: 321 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- 57.Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43: 1091. [DOI] [PubMed] [Google Scholar]
- 58.Oliver VF, Miles HL, Cutfield WS, Hofman PL, Ludgate JL, Morison IM. Defects in imprinting and genome-wide DNA methylation are not common in the in vitro fertilization population. Fertil Steril. 2012;97: 147–153. 10.1016/j.fertnstert.2011.10.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromosome, start and end coordinates for each differentially methylated region (DMR) are tabulated.
(DOCX)
Methylation levels at H19ICR, IGF2, SNRPN, PEG3, MEST1, NESPAS, KvDMR, ZAC1 and IGDMR1 imprints determined by pyrosequencing in blood samples from the ABC21 and ABC36 cohorts alongside cohort (ABCgroup), sex and analysis plate.
(XLSX)
Data Availability Statement
Anonymized data are available as Supporting Information files. Access to the full dataset may be requested by contacting the Aberdeen Birth Cohort steering committee (c.mcneil@abdn.ac.uk).