Abstract
Streptococcus pneumoniae (Spn) is an asymptomatic colonizer of the human nasopharynx but can also cause disease in the inner ear, meninges, lung and blood. Although various mechanisms contribute to the effective clearance of Spn, opsonophagocytosis by neutrophils is perhaps most critical. Upon phagocytosis, Spn is exposed to various degradative molecules, including a family of neutrophil serine proteases (NSPs) that are stored within intracellular granules. Despite the critical importance of NSPs in killing Spn, the bacterial proteins that are degraded by NSPs leading to Spn death are still unknown. In this report, we identify a 90kDa protein in a purified cell wall (CW) preparation, aminopeptidase N (PepN) that is degraded by the NSP neutrophil elastase (NE). Since PepN lacked a canonical signal sequence or LPxTG motif, we created a mutant expressing a FLAG tagged version of the protein and confirmed its localization to the CW compartment. We determined that not only is PepN a CW-localized protein, but also is a substrate of NE in the context of intact Spn cells. Furthermore, in comparison to wild-type TIGR4 Spn, a mutant strain lacking PepN demonstrated a significant hyper-resistance phenotype in vitro in the presence of purified NE as well as in opsonophagocytic assays with purified human neutrophils ex vivo. Taken together, this is the first study to demonstrate that PepN is a CW-localized protein and a substrate of NE that contributes to the effective killing of Spn by NSPs and human neutrophils.
Introduction
Streptococcus pneumoniae (Spn) is a Gram-positive bacterium that is a frequent, asymptomatic colonizer of the human upper respiratory tract. However, if it gains access to other anatomical sites in the human host, such as the lungs, inner ear, meninges or blood, it can cause a variety of diseases including pneumonia, otitis media, meningitis and sepsis, respectively [1–4]. Due to these invasive infections, about one million children die per year under the age of five, mostly in the developing world where access to healthcare is limited [5]. Neutrophils are the most abundant white blood cell in the body and are often the first immune cell type to migrate to the site of infection [6, 7]. Neutrophils play a critical role in the effective clearance of Spn via the process of opsonophagocytic killing. This multi-step process involves the tagging of Spn cells with complement proteins and subsequent internalization and degradation through the action of various factors including reactive oxygen and nitrogen species, antimicrobial peptides and a family of enzymes contained within the azurophilic granules, neutrophil serine proteases (NSPs) [8]. Of this repertoire of anti-microbial factors, previous work demonstrated that NSPs are the most important component for effectively killing Spn in vitro [9] and play a vital, protective role in murine models of pneumococcal pneumonia [10]. Furthermore, in individuals with Chediak-Higashi syndrome, a rare genetic disorder that impairs the mobilization of NSP-containing granules [11], neutrophils exhibited a reduced ability to kill Spn [12].
To date, four enzymes have been identified as members of the NSP family: neutrophil elastase (NE), cathepsin G (CG), proteinase 3 (PR3) and neutrophil serine protease 4 (NSP4) [13, 14]. NSPs are members of the chymotrypsin family of serine proteases and contain a His-Asp-Ser catalytic triad [14, 15]. NSPs become enzymatically active when NSP-containing granules fuse with the phagocytic compartment and can also be exocytosed as a component of neutrophil extracellular traps (NETs) to combat extracellular pathogens [16, 17]. However, previous studies demonstrated that Spn can persist within NETs and thus this mechanism is not believed to be responsible for killing during infection [18, 19]. Rather, the dominant means of clearing Spn seems to be opsonsophagocytosis. Several studies revealed that NSPs can reduce bacterial pathogenicity by degrading virulence factors produced by a range of pathogens, including Shigella flexneri, Salmonella enterica serovar Typhimurium, Yersinia enterocolitica and Staphylococcus aureus [20, 21]. In addition, NSPs have been shown to directly kill Pseudomonas aeruginosa, Escherichia coli and Klebsiella pneumoniae [22–25]. Specifically, in E. coli, it was revealed that NE degrades OmpA, which destabilized the cell and induced cell death [22, 23]. Additionally, NE-mediated degradation of OprF, a major outer membrane protein in P. aeruginosa, was demonstrated to be necessary for effective immune defense in a mouse model of lung infection [24]. These findings emphasize the importance of NSPs in anti-microbial defenses and that they achieve this, in part, by degrading specific bacterial proteins. Although the importance of NSPs in controlling Spn infection is well established [9, 10], the exact surface proteins on this pathogen that are degraded by NSPs leading to Spn death have yet to be identified.
In this study, we aimed to identify specific CW-localized Spn proteins that are degraded by NE and/or CG, since these two NSPs were shown to be important for killing Spn both in vitro and in vivo [9, 10]. In experiments using a purified CW preparation, we identified a ~90kDa protein that was specifically and significantly degraded by purified NE. Analysis by mass spectrometry revealed this protein to be aminopeptidase N (PepN), an annotated metalloproteinase predicted to cleave a variety of peptides from the N-terminus [26]. Since PepN lacked an obvious secretion signal or CW localization motif, we epitope-tagged the C-terminus and performed sub-cellular fractionation experiments that revealed that PepN did indeed localize to the CW compartment. Importantly, PepN was shown to be a substrate of purified NE both in experiments with purified CW and in intact Spn cells. Furthermore, a mutant strain of Spn lacking PepN (ΔpepN) was significantly more resistant to killing by purified NE and opsonophagocytic killing by human neutrophils. Taken together, these data identify the first Spn cell wall protein that is degraded by the NSP, neutrophil elastase, and demonstrate that the degradation of PepN contributes to the effective killing of Spn.
Materials and methods
Bacterial strains, growth conditions and growth curves
S. pneumoniae strain TIGR4 (serotype 4) was used throughout this study. S. pneumoniae was grown in Todd-Hewitt broth (BD) supplemented with 0.5% yeast extract (Fisher) (THY) and oxyrase (5μL/mL) or trypticase soy broth (TSB) (BD) supplemented with catalase (Sigma; 30U/mL). Alternatively, bacteria were grown on tryptic soy agar (TSA) supplemented with 200U/mL of catalase. Where appropriate during growth, antibiotics at the following concentrations were included: chloramphenicol (Cm; 4μg/mL) or streptomycin (Sm; 100μg/mL). All cells were grown at 37°C in a 5% CO2 incubator.
For growth curve experiments, strains of interest were grown to mid-log phase (OD600 ~0.6) in THY supplemented with oxyrase and then back-diluted to an OD600 ~0.003 in fresh media. Next, 200μL of cells were added to 96 well flat bottom plates in replicates of 8 wells per strain and were incubated at 37°C for 12h. OD600 measurements were taken every 30 minutes using a VersaMax plate reader.
Generation of mutant strains
All mutant strains of S. pneumoniae used in this study are described in Table 1 and were generated by allelic exchange using the DNA constructs shown in Fig 1A. Each allelic exchange construct was generated in vitro using splice by overlap extension PCR [27]. To create the ΔpepN mutant, ~1.5kb arms of homology flanking the pepN gene were PCR amplified from TIGR4 genomic DNA (gDNA). The CmR cassette was amplified from pAC100 [28] and incorporated into this construct to replace the pepN gene, which allowed for the direct selection of transformants. To generate the ΔpepNRevertant strain, the wild-type pepN gene plus ~3kb of flanking DNA sequence was PCR amplified from TIGR4 gDNA. Subsequently, using the ΔpepN mutant as the recipient strain, we performed co-transformation with the revertant DNA construct and the mutant rpsL allele (SmR) and screened for CmSSmR transformants. We also used co-transformation to make the PepNFLAG tag strain, which introduced a 1X FLAG epitope directly upstream of the stop codon in the pepN open reading frame [29, 30]. The constructs used to generate the ΔpepN and ΔpepNRev strains are illustrated in Fig 1A. Transformation of S. pneumoniae was performed as previously described [31]. PCR and Sanger sequencing was conducted on all mutant strains, including the flanking DNA regions, to confirm the presence of the correct DNA sequence (Eton Biosciences).
Table 1. Relevant strains and primers used in this study.
| Strain | Description | Source | |
|---|---|---|---|
| TIGR4 | Wild-type serotype 4 encapsulated strain, GentR | Laboratory Strain; Ingeborg Aaberge | |
| TIGR4Δcps | Serotype 4 acapsular strain. Cps locus replaced with SpecR gene | Laboratory Strain; Andrew Camilli | |
| TIGR4ΔpepN | Serotype 4 strain with the pepN gene (SP_0797) replaced with a CmR gene | This work | |
| TIGR4pepNFLAG | Serotype 4 strain with a FLAG epitope tag immediately upstream of the stop codon in the pepN gene, SmR | This work | |
| TIGR4pepNRevertant | Serotype 4 strain expresses the wild-type pepN gene in the ΔpepN genetic background, SmR | This work | |
| Primer Name | Sequence (5' to 3') | ||
| For generating ΔpepN mutant | |||
| F0 ΔpepN Sequencing primer |
GAGTTTTTTGACGAAGGG | ||
| R0 ΔpepN Sequencing primer |
CCTGCCCATAGCTATTAA | ||
| F1 ΔpepN For DNA construct |
GCACATGTCGTTACAGAA | ||
| R1 ΔpepN For DNA construct |
CATCAAGCTTATCGATACCGGCTTGCATAGTTTTCTCC | ||
| F2 ΔpepN For DNA construct |
GAAGGTTTTTATATTACAGCTCCAGTCGAAGCAGTTGTTCTT | ||
| R2 ΔpepN For DNA construct |
ATCCAAGCCCAAAAATCG | ||
| For generating pepNFLAG mutant | |||
| F0 pepNFLAG For sequencing |
TGATCGGAGATGAAATCG | ||
| R0 pepNFLAG For sequencing |
CAGCTGATGGAATTTCAC | ||
| F1 pepNFLAG For DNA construct |
CCTTTTAGCGGATTTGGT | ||
| R1 pepNFLAG For DNA construct |
TTTATCATCATCATCTTTATAATCTGCATTTCCGTATTGAAG | ||
| F2 pepNFLAG For DNA construct |
GATTATAAAGATGATGATGATAAATAAATAAGCCTAAAATAAAAAGAA | ||
| R2 pepNFLAG For DNA construct |
GTCCCCAGAAGTTAATCT | ||
GentR, gentamycin resistant; SpecR, spectinomycin resistant; CmR, chloramphenicol resistant; SmR, streptomycin resistant
Bolded text indicates sequences that are homologous to the CmR cassette
Underlined text indicate the FLAG epitope sequence
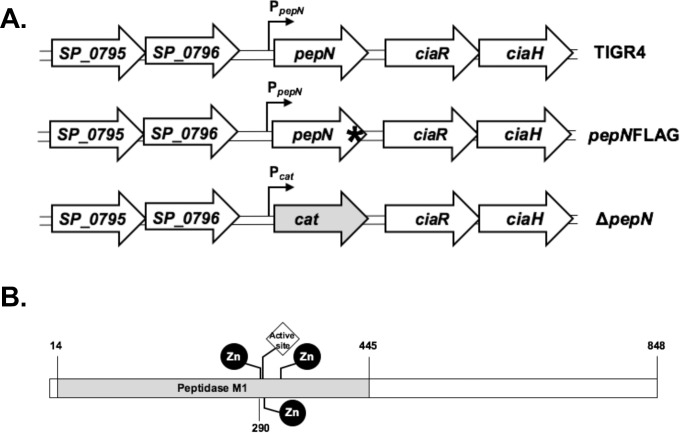
Fig 1. Schematic of DNA constructs used to generate Spn mutant strains and the domain structure of the PepN protein.
(A) The depicted DNA constructs were used to create mutant strains of S. pneumoniae via allelic exchange. In the ΔpepN strain, the pepN ORF was replaced by a Cm resistance cassette. In the pepNFLAG mutant a 1X FLAG tag (denoted by *) was added to the C terminus of the PepN protein, immediately upstream of the stop codon. (B) The domain structure of PepN was determined using BLASTP to be comprised of a peptidase M1 domain containing an active site coordinated by three zinc binding sites (residues 292, 296 and 315).
Sub-cellular fractionation of S. pneumoniae
To isolate purified cell walls and protoplasts, we followed an established protocol as described previously [32], with some modifications. Briefly, 10mL of mid-log cultures (OD600 ~0.6) were pelleted, washed once with 1mL 50 mM Tris-Cl, pH 7.5 and resuspended in 100 μL of cell wall digestion buffer (CWDB) containing 50 mM Tris-Cl, pH 7.5, 30% (w/v) sucrose, 1 mg/mL lysozyme, 300 U/μL mutanolysin (both Sigma-Aldrich), 1x protease inhibitor cocktail (Roche). Cells were incubated for at least 2 h at 37°C with rotation. Whole cell lysates (WCL) were prepared by directly adding SDS buffer to samples and boiling them for 10 mins at 100°C. For protoplast and cell wall (CW), samples were centrifuged at 13,000 x g for 10 min. The supernatant, containing the CW, was applied to SpinX 0.22μm spin columns (Sigma) and centrifuged for 1 min at 13,000 x g to remove any contaminating protoplasts. The pellet, containing the protoplasts, was resuspended in 100μL 50mM Tris-Cl pH 7.5. Samples were stored at -20°C until further use.
Neutrophil elastase-cell wall degradation assays and SDS-PAGE analysis
CW purified from 108 CFU were incubated with 68μM of neutrophil elastase (NE) (Elastin Products Company) or left untreated for 24 h at 37°C with rotation. Equal volumes of samples were boiled for 10 min in SDS sample buffer containing 50mM Tris-HCl, 10% glycerol, 2% SDS, 0.1% bromophenol blue, 2% β-mercaptoethanol and run on an AnykD gradient Tris-glycine polyacrylamide gel (Bio-Rad). Gels were stained with Colloidal Coomassie Blue (Bio-Rad) for 18h and were visualized using an Epson Perfection V550 Photo scanner.
Protein sequence analysis by LC-MS/MS
To determine the identity of proteins degraded by NE, bands of interest were excised from the gel and analyzed by the Taplin Mass Spectrometry Facility at Harvard University. Gel bands were cut into ~1 mm3 pieces and then subjected to a modified in-gel trypsin digestion procedure [33]. Gel pieces were washed and dehydrated with acetonitrile for 10 min, after which the acetonitrile was removed and the gel pieces were then completely dried in a speed-vac. Gel pieces were rehydrated with 50 mM ammonium bicarbonate solution containing 12.5 ng/μl modified sequencing-grade trypsin (Promega, Madison, WI) at 4°C. After 45 min, excess trypsin solution was removed and replaced with 50 mM ammonium bicarbonate solution. Samples were then incubated at 37°C overnight. Peptides were later extracted by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile and 1% formic acid. The extracts were then dried in a speed-vac (~1 hr) and the samples were then stored at 4°C until analysis.
For the analysis, samples were reconstituted in 5–10 μl of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reverse-phase HPLC capillary column was created by packing 2.6 μm C18 spherical silica beads into a fused silica capillary (100 μm inner diameter x ~30 cm length) with a flame-drawn tip [34]. After equilibrating the column, each sample was loaded onto the column via a Famos auto sampler (LC Packings, San Francisco CA). A gradient was formed and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid).
Eluted peptides were then subjected to electrospray ionization and then entered into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences, and hence protein identity, were determined by matching protein databases with the acquired fragmentation pattern by the software program, Sequest (Thermo Fisher Scientific, Waltham, MA) [35]. All databases include a reversed version of all the sequences and the data was filtered to between a one and two percent peptide false discovery rate.
Subcellular localization of PepN via Western blot analysis
CW was isolated from 107 CFU of the appropriate bacterial strains. Equal volumes of samples were analyzed via SDS-PAGE as described above, proteins were transferred to a nitrocellulose membrane (ThermoFisher) at 22V for 18h at 4°C and blocked for 1 h at room temperature with 5% skim milk in 1X PBS with 0.2% Tween-20 (PBST). Unconjugated primary mouse anti-FLAG antibody (Sigma-Aldrich) at 1:3,000 and rabbit anti-CodY serum (a generous gift from Dr. A. L. Sonenshein) at 1:10,000 diluted in 2.5% milk in 1X PBST were added to membranes and incubated for 1 h at room temperature with agitation. Membranes were then washed 3X with 10mL 1X PBST for 5 mins each. HRP-conjugated secondary antibodies (Jackson ImmunoResearch) were applied at a 1:10,000 dilution in 2.5% milk in 1X PBST and incubated for 1 h at room temperature, followed by three 45 min washes in 10mL 1X PBST. Membranes were then developed with SuperSignal West Dura Extended Duration Substrate (ThermoFisher).
Capsule immunodot blot assays
To quantify the amount of capsule present in wild-type TIGR4 and ΔpepN mutant strains, 1 ml of OD600-matched mid-exponential-growth-phase bacteria was pelleted and stored at −20°C until use. Samples were resuspended in 300 μl of CWDB, described above, and incubated at 37°C for 30 min. Samples were then sonicated for 4–10 second intervals while on ice using a probe sonic dismembrator (Fisher Scientific). Next, a 2-fold serial dilution of lysate was prepared in 1X PBS, and 5 μl was spotted on 0.2-μm-pore-size nitrocellulose membranes (Invitrogen, Inc.) with suction. Membranes were blocked with 10mL of 5% milk in 1X TBS with 0.1% Tween-20 (TBST) for 1 h at room temperature with shaking, then washed 1X with 10mL 1X TBST for 5 min. The membrane was then probed with an unconjugated rabbit anti-serotype 4 serum (Statens Serum Institut; 1:1000) in 5mL of 2.5% milk in 1X TBST for 1h at room temperature with shaking. After 3–5 min washes with 15mL 1X TBST, an HRP-conjugated goat-anti-rabbit secondary antibody (1:2500, Jackson ImmunoResearch) was applied to the membrane in 5mL of 2.5% milk in 1X TBST for 1h at room temperature with shaking. Finally, the membrane was washed 3X with 10mL 1X TBST, developed using ECL Blotting Substrate (Thermo Scientific), visualized using the C-Digit Western Blot Scanner, and quantified using ImageStudioLite (LI-COR Biosciences).
C3 deposition assay and FACS analysis
For C3 deposition assays, 1 ml of mid-exponential phase bacteria grown in THY were pelleted, washed in PBS and resuspended in 500 μl of Hanks buffer with Ca2+ and Mg2+ (Gibco, Corp.) supplemented with 0.1% gelatin (Fischer Scientific, Inc.). 107 CFU in 50 μl were added to a final concentration of 10% infant rabbit serum in 100 μl (AbD Serotec, Co.). Samples were incubated in a 37°C rolling incubator for 30 mins. Next, opsonization reactions were chilled for 3 mins on ice, quenched with 500 μl of Hanks buffer without Ca2+ and Mg2+ (Gibco) with 0.1% gelatin, and pelleted at 4000 rpm for 5 mins. Pellets were resuspended in 1:200 FITC-conjugated goat anti-rabbit C3 antibody (MP Biomedicals) in 100 μl Hanks buffer without Ca2+ and Mg2+ with 0.1% gelatin and incubated on ice in the dark for 30 mins. Staining reactions were quenched with 500 μl Hanks buffer with 0.1% gelatin but without Ca2+ and Mg2+, centrifuged at 4000 rpm for 5 mins, and pellets were resuspended in 300 μl of 1% paraformaldehyde (PFA; Sigma-Aldrich, Co.). Samples were analyzed on an Se3 Cell Sorter (Bio-Rad) and data were analyzed using FlowJo software.
In vitro bactericidal and PepN degradation assays with neutrophil elastase
Wild-type and mutant strains of S. pneumoniae were grown to early-exponential phase (OD600 ~0.3) in TSB supplemented with catalase, pelleted by centrifugation, washed in sterile 1X PBS, and resuspended in 10 μM sterile sodium phosphate reaction buffer to a concentration of ~107 CFU/mL. 50μL of cells were exposed to a two-fold dilution series of NE ranging from 3.4–13.6μM and incubated for 1h at 37°C with 5% CO2. These NE concentrations are within the documented physiological range observed in vivo [36] and are similar to concentrations tested by other groups [9, 37]. To determine viable counts, cells were then serially diluted, plated on TSA and incubated overnight at 37°C with 5% CO2.
To assess whether PepN is degraded by NE in the context of an intact Spn cell, ~5 x 106 CFU were exposed to 6.8μM NE, or left untreated, for 1h at 37°C with 5% CO2, followed by the isolation of CW and WCL fractions as described above. Samples were then subjected to SDS-PAGE and Western Blot analysis using anti-FLAG or anti-CodY antibodies. Data were compiled from at least three independent experiments.
Ex vivo neutrophil opsonophagocytic killing assay
These studies were approved by the Human Investigation Review Board (IRB) at both Tufts Medical Center and the University of Buffalo. Young, healthy human volunteers were recruited in accordance to University of Buffalo and Tufts Medical Center IRB and signed informed consent forms. Individuals taking medication, reporting symptoms of infection within the last two weeks, or who were pregnant were excluded from the study. 20mL of whole blood was obtained and anticoagulated with acid citrate/dextrose. PMNs were isolated using a 2% gelatin sedimentation technique as previously described [38] which allows for isolation of active PMNs with ~90% purity. Killing of opsonized Spn by human neutrophils was performed as previously described [39] based on a modified protocol described by Dalia et al. [40]. Strains of Spn were grown to mid-log and 103 CFU were opsonized in 10% (v/v) baby rabbit serum (Pel-Freeze) in 200μL of Hank’s Buffer supplemented with 0.1% gelatin for 30 mins. Pre-opsonized bacteria were then mixed with 5 x 105 PMNs for 45 mins at 37°C with rotation. Samples were then placed on ice to stop the process of opsonophagocytosis followed by serial dilution and plating to enumerate viable CFU. The percentage of bacterial killing was calculated relative to controls without PMNs. In some experiments, neutrophils were treated with 20μM cytochalasin D (Sigma-Aldrich) for 30 min at 37°C prior to incubation with opsonized Spn cells [9].
Statistical analysis
To determine if a statistically significant difference existed between wild-type TIGR4 and the mutant strain of interest across various concentrations of NE, two-way ANOVA statistical tests were performed [41]. In experiments that compared the phenotypes of wild-type TIGR4, ΔpepN and ΔpepNRev strains in the presence of only 3.4 μM NE, a one-way ANOVA followed by a multiple comparisons test was performed. Unpaired Student t-test was used for comparison of bacterial killing by PMNs. p values less than 0.05 were considered significant. All statistical analyses were performed using GraphPad Prism for Mac (GraphPad Software, Inc).
Results
Analysis of purified CW from TIGR4 treated with the NSPs, neutrophil elastase (NE) or cathepsin G (CG)
Although NSPs were demonstrated to be essential for the effective killing of Spn [9], the protein targets that are degraded by these enzymes remains unknown. To identify Spn CW proteins that are degraded by NSPs, CW was isolated from wild-type TIGR4 bacteria and were incubated with NE, CG or left untreated. Samples were then evaluated using SDS-PAGE analysis followed by staining with Coomassie blue. As depicted in Fig 2A and highlighted by the arrow heads, several protein species (~90kDa, ~70kDa, ~52kDa) were degraded by NE, but were still clearly visible in both the untreated and CG-treated samples. In particular, there was a significant reduction in the intensity of a ~90kDa band (Fig 2A, black arrow heads) in the NE-treated samples as compared to the untreated control (Fig 2B, P<0.0001). In order to determine the identity of this specific NE substrate, we excised the 90kDa band from the gel in both the untreated and NE-treated samples for mass spectrometry analysis. Based on its amino acid sequence, this protein was determined to be aminopeptidase N (PepN; SP_0797). Importantly, this analysis also revealed that the 90kDa band was vastly comprised of PepN, ranging from ~89–95% of the total band intensity (Fig 2C). Additionally, by comparing the relative abundance of the PepN protein in the untreated versus NE-treated samples, we observed that PepN was substantially degraded, with up to a 104-fold reduction in protein abundance (Fig 2C). Further confirming that the vast majority of the 90kDa band was comprised of PepN, CW isolated from a mutant strain of Spn that lacks PepN (ΔpepN) did not possess the 90kDa band (Fig 2A). Taken together, these data demonstrate that PepN is a 90kDa protein found in a purified Spn CW preparation that is specifically and markedly degraded by NE.
Fig 2. Neutrophil elastase degrades a 90kDa Spn CW protein in vitro.
CW was isolated from TIGR4 and ΔpepN cells of S. pneumoniae and were left untreated or incubated with 68μM NE or 10μM CG. (A) Samples were analyzed using an AnykD gradient Tris-glycine polyacrylamide gel followed by colloidal Coomassie blue staining. Arrow heads indicate bands degraded by NE. Black arrow heads highlight the 90kDa species degraded by NE. (B) The intensity of the 90 kDa band in the untreated control or in CW samples treated with NE was quantified using ImageStudioLite. The data are presented as relative band intensity normalized to a blank lane. Data shown are means ± SD from 5 independent experiments. P = 0.019 using a Student’s t-test. (C) To determine the identity of the 90kDa protein that is degraded by NE, we excised this band from both the untreated and NE-treated lanes and had it analyzed via mass spectrometry. Additionally, this analysis quantified the relative abundance of the 90kDa protein in both the untreated and NE-treated samples. The gel is from one experiment representative of 5 independent experiments. The mass spectrometry analysis is from 2 of those independent experiments. NE, neutrophil elastase; CG, cathepsin G.
Characterization of PepN as a CW-localized protein in Spn that is degraded by NE in vitro
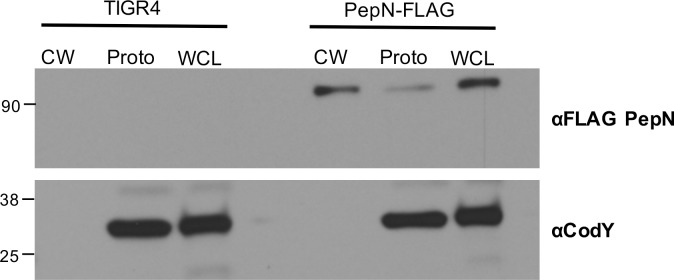
Based on its amino acid sequence, PepN is annotated as a member of the peptidase M1 family and possesses an active site that is coordinated by three zinc binding sites (Fig 1B). However, examination of the PepN amino acid sequence failed to reveal a canonical LPxTG motif, choline binding domain, Sec secretion signal peptide, or any other characterized export signal [42]. While it is not impossible for a protein lacking a classic export sequence to be secreted from the cell, such as the Pht proteins and those described as non-classical surface proteins [42, 43], this prompted a more thorough investigation of the sub-cellular localization of PepN. To do so, we generated a mutant strain of Spn via natural transformation using a DNA construct that added a FLAG epitope immediately upstream of the stop codon (Fig 1A; pepNFLAG). For these experiments, we prepared CW, protoplasts and whole cell lysates (WCL) from mid-exponential phase TIGR4 and pepNFLAG cells and analyzed these fractions via Western Blotting using anti-FLAG or anti-CodY (a cytoplasmic protein) antibodies. As expected, CodY was detected only in the protoplast and WCL fractions, and was completely absent in the CW samples, confirming that this fraction was free of cytoplasmic contaminants (Fig 3A, bottom panel). Importantly, the PepNFLAG protein was clearly detected in the both the WCL and CW fractions, with a fainter band present in the protoplast sample (Fig 3A, top panel). To confirm that observed variations in the PepNFLAG signal were not due to differences in the amount of total protein, we analyzed the CW, protoplast and WCL samples in parallel via SDS-PAGE and Coomassie blue staining. Indeed, similar amounts of protein were observed in the protoplast and WCL samples isolated from TIGR4 and PepNFLAG cells, while the CW fraction isolated from both strains contained the least amount of protein (S1 Fig). Thus, the intensity of the PepNFLAG signal in the CW fraction (Fig 3A, top panel) may possibly represent an underestimation of the amount of PepN protein in this compartment. Together, these data suggest that despite the absence of a canonical LPxTG motif, Sec-dependent signal peptide, or any known cell wall binding motif, PepN indeed localizes to the CW compartment.
Fig 3. PepN localizes to the cell wall compartment within Spn cells.
Whole cell lysates (WCL), CW and protoplast fractions were isolated from OD600-matched, mid-log phase TIGR4 and PepNFLAG cells. Samples were analyzed by Western blotting using anti-FLAG or anti-CodY (a cytoplasmic protein) antibodies. Data shown are from one experiment representative of three independent experiments.
Neutrophil elastase degrades PepN in intact Spn cells
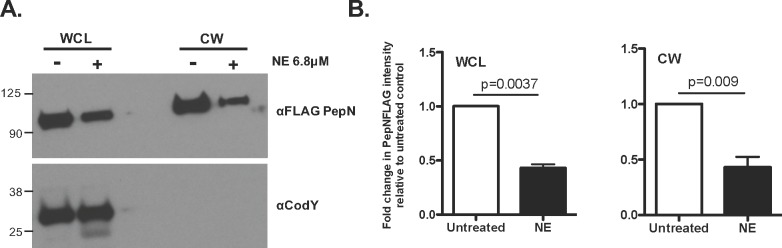
To validate PepN as the substrate for NE and confirm that NE can degrade PepN in the context of native cell wall architecture, we exposed PepNFLAG cells to NE, followed by subcellular fractionation and Western Blot analysis using anti-FLAG or anti-CodY antibodies. Again, the CodY signal was only present in the WCL samples and the intensity of this band was unchanged in the presence of NE (Fig 4A, bottom panel). Strikingly, analysis of both WCL and CW fractions revealed a strong PepNFLAG signal in the untreated samples (Fig 4A, top panel) that was diminished >2-fold upon exposure to NE (WCL P = 0.0037; CW P = 0.009) (Fig 4B). Importantly, quantification of total protein loaded in untreated or NE-treated WCL and CW samples were evaluated via SDS-PAGE analysis and Coomassie blue staining. These data confirm that, overall, there were no significant differences in protein content among these samples (S2 Fig). These data strongly suggest that in the context of an intact Spn cell, PepN is a CW-localized protein that is significantly degraded by NE.
Fig 4. NE degrades PepN in intact Spn cells.
Approximately 5 x 106 CFU of the PepNFLAG strain were grown to early-log phase and were either left untreated or were exposed to 6.8μM NE. (A) Cells were fractionated to isolate the CW and samples were analyzed via Western blotting using anti-FLAG or anti-CodY (a cytoplasmic protein) antibodies. Data shown are from one Western blot representative of four independent experiments. (B) Band intensity in CWL and CW samples were normalized to a blank lane, are expressed as fold change relative to the untreated control and were quantified using ImageStudioLite software. Data presented are the means ± SD from 4 independent experiments.
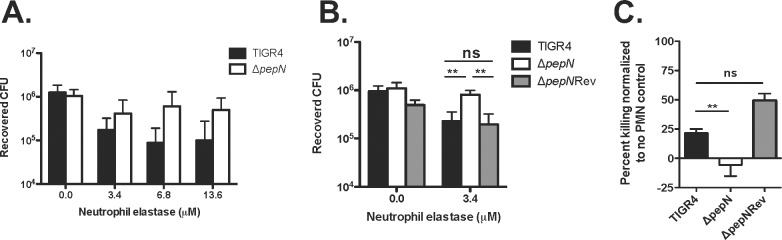
The ΔpepN mutant demonstrates enhanced resistance to killing by purified NE in vitro and by human neutrophils ex vivo
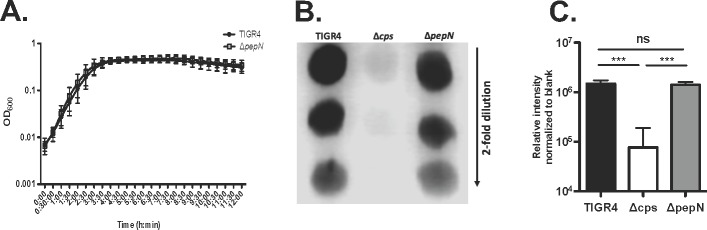
To assess the contribution of PepN to the effective killing of Spn in vitro, we generated a mutant strain lacking this protein (ΔpepN). To first characterize this mutant, we assessed its growth kinetic phenotype, and determined that it was similar to that of wild-type TIGR4 (Fig 5A). Additionally, to ensure that removal of the cell wall protein PepN did not have an unexpected impact on capsule level, a structure that was demonstrated to influence susceptibility to NSP-mediated killing [37], we quantified and compared capsule on wild-type and ΔpepN cells. As shown in Fig 5B and 5C, while wild-type TIGR4 and ΔpepN cells both express significantly more capsule than the acapsular control (Δcps; both P<0.001), there was no significant difference in capsule levels between wild-type TIGR4 and the ΔpepN strain.
Fig 5. Mutants lacking PepN (ΔpepN) demonstrate the same growth kinetics and capsule levels as wild-type TIGR4.
A) TIGR4 and ΔpepN cells were grown to mid-log phase, back-diluted in fresh media to OD600 0.003, incubated at 37°C and OD600 was measured every 30 minutes for 12h. Data presented are the means ± SD from 3 independent experiments. B and C) Capsule was isolated from OD600-matched, mid-log phase TIGR4, Δcps and ΔpepN cells. For each strain, 5μL spots of a 2-fold serial dilution were applied to a nitrocellulose membrane, which was developed using an unconjugated rabbit anti-serotype 4 serum, HRP-conjugated goat anti-rabbit antibody and ECL Blotting Substrate. B) Data shown are from one blot representative of 3 independent experiments. C) Data presented are the means ± SD from 3 independent experiments. ***, P<0.001; ns = no significant difference. Statistical analyses were conducted using a one-way ANOVA and Tukey’s Multiple Comparisons Test.
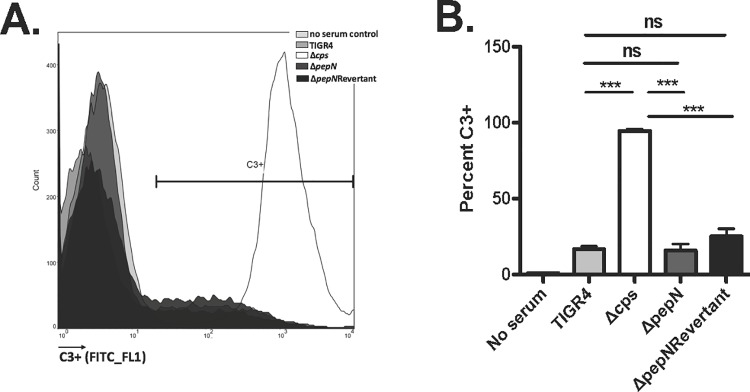
In subsequent experiments, we assessed the phenotype of the ΔpepN strain in the presence of various concentrations of purified NE. These data demonstrated that NE killed both wild-type TIGR4 and ΔpepN strains in a significant, dose-dependent fashion (P = 0.0035). Importantly, the ΔpepN mutant was significantly more resistant (P = 0.004) to NE-mediated killing as compared to wild-type TIGR4 across all tested concentrations of NE (Fig 6A and 6B). Furthermore, as shown in Fig 6B, when we reverted the ΔpepN mutant (ΔpepNRev) and exposed this strain to purified NE, it exhibited a sensitive phenotype similar to that of wild-type TIGR4. To test the ΔpepN mutant in a more physiologically relevant setting, we determined its survival phenotype using an opsonophagocytic assay with purified human neutrophils. These experiments revealed that, compared to wild-type Spn, the ΔpepN mutant was also significantly more resistant (P = 0.036) to extracellular phagocytic killing by human neutrophils (Fig 6C). In fact, while wild-type TIGR4 was killed in the presence of PMNs, the ΔpepN mutant was not and displayed a very slight overall increase in cell number (Fig 6C). Importantly, the resistance of the ΔpepN strain to neutrophil-mediated phagocytosis was not due to differences in neutrophil viability (S3 Fig) nor opsonization by complement C3, a protein vital to the uptake of Spn via the classical complement pathway [44, 45]. As shown in Fig 7A and 7B, there was no significant difference in the percent of C3+ Spn cells between the WT, ΔpepN and ΔpepNRev strains. Taken together, these data reveal that PepN is a nonessential, cell wall-localized protein that is a substrate of NE, and its degradation contributes to the effective killing of Spn by both purified NE and whole human neutrophils.
Fig 6. The ΔpepN mutant exhibits enhanced resistance to NE-mediated killing in vitro. and to killing by human neutrophils ex vivo.
A) Approximately 5 x 105 CFU of the TIGR4 or ΔpepN strain were left untreated or exposed to various concentrations of NE. Samples were diluted and plated to enumerate viable CFU. Data presented are the means ± SD from 3–4 independent experiments. P = 0.004 comparing TIGR4 to ΔpepN across all concentrations of NE; P = 0.0035 evaluating dose dependent effect of NE-mediated killing within each individual strain. Statistical analyses were calculated using two-way ANOVA. B) TIGR4, ΔpepN and ΔpepNRev cells were left untreated or were exposed to 3.4μM NE followed by serial dilution and plating to enumerate viable CFU. Data presented are the means ± SD from 3 independent experiments. **, P<0.01; ns = no significant difference. P values were calculated using one-way ANOVA. C) For opsonophagocytic killing experiments, PMNs were isolated from the blood of healthy donors and incubated with pre-opsonized Spn. Viable CFU were determined after serial dilution and plating. The percentage of bacteria killed was determined by comparing surviving CFU for each strain to a no PMN control. Positive percent killing indicates bacterial death while negative percent indicates bacterial growth. Data are shown as the means ± SD from 4 independent experiments with PMNs from four separate donors. P = 0.036 and was calculated using Student’s t-test.
Fig 7. Mutants lacking PepN demonstrate the same C3 deposition phenotype as wild-type TIGR4.
Spn cells were incubated with infant rabbit serum and stained with a FITC-conjugated anti-C3 antibody. Data are shown as A) a representative histogram plot from 1 experiment representative of 3 independent experiments and B) means ± SD from 3 independent experiments. ***, P<0.001; ns = no significant difference. P values were calculated using one-way ANOVA.
Discussion
In neutrophils, non-oxidative mechanisms of killing phagocytosed Spn were shown to be essential both in vitro and in vivo and primarily involve the activity of NSPs, such as NE and CG [9, 10]. Further supporting the importance of NSPs, individuals with impaired granule mobilization, which affects the fusion of NSP-containing granules with the phagolysosome, exhibit a defect in neutrophil-mediated killing of Spn [12]. In addition to playing a key role in controlling Spn infections, other studies demonstrated that NSPs have the capacity to directly kill or degrade virulence factors produced by a variety of pathogens including P. aeruginosa, E. coli, S. flexneri, K. pneumoniae and S. aureus [20–25]. Although previous reports highlight the importance of NSPs in bacterial clearance, little is known about the identity of the Spn proteins that are degraded by NSPs and thus facilitate effective killing.
In this study, we aimed to identify Spn CW proteins that are degraded by the NSPs NE and CG. To do so, we exposed a purified Spn CW preparation to NE and CG in vitro. These experiments revealed that a few bands, ~90kDa, ~70kDa and ~52kDa in size, were degraded only by NE, suggesting a difference in substrate specificity between these two NSP family members. Since over multiple independent experiments, the ~90kDa band was the most strikingly degraded by NE in multiple independent experiments, this study focused on further identifying and characterizing this CW protein as a putative NE substrate and determining its potential role in NSP-mediated killing of Spn. Subsequent analysis via mass spectrometry revealed this ~90kDa protein to be aminopeptidase N (PepN). Furthermore, these experiments demonstrated that the relative abundance of PepN was reduced 103−104-fold in purified CW samples treated with NE as compared to untreated controls, indicating that NE substantially degrades this Spn protein. Interestingly, in the context of an intact Spn cell, PepN abundance was more modestly reduced (~3-fold) in NE-treated samples, as compared to untreated controls. This discrepancy may be due, in part, to the masking effect provided by the capsular polysaccharide and/or the complex architecture of the cell wall, thus limiting the accessibility of PepN as a NE substrate. Nevertheless, degradation of PepN on intact Spn cells was still possible and thus may be sufficient to induce bacterial cell death.
Since analysis of the PepN sequence failed to reveal a canonical Sec-dependent secretion signal, LPxTG cell wall anchoring motif or other export signals [42, 43], we created a mutant strain of Spn harboring a FLAG-tagged version of the protein to more definitively assess its subcellular localization. From these experiments we visualized a faint PepN band in the protoplast fraction. It is still unclear how PepN is exported from the cell, therefore these data would suggest that the export mechanism is rate-limiting and may depend on there being a sufficient concentration of PepN in the cytoplasm. However, due to the absence of a Sec-dependent signal sequence, it seems unlikely that coupled translation and secretion is occurring.
Interestingly, despite the absence of an obvious secretory or CW localization motif, the majority of PepN was identified in the CW compartment. Although these findings are at odds with the reported cytoplasmic localization of PepN in S. mitis (97% sequence identity) and S. salivarius (67% sequence identity) [46, 47], the absence of the cytoplasmic control protein CodY in our CW fractions provides confidence that PepN is indeed CW localized in Spn. Additionally, in various other Gram-positive species including L. lactis, PepN (57% identity) is a CW anchored protein, indicating there is some variability in the localization of this protein [48]. Thus, akin to other Spn proteins including pneumolysin [32], PavA [49] and HtrA [42], our data indicate that PepN is yet another example of a non-classical protein that localizes to the CW despite the absence of an obvious export sequence.
These initial experiments strongly suggest that PepN is a CW-localized protein that serves as a substrate for NE. However, these conclusions were drawn from experiments using a purified preparation of CW proteins, which may affect the abundance, diversity or availability of substrates for NE-mediated degradation. More importantly, since the aim of this study was to identify Spn proteins that not only were degraded by NSPs, but were also involved in NSP-mediated killing of Spn, we conducted additional experiments in a more physiologically relevant system. Through experiments that exposed intact Spn cells expressing the FLAG-tagged version of PepN to NE, we confirmed that PepN does indeed localize to the CW and it is markedly degraded compared to untreated controls. Previous studies in E. coli and P. aeruginosa identified the outer membrane proteins OmpA and OprF, respectively, as targets that are degraded by NE that results in bacterial cell death [22, 24]. Both of these proteins are porins that contribute to virulence and isogenic mutants lacking either OmpA or OprF in the respective strain demonstrated enhanced resistance to NE-mediated killing in vitro and in relevant mouse models of infection [22, 24, 50, 51]. To directly assess the contribution of PepN to NSP-mediated killing of Spn cells, we created the ΔpepN strain and confirmed that its growth rate was indistinguishable from that of wild-type Spn. Importantly, since capsule level was shown to impact killing by NSPs [37], and it was feasible that the deletion of a CW-localized aminopeptidase could affect the attachment of capsule to the cell, we quantified capsule on wild-type TIGR4 and ΔpepN cells. Data from these experiments demonstrated that both strains possess similar amounts of capsule. In vitro bactericidal assays that exposed wild-type TIGR4 and ΔpepN cells to purified NE revealed that cells lacking PepN were significantly more resistant to killing. Additionally, ΔpepN cells were significantly more resistant to opsonophagocytic killing by whole human neutrophils ex vivo. Importantly, the differential killing phenotype was not due to variations in capsule level or C3 deposition. Rather, these data suggests that differential killing between wild-type and ΔpepN may be due to the absence of a NE substrate that is necessary for optimal killing of Spn once in the phagolysosome. Our observation that NE exhibits bactericidal activity with TIGR4 (serotype 4) strains in vitro are at odds with a study by Domon et al that report an absence of NE-mediated killing in D39, a serotype 2 strain of Spn [52]. These differences may be due, in part, to variations in the repertoire or expression level of surface-associated proteins between these two Spn serotypes [53]. However, another study by van der Windt and colleagues reported substantial NE-mediated killing of both TIGR4 and D39 strains in vitro [37]. Thus it may be possible that other factors, including slight technical variations in experimental parameters, might be responsible for the differences in outcome.
A key observation from these experiments was that NE-mediated degradation of a non-essential CW protein, PepN, may play an important role in Spn killing. One possible explanation for this observation is that degradation of PepN is sufficient to destabilize the cell envelope and induce cell lysis. Alternatively, if PepN is attached, in some fashion, to peptidoglycan, teichoic or lipotechoic acid, its degradation by NE may also damage these essential structures or impair their turnover such that normal Spn growth is disrupted. Another potential explanation may be that, via its aminopeptidase activity, PepN modifies other CW proteins and creates additional NE substrates. Thus in PepN-sufficient Spn cells, not only is PepN directly degraded, but also the modified CW proteins created via PepN aminopeptidase activity may also be degraded, which together is enough to cause cell death. It would be possible to test this last hypothesis by creating a mutant strain with an inactivated PepN catalytic site and assess its viability in the presence of NE. Additionally, since our initial experiments identified three protein bands that were notably degraded only by NE, it is feasible that NE may have additional substrates aside from PepN in the Spn CW that potentially contribute to bacterial cell death. These questions are beyond the scope of this current study, but emphasize the need for future experiments.
The role of PepN in Spn biology and in the context of host infection is not well understood. However, in other closely related species of Streptococcus, including S. thermophilus, PepN is characterized as a 95kDa monomeric, metallo-aminopeptidase that possesses three Zn2+ binding sites that coordinates its active site [26, 48]. Based on its sequence homology to other aminopeptidases, it is thought that PepN functions to degrade endogenous proteins and contributes to normal protein turnover as well as to provide free amino acids to be used for metabolic processes [48]. Interestingly, a recent study demonstrated that PepN present in Spn lysates dampens the effector function of cytotoxic T lymphocyte by modulating the intracellular TCR signaling cascade and ultimately dampens the production of the pro-inflammatory cytokine, IFN-γ [54]. Together with the findings in our study, these data begin to reveal a previously underappreciated role for PepN in host-pathogen interactions and Spn disease pathogenesis. In summary, this is the first report to identify a Spn protein, PepN, as a substrate that is degraded by NE and that also plays a key role in NSP-mediated killing of Spn both in vitro and ex vivo. Additionally, we determined that despite the absence of a canonical export sequence, PepN localizes to the CW compartment within Spn. We propose a model where Spn cells are opsonophagocytosed by neutrophils and are subsequently bombarded with an assortment of lysosome- and granule-derived anti-microbial factors. Based on our data, a key step in this process is the NE-mediated degradation of PepN, which contributes to the effective killing of Spn.
Supporting information
CW, protoplast and WCL fractions were isolated from TIGR4 and PepNFLAG cells. The samples analyzed in this experiment are the same as those presented in Fig 3. Samples were analyzed by (A) SDS-PAGE followed by Coomassie Blue staining. B) The band intensity in each lane was quantified using ImageStudioLite software and the data are expressed as fold change relative to the cell wall digestion buffer (CWDB) control lane. Data shown are from one experiment representative of three independent experiments.
(TIFF)
(A) Total protein content from three independent experiments in untreated and NE-treated WCL and CW samples were evaluated via SDS-PAGE and Coomassie blue staining. (B) Data are normalized to a blank lane, expressed as fold change relative to the respective untreated control and were quantified using ImageStudioLite software. Data shown are the means ± SD from three independent experiments. Student’s t-test revealed no significant differences.
(TIFF)
PMNs were isolated from the blood of two healthy donors and 5 x 105 cells were incubated with 103 CFU of WT or ΔpepN cells. Following a 45-minute incubation, neutrophil viability was determined via trypan blue exclusion and enumeration using a haemacytometer. Each sample was enumerated twice and by two independent individuals. Data shown are the means ± SD from two independent experiments with at least two technical replicates per strain per experiment. One-way ANOVA revealed no significant difference in neutrophil viability after exposure to WT or ΔpepN cells.
(TIFF)
Acknowledgments
This project was funded by Undergraduate Research Awards provided by the Jess and Mildred Fisher College of Science and Mathematics at Towson University and funds provided by the Department of Biological Sciences. We thank Dr. A. L. Sonenshein for providing the anti-CodY antibody and Dr. John Leong for sharing additional reagents needed for PMN experiments. We also thank Dr. Neil Greene for helpful discussions and for critically reading our manuscript and Ross Tomaino for help with the mass spectrometry analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was funded by Undergraduate Research Awards provided by the Jess and Mildred Fisher College of Science and Mathematics at Towson University and funds provided by the Department of Biological Sciences. We thank Dr. A. L. Sonenshein for providing the anti-CodY antibody and Dr. John Leong for sharing additional reagents needed for PMN experiments. We also thank Dr. Neil Greene for helpful discussions and for critically reading our manuscript and Ross Tomaino for help with the mass spectrometry analysis.
References
- 1.Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6(4):288–301. Epub 2008/03/15. nrmicro1871 [pii] 10.1038/nrmicro1871 . [DOI] [PubMed] [Google Scholar]
- 2.Mitchell TJ. Virulence factors and the pathogenesis of disease caused by Streptococcus pneumoniae. Res Microbiol. 2000;151(6):413–9. Epub 2000/08/29. S0923-2508(00)00175-3 [pii]. . [DOI] [PubMed] [Google Scholar]
- 3.Tuomanen EI, Austrian R, Masure HR. Pathogenesis of pneumococcal infection. N Engl J Med. 1995;332(19):1280–4. Epub 1995/05/11. 10.1056/NEJM199505113321907 . [DOI] [PubMed] [Google Scholar]
- 4.Weiser JN. The pneumococcus: why a commensal misbehaves. J Mol Med. 2009;88(2):97–102. Epub 2009/11/10. 10.1007/s00109-009-0557-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pneumococcal Disease www.who.int: World Health Organization; 2017. Available from: http://www.who.int/ith/diseases/pneumococcal/en/.
- 6.Dallaire F, Ouellet N, Bergeron Y, Turmel V, Gauthier MC, Simard M, et al. Microbiological and inflammatory factors associated with the development of pneumococcal pneumonia. J Infect Dis. 2001;184(3):292–300. 10.1086/322021 . [DOI] [PubMed] [Google Scholar]
- 7.van Rossum AM, Lysenko ES, Weiser JN. Host and bacterial factors contributing to the clearance of colonization by Streptococcus pneumoniae in a murine model. Infect Immun. 2005;73(11):7718–26. Epub 2005/10/22. 73/11/7718 [pii] 10.1128/IAI.73.11.7718-7726.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197–223. 10.1146/annurev.immunol.23.021704.115653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Standish AJ, Weiser J.N. Human Neutrophils Kill Streptococcus pneumoniae via Serine Proteases. Journal of Immunology. 2009;183(4):2602–9. [DOI] [PubMed] [Google Scholar]
- 10.Hahn I, Klaus A, Janze AK, Steinwede K, Ding N, Bohling J, et al. Cathepsin G and neutrophil elastase play critical and nonredundant roles in lung-protective immunity against Streptococcus pneumoniae in mice. Infect Immun. 2011;79(12):4893–901. 10.1128/IAI.05593-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganz T, Metcalf JA, Gallin JI, Boxer LA, Lehrer RI. Microbicidal/cytotoxic proteins of neutrophils are deficient in two disorders: Chediak-Higashi syndrome and "specific" granule deficiency. J Clin Invest. 1988;82(2):552–6. 10.1172/JCI113631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Root RK, Rosenthal AS, Balestra DJ. Abnormal bactericidal, metabolic, and lysosomal functions of Chediak-Higashi Syndrome leukocytes. J Clin Invest. 1972;51(3):649–65. 10.1172/JCI106854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perera NC, Schilling O, Kittel H, Back W, Kremmer E, Jenne DE. NSP4, an elastase-related protease in human neutrophils with arginine specificity. Proc Natl Acad Sci U S A. 2012;109(16):6229–34. 10.1073/pnas.1200470109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korkmaz B, Moreau T, Gauthier F. Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie. 2008;90(2):227–42. 10.1016/j.biochi.2007.10.009 . [DOI] [PubMed] [Google Scholar]
- 15.Hellman L, Thorpe M. Granule proteases of hematopoietic cells, a family of versatile inflammatory mediators—an update on their cleavage specificity, in vivo substrates, and evolution. Biol Chem. 2014;395(1):15–49. 10.1515/hsz-2013-0211 . [DOI] [PubMed] [Google Scholar]
- 16.Borregaard N, Cowland JB. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 1997;89(10):3503–21. . [PubMed] [Google Scholar]
- 17.Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol Rev. 2010;62(4):726–59. 10.1124/pr.110.002733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayana Moorthy A, Narasaraju T, Rai P, Perumalsamy R, Tan KB, Wang S, et al. In vivo and in vitro studies on the roles of neutrophil extracellular traps during secondary pneumococcal pneumonia after primary pulmonary influenza infection. Front Immunol. 2013;4:56 10.3389/fimmu.2013.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wartha F, Beiter K, Albiger B, Fernebro J, Zychlinsky A, Normark S, et al. Capsule and D-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell Microbiol. 2007;9(5):1162–71. Epub 2007/01/16. CMI857 [pii] 10.1111/j.1462-5822.2006.00857.x . [DOI] [PubMed] [Google Scholar]
- 20.Weinrauch Y, Drujan D, Shapiro SD, Weiss J, Zychlinsky A. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417(6884):91–4. 10.1038/417091a . [DOI] [PubMed] [Google Scholar]
- 21.Hazenbos WL, Kajihara KK, Vandlen R, Morisaki JH, Lehar SM, Kwakkenbos MJ, et al. Novel staphylococcal glycosyltransferases SdgA and SdgB mediate immunogenicity and protection of virulence-associated cell wall proteins. PLoS Pathog. 2013;9(10):e1003653 10.1371/journal.ppat.1003653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belaaouaj A, Kim KS, Shapiro SD. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. 2000;289(5482):1185–8. . [DOI] [PubMed] [Google Scholar]
- 23.Belaaouaj A, McCarthy R, Baumann M, Gao Z, Ley TJ, Abraham SN, et al. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4(5):615–8. . [DOI] [PubMed] [Google Scholar]
- 24.Hirche TO, Benabid R, Deslee G, Gangloff S, Achilefu S, Guenounou M, et al. Neutrophil elastase mediates innate host protection against Pseudomonas aeruginosa. J Immunol. 2008;181(7):4945–54. . [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Boado YS, Espinola M, Bahr S, Belaaouaj A. Neutrophil serine proteinases cleave bacterial flagellin, abrogating its host response-inducing activity. J Immunol. 2004;172(1):509–15. . [DOI] [PubMed] [Google Scholar]
- 26.Chavagnat F, Casey MG, Meyer J. Purification, characterization, gene cloning, sequencing, and overexpression of aminopeptidase N from Streptococcus thermophilus A. Appl Environ Microbiol. 1999;65(7):3001–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2(4):924–32. Epub 2007/04/21. nprot.2007.132 [pii] 10.1038/nprot.2007.132 . [DOI] [PubMed] [Google Scholar]
- 28.Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45(5):1389–406. Epub 2002/09/05. 3106 [pii]. [PMC free article] [PubMed] [Google Scholar]
- 29.Salles C, Creancier L, Claverys JP, Mejean V. The high level streptomycin resistance gene from Streptococcus pneumoniae is a homologue of the ribosomal protein S12 gene from Escherichia coli. Nucleic Acids Res. 1992;20(22):6103 Epub 1992/11/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalia AB, McDonough E, Camilli A. Multiplex genome editing by natural transformation. Proc Natl Acad Sci U S A. 2014;111(24):8937–42. 10.1073/pnas.1406478111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bricker AL, Camilli A. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett. 1999;172(2):131–5. Epub 1999/04/03. S0378-1097(99)00027-0 [pii]. 10.1111/j.1574-6968.1999.tb13460.x . [DOI] [PubMed] [Google Scholar]
- 32.Price KE, Camilli A. Pneumolysin localizes to the cell wall of Streptococcus pneumoniae. J Bacteriol. 2009;191(7):2163–8. Epub 2009/01/27. JB.01489-08 [pii] 10.1128/JB.01489-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–8. . [DOI] [PubMed] [Google Scholar]
- 34.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36(10):1083–91. 10.1002/jms.229 . [DOI] [PubMed] [Google Scholar]
- 35.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–89. 10.1016/1044-0305(94)80016-2 . [DOI] [PubMed] [Google Scholar]
- 36.Shapiro SD. Neutrophil elastase: path clearer, pathogen killer, or just pathologic? Am J Respir Cell Mol Biol. 2002;26(3):266–8. 10.1165/ajrcmb.26.3.f233 . [DOI] [PubMed] [Google Scholar]
- 37.van der Windt D, Bootsma HJ, Burghout P, van der Gaast-de Jongh CE, Hermans PW, van der Flier M. Nonencapsulated Streptococcus pneumoniae resists extracellular human neutrophil elastase- and cathepsin G-mediated killing. FEMS Immunol Med Microbiol. 2012;66(3):445–8. 10.1111/j.1574-695X.2012.01028.x . [DOI] [PubMed] [Google Scholar]
- 38.Hurley BP, Siccardi D, Mrsny RJ, McCormick BA. Polymorphonuclear cell transmigration induced by Pseudomonas aeruginosa requires the eicosanoid hepoxilin A3. J Immunol. 2004;173(9):5712–20. . [DOI] [PubMed] [Google Scholar]
- 39.Bou Ghanem EN, Lee JN, Joma BH, Meydani SN, Leong JM, Panda A. The Alpha-Tocopherol Form of Vitamin E Boosts Elastase Activity of Human PMNs and Their Ability to Kill Streptococcus pneumoniae. Front Cell Infect Microbiol. 2017;7:161 10.3389/fcimb.2017.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalia AB, Standish AJ, Weiser JN. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect Immun. 2010;78(5):2108–16. 10.1128/IAI.01125-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zar JH. Biostatistical Analysis. 5th ed: Pearson; 2010. 2010. [Google Scholar]
- 42.Perez-Dorado I, Galan-Bartual S, Hermoso JA. Pneumococcal surface proteins: when the whole is greater than the sum of its parts. Mol Oral Microbiol. 2012;27(4):221–45. 10.1111/j.2041-1014.2012.00655.x . [DOI] [PubMed] [Google Scholar]
- 43.Plumptre CD, Ogunniyi AD, Paton JC. Surface association of Pht proteins of Streptococcus pneumoniae. Infect Immun. 2013;81(10):3644–51. 10.1128/IAI.00562-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuste J, Sen A, Truedsson L, Jonsson G, Tay LS, Hyams C, et al. Impaired opsonization with C3b and phagocytosis of Streptococcus pneumoniae in sera from subjects with defects in the classical complement pathway. Infect Immun. 2008;76(8):3761–70. Epub 2008/06/11. IAI.00291-08 [pii] 10.1128/IAI.00291-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun. 2010;78(2):704–15. Epub 2009/12/02. IAI.00881-09 [pii] 10.1128/IAI.00881-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersson C, Linder LE, Sund ML, Lonnies H. Purification and characterization of an aminopeptidase from Streptococcus mitis ATCC 903. Curr Microbiol. 1992;25(5):261–7. . [DOI] [PubMed] [Google Scholar]
- 47.Midwinter RG, Pritchard GG. Aminopeptidase N from Streptococcus salivarius subsp. thermophilus NCDO 573: purification and properties. J Appl Bacteriol. 1994;77(3):288–95. . [DOI] [PubMed] [Google Scholar]
- 48.Gonzales T, Robert-Baudouy J. Bacterial aminopeptidases: properties and functions. FEMS Microbiol Rev. 1996;18(4):319–44. 10.1111/j.1574-6976.1996.tb00247.x . [DOI] [PubMed] [Google Scholar]
- 49.Holmes AR, McNab R, Millsap KW, Rohde M, Hammerschmidt S, Mawdsley JL, et al. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol Microbiol. 2001;41(6):1395–408. . [DOI] [PubMed] [Google Scholar]
- 50.Weiser JN, Gotschlich EC. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect Immun. 1991;59(7):2252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodruff WA, Parr TR Jr., Hancock RE, Hanne LF, Nicas TI, Iglewski BH. Expression in Escherichia coli and function of Pseudomonas aeruginosa outer membrane porin protein F. J Bacteriol. 1986;167(2):473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Domon H, Oda M, Maekawa T, Nagai K, Takeda W, Terao Y. Streptococcus pneumoniae disrupts pulmonary immune defence via elastase release following pneumolysin-dependent neutrophil lysis. Sci Rep. 2016;6:38013 10.1038/srep38013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergmann S, Hammerschmidt S. Versatility of pneumococcal surface proteins. Microbiology. 2006;152(Pt 2):295–303. Epub 2006/01/27. 152/2/295 [pii] 10.1099/mic.0.28610-0 . [DOI] [PubMed] [Google Scholar]
- 54.Blevins LK, Parsonage D, Oliver MB, Domzalski E, Swords WE, Alexander-Miller MA. A Novel Function for the Streptococcus pneumoniae Aminopeptidase N: Inhibition of T Cell Effector Function through Regulation of TCR Signaling. Front Immunol. 2017;8:1610 10.3389/fimmu.2017.01610 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CW, protoplast and WCL fractions were isolated from TIGR4 and PepNFLAG cells. The samples analyzed in this experiment are the same as those presented in Fig 3. Samples were analyzed by (A) SDS-PAGE followed by Coomassie Blue staining. B) The band intensity in each lane was quantified using ImageStudioLite software and the data are expressed as fold change relative to the cell wall digestion buffer (CWDB) control lane. Data shown are from one experiment representative of three independent experiments.
(TIFF)
(A) Total protein content from three independent experiments in untreated and NE-treated WCL and CW samples were evaluated via SDS-PAGE and Coomassie blue staining. (B) Data are normalized to a blank lane, expressed as fold change relative to the respective untreated control and were quantified using ImageStudioLite software. Data shown are the means ± SD from three independent experiments. Student’s t-test revealed no significant differences.
(TIFF)
PMNs were isolated from the blood of two healthy donors and 5 x 105 cells were incubated with 103 CFU of WT or ΔpepN cells. Following a 45-minute incubation, neutrophil viability was determined via trypan blue exclusion and enumeration using a haemacytometer. Each sample was enumerated twice and by two independent individuals. Data shown are the means ± SD from two independent experiments with at least two technical replicates per strain per experiment. One-way ANOVA revealed no significant difference in neutrophil viability after exposure to WT or ΔpepN cells.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.