SMYD3 is a member of the SMYD family of lysine methyltransferases (KMTs). In humans, there are five SMYD members (SMYD1–5) that are characterized by the presence of a split catalytic SET domain interrupted by a MYND zinc finger domain. The MYND domain is thought to potentially mediate DNA or protein binding but a clear function is yet to be established. SMYD homologs have been described in a wide range of organisms, such as zebrafish, drosophila and budding yeast (1,2). While methylation activity has been clearly established for SMYD2 and SMYD3 (3–6), the activities of SMYD1, 4 and 5 are not well understood.
A number of groups have focused their attention on SMYD3 in cancer because of its requirement for the growth of tumor cells in culture and its elevated levels in multiple cancer types, including cancers driven by activation of the Ras pathway (7–9). In particular, SMYD3 was shown for the first time to have a direct role in cancer development in vivo in mouse models of lung adenocarcinoma (LAC) and pancreatic ductal adenocarcinoma (PDAC) (4). In exciting new work, Sarris and colleagues also demonstrated an oncogenic role for SMYD3 in mouse models of colorectal carcinoma (CRC) and hepatocellular carcinoma (HCC) (10). Notably, SMYD3 may play some role in early development (11), in heart development (12), and in response to tissue injury (10), but organ-specific and whole-body SMYD3 deletion in mice does not result in any obvious pathology (4,10). These observations suggest that drugs inhibiting SMYD3 will have minimal side effects and therefore may hold great promise as therapeutics in the treatment of cancer.
To develop SMYD3-based therapeutics it is important to first understand the mechanisms of action of this enzyme in normal and cancer cells. SMYD3 was initially described to methylate histone H3 at lysine 4 (H3K4) (8); however subsequent work has shown that SMYD3 has virtually no activity on H3, particularly in a nucleosome context, and mutation of H3K4 to H3K4R has no impact on this trace activity [(4,6) and unpublished observations]. Thus, despite the persistence in the literature, SMYD3 is not an H3K4 methyltransferase. SMYD3 does methylate histone H4 at lysine 5, a non-canonical histone methylation site, and this site serves as a far more effective substrate than H3, particularly in a nucleosome context [Figure 1, (6)]. SMYD3 has been reported to regulate chromatin and transcriptional programs. Specifically, SMYD3 has been shown to directly bind to the regulatory regions of candidate target genes and to change the transcription of these genes [Figure 1, e.g., (7,9,13–15)]; whether this is related to the H4K5 methylation activity has not been determined.
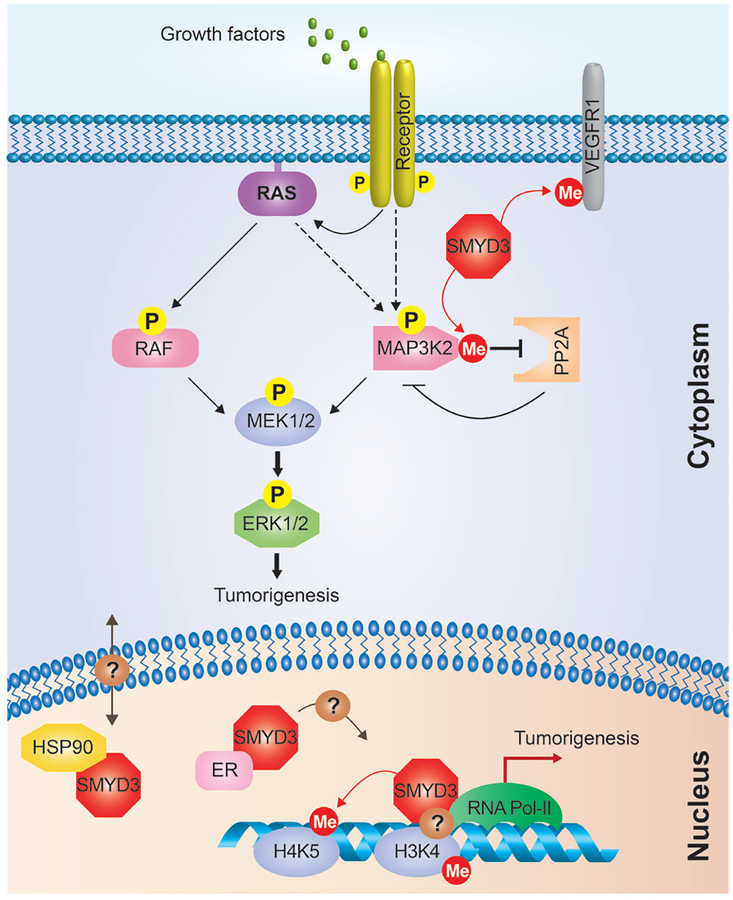
Figure 1.
Nuclear and cytoplasmic functions of SMYD3 in cancer cells. SMYD3 can be localized in the cytoplasm and the nucleus of cancer cells. In the cytoplasm, the methylation of MAP3K2 by SMYD3 inhibits the interaction between MAP3K2 and PP2A, which attenuates the inhibitory effects of this phosphatase on kinases in Ras signaling. SMYD3 may also affect the growth of cancer cells by methylation of VEGFR1. In the nucleus, SMYD3 acts as a transcription facilitator by binding to the DNA and RNA Pol-II, and methylates H4K5, which may affect other histone modifications and chromatin remodeling. SMYD3 interaction with HSP90 may regulate SMYD3 localization within the nucleus. SMYD3 binds to the estrogen receptor (ER) and has also been implicated in the regulation of its transcriptional targets. See text for details.
SMYD3 is primarily a cytoplasmic protein (4,16,17), including being virtually exclusively cytoplasmic in LAC and PDAC cells (4). Thus, it is likely that the primary substrates of SMYD3 reside in the cytoplasm. In this regard, we identified SMYD3 methylation of the cytoplasmic kinase MAP3K2 (also known as MEKK2) in LAC and PDAC cell lines; this methylation event inhibits the binding of the PP2A phosphatase to MAP3K2, which results in increased activity of the canonical RAS signaling pathway [Figure 1, (4)]. Notably, SMYD3 catalytic efficiency is orders of magnitude greater on MAP3K2 than on histones (4,18), supporting an important role for MAP3K2 as target of SMYD3 in PDAC and LAC tumorigenesis. SMYD3 also methylates the cytoplasmic portion of VEGFR1 (16), which may affect signaling downstream of this receptor (Figure 1). Together, these data support a role for the localization of SMYD3 activity in the cytoplasm in promoting the tumorigenic process.
The recent study from Sarris and colleagues adds another layer of complexity to the oncogenic functions of SMYD3 in cells (10). The authors used elegant chemical and genetic approaches to demonstrate that SMYD3 promotes the development of CRC and HCC. In these two cancer models, tumors also have active RAS signaling, which could suggest a role for MAP3K2 methylation by SMYD3. However, the authors did not observe significant differences in the phosphorylation levels of MEK1/2 [which are two of the kinases regulated by MAP3K2 (4)] between wild-type and SMYD3 mutant liver tumors. The authors did not experimentally address the possible role of the SMYD3-MAP3K2 axis regulating other pathways in these tumors, but overall their experiments suggested that the oncogenic activity of SMYD3 in CRC and HCC might be related to MAP3K2-independent events. Indeed, the authors found that SMYD3 is both nuclear and cytoplasmic in CRC and HCC tumors, suggesting that SMYD3 may have a nuclear function in these cancers. Sarris and colleagues observed that SMYD3 is involved in the activation of transcriptional programs related to oncogenic pathways (e.g., Myc, Ctnnb1, Jak1/2), including epithelial-mesenchymal transition (e.g., Snai1, Twist, Zeb1). While these observations would be consistent with a role for SMYD3 methylation of histones or unknown transcriptional regulators, the authors found that overexpression of a catalytically dead (H206G mutation) SMYD3 derivative still activates oncogenic targets such as Myc and Ctnnb1. These data argue for a non-catalytic function for SMYD3 in the nucleus of cancer cells tested by the authors. Since in Sarris et al., the analysis of the catalytic dead SMYD3 was limited to overexpression reporter assays, future experiments such as genetic complementation assays with the catalytic mutant will help more definitively rule out a role for SMYD3 catalytic activity in CRC and HCC.
In Sarris et al., intriguingly, SMYD3 was found to bind a large number of loci in the genome and its pattern of DNA binding is similar to that of RNA Pol-II next to the transcription start site (TSS); moreover, SMYD3 peaks in CHIP-seq analysis coincide with H3K4me3 peaks. Due to these observations, the authors speculated that SMYD3 directly binds to H3K4me3 and RNA Pol-II. The authors confirmed previous observations that ectopic SMYD3 interacts with RNA Pol-II (8), although the exact domain of interaction remains unknown. Preliminary results shown in the article suggest that the C-terminal domain of SMYD3, which promotes an interaction with HSP90 (17), also binds to H3K4me3 peptides. This observation raises the intriguing possibility that a yet to be identified domain in the C-terminus of SMYD3 folds into a novel H3K4me3-reader motif, an idea that will require rigorous biochemical, biophysical and biological testing. In addition to the candidate reader domain function, the authors also reported three possible DNA-binding consensus motifs for SMYD3 beyond the previously identified CCTCCC sequence (8). SMYD3 contains a conserved MYND-type domain, which might mediate either protein-protein interactions or bind to specific DNA sequences. If SMYD3 DNA-interaction is mediated by its MYND domain, and because of its localization within the catalytic SET domain, such DNA interaction could prevent SMYD3 lysine methyltransferase activity. However, no clear mutational analyses have been performed yet to identify the domain responsible for SMYD3 localization at specific gene promoters.
Together, the results presented in Sarris et al. identify a potentially new mechanism of action for SMYD3 in cancer cells beyond its ability to control signaling via lysine methylation. SMYD3 may be recruited to direct transcriptional targets in the nucleus by its interactions with H3K4me3 tails, RNA pol-II, and/or direct binding to DNA, where Sarris and colleagues propose that SMYD3 does not act as a classic transcription factor but rather as a potentiator of transcription at already active chromatin regions (Figure 1). Interestingly, by using SMYD3 transgenic mice, Sarris and colleagues provide the first evidence that SMYD3 overexpression is neither sufficient to promote tumorigenesis by itself nor to accentuate tumorigenesis progression, at least in the two models tested. However, if SMYD3 is indeed a transcription facilitator, one would expect that its overexpression in transgenic mice would lead to overexpression of subset of genes, which was not observed by the authors. These experiments suggest that SMYD3 may require unidentified co-factors for it to potentiate the transcription of its targets.
If SMYD3 directly regulates gene expression, it is not surprising that it can co-localize with RNA Pol-II and H3K4me3. As SMYD3 is not directly responsible of H3K4 methylation, past reports correlating SMYD3 binding at specific gene promoters with presence of H3K4me3 are intriguing (7,14). However, an indirect correlation between SMYD3 and H3K4me3 could explain such observations. Through H4K5 methylation, SMYD3 could promote an open chromatin environment subject to transcription, leading to H3K4me3 indirect deposition by other enzymes (Figure 1). A second hypothesis suggested by Sarris and colleagues is that SMYD3 could bind to euchromatin regions already modified for H3K4me3 and facilitate gene transcription by RNA Pol-II, independently of its lysine methyltransferase activity (Figure 1). Further studies are required to better understand SMYD3 activity in nucleus.
The new work by Sarris and colleagues raises a number of important and interesting questions. First, the mechanisms that control the induction of SMYD3 levels in cancer cells is yet not understood. Second, we still do not know what regulates the cellular localization of SMYD3 in normal and cancer cells, although one possibility is via interaction with the nuclear chaperone HSP90 [Figure 1, (17)]. It would be interesting to force the mislocalization of SMYD3 to the nucleus or the cytoplasm and to determine the effects of this misguided activity on gene expression programs and tumorigenic processes. Third, it is intriguing to imagine how the potential different oncogenic activities of SMYD3 are regulated in different cancer contexts. SMYD3 could promote cancer through regulation of cytoplasmic signaling (methylation of MAP3K2 and VEGFR), chromatin regulation (H4K5 methylation) or transcription facilitation independently of its methyltransferase activity (Figure 1). Current developed small molecules to inhibit SMYD3 target its lysine methyltransferase activity (18–20). But this new study suggests that SMYD3 may have pro-tumorigenic functions independent of its catalytic activity. If confirmed, this would open up the possibility of generating a different set of inhibitors that rather than targeting enzymatic activity suppress SMYD3’s capacity to bind gene promoters and regulate the transcription of oncogenic factors.
The analysis of SMYD3 levels in human cancer and functional studies in cell lines and mouse models all support the idea that SMYD3 plays a pivotal role in promoting a wide-range of tumor types. Because SMYD3 may promote cancer through multiple mechanisms, in the cytoplasm and in the nucleus, through its enzymatic activity and independent of it, it will be important to further characterize SMYD3 under specific physiologic and pathologic conditions. SMYD3 is a promising target in human cancer but more work will help guide use of future SMYD3 inhibitors in the treatment of cancer patients.
Acknowledgements
Funding: This work was supported by the National Institute of Health (R01CA172560 to Julien Sage and Or Gozani, K99CA19781601 to Pawel K. Mazur). Julien Sage is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases. Nicolas Reynoird is the recipient of an ERS-EU RESPIRE2 Marie Skłodowska-Curie Postdoctoral Research Fellowship (MCF 3776–2013).
Footnote
Provenance: This is a Guest Commentary commissioned by Section Editor Lei Huang, MD, MSc, MBBS, PhD candidate [Department of Gastrointestinal Surgery, the First Affiliated Hospital of Anhui Medical University, Hefei, China; German Cancer Research Center (DKFZ), Heidelberg, Germany].
Conflicts of Interest: Or Gozani is a co-founder of EpiCypher, Inc. The other authors have no conflicts of interest to declare.
References
- 1.Green EM, Mas G, Young NL, et al. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat Struct Mol Biol 2012;19:361–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calpena E, Palau F, Espinós C, et al. Evolutionary History of the Smyd Gene Family in Metazoans: A Framework to Identify the Orthologs of Human Smyd Genes in Drosophila and Other Animal Species. PLoS One 2015;10:e0134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang J, Perez-Burgos L, Placek BJ, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006;444:629–32. [DOI] [PubMed] [Google Scholar]
- 4.Mazur PK, Reynoird N, Khatri P, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 2014;510:283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynoird N, Mazur PK, Stellfeld T, et al. Coordination of stress signals by the lysine methyltransferase SMYD2 promotes pancreatic cancer. Genes Dev 2016;30:772–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Aller GS, Reynoird N, Barbash O, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics 2012;7:340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cock-Rada AM, Medjkane S, Janski N, et al. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res 2012;72:810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamamoto R, Furukawa Y, Morita M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol 2004;6:731–40. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Zhu MX, Zhang XD, et al. SMYD3 stimulates EZR and LOXL2 transcription to enhance proliferation, migration, and invasion in esophageal squamous cell carcinoma. Hum Pathol 2016;52:153–63. [DOI] [PubMed] [Google Scholar]
- 10.Sarris ME, Moulos P, Haroniti A, et al. Smyd3 Is a Transcriptional Potentiator of Multiple Cancer-Promoting Genes and Required for Liver and Colon Cancer Development. Cancer Cell 2016;29:354–66. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki S, Nozawa Y, Tsukamoto S, et al. Histone methyltransferase Smyd3 regulates early embryonic lineage commitment in mice. Reproduction 2015;150:21–30. [DOI] [PubMed] [Google Scholar]
- 12.Fujii T, Tsunesumi S, Yamaguchi K, et al. Smyd3 is required for the development of cardiac and skeletal muscle in zebrafish. PLoS One 2011;6:e23491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Heo K, Kim JH, et al. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J Biol Chem 2009;284:19867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Fang X, Ge Z, et al. The telomerase reverse transcriptase (hTERT) gene is a direct target of the histone methyltransferase SMYD3. Cancer Res 2007;67:2626–31. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Wang C, Wang K, et al. SMYD3 as an oncogenic driver in prostate cancer by stimulation of androgen receptor transcription. J Natl Cancer Inst 2013;105:1719–28. [DOI] [PubMed] [Google Scholar]
- 16.Kunizaki M, Hamamoto R, Silva FP, et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res 2007;67:10759–65. [DOI] [PubMed] [Google Scholar]
- 17.Brown MA, Foreman K, Harriss J, et al. C-terminal domain of SMYD3 serves as a unique HSP90-regulated motif in oncogenesis. Oncotarget 2015;6:4005–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Aller GS, Graves AP, Elkins PA, et al. Structure-Based Design of a Novel SMYD3 Inhibitor that Bridges the SAM-and MEKK2-Binding Pockets. Structure 2016;24:774–81. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell LH, Boriack-Sjodin PA, Smith S, et al. Novel Oxindole Sulfonamides and Sulfamides: EPZ031686, the First Orally Bioavailable Small Molecule SMYD3 Inhibitor. ACS Med Chem Lett 2015;7:134–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu W, Liu N, Qiao Q, et al. Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase. J Biol Chem 2016;291:9173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]