Abstract
Introduction:
CHD1 has been identified as a tumor suppressor gene in prostate cancer. Previous studies have shown strong associations between CHD1 deletion, prostate specific antigen [PSA] recurrence, and absence of ERG fusion. In this preliminary study we seek to find whether there is an independent correlation between CHD1 status and response to androgen deprivation therapy[ADT].
Materials and methods:
We identified 11 patients with prostate cancer who underwent prostatectomy and received at least 7 months of ADT at our institution. They were divided into undetectable [PSA < 0.2 ng/mL; n = 8] and detectable [PSA > 0.2 ng/mL; n = 3] according to their serum PSA nadir after 7 months of ADT. Tissue microarray was generated from their formalin-fixed paraffin-embedded prostatectomy and involved lymph node tissues. Fluorescence in situ hybridization [FISH] analysis for CHD1 and immunohistochemical stains for PSA, AR, PTEN, ERG and SPINK1 were performed.
Results:
Our results showed heterogeneity of FISH and immunostains expressions in different foci of tumor. Status of CHD1, ERG, PTEN, or SPINK1 did not correlate with one another or with response to ADT.
Conclusions:
Additional larger studies may be needed to further elucidate trends between these biomarkers and clinical outcomes in prostate cancer patients.
Keywords: Prostate cancer, CHD1, Androgen deprivation therapy, Genetics
INTRODUCTION
Prostate cancer remains the most commonly diagnosed non-cutaneous malignancy and the third most lethal malignancy in men with 161,360 new cases and 26,730 deaths expected in 2017 alone [1]. One of the significant challenges in the management of prostate cancer is understanding which factors portend a poor prognosis and require aggressive treatment [2,3].
Patient characteristics, cancer stage, Gleason score, and prostate specific antigen [PSA] level are currently used to risk stratify patients and inform the therapeutic recommendations of urologic, medical, and radiation oncologists. At this time, it is unclear how best to predict the natural history of these cancers. Therefore, many research efforts have focused on elucidating the molecular and genetic aberrancies underlying the aggressiveness of prostate cancer in order to help predict disease trajectory and guide treatment [2–5].
The chromosomal band 5q21 is a commonly deleted segment in prostate cancer. Several studies have localized CHD1, which encodes chromodomain helicase DNA-binding protein 1, to this region [2–4]. This deletion is estimated to underlie 10–26% of all prostate cancers [2,4]. CHD1 has been identified as a key tumor suppressor gene whose role involves chromatin remodeling [24], androgen receptor [AR]-dependent transcription regulation [2], recruiting homologous recombination repair proteins to double strand DNA breaks [6], and promoting cell invasiveness [3]. CHD1 may also be important in predicting sensitivity to androgen deprivation therapy [ADT] [7].
A variety of additional genetic alterations driving prostate cancer have been analyzed, although it is unclear if they represent distinct molecular subtypes of prostate cancer. Transcriptome analysis has revealed several of these gene modifications including tumor suppressor deletions, namely PTEN, RB1, and TP53, androgen receptor amplification and rearrangements, RTK-Ras-MAPK pathway aberrations such as SPINK1 overexpression [5,8], and ETS family transcription factor rearrangements, such as TMPRSS2:ERG fusion [3,5,9]. Interestingly, previous studies have shown strong associations between CHD1 deletion, prostate specific antigen [PSA] recurrence, and absence of ERG fusion, suggesting that CHD1 may play a role in ERG rearrangements [2,9,10]. In order to further define previously described, but partially understood, molecular subtypes of prostate cancer, this pilot study examines whether there is an independent correlation between CHD1, PTEN, ERG, and SPINK1 status and response to ADT.
MATERIALS AND METHODS
Patient classification
Nadir PSA after androgen deprivation has been shown to predict overall survival from metastatic prostate cancer [11]. For this IRB-approved pilot study, we retrospectively identified 11 patients with prostate cancer who underwent radical retropubic prostatectomy, subsequently developed advanced disease or a biochemical recurrence, and had received at least 7 months of ADT at our institution. We dichotomized the responses of this post-prostatectomy cohort into undetectable [PSA <0.2 ng/mL; n=8] and detectable [PSA >0.2 ng/mL; n=3] according to their serum PSA nadir after 7 months of ADT [11,12].
Tissue microarray [TMA]
TMA was generated from their formalin-fixed paraffin-embedded [FFPE] from the radical prostatectomy and lymphadenectomy [when available] specimens. The TMA includes normal prostatic glands and stroma, major tumor foci [up to 3 foci], high-grade prostatic intraepithelial neoplasia [HGPIN], intraductal carcinoma, and metastatic tumor from the lymph node.
Immunohistochemistry
Immunohistochemical stains for PSA, AR, PTEN, ERG and SPINK1 were performed on the TMA. To confirm gene expression in prostate cancer tissue samples, any significant staining [1+ to 3+ intensity] was considered positive for this study. The assessment for loss of PTEN was based on previously reported methods previously published by Lotan et al., which has been shown to be highly concordant to PTEN deletion by FISH [13]. The antibodies used included BOND prediluted Prostate Specific Antigen [35H9] mouse monoclonal antibody, Cell Marque prediluted Androgen Receptor [SP107] rabbit monoclonal antibody, Cell Signaling PTEN [D4.3] XP rabbit monoclonal antibody at 1:50 dilution, DAKO ERG [M7314] rabbit monoclonal antibody at 1:100 dilution, and Novusbio SPINK1 [4D4] mouse monoclonal antibody at 1:250 dilution.
Fluorescence in situ hybridization [FISH]
Fluorescence in situ hybridization [FISH] analysis for CHD1 and ERG rearrangements was performed using TMA. ERG gene rearrangement was assessed using break-apart fluorescence in situ hybridization assay as described previously [9,14,15]. Bacterial artificial chromosomes [BACs] were obtained from the BACPAC Resource Center [Oakland, CA, USA]. For detection of ERG rearrangement and TMPRSS2-ERG fusion, we used the following probes: RP11–95I21 [5’ to ERG] and RP11–476D17 [3’ to ERG] and RP11–35C4 [5’ to TMPRSS2]. We utilized the ARES™ Alexa Fluor® DNA Labeling Kit, which provides a two-step method DNA with Alexa Fluor® 555,488 and 647 dyes, respectively. For detection of CHD1 deletion we utilized a gene-specific DNA probe [RP11–432N19] and reporter probe that corresponded to pericentromeric sequence of Chr5q21 locus [RP11–929P16], Alexa Fluor® 647 and 555- labeled]. Gene mutation status was evaluated by counting spots for each probe in 50–100 non- overlapping, intact, interphase nuclei in which both chromosomes were identified.
Confocal microscopy
Labeled samples were scanned using a confocal microscope [Zeiss LSM 510 META, 100× objective]. Image stacks of 300 nm z-step size were captured and analyzed using Imaris Software [Bitplane]. Gene rearrangement and copy number were evaluated by scoring spots in morphologically intact, non-overlapping interphase nuclei as described previously.
Analysis of results
Fisher’s exact test was performed to test the association between biomarker loss and response to ADT.
RESULTS
PSA and tumor characteristics
In the 11 patients in this pilot study, the PSA at diagnosis ranged from 4 to 25.7 ng/mL with an average of 12.7 ng/mL. Gleason score on initial biopsy ranged from 6 to 9, whereas Gleason score after prostatectomy ranged from 7 to 9, as a result of upgrading in 3 patients and of downgrading in 2 patients. PSA prior to ADT initiation ranged from 0.14 to 24.55 ng/mL, and after 7 months of therapy, all patients achieved a decrease in PSA [Table 1]. Nine out of 11 patients had at least one positive margin identified in their prostatectomy tissue; no correlation was identified between margin status and response to ADT.
Table 1:
PSA, tumor, and ADT characteristics.
| Patient Number | PSA at Diagnosis [ng/mL] | Highest Gleason Score on Prostate Biopsy | Highest Gleason Score on Prostatectomy | Tumor Stage at ADT Initiation | PSA Prior to ADT [ng/mL] | PSA After Completion of >7 months of ADT [ng/mL] | Response | Hormone Therapy Type |
|---|---|---|---|---|---|---|---|---|
| 1 | 12.7 | 7 [3+4] | 7 [3+4] | pT3bNlMx | 0.14 | <0.01 | Undetectable | Leuprolide |
| 2 | 15 | 6 [3+3] | 7[3+4] | pT3bNxMx | 4.8 | 0.2 | Detectable | Leuprolide |
| 3 | 12 | unknown | 7 [3+4] | pT3aN0Ml | 1.3 | <0.1 | Undetectable | Leuprolide |
| 4 | 6 | 7 [3+4] | 7 [3+4 with tertiary 5] | pT3aNxMx | unknown | <0.01 | Undetectable | Degarelix |
| 5 | 5.1 | 8 [4+4] and 8 [3+5] | 9 [4+5] | pT3bN0Mx | 21.62 | <0.01 | Undetectable | Degarelix |
| 6 | 16.4 | 9 [4+5] | 8 [3+5] | pT3bNlMl | 12.3 | 0.6 | Detectable | Goserelin |
| 7 | 21 | 8 [4+4] | 8 [4+4] | pT2cNlMx | 1.03 | <0.01 | Undetectable | Leuprolide |
| 8 | 12 | 9 [4+5] | 8 [4+4] | pT2cNxMx | 21.4 | 0.24 | Detectable | Leuprolide |
| 9 | 4 | 6 [3+3] | 9 [4+5] | pT3aNlMx | 3.76 | <0.1 | Undetectable | Leuprolide |
| 10 | 4 | 7 [3+4] | 7 [3+4] | pT3aNxMx | 7.71 | <0.1 | Undetectable | Degarelix |
| 11 | 25.7 | 9 [4+5] | 9 [4+5] | pT3aNlMl | 24.55 | <0.01 | Undetectable | Leuprolide |
Abbreviations: PSA: prostate specific antigen; ADT: androgen deprivation therapy
FISH and immunostaining
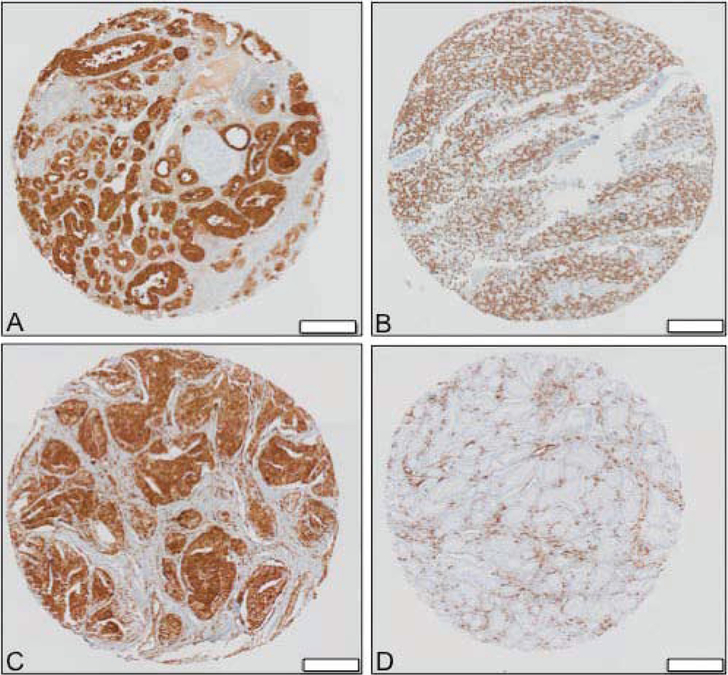
There was heterogeneity of FISH and marker detection by immunohistochemistry in different foci of the tumors. PSA was expressed in all normal tissue, high-grade prostatic intraepithelial neoplasia [HGPIN], and lymph node tumors; it was also expressed in all intraductal carcinoma and invasive foci, with the exception of one case [Figure 1A]. AR was expressed in all tissues except for one invasive focus in a single case [Figure 1B].
Figure 1.
Immunohistochemistry demonstrating [A] Positive PSA expression, [B] Positive AR expression, [C] Intact PTEN expression, and [D] PTEN loss in TMA analysis of prostate cancer specimens; white bar= 200 μg.
PTEN was expressed in all normal tissue and lymph node tumors [Figure 1C]. PTEN loss was seen in 1 of 3 HGPIN specimens, 2 out of 3 intraductal carcinomas, and 2 out of 25 invasive foci specimens. In the 2 specimens where PTEN loss is seen in intraductal component, PTEN loss is not seen in all invasive foci [Figure 1D]
SPINK1 was expressed in 1 of 3 HGPIN specimens and 2 of 25 invasive foci specimens. It was not expressed in any normal, intraductal, or lymph node tumor specimens.
ERG expression was not expressed in any normal tissue or HGPIN specimens. ERG was expressed in 2 out of 3 intraductal carcinomas, 6 out of 25 invasive foci, and 1 out of 4 lymph node tumors.
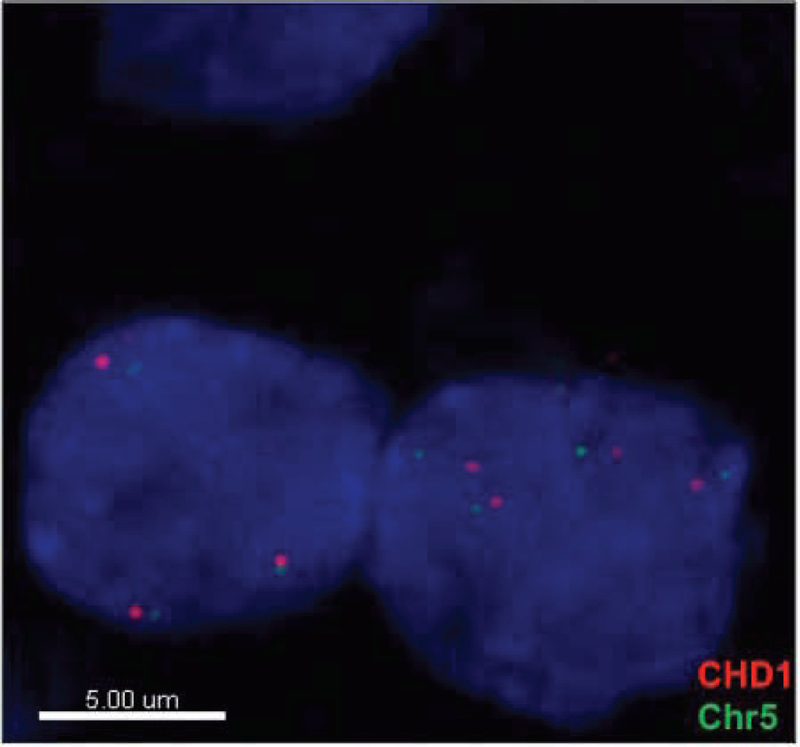
CHD1 deletion was detected in 11 out of 25 invasive foci and 1 out of 4 lymph node tumors. Copy number gain [CNG] was detected in 2 out of 25 invasive foci and 1 out of 4 lymph node tumors. All remaining tissues had no detectable copy number changes in CHD1 [Figure 2, Table 2].
Figure 2.
Representative sample of FISH demonstrating copy number variation [CNG] of CHD1.
Table 2:
Biomarkers in tumor tissue samples.
| Case | Prostate Tissue | PSA | AR | PTEN [√=LOSS] | ERG | SPINK1 | CHD1 [√=DELETED] | Response to ADT |
|---|---|---|---|---|---|---|---|---|
| 1 | Normal | √ | √ | 0 | 0 | 0 | 0 | Undetectable |
| Intraductal | √ | √ | √ | √ | 0 | 0 | ||
| Invasive focus1 | √ | √ | √ | √ | 0 | 0 | ||
| Invasive focus2 | √ | √ | 0 | √ | 0 | 0 | ||
| L.N. Tumor | √ | √ | 0 | √ | 0 | 0 | ||
| 2 | Normal | √ | √ | 0 | 0 | 0 | 0 | Detectable |
| HGPIN | √ | √ | 0 | 0 | 0 | 0 | ||
| Invasive focus1 | √ | √ | 0 | 0 | 0 | 0 | ||
| Invasive focus2 | √ | √ | 0 | 0 | 0 | 0 | ||
| Invasive focus3 | √ | √ | 0 | 0 | 0 | CNG | ||
| 3 | Normal | √ | √ | 0 | 0 | 0 | 0 | Undetectable |
| Intraductal | √ | √ | √ | √ | 0 | 0 | ||
| Invasive focus1 | √ | √ | 0 | √ | 0 | √ | ||
| Invasive focus2 | √ | √ | 0 | √ | 0 | 0 | ||
| Invasive focus3 | √ | √ | 0 | √ | 0 | 0 | ||
| 4 | Normal | √ | √ | 0 | 0 | 0 | 0 | Undetectable |
| HGPIN | √ | √ | √ | 0 | √ | 0 | ||
| Invasive focus1 | √ | √ | √ | 0 | 0 | 0 | ||
| 5 | Normal | √ | √ | 0 | 0 | 0 | 0 | Undetectable |
| Intraductal | 0 | √ | 0 | 0 | 0 | 0 | ||
| Invasive focus1 | 0 | √ | 0 | 0 | √ | 0 | ||
| Invasive focus2 | 0 | √ | 0 | 0 | 0 | 0 | ||
| 6 | Normal | √ | √ | 0 | 0 | 0 | 0 | Detectable |
| Invasive focus1 | √ | √ | 0 | 0 | 0 | 0 | ||
| Invasive focus2 | √ | √ | 0 | 0 | 0 | 0 | ||
| Invasive focus3 | √ | √ | 0 | 0 | 0 | 0 | ||
| L.N. Tumor | √ | √ | 0 | 0 | 0 | 0 | ||
| 7 | Normal | √ | √ | 0 | 0 | 0 | 0 | Undetectable |
| Invasive focus1 | √ | √ | 0 | 0 | 0 | CNG | ||
| Invasive focus2 | √ | √ | √ | 0 | √ | 0 | ||
| L.N. Tumor | √ | √ | 0 | 0 | 0 | CNG | ||
| 8 | Normal | √ | √ | 0 | 0 | 0 | 0 | Detectable |
| Invasive focus1 | √ | 0 | 0 | 0 | 0 | √ | ||
| Invasive focus2 | √ | √ | 0 | 0 | 0 | √ | ||
| 9 | Normal | √ | √ | 0 | 0 | 0 | 0 | Undetectable |
| Invasive focus1 | √ | √ | 0 | 0 | 0 | √ | ||
| Invasive focus2 | √ | √ | 0 | 0 | 0 | √ | ||
| Invasive focus3 | √ | √ | 0 | 0 | 0 | √ | ||
| 10 | Normal | √ | √ | 0 | 0 | 0 | 0 | Undetectable |
| Invasive focus1 | √ | √ | 0 | √ | 0 | √ | ||
| Invasive focus2 | √ | √ | 0 | 0 | 0 | √ | ||
| 11 | Normal | √ | √ | 0 | 0 | 0 | 0 | Undetectable |
| HGPIN | √ | √ | 0 | 0 | 0 | 0 | ||
| Invasive focus1 | √ | √ | 0 | 0 | 0 | √ | ||
| Invasive focus2 | √ | √ | 0 | 0 | 0 | √ | ||
| Invasive focus3 | √ | √ | 0 | 0 | 0 | √ |
Abbreviations: PSA: prostate specific antigen; AR: androgen receptor; ADT: androgen deprivation therapy; HGPIN: high grade prostate intraepithelial neoplasia
Biomarker correlation
No statistically significant correlations between CHD1, PTEN, ERG, and SPINK1 status were observed across prostate tissues [Table 3].
Table 3:
Biomarkers in tumor tissue samples.
| A | ERG | Fisher’s Exact Test | ||
| PTEN | 0 [intact] | 1 [loss] | p = 0.491 | |
| Intact [0] | 6 [54.6%] | 1 [9.1%] | ||
| Loss [1] | 2 [18.2%] | 2 [18.2%] | ||
| B | SPINK1 | |||
| PTEN | 0 [intact] | 1 [loss] | p = 0.491 | |
| Intact [0] | 6 [54.6%] | 1 [9.1%] | ||
| Loss [1] | 2 [18.2%] | 2 [18.2%] | ||
| C | CHD1 | |||
| PTEN | 0 [intact] | 1 [loss] | p = 0.546 | |
| Intact [0] | 3 [27.3%] | 4 [36.4%] | ||
| Loss [1] | 3 [27.3%] | 1 [9.1%] | ||
| D | SPINK1 | |||
| ERG | 0 [intact] | 1 [loss] | p = 0.491 | |
| Intact [0] | 5 [45.4%] | 3 [27.3%] | ||
| Loss [1] | 3 [27.3%] | 0 [0%] | ||
| E | CHD1 | |||
| ERG | 0 [intact] | 1 [loss] | p = 0.546 | |
| Intact [0] | 5 [45.4%] | 3 [27.3%] | ||
| Loss [1] | 1 [9.1%] | 2 [18.2%] | ||
| F | CHD1 | |||
| SPINK1 | 0 [intact] | 1 [loss] | p = 0.182 | |
| Intact [0] | 3 [27.3%] | 5 [45.4%] | ||
| Loss [1] | 3 [27.3%] | 0 [0%] | ||
Molecular subtype correlation with ADT Response after 7 Months
There were no statistically significant correlations between response to ADT after 7 months and PSA expression [p = 1], AR expression [p = 0.273], PTEN loss [p = 0.236], ERG overexpression [p = 0.491], SPINK overexpression [p = 0.491], and CHD1 status [p = 1] [Table 4].
Table 4:
Biomarkers and response to ADT.
| Biomarker | Response to ADT for 7 Months | Fisher’s exact test | ||
|---|---|---|---|---|
| Undetectable [N, %] | Detectable [N, %] | |||
| PSA | ||||
| Intact [0] | 1 [100] | 0 [0] | p = l | |
| Loss [1] | 7 [70] | 3 [30] | ||
| AR | ||||
| Intact [0] | 0 [0] | 1 [100] | p = 0.273 | |
| Loss [1] | 8 [80] | 2 [20] | ||
| PTEN | ||||
| Intact [0] | 4 [54.1] | 3 [42.9] | p = 0.236 | |
| Loss [1] | 4 [100] | 0 [0] | ||
| ERG | ||||
| Intact [0] | 5 [62.5] | 3 [37.5] | p = 0.491 | |
| Loss [1] | 3 [100] | 0 [0] | ||
| SPINK1 | ||||
| Intact [0] | 5 [62.5] | 3 [37.5] | p = 0.491 | |
| Loss [1] | 3 [100] | 0 [0] | ||
| CHD1 | ||||
| Intact [0] | 4 [66.7] | 2 [33.3] | p = l | |
| Loss [1] | 4 [80] | 1 [20] | ||
Abbreviations: ADT: androgen deprivation therapy; PSA: prostate specific antigen; AR: androgen receptor
DISCUSSION
In this era of personalized medicine, many research efforts in oncology have focused on creating targeted therapies based off of tumor genetics. In this pilot study, we analyzed the correlation between a variety of genetic alterations, including CHD1 deletions, with response to ADT after at least 7 months in men with prostate cancer. Using data from the Southwest Oncology Group Trial 9346, Hussain et al., demonstrated PSA after 7 months of ADT to be asurrogate for survival in patients with metastatic prostate cancer, with 4 ng/mL or less correlating with improved survival. When PSA is 0.2 ng/mL or less, risk of death was calculated to be one-fifth of that for those with a PSA greater than 4 ng/mL [11]. More recently Harshman et al., performed a retrospective survival analysis, which included 719 men randomized to receive either ADT and docetaxel or ADT alone. They report that PSA less than or equal to 0.2 ng/mL at 7 months after ADT, irrespective of docetaxel use, correlates with improved overall survival [16]. While we did not find any significant correlation between genetic status and response to ADT at 7 months, subsequent studies should evaluate long term survival in these patients to further validate this predictor of survival.
CHD1 deletions occur in a major subset of prostate cancers, but the clinical outcomes of patients with this molecular subtype are poorly defined. In our exploratory study, there was no significant association between response to ADT and CHD1 status. Unlike our results, Stein et al. observed a statistically insignificant but positive association demonstrating tumor sensitivity to ADT and AT-101, a small molecule Bcl-2 inhibitor, for CHD negative patients [7]. Moreover, findings by Kari et al. suggest that CHD1 is involved in recruitment of homologous recombination repair proteins to double stranded DNA breaks. Homologous repair in CHD1 negative prostate tissue is not optimal, leading to genomic instability and tumorigenesis [6,17]. These results indicate that PARP [poly ADP ribose polymerase] inhibitors, which target homologous repair proteins, may be an effective personalized therapeutic option for patients with CHD1 negative tumors [6]. In addition to correlating CHD1 status with response to first- and second-line ADT and other chemotherapies, futures studies should be directed at outlining natural disease progression of CHD1 negative tumors as well as clinical prognosis [18,19]. Furthermore, Huang et al., used comparative genomic hybridization to demonstrate that deletion is the most common molecular alteration in CHD1 in prostate cancer tissue samples [3]. In our study CHD1 deletion was the most common alteration observed, but we also detected CHD1 copy number gain in samples from two patients. While most research links CHD1 deletion with aggressive, castrate-resistant prostate cancers, it is possible that different CHD1 genetic alterations may play distinctive roles in other, less invasive molecular subtypes of prostate cancer. Future directions should be aimed at evaluating how these genetic alterations in CHD1 affect tumorigenesis and correspond to disease progression.
Gene rearrangements of the ETS transcription factor family are estimated to be present in 40–50% of all prostate cancers [20,21]. ERG mutations, specifically fusion between ERG and TMPRSS2, are some of the most common rearrangements involving the ETS family. When TMPRSS2:ERG fusion occurs, the ERG oncogene is overexpressed [ERG positive], leading to the over expression of androgen-driven genes and tumorigenesis. Our study did not show any statistically significant relationship between CHD1 status and ERG rearrangements. In contrast to our results, several prior studies have independently shown a negative association between CHD1 deletions and TMPRSS2: ERG fusion, indicating that absence of CHD1 may prevent ERG rearrangements [2,9,22]. Moreover, we did not find a significant link between ERG status and response to ADT. However, one study examining the effects of ADT on various molecular subtypes of prostate tumors showed that in TMPRSS2: ERG positive tumors, certain genes, namely CHGA, are upregulated after ADT. Such genetic changes may lead to neuroendocrine differentiation and progression to castrate-resistant prostate cancer. Therefore, the role of ADT in this molecular subset of patients is unclear [23].
Unlike CHD1, many groups have looked at clinical outcomes in the setting of ERG rearrangements. In a retrospective analysis by Berg et al., patients with ERG positive tumors were more likely than those with ERG negative tumors to progress on active surveillance within 2 years [58.6% [95% confidence interval (CI), 48.7–68.5] vs. 21.7% [95% CI, 14.3–29.1]]. They defined progression as stage >cT2b, PSA doubling time <3 years, Gleason score upgrade, or greater than 3 or bilaterally positive cores. Overall ERG positive tumors were identified as a predictive factor for progression from active surveillance [hazard ratio (HR), 2.45; 95% CI, 1.62–3.72; p < 0.0001] [25]. Furthermore, mouse models have demonstrated increased bone, limb, and spine metastases in individuals with TMPRSS2:ERG fusion, which interestingly correlate with sites of the most common prostate metastases in humans [26].
In contrast, a prospective study looking to correlate ERG status with clinical outcomes reported that after a median follow-up of 12.6 years no correlation was demonstrated between ERG over expression and lethal prostate cancer [HR, 0.93; 95% CI, 0.611.43]. In this case, lethal prostate cancer was defined as distant metastases or prostate cancer-specific mortality. There was also no association between ERG status and risk of biochemical recurrence [HR, 0.99; 95% CI, 0.78–1.26] [21]. Corroborating these findings, Johnson et al. proposed no correlation between ERG positive tumors and prostate cancer specific mortality in tumors untreated with any ADT or pharmacologic therapies [5].
Because of the prevalence of ETS-family fusions, some have suggested classifying tumors into “ETS fusion” and “ETS non-fusion” type cancers [27]. However, the aforementioned studies highlight the uncertain prognostic nature and clinical significance of ERG status in prostate cancer. To address these inconsistencies, Tsourlakis et al., analyzed all 1592 tumor blocks in 125 prostate cancer patients and found 89% ERG heterogeneity within tumors. The data suggests that although ERG fusions are extremely common, their expression varies, which may reflect clinical outcomes [28]. These findings underscore the need for randomized clinical trials to determine any definitive links between ETS-family fusion and response to various therapies, including ADT.
SPINK1, or serine peptidase inhibitor kazal type 1, is a protein in the RTK-Ras-MAPK pathway, which may facilitate cellular invasion via epidermal growth factor when expressed in high levels. Approximately 10% of TMPRSS2:ERG negative tumors overexpress SPINK1 [23]. Despite these previously described correlations, our results failed to show any significant association between SPINK1 and CHD1 or ERG status. A study by Johnson et al. reported that when compared to triple negative tumors [i.e., ERG negative, ETS negative, and SPINK1 negative], SPINK1 overexpression was predictive of prostate cancer specific mortality, as indicated by biochemical recurrence [HR 3; 95% CI, 1.1–8] and distant metastases [HR 2.48; 95% CI, 0.96–6.4]. An important consideration is that the tissue samples in this report had not been treated with any form of ADT or pharmacologic therapy, unlike those in our study, underscoring the need for larger studies to elaborate upon our results [5]. We also found no correlation between SPINK1 and response to ADT. Prior work concerning the effects of SPINK1 overexpression on response to ADT is largely indeterminate as well [23].
Deletion of phosphatase and tensin homolog [PTEN], a tumor suppressor gene, has been associated with castrate-resistant metastatic prostate cancer and other markers of poorer prognosis, such as higher Gleason score and seminal vesicle infiltration [29]. PTEN deletions have been reported to underlie approximately 50% of castrate resistant prostate cancers [30]. Our results support these prior studies, as the patients in 3 out of the 4 samples with PTEN aberrations had either metastatic or nodal disease. PTEN deletion has a high correlation with TMPRSS2: ERG rearrangements, and when found together, correlate with shorter progression-free survival in prostate cancer patients post-prostatectomy [31]. To the contrary, our findings did not identify any significant association between PTEN deletion and the status of other biomarkers.
PTEN deletions have been found more frequently in intraductal carcinomas [IDC] than other aggressive prostate cancers [32]. IDC in localized prostate cancer is suggestive for development of high grade and advanced stage disease [33]. IDC on surgical pathology in patients with a Gleason score of 6 has been shown to be prognostic for aggressive and invasive cancer, despite the usual dogma that Gleason 6 disease is unlikely to progress [34–36]. Furthermore, patients with ICDs have been shown to be poor responders to ADT [37]. In our study, we did observe two specimens where PTEN deletion was observed in an intraductal component but not in accompanying invasive foci. However, these results did not correspond with response to ADT.
There are several limitations to our pilot study. Most notably, this study was exploratory in character with the goal of establishing proof of principle. With a preliminary sample size of 11 patients, it is certainly possible that significant correlations may be revealed if our study was larger. In addition, prostatectomies were performed at various locations without standardization of technique or performance of lymph node dissection. In terms of our pathologic analysis, our AR staining was unable to be stratified by percentage and staining intensity because our sample size was limited. Additionally, the punch cores used for our TMA are small and are subject to sampling error. Lastly, our TMA was created using radical prostatectomy specimens, and CHD1 expression could vary between primary and metastatic tumors. To the best of our knowledge, our study is the only one to specifically ascertain associations between CHD1 status and response to ADT after 7 months. Our findings highlight the need to design future studies to determine how alterations in the prostate cancer genome allow for personalized therapeutics and influence disease prognosis. Ultimately, future directions may include analyzing the correlation between genomic alterations and RNA expression levels to gain more insight into the molecular underpinnings behind various prostate cancer subtypes.
CONCLUSION
The natural disease history of prostate cancer is heterogeneous, but recently many research efforts have concentrated on defining tumor molecular profiles to better personalize treatment plans. Our study does not demonstrate any correlation between CHD1 status and response to ADT after at least 7 months of therapy. We also did not find any significant association in co-occurrence between tumor markers. Additional studies are required to delineate how CHD1 status, as well as other potential biomarkers, influence response to ADT.
ACKNOWLEDGEMENTS
This work is supported by funding from the National Cancer Institute [P30CA072720], C.R. Bard Foundation, and the Walter and Louise Sutcliffe Foundation.
ABBREVIATIONS
- PSA
Prostate Specific Antigen
- AR
Androgen Receptor
- ADT
Androgen Deprivation Therapy
- FISH
Fluorescence in Situ Hybridization
- TMA
Tissue Microarray
- HGPIN
High-Grade Prostatic Intraepithelial Neoplasia
REFERENCES
- 1.National Cancer Institute Surveillance E, and End Results Program. Cancer Stat Facts: Prostate Cancer: National Cancer Institute Surveillance, Epidemiology, and End Results Program; 2017. [Google Scholar]
- 2.Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013; 73: 2795–2805. [DOI] [PubMed] [Google Scholar]
- 3.Huang S, Gulzar ZG, Salari K, Lapointe J, Brooks JD, Pollack JR. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene. 2012; 31: 4164–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri CE, Bangma CH, Bjartell A, Catto JW, Culig Z, Gronberg H, et al. The mutational landscape of prostate cancer. Eur Urol. 2013; 64: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson MH, Ross AE, Alshalalfa M, Erho N, Yousefi K, Glavaris S, et al. SPINK1 Defines a Molecular Subtype of Prostate Cancer in Men with More Rapid Progression in an at Risk, Natural History Radical Prostatectomy Cohort. J Urol. 2016; 196: 1436–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kari V, Mansour WY, Raul SK, Baumgart SJ, Mund A, Grade M, et al. Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 2016; 17: 1609–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein MN, Hussain M, Stadler WM, Liu G, Tereshchenko IV, Goodin S, et al. A Phase II Study of AT-101 to Overcome Bcl-2--Mediated Resistance to Androgen Deprivation Therapy in Patients With Newly Diagnosed Castration-Sensitive Metastatic Prostate Cancer. Clin Genitourin Cancer. 2016; 14: 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yadav SS, Li J, Lavery HJ, Yadav KK, Tewari AK. Next-generation sequencing technology in prostate cancer diagnosis, prognosis, and personalized treatment. Urol Oncol. 2015; 33: 267. [DOI] [PubMed] [Google Scholar]
- 9.Tereshchenko IV, Zhong H, Chekmareva MA, Kane-Goldsmith N, Santanam U, Petrosky W, et al. ERG and CHD1 heterogeneity in prostate cancer: use of confocal microscopy in assessment of microscopic foci. Prostate. 2014; 74: 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012; 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussain M, Tangen CM, Higano C, Schelhammer PF, Faulkner J, Crawford ED, et al. Absolute prostate-specific antigen value after androgen deprivation is a strong independent predictor of survival in new metastatic prostate cancer: data from Southwest Oncology Group Trial 9346 [INT-0162]. J Clin Oncol. 2006; 24: 3984–3890. [DOI] [PubMed] [Google Scholar]
- 12.Cookson MS, Aus G, Burnett AL, Canby-Hagino ED, D’Amico AV, Dmochowski RR, et al. Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: the American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel report and recommendations for a standard in the reporting of surgical outcomes. J Urol. 2007; 177: 540545. [DOI] [PubMed] [Google Scholar]
- 13.Lotan TL, Heumann A, Rico SD, Hicks J, Lecksell K, Koop C, et al. PTEN loss detection in prostate cancer: comparison of PTEN immunohistochemistry and PTEN FISH in a large retrospective prostatectomy cohort. Oncotarget. 2017; 8: 65566–65576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007; 20: 538–544. [DOI] [PubMed] [Google Scholar]
- 15.Clark J, Attard G, Jhavar S, Flohr P, Reid A, De-Bono J, et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2008; 27: 1993–2003. [DOI] [PubMed] [Google Scholar]
- 16.Harshman LC, Chen YH, Liu G, Carducci MA, Jarrard D, Dreicer R, et al. Seven-Month Prostate-Specific Antigen Is Prognostic in Metastatic Hormone-Sensitive Prostate Cancer Treated With Androgen Deprivation With or Without Docetaxel. J Clin Oncol. 2018; 36: 376382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shenoy TR, Boysen G, Wang MY, Xu QZ, Guo W, Koh FM, et al. CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann Oncol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virgo KS, Basch E, Loblaw DA, Oliver TK, Rumble RB, Carducci MA, et al. Second-Line Hormonal Therapy for Men With Chemotherapy-Naive, Castration-Resistant Prostate Cancer: American Society of Clinical Oncology Provisional Clinical Opinion. J Clin Oncol. 2017; 35: 1952–1964. [DOI] [PubMed] [Google Scholar]
- 19.Basch E, Loblaw DA, Oliver TK, Carducci M, Chen RC, Frame JN, et al. Systemic therapy in men with metastatic castration-resistant prostate cancer: American Society of Clinical Oncology and Cancer Care Ontario clinical practice guideline. J Clin Oncol. 2014; 32: 3436–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, et al. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009; 56: 275–286. [DOI] [PubMed] [Google Scholar]
- 21.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, et al. The TMPRSS2: ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012; 21: 1497–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SL, Yu D, Wang C, Saba R, Liu S, Trpkov K, et al. ERG Expression in Prostate Needle Biopsy: Potential Diagnostic and Prognostic Implications. Appl Immunohistochem Mol Morphol. 2015; 23: 499505. [DOI] [PubMed] [Google Scholar]
- 23.Volante M, Tota D, Giorcelli J, Bollito E, Napoli F, Vatrano S, et al. Androgen deprivation modulates gene expression profile along prostate cancer progression. Hum Pathol. 2016; 56: 81–88. [DOI] [PubMed] [Google Scholar]
- 24.World Medical A World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013; 310: 2191–2194. [DOI] [PubMed] [Google Scholar]
- 25.Berg KD, Vainer B, Thomsen FB, Roder MA, Gerds TA, Toft BG, et al. ERG protein expression in diagnostic specimens is associated with increased risk of progression during active surveillance for prostate cancer. Eur Urol. 2014; 66: 851–860. [DOI] [PubMed] [Google Scholar]
- 26.Deplus R, Delliaux C, Marchand N, Flourens A, Vanpouille N, Leroy X, et al. TMPRSS2-ERG fusion promotes prostate cancer metastases in bone. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011; 17: 5878–5888. [DOI] [PubMed] [Google Scholar]
- 28.Tsourlakis MC, Stender A, Quaas A, Kluth M, Wittmer C, Haese A, et al. Heterogeneity of ERG expression in prostate cancer: a large section mapping study of entire prostatectomy specimens from 125 patients. BMC Cancer. 2016; 16: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva MP, Barros-Silva JD, Ersvaer E, Kildal W, Hveem TS, Pradhan M, et al. Cancer Prognosis Defined by the Combined Analysis of 8q, PTEN and ERG. Transl Oncol. 2016; 9: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018; 15: 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leinonen KA, Saramaki OR, Furusato B, Kimura T, Takahashi H, Egawa S, et al. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Cancer Epidemiol Biomarkers Prev. 2013; 22: 2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura K, Tsuzuki T, Kato M, Saito AM, Sassa N, Ishida R, et al. Prognostic value of intraductal carcinoma of the prostate in radical prostatectomy specimens. Prostate. 2014; 74: 680–687. [DOI] [PubMed] [Google Scholar]
- 33.Korsten H, Ziel-van der Made AC, van W eerden WM, van der Kwast T, Trapman J, Van Duijn PW. Characterization of Heterogeneous Prostate Tumors in Targeted Pten Knockout Mice. PLoS One. 2016; 11: e0147500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khani F, Epstein JI. Prostate Biopsy Specimens With Gleason 3+3=6 and Intraductal Carcinoma: Radical Prostatectomy Findings and Clinical Outcomes. Am J Surg Pathol. 2015; 39: 1383–1389. [DOI] [PubMed] [Google Scholar]
- 35.Eggener SE, Badani K, Barocas DA, Barrisford GW, Cheng JS, Chin AI, et al. Gleason 6 Prostate Cancer: Translating Biology into Population Health. J Urol. 2015; 194: 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter HB, Partin AW, Walsh PC, Trock BJ, Veltri RW, Nelson WG, et al. Gleason score 6 adenocarcinoma: should it be labeled as cancer? J Clin Oncol. 2012; 30: 4294–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, Chen N, Shen P, Gong J, Li X, Zhao T, et al. The presence and clinical implication of intraductal carcinoma of prostate in metastatic castration resistant prostate cancer. Prostate. 2015; 75: 1247–1254. [DOI] [PubMed] [Google Scholar]