Abstract
ABSTRACT: Porcine reproductive and respiratory syndrome (PRRS) is one of the most important global swine diseases from both an economic and animal welfare standpoint. PRRS has plagued the US swine industry for over 25 yr, and containment of PRRS virus (PRRSV) has been unsuccessful to date. The primary phase of PRRS, tracked by serum viremia, typically clears between 21 and 42 d postinfection (dpi) but tonsils are a main site of PRRSV persistence and PRRSV can be detected in tonsils in excess of 150 dpi. Measuring tonsil virus (TV) levels at late stages of infection (6 to 7 wk postinfection) can be used to assess tonsil persistence, as levels of virus in tonsil at this time likely influence how long the virus will remain in the tissue. TV levels were measured on pigs experimentally infected with either the NVSL-97-7895 (NVSL; n = 524) or KS-2006-72109 (KS06; n = 328) PRRSV type 2 isolates across five trials. The objectives of this study were to (i) estimate the heritability of TV levels at 35 or 42 dpi; (ii) identify factors the affect TV level, including serum viremia; (iii) identify genomic regions associated with TV level; and (iv) compare results for the two PRRSV isolates. TV level was lowly heritable for both isolates (NVSL: 0.05 ± 0.06; KS06: 0.11 ± 0.10). Level of TV was phenotypically associated with traits related to viral clearance from serum: pigs with low TV levels had an earlier and faster rate of maximal serum viral clearance, lower total serum viral load, and lower viremia level at 35 or 42 dpi. Although no genomic regions with major effects on TV level were identified, several showed some association (>0.1% of total genetic variance in the NVSL-infected dataset, the KS06-infected dataset, and the combined dataset). These regions contained the genes CCL1, CCL2, CCL8, HS3ST3B1, GALNT10, TCF7, C1QA/B/C, HPSE, G0S2, and CD34, which are involved in viral infiltration or replication, immune cell migration, and viral clearance from tissue. Results were similar between the two PRRSV isolates. In conclusion, selection for viral clearance traits in serum may reduce PRRSV persistence in the tonsil across PRRSV isolates. However, genetic correlations need to be estimated to determine whether this will be successful.
Keywords: genetics, pigs, porcine reproductive and respiratory syndrome, tonsils
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS) is an economically devastating, persistent viral swine disease (Holtkamp et al., 2013). Painstaking efforts are necessary to return an infected herd to negative status. Pigs are expected to clear PRRS virus (PRRSV) from serum 3 to 8 wk postinfection (Wills et al., 2003; Hess et al., 2016), but some pigs can be PRRSV positive in tissues for 150 to 250 d post infection (dpi) (Allende et al., 2000; Wills et al., 2003). The tonsil is an active site of PRRSV replication (Rowland et al., 2003). Pigs with PRRSV present in tonsils after the acute phase of infection are classified as persistently infected (Benfield et al., 1998). They can experience a resurgence of circulating virus, triggering a secondary outbreak (Rowland and Morrison, 2012)—a major concern to naïve pigs introduced into the herd after the initial PRRSV outbreak. Identification and removal of persistently infected pigs could substantially help with containment of PRRS and improve herd health, but persistently infected pigs are often asymptomatic, hampering efforts to identify them despite high levels of PRRSV in tissues (Wills et al., 2003). Genetic selection of pigs that are more resistant to PRRS is one method to improve the health status of the swine herd (Lewis et al., 2007). Serum viremia was previously found to be under genetic control, with a major QTL, marked by the SNP WUR10000125 (WUR) (Boddicker et al., 2014b; Hess et al., 2016). We hypothesized that animals that clear serum viremia levels faster, and thus have lower serum viremia at 35 or 42 dpi, will also have a lower tonsil virus (TV) level at that time, and that TV has a heritable genetic component, similar to serum viremia level. The objectives of this study were to (i) estimate the heritability of TV; (ii) identify factors the affect TV, including serum viremia; (iii) conduct a genome-wide association study (GWAS) to identify genomic regions associated with TV, including the WUR region; and (iv) compare the results for the two type 2 PRRSV isolates, NVSL, and KS06.
MATERIALS AND METHODS
Study Design
Pigs used for this study were from five experimental PRRS trials (3, 5, 7, 11, and 14) of the PRRS host genetics consortium (PHGC) (Lunney et al. 2011), whose objective is to identify host genes or genomic regions associated with increased resistance or reduced susceptibility of pigs to PRRSV infection. The Kansas State University Institutional Animal Care and Use Committee approved all experimental protocols for the animal trials used herein. A detailed description of the design, data collection and molecular techniques used in the PHGC trials is in Lunney et al. (2011), with additional information provided in Hess et al. (2016). Briefly, for each trial, ~200 crossbred weaner pigs were provided from a commercial breeding program in the United States and Canada. Trials 3 and 11, as well as trials 5 and 14 had the same genetic background. Within each trial, pigs were from a single high health farm and genetic background, except for trial 5, which included pigs from one genetic background but two farms. All source farms were free of PRRS, Mycoplasma hyopneumoniae, and swine influenza. Animals were transported to Kansas State University at weaning (average age 21 d) and randomly placed into pens of 10 to 15 pigs as they were unloaded. After a 7-d acclimation period, pigs were experimentally infected, both intramuscularly and intranasally (see Lunney et al. 2011), with 105 (TCID50) of NVSL-97-7985 (trials 3, 5, and 7), a highly virulent PRRSV isolate (Truong et al., 2004), or KS-2006-72109 (trials 11 and 14), a more contemporary PRRSV isolate. NVSL and KS06 are 89% similar at the GP5 nucleotide sequence level (Ladinig et al., 2015). Blood samples were collected at −6, 0, 4, 7, 11, 14, 21, 28, 35, and 42 d postinfection (dpi). BW was measured at 0, 7, 14, 21, 28, 35, and 42 dpi. Pigs were euthanized at 42 dpi. Trial 7 ended at 35 dpi due to limits on facility availability. Tonsil samples were collected immediately after pigs were euthanized. Across the NVSL trials, 5% of pigs died or were euthanized for humane reasons before 35/42 dpi. Mortality rate was similar in the KS06 trials, with 7% pigs dying or euthanized before 42 dpi across the two trials. Dead pigs were necropsied and subsequent gross and microscopic pathology by a board-certified pathologist identified PRRS associated disease as the major source of mortality.
Serum Viremia and TV Levels
Serum viremia and TV levels were measured using a semi-quantitative TaqMan PCR assay for PRRSV RNA, as described in Boddicker et al. (2012), with samples run in duplicate using 96-well plates. Serum and tonsil assay results were reported as the log10 of PRRSV RNA copies per milliliter of serum and per milligram of tissue, respectively. For the NVSL trials (3, 5, and 7), the Applied Biosystems AgPath ID NA and EU PRRSV reagents (AB assay) were used to measure TV, while the Tetracore US and EURO PRRSV Master Mix reagents (Tetracore assay) were used for the KS06 trials (11 and 14) because the AB primers failed to amplify cDNA of the KS06 isolate.
TV level data were available on 524 NVSL-infected animals and on 328 KS06-infected animals. The RNA integrity number (RIN) score of RNA extracted from the tonsil samples was determined by an Agilent bioanalyzer, which serves as a measurement of the quality of the RNA in a sample. Fourteen animals were removed due to having an unreadable RIN, which is interpreted as a low quality RNA sample (Schroeder et al., 2006). Virus levels were assigned to a sample using a standard curve when performing the TaqMan qPCR assay. This standard curve has a linear portion for which virus levels can be accurately measured, while samples falling in the nonlinear range are not accurately measured at the dilution used. Based on this and the 14 animals without RIN, in total 30 NVSL-infected animals and 26 KS06-infected animals were identified as having low-confidence measurements, including samples whose virus levels fell outside the standard curve (too high (28) or too low (19)) or for which no virus was detected (9) due to either assay failure or lack of virus in the sample. These animals were removed from the analysis.
To evaluate the consistency between the AB and Tetracore assays, tonsil samples from a subset of 35 NVSL-infected animals were also evaluated using the Tetracore kit. Results showed that TV from the Tetracore assay had a lower mean and smaller standard deviation between samples than TV from the AB assay but they were highly correlated at 0.87 (Appendix 2, Supplementary material). To convert TV from the Tetracore assay to the scale of the AB assay, Tetracore results were regressed on the AB results (Appendix 2, Supplementary material) (R2 = 0.76), and the resulting linear regression equation was used to adjust the NVSL TV data for differences between the kits, in order to obtain the expected TV based on the Tetracore kit.
Genotypes
Ear tissue was collected from all pigs at death or euthanasia for DNA isolation. Tissue or DNA samples for trials 3, 5, and 7 were genotyped with the Illumina Porcine SNP60 Beadchip (Ramos et al., 2009) v1 (San Diego, California) at GeneSeek Inc. (Lincoln, Nebraska) and samples from trials 11 and 14 were genotyped with the Illumina Porcine SNP60 Beadchip v2 (San Diego, CA) at Delta Genomics (Edmonton, AB). Only SNPs that were included on both versions of the Illumina’s Porcine SNP60 Beadchip were used for analysis. SNPs were also removed if they were uninformative or would give unreliable results, i.e. fixed within trial, unmapped, or mapped to a sex chromosome in build 10.2 of the swine genome (http://www.ncbi.nlm.nih.gov/genome/guide/pig/, accessed August 13, 2015); this left 48,164 SNPs. For these SNPs, missing genotypes (0.19%) were assigned the average genotype (on a 0/1/2 scale) for animals in that trial for that SNP. This set of SNPs will be referred to as the 60k SNPs.
Modeling Viremia Profiles with the Wood’s Function
Previous studies utilized the Wood’s curve to model serum viremia profiles in the PHGC trials (Hess et al., 2016, 2018; Islam et al., 2013):
where V(t) is serum viremia on the log10 scale at t. i, a1 impact the overall viremia level across time, b1 is an indicator of the initial rate of increase to peak viremia, c1 is an indicator of the rate of decline after the peak and dominates the viremia profile at the later stages of infection. These three function parameters were estimated for each individual that had at least five time points using Bayesian inference with a likelihood framework, implemented using a Markov chain Monte-Carlo method (Islam et al., 2013). The raw viremia profiles of some pigs appeared bi-modal, so an extended Wood’s curve was also fitted for each pig using the same methodology:
where t0 denotes the onset of the second phase of the profile, which is assumed to follow the same Wood’s curve shape as the primary phase and is thus defined by a second set of Wood’s model parameters. A pig was classified as experiencing viremia rebound based on the Akaike’s information criterion (AIC) if the extended Wood’s curve had an AIC that was more than 1.488 smaller than the single Wood’s curve, which corresponds to the 95% significance level of the likelihood ratio test between these models (Islam et al., 2013). In total, 15.5% of NVSL-infected pigs and 4.6% of KS06-infected pigs were classified as rebound pigs. Previously, NVSL was determined to be more virulent, while KS06 was characterized as being more persistent in serum (Hess et al., 2016). A total of 45.2% of NVSL-infected pigs and 65.2% of KS06-infected pigs were classified as having persistent PRRSV levels in serum, defined as having an estimated log10 serum value that exceeded 1 at the end of the trial (Islam et al., 2013), as determined by the most appropriate fitted Wood’s curve (single or extended) for that individual. The remaining animals were assumed to have cleared viremia from serum by the end of the trial.
Five curve characteristics were derived to describe individual viremia profiles using the estimates of the curve parameters ( of the single or the extended Wood’s curve in order to elucidate the relationship between serum and TV levels, as described in Hess et al. (2016):
VLTotal = area under the Wood’s curve from infection until euthanasia, i.e. 35 (trial 7) or 42 dpi (trials 3, 5, 11, and 14). VLTotal includes both the level of viremia and the extent to which viremia is maintained.
Other Response Phenotypes
BW was collected weekly on all pigs and used to interpolate daily weights as described in Hess et al. (2016). Fitted weights from 0 to 7, 7 to 14, 14 to 21, 21 to 28, 28 to 35, and 35 to 42 dpi (when available) were used for analysis. Total PRRSV N-protein IgG (measured as an S:P ratio) was also available on 207 NVSL (trials 3 and 5) and 156 KS06 (trial 11)-infected pigs that had TV level data, as detailed in Hess et al. (2018). Neutralizing antibody (NAb) titer data were available on 360 NVSL (trials 3, 5, and 7) and 157 KS06 (trial 11)-infected animals that had sufficient serum available at the end of the trial. To determine NAb, collected serum samples were assayed as described in Trible et al. (2015) by incubating a 200 50% tissue culture infectious dose (TCID50) of the virus with 1:2 dilutions of serum and then transferring to tissue culture plates with confluent MARC-145 cells; PRRSV cytopathic effects were assayed 4 d later. The inverse of the highest serum dilution without cytopathic effects was recorded as the NAb titer, which ranged from <8 to >1,024. For statistical analysis, NAb response was converted to an adjusted log2 scale (0 to 8).
Statistical Analyses
Factors Affecting TV Levels.
TV assays were run on 96-well plates, and plate was partially confounded with both pen and family—with individuals from the same pen tending to be run on the same plate and half or full sibs also often run on the same plate. To evaluate the effect of including plate in the model, the following model was fitted to the TV data with or without plate and separately for each PRRSV isolate, using ASReml 3.0 (Gilmour et al., 2009):
where Y is the dependent variable of TV level. Parity of the dam (P), classified as first, second, or later parity, sex of the pig (S), and trial (Tr) were fitted as a fixed class effects. To account for potential differences between rebound and nonrebound pigs based on serum viremia, the fixed class effect of rebound (R) was also included in the model. Age (A) and weight (W) of the piglet at infection, and the RIN of the tonsil sample were fitted as linear covariates. Random effects included animal genetic effects (An), litter (Li), Pen nested within trial (Pen(Tr)), Plate (Pl), and ε as the residual. Due to the limited pedigree information and availability of genotypes on almost all animals, the 60k SNP genotype data were used to construct a genomic relationship matrix (G-matrix) among all animals, using the method of VanRaden (2008). The G-matrix also included relationships between animals from different trials and genetic backgrounds.
Forty-three animals that did not have plate information were not used in this analysis. Results showed that plate explained a large part of the variance when it was included (58% for NVSL and 15% for KS06) and that this variance was partitioned between animal, pen, and residual when plate was not included in the model (Appendix 1, Supplementary material). The effect of plate that is partitioned to the animal component when plate is not included in the model is expected to be a combination of genetic and plate effects due to confounding between plate and family, but it is not known in what proportions. Thus, estimates of heritability are expected to be biased upward when excluding plate and downward when including plate. We decided to exclude plate from the model because including plate would remove genetic effects from TV and from genetic correlations of TV with other traits.
Estimates of the heritability of TV and of phenotypic associations of TV with other PRRS response traits were obtained from the model described above without plate effects. The relationships of TV with serum viremia characteristics, weekly weight gains, and antibody response were estimated by separately including each of these traits as a fixed covariate in the model. When estimating the association between TV and WUR genotype, WUR genotype was added as a fixed effect. The significance threshold for the aforementioned analyses was set to P ≤ 0.05, and the coefficient of determination of the independent variable was used to assess the proportion of TV explained by the covariate.
Genome-Wide Association Study
The Bayes C method implemented in GenSel 4.73 (Fernando and Garrick, 2012) was used to test associations of 60k SNPs with TV based on the following model:
where Y is the dependent variable of adjusted TV; P, S, A, W, R, RIN, Tr, and Pen(Tr) are as defined above but all fitted as fixed effects because GenSel does not accommodate random effects other than SNP effects; s is the number of SNPs fitted; zr is the vector of genotypes of SNP r for each animal (t) coded as −10, 0, 10, αr is the allele substitution effect for SNP r, and δr indicates whether SNP r was included (δr = 1) or excluded (δr = 0) in the model for a given iteration of the Monte-Carlo Markov Chain. The prior probability of δr = 0 was set equal to π = 0.995. A total of 500,000 iterations were run, including a burn-in of 50,000. Breeding values were calculated for each nonoverlapping 1 Mb region (window) for each iteration to obtain a sample of the breeding value for each animal for each window. The total breeding value of an animal was computed as the sum of the animal’s window breeding values across the genome. The percent genetic variance explained by a window in a given iteration was calculated as the variance of the breeding values for that window across all animals divided by the variance of total breeding values, multiplied by 100. The estimate of the percent genetic variance explained by a window was the posterior mean percent genetic variance explained by that window across all iterations (excluding 50,000 burn-in) (Garrick and Fernando, 2013).
RESULTS
Factors Affecting TV Level
Genetic Parameters.
TV level was estimated to be lowly heritable for both isolates, with both heritabilities not significantly different from zero (Table 1). Genetic correlation estimates could not be obtained due to lack of model convergence.
Table 1.
Estimates of heritability for the log of tonsil virus level for two PRRSV isolates
| Isolate | Heritability (s.e.) | Phenotypic SD |
|---|---|---|
| NVSL | 0.05 (0.06) | 1.03 |
| KS06 | 0.11 (0.10) | 0.89 |
| Both | 0.09 (0.06) | 0.98 |
Litter variance was zero for all analyses.
Phenotypic SD includes the effects of trial, pen, animal, and residual.
Isolate and Serum Viremia Status.
NVSL-infected pigs had 43% higher TV than KS06-infected pigs (P < 0.001); however, after adjusting TV for NVSL to the Tetracore scale, NVSL-infected pigs had only 2% higher TV than KS06-infected pigs, and this difference was not significant (P > 0.1; Table 2). For subsequent analyses, the unadjusted TV was used for NVSL-infected animals but for comparisons between isolates, estimates can be rescaled to the Tetracore scale by multiplying the least square means (Table 2) by 0.6753 and adding 0.2157 and multiplying regression coefficients (Table 3) by 0.6753.
Table 2.
LS means (s.e.; number of animals) of tonsil virus levels by isolate, serum viremia status, and WUR genotype
| Trait | Level | NVSL | KS06 |
|---|---|---|---|
| Isolate | Unadjusted* | 4.69 (0.09; 480) |
3.27 (0.13; 302) |
| Adjusted | 3.38 (0.07; 480) |
3.30 (0.10; 302) |
|
| Serum viremia status | Cleared | 4.45a (0.09; 203) |
3.35a (0.17; 94) |
| Rebound | 4.62a,b (0.13; 71) |
3.48a (0.27; 13) |
|
| Persistent | 4.88b (0.09; 206) |
3.54a (0.16; 195) |
|
| WUR genotype | AA | 4.63 (0.09; 270) |
3.47 (0.15; 237) |
| AB | 4.71 (0.10; 181) |
3.43 (0.18; 59) |
|
| BB | 4.65 (0.21; 29) |
3.19 (0.38; 6) |
Unadjusted: original tonsil virus levels were analyzed as measured.
Adjusted: tonsil virus levels for NVSL were adjusted to their Tetracore equivalent.
Estimates within the same isolate and dataset that have different letters are significantly different from each other.
*Estimates for NVSL and KS06 were significantly different from each other (P < 0.001).
Table 3.
Associations of tonsil virus levels with serum viremia characteristics, weight gain, and antibody levels for the NVSL and KS06 isolate data
| Statistic | TP | PV | T max | V max | VLTotal | v END | W7 | W14 | W21 | W28 | W35 | W42 | WGEND | S:P | NAb | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NVSL | N | 480 | 480 | 480 | 480 | 480 | 471 | 480 | 480 | 480 | 480 | 480 | 339 | 480 | 207 | 332 |
| Reg. coeff. | 0.043 | −0.183 | 0.094 | −3.244 | 0.013 | 0.164 | 0.151 | 0.185 | 0.174 | 0.108 | 0.05 | −0.022 | 0.117 | −0.09 | −0.006 | |
| s.e. | 0.038 | 0.14 | 0.021 | 0.64 | 0.003 | 0.036 | 0.087 | 0.123 | 0.137 | 0.121 | 0.102 | 0.128 | 0.055 | 0.31 | 0.038 | |
| P-value | 0.26 | 0.19 | <0.001 | <0.001 | <0.001 | <0.001 | 0.09 | 0.14 | 0.21 | 0.38 | 0.62 | 0.86 | 0.04 | 0.78 | 0.86 | |
| R 2 | 0 | 0 | 0.05 | 0.05 | 0.06 | 0.04 | 0.01 | 0.01 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0 | |
| KS06 | N | 302 | 302 | 302 | 302 | 302 | 281 | 302 | 302 | 302 | 302 | 302 | 302 | 302 | 137 | 132 |
| Reg. coeff. | 0.043 | −0.145 | 0.048 | −2.362 | 0.006 | 0.215 | 0.108 | 0.102 | 0.016 | −0.035 | −0.05 | −0.054 | −0.054 | −0.93 | 0.103 | |
| s.e. | 0.035 | 0.125 | 0.02 | 1.004 | 0.003 | 0.055 | 0.122 | 0.156 | 0.154 | 0.132 | 0.115 | 0.104 | 0.104 | 0.40 | 0.044 | |
| P-value | 0.23 | 0.25 | 0.02 | 0.02 | 0.07 | <0.001 | 0.37 | 0.51 | 0.91 | 0.79 | 0.66 | 0.6 | 0.6 | 0.02 | 0.02 | |
| R 2 | 0.01 | 0 | 0.02 | 0.02 | 0.01 | 0.05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.05 | 0.05 | |
TP = time to peak serum viremia (days); PV = peak serum viremia (log10 serum viremia); Tmax = time to maximal decay rate of serum viremia (days); Vmax = rate of maximal decay rate of serum viremia (log10 serum viremia/d); VLTotal = total serum viral load (log10 serum viremia*day), vEND = measured serum viremia at the end of the trial (log10 serum viremia); W7 = weight gain from 0 to 7 dpi (kg); W14 = weight gain from 7 to 14 dpi (kg); W21 = weight gain from 14 to 21 dpi (kg); W28 = weight gain from 21 to 28 dpi (kg); W35 = weight gain from 28 to 35 dpi (kg); W42 = weight gain from 35 to 42 dpi (kg); WGEND = weight gain in the last week of the trial (kg); S:P = virus N-protein-specific IgG response at 42 dpi (S:P); Nab = neutralizing antibody response at 42 dpi (log2 endpoint titer).
For NVSL, pigs that were classified as having persistent serum viremia levels had significantly higher TV than pigs that had cleared serum viremia (P < 0.001). Rebound animals had intermediate average TV, which was not significantly different from the average TV of cleared or persistent animals (P > 0.1). For KS06, the direction of the effect of serum viremia status on TV was the same as for NVSL but differences were not as significant (Table 2). Adjusting NVSL to their Tetracore equivalent resulted in differences between cleared and persistent animals that were similar to those for KS06-infected animals.
Serum Viremia.
Associations of different serum viremia curve characteristics with TV were assessed based on regression coefficients and coefficients of determination (R2; Table 3). Neither TP nor PV were significantly associated with TV for either the NVSL or KS06 data. An earlier and faster virus clearance (Tmax and Vmax) was, however, associated with lower TV for both NVSL- and KS06-infected pigs. After adjustment of NVSL to their Tetracore equivalents, the regression coefficients for Tmax and Vmax were more similar for NVSL- and KS06-infected pigs. Animals that had lower levels of serum viremia throughout the trial (VLTotal) or on the day that the tonsils were collected (vEND) had lower TV for both NVSL- and KS06-infected pigs (Table 3).
Weight Gain.
Analysis of weekly weight gain did not reveal any associations with TV (Table 3). However, there was a marginally significant (P = 0.04) positive association of weight gain during the last week of the trial with TV, suggesting that pigs that gained more weight at the end of the trial had higher TV. However, this was only evident for NVSL-infected pigs.
Antibody Response.
Both S:P and NAb response had significant associations with TV in KS06-infected pigs but not for NVSL-infected pigs (Table 3). The estimates of the regression coefficients suggest that animals that had a higher antibody levels at the end of the trial had lower TV, while animals with higher neutralizing antibody titers had higher TV.
Genome-Wide Association Studies.
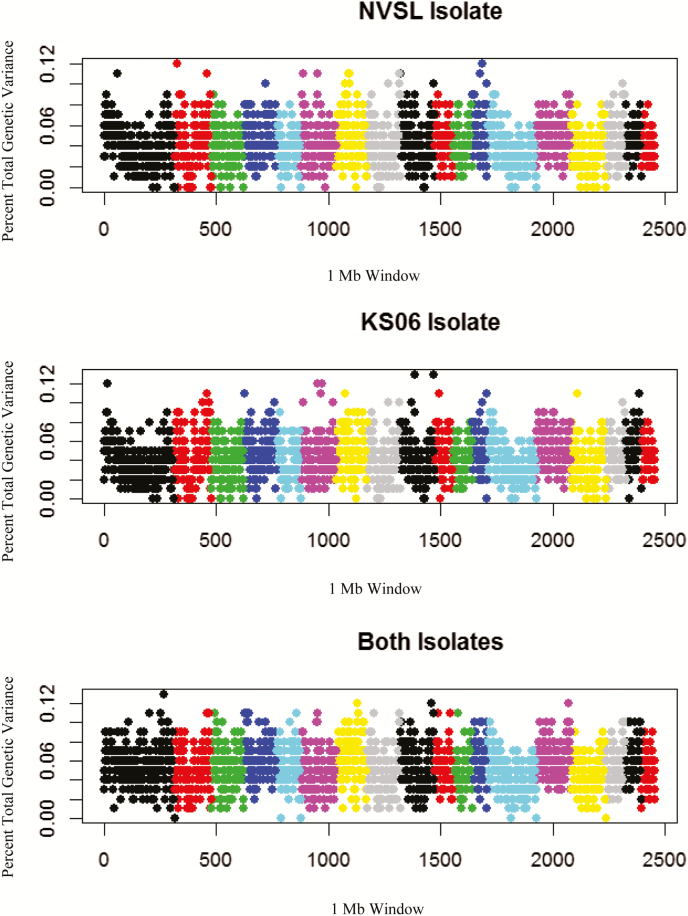
Genotype at the WUR locus did not have a significant association with TV for either isolate (P > 0.6; Table 2). The full GWAS identified no major QTL for either isolate separately or jointly (Figure 1). The 1 Mb window with the greatest association explained only 0.14% of the genetic variance. One Mb regions that explained >0.1% of genetic variance when analyzing each isolate separately and combined were further investigated for their biological relevance to PRRSV infection because they were identified in independent analyses (NVSL and KS06). In the conservative dataset, 21, 24, and 62 regions were identified that explained at least 0.1% of genetic variance in the NVSL, KS06, and combined data sets, respectively. Of these, eight regions were identified in all three analyses, of which seven had functionally relevant genes within 1 Mb in either direction of the identified 1 Mb window (Table 4).
Figure 1.
Manhattan plots for tonsil virus levels in pigs infected with NVSL or KS06 PRRSV isolates.
Table 4.
Genomic regions and candidate genes associated with tonsil virus level identified by genome-wide association study (GWAS)
| GWAS region (Chr_Mb) |
Candidate genes | Relative location (Chr_Mb) |
Function |
|---|---|---|---|
| 12_42 | CCL2 | CCL2: displays chemotactic activity for monocytes and basophils | |
| CCL1 | 12_42.6 | CCL1: secreted by activated T cells and displays chemotactic activity for monocytes | |
| CCL8 | CCL8: chemotactic factor that attracts monocytes, lymphocytes, basophils, and eosinophils; this protein can bind heparin | ||
| 12_60 | HS3ST3B1 | 12_61.0 | Catalyzes the addition of sulfate groups at the 3-OH position of glucosamine in heparan sulfate |
| 16_75 | GALNT10 | 16_74.6 | Membrane-bound polypeptide that catalyzes the first step in mucin-type O-glycosylation of peptides in the Golgi apparatus |
| 2_141 | TCF7 | 2_142.0 | Transcriptional activator—T-cell lymphocyte differentiation |
| 6_74 | C1QA/B/C | 6_74.6 | C1q associates with C1r and C1s in order to yield the first component of the serum complement system |
| 7_39 | ? | 7_39 | |
| 8_145 | HPSE | 8_144.3 | Cleaves heparan sulfate proteoglycans to permit cell movement through remodeling of the extracellular matrix |
| 9_147 | G0S2 | 9_146.5 | G0S2: promotes apoptosis by binding to BCL2 |
| CD34 | 9_148.4 | CD34: important adhesion molecule and is required for T cells to enter lymph nodes; It also interacts with L-selectin, important in inflammation |
All windows explained between 0.10% and 0.14% of genetic variance for GWAS including NVSL-infected animals, KS06-infected animals, or both
DISCUSSION
Effect of Assay on TV Level Data
In this study, TV of NVSL-infected pigs was based on the AB assay and TV for the KS06-infected pigs on the Tetracore assay. Ideally, the reported level of virus in a sample is invariant to the probe set that is used for measurement, i.e., a regression coefficient and an R2 near 1 when comparing results from the two probe sets on the same animals. However, here, the regression coefficient and R2 were 0.63 and 0.76, respectively, for results from the AB and Tetracore assays on the set of 35 animals. The resulting regression equation was used to put results from the AB assay on the scale of the Tetracore assay, and this was used to compare results from the NVSL- and KS06-infected pigs.
Genetic Parameter Estimates
Heritability estimates for TV were low and not significantly different from zero (Table 1). Bishop and Woolliams (2010) reviewed potential sources of variation in the macro environment that contribute to phenotypic variation, such as incomplete recording, incomplete exposure, and imperfect sensitivity and specificity of diagnosis, and showed that, if not controlled for, these factors have the potential to underestimate heritability. This may also be occurring with regard to the microenvironment when measuring virus levels in tonsils. Tonsils are a site of PRRSV replication and PRRSV preferentially replicates in macrophages. Thus, potential sources of variation that impact TV levels include the size of tonsils and the site at which the biopsy was taken (partial tonsil biopsies were taken in this study rather than whole tonsils), as well as macrophage abundance in the tonsils. If available, accounting for these factors as covariates in the model is expected to increase heritability estimates.
Our measures of TV capture both genomic and subgenomic RNA because the probes used in the TaqMan assays measure the amount of particular segments of RNA, rather than the whole strand of genomic RNA. Estimates of genetic correlations between traits give an indication of the genetic progress that can be made in a more difficult to measure trait (e.g. TV levels) by selecting on an easier trait to measure (e.g., serum viremia). Unfortunately, genetic correlations could not be calculated for these data due to convergence issues, likely because of the low heritability of TV. In studies involving the same animals used in this study, heritability estimates of curve characteristics of serum viremia curve were moderate–high (Hess et al., 2016), so if serum viremia clearance characteristics are genetically correlated with TV levels, selection on those serum viremia characteristics may decrease viral persistence in tonsils.
Association of TV Levels with Host Response Traits
Estimates of regression coefficients of serum viremia traits on TV provided evidence that there is a relationship between serum viremia and TV levels, although these relationships were weak (low R2) (Table 3). The results, however, suggest that faster clearance of virus from serum was associated with a lower level of virus that localizes to the tonsil, most likely because a lower level of circulating virus in serum affords the virus less of an opportunity to localize to tissues in which it persists. To our knowledge, this is the first time that such a relationship has been assessed for PRRSV infection. The relationship of TV with clearance of PRRSV from serum is of particular interest because these serum viremia characteristics explained the most variation in TV levels of all PRRSV response traits investigated when considering both isolates (Table 3).
Weight gain (WG) was not significantly associated with TV levels, with the exception of final WG in NVSL-infected pigs (Table 3). This is consistent with the observation that persistently infected pigs are generally asymptomatic (Benfield et al., 1998; Wills et al., 2003).
A dynamic relationship has been observed between antibody response measured by S:P at 42 dpi and serum viremia levels throughout the course of infection (Hess et al., 2018). The phenotypic relationship between S:P and TV, both at 42 dpi, was negative following infection with either NVSL or KS06 but the association was only significant for KS06-infected animals. The stage of infection at 42 dpi may be different between NVSL- and KS06-infected pigs, because a greater proportion of NVSL-infected pigs cleared serum viremia before 42 dpi compared to KS06-infected pigs. To gain a deeper understanding of the relationship between TV and antibody levels, the relationship between these two traits needs to be investigated at multiple time points during infection.
A positive association was identified between NAb and TV for KS06-infected pigs but not for NVSL-infected pigs. This positive association may be due to antibody-dependent enhancement, a phenomenon in which virus-specific antibodies enhance entry and in some cases replication of virus in monocytes/macrophages and granulocytic cells through interaction with Fc and/or complement receptors (Yang et al., 2000). Antibody-dependent enhancement has been demonstrated for PRRS and has been shown to differ between virus isolates (Yoon et al., 1997). Neutralizing antibody titer was assessed using the inoculum but it is possible that the PRRS virions that are represented in the tonsil are immune escape variants (Evans et al., 2017) and that the antibodies that were previously neutralizing aid the population of PRRSV in the tonsil. Therefore, in addition to differences in results between NVSL and KS06 due to the timing of TV collection in relation to clearance of virus from the serum, these two virus strains may differ in their ability to enhance viral replication in the tonsils via antibody dependent enhancement. The negative (favorable) relationship observed for S:P with TV and the positive (unfavorable) relationship for NAb and TV could represent different levels of host-pathogen interaction; non-neutralizing antibodies could reduce viremia through C1q complement mediated responses (Lu et al., 2018) as discussed further below, whereas NAb could exacerbate virus levels due to antibody-dependent enhancement (Tirado and Yoon, 2003).
Genome-Wide Association Studies
In previous studies using animals included in this study, a major QTL was identified by GWAS, which is tracked by the SNP WUR10000125 (WUR), for which animals that carry at least one copy of the B allele had reduced serum viremia and increased weight gain during PRRSV infection (Boddicker et al., 2012; Hess et al., 2016). A mutation in the GBP5 gene was identified as the putative causal for this QTL by Koltes et al. (2015), which is a gene that is involved with formation of the inflammasome, mediated by NLRP3 (Shenoy et al., 2012). This QTL was estimated to explain 13.4% and 7.5% of the genetic variation for serum viral load in NVSL- and KS06-infected pigs, respectively (Waide et al., 2018). However, genotype at the WUR locus did not have a significant association with TV for either isolate (P > 0.6; Table 2), suggesting a different biological mechanism for the prevalence of PRRSV in the tonsils.
The GWAS did not identify QTL with large effects. Nevertheless, eight genomic regions explained at least 0.1% of the genetic variance in the GWAS for each isolate and in the combined dataset. Genes that encode proteins that are responsible for modification of heparan sulfate, HPSE and HS3ST3B1, were located in the identified regions (Table 2). Heparan sulfate is a proteoglycan with covalently attached glycosaminogylcans (GAGs) and was the first identified mediator of PRRSV infection (Delputte et al., 2002). Virions are thought to concentrate on the surface of macrophages by attaching to GAGs, which stabilizes them and permits more efficient infection of the cell (Delputte et al., 2005). Heparanase is a hydrolase that cleaves between the saccharide subunits of the glycosaminoglycan chain, is expressed in immune-related tissues, and is upregulated in pigs infected with PRRSV (Tao et al., 2013). In vitro treatment of cells with HPSE has been shown to reduce PRRSV levels in infected porcine alveolar macrophages (Delputte et al., 2002). Heparanase activity is present in both endosomal and lysosomal compartments but can also occur at the cell surface, where it might digest adjacent heparin sulfate proteoglycans (Iozzo, 2001). The HS3ST3B1 gene is involved in GAG sulfation of the 3-OH position of glucosamine heparan sulfate, one of the final steps in biosynthesis of HS (Liu et al., 1999). Thus, the ability of the animal to modify heparan sulfate may be an area of interest for future studies that focus on susceptibility of the host to succumb to PRRSV persistence in the tonsil.
GALNT10 is another gene that is located in a region that was identified in the GWAS (Table 2). The short cytoplasmic tails of all GalNAc-Ts contain basic residues that may interact with peripheral Golgi membrane protein tethering complexes (Smith and Lupashin, 2008). Upon entry into the macrophage, PRRSV particles migrate to the vesicles of the endoplasmic reticulum and the Golgi apparatus (Thanawongnuwech et al., 1997), where they then rearrange host cell membranes to establish a viral replication complex. The latter plays a structural and/or functional role by offering a suitable microenvironment for viral RNA synthesis, or facilitates recruitment of membrane associated host cell proteins for the purpose of viral replication and transcription (Pedersen et al., 1999). Thus, GALNT10 may be involved in establishment of the replication complex.
Other genes that are located in regions that were identified in the GWAS (Table 2) play a role in recruitment and activation of cells involved in immune response related to T-cell responses, including transcriptional activation of T-cell differentiation (TCF7), recruitment of monocytes (which differentiate into macrophages) by chemotactic cytokines such as CCL1 (secreted by activated T-cells), CCL2, and CCL8, and T-cell migration into lymph nodes (CD34). The genes C1QA/B/C are also located in one of the eight regions identified in the GWAS. C1q is the first subcomponent of the classical pathway of activation of complement. Both immunoglobulin (IgG and IgM) and viruses are known to interact with C1q (Kishore and Reid, 2000), which has been identified as a mediator of the clearance of apoptotic cells by macrophages (Taylor et al., 2000). Therefore, the ability of an animal to recruit T-cells to the site of infection and to signal uninfected macrophages to the site of infection may play a crucial role in containing PRRSV infection in the tonsils by targeting infected cells for clearance by macrophages via C1q.
CONCLUSIONS
This study evaluated factors that could be used to reduce PRRSV persistence in infected swine herds. TV levels at 35 or 42 dpi were estimated to be lowly heritable following infection with either NVSL or KS06 and no major QTL were identified for either PRRSV isolate. However, several genomic regions that explained a proportion of genetic variance and that harbored strong candidate genes were identified. Identified genomic regions included genes involved with the host’s ability to control viral infiltration/replication, as well as the ability to clear infected cells from tissue. These genes may be useful targets for future gene expression analyses on tonsil tissue in order to gain insight into the host genetic control of TV levels and viral persistence. Clearance of viremia from serum and serum viremia levels at 35 or 42 dpi was phenotypically associated with TV levels. The results from the GWAS analyses suggest that TV levels are likely regulated by both the host’s ability to control viral infiltration/replication, as well as its ability to clear infected cells from tissue, which may be associated with serum viremia levels. This suggests that there may be a genetic correlation of TV level with Tmax, Vmax, or vEND, in which case selection that reduces serum viremia late in infection may also decrease TV levels and persistence. If serum viremia clearance characteristics are genetically correlated with TV levels, selection on those serum viremia characteristics could decrease viral persistence in tonsils.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge contributions from members of the PRRS Host Genetics Consortium. Students involved in the Hispanic Serving Institutions summer program, Wilbethsie Vazquez Degro and Gabriela Solis, assisted in preparation of tonsil RNAs.
Footnotes
This project was funded by National Pork Board (grant nos. #12-061 and #14-223), USDA-NIFA (grant no. 2013-68004-20362), and Genome Canada.
LITERATURE CITED
- Allende R., W. W. Laegreid G. F. Kutish J. A. Galeota R. W. Wills, and Osorio F. A.. 2000. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J. Virol. 74:10834–10837. doi:10.1128/JVI.74.22.10834-10837.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfield D., Nelson J., Rossow K., Rowland R., Lawson S., Steffen M., and Collins J.. 1998. Pathogenesis and persistence of PRRS. Proceedings of Allen D. Leman Swine Conference, Minneapolis, Minnesota: University of Minnesota. vol. 25; pp. 169–171. Retrieved from the University of Minnesota Digital Conservancy, http://hdl.handle.net/11299/148428 [Google Scholar]
- Bishop S. C. and Woolliams J. A.. 2010. On the genetic interpretation of disease data. PLoS One. 5:e8940. doi: 10.1371/journal.pone.0008940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker N. J., A. Bjorkquist R. R. Rowland J. K. Lunney J. M. Reecy, and Dekkers J. C.. 2014a. Genome-wide association and genomic prediction for host response to porcine reproductive and respiratory syndrome virus infection. Genet. Sel. Evol. 46:18. doi: 10.1186/1297-9686-46-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker N. J., D. J. Garrick R. R. Rowland J. K. Lunney J. M. Reecy, and Dekkers J. C.. 2014b. Validation and further characterization of a major quantitative trait locus associated with host response to experimental infection with porcine reproductive and respiratory syndrome virus. Anim. Genet. 45:48–58. doi: 10.1111/age.12079 [DOI] [PubMed] [Google Scholar]
- Boddicker N., Waide E. H., Rowland R. R. R., Lunney J. K., Garrick D. J., Reecy J. M., and Dekkers J. C. M.. 2012. Evidence for a major QTL associated with host response to porcine reproductive and respiratory syndrome virus challenge. J. Anim. Sci. 90:1733–1746. doi:10.2527/jas.2011-4464 [DOI] [PubMed] [Google Scholar]
- Delputte P. L., S. Costers, and Nauwynck H. J.. 2005. Analysis of porcine reproductive and respiratory syndrome virus attachment and internalization: distinctive roles for heparan sulphate and sialoadhesin. J. Gen. Virol. 86(Pt 5):1441–1445. doi: 10.1099/vir.0.80675-0 [DOI] [PubMed] [Google Scholar]
- Delputte P. L., N. Vanderheijden H. J. Nauwynck, and Pensaert M. B.. 2002. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 76:4312–4320. doi:10.1128/JVI.76.9.4312-4320.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A. B., Loyd H., Dunkelberger J. R., van Tol S., Bolton M. J., Dorman K. S., Dekkers J. and Carpenter S.. 2017. Antigenic and biological characterization of ORF2–6 variants at early times following PRRSV infection. Viruses. 9(5):p.113. doi:10.3390/v9050113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando R., and Garrick D.. 2012. GenSel – user manual for a portfolio of genomic selection related analyses. Ames, IA: Iowa State University. [Google Scholar]
- Garrick D. J. and Fernando R. L.. 2013. Implementing a QTL detection study (GWAS) using genomic prediction methodology. Methods Mol. Biol. 1019:275–298. doi: 10.1007/978-1-62703-447-0_11 [DOI] [PubMed] [Google Scholar]
- Gilmour A. R., Gogel B., Cullis B., and Thompson R.. 2009. ASReml user guide release 3.0.Hemel Hempstead, UK:VSN International Ltd. [Google Scholar]
- Hess A. S., Z. Islam M. K. Hess R. R. Rowland J. K. Lunney A. Doeschl-Wilson G. S. Plastow, and Dekkers J. C.. 2016. Comparison of host genetic factors influencing pig response to infection with two North American isolates of porcine reproductive and respiratory syndrome virus. Genet. Sel. Evol. 48:43. doi: 10.1186/s12711-016-0222-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, A. S., Trible, B. R., Hess, M. K., Rowland, R. R. R., Lunney, J. K., Plastow, G. S., & Dekkers, J. C. M. (2018). Genetic relationships of antibody response, viremia level and weight gain in pigs experimentally infected with porcine reproductive and respiratory syndrome virus. J. Anim. Sci. 96(9):3565–3581. doi:10.1093/jas/sky229. doi: 10.1093/jas/sky229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp D. J., Kliebenstein J. B., Neumann E. J., Zimmerman J. J., Rotto H. F., Yoder T. K., Wang C., Yeske P. E., Mowrer C. L., and Haley C. A.. 2013. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health Prod. 21(2):72–84. [Google Scholar]
- Iozzo R. V. 2001. Heparan sulfate proteoglycans: intricate molecules with intriguing functions. J. Clin. Invest. 108:165–167. doi: 10.1172/JCI13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam Z. U., S. C. Bishop N. J. Savill R. R. Rowland J. K. Lunney B. Trible, and Doeschl-Wilson A. B.. 2013. Quantitative analysis of porcine reproductive and respiratory syndrome (PRRS) viremia profiles from experimental infection: a statistical modelling approach. PLoS One. 8:e83567. doi: 10.1371/journal.pone.0083567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore U. and Reid K. B.. 2000. C1q: structure, function, and receptors. Immunopharmacology. 49:159–170. doi:10.1016/S0162-3109(00)80301-X [DOI] [PubMed] [Google Scholar]
- Koltes J. E., E., Fritz-Waters C. J., Eisley I., Choi H., Bao A., Kommadath N. V., Serão N. J., Boddicker S. M., Abrams M., Schroyen, et al. 2015. Identification of a putative quantitative trait nucleotide in guanylate binding protein 5 for host response to PRRS virus infection. BMC Genomics. 16:412. doi: 10.1186/s12864-015-1635-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinig A., S. E. Detmer K. Clarke C. Ashley R. R. Rowland J. K. Lunney, and Harding J. C.. 2015. Pathogenicity of three type 2 porcine reproductive and respiratory syndrome virus strains in experimentally inoculated pregnant gilts. Virus Res. 203:24–35. doi: 10.1016/j.virusres.2015.03.005 [DOI] [PubMed] [Google Scholar]
- Lewis C. R., T. Ait-Ali M. Clapperton A. L. Archibald, and Bishop S.. 2007. Genetic perspectives on host responses to porcine reproductive and respiratory syndrome (PRRS). Viral Immunol. 20:343–358. doi: 10.1089/vim.2007.0024 [DOI] [PubMed] [Google Scholar]
- Liu J., Z. Shriver P. Blaiklock K. Yoshida R. Sasisekharan, and Rosenberg R. D.. 1999. Heparan sulfate D-glucosaminyl 3-O-sulfotransferase-3A sulfates N-unsubstituted glucosamine residues. J. Biol. Chem. 274:38155–38162. doi:10.1074/jbc.274.53.38155 [DOI] [PubMed] [Google Scholar]
- Lu L. L., T. J. Suscovich S. M. Fortune, and Alter G.. 2018. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 18:46–61. doi: 10.1038/nri.2017.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney J. K., Steibel J. P., Reecy J. M., Fritz E., Rothschild M. F., Kerrigan M., Trible B., and Rowland R. R.. 2011. Probing genetic control of swine responses to PRRSV infection: current progress of the PRRS host genetics consortium. BMC Proc. 5.4:S30–S30. doi:10.1186/1753-6561-5-S4-S30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K. W., Y. van der Meer N. Roos, and Snijder E. J.. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, A. M., R. P. M. A. Crooijmans, N. A. Affara, A. J. Amaral, A. L. Archibald, J. E. Beever, C. Bendixen, C. Churcher, R. Clark, P. Dehais, et al. 2009. Design of a High Density SNP Genotyping Assay in the Pig Using SNPs Identified and Characterized by Next Generation Sequencing Technology. PLoS One. 4(8). doi:10.1371/journal.pone.0006524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R. R., S. Lawson K. Rossow, and Benfield D. A.. 2003. Lymphoid tissue tropism of porcine reproductive and respiratory syndrome virus replication during persistent infection of pigs originally exposed to virus in utero. Vet. Microbiol. 96:219–235. doi:10.1016/j.vetmic.2003.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R. R., J. Lunney, and Dekkers J.. 2012. Control of porcine reproductive and respiratory syndrome (PRRS) through genetic improvements in disease resistance and tolerance. Front. Genet. 3:260. doi: 10.3389/fgene.2012.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A., O. Mueller S. Stocker R. Salowsky M. Leiber M. Gassmann S. Lightfoot W. Menzel M. Granzow, and Ragg T.. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7:3. doi: 10.1186/1471-2199-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A. R., D. A. Wellington P. Kumar H. Kassa C. J. Booth P. Cresswell, and MacMicking J. D.. 2012. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 336:481–485. doi: 10.1126/science.1217141 [DOI] [PubMed] [Google Scholar]
- Smith R. D. and Lupashin V. V.. 2008. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 343:2024–2031. doi: 10.1016/j.carres.2008.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao C., W. Wang P. Zhou T. Xia X. Zhou C. Zeng Q. Zhang, and Liu B.. 2013. Molecular characterization, expression profiles, and association analysis with hematologic parameters of the porcine HPSE and HPSE2 genes. J. Appl. Genet. 54:71–78. doi: 10.1007/s13353-012-0119-8 [DOI] [PubMed] [Google Scholar]
- Taylor P. R., A. Carugati V. A. Fadok H. T. Cook M. Andrews M. C. Carroll J. S. Savill P. M. Henson M. Botto, and Walport M. J.. 2000. A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J. Exp. Med. 192:359–366. doi:10.1084/jem.192.3.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawongnuwech R., E. L. Thacker, and Halbur P. G.. 1997. Effect of porcine reproductive and respiratory syndrome virus (PRRSV) (isolate ATCC VR-2385) infection on bactericidal activity of porcine pulmonary intravascular macrophages (PIMS): in vitro comparisons with pulmonary alveolar macrophages (PAMS). Vet. Immunol. Immunopathol. 59:323–335. doi:10.1016/S0165-2427(97)00078-0 [DOI] [PubMed] [Google Scholar]
- Tirado S. M. and Yoon K. J.. 2003. Antibody-dependent enhancement of virus infection and disease. Viral Immunol. 16:69–86. doi: 10.1089/088282403763635465 [DOI] [PubMed] [Google Scholar]
- Trible B. R., L. N. Popescu N. Monday J. G. Calvert, and Rowland R. R.. 2015. A single amino acid deletion in the matrix protein of porcine reproductive and respiratory syndrome virus confers resistance to a polyclonal swine antibody with broadly neutralizing activity. J. Virol. 89:6515–6520. doi: 10.1128/JVI.03287-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong H. M., Z. Lu G. F. Kutish J. Galeota F. A. Osorio, and Pattnaik A. K.. 2004. A highly pathogenic porcine reproductive and respiratory syndrome virus generated from an infectious cDNA clone retains the in vivo virulence and transmissibility properties of the parental virus. Virology. 325:308–319. doi: 10.1016/j.virol.2004.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Waide E. H., C. K. Tuggle N. V. L. Serão M. Schroyen A. Hess R. R. R. Rowland J. K. Lunney G. Plastow, and Dekkers J. C. M.. 2018. Genomic prediction of piglet response to infection with one of two porcine reproductive and respiratory syndrome virus isolates. Genet. Sel. Evol. 50:3. doi: 10.1186/s12711-018-0371-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills R. W., A. R. Doster J. A. Galeota J. H. Sur, and Osorio F. A.. 2003. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J. Clin. Microbiol. 41:58–62. doi:10.1128/JCM.41.1.58-62.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., M. L. Frey K. J. Yoon J. J. Zimmerman, and Platt K. B.. 2000. Categorization of north american porcine reproductive and respiratory syndrome viruses: epitopic profiles of the N, M, GP5 and GP3 proteins and susceptibility to neutralization. Arch. Virol. 145:1599–1619. doi:10.1007/s007050070079 [DOI] [PubMed] [Google Scholar]
- Yoon K. J., L. L. Wu J. J. Zimmerman, and Platt K. B.. 1997. Field isolates of porcine reproductive and respiratory syndrome virus (PRRSV) vary in their susceptibility to antibody dependent enhancement (ADE) of infection. Vet. Microbiol. 55:277–287. doi:10.1016/S0378-1135(96)01338-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.