Abstract
The goal of this study was to test the hypothesis that sodium selenite (inorganic Se, ISe), SEL-PLEX (organic forms of Se, OSe), vs. a 1:1 blend (MIX) of ISe and OSe in a basal vitamin–mineral (VM) mix would differentially alter pituitary transcriptome profiles in growing beef steers grazing an endophyte-infected tall fescue (E+) pasture. Predominately Angus steers (BW = 183 ± 34 kg) were randomly selected from fall-calving cows grazing E+ pasture and consuming VM mixes that contained 35 ppm Se as ISe, OSe, or MIX forms. Steers were weaned, depleted of Se for 98 d, and subjected to summer-long common grazing of a 10.1 ha E+ pasture containing 0.51 ppm ergot alkaloids. Steers were assigned (n = 8 per treatment) to the same Se-form treatments on which they were raised. Selenium treatments were administered by daily top-dressing 85 g of VM mix onto 0.23 kg soyhulls, using in-pasture Calan gates. As previously reported, serum prolactin was greater for MIX (52%) and OSe (59%) steers vs. ISe. Pituitaries were collected at slaughter and changes in global and selected mRNA expression patterns determined by microarray and real-time reverse transcription PCR analyses, respectively. The effects of Se treatment on relative gene expression were subjected to one-way ANOVA. The form of Se affected the expression of 542 annotated genes (P < 0.005). Integrated pathway analysis found a canonical pathway network between prolactin and pro-opiomelanocortin (POMC)/ACTH/α-melanocyte-stimulating hormone (α-MSH) synthesis-related proteins and that mitochondrial dysfunction was a top-affected canonical pathway. Targeted reverse transcription-PCR analysis found that the relative abundance of mRNA encoding prolactin and POMC/ACTH/α-MSH synthesis-related proteins was affected (P < 0.05) by the form of Se, as were (P ≤ 0.05) mitochondrial dysfunction-related proteins (CYB5A, FURIN, GPX4, and PSENEN). OSe steers appeared to have a greater prolactin synthesis capacity (more PRL mRNA) vs. ISe steers through decreased dopamine type two receptor signaling (more DRD2 mRNA), whereas MIX steers had a greater prolactin synthesis capacity (more PRL mRNA) and release potential by increasing thyrotropin-releasing hormone concentrations (less TRH receptor mRNA) than ISe steers. OSe steers also had a greater ACTH and α-MSH synthesis potential (more POMC, PCSK2, CPE, and PAM mRNA) than ISe steers. We conclude that form of Se in VM mixes altered expression of genes responsible for prolactin and POMC/ACTH/α-MSH synthesis, and mitochondrial function, in pituitaries of growing beef steers subjected to summer-long grazing an E+ pasture.
Keywords: ACTH, cattle, fescue toxicosis, mitochondria, prolactin, selenium supplementation
INTRODUCTION
Two simultaneous challenges faced by many south-eastern United States cattle producers are fescue toxicosis and Se deficiency. Fescue toxicosis results from consumption of ergot alkaloids found in Epichloe coenophialum-infected tall fescue (Lolium arundinaceum) pastures and is a clinical condition consisting of impaired metabolic, vascular, growth, and reproductive processes in cattle (Strickland et al., 2011). Reduced serum prolactin is a recognized marker of fescue toxicosis (Goetsch et al., 1987; Davenport et al., 1993). Selenium-poor soils in this same geographic region result in Se-deficient forages necessitating the need to provide supplemental Se (Dargatz and Ross, 1996). Inorganic Se (ISe, sodium selenite) is the most common form of Se supplemented in cattle diets, whereas organic forms of Se (OSe) derived from specially cultivated Saccharomyces cerevisiae also are available and approved for use in beef cattle diets.
Serendipitously, it was found that expression of several genes downregulated in the liver (Liao et al., 2015) and pituitary (Li et al., 2017) of steers grazing high vs. low endophyte-infected forages were upregulated in cattle by consumption of a 1:1 blend of ISe:OSe (MIX) in vitamin–mineral (VM) mixes (Matthews et al., 2014; Matthews and Bridges, 2014). Moreover, it was determined subsequently that steers subjected to summer-long grazing of endophyte-infected pasture and supplemented (3 mg/d) with MIX or OSe forms of Se had greater serum prolactin concentrations than ISe-supplemented steers (Jia et al., 2018). The first goal of the present study was to test the specific hypothesis that the amount of prolactin mRNA would be greater in the pituitary tissue of the same MIX and OSe vs. ISe steers, whereas the second goal was to test the general hypothesis that the form of supplemental Se would alter pituitary transcriptome profiles.
MATERIALS AND METHODS
All experimental procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee.
Animal Model
The animal management regimen and model for steers that yielded the pituitary tissue of the present experiment have been reported (Jia et al., 2018). Briefly, 24 predominantly Angus beef steers (BW, 182.6 ± 33.9 kg; age, 165.5 ± 14.2 d) were randomly selected from three Se phenotypic herds (eight steers per herd), which were managed under a typical forage based (predominately endophyte-infected tall fescue), fall-calving, and cow-calf production regimen. The three Se phenotypic herds had free access to VM premixes (UK Beef IRM Cow-Calf Mineral, Burkmann Feeds, Danville, KY) containing 35 ppm of ISe (sodium selenite, Prince Se Concentrate; Prince Agri Products, Inc., Quincy, IL), OSe (SEL-PLEX, Alltech Inc., Nicholasville, KY), and 1:1 mix of ISe:OSe (MIX). After adapted to consuming VM premixes from in-pasture Calan gate feeders, 24 steers with three Se phenotypes (n = 8) started (day 0) summer-long grazing of a 10.1-ha predominately endophyte-infected tall fescue-mixed pasture (0.51 μg/g total ergot alkaloids) (Jia et al., 2018). Three Se form treatments were administered using in-pasture Calan gate feeders to steers with the same Se phenotypes. All three Se form treatments contained a common basal VM premix that also contained 35 ppm Se as either ISe, OSe, or MIX. After the common 86-d grazing period on pastures, steers were slaughtered in the University of Kentucky Meat Laboratory (Lexington, KY) over a 26-d period. Throughout the slaughter period, steers continued on their respective Se treatment. Details of the slaughter period and process have been reported (Jia et al., 2018).
Sample Collection and RNA Preparation
Steers were stunned by captive bolt pistol and exsanguinated. Within 10 to 12 min of death, the whole pituitary was collected from each animal, placed in a foil pack, flash-frozen in liquid nitrogen, and stored at −80 °C. Four pituitary glands (two ISe, one OSe, and one MIX) were not used in the microarray analysis because of tissue damage incurred during the collection process. As a result, six pituitaries for ISe and seven pituitaries for both OSe and MIX treatment groups were subjected to RNA analyses.
Total RNA was extracted from the whole frozen pituitary tissue and its purity and integrity determined as described (Li et al., 2017). Extracted RNA samples had an average concentration of 706 ng/µL and were of high purity with 260:280 nm absorbance ratios ranging from 1.85 to 2.05 and 260:230 nm absorbance ratios ranging from 2.09 to 2.50. All RNA samples had 28S:18S rRNA absorbance ratios >1.8 and RNA integrity numbers >8.9.
Microarray Analysis
The GeneChip Bovine Gene 1.0 ST Array (Affymetrix, Inc., Santa Clara, CA) was used to investigate the effect of Se treatment on bovine pituitary gene expression profiles. Microarray analysis was conducted according to the manufacturer’s standard protocol at the University of Kentucky Microarray Core Facility as described (Li et al., 2017), using 3 μg of RNA per sample (chip). All the GeneChip transcripts were annotated using the NetAffx annotation database for gene expression on Bovine GeneChip Array ST 1.0, provided by the manufacturer (http://www.affymetrix.com/analysis/index.affx, accessed January 2018, annotation file last updated in May 2016). Quality control of the microarray hybridization and data presentation was performed by MA plot on all the gene expression values and by box plot on the control probe sets on the Affymetrix chips (data not shown). Pearson (linear) correlation generated the similarity matrix (accessed January 2018, PGS 7.17.0918). The average correlation between any pair of the 20 GeneChips was 0.96 (Supplementary Figure S1), and all GeneChips were further analyzed. Principal component analysis (PCA) was performed to elucidate the quality of the microarray hybridization and visualize the general data variation among the chips (Partek, 2009). To assess treatment effects (ISe vs. OSe vs. MIX) on the relative expression of the pituitary gene transcripts, qualified microarray data were subjected to one-way ANOVA using the same PGS software. To achieve a greater degree of confidence (i.e., a more conservative approach), transcripts showing treatment effects at the significance level of P < 0.005 (false discovery rate of ≤ 18.8%) were defined as differentially expressed. These differentially expressed genes/gene transcripts (DEG) were subjected to hierarchical clustering analysis using PGS software and to canonical, functional, and network pathway analyses using the core analysis program of Ingenuity Pathway Analysis online software [IPA, Build version 470319M, Content version 43605602; http://www.ingenuity.com (accessed in March, 2018); Ingenuity Systems, Inc., Redwood City, CA].
All the microarray *.cel files collected by Command Console plus the GC Robust Multichip Averaging-corrected data processed by PGS software of this manuscript have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/, released May 25, 2018), are compliant with the minimum information about a microarray experiment guidelines (Brazma et al., 2001), and are accessible through GEO series accession number GSE114893.
Real-Time Reverse Transcription (RT)-PCR Analysis
Primer sets for genes selected for real-time RT-PCR analysis (Supplementary Table S1) were designed using the NCBI Pick Primers online program against RefSeq sequences (accessed March to November 2017), except for s-PRLR and l-PRLR, which have been reported (Thompson et al., 2011). Real-time RT-PCR was performed as described (Li et al., 2017) using 1 μg of RNA used for each reverse transcription reaction. Gene expression was analyzed by the 2−ΔΔCT method (Livak and Schmittgen, 2001).
The resulting real-time RT-PCR products were purified using a PureLink Quick Gel Extraction Kit (Invitrogen) and sequenced at Eurofins Scientific (Eurofins, Louisville, KY). Sequences were compared with the corresponding RefSeq mRNA sequences used as the templates for primer set design. The sequences of the primers and the resulting sequence-validated real-time RT-PCR reaction amplicons for selected DEG and the endogenous control genes RPS11, TFRC, and UBC are presented in Supplementary Table S1 and Supplementary Figure S2, respectively. All sequenced amplicons had at least 98% identity with their template sequences. Three constitutively expressed genes (RPS11, TFRC, and UBC) were used and their CT values were not affected (P = 0.59, 0.51, 0.66. respectively) by Se-form treatments. Thus, the geometric mean expression of RPS11, TFRC, and UBC was used to normalize the relative quantities of the selected DEG mRNA expression, and all RT-PCR reactions were conducted in triplicate.
Statistical Analysis
Steers were the experimental units. To test for Se treatment effects on the relative expression of the pituitary gene transcripts, microarray hybridization data were subjected to one-way ANOVA using the PGS software as described in the “Microarray Analysis”. To determine the effects of treatment, the relative expression levels of selected DEG analyzed by real-time RT-PCR were subjected to one-way ANOVA using the GLM procedure of the SAS statistical software package (version 9.4; SAS Inst., Inc., Cary, NC), with the Se treatment as the fixed effect. For these data, significance was declared when P ≤ 0.05, and a tendency to differ was declared when 0.10 > P > 0.05. When P < 0.10, means were separated using Fisher’s LSD procedure.
RESULTS
Differentially Expressed Genes
PCA of all microarray data was performed to examine the correlation and variation among the chips, revealing a total variance of 25.53% (Supplementary Figure S3). The first principal component (PC #1, x-axis) explained a median degree of variance (10%), whereas PC #2 (y-axis) and PC #3 (z-axis) explained low degrees of variance (9.1% and 6.43%, respectively). Overall, PCA clearly demonstrated that the chips within each treatment group were clustered closely together.
Individual ANOVA was conducted to identify altered expression of RNA transcripts in the pituitary tissue across Se form treatments. At the P < 0.01 level and a false discovery rate of <21.5%, 948 annotated gene transcripts were identified. To refine this analysis, 542 genes with the criteria of a false discovery rate of <18.8% and P < 0.005 were considered to be differentially expressed (Supplementary Table S2).
Hierarchical cluster analysis of the 542 DEG revealed all steers segregated within their treatment group, except for one ISe steer, which displayed a DEG pattern similar to OSe and MIX steer groups (Supplementary Figure S4).
Pathways and Gene Network Analyses
To determine the physiological significance of Se treatment-induced DEG (Supplementary Table S2), bioinformatic analysis of canonical, functional, and network pathway analyses was performed. Canonical pathway analysis (Table 1) revealed (P < 0.005) that the top six pathways were ephrin receptor signaling (12 genes), Th1 and Th2 activation pathway (11 genes), Th1 pathway (nine genes), breast cancer regulation by stathmin1 (11 genes), ephrin B signaling (six genes), and mitochondrial dysfunction (10 genes).
Table 1.
Top six IPA-identified canonical pathways of genes differentially expressed by pituitary tissue of steers grazing endophyte-infected tall fescue and supplemented with 3 mg Se/d in vitamin–mineral mixes as either sodium selenite (ISe), SEL-PLEX (OSe), or a 1:1 mix of ISe and OSe (MIX)
| Canonical pathway | Number1 | Gene symbol | Ratio2 | Log (P-value) |
|---|---|---|---|---|
| Ephrin receptor signalling | 12 | ROCK2,EPHB6,ITGA3,SDC2,RACK1,LIMK2,EFNB3,STAT3,RAP1A,EPHA2,GRINA,LIMK1 | 0.068 | 3.57 |
| Th1 and Th2 activation pathway | 11 | PSENEN,TGFB1,IL1RL1,LTA,IL6R,mir-155,VAV1,IL27RA,STAT3,IFNAR1,IL18R1 | 0.060 | 2.83 |
| Th1 pathway | 9 | PSENEN,LTA,IL6R,mir-155,VAV1,IL27RA,STAT3,IFNAR1,IL18R1 | 0.066 | 2.74 |
| Breast Cancer Regulation by Stathmin1 | 11 | ROCK2,TUBB4B,PPP2R3A,PPP1R14D,PRKCD,RACK1,ARHGEF1,LIMK2,ARHGEF3,PPP1CA,LIMK1 | 0.053 | 2.43 |
| Ephrin B Signalling | 6 | ROCK2,EPHB6,RACK1,VAV1,EFNB3,LIMK1 | 0.080 | 2.38 |
| Mitochondrial dysfunction | 10 | FURIN,SDHB,ATP5G1,PSENEN,COX7A2,LRRK2,CYB5A,GPX4,NDUFAB1,NDUFA2 | 0.053 | 2.29 |
1The number of genes (listed in the “Symbol” column) associated with the particular canonical pathway.
2The ratio is calculated as the number of genes in a given pathway that meet cutoff criteria (e.g., the ANOVA P-value for the differential expression among Se groups is <0.005) divided by the total number of genes that make up that pathway.
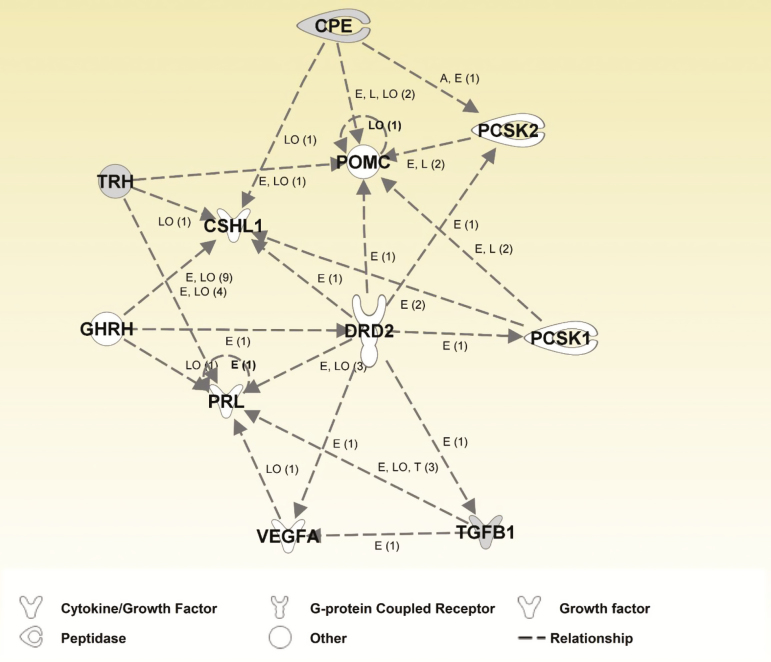
To gain insight into potentially interacting canonical pathways, canonical pathway network analysis (Figure 1) revealed one network that included four DEG [CPE, CSHL1, Transforming growth factor-β1 (TGFB1), and thyrotropin-releasing hormone (TRH)] and seven other affected (P < 0.10) genes [DRD2, PCSK1, PCSK2, pro-opiomelanocortin (POMC), PRL, VEGFA, and TRH receptors (TRHR)], all of which are related to either prolactin or POMC/ACTH/α-melanocyte-stimulating hormone (α-MSH) synthesis or release.
Figure 1.
Canonical pathway network analysis. Shaded color indicates differentially expressed genes (P < 0.005). Non-shaded color indicates genes added from the Ingenuity Knowledge Base (Ingenuity Pathway, Ingenuity Systems, Inc., Redwood City, CA). Arrowheads symbolize action-on. Labels of interaction or relationship: A = Activation, E = Expression (includes metabolism or synthesis for chemicals), I = Inhibition, L = Molecular Cleavage, LO = Localization, T= Transcription. The number in parenthesis for each interaction indicates the number of published references in the Ingenuity Knowledge Base that support the particular interaction.
Real-Time RT-PCR Analysis of Selected mRNA
Real-time RT-PCR analysis was used to corroborate the microarray analysis-identified DEG responsible for prolactin synthesis and secretion and POMC/ACTH production in Se treatment steers (Table 2). For the prolactin receptor (PRLR), unlike the microarray analysis, the RT-PCR analysis was designed to delineate the long (l-PRLR) and short (s-PRLR) forms. With the exception of VEGFA, the ANOVA P-values for Se treatment effect were consistent between the two analytical techniques. For VEGFA, microarray analysis indicated that MIX steer expression of VEGFA tended to be greater (P = 0.093), whereas RT-PCR analysis found no difference (P = 0.250). With regard to fold-changes, the direction of Se treatment-induced change was the same between microarray and RT-PCR analyses while the magnitude of the determined fold-changes typically was greater by RT-PCR analysis (Table 2).
Table 2.
Comparison of microarray- and real-time RT-PCR (RT-PCR)-determined relative expression of prolactin and POMC/ACTH synthesis related genes in pituitary tissue of steers grazing endophyte-infected tall fescue and supplemented with 3 mg Se/d in vitamin–mineral mixes as either sodium selenite (ISe), SEL-PLEX (OSe), or a 1:1 mix of ISe and OSe (MIX)
| Gene | Gene name | Microarray1 | RT-PCR2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Treatment3,4 | P-value | Treatment3,4 | P-value | ||||||
| ISe | MIX | OSe | ISe | MIX | OSe | ||||
| Item | |||||||||
| Prolactin synthesis-related genes | |||||||||
| DRD2 | Dopamine receptor D2 | 1.00a | 1.15ab | 1.27b | 0.027 | 1.13a | 1.64ab | 2.48b | 0.039 |
| POU1F1 | POU class 1 homeobox 1 | 1.00 | 1.06 | 1.05 | 0.530 | 1.02 | 1.11 | 0.87 | 0.277 |
| PRL | Prolactin | 1.00a | 1.12ab | 1.18b | 0.022 | 1.02a | 2.08b | 3.67b | 0.007 |
| TRHR | Thyrotropin releasing hormone receptor | 1.00a | 0.76b | 0.83ab | 0.045 | 1.00a | 0.71b | 0.81ab | 0.039 |
| VIP | Vasoactive intestinal peptide | 1.00 | 1.64 | 1.43 | 0.331 | 1.21 | 1.74 | 2.31 | 0.311 |
| GAL | Galanin/GMAP prepropeptide | 1.00 | 1.03 | 1.00 | 0.821 | 1.32 | 2.10 | 2.35 | 0.182 |
| VEGFA | Vascular endothelial growth factor A | 1.00a | 1.15b | 1.05ab | 0.093 | 1.01 | 1.18 | 1.03 | 0.250 |
| TGFB1 | Transforming growth factor beta 1 | 1.00a | 1.46b | 1.30b | 0.001 | 1.02a | 1.78b | 2.06c | 0.005 |
| GHRHR | Growth hormone releasing hormone receptor | 1.00 | 0.93 | 1.00 | 0.532 | 1.01 | 1.07 | 1.29 | 0.166 |
| CSH2 | Chorionic somatomammotropin hormone 2 | 1.00 | 1.09 | 0.97 | 0.173 | 1.10 | 0.74 | 0.87 | 0.279 |
| PRLR | Prolactin receptor | 1.00 | 1.13 | 1.23 | 0.141 | NA | NA | NA | NA |
| L-PRLR | Prolactin receptor long isoform | NA | NA | NA | NA | 1.01 | 0.95 | 1.17 | 0.376 |
| S-PRLR | Prolactin receptor short isoform | NA | NA | NA | NA | 1.01 | 1.01 | 1.09 | 0.761 |
| POMC/ACTH/α-MSH synthesis related gene | |||||||||
| POMC | Proopiomelanocortin | 1.00a | 1.10ab | 1.23b | 0.045 | 1.14a | 1.91a | 4.06b | 0.002 |
| PCSK1 | Proprotein convertase subtilisin/kexin type 1 | 1.00a | 1.09ab | 1.44b | 0.059 | 1.03a | 1.48ab | 1.68b | 0.076 |
| PCSK2 | Proprotein convertase subtilisin/kexin type 2 | 1.00a | 1.13ab | 1.33b | 0.074 | 1.03a | 1.14ab | 1.41b | 0.048 |
| CPE | Carboxypeptidase E | 1.00a | 0.98a | 1.04b | 0.002 | 1.01a | 1.01a | 1.31b | 0.003 |
| PAM | Peptidylglycine α-amidating monooxygenase | 1.00a | 1.04a | 1.29b | 0.044 | 1.06a | 1.28a | 1.96b | 0.008 |
1The abundance of gene transcripts is reported relative to the mean expression of the ISe treatment group and is expressed as fold-change of the untransformed intensity value.
2The abundance of gene transcripts is reported relative to the geometric mean expression of the reference genes.
3Values are least-squares means (n = 6 for ISe, n = 7 for OSe and MIX).
4Means within a row that lack a common letter differ (P < 0.05).
The relative expression of the nine genes that constituted the mitochondrial dysfunction pathway was analyzed by RT-PCR to corroborate the microarray analysis (Table 3). The trend of the numeric values of the two analyses was consistent for eight of nine evaluated genes, and statistically different for five of nine genes. Specifically, both analyses revealed that the content of CYB5A, FURIN, GPX4, and PSENEN mRNA was greater (P ≤ 0.052), or tended (P = 0.096) to be greater (COX7A2), in OSe vs. ISe steer pituitaries. In contrast, the contents of ATP5G1, LRRK2, NDUFA2, and SDHB mRNA did not differ (P > 0.130), as assessed by RT-PCR analysis.
Table 3.
Comparison of microarray and real-time RT-PCR (RT-PCR) identification of mitochondrial dysfunction related genes by pituitary tissue of steers grazing endophyte-infected tall fescue and supplemented with 3 mg Se/d in vitamin–mineral mixes as either sodium selenite (ISe), SEL-PLEX (OSe), or a 1:1 mix of ISe and OSe (MIX)
| Gene | Gene name | Microarray1 | RT-PCR2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Treatment3,4 | P-value | Treatment3,4 | P-value | ||||||
| ISe | MIX | OSe | ISe | MIX | OSe | ||||
| Item | |||||||||
| Mitochondrial dysfunction related genes | |||||||||
| ATP5G1 | ATP synthase, H+ transporting, mitochondrial Fo complex subunit C1 (subunit 9) | 1.00a | 1.22b | 1.26b | 0.004 | 1.04 | 1.24 | 1.31 | 0.266 |
| COX7A2 | Cytochrome C oxidase subunit 7A2 | 1.00a | 1.05a | 1.24b | 0.001 | 1.00a | 1.20ab | 1.29b | 0.096 |
| CYB5A | Cytochrome B5 Type A | 1.00a | 1.00a | 1.13b | 0.002 | 1.02a | 1.02a | 1.27b | 0.024 |
| FURIN | Paired basic AA cleaving enzyme | 1.00a | 1.04a | 1.12b | 0.002 | 1.04a | 1.33a | 1.81b | 0.011 |
| GPX4 | Glutathione peroxidase 4 | 1.00a | 1.21b | 1.23b | 0.004 | 1.03a | 1.32ab | 1.56b | 0.052 |
| LRRK2 | Leucine rich repeat kinase 2 | 1.00a | 0.88b | 0.79c | 0.001 | 1.00 | 0.88 | 0.85 | 0.130 |
| NDUFA2 | NADH:ubiquinone oxidoreductase subunit A2 | 1.00a | 1.18b | 1.16b | 0.003 | 1.01 | 1.16 | 1.23 | 0.142 |
| PSENEN | Presenilin enhancer gamma-secretase subunit | 1.00a | 1.13b | 1.24c | 0.001 | 1.01a | 1.17ab | 1.32b | 0.032 |
| SDHB | Succinate dehydrogenase complex iron sulphur subunit B | 1.00a | 1.03a | 1.12b | 0.003 | 1.23 | 1.71 | 1.46 | 0.573 |
| Antioxidant enzyme-encoding genes | |||||||||
| CAT | Catalase | 1.00 | 1.00 | 1.04 | 0.127 | 1.02 | 1.05 | 1.24 | 0.138 |
| SOD1 | Superoxide dismutase 1 | 1.00 | 1.06 | 1.04 | 0.115 | 1.05a | 1.35ab | 1.80b | 0.018 |
1The abundance of gene transcripts is reported relative to the mean expression of the ISe treatment group and is expressed as fold-change of the untransformed intensity value.
2The abundance of gene transcripts is reported relative to the geometric mean expression of the reference genes.
3Values are least squares means (n = 6 for ISe, n = 7 for OSe and MIX).
4Means within a row that lack a common letter differ (P < 0.05).
Because it has been reported that mitochondrial dysfunction is highly correlated to increased oxidative stress (Prabakaran et al., 2004; Calabrese et al., 2005; Lin and Beal, 2006), to evaluate the antioxidant response to oxidative stress, even though they were not identified as DEG by microarray analysis (Supplementary Table 2), the content of catalase (CAT) and superoxide dismutase 1 (SOD1) mRNA was evaluated by RT-PCR (Table 3). The amount of SOD1 in OSe pituitaries was greater (P = 0.018) than in ISe steers, whereas the amount of CAT mRNA did not differ (P = 0.138).
DISCUSSION
Animal Model
The reduction of serum prolactin by cattle consuming endophyte-infected tall fescue is a physiologic hallmark of fescue toxicosis. For example, the serum prolactin concentrations in growing beef steers subjected to summer-long grazing of high endophyte-infected tall fescue were decreased 85% to 90% relative to steers grazing low endophyte-infected forage (Brown et al., 2009; Jackson et al., 2015). Unlike the well-described suppression of serum prolactin in cattle consuming ergot alkaloids, the potential effect of supplemental Se form on serum prolactin and other indicators of fescue toxicosis has not been well characterized. For reasons (Matthews and Bridges, 2014; Matthews et al., 2014) outlined in Introduction, we conducted a trial comparing the potential ability of the form of Se in VM mixes (35 ppm) to ameliorate some of the characteristic effects of fescue toxicosis on growing beef steers. The results (Jia et al., 2018) showed that OSe and MIX steers subjected to grazing of endophyte-infected pasture had 59% (P < 0.03) and 52% (P < 0.05) more serum prolactin than ISe steers, respectively. Using the pituitaries from the same animals, the overall goal of the present study was to determine the effect of the form of supplemental Se in VM mix on expression of pituitary targeted mRNA content transcriptome profiles to gain insight into mechanisms responsible for Se-form-specific concentrations of serum prolactin.
The Content of Prolactin mRNA Is Greater in OSe and MIX Steer Pituitaries
The first goal of the present study was to test the specific hypothesis that the amount of prolactin mRNA would be greater in the pituitary tissue of the same (Jia et al., 2018) MIX and OSe vs. ISe steers. As shown in Table 2, the content of prolactin mRNA transcripts did not differ between MIX and ISe steers according to microarray analysis, whereas RT-PCR analysis found that MIX had 100% greater content of prolactin mRNA than ISe steers. In addition, OSe steers had 18% (microarray analysis) and 250% (RT-PCR analysis) greater content of prolactin mRNA than ISe steers (Table 2). Thus, we accept the original hypothesis that the amount of prolactin mRNA would be greater in the pituitary tissue of MIX and OSe vs. ISe steers.
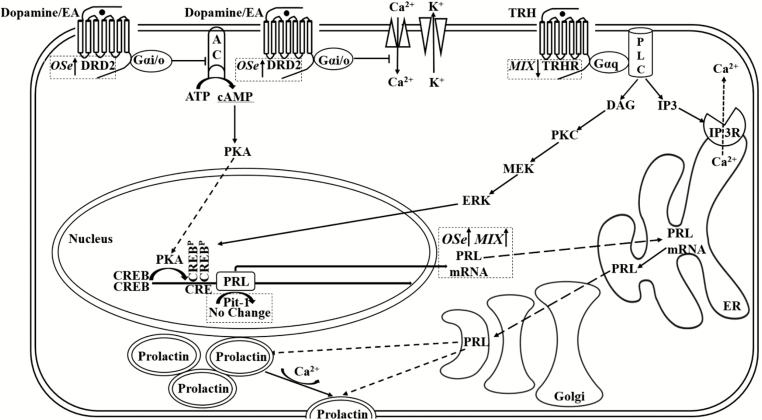
To gain insight into the mechanisms by which MIX and OSe steers had greater amounts of serum prolactin, the second goal of this experiment was to identify candidate molecules and signaling pathways in pituitary tissue known to be associated with prolactin synthesis (Figure 2) using microarray and RT-PCR transcript analyses (Table 2).
Figure 2.
Mechanisms, and mRNA expression responses to Se form treatments, by which dopamine and TRH affect prolactin synthesis and release. AC, adenylyl cyclase; CRE, cAMP response element; CREB, cAMP response element-binding protein; DAG, diacylglycerol; DRD2, dopamine receptor D2; EA, ergot alkaloid; ERK, extracellular signal-regulated kinase; IP3, inositol trisphosphate; ISe, sodium selenite; MEK, mitogen-activated protein kinase kinase; MIX, 1:1 mix of ISe and OSe; OSe, SEL-PLEX; Pit-1, pituitary-specific positive transcription factor 1; PKA, protein kinase A; PKC, protein kinase C; PLC, phospholipase C; PRL, prolactin; TRH, thyrotropin-releasing hormone; TRHR, thyrotropin-releasing hormone receptor. A line with arrowhead signifies interaction. A crosshead bar signifies inhibition. A dash line with arrowhead signifies transportation between cellular organelles. Adapted from Ben-Jonathan and Hnasko (2001) and Kanasaki et al. (2015). Sodium selenite (ISe), SEL-PLEX (OSe), or a 1:1 mix of ISe and OSe (MIX).
OSe Form of Se Supplementation Had Greater Prolactin Synthesis Capacity.
Dopamine is one of the most influential regulators of prolactin secretion. Activation of the dopamine type two receptor (DRD2) signaling by dopamine suppresses prolactin gene (PRL) expression via the inhibition of adenylyl cyclase and prolactin exocytosis through modification of several potassium and calcium channels (Fitzgerald and Dinan, 2008; Figure 2). Ergot alkaloids contained in endophyte-infected tall fescue resemble dopamine and trigger DRD2 signaling (Larson et al., 1999), resulting in decreased PRL transcription and serum prolactin concentrations (Strickland et al., 2011). In a previous summer-long grazing trial, the abundance of DRD2 mRNA was reduced in the pituitaries of steers that had decreased serum PRL as a result of grazing high versus low endophyte-infected tall fescue (Li et al., 2017). Consistent with this observation, consumption of endophyte-infected fescue seed reduced DRD2 mRNA and density in rat brain (Larson et al., 1994). Moreover, DRD2 mRNA and protein levels were down-regulated under constitutive hyperdopaminergia (Fauchey et al., 2000). Hence, agonists such as dopamine and ergot alkaloids negatively regulate DRD2 mRNA expression. Because OSe steers had more serum prolactin (Jia et al., 2018) and a greater pituitary content of PRL mRNA (Table 2) than ISe steers, we expected to find a greater pituitary DRD2 mRNA content in OSe steers. As expected, OSe steers did have greater pituitary DRD2 mRNA content than did ISe steers. Thus, it is possible that the serum prolactin difference between OSe and ISe steers (OSe > ISe) was caused by differential activation of DRD2 signaling (ISe > OSe) which was derived from different dopamine/ergot alkaloids concentrations (ISe > OSe) induced by different Se forms. This possibility also is consistent with the observation that consumption of ISe (selenite), but not OSe, resulted in increased dopamine concentrations in murine striatum (Tsunoda et al., 2000), where DRD2 mRNA is down-regulated by persistent stimulation of DRD2 (Chen et al., 1993).
Pituitary transcription factor Pit-1 (encoded by POU1F1) plays a pivotal role in PRL expression by binding to specific sites of promoter elements in the PRL gene and stimulating expression of prolactin mRNA (Fox et al., 1990). However, Pit-1 mRNA was not affected by Se treatment (Table 2, Figure 2). One explanation for the finding of no difference of Pit-1 mRNA but greater pituitary PRL mRNA in OSe vs. ISe steers could be that although Pit-1 is indispensable for prolactin production, an increase in Pit-1 mRNA is not necessary to promote PRL gene expression. For example, in estrogen-treated rats, lactotroph proliferation and enhanced expression of PRL mRNA but not an increase in Pit-1 mRNA have been observed (Tsukahara et al., 1994).
During DRD2-dependent inhibition of PRL gene expression, rapid histone deacetylation of prolactin promoter occurs after activation of DRD2 signaling, followed by inhibition of extracellular signal-regulated kinase (ERK)1/2 activity, and an unchanged association between Pit-1 and the prolactin promoter (Liu et al., 2005). Hence, a Pit-1-independent, epigenetic mechanism of DRD2 signaling also may be responsible for the difference between prolactin mRNA expression levels of OSe and ISe steers.
MIX Form Increased Prolactin Synthesis and Release Potential.
As found for OSe steers, MIX steers had a greater content of pituitary PRL mRNA (Table 2) and a greater serum prolactin concentration than ISe throughout the grazing period (Jia et al., 2018). However, in contrast to OSe steers, DRD2 mRNA content in MIX steers did not differ from ISe steers (Table 2), indicating the mechanisms by which both MIX and OSe steers had greater content of prolactin mRNA and serum prolactin levels likely differed. We have examined several other genes associated with prolactin secretion, and among them is TRH the principle prolactin secretagogue, which has been reported to stimulate prolactin production in both rat pituitary cells and cow pituitary tissue (Tashjian et al., 1971; Kelly et al., 1973). TRH induces prolactin mRNA levels via activation of ERK signaling pathway with synergistic increase in intracellular Ca2+ (White and Bancroft, 1983; Kanasaki et al., 2002) (Figure 2). TRH also was found to induce prolactin release from lactotrophs in a dose-dependent manner (Sheward et al., 1983; Lamberts and Macleod, 1990; Freeman et al., 2000). The way TRH stimulates prolactin release is via stimulation of Ca2+-dependent exocytosis in lactotrophs (Sikdar et al., 1989; Christian et al., 2007). It is known that TRHR mRNA expression is negatively regulated by TRH in rat pituitary (Oron et al., 1987; Narayanan et al., 1992). Hence, we tested mRNA expression of TRHR which have been detected in rat lactotrophs (Hinkle and Tashjian, 1973; Figure 2). Expression of TRHR decreased in MIX vs. ISe steers according to both microarray and real-time RT-PCR analyses (Table 2). Hence, the greater serum prolactin concentrations of MIX vs. ISe steers might be due to greater TRH concentrations available in pituitaries of MIX vs. ISe steers, which may have stimulated more prolactin synthesis and release in MIX vs. ISe steers.
TGFB1 has been shown present in lactotrophs and is capable of inhibiting prolactin release and lactotroph proliferation (Minami and Sarkar, 1997). Selenite was reported to inhibit the expression of TGFB1 induced by LPS (Pei et al., 2010). In agreement with the above study, we found that both OSe and MIX steers had more TGFB1 mRNA than ISe steers (Table 2). This finding is contradictive to the observation that serum prolactin levels were greater in OSe and MIX vs. ISe steers. One explanation to the contradiction is that the low level of TGFB1 expression limited its inhibitory potential over prolactin release, as the magnitude of TGFB1 mRNA expression appears to be lower than other genes in this network (e.g. 64-fold less than POU1F1 mRNA based on raw CT value, data not shown).
As mentioned above, we conducted RT-PCR analysis of several other genes with regard to prolactin synthesis and release, including VIP, GAL, GHRHR, VEGFA, and CSH2 based on IPA network analysis (Figure 1) and a previous study (Li et al., 2017). We also evaluated PRLR mRNA expression of both short form and long form. However, neither microarray nor RT-PCR showed mRNA expression of these genes above affected by Se treatment.
Besides synthesis and release, metabolic clearance of prolactin may contribute to the differences in serum prolactin concentrations. That is, the kidney has been reported to metabolize two-thirds of circulating prolactin (Emmanouel et al., 1981). Hence, future studies need to examine potential Se treatment-induced differences in prolactin clearance using the same steer model.
OSe Form of Se Supplementation Increased POMC/ACTH/α-MSH Synthesis Potential
POMC is a precursor polypeptide encoded by gene POMC and is synthesized mainly by corticotrophs of the anterior pituitary. Adrenocorticotropic hormone and α-MSH are two important hormones derived from POMC and secreted by the anterior pituitary and intermediate lobe of the pituitary, respectively (Figure 3). Adrenocorticotropic hormone induces the adrenal cortex to secrete glucocorticoids (Schwyzer, 1977), whereas α-MSH affects feeding behavior, energy homeostasis, and inflammation (Gantz and Fong, 2003). Previous research found that the liver tissue of beef steers grazing high vs. low endophyte-infected tall fescue and consuming ad libitum amounts of ISe-containing VM mix had increased amounts of mitochondrial mass, capacity for ATP synthesis, and AA-derived gluconeogenesis (Brown et al., 2009; Liao et al., 2015). These processes may have been coordinated through the glucocorticoid receptor-mediated pathway (Liao et al., 2015). A subsequent gene expression study of the pituitaries from these same steers (Li et al., 2017) found that the potential for pituitary POMC/ACTH synthesis was reduced in steers consuming forage with the high amounts of endophyte-infected tall fescue. This understanding, plus the finding that selenite inhibited glucocorticoid receptor hormone binding (Tashima et al., 1989), led to the general hypothesis of the present study that the form of supplemental Se would differentially affect mRNA content of pituitary genes related to POMC/ACTH synthesis in steers grazing endophyte-infected tall fescue.
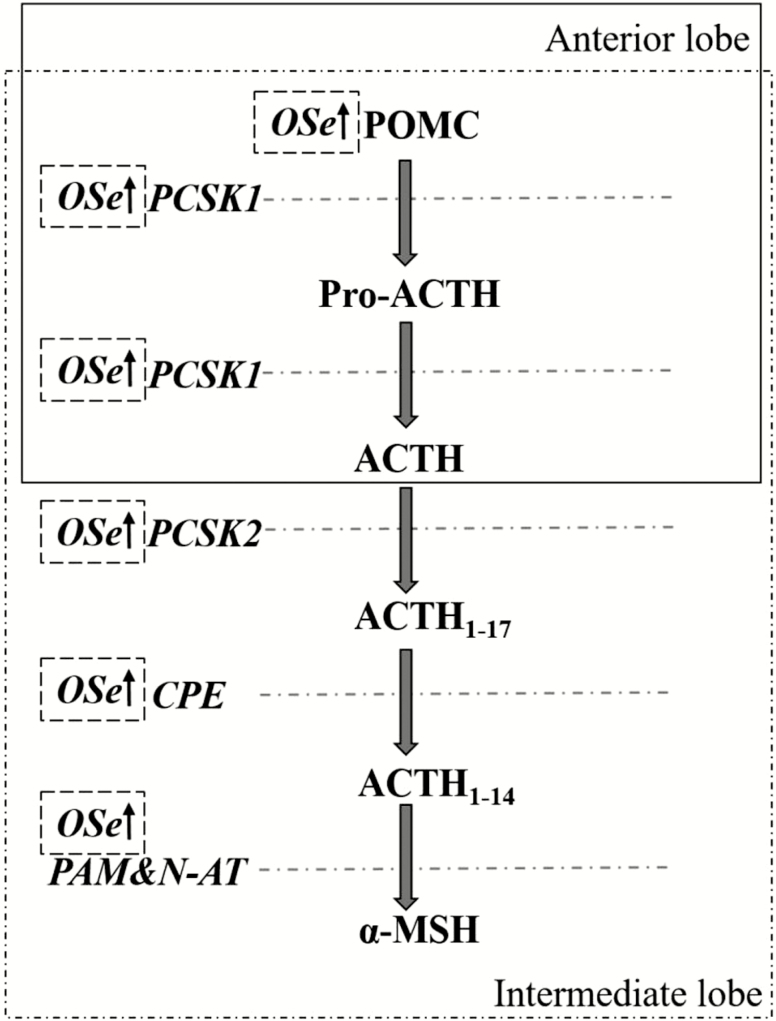
Figure 3.
Regional biosynthesis of ACTH and α-MSH from POMC in the pituitary. CPE, carboxypeptidase E; MSH, melanocyte-stimulating hormone; N-AT, N-acetyltransferase; PAM, peptidylglycine α-amidating monooxygenase; PCSK: proprotein convertase subtilisin/kexin. Adapted from Getting (2006) and Cawley et al. (2016).
In pituitary corticotrophs, proprotein convertase 1 (encoded by the PCSK1 gene) is expressed and cleaves POMC, producing ACTH1-39, β-endorphin, β-lipotrophin, amino-terminal peptide, and joining peptide (Millington, 2007). That the abundance of both POMC and PCSK1 mRNA was increased in pituitaries of OSe vs. ISe steers (Table 2) indicates that OSe steers possessed a greater POMC/ACTH synthesis capacity in OSe steers. To complete the assessment of Se treatment effects on the POMC/ACTH/α-MSH synthesis pathway (Figures 1 and 3), the expression of PCSK2, CPE, and PAM was evaluated (Table 2). Collectively, the results indicate that OSe steers possess greater POMC, ACTH, and α-MSH synthesis potential than ISe steers, and a greater α-MSH synthesis potential than MIX steers. As for prolactin, the exact physiological consequences of Se form-altered expression of ACTH and α-MSH remains to be determined.
Functional Analysis of the Genes Involved in Mitochondrial Dysfunction and Antioxidant Defense
As noted above, gene expression profiling indicated that the liver of steers grazing high vs. low endophyte-infected tall fescue and consuming ISe as an Se source had increased mitochondrial mass and respiratory chain mediated ATP synthetic capacity (Liao et al., 2015). The role that Se plays in preserving mitochondrial function is controversial. Whereas Se induces apoptosis associated with ROS accumulation and mitochondrial dysfunction (Guan et al., 2009), and selenite is detrimental to mitochondrial membrane potential by induction of mitochondrial permeability transition through thiol-oxidation (Kim et al., 2002), Se also is known to attenuate apoptosis (of at least damaged spinal cord tissue) through protection of mitochondrial function (Yeo et al., 2008) and shows a protective effect on cadmium-induced apoptosis in mice kidney (Wang et al., 2013). Because canonical pathway analysis of pituitary DEG identified “mitochondrial dysfunction” as one of the top pathways affected by Se treatment (Table 1), expression of DEG involved in mitochondrial dysfunction pathways was further examined by RT-PCR analysis, along with two genes (SOD1 and CAT) encoding key antioxidant enzymes.
Although microarray analysis showed that OSe steers expressed more NDUFA2, COX7A2, SDHB, and ATP5G1 mRNA content than ISe steers, RT-PCR analysis (Table 3) corroborated increased expression of NDUFA2 and COX7A2. Collectively, these data indicate that ISe had a reduced electron transport chain capacity than OSe steers, thereby likely less ATP generation and more damaging ROS in mitochondria (Bosetti et al., 2002; Musatov and Robinson, 2012; Saito et al., 2016). In terms of mitigating oxidative stress, genes involved with control of ROS production (LRRK2, CYB5A, and PSNENEN) and antioxidant production and use (FURIN, CAT, SOD1, and GPx4) were evaluated. OSe pituitaries expressed greater levels of SOD1, GPx4, and PSENEN than ISe steers and CYB5A and FURIN than ISe and MIX steers. Collectively, these findings strongly indicate that the pituitaries of OSe steers had a greater capacity to manage oxidative stress vs. ISe steers (Mates et al., 1999; Zangar et al., 2004; Heo et al., 2010).
In summary, consumption of 3 mg Se/d in VM mixes as OSe, MIX, or ISe differentially affected the expression of genes responsible for the synthesis or release of prolactin and POMC/ACTH/α-MSH, and for mitochondrial function, in the pituitaries of growing beef steers grazing an endophyte-infected tall fescue pasture. Consumption of OSe resulted in a greater prolactin synthesis capacity, whereas consumption of MIX resulted in increased prolactin synthesis and release potential, both of which resulted in greater serum prolactin concentrations in OSe and MIX steers vs. ISe steers, respectively. In addition, consumption of OSe resulted in greater POMC/ACTH/α-MSH synthesis potential than consumption of ISe and MIX forms of Se, and a better capacity to manage against mitochondrial dysfunction and oxidative stress, than consumption of ISe. The implications from these findings are that the inclusion of an organic form of Se in free-choice VM mixes can partially ameliorate the negative impact of fescue toxicosis on growing beef steers by restoration of both prolactin and POMC/ACTH synthesis capacities. In addition, because the role of prolactin is best understood in regulating lactation (Lamberts and Macleod, 1990; Freeman et al., 2000), it may be of especial commercial importance to evaluate the potential effect of MIX and OSe forms of Se in VM mixes to ameliorate the negative effects of grazing endophyte-infected tall fescue in lactating/sucking cow/calf pairs.
Supplementary Material
LITERATURE CITED
- Ben-Jonathan N., and Hnasko R.. 2001. Dopamine as a prolactin (PRL) inhibitor. Endocr. Rev. 22:724–763. doi: 10.1210/edrv.22.6.0451 [DOI] [PubMed] [Google Scholar]
- Bosetti F., Brizzi F., Barogi S., Mancuso M., Siciliano G., Tendi E. A., Murri L., Rapoport S. I., and Solaini G.. 2002. Cytochrome c oxidase and mitochondrial F1F0-atpase (ATP synthase) activities in platelets and brain from patients with Alzheimer’s disease. Neurobiol. Aging 23:371–376. doi: 10.1016/S0197-4580(01)00314-1 [DOI] [PubMed] [Google Scholar]
- Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C. A., Causton H. C.,. et al. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29:365–371. doi: 10.1038/ng1201-365 [DOI] [PubMed] [Google Scholar]
- Brown K. R., Anderson G. A., Son K., Rentfrow G., Bush L. P., Klotz J. L., Strickland J. R., Boling J. A., and Matthews J. C.. 2009. Growing steers grazing high versus low endophyte (neotyphodium coenophialum)-infected tall fescue have reduced serum enzymes, increased hepatic glucogenic enzymes, and reduced liver and carcass mass. J. Anim. Sci. 87:748–760. doi: 10.2527/jas.2008-1108 [DOI] [PubMed] [Google Scholar]
- Calabrese V., Lodi R., Tonon C., D’Agata V., Sapienza M., Scapagnini G., Mangiameli A., Pennisi G., Stella A. M., and Butterfield D. A.. 2005. Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich’s ataxia. J. Neurol. Sci. 233:145–162. doi: 10.1016/j.jns.2005.03.012 [DOI] [PubMed] [Google Scholar]
- Cawley N. X., Li Z., and Loh Y. P.. 2016. 60 years of POMC: biosynthesis, trafficking, and secretion of pro-opiomelanocortin-derived peptides. J. Mol. Endocrinol. 56:T77–T97. doi: 10.1530/JME-15-0323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. F., Aloyo V. J., and Weiss B.. 1993. Continuous treatment with the D2 dopamine receptor agonist quinpirole decreases D2 dopamine receptors, D2 dopamine receptor messenger RNA and proenkephalin messenger RNA, and increases mu opioid receptors in mouse striatum. Neuroscience 54:669–680. doi: 10.1016/0306-4522(93)90238-B [DOI] [PubMed] [Google Scholar]
- Christian H. C., Chapman L. P., and Morris J. F.. 2007. Thyrotrophin-releasing hormone, vasoactive intestinal peptide, prolactin-releasing peptide and dopamine regulation of prolactin secretion by different lactotroph morphological subtypes in the rat. J. Neuroendocrinol. 19:605–613. doi: 10.1111/j.1365-2826.2007.01567.x [DOI] [PubMed] [Google Scholar]
- Dargatz D. A., and Ross P. F.. 1996. Blood selenium concentrations in cows and heifers on 253 cow-calf operations in 18 states. J. Anim. Sci. 74:2891–2895. doi: 10.2527/1996.74122891x [DOI] [PubMed] [Google Scholar]
- Davenport G. M., Boling J. A., and Rahe C. H.. 1993. Growth and endocrine responses of cattle to implantation of estradiol-17 beta during continuous or discontinuous grazing of high- and low-endophyte-infected tall fescue. J. Anim. Sci. 71:757–764. doi:10.2527/1993.713757x [DOI] [PubMed] [Google Scholar]
- Emmanouel D. S., Fang V. S., and Katz A. I.. 1981. Prolactin metabolism in the rat: role of the kidney in degradation of the hormone. Am. J. Physiol. 240:F437–F445. doi: 10.1152/ajprenal.1981.240.5.F437 [DOI] [PubMed] [Google Scholar]
- Fauchey V., Jaber M., Caron M. G., Bloch B., and Le Moine C.. 2000. Differential regulation of the dopamine D1, D2 and D3 receptor gene expression and changes in the phenotype of the striatal neurons in mice lacking the dopamine transporter. Eur. J. Neurosci. 12:19–26. doi: 10.1046/j.1460-9568.2000.00876.x [DOI] [PubMed] [Google Scholar]
- Fitzgerald P., and Dinan T. G.. 2008. Prolactin and dopamine: what is the connection? A review article. J. Psychopharmacol. 22(2 Suppl):12–19. doi: 10.1177/0269216307087148 [DOI] [PubMed] [Google Scholar]
- Fox S. R., Jong M. T., Casanova J., Ye Z. S., Stanley F., and Samuels H. H.. 1990. The homeodomain protein, pit-1/GHF-1, is capable of binding to and activating cell-specific elements of both the growth hormone and prolactin gene promoters. Mol. Endocrinol. 4:1069–1080. doi: 10.1210/mend-4-7-1069 [DOI] [PubMed] [Google Scholar]
- Freeman M. E., Kanyicska B., Lerant A., and Nagy G.. 2000. Prolactin: structure, function, and regulation of secretion. Physiol. Rev. 80:1523–1631. doi: 10.1152/physrev.2000.80.4.1523 [DOI] [PubMed] [Google Scholar]
- Gantz I., and Fong T. M.. 2003. The melanocortin system. Am. J. Physiol. Endocrinol. Metab. 284:E468–E474. doi: 10.1152/ajpendo.00434.2002 [DOI] [PubMed] [Google Scholar]
- Getting S. J. 2006. Targeting melanocortin receptors as potential novel therapeutics. Pharmacol. Ther. 111:1–15. doi: 10.1016/j.pharmthera.2005.06.022 [DOI] [PubMed] [Google Scholar]
- Goetsch A. L., Jones A. L., Stokes S. R., Beers K. W., and Piper E. L.. 1987. Intake, digestion, passage rate and serum prolactin in growing dairy steers fed endophyte-infected fescue with noninfected fescue, clover or wheat straw. J. Anim. Sci. 64:1759–1768. doi: 10.2527/jas1987.6461759x [DOI] [PubMed] [Google Scholar]
- Guan L., Han B., Li Z., Hua F., Huang F., Wei W., Yang Y., and Xu C.. 2009. Sodium selenite induces apoptosis by ROS-mediated endoplasmic reticulum stress and mitochondrial dysfunction in human acute promyelocytic leukemia NB4 cells. Apoptosis 14:218–225. doi: 10.1007/s10495-008-0295-5 [DOI] [PubMed] [Google Scholar]
- Heo H. Y., Park J. M., Kim C. H., Han B. S., Kim K. S., and Seol W.. 2010. LRRK2 enhances oxidative stress-induced neurotoxicity via its kinase activity. Exp. Cell Res. 316:649–656. doi: 10.1016/j.yexcr.2009.09.014 [DOI] [PubMed] [Google Scholar]
- Hinkle P. M., and Tashjian A. H. Jr. 1973. Receptors for thyrotropin-releasing hormone in prolactin producing rat pituitary cells in culture. J. Biol. Chem. 248:6180–6186. [PubMed] [Google Scholar]
- Jackson J. J., Lindemann M. D., Boling J. A., and Matthews J. C.. 2015. Summer-long grazing of high vs. low endophyte (neotyphodium coenophialum)-infected tall fescue by growing beef steers results in distinct temporal blood analyte response patterns, with poor correlation to serum prolactin levels. Front. Vet. Sci. 2:77. doi: 10.3389/fvets.2015.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Li Q., Burris W. R., Aiken G. E., Bridges P. J., and Matthews J. C.. 2018. Forms of selenium in vitamin-mineral mixes differentially affect serum prolactin concentration and hepatic glutamine synthetase activity of steers grazing endophyte-infected tall fescue. J. Anim. Sci. 96:715–727. doi: 10.1093/jas/skx068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasaki H., Oride A., Mijiddorj T., and Kyo S.. 2015. Role of thyrotropin-releasing hormone in prolactin-producing cell models. Neuropeptides 54:73–77. doi: 10.1016/j.npep.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Kanasaki H., Yonehara T., Yamamoto H., Takeuchi Y., Fukunaga K., Takahashi K., Miyazaki K., and Miyamoto E.. 2002. Differential regulation of pituitary hormone secretion and gene expression by thyrotropin-releasing hormone. A role for mitogen-activated protein kinase signaling cascade in rat pituitary GH3 cells. Biol. Reprod. 67: 107–113. doi: 10.1095/biolreprod67.1.107 [DOI] [PubMed] [Google Scholar]
- Kelly P. A., Bedirian K. N., Baker R. D., and Friesen H. G.. 1973. Effect of synthetic TRH on serum prolactin, TSH and milk production in the cow. Endocrinology 92:1289–1293. doi: 10.1210/endo-92-4-1289 [DOI] [PubMed] [Google Scholar]
- Kim T. S., Jeong D. W., Yun B. Y., and Kim I. Y.. 2002. Dysfunction of rat liver mitochondria by selenite: induction of mitochondrial permeability transition through thiol-oxidation. Biochem. Biophys. Res. Commun. 294:1130–1137. doi: 10.1016/S0006-291X(02)00612-5 [DOI] [PubMed] [Google Scholar]
- Lamberts S. W., and Macleod R. M.. 1990. Regulation of prolactin secretion at the level of the lactotroph. Physiol. Rev. 70:279–318. doi: 10.1152/physrev.1990.70.2.279 [DOI] [PubMed] [Google Scholar]
- Larson B. T., Harmon D. L., Piper E. L., Griffis L. M., and Bush L. P.. 1999. Alkaloid binding and activation of D2 dopamine receptors in cell culture. J. Anim. Sci. 77:942–947. doi: 10.2527/1999.774942x [DOI] [PubMed] [Google Scholar]
- Larson B. T., Sullivan D. M., Samford M. D., Kerley M. S., Paterson J. A., and Turner J. T.. 1994. D2 dopamine receptor response to endophyte-infected tall fescue and an antagonist in the rat. J. Anim. Sci. 72:2905–2910. doi: 10.2527/1994.72112905x [DOI] [PubMed] [Google Scholar]
- Li Q., Hegge R., Bridges P. J., and Matthews J. C.. 2017. Pituitary genomic expression profiles of steers are altered by grazing of high vs. Low endophyte-infected tall fescue forages. PLoS One 12:e0184612. doi: 10.1371/journal.pone.0184612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. F., Boling J. A., and Matthews J. C.. 2015. Gene expression profiling indicates an increased capacity for proline, serine, and ATP synthesis and mitochondrial mass by the liver of steers grazing high vs. low endophyte-infected tall fescue. J. Anim. Sci. 93:5659–5671. doi: 10.2527/jas.2015-9193 [DOI] [PubMed] [Google Scholar]
- Lin M. T., and Beal M. F.. 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795. doi: 10.1038/nature05292 [DOI] [PubMed] [Google Scholar]
- Liu J. C., Baker R. E., Chow W., Sun C. K., and Elsholtz H. P.. 2005. Epigenetic mechanisms in the dopamine D2 receptor-dependent inhibition of the prolactin gene. Mol. Endocrinol. 19:1904–1917. doi: 10.1210/me.2004-0111 [DOI] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Matés J. M., Pérez-Gómez C., and Núñez de Castro I.. 1999. Antioxidant enzymes and human diseases. Clin. Biochem. 32:595–603. [DOI] [PubMed] [Google Scholar]
- Matthews J.C., and Bridges P.J.. 2014. NutriPhysioGenomics applications to identify adaptations of cattle to consumption of ergot alkaloids and inorganic versus organic forms of selenium: altered nutritional, physiological and health states?Anim. Prod. Sci. 54:1594–1604. doi: 10.1071/AN14274 [DOI] [Google Scholar]
- Matthews J. C., Zhang Z., Patterson J. D., Bridges P. J., Stromberg A. J., and Boling J. A.. 2014. Hepatic transcriptome profiles differ among maturing beef heifers supplemented with inorganic, organic, or mixed (50% inorganic:50% organic) forms of dietary selenium. Biol. Trace Elem. Res. 160:321–339. doi: 10.1007/s12011-014-0050-4 [DOI] [PubMed] [Google Scholar]
- Millington G. W. 2007. The role of proopiomelanocortin (POMC) neurones in feeding behaviour. Nutr. Metab. (Lond.). 4:18. doi: 10.1186/1743-7075-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami S., and Sarkar D. K.. 1997. Transforming growth factor-beta 1 inhibits prolactin secretion and lactotropic cell proliferation in the pituitary of oestrogen-treated Fischer 344 rats. Neurochem. Int. 30:499–506. doi: 10.1016/S0197-0186(96)00087-3 [DOI] [PubMed] [Google Scholar]
- Musatov A., and Robinson N. C.. 2012. Susceptibility of mitochondrial electron-transport complexes to oxidative damage. Focus on cytochrome c oxidase. Free Radic. Res. 46:1313–1326. doi: 10.3109/10715762.2012.717273 [DOI] [PubMed] [Google Scholar]
- Narayanan C. S., Fujimoto J., Geras-Raaka E., and Gershengorn M. C.. 1992. Regulation by thyrotropin-releasing hormone (TRH) of TRH receptor mRNA degradation in rat pituitary GH3 cells. J. Biol. Chem. 267:17296–17303. [PubMed] [Google Scholar]
- Oron Y., Straub R. E., Traktman P., and Gershengorn M. C.. 1987. Decreased TRH receptor mRNA activity precedes homologous downregulation: assay in oocytes. Science 238:1406–1408. [DOI] [PubMed] [Google Scholar]
- Partek D. 2009. Partek documentation: turning data into discovery. St. Louis:Partek Incorporated. [Google Scholar]
- Pei Z., Li H., Guo Y., Jin Y., and Lin D.. 2010. Sodium selenite inhibits the expression of VEGF, TGFbeta(1) and IL-6 induced by LPS in human PC3 cells via TLR4-NF-(K)B signaling blockage. Int. Immunopharmacol. 10:50–56. doi: 10.1016/j.intimp.2009.09.020 [DOI] [PubMed] [Google Scholar]
- Prabakaran S., Swatton J. E., Ryan M. M., Huffaker S. J., Huang J. T., Griffin J. L., Wayland M., Freeman T., Dudbridge F., Lilley K. S.,. et al. 2004. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol. Psychiatry 9:684–697, 643. doi: 10.1038/sj.mp.4001511 [DOI] [PubMed] [Google Scholar]
- Saito Y., Ishii K. A., Aita Y., Ikeda T., Kawakami Y., Shimano H., Hara H., and Takekoshi K.. 2016. Loss of SDHB elevates catecholamine synthesis and secretion depending on ROS production and HIF stabilization. Neurochem. Res. 41:696–706. doi: 10.1007/s11064-015-1738-3 [DOI] [PubMed] [Google Scholar]
- Schwyzer R. 1977. ACTH: a short introductory review. Ann. N. Y. Acad. Sci. 297:3–26. doi: 10.1111/j.1749-6632.1977.tb41843.x [DOI] [PubMed] [Google Scholar]
- Sheward W. J., Harmar A. J., Fraser H. M., and Fink G.. 1983. Thyrotropin-releasing hormone in rat pituitary stalk blood and hypothalamus: studies with high performance liquid chromatography. Endocrinology 113:1865–1869. doi: 10.1210/endo-113-5-1865 [DOI] [PubMed] [Google Scholar]
- Sikdar S. K., Zorec R., Brown D., and Mason W. T.. 1989. Dual effects of G-protein activation on ca-dependent exocytosis in bovine lactotrophs. FEBS Lett. 253:88–92. doi: 10.1016/0014-5793(89)80936-6 [DOI] [PubMed] [Google Scholar]
- Strickland J. R., Looper M. L., Matthews J. C., Rosenkrans C. F. Jr, Flythe M. D., and Brown K. R.. 2011. Board-invited review: st. Anthony’s fire in livestock: causes, mechanisms, and potential solutions. J. Anim. Sci. 89:1603–1626. doi: 10.2527/jas.2010-3478 [DOI] [PubMed] [Google Scholar]
- Tashima Y., Terui M., Itoh H., Mizunuma H., Kobayashi R., and Marumo F.. 1989. Effect of selenite on glucocorticoid receptor. J. Biochem. 105:358–361. [DOI] [PubMed] [Google Scholar]
- Tashjian A. H. Jr, Barowsky N. J., and Jensen D. K.. 1971. Thyrotropin releasing hormone: direct evidence for stimulation of prolactin production by pituitary cells in culture. Biochem. Biophys. Res. Commun. 43:516–523. doi: 10.1016/0006-291X(71)90644-9 [DOI] [PubMed] [Google Scholar]
- Thompson I. M., Ozawa M., Bubolz J. W., Yang Q., and Dahl G. E.. 2011. Bovine luteal prolactin receptor expression: potential involvement in regulation of progesterone during the estrous cycle and pregnancy. J. Anim. Sci. 89:1338–1346. doi: 10.2527/jas.2010-3559 [DOI] [PubMed] [Google Scholar]
- Tsukahara S., Kambe F., Suganuma N., Tomoda Y., and Seo H.. 1994. Increase in pit-1 mRNA is not required for the estrogen-induced expression of prolactin gene and lactotroph proliferation. Endocr. J. 41:579–584. doi: 10.1507/endocrj.41.579 [DOI] [PubMed] [Google Scholar]
- Tsunoda M., Johnson V. J., and Sharma R. P.. 2000. Increase in dopamine metabolites in murine striatum after oral exposure to inorganic but not organic form of selenium. Arch. Environ. Contam. Toxicol. 39:32–37. doi: 10.1007/s002440010076 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wu Y., Luo K., Liu Y., Zhou M., Yan S., Shi H., and Cai Y.. 2013. The protective effects of selenium on cadmium-induced oxidative stress and apoptosis via mitochondria pathway in mice kidney. Food Chem. Toxicol. 58:61–67. doi: 10.1016/j.fct.2013.04.013 [DOI] [PubMed] [Google Scholar]
- White B. A., and Bancroft F. C.. 1983. Epidermal growth factor and thyrotropin-releasing hormone interact synergistically with calcium to regulate prolactin mRNA levels. J. Biol. Chem. 258:4618–4622. [PubMed] [Google Scholar]
- Yeo J. E., Kim J. H., and Kang S. K.. 2008. Selenium attenuates ROS-mediated apoptotic cell death of injured spinal cord through prevention of mitochondria dysfunction; in vitro and in vivo study. Cell. Physiol. Biochem. 21:225–238. doi: 10.1159/000113764 [DOI] [PubMed] [Google Scholar]
- Zangar R. C., Davydov D. R., and Verma S.. 2004. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol. Appl. Pharmacol. 199:316–331. doi: 10.1016/j.taap.2004.01.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.