Abstract
Interactions between regulatory pathways allow organisms to adapt to their environment and respond to stress. One interaction that has been recently identified occurs between the aryl hydrocarbon receptor (AHR) and the nuclear factor erythroid-2 related factor (NRF) family. Each transcription factor regulates numerous downstream genes involved in the cellular response to toxicants and oxidative stress; they are also implicated in normal developmental pathways. The zebrafish model was used to explore the role of AHR regulation of nrf genes during development and in response to toxicant exposure. To determine if AHR1b is responsible for transcriptional regulation of 6 nrf genes during development, a loss-of-function experiment using morpholino-modified oligonucleotides was conducted followed by a chromatin immunoprecipitation study at the beginning of the pharyngula period (24 h postfertilization). The expression of nrf1a was AHR1b dependent and its expression was directly regulated through specific XREs in its cis-promoter. However, nrf1a expression was not altered by exposure to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD), a toxicant and prototypic AHR agonist. The expression of nrf1b, nrf2a, and nfe2 was induced by TCDD, and AHR1b directly regulated their expression by binding to cis-XRE promoter elements. Last, nrf2b and nrf3 were neither induced by TCDD nor regulated by AHR1b. These results show that AHR1b transcriptionally regulates nrf genes under toxicant modulation via binding to specific XREs. These data provide a better understanding of how combinatorial molecular signaling potentially protects embryos from embryotoxic events following toxicant exposure.
Keywords: AHR, ChIP, development, nrf, TCDD, zebrafish
The expression of most genes is carefully regulated by multiple transcription factors. The crosstalk between these transcription factors allows biological systems to respond to endogenous and exogenous signals, and adapt to stress. Recently, the aryl hydrocarbon receptor (AHR) and the nuclear factor erythroid-2 related factor-2 (NRF2) were discovered to engage in crosstalk in multiple biological systems (Kalthoff et al., 2010; Ma et al., 2004; Miao et al., 2005; Shin et al., 2007; Timme-Laragy et al., 2012; Yeager et al., 2009). Because AHR and NRF2 are already known to each regulate a large and diverse set of target genes (Boutros et al., 2009; Ma, 2013), crosstalk could also play a significant role.
AHR is a highly conserved ligand-activated transcription factor (Denison and Nagy, 2003) that binds to dioxin response elements (DREs)/xenobiotic response elements (XREs) (Denison et al., 1988) on the promoter of target genes (Beischlag et al., 2008). The AHR transcriptional program is extensively implicated in toxicology, carcinogenesis, physiology, and development, which highlights the importance of understanding how it interacts with other signaling pathways, including those controlled by NRF2 and related proteins.
In mammals, 4 Cap’n’collar (CNC) basic leucine zipper (bZIP) transcription factors have been characterized: nuclear factor erythroid-2 (NF-E2), NF-E2-related factor-1 (NRF1; also called NFE2L1), NF-E2-related factor- 2 (NRF2 or NFE2L2), and NF-E2-related factor- 3 (NRF3 or NFE2L3). CNC-bZIP transcription factors are responsible for the cellular transcriptional response to oxidative stress (Ma and He, 2012; Sykiotis and Bohmann, 2010). Although the expression patterns and cellular functions of various NRF proteins vary widely, most have been found to play a role in basal cellular functions and response to imbalances in cellular redox conditions (Chevillard and Blank, 2011; Gasiorek and Blank, 2015; Hahn et al., 2015; Ma, 2013; Zhang and Xiang, 2016). The many and diverse cellular roles make understanding NRF regulation and intrapathway interactions essential.
Genes that are transcriptionally activated through either xenobiotic or antioxidant response elements (XRE or ARE), and thus by AHR or NRF proteins, respectively, were originally thought to be independently regulated. The first gene found to be dependent on both proteins was NAD(P)H: quinone oxidoreductase 1 (NQO1). Induction of NQO1 by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) requires XRE (Favreau and Pickett, 1991), whereas induction by the prototypical oxidant tert-butylhydroquinone (tBHQ) proceeds through protein binding to AREs (Radjendirane and Jaiswal, 1999). A subsequent study showed that basal and inducible expression of NQO1 by either TCDD or tBHQ required the interaction of AHR and NRF2 (Ma et al., 2004). The promoter region of NQO1 as well as GSTA1 and UGT1A10 contain XRE and ARE in close proximity to each other, suggesting that AHR and NRF2 may physically interact to drive transcription of the genes (Kalthoff et al., 2010; Vasiliou et al., 1995). In addition, the induction by TCDD of AHR-regulated genes, including UGT1A6, GSTA1, and other UDP glucuronosyltransferases and glutathione S-transferase isoforms was then shown to require NRF2 (Yeager et al., 2009). These studies presented a new paradigm in gene regulation, that of the “TCDD-inducible AHR-NRF2 gene battery” (Yeager et al., 2009), raising important questions about which other genes may be regulated through this mechanism, especially during the most sensitive and consequential life stage, the embryo.
Zebrafish have emerged as a powerful model for studying molecular mechanisms of vertebrate development and developmental toxicology (Horzmann and Freeman, 2018; Shin and Fishman, 2002; Tanguay, 2018; Ward and Lieschke, 2002). In zebrafish, 3 ahr genes (Hahn et al., 2017) and 6 nrf genes (Timme-Laragy et al., 2012) have been identified, arising from a whole genome duplication event (Amores et al., 1998; Taylor et al., 2001). Subsequently, the genes (single copies of nfe2 and nrf3 as well paralogs nrf1a, nrf1b, nrf2a, and nrf2b) and their protein products have subfunctionalized in their temporal and spatial expression patterns as well as in their transcriptional roles (Kobayashi et al., 2009; Pratt et al., 2002; Timme-Laragy et al., 2012; Williams et al., 2013). All nrf genes have putative XREs in their cis-promoters (Williams et al., 2013), making them potential targets of Ahr regulation.
Of particular interest to this study was ahr1b due to its expression throughout development (Karchner et al., 2005) and spatial expression in the developing eye (Karchner et al., 2017; Sugden et al., 2017), an organ that at 24 h postfertilization (hpf) makes up a large percentage of the larval body mass and is critical to later life behavior such as feeding and predator evasion (Glass and Dahm, 2004). Although ahr1b’s transcription is unaffected by chemical exposure to TCDD (Karchner et al., 2005) or β-naphthoflavone (Sugden et al., 2017), AHR1b has the ability to bind TCDD and is able to activate transcription of a reporter gene under the control of XREs (Karchner et al., 2005). The goal of this study was to assess whether AHR1b transcriptionally regulates the expression of nrf gene family members through binding to XREs.
MATERIALS AND METHODS
Chemicals
TCDD was purchased from Ultra Scientific (N. Kingston, Rhode Island) and dimethyl sulfoxide (DMSO) was purchased from Sigma-Aldrich (St Louis, Missouri).
Fish husbandry and strains
For all experiments, the Tupfel/Longfin mutation wild-type strain was used. Adults and embryos were maintained and used as previously described in Jönsson et al. (2007). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Woods Hole Oceanographic Institution Animal Care and Use Committee (Animal Welfare Assurance Number A3630-01) and the Bates College Animal Care and Use Committee (Animal Welfare Assurance Number A3320-01).
Morpholino oligonucleotides
A morpholino antisense oligonucleotide (MO), designed to block translation of ahr1b (AGGCACCCAAAATCTCAATATCACA) by binding upstream of the translational start site, was obtained from Gene Tools, LLC (Philomath, Oregon). The standard control-MO from Gene Tools (CCTCTTACCTCAGTTACAATTTATA) was used as an injection and MO control.
The efficacy of the translational knockdown of AHR1b was assessed via the TNT T7 Quick Coupled Reticulocyte Lysate system (Promega, Madison, Wisconsin). [35S]methionine-labeled AHR1b was synthesized per manufacturer’s instructions in the presence of ahr1b-MO or control-MO (0.05 μM final concentration) and synthesis of AHR1b was analyzed by gel electrophoresis and fluorography as previously described (Jenny et al., 2009). Efficacy of knockdown was quantified for a single experiment by comparing the densitometric signals of the ahr1b- and control-MO samples using ImageJ (Schneider et al., 2012).
Microinjection of zebrafish embryos with morpholinos and chemical exposure
Embryos between the 2- and 4-cell stage were injected with 2–2.5 nL of a 0.1 mM solution of ahr1b MO as previously described in Jenny et al. (2009) and Williams et al. (2013).
At 6 hpf, control MO and ahr1b injected embryos (3 pools of 30 embryos) were placed in glass scintillation vials containing no more than 3 embryos per milliliter of 0.3× Danieau’s and then exposed to either 0.1% DMSO (vehicle control) or 2 nM TCDD (dissolved in DMSO) for 1 h. The timing, duration, and TCDD concentration were chosen because they did not cause severe toxicity but did elicit strong induction of cyp1a (Jenny et al., 2009), a prototypic AHR-regulated gene (Whitlock, 1999). After the exposure, embryos were washed in 0.3× Danieau’s and then placed in petri dishes containing fresh 0.3× Danieau’s and held in an incubator with a 14/10-light/dark cycle until 24 hpf (Jenny et al., 2009).
RNA extraction, cDNA synthesis, and real-time quantitative RT-PCR
Using RNA STAT-60 (AMS Biotechnology, Abingdon, UK), total RNA was isolated from pooled embryos following manufacturer’s instructions. Following analysis of the isolated RNA with a Nanodrop 2000 (Thermo Fisher Scientific, Waltham, Massachusetts), 1 μg of total RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, California).
Quantitative RT-PCR (qPCR) on all nrf transcripts (nfe2, nrf1a, nrf1b, nrf2a, nrf2b, and nrf3) and a housekeeping gene (β-actin) in control MO and ahr1b MO samples was conducted using the MyiQ Single-Color Real-Time PCR Detection system (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad) with the protocol and primers described previously (Evans et al., 2005; Timme-Laragy et al., 2012; Williams et al., 2013). Relative expression of each gene was analyzed using the ΔΔCT method (Livak and Schmittgen, 2001). Statistical differences in expression were determined using a 2-way ANOVA followed by a Sidak’s multiple comparisons test in Graphpad Prism 7 software (La Jolla, California).
In silico promoter analysis of XREs
An in silico promoter analysis was carried out on 10 000 bp upstream of the transcriptional start site to identify potential XREs [KNGCGTG] (Lusska et al., 1993; ZeRuth and Pollenz, 2007) in the cis-promoter of nrf genes where AHR1b could potentially bind. Both strands of the DNA were searched using a fuzzy search algorithm, fuzznuc (Rice et al., 2000), as previously described in Williams et al. (2013).
Antibody against AHR1b and confirmation of antibody specificity
A rabbit polyclonal antibody targeting AHR1b protein was raised against the peptide LENQTEDPAESQKPSTA (amino acids 592-608 of AHR1b) by 21st Century Biochemicals (Marlboro, Massachusetts) and affinity-purified. Antibody specificity was assessed by Western blotting of COS-7 lysates expressing AHR1b from transfected cDNA (data not shown) and zebrafish embryo homogenates. In 3 independent experiments, 200 embryos were exposed to DMSO or 2 nM TCDD starting at 6 hpf for 1 hour and prepared for Western blotting at 24 hpf using methods that are previously described in Westerfield (2007). Following electrophoresis of 10 μL of homogenate from each sample on a 10% Mini-PROTEAN TGX Stain-Free gel (Bio-Rad), total protein was quantified with a Biorad Chemidoc imaging system by measuring the total density of protein in each lane using Biorad ImageLab 6.0 (Gilda and Gomes, 2013). This step ensured equal loading of total protein in each well. Protein was then transferred to an Immuno-Blot LF PVDF membrane (Bio-Rad) and preblocked in 5% milk in 1× Tris-buffered saline Tween (TBST) for 1 hour at room temperature. After blocking, the membrane was incubated with AHR1b (1 μg/ml) antibody in Tris-buffered saline for 1 h at room temperature. Following 3 washes with 1× TBST, a goat antirabbit IgG (H + L)-HRP conjugate secondary antibody (Bio-Rad) was used at a dilution of 1:3000 and incubated for 1 h. Following three 1× TBST washes, Clarity Max Western ECL substrate (Bio-Rad) was added to the blot and the blot was exposed for 60 s on a Biorad Chemidoc imaging system. ImageJ (Schneider et al., 2012) was used to quantify and compare the AHR1b protein between DMSO and TCDD samples. A 1-way ANOVA was used to determine statistical differences in the relative density of the AHR1b specific band between DMSO and TCDD treated embryos in GraphPad Prism 7 software, with the DMSO-treated samples serving as the control in the density calculations (Taylor et al., 2013).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was carried out in triplicate on 24 hpf embryos as described previously in Aday et al. (2011) with slight modifications. For each independent sample, 300 embryos were used. Embryos were manually dechorionated. Samples were sonicated with a Diagnode Bioruptor Pico with a water cooler set to 4°C (Denville, New Jersey) for 13 cycles of 30 s on and 30 s off. Shearing was verified with gel electrophoresis to ensure a fragment size between 200 and 300 bp. Samples were not precleared or preblocked. A total of 8 μg of AHR1b antibody was used in the immunoprecipitation.
Immunoprecipitated and input samples were amplified for quantification with qPCR using a Stratagene Mx3000P qPCR Machine and Agilent Brilliant II SYBR Green dye. Primers for each XRE in the cis-promoter of all nrf genes were designed (Table 1) and used. For the positive control, primers were designed to amplify XRE 4 from cyp1a1 (ZeRuth et al., 2007), and the negative control primers amplified a segment of nrf1a cis-promoter that was at least 5800 bp upstream from any of the 2 identified XREs. Promoter content was calculated as previously described in Hestermann and Brown (2003).
Table 1.
Quantitative Real-Time PCR Primers Used to Amplify XRE Containing Regions of nrf cis-Promoters Following ChIP in 24 hpf zebrafish
| Gene and XRE | Forward Primer | Reverse Primer | XRE Sequence | Amplicon Length |
|---|---|---|---|---|
| nrf1a XRE1 | TGTTGTTTA TATAGGTCA ACTCATAAG | TTGGAGGAG ATCAGGGTT ACT | CACGCGC | 76 |
| nrf1a XRE2 | GGCACACAT AACCTACCA GAG | CGCTACGGT GAGGTTGAG CC | GGGCGTG | 166 |
| nrf1a XRE3 | ACACTGATG AACGGAGTG AG | GAGTCG GTG AGTGCTGGA AAGC | CACGCGC | 104 |
| nrf1a Negative Control | CTCATCTGC ATACAGGAC TCAC | GTCTACAAG AGCAGCATA GACC | — | 82 |
| nrf1b XRE1 | GACGCAGTG CTCTAATAT GGC | CTTGTCCTG AAAGAACTC GG | TAGCGTG | 73 |
| nrf2a XRE1 | CTGCTCTCC GCCTGTTTA C | AGTGCTCTG CTCCACATT TG | GCGCGTG | 108 |
| nrf2a XRE2 | CATGCACGC ACAGTCTAA TC | CAGTGTTCC CAGTGCTTT AC | CACGCAC | 84 |
| nrf2a XRE3 | AGCTTCAGC TGGAGAATG TC | GACTAGGAG AACAGTGAA GTGC | CACGCACGCGC | 115 |
| nrf2a XRE4 | GCACTTCAC TGTTCTCCT AGTC | CAGACACAC ATCTCCAGC AC | GTGCGTG | 147 |
| nrf2b XRE1 | GTCCAGCAG TTCTTATCA GC | GACCATCTG TGTAGTCTT CTGC | CACGCCA | 115 |
| nrf2b XRE2 | GTGACGCAG TGCTCTAAT ATG | TTTCTTGTC CTGAAAGAA CTCG | CACGCGA | 89 |
| nfe2 XRE1 | CCTCTTTGG ACGGTGTCA GCC | CGCTCCATC AAGCCTCAT GCC | CACGCCA | 146 |
| nfe2 XRE2 | CAAAGCCTC CACTGGTCA TGG | GCCACATTG TTCCTGTAT CTGC | CACGCTC | 79 |
| nrf3 XRE1 | AACCTTATT GTAAAGTGT GACCATC | ATAGCTCCA GTCCCCCTA GC | CACGCCA | 125 |
| nrf3 XRE2 | CTCGGACAG CAATATCTC CTTG | CGACCTTGC CATTCCTAT AAC | CACGCAA | 89 |
| cyp1a1 XRE4 Positive Control | CATTCCGCC AGCTCTTCC TG | GCCTGCATG TTTGAGTCT CTGC | CGCGTG | 114 |
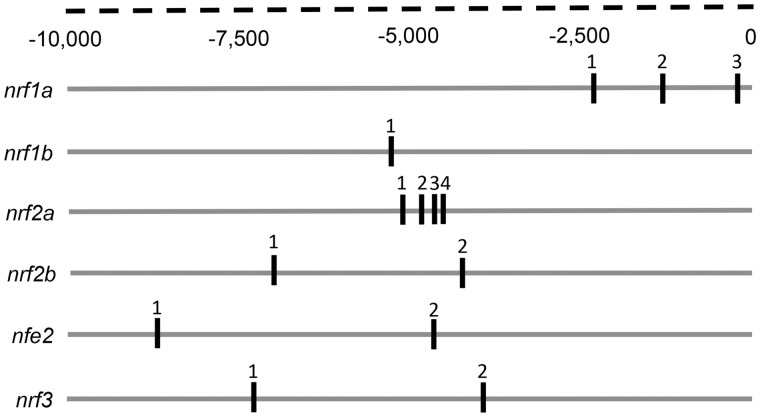
Primers are written 5’ to 3’ and amplicon length is given. Putative XRE sequence locations are mapped in Figure 1.
RESULTS
Given that there was at least 1 well-conserved putative XRE in the cis-promoter of each nrf gene (Figure 1, Table 1) (Williams et al., 2013), we hypothesized that the temporary suppression of AHR1b by a MO could affect the transcription of these genes during the pharyngula period. In vitro, the MO knocked down the expression of AHR1b by 66% (Figure 2) and thus was considered to be effective at reducing AHR1b in vivo. It is advisable that future studies test the knockdown of AHR1b via MO in embryos using the AHR1b antibody.
Figure 1.
Putative XREs in the proximal regulatory region of zebrafish nrf genes. Specific XREs were identified using a fuzzy search for canonical zebrafish XREs within each cis-nrf promoter 10, 000 bp upstream of the transcriptional start site. Exon positions were exported from the Ensembl database (Zv9).
Figure 2.
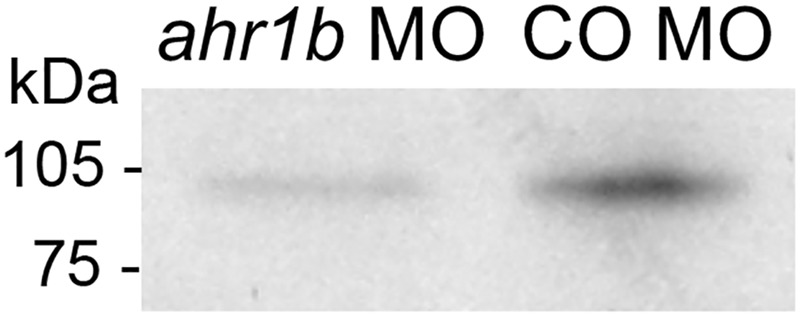
Morpholino efficacy as assessed by in vitro translation. The in vitro translation of AHR1b was measured using incorporation of [35S]methionine in the presence of ahr1b MO or control MO. The ahr1b MO decreased the expression of AHR1b by 66%.
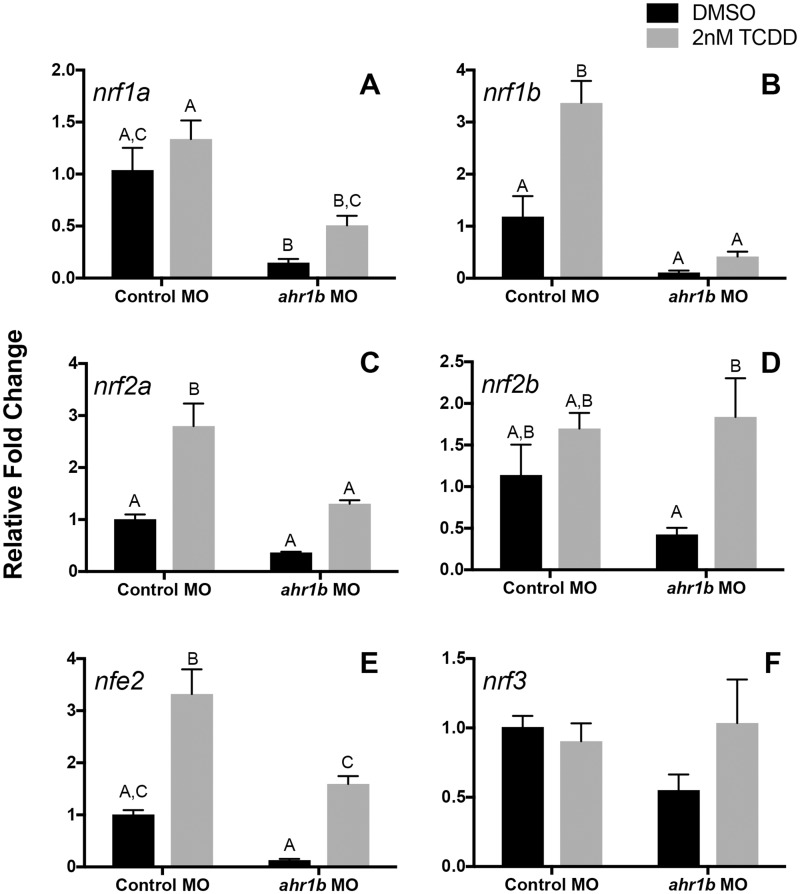
Upon exposure to TCDD, nrf1b, nrf2a, and nfe2 transcripts were induced and the relative fold change values of nrf1b and nrf2a were significantly decreased upon knockdown of AHR1b (Figs. 3B and 3C). Interestingly, there was little difference in the TCDD-induced fold induction ratio of nrf1b in the control morphants (2.8) as compared with ahr1b morphants (3.7). The same was true for nrf2a, which had a ratio of 2.7 for control morphants and 3.5 for ahr1b morphants. For nfe2, the knockdown of AHR1b did not block the induction by TCDD (Figure 3E); moreover, the fold induction ratio caused by TCDD in the ahr1b morphants (12.1) was much greater than that in the control morphants (3.3). The constitutive expression of nrf1b, nrf2a, and nfe2 (in DMSO-treated samples) was not significantly affected by knockdown of AHR1b. nrf1a was down-regulated upon knockdown of AHR1b in the vehicle controls, but its expression was not affected by exposure to TCDD (Figure 3A). Although the relative fold change values of this gene were not significant, there was an induction difference due to TCDD treatment in control morphant (1.3) versus ahr1b morphant samples (3.4). The expression of nrf2b was not affected by TCDD in the embryos injected with the control MO, but in the AHR1b morphants nrf2b was induced by TCDD (Figure 3D). TCDD had an effect on the fold induction of this gene in ahr1b morphants (4.3) as compared with control morphants (1.4). Under both vehicle control and TCDD conditions, the expression of nrf3 remained the same and the loss of AHR1b had no effect (Figure 3F). There was no difference in TCDD induction ratios between control morphants (0.9) and ahr1b morphants (1.7).
Figure 3.
AHR1b knockdown resulted in decreased expression of nrf genes in zebrafish embryos. Relative fold change of nuclear factor erythroid 2 (nfe2)-related factors (nrf) genes in 24 hpf zebrafish embryos is shown for control and ahr1b morphants. Zebrafish were dosed with DMSO or 2 nM TCDD for 1 h starting at 6hpf. Data are presented as mean + standard deviation, where different letters over bars indicate significant difference (2-way ANOVA with Sidak’s multiple comparisons test, p = .05). β-actin, whose expression was consistent across treatments, was used as a housekeeping gene and relative expression was calculated using the ΔΔCT method.
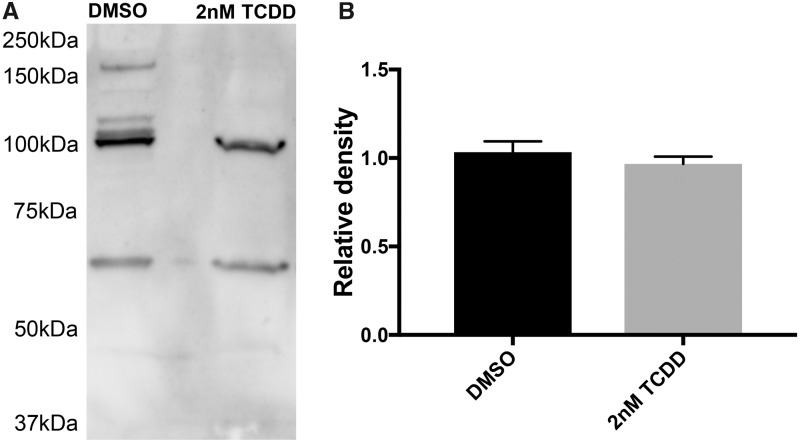
Prior to using the AHR1b antibody in the ChIP assay, its efficacy in recognizing AHR1b in 24 hpf embryos was verified using a Western blot. In both the DMSO and TCDD-treated embryos, the antibody recognized a major band of approximately 105 kDa, very close to the predicted size of 104.8 kDa (Karchner et al., 2005) (Figure 4A). There were a few nonspecific bands seen in the blots, some of which were of high molecular weight and only found in the DMSO sample. Total immunodetectable AHR1b protein was quantified by densitometry across 3 blots from independent samples and the average relative density of the 105-kDa band did not differ between DMSO and TCDD 24 hpf samples as determined by a 1-way ANOVA (Figure 4B).
Figure 4.
Western blot analysis of 24 hpf embryos with AHR1b antibody. A, Total protein from 24 hpf embryos was electrophoresed and blotted with a custom AHR1b antibody in 3 independent experiments and a representative image from 1 experiment is shown. B, Relative density of AHR1b concentration (band at approximately 105 kDa) was determined across 3 independent experiments by ImageJ and mean + SD is shown. No statistical difference was found between treatments (1-way ANOVA, p = .05).
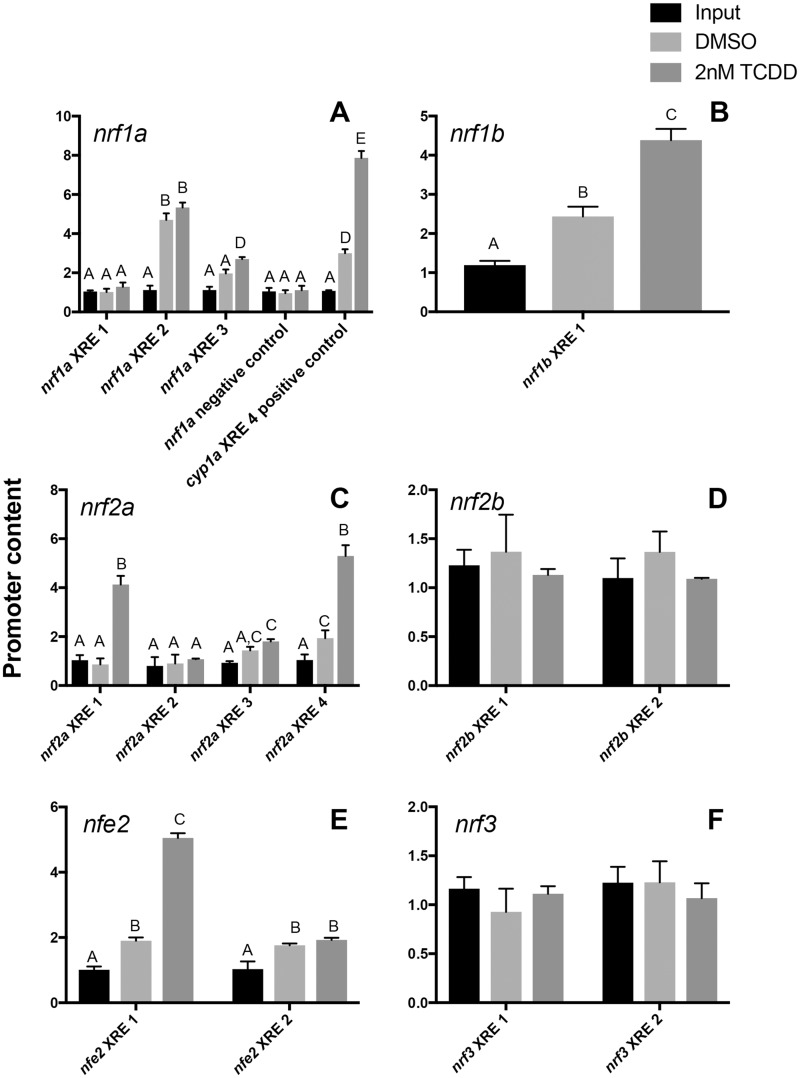
The antibody was subsequently used to immunoprecipitate the AHR1b-bound regions of the chromatin. PCR amplification of the cyp1a cis-promoter, containing XRE4 (TGGCGTGCAAAG), which has been shown to drive expression of luciferase upon exposure to TCDD (ZeRuth et al., 2007), was used as a positive control. Indeed, AHR1b directly bound to XRE4 under conditions of TCDD exposure (Figure 5A), serving as a positive control for the assay. For our negative control, we tested the promoter content of a region of the nrf1a cis-promoter that was located at least 5800 bp upstream from any of the 3 identified XREs. This negative control showed very little promoter content after immunoprecipitation (Figure 5A). For nrf1b, nrf2a, and nfe2, which showed transcriptional activation via TCDD exposure, the ChIP assay verified that there was direct binding of AHR1b in a XRE-specific manner (Figs. 5B, C, and E). Despite the presence of only a single XRE (TAGCGTG) in the nrf1b proximal promoter (Figure 1), this putative element was able to bind AHR1b (Figure 5B). Although the 4 XREs for nrf2a are all clustered together (within 1000 bp), only 2 seem to play a major role in binding AHR1b. XRE1 (GCGCGTG) and XRE4 (GTGCGTG) were both bound by AHR1b after treatment with TCDD, and XRE4 also bound AHR1b under vehicle control conditions. XRE3 (CACGCACGCGC) also bound some AHR1b weakly under control and TCDD treatments. There are 2 XREs in the proximal promoter for nfe2; however, only XRE1 (CACGCCA) bound AHR1b under TCDD conditions; in the control samples, the binding of AHR1b was weak for both XRE1 and XRE2 (CACGCTC). For nrf1a, XRE2 (GGGCGTG) bound AHR1b under control and TCDD conditions, whereas XRE3 (CACGCGC) participated in binding AHR1b following TCDD treatment (Figure 5A). Both of these XREs were located more proximal to the transcriptional start site (Figure 1) as compared with XRE1, which was more distal and did not show binding of AHR1b. AHR1b did not bind to the XREs in either of the cis-promoters of nrf2b or nrf3 (Figs. 5D and 5F).
Figure 5.
AHR is bound specifically to cis-XREs in the promoters of nrf genes in zebrafish embryos. ChIP assays were performed on 24 hpf zebrafish treated with either DMSO or 2 nM TCDD using an antibody specific for AHR1b followed by qPCR of individual XREs. qPCR of DNA from input fraction and AHR1b immunoprecipitation fraction are shown for samples treated with the control DMSO vehicle or TCDD. Shown in the nrf1a panel is the negative control which was an AHR1b IP followed by an amplification of a cis region of the nrf1a promoter lacking an XRE, and the positive control involved an AHR1b IP followed by an amplification of a cis region of the cyp1a promoter with XRE4 that has been shown to bind AHR2 (ZeRuth and Pollenz, 2007). Data are pooled from 3 experiments and presented as mean + SD where different letters over bars indicate significant difference (2-way ANOVA with Sidak’s multiple comparisons test, p = .05).
DISCUSSION
Since its discovery in the 1970s (Okey et al., 1979; Poland et al., 1976), the direct transcriptional role of AHR in cellular signaling has been well studied in the context of both its toxicological and physiological functions (Hahn et al., 2017; Jan et al., 2011; Lindsey and Papoutsakis, 2012; Mulero-Navarro and Fernandez-Salguero, 2016; Okey, 2007; Zhang, 2011). What has been less studied is its effects on cell signaling through crosstalk with other transcription factors (Puga et al., 2009). Due to genome duplication in the teleost lineage (Amores et al., 1998; Taylor et al., 2001), there are multiple Ahr and Nrf proteins for which this crosstalk has not been evaluated. This study is the first to demonstrate direct regulation of nrf genes by 1 AHR paralog, AHR1b, during early embryonic development.
At the beginning of the pharyngula period (24 hpf), a phylotypic stage, all of the nrf genes and ahr1b are expressed (Karchner et al., 2005; Mukaigasa et al., 2012; Pratt et al., 2002; Timme-Laragy et al., 2012; Williams et al., 2013). At 24 hpf, embryos are differentiating, utilizing endogenous ROS for cellular signaling (Thannickal and Fanburg, 2000). Yet, these embryos have low concentrations of total glutathione and oxidizing redox potential (Timme-Laragy et al., 2013), leaving them highly susceptible to imbalances in redox status that can be brought about by chemical exposure to compounds such as TCDD (Lin et al., 2007).
Exposure to TCDD can cause oxidative stress (Reichard et al., 2006). The AHR1b-dependent up-regulation of nrf genes, namely nrf1b, nrf2a, and nfe2 (Figs. 3 and 5), which would likely enhance the transcription of antioxidant defenses, may help the embryos to maintain redox balance upon exposure to TCDD. The up-regulation of nrf2a is similar to results obtained in mammalian systems, in which AHR regulated NRF2 expression in mouse cells (Miao et al., 2005). However, Hahn et al. (2015) did not find acute induction of nrf2a following a 6-h TCDD exposure in 24 hpf zebrafish with immediate sampling nor did a study that exposed 4 hpf embryos for 1-h to 1 nM TCDD with sampling at 24 hpf (Alexeyenko et al., 2010). Disparities in nrf2a gene expression patterns between the studies are likely due to differences in exposure (time and concentration) and collection regimes. Induction results may also vary by embryonic stage. Timme-Laragy et al. (2012) found that at 48 hpf nrf2a was not induced by TCDD as compared with DMSO. We also collected 48 hpf expression data for nrf2a following the protocol used in this study and in Timme-Laragy et al. (2012) and obtained results similar to those of Timme-Laragy et al. (Supplementary Figure 1), whereas Hahn et al. (2015) and Alexeyenko et al. (2010) found induction of nrf2a at 48hpf. Again, the differences in experimental design may explain the different results of these studies. Thus, it is imperative to follow the same exposure regime in order to compare gene expression profiles between experiments. Furthermore, these additional data point to temporal differences in gene expression patterns that are worthy of future experiments, which may include the use of qPCR, RNA-sequencing, whole mount in situ hybridization, and ChIP.
The AHR-dependent up-regulation of nrf1b and nfe2 in our study is a novel finding that has not previously been observed in mammalian systems. This result suggests that the regulation of genes involved in redox signaling and glutathione synthesis after AHR activation may not be carried out solely by Nrf2a. One possibility, which remains to be tested, is that AHR1b up-regulates nrf genes and then physically interacts or otherwise cooperates with their protein products to drive the transcription of shared antioxidant targets that contain XREs and AREs, such as nqo1 and gsta1 (Rousseau et al., 2015; Wang et al., 2013).
An alternative idea is that TCDD may not be causing oxidative stress, as suggested by several studies (Alexeyenko et al., 2010; Wang et al., 2013). The AHR1b-dependent induction of nrfs may regulate additional downstream targets that are distinct from those involved in antioxidant defense. The identity of genes that are regulated through AHR1b-NRF crosstalk could be ascertained through a RNA-seq experiment following knockdown or knockout of ahr1b and nrf genes.
Although much of the research on AHR has centered around exogenous ligands, AHR is also constitutively active in the absence of such compounds (Chang and Puga, 1998). We showed that the basal expression of nrf1a is AHR1b dependent (Figure 3A) but that nrf1a is not inducible by TCDD. To date, there are no known targets of nrf1a, although its ortholog in mammals is critical to both regulating genes involved in the oxidative stress response as well as basal functions like proteostasis and metabolism (Kim et al., 2016). Further determination of nrf1 paralog targets in zebrafish may elucidate the importance of this crosstalk during development.
Gene expression data reported in this study relied on the use of MO technology. This technology has been shown to be effective at temporally knocking down the expression of protein in developing zebrafish (Corey and Abrams, 2001), and in particular AHR2 (Massarsky et al., 2016; Mathew et al., 2006; Prasch et al., 2003). Although morpholinos can induce off-target effects (Bedell et al., 2011), their use avoids genetic compensation, which can occur in null mutants (Rossi et al., 2015). This is especially powerful when dealing with paralogous genes that have arisen through genome duplication, such as the AHR family (Hahn, 2002), which may have similar biological functions including promoter binding. However, future experiments should consider using an ahr1b germ line knockout (Karchner et al., 2017; Sugden et al., 2017) as important differences have been reported between mutants and morphants in zebrafish (Kok et al., 2015).
Although it is important to understand the cellular pathways that may be affected by AHR1b-NRF crosstalk, this study is also the first to demonstrate binding specificity of the AHR1b protein to cis-promoter elements in vivo. In the presence of its binding partner ARNT, AHR is known to bind to the DRE consensus sequences 5’TNGCGTG-3’ (Li et al., 2014; Lusska et al., 1993). In cell culture, AHR1b/ARNT2b has been shown to drive the expression of luciferase (pGudLuc6.1) in vehicle control conditions and upon exposure to TCDD (Karchner et al., 2005). In this plasmid, the expression of luciferase is under the control of four 5’GCGTG-3′ DRE core sequences (Han et al., 2004). In this study, those genes whose expression was altered by the knockdown of AHR1b, namely nrf1a, nrf1b, nrf2a, and nfe2 (Figs. 3A–C and E), directly bound AHR1b to XRE sequences (Figs. 5A–C and E). The expression of nrf1a via AHR1b is most dependent upon XRE2 with a core sequence of GCGTG. This core sequence has been shown to bind AHR efficiently in both murine and zebrafish assays (Li et al., 2014; ZeRuth et al., 2007). Interestingly, however, the flanking sequence around XRE2 (GGGGGCGTGTCTGC) more closely matches zebrafish cyp1a XRE 1, 3, and 6 at position 4 (T vs the consensus nucleotides C/A), and XRE 1, 2, and 6 at position 6 (T vs the consensus nucleotides A/C) (ZeRuth et al., 2007), all of which do not have in vivo activity. It does, however, match the consensus sequence at position 8, which was hypothesized to be an important residue in AHR binding (ZeRuth et al., 2007). The difference in binding could be due to differences between AHR/AHR2 and AHR1b.
With respect to nrf1b, there was a single XRE (TTAGCGTGCCGAGT) that closely matched the XRE consensus sequence across species (Li et al., 2014; Lusska et al., 1993; ZeRuth et al., 2007). This sequence bound AHR1b under both DMSO and TCDD treatment (Figure 5B). In cell culture, a single murine XRE of this same core sequence has been shown to mediate the activation of transcription (Li et al., 2014). Combined with our data, this finding points to AHR orthologs and paralogs being able to regulate transcription with a single XRE. Multiple XREs were shown to bind AHR1b in the cis-promoter of nrf2a, with the strongest binding in XRE1 (GCGCGTGCAGACG) and XRE4 (GCGCGTGCTATTA), with the greatest occupancy on XRE4, the more proximal element to the transcriptional start site (Figs. 1 and 5C). In the cyp1a promoter region, positions 4, 5, 6, and 8 were important for in vivo activity; for nrf2a these XREs had conserved residues at position 4 with the consensus sequence were not conserved at position 5, and XRE4 was conserved at position 6 and XRE1 at position 8. A mutation study similar to that of ZeRuth and Pollenz (2007) for the nrf2a promoter would elucidate the importance of particular residues and their positions for activation of transcription by AHR1b in vivo.
The cis-promoter of nfe2 also bound AHR1b in 24 hpf embryos (Figure 5E). Of those XREs (Figure 1), XRE1 for nfe2 (CACGCCA) had the strongest binding of AHR1b as compared with XRE2 (CACGCTC). In the murine cyp1a1 promoter, where the consensus sequence is CACGCNA, T or C residues at the “N” position increase transcriptional efficiency (Li et al., 2014); nfe2 XREs both have a T or C at this “Nth” position but XRE2 deviates from the consensus sequence where it has a C rather than an A at the next position which may explain the difference in promoter occupancy.
It is unclear why XREs in nrf2b and nrf3 do not bind AHR1b at 24 hpf. One possibility is that there may be temporal control of binding during development. Extending the ChIP assay to other time points would reveal whether AHR1b binds similarly to nrf genes during different developmental stages. Since nrf2b is transcriptionally activated by TCDD at 48 hpf (Timme-Laragy et al., 2012), it is possible that AHR1b is involved in regulation at this point in development, where it is not at 24 hpf.
This study is the first to show both direct binding of AHR1b to promoters in vivo and provide evidence of AHR1b-NRFcrosstalk during development. It is also the first evidence for AHR crosstalk with Nfe2, Nrf1, and Nrf2, extending the previously identified AHR-NRF2 interaction to other NRF family members. Future studies should focus on determining cell- or tissue-specific effects of AHR1b-nrf interactions. Since the whole embryo was used to determine changes in gene expression and AHR1b binding, we may have missed cell-to-cell variation, especially since the majority of AHR1b expression is in the eye (Karchner et al., 2017; Sugden et al., 2017). Methods that tag single-cell types such as the translating ribosome affinity purification approach (TRAP) (Doyle et al., 2008; Heiman et al., 2008; Tryon et al., 2013) or laser capture microdissection (Bonner et al., 1997) may be able to isolate enough embryonic material for gene expression and binding analyses. In order to determine the role of AHR1b as compared with AHR2, the AHR form most responsible in mediating the developmental toxicity of TCDD in zebrafish (Antkiewicz et al., 2006; Dong et al., 2003; Jönsson et al., 2007; Prasch et al., 2003), nrf gene expression could be ascertained in AHR2 knockouts as well as in AHR1b/AHR2 double knockouts (Chlebowski et al., 2017; Sugden et al., 2017). In order to complete ChIP assays, though, an effective AHR2 antibody must first be generated. Furthermore, by expanding this work to understand the downstream effects of this transcriptional regulation, the importance of AHR-NRF crosstalk will be better understood.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
This work was supported by the National Institutes of Health (F32ES019832 and P20GM103423 to L.M.W.; R01ES016366, R01ES006272, and R56ES028728 to M.E.H.). This work was also supported by Walter A. and Hope Noyes Smith Fund, the J. Seward Johnson Fund, and the Bates College Departments of Biology as well as Chemistry and Biochemistry.
Supplementary Material
ACKNOWLEDGMENTS
Excellent fish care was provided by Gale Clark, Abigail Haslett, Brandy Joyce, Erol Karchner, and Bruce Woodin at the Woods Hole Oceanographic Institution as well as Mary Hughes at Bates College. Dr Matthew Jenny designed the morpholino and was pivotal in the initial characterization of the antibody.
REFERENCES
- Aday A. W., Zhu L. J., Lakshmanan A., Wang J., Lawson N. D. (2011). Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev. Biol. 357, 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyenko A., Wassenberg D. M., Lobenhofer E. K., Yen J., Linney E., Sonnhammer E. L. L., Meyer J. N. (2010). Dynamic zebrafish interactome reveals transcriptional mechanisms of dioxin toxicity. PLoS One 5, e10465.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A., Force A., Yan Y. L., Joly L., Amemiya C., Fritz A., Ho R. K., Langeland J., Prince V., Wang Y. L, et al. (1998). Zebrafish hox clusters and vertebrate genome evolution. Science 282, 1711–1714. [DOI] [PubMed] [Google Scholar]
- Antkiewicz D. S., Peterson R. E., Heideman W. (2006). Blocking expression of AHR2 and ARNT1 in zebrafish larvae protects against cardiac toxicity of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 94, 175–182. [DOI] [PubMed] [Google Scholar]
- Bedell V. M., Westcot S. E., Ekker S. C. (2011). Lessons from morpholino-based screening in zebrafish. Brief. Funct. Genomics 10, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag T. V., Morales J. L., Hollingshead B. D., Perdew G. H. (2008). The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 18, 207–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner R. F., Emmert-Buck M., Cole K., Pohida T., Chuaqui R., Goldstein S., Liotta L. A. (1997). Laser capture microdissection: Molecular analysis of tissue. Science 278, 1481–1483. [DOI] [PubMed] [Google Scholar]
- Boutros P. C., Bielefeld K. A., Pohjanvirta R., Harper P. A. (2009). Dioxin-dependent and dioxin-independent gene batteries: Comparison of liver and kidney in AHR-null mice. Toxicol. Sci. 112, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-Y., Puga A. (1998). Constitutive activation of the aromatic hydrocarbon receptor. Mol. Cell. Biol. 18, 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillard G., Blank V. (2011). NFE2L3 (NRF3): The Cinderella of the Cap'n'Collar transcription factors. Cell. Mol. Life Sci. 68, 3337–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A. C., Garcia G. R., La Du J. K., Bisson W. H., Truong L., Massey Simonich S. L., Tanguay R. L. (2017). Mechanistic investigations into the developmental toxicity of nitrated and heterocyclic PAHs. Toxicol. Sci. 157, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey D. R., Abrams J. M. (2001). Morpholino antisense oligonucleotides: Tools for investigating vertebrate development. Genome Biol. 2, reviews1015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. S., Fisher J. M., Whitlock J. P. (1988). The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. J. Biol. Chem. 263, 17221–17224. [PubMed] [Google Scholar]
- Denison M. S., Nagy S. R. (2003). Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334. [DOI] [PubMed] [Google Scholar]
- Dong W., Teraoka H., Tsujimoto Y., Stegeman J. J., Hiraga T. (2003). Role of aryl hydrocarbon receptor in mesencephalic circulation failure and apoptosis in zebrafish embryos exposed to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 77, 109–116. [DOI] [PubMed] [Google Scholar]
- Doyle J. P., Dougherty J. D., Heiman M., Schmidt E. F., Stevens T. R., Ma G., Bupp S., Shrestha P., Shah R. D., Doughty M. L, et al. (2008). Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B. R., Karchner S. I., Franks D. G., Hahn M. E. (2005). Duplicate aryl hydrocarbon receptor repressor genes (ahrr1 and ahrr2) in the zebrafish Danio rerio: Structure, function, evolution, and AHR-dependent regulation in vivo. Arch. Biochem. Biophys. 441, 151–167. [DOI] [PubMed] [Google Scholar]
- Favreau L. V., Pickett C. B. (1991). Transcriptional regulation of the rat nad(P)H-quinone reductase gene - Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic-compounds and phenolic antioxidants. J. Biol. Chem. 266, 4556–4561. [PubMed] [Google Scholar]
- Gasiorek J. J., Blank V. (2015). Regulation and function of the NFE2 transcription factor in hematopoietic and non-hematopoietic cells. Cell. Mol. Life Sci. 72, 2323–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilda J. E., Gomes A. V. (2013). Stain-Free total protein staining is a superior loading control to beta-actin for Western blots. Anal. Biochem. 440, 186–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass A. S., Dahm R. (2004). The zebrafish as a model organism for eye development. Ophthalmic Res. 36, 4–24. [DOI] [PubMed] [Google Scholar]
- Hahn M. E. (2002). Aryl hydrocarbon receptors: Diversity and evolution11Invited review for Chemico-Biological Interactions. Chem. Biol. Interact. 141, 131–160. [DOI] [PubMed] [Google Scholar]
- Hahn M. E., Karchner S. I., Merson R. R. (2017). Diversity as opportunity: Insights from 600 million years of AHR evolution. Curr. Opin. Toxicol. 2, 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. E., Timme-Laragy A. R., Karchner S. I., Stegeman J. J. (2015). Nrf2 and Nrf2-related proteins in development and developmental toxicity: Insights from studies in zebrafish (Danio rerio). Free Radic. Biol. Med. 88(Pt B), 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Nagy S. R., Denison M. S. (2004). Comparison of recombinant cell bioassays for the detection of Ah receptor agonists. BioFactors 20, 11–22. [DOI] [PubMed] [Google Scholar]
- Heiman M., Schaefer A., Gong S., Peterson J., Day M., Ramsey K. E., Suárez-Fariñas M., Schwarz C., Stephan D. A., Surmeier D. J.. et al. (2008). Development of a BACarray translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hestermann E. V., Brown M. (2003). Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol. Cell. Biol. 23, 7920–7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzmann K. A., Freeman J. L. (2018). Making waves: New developments in toxicology with the zebrafish. Toxicol. Sci. 163, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan V., Lenka U., Miroslav M. (2011). Interactions of the aryl hydrocarbon receptor with inflammatory mediators: Beyond CYP1A Regulation. Curr. Drug Metab. 12, 89–103. [DOI] [PubMed] [Google Scholar]
- Jenny M. J., Karchner S. I., Franks D. G., Woodin B. R., Stegeman J. J., Hahn M. E. (2009). Distinct roles of two zebrafish AHR repressors (AHRRa and AHRRb) in embryonic development and regulating the response to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 110, 426–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson M. E., Jenny M. J., Woodin B. R., Hahn M. E., Stegeman J. J. (2007). Role of AHR2 in the expression of novel cytochrome P450 1 family genes, cell cycle genes, and morphological defects in developing zebra fish exposed to 3, 3′, 4, 4′, 5-pentachlorobiphenyl or 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 100, 180–193. [DOI] [PubMed] [Google Scholar]
- Kalthoff S., Ehmer U., Freiberg N., Manns M. P., Strassburg C. P. (2010). Interaction between oxidative stress sensor nrf2 and xenobiotic-activated aryl hydrocarbon receptor in the regulation of the human phase II detoxifying UDP-glucuronosyltransferase 1A10. J. Biol. Chem. 285, 5993–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner S. I., Franks D. G., Hahn M. E. (2005). AHR1B, a new functional aryl hydrocarbon receptor in zebrafish: Tandem arrangement of ahr1b and ahr2 genes. Biochem. J. 392, (Pt 1) 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner S. I., Jenny M. J., Aluru N., Franks D. G., Laub L. B., Linney E., Williams L. M., Teraoka H., Hahn M. E. (2017). Evidence for Developmental Versus Toxicological Roles for Zebrafish AHR1b, Vol. 156, p. s39, Abstract #1165. Society of Toxicology, Baltimore, MD. [Google Scholar]
- Kim H. M., Han J. W., Chan J. Y. (2016). Nuclear factor erythroid-2 like 1 (NFE2L1): Structure, function and regulation. Gene 584, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. (2009). The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 29, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok F. O., Shin M., Ni C. W., Gupta A., Grosse A. S., van Impel A., Kirchmaier B. C., Peterson-Maduro J., Kourkoulis G., Male I.. et al. (2015). Reverse genetic screening reveals poor correlation between Morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Pei X., Zhang W., Xie H. Q., Zhao B. (2014). Functional analysis of the dioxin response elements (DREs) of the murine CYP1A1 gene promoter: Beyond the core DRE sequence. Int. J. Mol. Sci. 15, 6475–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.-H., Lin C.-H., Huang C.-C., Chuang M.-C., Lin P. (2007). 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin (TCDD) induces oxidative stress, DNA strand breaks, and poly(ADP-ribose) polymerase-1 activation in human breast carcinoma cell lines. Toxicol. Lett. 172, 146–158. [DOI] [PubMed] [Google Scholar]
- Lindsey S., Papoutsakis E. T. (2012). The evolving role of the aryl hydrocarbon receptor (AHR) in the normophysiology of hematopoiesis. Stem Cell Rev. 8, 1223–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lusska A., Shen E., Whitlock J. P. (1993). Protein-DNA interactions at a dioxin-responsive enhancer. Analysis of six bona fide DNA-binding sites for the liganded Ah receptor. J. Biol. Chem. 268, 6575–6580. [PubMed] [Google Scholar]
- Ma Q. (2013). Role of Nrf2 in Oxidative Stress and Toxicity. Annu. Rev. Pharmacol. Toxicol. 53, 401–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., He X. (2012). Molecular basis of electrophilic and oxidative defense: Promises and Perils of Nrf2. Pharmacol. Rev. 64, 1055–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Kinneer K., Bi Y. Y., Chan J. Y., Kan Y. W. (2004). Induction of murine NAD(P)H: Quinone oxidoreductase by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin requires the CNC (cap ‘n’ collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): Cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. Biochem. J. 377, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarsky A., Bone A. J., Dong W., Hinton D. E., Prasad G. L., Di Giulio R. T. (2016). AHR2 morpholino knockdown reduces the toxicity of total particulate matter to zebrafish embryos. Toxicol. Appl. Pharmacol. 309, 63–76. [DOI] [PubMed] [Google Scholar]
- Mathew L. K., Andreasen E. A., Tanguay R. L. (2006). Aryl hydrocarbon receptor activation inhibits regenerative growth. Mol. Pharmacol. 69, 257. [DOI] [PubMed] [Google Scholar]
- Miao W., Hu L., Scrivens P. J., Batist G. (2005). Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: Direct cross-talk between phase I and II drug-metabolizing enzymes. J. Biol. Chem. 280, 20340–20348. [DOI] [PubMed] [Google Scholar]
- Mukaigasa K., Nguyen L. T. P., Li L., Nakajima H., Yamamoto M., Kobayashi M. (2012). Genetic evidence of an evolutionarily conserved role for Nrf2 in the protection against oxidative stress. Mol. Cell. Biol. 32, 4455–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Navarro S., Fernandez-Salguero P. M. (2016). New trends in aryl hydrocarbon receptor biology. Front. Cell Dev. Biol. 4, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okey A. B. (2007). An aryl hydrocarbon receptor odyssey to the shores of toxicology: The deichmann lecture, international congress of toxicology-XI. Toxicol. Sci. 98, 5–38. [DOI] [PubMed] [Google Scholar]
- Okey A. B., Bondy G. P., Mason M. E., Kahl G. F., Eisen H. J., Guenthner T. M., Nebert D. W. (1979). Regulatory gene product of the Ah locus. Characterization of the cytosolic inducer-receptor complex and evidence for its nuclear translocation. J. Biol. Chem. 254, 11636–11648. [PubMed] [Google Scholar]
- Poland A., Glover E., Kende A. S. (1976). Stereospecific, high affinity binding of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J. Biol. Chem. 251, 4936–4946. [PubMed] [Google Scholar]
- Prasch A. L., Teraoka H., Carney S. A., Dong W., Hiraga T., Stegeman J. J., Heideman W., Peterson R. E. (2003). Aryl hydrocarbon receptor 2 mediates 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol. Sci. 76, 138–150. [DOI] [PubMed] [Google Scholar]
- Pratt S. J., Drejer A., Foott H., Barut B., Brownlie A., Postlethwait J., Kato Y., Yamamoto M., Zon L. I. (2002). Isolation and characterization of zebrafish NFE2. Physiol. Genomics 11, 91–98. [DOI] [PubMed] [Google Scholar]
- Puga A., Ma C., Marlowe J. L. (2009). The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem. Pharmacol. 77, 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radjendirane V., Jaiswal A. K. (1999). Antioxidant response element-mediated 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) induction of human NAD(P)H: Quinone oxidoreductase 1 gene expression. Biochem. Pharmacol. 58, 1649–1655. [DOI] [PubMed] [Google Scholar]
- Reichard J. F., Dalton T. P., Shertzer H. G., Puga A. (2006). Induction of oxidative stress responses by dioxin and other ligands of the aryl hydrocarbon receptor. Dose Response 3, 306–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: The european molecular biology open software suite. Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Holper S., Kruger M., Stainier D. Y. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233. [DOI] [PubMed] [Google Scholar]
- Rousseau M. E., Sant K. E., Borden L. R., Franks D. G., Hahn M. E., Timme-Laragy A. R. (2015). Regulation of Ahr signaling by Nrf2 during development: Effects of Nrf2a deficiency on PCB126 embryotoxicity in zebrafish (Danio rerio). Aquat. Toxicol. 167, 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. T., Fishman M. C. (2002). From zebrafish to human: Modular medical models. Annu. Rev. Genomics Hum. Genet. 3, 311–340. [DOI] [PubMed] [Google Scholar]
- Shin S., Wakabayashi N., Misra V., Biswal S., Lee G. H., Agoston E. S., Yamamoto M., Kensler T. W. (2007). NRF2 modulates aryl hydrocarbon receptor signaling: Influence on adipogenesis. Mol. Cell. Biol. 27, 7188–7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden W. W., Leonardo-Mendonca R. C., Acuna-Castroviejo D., Siekmann A. F. (2017). Genetic dissection of endothelial transcriptional activity of zebrafish aryl hydrocarbon receptors (AHRs). PLoS One 12, e0183433.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis G. P., Bohmann D. (2010). Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci. Signal. 3, re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguay R. L. (2018). The rise of zebrafish as a model for toxicology. Toxicol. Sci. 163, 3–4. [DOI] [PubMed] [Google Scholar]
- Taylor J. S., Van de Peer Y., Braasch I., Meyer A. (2001). Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1661–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. C., Berkelman T., Yadav G., Hammond M. (2013). A defined methodology for reliable quantification of Western blot data. Mol. Biotechnol. 55, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal V. J., Fanburg B. L. (2000). Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 279, L1005–L1028. [DOI] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Goldstone J. V., Imhoff B. R., Stegeman J. J., Hahn M. E., Hansen J. M. (2013). Glutathione redox dynamics and expression of glutathione-related genes in the developing embryo. Free Radic. Biol. Med. 65, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Karchner S. I., Franks D. G., Jenny M. J., Harbeitner R. C., Goldstone J. V., McArthur A. G., Hahn M. E. (2012). Nrf2b, novel zebrafish paralog of oxidant-responsive transcription factor NF-E2-related factor 2 (NRF2). J. Biol. Chem. 287, 4609–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryon R. C., Pisat N., Johnson S. L., Dougherty J. D. (2013). Development of translating ribosome affinity purification for zebrafish. Genesis 51, 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliou V., Puga A., Chang C. Y., Tabor M. W., Nebert D. W. (1995). Interaction between the Ah receptor and proteins binding to the AP-1-like electrophile response element (EpRE) during murine phase II [Ah] battery gene expression. Biochem. Pharmacol. 50, 2057–2068. [DOI] [PubMed] [Google Scholar]
- Wang L., He X., Szklarz G. D., Bi Y., Rojanasakul Y., Ma Q. (2013). The aryl hydrocarbon receptor interacts with nuclear factor erythroid 2-related factor 2 to mediate induction of NAD(P)H: Quinoneoxidoreductase 1 by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Arch. Biochem. Biophys. 537, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A. C., Lieschke G. J. (2002). The zebrafish as a model system for human disease. Front. Biosci. 7, D827–D833. [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2007). The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio), 5th ed University of Oregon Press, Eugene, OR. [Google Scholar]
- Whitlock J. P. (1999). Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 39, 103–125. [DOI] [PubMed] [Google Scholar]
- Williams L. M., Timme-Laragy A. R., Goldstone J. V., McArthur A. G., Stegeman J. J., Smolowitz R. M., Hahn M. E. (2013). Developmental expression of the Nfe2-related factor (Nrf) transcription factor family in the zebrafish, Danio rerio. PLoS One 8, e79574.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager R. L., Reisman S. A., Aleksunes L. M., Klaassen C. D. (2009). Introducing the “TCDD-Inducible AhR-Nrf2 Gene Battery”. Toxicol. Sci. 111, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZeRuth G., Pollenz R. S. (2007). Functional analysis of cis-regulatory regions within the dioxin-inducible CYP1A promoter/enhancer region from zebrafish (Danio rerio). Chem. Biol. Interact. 170, 100–113. [DOI] [PubMed] [Google Scholar]
- Zhang N. (2011). The role of endogenous aryl hydrocarbon receptor signaling in cardiovascular physiology. J. Cardiovasc. Dis. Res. 2, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiang Y. (2016). Molecular and cellular basis for the unique functioning of Nrf1, an indispensable transcription factor for maintaining cell homoeostasis and organ integrity. Biochem. J. 473, 961–910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.