Abstract
Detection of developmental phenotypes in zebrafish embryos typically involves a visual assessment and scoring of morphological features by an individual researcher. Subjective scoring could impact results and be of particular concern when phenotypic effect patterns are also used as a diagnostic tool to classify compounds. Here we introduce a quantitative morphometric approach based on image analysis of zebrafish embryos. A software called FishInspector was developed to detect morphological features from images collected using an automated system to position zebrafish embryos. The analysis was verified and compared with visual assessments of 3 participating laboratories using 3 known developmental toxicants (methotrexate, dexamethasone, and topiramate) and 2 negative compounds (loratadine and glibenclamide). The quantitative approach exhibited higher sensitivity and made it possible to compare patterns of effects with the potential to establish a grouping and classification of developmental toxicants. Our approach improves the robustness of phenotype scoring and reliability of assay performance and, hence, is anticipated to improve the predictivity of developmental toxicity screening using the zebrafish embryo.
Keywords: developmental toxicity, zebrafish embryo, alternatives to animal testing, image analysis
Zebrafish (Danio rerio) exhibit 70–80% gene sequence homology with humans and share structural similarities with vertebrates (Dooley and Zon, 2000; Gunnarsson et al., 2008). Therefore, their embryos are used as an alternative model for developmental toxicity screening of drugs and chemicals (Brannen et al., 2010; Selderslaghs et al., 2009). The possibility of holistic assessment in a small-scale system, the ability to produce large numbers of progeny, and the transparency of the embryos and their rapid development have made the model particularly attractive and led to the development of high-throughput assays (Padilla et al., 2012; Truong et al., 2014).
Results from small-scale pilot studies have demonstrated a high concordance between zebrafish and mammalian developmental toxicity with an overall concordance of 72–92% (Brannen et al., 2010; Hermsen et al., 2011; Krupp, 2016; Selderslaghs et al., 2009; Van den Bulck et al., 2011). However, in interlaboratory variability studies (Ball et al., 2014; Gustafson et al., 2012), some inconsistencies with respect to concordance analysis were also observed. The concordance of individual laboratories for developmental toxicity or teratogenic classification ranged between 60% and 70% when compared with mammalian data, but only 5 of 20 compounds were similarly classified (ie, teratogenic or nonteratogenic) by all 4 participating laboratories (Gustafson et al., 2012). In a subsequent study with 37 compounds and 2 laboratories, a concordance of 71% for teratogen classification was observed (Ball et al., 2014). This variability between laboratories may have been partly caused by the visual observation and classification of developmental alterations by an individual technician or researcher and limited standardization. Hence, the approach currently used for developmental toxicity screening in zebrafish embryos might be biased by the experience and accuracy of the observer. Furthermore, observations are often not documented by storing the relevant images, thus making verification and reanalysis of data difficult.
Previous phenotypic image analyses have focused on fluorescent imaging for measuring, that is, cardiovascular development (Leet et al., 2014), cardiovascular function (Burns et al., 2005; Leet et al., 2014; Letamendia et al., 2012; Yozzo et al., 2013), and angiogenesis (Letamendia et al., 2012; Vogt et al., 2009). There are few published studies using automatic phenotypic image analysis for bright-field microscope images without fluorescent markers or staining (Arslanova et al., 2010; Deal et al., 2016; Jeanray et al., 2015; Liu et al., 2012; Schutera et al., 2016). Some of these studies were limited to the identification of specific phenotypes such as lethality (Alshut et al., 2010; Liu et al., 2012), hatching status (Liu et al., 2012), changes in pigmentation (Arslanova et al., 2010; Schutera et al., 2016), or lack of eyes (Schutera et al., 2016). One study aimed at developing a computational malformation index through the use of morphometric parameters (eg, total body area, convexity) in combination with a very brief human visual assessment (Deal et al., 2016). That method was more objective as user scoring was based on microscopic observations and the cumulative degree of abnormality could be described, but the different phenotypes (eg, edema, small eyes) were not resolved. A different approach was developed by Jeanray et al. (2015) using supervised machine learning to identify developmental phenotypes. This approach is based on an initial expert classification of phenotypes and requires several rounds of classification and learning but can be used to establish concentration-response curves for cumulative phenotypic assessment. However, the same or similar instrumentation and settings would be required to apply their established models directly.
Crucial for a quantitative, unbiased approach to phenotype assessment using 2D images is a proper orientation of the fish embryos. Slight differences in the orientation and the subsequent 2D projection could lead to changes in feature detection. Therefore, in this study, an image-based detection and quantification of morphological features in zebrafish embryos was developed based on an automated system for positioning of the embryos in a capillary. Multiple morphological features were automatically extracted from zebrafish images using a custom MATLAB-based software called FishInspector. Although our workflow was developed for automated positioning in a capillary, it can also be applied to manually positioned embryos as conducted in other studies (eg, Peravali et al., 2011). However, this may be more time consuming and may introduce additional variability. In a second step, we used the analytical platform KNIME and R scripts for morphometric analysis and quantification using the coordinates of each feature detected by FishInspector.
Morphological features were complemented by video-based measurements of heart rate and behavioral effects (locomotor response at 96 h post-fertilization [hpf]). These two functional parameters provide further endpoints relevant for safety areas assessment and potentially linked to developmental toxicity. For instance, a comparative endpoint analysis (Ducharme et al., 2013) has revealed a high correlation of behavioral endpoints with (gross) malformations of fish embryos and hence may support quantitation of overall assessment of teratogenic effects.
To demonstrate the capacity of the software for the multi-endpoint analysis, it was applied to a set of 5 model compounds representing diverse drug classes. Three compounds (methotrexate, topiramate, and dexamethasone) known to cause developmental toxicity in mammals and two compounds (glibenclamide and loratadine) as non-developmental toxicants. The performance of this method was also analyzed in the context of sensitivity differences between 3 laboratories experienced with conventional visual assessment and scoring of developmental anomalies in the zebrafish embryo. The intention was, for example, to understand whether the automatic assessment provides increased sensitivity compared with conventional assessments in other laboratories.
MATERIALS AND METHODS
Chemicals
The following chemicals were used: loratadine (CAS-RN 79794-75-5, purity ≥ 98%, Sigma-Aldrich), metothrexate (CAS-RN 59-05-2, purity ≥ 98.5%, AppliChem), glibenclamide (CAS-RN 10238-21-8, purity ≥ 99%, Sigma-Aldrich), dexamethasone (CAS-RN 50-02-2, purity ≥ 97%, Fluka), topiramate CAS-RN 97240-79-4, purity ≥ 98%, Sigma-Aldrich), all-trans retinoic acid (CAS-RN 302-79-4, purity≥ 98%, AppliChem Panreac), and N-phenylthiourea (PTU, CAS-RN 103-85-5, purity ≥ 98%, Sigma-Aldrich). Loratadine, glibenclamide, dexamethasonse, and all-trans retinoic acid were dissolved in dimethyl sulfoxide (DMSO). Test solutions were obtained by dilution of the stock solutions in embryo test medium according to the OECD testing guideline 236 (OECD, 2013, pH = 7.4–7.5) resulting in final DMSO concentrations of 0.01% (all-trans retinoic acid), 0.5% (loratadine and glibenclamide), and 1% (dexamethasone). The different DMSO concentrations reflect the different solubility in DMSO, that is, the concentration of DMSO was kept as low as possible to obtain full concentration-response curves for mortality and sublethal phenotypes.
Zebrafish Developmental Toxicity Assay Overview
Adult, healthy, and unexposed zebrafish were used for the production of fertilized eggs. We used the UFZ-OBI strain (generation F14–15), obtained originally from a local breeder and kept for several generations at the UFZ. Fish were cultured and used according to German and European animal protection standards and approved by the Government of Saxony, Landesdirektion Leipzig, Germany (Aktenzeichen 75-9185.64). Just after fertilization eggs were treated against fungal infection with a diluted Chloramine-T bleaching solution (0.5% w/v) for 60 s with gentle periodic agitation, washed twice with embryo medium and transferred into a petri dish for egg selection. Bleaching did not affect the hatching of embryos at later stages. All control embryos were hatched at 96 hpf. The bleaching was conducted to avoid carry over of fungi or microbes from the tanks. Embryos were exposed to the test compound, a solvent control and a positive control (all-trans retinoic acid at 12.5 nM) from 2 to 48 hpf and from 2 to 96 hpf, at a temperature of 28 (±1)°C (14:10 light: dark cycle). Forty eight-hour exposures were conducted in crystallization dishes covered with watchmaker glasses with a test volume of 16 ml and 16 embryos per dish. Ninety six-hour exposures were conducted in rectangular 96-well microplates (Clear Polystyrene, flat bottom, Uniplate, Whatman, GE Healthcare, Little Chalfont, UK) covered by a lid with a test volume of 400 µl (one embryo per well, 16 wells per concentration tested). No evaporation was observed during the exposure period. The different protocols were used since manual dechorionation is difficult to conduct in 96-well plates. For hydrophobic compounds (log P > 4) low exposure volumes in 96-well microplates (400 µl exposure volume per embryo) may result in a (pronounced) decline in exposure concentration when compared with exposure in crystallization dishes (1000 µl volume per embryo). Therefore, for hydrophobic compounds (loratadine and glibenclamide) exposure was conducted in crystallization dishes for both the 48 and 96 h exposure in order to compensate for a potential loss of exposure concentration due to absorption in embryos and to the wells of the microplate. Tests were performed with at least 2 replicates. Renewal of the exposure solutions were performed every 24 h, except for methotrexate, for which, due to confirmation of stable exposure concentration, a 48 h renewal interval was selected (see Supplementary Table 2), and for topiramate, for which stability was assumed (Micheel et al., 1998) and no renewal was done. Phenotypic assessment by automated imaging (“Image-Based Quantification of Morphological Features” section) was conducted after assessment of lethality, behavioral effects (at 96 hpf), and visual assessment using a stereomicroscope (Olympus SZX10, Massachusetts). Visual and automatic image-based assessment of phenotypes at the UFZ was conducted for the same experiment and same fish. Supplementary Table 1 shows the endpoints evaluated by visual observation. More details on the test protocol can be found in Supplementary Table 2.
Developmental Staging Analysis
Comparison of developmental stages of zebrafish incubated at 28 (±1)°C was done using untreated embryos from 5 different stages from 32 to 96 hpf (32, 48, 72, 82, and 96 hpf). Linear regression analysis was performed to determine which of the features quantified using the FishInspector exhibit a significant correlation during normal development.
Image-Based Quantification of Morphological Features
Automated imaging of zebrafish embryos
Images of zebrafish embryos were obtained using the VAST Bioimager (Union Biometrica, Gees, Belgium) (Pardo-Martin et al., 2010; Pulak, 2016) with the on-board camera of 10 µm resolution. Beforehand imaging embryos were dechorionated (required for 48 hpf stages only) and anesthetized with a tricaine solution (150 mg/l, TRIS 26 mM, pH 7.5). Embryos exposed in crystallization dishes were transferred to a 96-well microplate with rectangular wells. Loading of each fish from rectangular 96-well plates was done using the LP sampler (Union Biometrica, Gees, Belgium) and 4 pictures were automatically collected (two laterals, one dorsal, and one ventral image). Additionally, a video of 15 s at 30 frames per second was recorded of each embryo in lateral position for later video-based determination of the heart frequency. For the analysis, fish embryos were removed from the microtiter plates such that individuals from different concentrations were analyzed alternately. This was done to avoid time bias. The concentration of tricaine used here has been shown not to affect the heart rate frequency within the time frame (2 h) that was used for analysis (Yozzo et al., 2013).
Feature detection using the FishInspector software
Lateral control images of embryos at 48 and 96 hpf were used initially for software development. FishInspector was developed within MATLAB environment and the source code and an executable version for windows operation system is freely available (last updated version is available via Zenodo [Kießling et al., 2018]). The detection of various features is organized hierarchically, that is, in order to locate a certain feature the locations of previously detected features are included. For example, detection of the contour of the embryo is guided by the capillary boundaries, since the embryo definitely will be located inside the capillary. Subsequently, other features are identified in a stepwise manner (Supplementary Figure 1). Hence, the detection of specific morphological features is dependent on the detection of other features and is facilitated by excluding regions that may interfere. The identification of the regions of interest was driven by visual observation and measurement of generic object properties. For example, once the contour of the fish was localized, the eye was detected by searching for a dark object either in the right or left half of the zebrafish. The detection algorithms were successively improved by using images of embryos treated with all-trans retinoic acid (used as the positive control for gross changes in body morphology). Given that establishment of a 100% correct automated feature detection would be very challenging and to allow improvement by the user, the software permits modification of the parameters used for the automated feature detection, and also manual correction if the feature is not sufficiently detected. At present, jaw morphology analysis cannot be detected automatically with the FishInspector and requires a manual annotation step, that is, label of the tip of the lower part of the mouth. The resulting output of the FishInspector is a set of xy coordinates of the morphological feature detected. For each image analyzed, data are exported to a single JSON file, which is a language independent open-standard file format typically used for transmitting data between applications. The boundary coordinates of multiple detected features can then be stored in a structured text file. This allows the seamless integration of the FishInspector output into custom post-processing algorithms, which can be implemented in any programming language.
Quantification of phenotypic features
The JSON data files were used as input in a customized KNIME workflow with R scripts (Berthold et al., 2008, R Core Team 2017). The phenotypic features analyzed are described in Table 1. Shape information (mainly length and surface area) was extracted using the “Momocs” R package (Bonhomme et al., 2013; Claude et al., 2008). For extraction of the fish tail curvature, only the notochord coordinates from the tail of the fish were considered (Supplementary Figure 2). Curvatures along the tail were calculated by extracting from the smoothed notochord line the value of the second derivative when the first derivative is 0. The maximum curvature value along the tail was used for the analysis. Tail curvature was calculated using R with the package “features” (Varadhan et al., 2015) using as smoother the function “smooth.spline” with a spar value of 0.9. Head size was quantified by drawing a line between the eye and otolith centroid, then an angle was taken from the otolith to the upper contour of the fish, also from the eye to the bottom contour of the fish to enclose the head region (see Supplementary Figure 3). Lower jaw position was evaluated at 96 hpf by using the manual selection on the FishInspector. To quantify the effects, the distance in the x coordinate between the eye centroid and the lower jaw tip was calculated (see Supplementary Figure 4).
Table 1.
Morphological Features Measured in the Zebrafish Using the FishInspector Software
| Phenotypic Feature | Data Exported as Json Format | Parameter or Metric | Corresponding Endpoint in Visual Assessment |
|---|---|---|---|
| Eye size | Eye xy coordinates | Surface area (mm2) | Reduced eye size |
| Body length | Fish contour xy coordinates | Length (mm) | Not assessed |
| Yolk sac size | Yolk sac contour xy coordinates | Surface area (mm2) | Increased yolk sac size or abnormal morphology |
| Otolith-eye distance | Otolith xy centroid (saccule, the largest otolith) | Length (mm) | Not assessed |
| Pericard size | Pericard contour xy coordinates | Surface area (mm2) | Increased pericard size |
| Tail malformations | Notochord xy coordinates | Curvature | Tail curvature |
| Swim bladder inflation | Swim bladder contour xy coordinates | Surface area (mm2) | Failure to inflate the swim bladder |
| Head size | Fish contour xy coordinates, otolith, and eye centroid | Surface area (mm2) | Reduced or abnormal head size |
| Pigmentation | Area (in pixels) of pigment cells from lateral line | Sum surface area (mm2) | Not assessed |
| Lower jaw position | Distance in the x coordinate between eye centroid and lower jaw tip | Distance (mm) | Underdeveloped or abnormal jaw |
Notes: The data are exported in Json file format and used to quantify different metrics by the use of a customized KNIME workflow. The corresponding assessment using the conventional visual assessment is also shown in the table.
Application of the workflow does not require knowledge of computer programing languages. The complete workflow only requires the use of the standard open source tools (KNIME, R, and ImageJ. The workflow is provided in Dryad, Teixido et al., 2018). Pigmentation was quantified by measuring the sum area of pigment cells along the lateral line, using the area covered by the notochord as the enclosure region. In order to validate the pigmentation analysis, embryos were exposed to increasing concentrations (0−150 µM) of N-phenylthiourea (PTU) (Supplementary Figure 5), a model compound that is known to inhibit melanization (Karlsson et al., 2001).
Heart Rate Quantification
An automated image workflow was developed using the KNIME Analytics Platform (workflow available in Dryad, Teixido et al., 2018). The zebrafish heart as the region of interest (ROI) is detected by comparing the absolute difference in pixel intensity between two consecutive frames. By using a threshold method and morphological operations, irrelevant areas were removed from the analysis. Then the pixel variance of the ROI in each frame was used to determine the heart frequency using a Fast Fourier transform with the spectrum function included in the base package of R.
Locomotor Response (LMR)
The locomotor response was assessed at 96 hpf prior to the analysis with the VAST Bioimager system. Embryonic movement was tracked using the ZebraBox video tracking system (Viewpoint, Lyon, France) for 40 min in a series of light and dark periods to stimulate movement (10 min equilibration in light, followed by 20 min in dark, and a final 10 min light phase) as described in Irons et al. (2010). The movement in the light periods was recorded using maximum intensity (1200 lux). Movement in light and dark periods was recorded using an infrared camera and the video tracking mode with a detection threshold set to 20. The temperature was continuously maintained at 28(±1) °C. Live embryos, including malformed embryos and embryos showing no inflation of the swim bladder, were considered for the analysis of the locomotor response. The percentage of effects (EC50) was calculated on the basis of the mean travelled distance as described in Klüver et al. (2015) using the dark phase interval (10–20 min).
Inter-Laboratory Study Design
Three laboratories participated in this study. They were: Department of Bioanalytical Ecotoxicology, Helmholtz Center for Environmental Research (UFZ), R&D Preclinical Safety, Sanofi-Aventis Deutschland GmbH, and BBD BioPhenix-Biobide. The laboratories used an agreed test protocol (described in “Zebrafish Developmental Toxicity Assay Overview” section) with minor differences between laboratories as shown in Supplementary Table 2. The UFZ was the only laboratory to include an image-based quantification of morphological features using the FishInspector (as described in “Image-Based Quantification of Morphological Features” section), heart rate quantification (“Heart rate quantification” section), and behavior analysis (“Locomotor Response (LMR)” section). Testing of the compounds was done in a blind manner at 2 of the 3 laboratories (Biobide and UFZ), that is, identity of the compounds was only released after completion of the effect assessment. The test concentrations were not harmonized between the different laboratories and were individually adjusted based on range findings or to improve the description of the concentration response curves in replicates.
Data Evaluation
Two approaches were used for the concentration-response analysis: (a) effect quantification with continuous data normalized to the mean control value and, (b) threshold-based quantal effect data. The first approach was used for endpoints with high variability between controls of replicates, observed for heart rate, behavior, and pigmentation. For these endpoints, data were normalized to the mean control of each replicate and concentration-response curves were derived from these data. For all other endpoints (eye size, body length, yolk sac size, head size, swim bladder, jaw-eye distance, and otolith-eye distance), similar to the method proposed for obtaining benchmark responses with dichotomized continuous data (U.S. EPA 2012), a threshold value was established by analysis of the variability of about 130 control embryos of different replicates (Supplementary Table 3). Values deviating by ±2 SD were considered as indicating a deviation from the control and were used to calculate the fraction of embryos for which the appropriate endpoint was affected. For the overall cumulative effect assessment, a threshold of 2.5 SD was used given the higher likelihood that one of the features was affected randomly. Concentration-response curves were derived for all the morphological features and also for lethality and abnormalities (visual assessment) only when a clear concentration-response was observed and more than 30% of embryos were affected. To characterize responses for each chemical we derived an EC50 as the concentration at which 50% of the embryos were deviating from the feature as it was observed in controls. Lethal concentrations (LC50) and effect concentrations (EC50) for each endpoint were obtained with the sigmoidal dose-response (Hill-slope) equation (equation 1) calculated in SigmaPlot (version 13.0).
| (1) |
Constraints for max and min were set to 100 and 0.
In order to rank the capability of an agent to produce developmental toxicity in relation to lethal effects we calculated the teratogenic index (TI), which is defined as the ratio between the LC50/EC50 and was successfully established in the Xenopus frog embryo’s developmental toxicity screening assay (Mouche et al., 2017). A chemical was classified as developmentally toxic if the teratogenic index was greater than 1.2 in either developmental stage based on previous internal results obtained in the Sanofi lab (data not shown). If no mortality was observed, the chemical was considered developmentally toxic if morphological alterations were concentration-dependent reaching more than the 30% effect level. For the automatic image-based assessment, effect concentrations (EC50) for all endpoints were calculated based on a log-logistic model in R (LL.4 model from package drc [Ritz et al., 2015]). To reduce uncertainty, treatment groups with less than 4 surviving individuals were excluded from the analysis. Effect signatures of visual and image-based assessment were obtained by normalizing each effect concentration to the most sensitive feature (EC50 most sensitive feature/EC50 specific feature) for each time point (48 and 96 hpf). This allows for comparison of all features at the same scale. Hierarchical clustering was performed based on the “Manhattan” distance using the hclust function in R and “Ward.D2” method.
RESULTS
The FishInspector Software and Phenotype Characterization
A user-friendly platform for feature detection based on two-dimensional projection of fish embryos called FishInspector was developed. The graphical user interface of the software is illustrated in Figure 1. FishInspector is written in MATLAB and an executable version for Windows is freely available (latest software update available at Zenodo [Kießling et al., 2018]). The software has a modular structure and the MATLAB code can, in principle, be extended to include more features by programming appropriate plugins. In order to compensate for potential errors of the automated image analysis, particularly during the development of the software or in cases where it is difficult to establish error-free automated detection, the software allows user interaction and correction. Variability of image qualities depending on the source (camera and microscope settings, resolution, contrast, intensity) may impact on feature detection. Therefore, adjustable parameters were included in the software, making it possible to compensate for camera or microscope-dependent differences. In its current version the FishInspector is able to locate up to 10 different morphological features (Table 1), and export their coordinates to an open format (JSON—JavaScript Object Notation—file). The average processing time was 3 h per plate (2 h unsupervised for the image acquisition and 1 h for the FishInspector analysis). It should be noted that FishInspector is not intended to detect deviations from normal phenotypes. This is done by subsequent analysis using the identified feature coordinates and existing analysis routines. The identified feature coordinates are processed subsequently in a KNIME workflow to derive their metrics (Table 1, see Material and Methods, supplementary KNIME workflow in Dryad, Teixido et al., 2018). The features were chosen because of their relevance in zebrafish embryo development and the observed phenotypes of the model compound exposures.
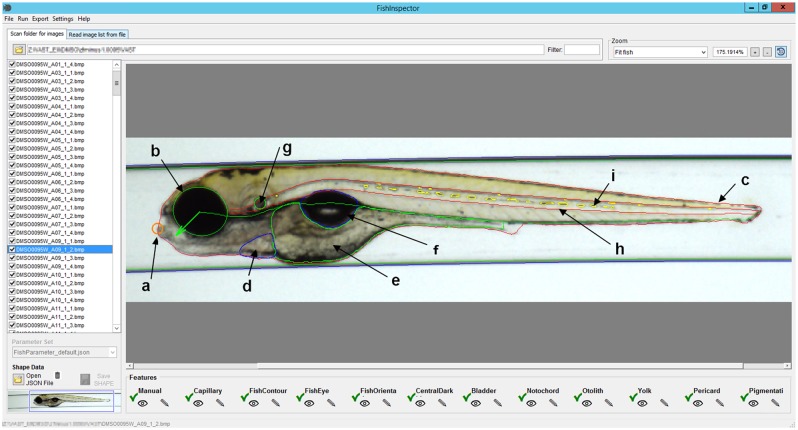
Figure 1.
Screenshot of the FishInspector graphical user interface showing an image with detected regions of interest (ROIs) for each feature. The interface allows users to adjust and correct detected ROIs manually. The image shows the final corrected ROIs and the detected features are the following: a, lower jaw tip (orange); b, eye contour (green); c, fish contour (red); d, pericard (blue); e, yolk sac (green); f, swim bladder (blue); g, otolith (green); h, notochord (green); i, pigmentation (yellow). Colours refer to the online version.
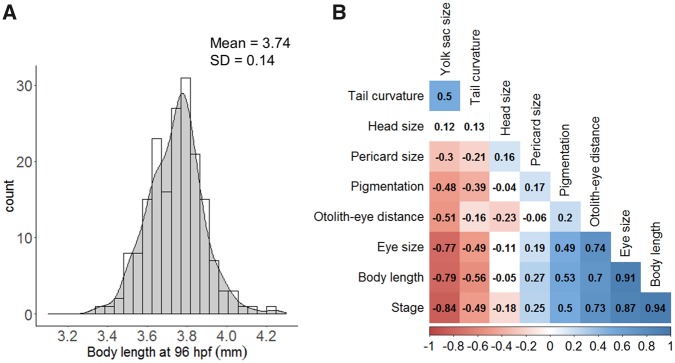
Some features can be expected to change during the course of development. So, developmental retardation would lead to changes in those parameters in particular. If several features that correlate during the course of normal development change in a consistent manner, this could serve as an indicator for developmental retardation. Therefore, cross-correlation of the different features was analyzed in untreated embryos from 32 to 96 hpf (Figure 2b). Body length and eye size were the most highly correlated features (r = 0.94 and 0.87, respectively) following by yolk sac size (r = −0.84). The eye-ear distance, a common morphological marker used to stage zebrafish embryos (Beasley et al., 2012; Kimmel et al., 1995), showed a slight correlation (r = 0.7). However, if restricted to stages between 48 and 96 hpf, the correlation increased (r = 0.92, Supplementary Figure 6) and was therefore used to asses growth retardation at 96 hpf. The lower jaw position was analyzed between 72 and 96 hpf and also showed a positive correlation (Supplementary Figure 7).
Figure 2.
Control variability and cross-correlation of morphological features. A, Example of distribution plot for total body length obtained for control population at 96 hpf (n = 183). The mean and standard deviation (SD) were used to derive a threshold to detect the fraction of treated embryos that deviate from controls (see Materials and Methods). B, Cross-correlation of the morphological features over zebrafish development (from 32 to 96 hpf). The plot displays Pearson’s linear correlation coefficient for every pair of variables. Correlation was based using the individual metric of each embryo (N = 44–79). Jaw-eye distance correlation was not included as it was only analyzed between 72 and 96 hpf (see Supplementary Figure 7).
In fish embryo toxicity assays, DMSO is often used as carrier solvent to accelerate solubilization of hydrophobic chemicals, up to concentrations of around 1%. Therefore, effects of DMSO were also evaluated using the FishInspector software and KNIME workflows. Most of the affected endpoints exhibited EC50 ≥ 2% (v/v) DMSO, except for noninflation of the swim bladder and locomotor response. Both showed an EC50 value of around 1% DMSO (Supplementary Table 4) representing the maximum solvent concentration that was used for analyzing the effects of dexamethasone (for loratadine, glibenclamide, and all-trans retinoic maximum DMSO concentrations of 0.5%, 0.5%, and 0.01%, respectively, were used).
Comparison of the Automated Quantitative versus Visual Analysis
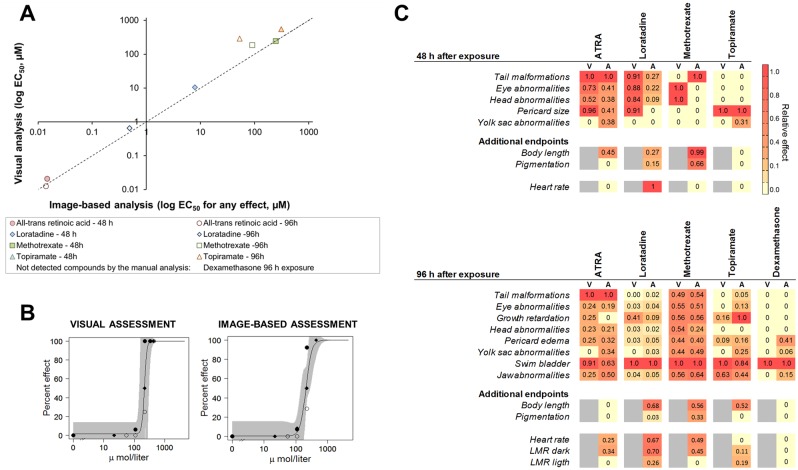
To illustrate the performance of the software, we analyzed the phenotypic effects of six model compounds previously characterized in the zebrafish and mammalian models for developmental toxicity (Supplementary Table 5). First, the visual assessment and the automated quantitative assessment with the FishInspector were compared by calculating a cumulative EC50 representing the concentration where 50% of the embryos were affected by any of the quantified individual endpoints (swim bladder effects were excluded for this analysis). The two assessments revealed very similar effect levels (Figure 3a). However, the visual assessment did not reach an EC50 for dexamethasone, whereas the automated assessment—based mainly on morphological changes of pericard size, yolk sac size, and lower jaw position—was able to reveal an EC50 of 5 µM after 96 h of exposure.
Figure 3.
Comparison of quantitative versus the visual assessment of zebrafish embryo phenotypes. A, Correlation between aggregated EC50 values derived from the visual and the image-based quantitative automatic analysis. Dashed line indicates the line of unity. B, Concentration-response curves for decreased eye size in zebrafish embryos at 96 hpf after exposure to methotrexate obtained by visual and image-based analysis. Different symbols refer to independent replicates. C, Effect signatures obtained using visual (V) and image-based (A) assessment. The relative effects are shown by a color code from the most sensitive effect (red, 1) to no effect (yellow, 0). Areas without a value (in gray) indicate that the endpoint was not assessed. Endpoint terminology was adapted for a better comparison, as manual analysis is a subjective measure and the automatic image-based analysis gives a quantitative measure of a detailed effect (eg, eye abnormalities vs eye size, growth retardation vs otolith-eye distance). Glibenclamide at 48 hpf/96 hpf and dexamethasone at 48 hpf did not provoke any effects. Abbreviations: V, visual assessment; A, automatic image-based assessment using the FishInspector. ATRA, all-trans retinoic acid; LMR, locomotor response.
EC50 values were also derived for each individual endpoint analyzed with the visual and automatic image-based method (see Figure 3b for an example of concentration-response curve).
Figure 3c shows the comparison between visual and image-based specific altered endpoints using a color scale that represents the EC50 normalized to the most sensitive endpoint for each of the time points (48 and 96 hpf).
In addition to the morphological endpoints analyzed with the FishInspector, two functional endpoints, heart rate and locomotor response for behavior analysis, were added to our analysis to increase the diagnostic power of the phenotype assessment. Loratadine showed a strong reduction in heart rate at both measurement time points. Topiramate exposure was found to alter heart rate at 96 hpf. Methotrexate and all-trans retinoic acid showed reduced locomotion in the dark phase, in contrast to topiramate and loratadine, which showed increased locomotion during light phase.
Chemical Signatures
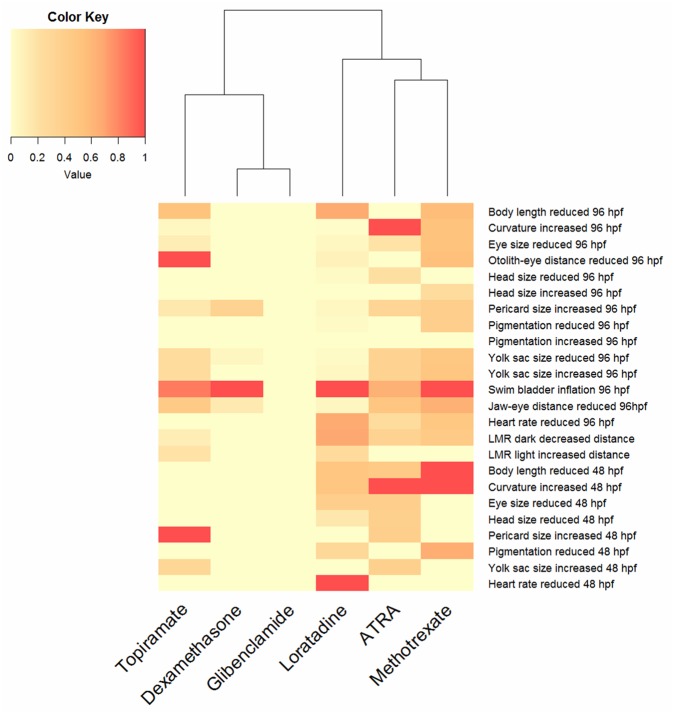
The measurement of each individual endpoint enabled the construction of a phenotypic signature for each compound according to the most affected endpoint. Figure 4 shows these signatures with a color code scaled from no effect (yellow, 0) to specific effect (red, 1).
Figure 4.
Heat map of phenotypes and functional endpoints observed after chemical exposure of zebrafish embryos. The color code refers to normalized effect concentrations at the appropriate time point (48 hpf and 96 hpf). The scale ranges from no effect (0, yellow) to most sensitive endpoints (1, red). Abbreviation: ATRA, all-trans retinoic acid.
Inter-Laboratory Assessment of the Zebrafish Developmental Toxicity Assay
The five selected compounds were also evaluated in two other laboratories that are currently using visual assessment to score for developmental toxic effect in zebrafish (Sanofi and Biobide).The overall results (LC50, EC50 values) are shown in Table 2.
Table 2.
Interlaboratory Comparison of Effect Concentrations, NOAEL, and Teratogenic Index after Embryo Exposure to the Selected Compounds at 48 and 96 hpf
| Substance | Type of Assessment | Laboratory | EC50 (µM) |
LC50 (µM) |
TI (LC50/EC50) |
Highest Tested Concentration (µM) | |||
|---|---|---|---|---|---|---|---|---|---|
| 48 hpf | 96 hpf | 48 hpf | 96 hpf | 48 hpf | 96 hpf | ||||
| Loratadine | V | Biobide | 10.78 | 1.64 | >30 | 11.51 | >2.8 | 7.1 | 30 |
| V | Sanofi | 9.31 | 7.1 | 13.9 | 9.25 | 1.5 | 1.3 | 30 | |
| V | BIOTOX-UFZ | 10.34 | 0.65 | 19.14 | 12.82 | 1.8 | 19.7 | 26 | |
| A | BIOTOX-UFZ | 7.9 | 0.38 | — | — | 2.4 | 33.7 | ||
| Methotrexate | V | Biobide | 337.3 | 216.1 | >1000 | 351.2 | >3 | 1.6 | 1000 |
| V | Sanofi | 260 | 75.4 | 321 | 101 | 1.2 | 1.3 | 500 | |
| V | BIOTOX-UFZ | 244.48 | 184.4 | 357.8 | 304.8 | 1.5 | 1.6 | 550 | |
| A | BIOTOX-UFZ | 247.6 | 90.8 | — | — | 1.4 | 3.4 | ||
| Dexamethasone | V | Biobidea | >300 | >300 | >300 | >300 | — | — | 600 |
| V | Sanofi | >500 | 559b | >500 | >500 | — | — | 500 | |
| V | BIOTOX-UFZ | >255 | >255 | >255 | >255 | — | — | 255 | |
| A | BIOTOX-UFZ | >255 | 5 | >255 | >255 | — | >51 | ||
| Topiramate | V | Biobide | 863.5 | 198.6 | >1500 | 671.7 | >1.7 | 3.4 | 1500 |
| V | Sanofi | 767 | 325 | 1279 | 678 | 1.7 | 2.1 | 1000 | |
| V | BIOTOX-UFZ | 551.2 | 284.2 | 1224.1 | 937.9 | 2.2 | 3.2 | 1500 | |
| A | BIOTOX-UFZ | 311.7 | 58.8 | — | — | 3.9 | 15.9 | ||
| Glibenclamide | V | Biobide | >500 | >500 | >500 | >500 | — | – | 500 |
| V | Sanofi | >200 | >200 | >200 | >200 | — | — | 200 | |
| V | BIOTOX-UFZ | >101.2 | >101.2 | >101.2 | >101.2 | — | — | 101.2 | |
| A | BIOTOX-UFZ | >101.2 | >101.2 | >101.2 | >101.2 | — | — | ||
Abbreviations: V, visual assessment; A, automatic image-based assessment using the FishInspector.
Precipitation was observed from 350 μM.
Effect concentration was extrapolated.
Only in 1 laboratory (Sanofi), dexamethasone showed a concentration-dependent increase in effects and an EC50 could be extrapolated. Based on the teratogenic index with individually set laboratory thresholds (Sanofi threshold for developmental toxicity liability of TI > 1.2), there were 4 compounds classified as developmentally toxic compounds (loratadine, methotrexate, topiramate, and dexamethasone) and 1 (glibenclamide) classified as nondevelopmentally toxic. Glibenclamide is not reported to cause developmental toxicity in mammals.
DISCUSSION
The FishInspector as a Flexible Platform for Detecting Morphological Features
Although large-scale toxicity screens have been carried out with zebrafish (Gustafson et al., 2012; Padilla et al., 2012; Truong et al., 2014), the phenotypic assessments are typically nonquantitative or semiquantitative at best. Morphological phenotyping remains a subjective process that may vary greatly between laboratories and could be affected by the fatigue, training, and expertise of those who perform the analysis and scoring. The use of a more unbiased, quantitative phenotypic assessment using image analysis, such as the one presented in this manuscript, can mitigate the subjectivity inherent in tests that rely on phenotype observations. Aiming to reduce this potential subjective bias from zebrafish embryo morphological analysis and to potentially link phenotype patterns to mode of action in subsequent analyses, we developed the software FishInspector. It provides an integrated and user-friendly platform for feature detection based on a two-dimensional projection of fish embryos. A crucial prerequisite is that embryos are analyzed out of their chorion (requiring manual dechorionation for stages <72 hpf) and that images are obtained after precise orientation of embryos. For instance, more than 75% eye overlap of the left and right eye in lateral two-dimensional projections was reported to be required for ear-eye distance analyses with less than 5% error (Beasley et al., 2012).
Correction of feature detection with the FishInspector is frequently required, but not for all features. For example, eye size, body length, notochord, and yolk are robust parameters that rarely need interaction or require only little correction. Other features like the jaw or pericard mostly require user correction. However, user interaction in the FishInspector is required only for the detection of the features and can also be conducted blind. Assessment of whether the chemical is provoking a certain phenotype or deviation from controls is made via concentrations-response modeling. This greatly reduces the bias if compared with visual microscopic observation and scoring. Furthermore, with the FishInspector one has an improved documentation of the analysis given that assessments can always be traced back to the original images.
Existing image analysis platforms (Molecular Devices ImageXpress, Definiens Developer software, Noldus Danioscope, Thermo Scientific Cellomics Zebrabox, or GE Healthcare Lifesciences Cell Investigator Zebrafish Analysis) do not at present allow feature annotation to the same extent or with the same flexibility or future development potential as our approach. Moreover they are not freely available as open source software, and some of them require co-purchase of certain equipment and/or have been discontinued. The FishInspector software in our study has been used in conjunction with the VAST bioimager system which automatically positions embryos in a glass capillary prior to imaging (Pardo-Martin et al., 2010). However, in principle, it is possible to use conventional pictures obtained with a bright-field microscope (Supplementary Figure 8). Therefore, we provide a simple workflow that automatically rotates the images and draws a virtual capillary. The user-friendly workflow processes multiple images simultaneously based on an imageJ macro embedded in a KNIME workflow (Teixido et al., 2018). Hence, it uses established and open source software. The workflow can easily be adapted to accommodate different image properties depending on the source of the image (eg, intensity, contrast). As for any type of image analysis, the quality of the images is critical even if manually positioned embryo images are used.
A limited number of features can be detected at present (Table 1). Due to the modular architecture of the FishInspector, the plan is to increase the number of detected morphological features, including support for dorsal and ventral images. Future versions may also implement self-learning algorithms to make automatic feature detection more robust. Manually approved feature contours could be used, for example, to train Active Shape/Appearance models (Cootes et al., 1998; Cootes and Taylor, 1992) and to minimize the need for manual correction.
Cross-correlation analysis of all the features with progressing development indicated that a subset of the morphological endpoints exhibit a high correlation and enable improved identification of growth retardation (Figure 2b), a common parameter evaluated in mammalian developmental toxicity studies. The potential confounding effects of DMSO on phenotypes and behavior was also revealed in this study. DMSO was used up to a concentration of 1%, representing the EC50 for nonswim bladder inflation and reduced locomotor activity. Effects of DMSO on locomotion have been previously reported in other studies at a concentration as low as 0.01% (Chen et al., 2011). The effect on these parameters should be carefully interpreted (eg, reduce locomotion in dexamethasone-treated embryos in combination with 1% DMSO in our study). Hence, we suggest, in general, minimizing the amount of DMSO especially for specific examinations or considering potential interference by solvents in the interpretation of results. However, for screening purposes, maximization of the compound solubility and uptake through standardized DMSO concentrations (eg, 1%) have been used effectively with good predictivity (Krupp, 2016).
Software Performance and Differences between Visual and Automated Assessment
The ability of our approach to detect developmental toxicity was demonstrated by using six compounds previously assessed by other laboratories for the optimization and performance evaluation of the zebrafish developmental toxicity assay (Gustafson et al., 2012). Our image-based quantitative approach eliminates possible observation bias whereas demonstrating consistency with the overall effect assessment by visual analysis of an experienced researcher. Furthermore, automated assessment included the evaluation of two additional endpoints, body length, and pigmentation, which could not be properly evaluated by visual analysis due to its inherent subjectivity. Our approach slightly increases throughput given that the imaging is conducted unsupervised. However, the amount of data generated also increases the subsequent analysis workload. Indeed we did not primarily aim or expect to increase throughput, rather to increase content and accuracy in the morphological assessment.
Comparison between visual and automatic specific altered endpoints reveals in general good agreement, with 3 major exceptions (Figure 3c): (1) Methotrexate exposure resulted in increased incidence of embryos showing bending of the tail after 48 h of exposure. However the visual analysis was not sensitive enough to capture this effect. (2) Using visual assessment we were only able to observe a concentration-dependent effect on swim bladder inflation for dexamethasone after 96 h of exposure, but the automatic assessment revealed also a concentration-dependent increase of pericard size, reduction of yolk sac size, and reduced jaw-eye distance. (3) For loratadine exposure after 96 h, the visual assessment indicated swim bladder inflation and growth retardation as the most sensitive endpoints. However, the measurement of body length revealed that loratadine specifically affects body length of the embryo at much lower concentrations than other indicators of growth retardation. Failure to inflate the swim bladder at 96 hpf represented the most sensitive endpoint in almost all the chemical exposures and could be related to a developmental delay as untreated embryos at 96 hpf often do not have a fully inflated swim bladder (Supplementary Figure 9). The swim bladder in the developing zebrafish has been shown to be evolutionarily homologous to the mammalian lung (Zheng et al., 2011). However, it is not known whether swim bladder malformations could relate to developmental toxicity in higher vertebrates. Moreover, swim bladder development depends on blood circulation and, hence, may represent a secondary effect of disturbed vascularization (Yue et al., 2015). Chemicals affecting heart rate (eg, β-blockers, Bittner et al., 2018) displayed a co-occurrence of missing swim bladder inflation and heart rate decrease. Therefore, as swim bladder inflation seems to be affected by many chemicals, it may have a limited diagnostic value at the 96 hpf stage. Two functional endpoints, heart rate and locomotor response, allowed us to discover potential off-target effects of drugs, like reduced heart rate after loratadine exposure. Heart and jaw abnormalities are frequently analyzed as teratogenic indicators, using transgenic or stained fish embryos. Heart morphology has not yet been included in the automatic assessment with the FishInspector, but heart rate quantification may partially capture heart malformations.
Comparative Effect Analysis
Using the different morphological and functional endpoints quantified in our study, phenotypic signatures were derived for each chemical and scaled by normalization to the effect concentration of the most sensitive endpoint. Our data suggests that observed differences in phenotype patterns could reflect the differences in the underlying mechanism of action (Figure 4). Using the FishInspector software, a larger amount of chemicals with similar mechanisms could now be analyzed to reveal whether commonalities between compound effect patterns could be derived and linked to modes of action or common key events. In the present analysis, embryos exposed to all-trans retinoic acid and methotrexate both showed tail or body axis curvature as the most sensitive morphological feature. Both compounds are associated with neural tube defects in mammals. All-trans retinoic acid interferes with the retinoic pathway, which is especially important for anterior-posterior patterning of the spinal cord and hindbrain, neuronal differentiation, and axis elongation (Tonk et al., 2015). Methotrexate is a folate analog that acts by competitively inhibiting dihydrofolate reductase, an enzyme involved in DNA biosynthesis. This impairment in nucleotide biosynthesis can decrease mitotic rates during critical morphogenetic windows (Lee et al., 2012). Hence, similarities in effect patterns may reflect the conversion of both pathways at neural tube organogenesis.
Our study also supports evidence for the known side effects of the antihistaminic loratadine. The most affected endpoint for loratidine exposure was reduced heart rate and body length of the embryos. Some antihistaminic compounds have been shown to reduce the heart rate by competitive inhibition of the muscarinic receptors in mammals (Liu et al., 2006). In zebrafish, knock-down of muscarinic receptors has been demonstrated to alter cardiac β-adrenergic receptor activity (Steele et al., 2009).
The phenotypic effect observed after exposure to the antiepileptic drug topiramate revealed growth retardation as the most affected endpoint after 48 and 96 h exposure. The use of antiepileptic drugs during pregnancy has been associated with congenital defects and developmental delay in humans (Campbell et al., 2013), however the underlying mechanism is still unknown. Our approach allowed us to identify growth retardation as the main endpoint of topiramate exposure, rather than teratogenic effects. Antiepileptic drugs are also capable of inducing neurodevelopmental effects (Ornoy, 2006) and interfere with the GABA and AMPA/kainate glutamate receptor and block voltage-dependent sodium channels (Schneiderman, 1998). In our study we observed increased locomotion during the light phase of the locomotor response analysis, which may potentially relate to the MoA of topiramate.
Dexamethasone exposure caused reduced yolk sac size in zebrafish embryos, potentially related to the role of glucocorticoid in energy metabolism by mobilizing and relocating energy substrate stores (Nesan and Vijayan, 2013). Mammalian studies have demonstrated that glucocorticoids cause cleft palate and some studies have shown that glucocorticoids alter craniofacial development in zebrafish as well (Hillegass et al., 2008). Our study also revealed an alteration in jaw development by a reduced jaw-eye distance (Figure 3c).
Interlaboratory Assessment
The performance of our method was verified by comparing it with the visual assessments of 3 different laboratories experienced with conventional visual assessment of the zebrafish embryos. A previous interlaboratory assessment study showed that technical differences were the primary contributor to interlaboratory differences in classification of a compound as developmentally toxic using zebrafish embryos (Ball et al., 2014). Our approach avoids score assignment based on qualitative measures of effect. The interlaboratory study showed good agreement; however dexamethasone was classified as developmentally toxic by only one laboratory (Sanofi) using the visual inspection method. The quantitative approach showed a higher sensitivity for the detection of chemical effects and the sensitivity of effect assessment for dexamethasone was increased (Table 2). The overall weak effects caused by dexamethasone, however, could also be due to reduced bioavailability of the compound. It has been reported that embryonic concentrations reached only 20% of the exposure concentrations indicating a potential slow uptake and internal concentration not in equilibrium (Steenbergen et al., 2017). Uptake of the chemicals by zebrafish embryos was not analyzed in our study, as the focus was on feature detection and quantification of developmental toxicity. However, we consider it important that this be included in routine screens, either via appropriate TK models or by internal concentration analysis (Brox et al., 2014) since a slow and/or limited uptake of a substance by an embryo could represent a confounding factor in the assessment of effects. Loratadine was classified as a false positive in all laboratories including the automatic image-based assessment. This compound demonstrated a high uptake in previous studies, which may have contributed to the false positive results in the assay (Gustafson et al., 2012). Whether analysis with the FishInspector would lead to a higher number of false positives, however, requires a more thorough analysis of a greater number of chemicals.
CONCLUSIONS
This study has demonstrated the value of the FishInspector software and quantitative analysis has been demonstrated. The FishInspector software allows an unbiased and automated quantitative assessment of morphological changes in zebrafish embryos after chemical treatment, particularly for embryos positioned to a precise orientation. Its modular architecture allows users to implement the detection of additional features. Furthermore, to facilitate automatic recognition of features and reduce user interaction, self-learning algorithms for feature detection could be considered.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Leuthold of the Bioanalytical Ecotoxicology Department (UFZ) for his support in the laboratory and Nicole Schweiger for help with the fish care. We kindly acknowledge Dr Benjamin Piña and Rubén Martínez from IDAEA-CSIC (Spain) for providing zebrafish images without capillary boundaries, enabling us to develop a workflow to process images from conventional microscopic analysis with the FishInspector. Anne Carney, Berlin, is thanked for professional English language editing. The authors declare that there are no conflicts of interest, except for C.Q. and A.M., who are affiliated with BBD Biophenix-Biobide and have a financial or nonfinancial interest in the subject matter or materials discussed in the manuscript. The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the companies with which the authors are affiliated.
FUNDING
This work was supported by a grant from the German Ministry of Education and Research (BMBF) to the project ZFminus1 (grant number 031A582). We gratefully acknowledge access to the platform CITEPro (Chemicals in the Terrestrial Environment Profiler) funded by the Helmholtz Association.
REFERENCES
- Alshut R., Legradi J., Liebel U., Yang L., Van W. J., Strähle U., Mikut R., Reischl M. (2010) Methods for automated high-throughput toxicity testing using zebrafish embryos. In KI 2010: Advances in Artificial Intelligence (Dillmann R., Beyerer J., Hanebeck U. D., Schultz T., Eds) Lecture Notes in Computer Science, 6359 Springer, Berlin, Heidelberg. [Google Scholar]
- Arslanova D., Yang T., Xu X., Wong S. T., Augelli-Szafran C. E., Xia W. (2010). Phenotypic analysis of images of zebrafish treated with Alzheimer’s gamma-secretase inhibitors. BMC Biotechnol. 10, 24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball J. S., Stedman D. B., Hillegass J. M., Zhang C. X., Panzica-Kelly J., Coburn A., Enright B. P., Tornesi B., Amouzadeh H. R., Hetheridge M., et al. (2014). Fishing for teratogens: A consortium effort for a harmonized zebrafish developmental toxicology assay. Toxicol. Sci. 139, 210–219. [DOI] [PubMed] [Google Scholar]
- Beasley A., Elrod-Erickson M., Otter R. R. (2012). Consistency of morphological endpoints used to assess developmental timing in zebrafish (Danio rerio) across a temperature gradient. Reprod. Toxicol. 34, 561–567. [DOI] [PubMed] [Google Scholar]
- Berthold M. R., Cebron N., Dill F., Gabriel T. R., Kötter T., Meinl T., Ohl P., Sieb C., Thiel K., Wiswedel B., (2008) KNIME: The Konstanz Information Miner In Data Analysis, Machine Learning and Applications (Preisach C., et al. , Eds.), Studies in Classification, Data Analysis, and Knowledge Organization, pp. 319–326. Springer, Berlin, Heidelberg. [Google Scholar]
- Bittner L., Teixido E., Seiwert B., Escher B. I., Klüver N. (2018). Influence of pH on the uptake and toxicity of β-blockers in embryos of zebrafish, Danio rerio. Aquat. Toxicol. 201, 129–137. [DOI] [PubMed] [Google Scholar]
- Bonhomme V., Picq S., Gaucherel C., Claude J. (2013). Momocs: Outline analysis using R. J. Stat. Softw. 56, 1–24. [Google Scholar]
- Brannen K. C., Panzica-Kelly J. M., Danberry T. L., Augustine-Rauch K. A. (2010). Development of a zebrafish embryo teratogenicity assay and quantitative prediction model. Birth Defects Res. B Dev. Reprod. Toxicol. 89, 66–77. [DOI] [PubMed] [Google Scholar]
- Brox S., Ritter A. P., Küster E., Reemtsma T. (2014). A quantitative HPLC–MS/MS method for studying internal concentrations and toxicokinetics of 34 polar analytes in zebrafish (Danio rerio) embryos. Anal. Bioanal. Chem. 406, 4831–4840. [DOI] [PubMed] [Google Scholar]
- Burns G. C., Milan D. J., Grande E. J., Rottbauer W., Macrae C. A., Fishman M. C. (2005). High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 1, 263–264. [DOI] [PubMed] [Google Scholar]
- Campbell E., Devenney E., Morrow J., Russell A., Smithson W. H., Parsons L., Robertson I., Irwin B., Morrison P. J., Hunt S., et al. (2013). Recurrence risk of congenital malformations in infants exposed to antiepileptic drugs in utero. Epilepsia 54, 165–171. [DOI] [PubMed] [Google Scholar]
- Chen T.-H. H., Wang Y.-H. H., Wu Y.-H. H. (2011). Developmental exposures to ethanol or dimethylsulfoxide at low concentrations alter locomotor activity in larval zebrafish: Implications for behavioral toxicity bioassays. Aquat. Toxicol. 102, 162–166. [DOI] [PubMed] [Google Scholar]
- Claude J., Baylac M., Stayton T. (2008) Traditional statistics for morphometrics. In Morphometric with R. pp. 70–131. Springer, Berlin, Heidelberg. [Google Scholar]
- Cootes T. F., Edwards G. J., Taylor C. J. (1998) Active Appearance Models. Springer, Berlin, Heidelberg, pp. 484–498. [Google Scholar]
- Cootes T. F., Taylor C. J. (1992) Active shape models—‘Smart snakes’ In BMVC92, pp. 266–275. Springer, London. [Google Scholar]
- Deal S., Wambaugh J., Judson R., Mosher S., Radio N., Houck K., Padilla S. (2016). Development of a quantitative morphological assessment of toxicant-treated zebrafish larvae using brightfield imaging and high-content analysis. J. Appl. Toxicol. 36, 1214–1222. [DOI] [PubMed] [Google Scholar]
- Dooley K., Zon L. I. (2000). Zebrafish: A model system for the study of human disease. Curr. Opin. Genet. Dev .10, 252–256. [DOI] [PubMed] [Google Scholar]
- Ducharme N. a., Peterson L. E., Benfenati E., Reif D., McCollum C. W., Gustafsson J.-Å., Bondesson M. (2013). Meta-analysis of toxicity and teratogenicity of 133 chemicals from zebrafish developmental toxicity studies. Reprod. Toxicol. 41, 98–108. [DOI] [PubMed] [Google Scholar]
- Gunnarsson L., Jauhiainen A., Kristiansson E., Nerman O., Larsson D. G. (2008). Evolutionary conservation of human drug targets in organisms used for environmental risk assessments. Environ. Sci Technol. 42, 5807–5813. [DOI] [PubMed] [Google Scholar]
- Gustafson A.-L. L., Stedman D. B., Ball J., Hillegass J. M., Flood A., Zhang C. X., Panzica-Kelly J., Cao J., Coburn A., Enright B. P., et al. (2012). Inter-laboratory assessment of a harmonized zebrafish developmental toxicology assay—Progress report on phase I. Reprod. Toxicol. 33, 155–164. [DOI] [PubMed] [Google Scholar]
- Hermsen S. A. B., van den Brandhof E.-J., van der Ven L. T. M., Piersma A. H. (2011). Relative embryotoxicity of two classes of chemicals in a modified zebrafish embryotoxicity test and comparison with their in vivo potencies. Toxicol. In Vitro 25, 745–753. [DOI] [PubMed] [Google Scholar]
- Hillegass J. M., Villano C. M., Cooper K. R., White L. A. (2008). Glucocorticoids alter craniofacial development and increase expression and activity of matrix metalloproteinases in developing zebrafish (Danio rerio). Toxicol. Sci. 102, 413–424. [DOI] [PubMed] [Google Scholar]
- Irons T. D., MacPhail R. C., Hunter D. L., Padilla S. (2010). Acute neuroactive drug exposures alter locomotor activity in larval zebrafish. Neurotoxicol. Teratol. 32, 84–90. [DOI] [PubMed] [Google Scholar]
- Jeanray N., Marée R., Pruvot B., Stern O., Geurts P., Wehenkel L., Muller M. (2015). Phenotype classification of zebrafish embryos by supervised learning. PLoS One 10, e0116989.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson J., von Hofsten J., Olsson P. E. (2001). Generating transparent zebrafish: A refined method to improve detection of gene expression during embryonic development. Mar. Biotechnol. 3, 522–527. [DOI] [PubMed] [Google Scholar]
- Kießling T. R., Teixidó E., Scholz S. (2018). FishInspector v1.03 - Annotation of features from zebrafish embryo images (Version v1.03). Zenodo. 10.5281/zenodo.1436689. [DOI]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Klüver N., König M., Ortmann J., Massei R., Paschke A., Kühne R., Scholz S. (2015). Fish embryo toxicity test: Identification of compounds with weak toxicity and analysis of behavioral effects to improve prediction of acute toxicity for neurotoxic compounds. Environ. Sci. Technol. 49, 7002–7011. [DOI] [PubMed] [Google Scholar]
- Krupp E. (2016). Screening of developmental toxicity—Validation and predictivity of the zebrafish embryotoxicity assay (ZETA) and strategies to optimize de-risking developmental toxicity of drug candidates. Toxicol. Lett. 258, S39. [Google Scholar]
- Lee M. S., Bonner J. R., Bernard D. J., Sanchez E. L., Sause E. T., Prentice R. R., Burgess S. M., Brody L. C. (2012). Disruption of the folate pathway in zebrafish causes developmental defects. BMC Dev. Biol. 12, 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leet J. K., Lindberg C. D., Bassett L. A., Isales G. M., Yozzo K. L., Raftery T. D., Volz D. C. (2014). High-content screening in zebrafish embryos identifies butafenacil as a potent inducer of anemia. PLoS One 9, e104190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letamendia A., Quevedo C., Ibarbia I., Virto J. M., Holgado O., Diez M., Izpisua Belmonte J. C., Callol-Massot C. (2012). Development and validation of an automated high-throughput system for zebrafish in vivo screenings. PLoS One 7, e36690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zheng Q., Farley J. M. (2006). Antimuscarinic actions of antihistamines on the heart. J. Biomed. Sci. 13, 395–401. [DOI] [PubMed] [Google Scholar]
- Liu R., Lin S., Rallo R., Zhao Y., Damoiseaux R., Xia T., Lin S., Nel A., Cohen Y. (2012). Automated phenotype recognition for zebrafish embryo based in vivo high throughput toxicity screening of engineered nano-materials. PLoS One 7, e35014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheel A. P., Ko C. Y., Guh H. Y. (1998). Ion chromatography method and validation for the determination of sulfate and sulfamate ions in topiramate drug substance and finished product. J. Chromatogr. B Biomed. Appl. 709, 166–172. [DOI] [PubMed] [Google Scholar]
- Mouche I., Malésic L., Gillardeaux O. (2017). FETAX assay for evaluation of developmental toxicity. In Drug Safety Evaluation (Gautier J. C., Eds). Methods in Molecular Biology, 1641 New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- Nesan D., Vijayan M. M. (2013). Role of glucocorticoid in developmental programming: Evidence from zebrafish. Gen. Comp. Endocrinol. 181, 35–44. [DOI] [PubMed] [Google Scholar]
- OECD (2013) Test no. 236: Fish embryo acute toxicity (FET) test. Paris, France. OECD Guidel. Test. Chem. Sect. 2, OECD Publ., pp. 1–22.
- Ornoy A. (2006). Neuroteratogens in man: An overview with special emphasis on the teratogenicity of antiepileptic drugs in pregnancy. Reprod. Toxicol. 22, 214–226. [DOI] [PubMed] [Google Scholar]
- Padilla S., Corum D., Padnos B., Hunter D. L., Beam A., Houck K. A., Sipes N., Kleinstreuer N., Knudsen T., Dix D. J., et al. (2012). Zebrafish developmental screening of the ToxCastTM Phase I chemical library. Reprod. Toxicol. 33, 174–187. [DOI] [PubMed] [Google Scholar]
- Pardo-Martin C., Chang T.-Y., Koo B. K., Gilleland C. L., Wasserman S. C., Yanik M. F. (2010). High-throughput in vivo vertebrate screening. Nat. Methods 7, 634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peravali R., Gehrig J., Giselbrecht S., Lütjohann D. S., Hadzhiev Y., Müller F., Liebel U. (2011). Automated feature detection and imaging for high-resolution screening of zebrafish embryos. Biotechniques 50, 319–324. [DOI] [PubMed] [Google Scholar]
- Pulak R. (2016). Tools for automating the imaging of zebrafish larvae. Methods 96, 118–126. [DOI] [PubMed] [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: ISBN 3-900051-07-0. http://www.R-project.org. [Google Scholar]
- Ritz C., Baty F., Streibig J. C., Gerhard D. (2015). Dose-response analysis using R. PLoS One 10, e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiderman J. H. (1998). Topiramate: Pharmacokinetics and pharmacodynamics. Can. J. Neurol. Sci. 25, S3–S5. [DOI] [PubMed] [Google Scholar]
- Schutera M., Dickmeis T., Mione M., Peravali R., Marcato D., Reischl M., Mikut R., Pylatiuk C. (2016). Automated phenotype pattern recognition of zebrafish for high-throughput screening. Bioengineered 7, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selderslaghs I. W. T., Van Rompay A. R., De Coen W., Witters H. E. (2009). Development of a screening assay to identify teratogenic and embryotoxic chemicals using the zebrafish embryo. Reprod. Toxicol. 28, 308–320. [DOI] [PubMed] [Google Scholar]
- Steele S. L., Lo K. H., Li V. W., Cheng S. H., Ekker M., Perry S. F. (2009). Loss of M2 muscarinic receptor function inhibits development of hypoxic bradycardia and alters cardiac-adrenergic sensitivity in larval zebrafish (Danio rerio). Am. J. Physiol. Regul. Integr. Comp. Physiol 297, R412–R420. [DOI] [PubMed] [Google Scholar]
- Steenbergen P. J., Bardine N., Sharif F. (2017). Kinetics of glucocorticoid exposure in developing zebrafish: A tracer study. Chemosphere 183, 147–155. [DOI] [PubMed] [Google Scholar]
- Teixido E., Kießling T. R., Krupp E., Quevedo C., Muriana A., Scholz S. (2018) Data from: Automated morphological feature assessment for zebrafish embryo developmental toxicity screens. doi: 10.5061/dryad.gv144d5. [DOI] [PMC free article] [PubMed]
- Tonk E. C. M., Pennings J. L. A., Piersma A. H. (2015). An adverse outcome pathway framework for neural tube and axial defects mediated by modulation of retinoic acid homeostasis. Reprod. Toxicol. 55, 104–113. [DOI] [PubMed] [Google Scholar]
- Truong L., Reif D. M., St Mary L., Geier M. C., Truong H. D., Tanguay R. L. (2014). Multidimensional in vivo hazard assessment using zebrafish. Toxicol. Sci. 137, 212–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA. (2012) Benchmark dose technical guidance, EPA/100/R-12/001, June 2012. Risk Assessment Forum, US Environmental Protection Agency (EPA), Washington, DC. https://www.epa.gov/sites/production/files/2015-01/documents/benchmark_dose_guidance.pdf. Last accessed: August 2018.
- Van den Bulck K., Hill A., Mesens N., Diekman H., De Schaepdrijver L., Lammens L. (2011). Zebrafish developmental toxicity assay: A fishy solution to reproductive toxicity screening, or just a red herring? Reprod. Toxicol. 32, 213–219. [DOI] [PubMed] [Google Scholar]
- Varadhan R., Johns Hopkins University, MKG Subramaniam and AT&T Reserach Labs. (2015) features: Feature Extraction for Discretely-Sampled Functional Data. R package version 2015.12-1. https://CRAN.R-project.org/package=features.
- Vogt A., Cholewinski A., Shen X., Nelson S. G., Lazo J. S., Tsang M., Hukriede N. A. (2009). Automated image-based phenotypic analysis in zebrafish embryos. Dev. Dyn. 238, 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yozzo K. L., Isales G. M., Raftery T. D., Volz D. C. (2013). High-content screening assay for identification of chemicals impacting cardiovascular function in zebrafish embryos. Environ. Sci. Technol. 47, 11302–11310. [DOI] [PubMed] [Google Scholar]
- Yue M. S., Peterson R. E., Heideman W. (2015). Dioxin inhibition of swim bladder development in zebrafish: Is it secondary to heart failure? Aquat. Toxicol. 162, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Wang Z., Collins J. E., Andrews R. M., Stemple D., Gong Z. (2011). Comparative transcriptome analyses indicate molecular homology of zebrafish swimbladder and mammalian lung. PLoS One 6, e24019.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.