Abstract
The objective of this study was to enhance estimates of additive, dominance, and epistatic effects of marker polymorphisms on beef carcass and quality traits. Myostatin (MSTN) F94L SNP and the µ-calpain (CAPN1) 316 and 4751 SNP haplotype have previously been associated with fat and muscle traits in beef cattle. Multiyear selection in a composite population segregating these polymorphisms increased minor allele (F94L L) and chosen haplotype (CAPN1 CC and GT) frequencies to intermediate levels resulting in more precise estimates of additive and nonadditive genetic effects. During the 3 yr after selection, 176 steers were evaluated for growth, carcass, meat quality, tenderness (n = 103), and meat color traits. The statistical model included year, age of dam, age of the steer, and genotype in a random animal model. The 9 genotypes (3 CAPN1 diplotypes × 3 F94L genotypes) affected marbling score, ribeye area, adjusted fat thickness, vision yield grade (all P < 0.001), slice shear force (P = 0.03), and CIE L* reflectance (P = 0.01). Linear contrasts of the 9 genotypes estimated additive, recessive, and epistatic genetic effects. Significant additive effects of the F94L L allele decreased marbling score, adjusted fat thickness, vision yield grade, and slice shear force; and increased ribeye area and CIE L* reflectance. The homozygous F94L FF and LL genotypes differed by 1.3 to 1.9 phenotypic SD for most carcass traits and by 0.8 to 0.9 SD for slice shear force and CIE L* reflectance but carcass weight differed by only 3 kg (0.1 SD). The L allele was partially recessive to F for ribeye area (P = 0.02) and the heterozygous FL means tended to be closer to the FF genotype than the LL genotype for other carcass traits but differences from additive were not significant. The CAPN1 additive × F94L additive effect on slice shear force was the only significant epistatic estimate. The F94L L allele is prevalent in Limousin but nearly absent in other U.S. purebreds. This allele had about half of the effects on birth weight, muscle, and fat traits reported for severe MSTN mutations in Belgian Blue and Piedmontese breeds. The interaction between MSTN and CAPN1 genotypes may reflect the strong additive effects of MSTN F94L L allele on fat and muscle traits interfering with the phenotypic effect of CAPN1 genotype on meat tenderness.
Keywords: calpain, cattle, epistasis, F94L, myostatin, selection
INTRODUCTION
Some polymorphisms in the myostatin (MSTN) gene in cattle reduce functionally effective myostatin resulting in muscle hypertrophy. Before breeders knew what caused muscle hypertrophy and before DNA tests were developed, some breeders selected their cattle for increased muscling (Arthur, 1995). Selection greatly increased frequencies of some MSTN alleles in breeds such as Piedmontese and Belgian Blue (Grobet et al., 1997; Kambadur et al., 1997).
Surveys of some European cattle breeds for DNA variants in MSTN revealed many polymorphisms (Grobet et al., 1998; Dunner et al., 2003). Among those was F94L, a phenylalanine to leucine substitution at amino acid position 94 in myostatin (Grobet et al., 1998). The leucine allele frequency was high in Limousin and low or absent in many other breeds. Subsequently, this allele was associated with increased muscling and reduced fatness in Limousin crossbreds (Esmailizadeh et al., 2008; Alexander et al., 2009). However, mating designs limited statistical power for estimating dominance effects (Esmailizadeh et al., 2008).
Polymorphisms in µ-calpain (CAPN1) are associated with postmortem proteolysis of muscle leading to increased meat tenderness (Page et al., 2004; White et al., 2005; Casas et al., 2006; Robinson et al., 2012). Also, some MSTN polymorphisms suppressing production of functional myostatin increase meat tenderness (Wheeler et al., 2001). Casas et al. (2001) identified an interaction of a QTL affecting meat tenderness on BTA 4 with heterozygous MSTN and homozygous normal progeny of a Belgian Blue crossbred bull. Epistasis between MSTN and CAPN1 is possible because both genes are thought to affect protein turnover of muscle.
The objective of this study was to estimate additive and nonadditive effects associated with the F94L polymorphism in MSTN and SNP 316 and 4751 haplotypes in CAPN1. Estimates were enhanced by increasing minor allele frequency of F94L and increasing frequencies of chosen CAPN1 haplotypes through selection and by using bulls heterozygous for F94L and for CAPN1.
MATERIALS AND METHODS
The U.S. Meat Animal Research Center (USMARC) Institutional Animal Care and Use Committee approved the experiment following recommendations by FASS (1999).
Composite Population
A composite cattle population known as MARC I was formed beginning in 1978 and consisted of 0.125 Angus, 0.125 Hereford, 0.25 Braunvieh, 0.25 Charolais, and 0.25 Limousin (Gregory et al., 1991). From 1992 through 1999, the composite was divided into 2 lines: a calving ease selection line and a control line (Bennett, 2008). After completing the selection experiment, cows from both lines were bred to the same bulls and their progeny treated as a single population. During this period from 2000 through 2006, the MARC I population was continued with about 205 calves from 18 sires born each year. Approximately half of sires were replaced each year resulting in the use of 68 bulls selected from within the herd.
Genetic Markers
Selected markers were from 2 genes thought to affect muscles, especially protein turnover and proteolysis. They were the large subunit of micromolar activated calpain (CAPN1) and myostatin (MSTN). The SNP marker chosen for MSTN on BTA 2 was a phenylalanine to leucine substitution at amino acid position 94 in myostatin (F94L; rs110065568, Grobet et al., 1998). Two SNP markers, CAPN1_316 (BTA29; rs17872000) and CAPN1_4751 (BTA29; rs17872050), were used for the CAPN1 gene located on BTA 29. CAPN1_316 segregates C and G alleles, while CAPN1_4751 segregates C and T alleles. Initial findings associated the C allele of CAPN1_316 with tender beef (Page et al., 2002). Common haplotypes determined from CAPN1_316 and CAPN1_4751 SNP are CC (CAPN1hCC), GT (CAPN1hGT), and GC (CAPN1hGC). A fourth haplotype, CT (CAPN1hCT), is rare. White et al. (2005) found the largest difference for 14 d Warner-Bratzler shear force between CAPN1hCC and CAPN1hGT. The most frequent haplotype in that study (CAPN1hGC) was intermediate in tenderness to CAPN1hCC and CAPN1hGT.
Samples of DNA were extracted from blood or semen. Extraction of DNA was done using a Qiagen QIAmp DNA mini blood kit (Qiagen, Valencia, CA). Blood samples were collected in 10 mL syringes with 4% EDTA. Blood was frozen until DNA was extracted. Genotyping was performed using a primer extension method with mass spectrometry-based analysis of the extension products on a MassArray system as suggested by the manufacturer (Sequenom, Inc., San Diego, CA) and described by Stone et al. (2002). When necessary, genotype assays were repeated to reduce missing genotypes.
Base, Selection, and Evaluation Phases
The experiment was conducted in 3 phases: base, selection, and evaluation. The base phase surveyed live animals and some frozen semen from 4 populations (Angus and 3 composites; MARC I, MARC II, and MARC III) completing the calving ease selection experiment (Bennett, 2008), for CAPN1 allele and haplotype frequencies. Frequencies of CAPN1hCC, CAPN1hGT, and CAPN1hGC were 0.47, 0.28, and 0.25 for Angus; 0.20, 0.21, and 0.58 for MARC I; 0.20, 0.39, and 0.40 for MARC II; and 0.22, 0.46, and 0.32 for MARC III. Allele L (F94LaL) is more frequent than allele F (F94LaF) in the Limousin breed but absent or near zero in most other breeds in the United States (Dunner et al., 2003). Limousin makes up 0.25 of MARC I and it was assumed to be the only population segregating F94L. The estimated frequency of F94LaL in MARC I was 20.7% for 363 animals born before 2004. The Limousin specificity was verified later by genotyping Angus (n = 564), MARC II (Angus, Hereford, Gelbvieh, and Simmental; n = 538), and MARC III (Angus, Hereford, Red Poll, and Pinzgauer; n = 747) populations born in 2009, 2010, and 2011 for F94L. All were homozygous for the phenylalanine allele (F94LaF) except for 1 Angus, 1 MARC II, and 2 unrelated MARC III heterozygotes, a frequency less than assumed genotyping error rate. MARC I was chosen for this experiment, because F94LaL was present in the population.
Selection was applied from 2004 through 2006 with the goal of increasing frequencies of F94LaL, CAPN1hCC, and CAPN1hGT to 0.5 and eliminating CAPN1hGC. Calves were bled before weaning, the DNA was extracted, and then genotyped. Marker genotypes were used to select replacement bulls and heifers soon after weaning. Selection of replacement animals in this phase was based on the presence of F94LaL, CAPN1hCC, and CAPN1hGT and absence of CAPN1hGC.
The evaluation phase (birth years 2007, 2008, and 2009) increased the number of animals evaluated for carcass traits. This phase also used bulls heterozygous for both F94L and CAPN1 if available. These bulls increase the number of within sire comparisons across any combination of progeny genotypes. A heterozygous bull can sire progeny with heterozygous or either homozygous genotype. Thirty bulls sired calves in the evaluation phase. Bulls heterozygous for CAPN1hCC/CAPN1hGT (n = 15) sired 70% of calves. Bulls heterozygous for F94L (n = 23) sired 87% of calves. Bulls heterozygous for both CAPN1 and F94L (n = 10) sired 59% of calves.
Blood samples from spring-born progeny were collected before weaning and genotyped. Calves with incomplete genotypes were removed from the experiment. Replacement bulls were randomly sampled within sire from among males heterozygous for both F94L and the chosen CAPN1 haplotypes. The remaining males were castrated by banding soon after weaning and genotyping. Steers consumed corn and corn silage-based diets until harvest. Weights were taken at birth (mean date = April 13), at weaning (mean age = 159 d, SD = 18 d), and as yearlings (mean age = 344 d, SD = 22 d).
All experimental steers were harvested on a single day each year at a commercial abattoir at an average age of 487 d. In 2010 (born 2009), 29 steers were removed from the experiment based on either having unselected haplotypes (CAPN1hGC or CAPN1hCT) or having genotypes in common with many steers (reduced at random). Carcasses were weighed hot, electrically-stimulated, and chilled using the commercial facility’s proprietary system. At 36 h postmortem, carcasses were ribbed between the 12th and 13th ribs and an image analysis based (VBG2000) grading system (Shackelford et al., 2003) assessed adjusted fat thickness, ribeye area, USDA marbling score, CIE L* of the longissimus muscle, and calculated vision yield grade. A longissimus steak from the 13th rib region was returned to USMARC to evaluate slice shear force at 14 d postmortem (Shackelford et al., 1999).
Statistical Analysis
Either trait measurements or logarithms of measurements (marbling score; slice shear force) were analyzed with a mixed model using MTDFREML (Boldman et al., 1995). The model was:
where Yi,j,k,l is the observation or its logarithm for the i, j, k, l-th animal, µ is the mean, Yeari is birth year 2007, 2008, or 2009, Aod5j is age of dam (2, 3, 4, or ≥ 5 yr), b is a linear regression coefficient on the i, j, k, l-th animal’s age (Agei,j,k,l) in days, Genotypek is 1 of the 9 combinations of 3 CAPN1 diplotypes and 3 F94L genotypes, ai,j,k,l is the additive polygenic animal effect, and ei,j,k,l is of the residual effect of the i, j, k, l-th observation. Covariances of polygenic effects were assumed proportional to the pedigree relationship matrix. Residual effects were assumed independent with constant variance. The pedigree used to calculate relationships included more than 6,600 animals. Heritabilities were constrained between 0.20 and 0.70 because of few observations and imprecise genetic variance estimates. Similar ranges in heritabilities for these traits were estimated from larger, related populations (Gregory et al., 1994; Bennett and Gregory, 1996). Skewed distributions of marbling scores and meat tenderness values were transformed using base 10 logarithms to determine P values but reported means and contrasts are from analyses of untransformed data.
A P < 0.10 for the genotype effect (9 genotypes) was used as a guideline for then calculating linear contrasts for additive, dominance, and epistasis effects associated with CAPN1 haplotypes and F94L alleles similar to Tait et al. (2016). Linear contrast coefficients used to estimate genetic effects (Table 1) which were divided by their SE to determine significance based on a t-test. Only testing genetic effect contrasts after meeting an overall genotype probability test (e.g., P < 0.10 guideline) protects against probability inflation due to multiple testing. A significance level of P < 0.05 was used for individual contrasts.
Table 1.
Linear contrast coefficients used to estimate additive, dominance, and epistasis effects for µ-calpain (CAPN1) haplotype and F94L SNP
| Genotype mean1 | F94L2 | CAPN1 2 | F94L × CAPN13 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| F94L | CAPN1 | A | D | A | D | AA | AD | DA | DD |
| FF | NN-NN | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 |
| FF | CC-NN | −1 | −1 | 0 | 2 | 0 | −2 | 0 | −2 |
| FF | CC-CC | −1 | −1 | 1 | −1 | −1 | 1 | −1 | 1 |
| FL | NN-NN | 0 | 2 | −1 | −1 | 0 | 0 | −2 | −2 |
| FL | CC-NN | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 4 |
| FL | CC-CC | 0 | 2 | 1 | −1 | 0 | 0 | 2 | −2 |
| LL | NN-NN | 1 | −1 | −1 | −1 | −1 | −1 | 1 | 1 |
| LL | CC-NN | 1 | −1 | 0 | 2 | 0 | 2 | 0 | −2 |
| LL | CC-CC | 1 | −1 | 1 | −1 | 1 | −1 | −1 | 1 |
| Divisor4 | 6 | 6 | 6 | 6 | 1 | 2 | 2 | 4 | |
1Estimated genotype means are identified by the 9 combinations of 3 MSTN F94L genotypes and 3 µ-calpain (CAPN1) diplotypes. F94L genotypes are designated by FF (homozygous F94LaF), FL (heterozygotes), and LL (homozygous F94LaL). CAPN1 diplotypes are designated NN-NN (homozygous CAPN1hNN), CC-NN (heterozygotes) and CC-CC (homozygous CAPN1hCC).
2Linear contrast coefficients multiplied by genotype means to estimate F94LaL and CAPNhCC haplotype additive (A) and dominance (D) effects.
3Linear contrast coefficients for 2-factor epistatic effects identified with 2 letters. The first letter is the F94LaL effect and the second letter is the CAPN1hCC haplotype effect, e.g., AD is additive F94L × CAPN1 dominance epistatic effect.
4Actual coefficients used were the whole numbers in table divided by this number.
RESULTS AND DISCUSSION
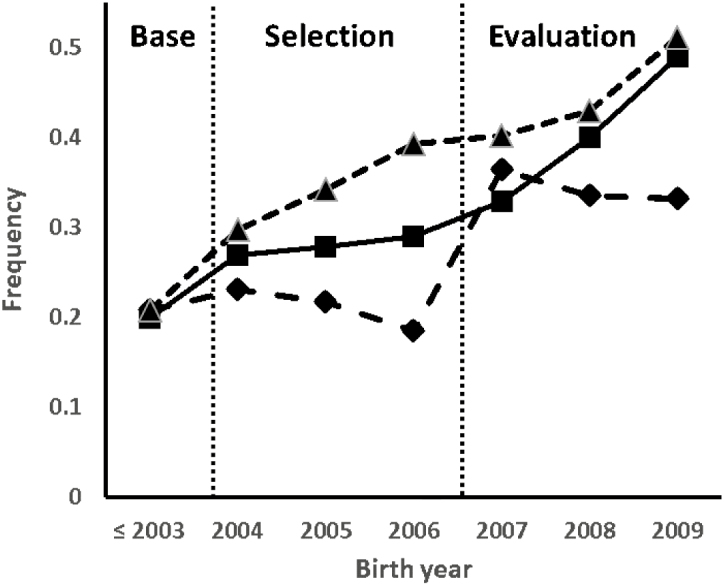
Frequencies of chosen CAPN1 haplotypes and F94LaL changed during the 3 phases of the experiment (Fig. 1). The frequency of F94LaL and CAPN1hCC approached 0.5 during the evaluation phase. The combined frequencies of the 2 selected CAPN1 haplotypes were about 0.4 during the base phase and increased to 0.75 during the evaluation phase (Fig. 1). About 40% of animals still had at least one CAPN1hGC haplotype during the evaluation phase. Because CAPN1hCC was more frequent than CAPN1hGT, analyses of all traits except slice shear force used CAPN1hCC and all other haplotypes (CAPN1hNN) instead of CAPN1hGT specifically. Only 103 animals with unambiguous diplotypes consisting of only CAPN1hGT and (or) CAPN1hCC haplotypes were used to analyze slice shear force. This method of addressing the CAPN1hGC haplotypes was chosen for slice shear force because a direct effect of CAPN1 on this trait was expected. This method should have no bias and a straight forward interpretation of CAPN1hCC and CAPN1hGT associations but does result in fewer animals and less power. The CAPN1hNN method was used for all other traits because CAPN1 effects, if any, are expected to be less direct. The CAPN1hNN method maximized the number of steers that could be used to estimate F94L effects for traits other than slice shear force while still accounting for the possibility of CAPN1 influence.
Figure 1.
Frequencies for MSTN F94L allele L (▲) and µ-calpain 316–4751 haplotypes CC (■) and GT (♦) by birth year. Base, selection, and evaluation phases of the experiment are identified by vertical dashed lines.
Table 2 shows the combinations of CAPN1 diplotypes and F94L genotypes for the 176 harvested steers and the 103 used in slice shear force analyses. Table 3 characterizes the opportunity for increasing power through within sire comparisons across genotypes. Median values for 28 sires used were 4.5 progeny distributed among 3 of the 9 possible genotypes (Table 3). Four sires had progeny in 7, 8, or 9 genotype classes making especially strong contributions to increasing power. For example, the 2 steers in the smallest CAPN1 × MSTN combination for slice shear force had 18 half-sibs among 7 or the other 8 CAPN1 × MSTN combinations.
Table 2.
Number of harvested MARC I steers by genotype
| CAPN1 diplotype1 | F94L genotype2 | Total | ||
|---|---|---|---|---|
| FF | FL | LL | ||
| CAPN1hNN, CAPN1hNN | 26 | 26 | 9 | 61 |
| CAPN1hGT, CAPN1hGT | 9 | 12 | 2 | 23 |
| CAPN1hCC, CAPN1hNN | 24 | 40 | 16 | 80 |
| CAPN1hCC, CAPN1hGT | 17 | 18 | 10 | 45 |
| CAPN1hCC, CAPN1hCC | 8 | 17 | 10 | 35 |
| Total | 58 | 83 | 35 | 176 |
1Diplotypes composed of µ-Calpain (CAPN1) haplotypes. Any haplotype other than CAPN1hCC is represented by CAPN1hNN and these animals were used for analyses of most traits. Only animals with diplotypes consisting of CAPN1hGT and CAPN1hCC were used to analyze slice shear force (n = 103) and are a subset of the 176 steers and indicated by italics.
2 MSTN F94L homozygous F94LaF (FF), heterozygous (FL), and homozygous F94LaL (LL) genotypes.
Table 3.
Characterization of potential for within sire comparisons among genotypes
| Progeny distribution measures for 28 sires | Value |
|---|---|
| Median number of progeny per sire | 4.5 |
| Sires with 3 progeny F94L genotypes1 | 12 |
| Average F94L progeny genotypes per sire | 2.04 |
| Sires with 3 progeny CAPN1 diplotypes2 | 7 |
| Average CAPN1 progeny diplotypes per sire | 2.11 |
| Sires with 7 to 9 CAPN1 × F94L progeny genotypes | 4 |
| Sires with 1 to 3 CAPN1 × F94L progeny genotypes | 16 |
| Median CAPN1 × MSTN progeny genotypes per sire | 3 |
1Myostatin (MSTN) F94L genotypes were homozygous F94LaF, heterozygous F94L, and homozygous F94LaL.
2µ-Calpain (CAPN1) 316–4751 diplotypes were homozygous CAPN1hCC, the heterozygotes, and homozygous CAPN1hNN.
Averages for steer traits, their estimated heritabilities, SD, and phenotypic SD are shown in Tables 4 and 5. The P-values for genotypes (8 df) were not significant for any weights from birth through harvest (Table 5). Genotype effects were significant (P < 0.05) for all carcass and meat traits except hot carcass weight. Estimated genotype and diplotype means for all traits are shown in Table 6.
Table 4.
Averages and SD of unadjusted measurements on 176 harvested steers
| Trait | Average | SD |
|---|---|---|
| Birth weight, kg | 41.0 | 5.5 |
| Weaning weight, kg | 202 | 25 |
| Yearling weight, kg | 409 | 44 |
| Final weight, kg | 604 | 55 |
| Hot carcass weight, kg | 376 | 37 |
| Marbling score1 | 329 | 38 |
| Ribeye area, cm2 | 94.0 | 9.2 |
| Adjusted fat thickness, mm | 9.3 | 3.6 |
| Vision yield grade2 | 2.26 | 0.73 |
| Slice shear force3, kg | 14.8 | 4.0 |
| CIE L*4 | 35.6 | 2.0 |
1300 = Slight00; 400 = Small00 (USDA, 1997).
2Prediction of USDA Yield Grade. Smaller numbers indicate greater yield of boneless, closely trimmed retail cuts.
3Values for 103 steers having only diplotypes consisting of CAPN1hCC and (or) CAPN1hGT and analyzed for slice shear force.
4CIE L* measure of lightness. Greater values indicate lighter lean color.
Table 5.
Heritability and phenotypic SD estimates and P-values for sources of variation
| Trait | Year | Dam age | Calf age1 | Genotype | h2 | σP |
|---|---|---|---|---|---|---|
| Birth weight, kg | 0.10 | 0.001 | 0.09 | 0.38 | 0.68 | 5.3 |
| Weaning weight, kg | 0.91 | <0.001 | <0.001 | 0.73 | 0.30 | 19.0 |
| Yearling weight, kg | <0.001 | <0.001 | <0.001 | 0.99 | 0.31 | 24.4 |
| Final weight, kg | 0.005 | 0.001 | <0.001 | 0.85 | 0.50 | 48.5 |
| Hot carcass weight, kg | <0.001 | 0.001 | <0.001 | 0.92 | 0.42 | 31.4 |
| Marbling score2,3 | 0.13 | 0.60 | 0.53 | <0.001 | 0.47 | 34 |
| Ribeye area, cm2 | 0.004 | 0.04 | 0.16 | <0.001 | 0.39 | 7.1 |
| Adjusted fat thickness, mm | 0.15 | 0.61 | 0.23 | <0.001 | 0.51 | 3.2 |
| Vision yield grade4 | 0.22 | 0.35 | 0.10 | <0.001 | 0.52 | 0.62 |
| Slice shear force3, kg | 0.08 | 0.94 | 0.34 | 0.03 | 0.205 | 4.0 |
| CIE L* reflectance6 | 0.03 | 0.86 | 0.05 | 0.01 | 0.44 | 1.9 |
1Julian birthday linear covariate was used for all traits and is a proxy for age for all traits (except birth weight) because a single harvest date was used each year.
2300 = Slight00; 400 = Small00 (USDA, 1997).
3Logarithm (base 10) values of traits were analyzed.
4Prediction of USDA Yield Grade. Smaller numbers indicate greater yield of boneless, closely trimmed retail cuts.
5Constrained to 0.20 ≤ h2 ≤ 0.70.
6CIE L* measure of lightness. Greater values mean lighter lean color.
Table 6.
Means of traits by myostatin F94L genotypes and µ-calpain (CAPN1) diplotypes
| Trait | F94L1 | CAPN1 2 | SED3 | |||||
|---|---|---|---|---|---|---|---|---|
| FF | FL | LL | NN-NN | NN-CC | CC-CC | SEDhom | SEDhet | |
| Birth weight, kg | 40.7 | 41.4 | 43.3 | 41.7 | 41.3 | 42.4 | 1.2 | 1.0 |
| Weaning weight, kg | 202 | 199 | 206 | 204 | 202 | 201 | 4.6 | 3.9 |
| Yearling weight, kg | 409 | 410 | 412 | 408 | 409 | 413 | 8.5 | 7.1 |
| Final weight, kg | 608 | 603 | 598 | 599 | 604 | 607 | 11.6 | 9.7 |
| Hot carcass weight, kg | 375 | 375 | 378 | 374 | 377 | 377 | 7.6 | 6.3 |
| Marbling score4 | 345 | 329 | 294 | 325 | 327 | 316 | 8.2 | 6.8 |
| Ribeye area, cm2 | 89 | 93 | 103 | 95 | 95 | 95 | 1.7 | 1.4 |
| Adjusted fat thickness, mm | 10.5 | 9.1 | 6.4 | 8.9 | 8.9 | 8.3 | 0.8 | 0.6 |
| Vision Yield Grade5 | 2.59 | 2.26 | 1.57 | 2.18 | 2.18 | 2.05 | 0.15 | 0.12 |
| Slice shear force, kg | 16.2 | 14.5 | 12.8 | 15.1 | 13.7 | 14.8 | 0.96 | 0.81 |
| CIE L* reflectance6 | 34.65 | 35.58 | 36.20 | 35.40 | 35.58 | 35.44 | 0.45 | 0.38 |
1 MSTN F94L homozygous F94LaF (FF), heterozygous (FL), and homozygous F94LaL (LL) genotypes.
2 CAPN1 diplotypes are designated NN-NN (homozygous CAPN1hNN), CC-NN (heterozygotes) and CC-CC (homozygous CAPN1hCC).
3Approximate within gene SED for the difference between homozygotes (SEDhom) and between the heterozygote and either of the homozygotes (SEDhet).
4300 = Slight00; 400 = Small00 (USDA, 1997).
5USDA vision yield grade. Smaller numbers indicate greater yield of boneless, closely trimmed retail cuts.
6CIE L* measure of lightness. Lighter lean color results in greater values.
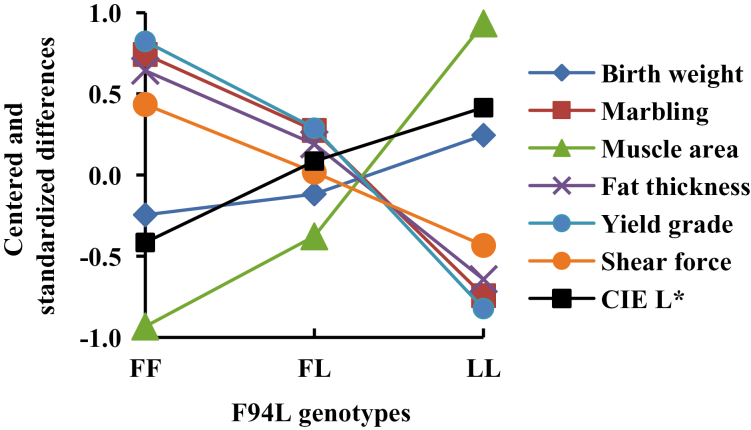
Linear contrasts for genetic effects were estimated for significant carcass and meat traits (Table 7). Additive effects of MSTN F94L were significant (P < 0.01) for all these traits. Reduced fat thickness, larger ribeye area, and better yield grade were associated with F94LaL. It was also associated with lower marbling scores, more tender meat, and lighter meat color. The F94LaL was partially recessive to F94LaF for ribeye area resulting in the heterozygote being closer to the F94LaF homozygote (Tables 6 and 7). The estimated differences of homozygous F94LaF, heterozygous, and homozygous F94LaL on traits with significant additive and dominance effects are shown in Fig. 2. Differences are standardized by subtracting the average of the homozygotes and dividing by phenotypic SD. Although genotype effect (8 df) was not significant in this study (P = 0.38; Table 5), birth weight is also shown because it could affect use of F94LaL in mating systems, approached significance (P = 0.06) in heifers that were sibs to these steers (Cushman et al., 2015), and has shown significant increases in other homozygous MSTN mutations (e.g., Casas et al., 2004).
Table 7.
Estimated marker associated additive and nonadditive effects for traits with overall P < 0.10 for genotype
| Marker effect1 | Marbling score2 | Ribeye area, cm2 | Adjusted fat thickness, mm | Vision Yield Grade | Slice shear force2, kg | L* reflectance |
|---|---|---|---|---|---|---|
| Value ± SE | Value ± SE | Value ± SE | Value ± SE | Value ± SE | Value ± SE | |
| F94L A | −25.4*** ± 3.9 | 6.6*** ± 0.8 | −2.06*** ± 0.36 | −0.57*** ± 0.07 | −1.7** ± 0.7 | 0.78*** ± 0.21 |
| F94L D | 9.4 ± 5.6 | −2.7* ± 1.2 | 0.61 ± 0.52 | 0.18 ± 0.10 | 0.0 ± 0.9 | 0.16 ± 0.30 |
| CAPN1 A | −4.7 ± 4.3 | 0.3 ± 0.9 | −0.32 ± 0.40 | 0.07 ± 0.08 | −0.1 ± 0.7 | 0.02 ± 0.24 |
| CAPN1 D | 5.8 ± 5.5 | 0.0 ± 1.1 | 0.30 ± 0.51 | 0.07 ± 0.10 | −1.3 ± 0.9 | 0.16 ± 0.30 |
| A × A | 13.4 ± 20.4 | −2.3 ± 4.3 | 0.61 ± 1.90 | 0.06 ± 0.37 | 7.3* ± 3.7 | 0.49 ± 1.12 |
| A × D | 9.2 ± 15.0 | 5.2 ± 3.1 | −1.03 ± 1.40 | −0.23 ± 0.27 | −0.8 ± 2.5 | 0.80 ± 0.82 |
| D × A | 13.2 ± 14.4 | −2.6 ± 3.0 | −0.01 ± 1.37 | 0.21 ± 0.26 | −0.3 ± 2.4 | 0.96 ± 0.79 |
| D × D | 4.1 ± 10.3 | 2.3 ± 2.2 | 1.06 ± 1.02 | 0.05 ± 0.19 | −0.1 ± 1.7 | −0.51 ± 0.56 |
1Epistatic effects are listed as myostatin F94L effect × µ-Calpain (CAPN1) haplotype effect. Linear contrast coefficient used to estimate additive (A), dominance (D), and epistatic (A × A, D × A, A × D, D × D) effects are shown in Table 1.
2Means and SE estimates from actual values. Significance determined from analyses of logarithms of data.
* P < 0.05; **P < 0.01; ***P < 0.001.
Figure 2.
Means for MSTN F94L genotypes divided by their phenotypic SD and deviated from the average of F94LaF (FF) and F94LaL (LL) homozygotes. All differences between divergent homozygotes are significant except birth weight. Heterozygotes were different from the average of homozygotes for ribeye muscle area (P < 0.05). Birth weight is included for comparison with previous literature reports of significant differences.
Multiple variants in MSTN that decrease functional activity and cause muscular hypertrophy in cattle have been found (Dunner et al., 2003). An 11 base pair deletion in the Belgian Blue breed causes a frame shift in the translation frame that prevents translation of the active signaling domain of the protein. In Piedmontese, there is a single base change that eliminates a proteolytic self-cleavage site immediately proximal to the signaling domain that prevents process to the active form. These 2 highly disruptive variants contrast with the relatively minor substitution of an aliphatic side chain (leucine) for an aromatic (phenylalanine) in the propeptide domain resulting from the F94L variant, because both are nonpolar residues. However, it is possible this substitution affects protein folding, stability, or trafficking of the myostatin protein with the primary evidence for this being the effect on muscle growth in cattle (Alexander et al., 2007). Differences between F94L homozygote means in this study ranged from 1.3 to 1.9 phenotypic SD for fat and muscle traits. Most carcass traits show a nonsignificant tendency for the heterozygote to be partially recessive to the F94LaF allele (the mean being closer to the homozygous F94LaF mean than the homozygous F94LaL mean), although the effect did reach significance for ribeye area. Esmailizadeh et al. (2008) reported on Limousin-Jersey back-cross families in Australia and New Zealand. They found no significant effects of the F94LaL SNP on birth and live weights. Additive and dominance effects on Longissimus muscle area were significant but not as large as the additive effects in this study. Several measures of fatness were also decreased, and meat weights increased. Alexander et al. (2009) found increased muscle area and reduced marbling in a Wagyu-Limousin F2 family.
Differences in birth weight, fat, and muscle traits between homozygotes for some of the severe MSTN variants estimated in 2 other experiments exceed the values for F94L estimated in this experiment. Casas et al. (2004) compared homozygous active and inactive F2 progeny from Belgian Blue F1 × F1 matings in the same location and under similar management as this experiment. Short et al. (2002) compared F2 Piedmontese under similar management but in a different location. Both experiments found significant differences between divergent homozygotes for birth weight, ribeye area, marbling score, fat thickness, and yield grade. The average percentage differences from the active MSTN homozygotes were 15%, 32%, −32%, −62%, and −78%, respectively, compared with 7%, 15%, −14%, −39%, and −38% for F94L in this experiment. The effects of the F94L mutation were about half the percentage differences of the Belgian Blue and Piedmontese mutations.
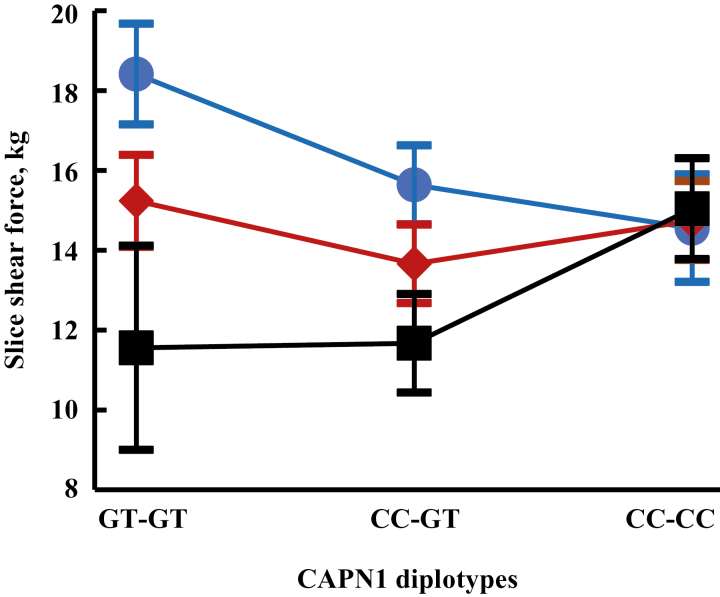
The CAPN1 haplotypes had no significant additive or dominance associations with any measured trait including meat tenderness. A significant CAPN1 additive by F94L additive epistatic effect is illustrated in Fig. 3. One way of characterizing this epistasis is that CAPN1hCC reduced slice shear force (increased tenderness) in animals homozygous for the common F94LaF allele, had no effect in F94L heterozygotes, and decreased tenderness in F94LaL homozygotes. Increased tenderness is often observed in meat from animals with heterozygous and homozygous MSTN mutations resulting in nonfunctional myostatin (e.g., Wheeler et al., 2001). Because few animals are homozygous CAPN1hCC in most common populations surveyed (White et al., 2005), F94LaL would be associated with increased tenderness in most populations using the epistatic estimates. Frequency of CAPN1hCC was increased in 2 similar experiments. In an Angus population (Tait et al., 2014a), the estimated additive effect of CAPN1aCC (compared to CAPN1aGT) on slice shear force was −1.05 ± 0.25 kg and in the composite MARC III population (Tait et al., 2014b) was −1.15 ± 0.48 kg. Using only estimated means for homozygous F94LaF, the equivalent additive estimate was −1.93 ± 0.97 kg in the current study. The usual relationship between CAPN1 haplotypes and meat tenderness appears to be disrupted by the F94LaL allele. Previous QTL discovery in progeny of an F1 Piedmontese sire and an F1 Belgian Blue sire heterozygous for active and inactive myostatin found interactions of the MSTN variants with other QTL located on BTA 4 (Casas et al., 2001) and BTA 5 (Casas et al., 2000) for meat tenderness.
Figure 3.
MSTN F94L × µ-calpain genotypic means for slice shear force. F94L homozygous F94LaF, heterozygous, and homozygous F94LaL genotypes are designated by FF, FL, and LL, respectively. CAPN1 diplotypes are designated GT-GT (homozygous CAPN1hGT), CC-GT (heterozygotes) and CC-CC (homozygous CAPN1hCC). The additive effect is significant for F94L (P < 0.01) and the additive F94L × additive µ-calpain effect is significant (P < 0.05). Variation is shown as ±1 SEM.
Research using heifer half-sibs from this population showed delayed age of puberty due to F94LaL alleles (Cushman et al., 2015). However, conception was not reduced or delayed in this population and management system. This moderate form of MSTN mutation with high frequency in the Limousin breed shows the potential for variation among the effects of mutations in some genes. In this case, the resulting increase in lean meat yield, moderate birth weight increase, and limited effect on heifer conception is a useful resource for improving efficiency of lean beef production in conventional production systems. Because F94L effects are mostly additive, 0, 1, or 2 copies of F94LaL can be used to create 3 product types with increasing lean meat yield and decreasing marbling. Terminal cross bulls with 1 or 2 copies of F94LaL mated to cows with 0 copies would produce 0 and 1 copy progeny (1 copy bulls) or 1 copy progeny (2 copy bulls). Based on the epistatic estimates for slice shear force, CAPN1 selection would be less effective for progeny with 1 copy but effective for progeny with 0 copies.
Footnotes
Mention of trade name, proprietary product, or specified equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable.
The USDA is an equal opportunity provider and employer.
The authors acknowledge the important contributions of technicians and cattle operations staff to the conduct and completion of this study.
LITERATURE CITED
- Alexander L. J., Geary T. W., Snelling W. M., and Macneil M. D.. 2007. Quantitative trait loci with additive effects on growth and carcass traits in a wagyu-limousin F2 population. Anim. Genet. 38:413–416. doi: 10.1111/j.1365-2052.2007.01616.x [DOI] [PubMed] [Google Scholar]
- Alexander L. J., Kuehn L. A., Smith T. P., Matukumalli L. K., Mote B., Koltes J. E., Reecy J., Geary T. W., Rule D. C., and Macneil M. D.. 2009. A limousin specific myostatin allele affects longissimus muscle area and fatty acid profiles in a wagyu-limousin F2 population. J. Anim. Sci. 87:1576–1581. doi: 10.2527/jas.2008-1531 [DOI] [PubMed] [Google Scholar]
- Arthur P. F. 1995. Double muscling in cattle: a review. Aust. J. Agric. Res. 46:1493–1515. doi: 10.1071/AR9951493 [DOI] [Google Scholar]
- Bennett G. L. 2008. Experimental selection for calving ease and postnatal growth in seven cattle populations. I. Changes in estimated breeding values. J. Anim. Sci. 86:2093–2102. doi: 10.2527/jas.2007-0767 [DOI] [PubMed] [Google Scholar]
- Bennett G. L., and Gregory K. E.. 1996. Genetic (co)variances among birth weight, 200-day weight, and postweaning gain in composites and parental breeds of beef cattle. J. Anim. Sci. 74:2598–2611. doi: 10.2527/1996.74112598x [DOI] [PubMed] [Google Scholar]
- Boldman K. G., Kriese L. A., Van Vleck L. D., Van Tassell C. P., and Kachman S. D.. 1995. A manual for use of MTDFREML. A set of programs to obtain estimates of variance and covariances. USDA, Agricultural Research Service, Washington, DC. [Google Scholar]
- Casas E., Bennett G. L., Smith T. P., and Cundiff L. V.. 2004. Association of myostatin on early calf mortality, growth, and carcass composition traits in crossbred cattle. J. Anim. Sci. 82:2913–2918. doi: 10.2527/2004.82102913x [DOI] [PubMed] [Google Scholar]
- Casas E., Shackelford S. D., Keele J. W., Stone R. T., Kappes S. M., and Koohmaraie M.. 2000. Quantitative trait loci affecting growth and carcass composition of cattle segregating alternate forms of myostatin. J. Anim. Sci. 78:560–569. doi: 10.2527/2000.783560x [DOI] [PubMed] [Google Scholar]
- Casas E., Stone R. T., Keele J. W., Shackelford S. D., Kappes S. M., and Koohmaraie M.. 2001. A comprehensive search for quantitative trait loci affecting growth and carcass composition of cattle segregating alternative forms of the myostatin gene. J. Anim. Sci. 79:854–860. doi: 10.2527/2001.794854x [DOI] [PubMed] [Google Scholar]
- Casas E., White S. N., Wheeler T. L., Shackelford S. D., Koohmaraie M., Riley D. G., Chase C. C. Jr, Johnson D. D., and Smith T. P.. 2006. Effects of calpastatin and micro-calpain markers in beef cattle on tenderness traits. J. Anim. Sci. 84:520–525. doi: 10.2527/2006.843520x [DOI] [PubMed] [Google Scholar]
- Cushman R. A., Tait R. G. Jr, McNeel A. K., Forbes E. D., Amundson O. L., Lents C. A., Lindholm-Perry A. K., Perry G. A., Wood J. R., Cupp A. S.,. et al. 2015. A polymorphism in myostatin influences puberty but not fertility in beef heifers, whereas µ-calpain affects first calf birth weight. J. Anim. Sci. 93:117–126. doi: 10.2527/jas.2014-8505 [DOI] [PubMed] [Google Scholar]
- Dunner S., Miranda M. E., Amigues Y., Cañón J., Georges M., Hanset R., Williams J., and Ménissier F.. 2003. Haplotype diversity of the myostatin gene among beef cattle breeds. Genet. Sel. Evol. 35:103–118. doi: 10.1051/gse:2002038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmailizadeh A. K., Bottema C. D., Sellick G. S., Verbyla A. P., Morris C. A., Cullen N. G., and Pitchford W. S.. 2008. Effects of the myostatin F94L substitution on beef traits. J. Anim. Sci. 86:1038–1046. doi: 10.2527/jas.2007-0589 [DOI] [PubMed] [Google Scholar]
- FASS 1999. Guide for the care and use of agricultural animals in agricultural research and teaching. 1st rev. ed. Fed. Anim. Sci. Soc, Savoy, IL. [Google Scholar]
- Gregory K. E., Cundiff L. V., and Koch R. M.. 1991. Breed effects and heterosis in advanced generations of composite populations for preweaning traits of beef cattle. J. Anim. Sci. 69:947–960. doi: 10.2527/1991.693947x [DOI] [PubMed] [Google Scholar]
- Gregory K. E., Cundiff L. V., Koch R. M., Dikeman M. E., and Koohmaraie M.. 1994. Breed effects, retained heterosis, and estimates of genetic and phenotypic parameters for carcass and meat traits of beef cattle. J. Anim. Sci. 72:1174–1183. doi: 10.2527/1994.7251174x [DOI] [PubMed] [Google Scholar]
- Grobet L., Martin L. J., Poncelet D., Pirottin D., Brouwers B., Riquet J., Schoeberlein A., Dunner S., Ménissier F., Massabanda J.,. et al. 1997. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 17:71–74. doi: 10.1038/ng0997-71 [DOI] [PubMed] [Google Scholar]
- Grobet L., Poncelet D., Royo L. J., Brouwers B., Pirottin D., Michaux C., Ménissier F., Zanotti M., Dunner S., and Georges M.. 1998. Molecular definition of an allelic series of mutations disrupting the myostatin function and causing double-muscling in cattle. Mamm. Genome 9:210–213. doi: 10.1007/s003359900727 [DOI] [PubMed] [Google Scholar]
- Kambadur R., Sharma M., Smith T. P., and Bass J. J.. 1997. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 7:910–916. doi: 10.1101/gr.7.9.910 [DOI] [PubMed] [Google Scholar]
- Page B. T., Casas E., Heaton M. P., Cullen N. G., Hyndman D. L., Morris C. A., Crawford A. M., Wheeler T. L., Koohmaraie M., Keele J. W.,. et al. 2002. Evaluation of single-nucleotide polymorphisms in CAPN1 for association with meat tenderness in cattle. J. Anim. Sci. 80:3077–3085. doi: 10.2527/2002.80123077x [DOI] [PubMed] [Google Scholar]
- Page B. T., Casas E., Quaas R. L., Thallman R. M., Wheeler T. L., Shackelford S. D., Koohmaraie M., White S. N., Bennett G. L., Keele J. W.,. et al. 2004. Association of markers in the bovine CAPN1 gene with meat tenderness in large crossbred populations that sample influential industry sires. J. Anim. Sci. 82:3474–3481. doi: 10.2527/2004.82123474x [DOI] [PubMed] [Google Scholar]
- Robinson D. L., Cafe L. M., McIntyre B. L., Geesink G. H., Barendse W., Pethick D. W., Thompson J. M., Polkinghorne R., and Greenwood P. L.. 2012. Production and processing studies on calpain-system gene markers for beef tenderness: consumer assessments of eating quality. J. Anim. Sci. 90:2850–2860. doi: 10.2527/jas.2011-4928 [DOI] [PubMed] [Google Scholar]
- Shackelford S. D., Wheeler T. L., and Koohmaraie M.. 1999. Evaluation of slice shear force as an objective method of assessing beef longissimus tenderness. J. Anim. Sci. 77:2693–2699. doi: 10.2527/1999.77102693x [DOI] [PubMed] [Google Scholar]
- Shackelford S. D., Wheeler T. L., and Koohmaraie M.. 2003. On-line prediction of yield grade, longissimus muscle area, preliminary yield grade, adjusted preliminary yield grade, and marbling score using the MARC beef carcass image analysis system. J. Anim. Sci. 81:150–155. doi: 10.2527/2003.811150x [DOI] [PubMed] [Google Scholar]
- Short R. E., MacNeil M. D., Grosz M. D., Gerrard D. E., and Grings E. E.. 2002. Pleiotropic effects in hereford, limousin, and piedmontese F2 crossbred calves of genes controlling muscularity including the piedmontese myostatin allele. J. Anim. Sci. 80:1–11. doi:10.2527/2002.8011 [DOI] [PubMed] [Google Scholar]
- Stone R. T., Grosse W. M., Casas E., Smith T. P., Keele J. W., and Bennett G. L.. 2002. Use of bovine EST data and human genomic sequences to map 100 gene-specific bovine markers. Mamm. Genome 13:211–215. doi: 10.1007/s00335-001-2124-9 [DOI] [PubMed] [Google Scholar]
- Tait R. G. Jr, Cushman R. A., McNeel A. K., Casas E., Smith T. P., Freetly H. C., and Bennett G. L.. 2016. Estimates of epistatic and pleiotropic effects of casein alpha s1 (CSN1S1) and thyroglobulin (TG) genetic markers on beef heifer performance traits enhanced by selection. J. Anim. Sci. 94:920–926. doi: 10.2527/jas.2015-9860 [DOI] [PubMed] [Google Scholar]
- Tait R. G. Jr, Shackelford S. D., Wheeler T. L., King D. A., Casas E., Thallman R. M., Smith T. P. L., and Bennett G. L.. 2014a. µ-Calpain, calpastatin, and growth hormone receptor genetic effects on preweaning performance, carcass quality traits, and residual variance of tenderness in Angus cattle selected to increase minor haplotype and allele frequencies. J. Anim. Sci. 92:456–466. doi: 10.2527/jas.2013-7075 [DOI] [PubMed] [Google Scholar]
- Tait R. G. Jr, Shackelford S. D., Wheeler T. L., King D. A., Keele J. W., Casas E., Smith T. P., and Bennett G. L.. 2014b. CAPN1, CAST, and DGAT1 genetic effects on preweaning performance, carcass quality traits, and residual variance of tenderness in a beef cattle population selected for haplotype and allele equalization. J. Anim. Sci. 92:5382–5393. doi: 10.2527/jas.2014-8211 [DOI] [PubMed] [Google Scholar]
- USDA 1997. Official United States standards for grades of carcass beef. Agric. Market. Serv., USDA, Washington, DC. [Google Scholar]
- Wheeler T. L., Shackelford S. D., Casas E., Cundiff L. V., and Koohmaraie M.. 2001. The effects of piedmontese inheritance and myostatin genotype on the palatability of longissimus thoracis, gluteus medius, semimembranosus, and biceps femoris. J. Anim. Sci. 79:3069–3074. doi: 10.2527/2001.79123069x [DOI] [PubMed] [Google Scholar]
- White S. N., Casas E., Wheeler T. L., Shackelford S. D., Koohmaraie M., Riley D. G., Chase C. C. Jr, Johnson D. D., Keele J. W., and Smith T. P.. 2005. A new single nucleotide polymorphism in CAPN1 extends the current tenderness marker test to include cattle of Bos indicus, Bos taurus, and crossbred descent. J. Anim. Sci. 83:2001–2008. doi: 10.2527/2005.8392001x [DOI] [PubMed] [Google Scholar]