Abstract
Osteosarcoma (OS) is the most common form of primary bone cancer in humans. The early detection and subsequent control of metastasis has been challenging in OS. Lipids are important constituents of cells that maintain structural integrity that can be converted into lipid-signaling molecules and are reprogrammed in cancerous states. Here, we investigate the global lipidomic differences in metastatic (143B) and nonmetastatic (HOS) human OS cells as compared with normal fetal osteoblast cells (FOB) using lipidomics. We detect 15 distinct lipid classes in all three cell lines that included over 1,000 lipid species across various classes including phospholipids, sphingolipids and ceramides, glycolipids, and cholesterol. We identify a key class of lipids, diacylglycerols, which are overexpressed in metastatic OS cells as compared with their nonmetastatic or nontumorigenic counterparts. As a proof of concept, we show that blocking diacylglycerol synthesis reduces cellular viability and reduces cell migration in metastatic OS cells. Thus, the differentially regulated lipids identified in this study might aid in biomarker discovery, and the synthesis and metabolism of specific lipids could serve as future targets for therapeutic development.
Keywords: lipidomics, mass spectrometry, diacylglycerol, cholesterol
Osteosarcoma (OS) is the most common form of primary bone cancer in humans. The treatment options for OS consists of multiagent induction chemotherapy, tumor excision, and adjuvant chemotherapy. The survival rates remain poor, despite aggressive treatment. In patients with localized disease, 5-year survival rates are approximately 65%; however, in the case of metastatic disease, the 5-year survival rates have plateaued to about 20% (1, 2). Although progress has been made toward improving treatment options in OS, the early detection and subsequent control of metastasis has been challenging. Lipids are important constituents of cells that act as signaling molecules, maintain structural integrity as well as storage of energy, and are often reprogrammed in cancerous states (3). The discovery of metabolically altered pathways, especially those related to lipid synthesis, can aid in understanding the mechanism of tumor formation and metastasis and might support in the development of novel OS therapeutics.
Lipids are an important component of the cellular machinery. They encompass a large class of biomolecules, including diversity in backbone and chain length, as well as number and position of unsaturated bonds (4, 5). Although the functional consequences of lipid diversity are not fully understood, it is well characterized that lipids help regulate a variety of cellular functions, including protein signaling in the membrane as well as homeostasis (6, 7). Thus, dysregulation in lipid metabolism is linked to a variety of pathological conditions, including immune disorders and cancer (8).
Cancer cells are metabolically reprogrammed to sustain their uncontrolled growth and proliferation. An increased production of lipids, nucleic acids, and proteins is required to drive downstream oncogenic processes (3). Although significant research has delved into the transcriptomics and proteomics of various cancers, the lipidomic profiles of most cancers remain poorly understood. Lipids form a major component of the plasma membrane and various cellular membranes, as well as in vesicles for intercellular and intracellular transport. Altered lipid composition leads to changes in membrane fluidity and cellular polarization (9). Thus, the lipidomic profiles of cancer cells can lead to the development of biomarkers, identify potential therapeutic targets, and help understand the underlying mechanisms of various malignant pathologies.
Various analytical approaches have been taken toward understanding lipid profiles of cancer cells. The expression level and activity profiling of various lipid-synthesis enzymes have been performed to identify increased fatty acid synthase activity and ATP citrate lyase activity in tumors (10, 11). Microarray profiling of metabolic genes has revealed altered cholesterol metabolism and LDL profiles (12). Raman spectroscopy has also been used for understanding cellular lipid profiles at the single-cell level (13), and 31P NMR spectroscopy has been used toward lipidomic profiling (14).
The complexity in study of lipids arises from the similarity in structures of lipids accompanied with overlapping mass. Lipids are categorized in various ways, including classifications based on their chain length, backbone, or saturation levels. Based on their backbone, lipids are classified mainly into phospholipids, glycolipids, sphingolipids, and sterols (Fig. 1). The saponifiable part of phospholipids, glycolipids, and sphingolipids are free fatty acids. Thus, mixtures of lipids are highly diverse and numerous, based on combinations of fatty acids and the backbone. The development of mass-spectrometric techniques has enabled the separation and identification of complex lipid mixtures. Soft ionization techniques including ESI and MALDI allow lipids to remain intact for mass-spectrometric analyses. These techniques, in combination with highly specialized software programs, are also quantitative through the use of internal standards, allowing a thorough and high-throughput analysis of lipid mixtures.
Fig. 1.
The various classes of lipids. The major classes of biological lipids are phospholipids, sterols, acylglycerides, and sphingolipids. Phospholipids are further classified into PCs, PEs, PSs, PGs, CLs, and phosphatidylinositols (PIs). The major human sterol is cholesterol, which is often stored as ChE. Acylglycerols can be MGs, DGs, and TGs based on the number of acyl groups attached. Sphingolipids are mainly classified into SMs and Cers.
The use of mass spectrometry to study cancer lipidomics has precedence in breast cancer, prostate cancer, and adenocarcinoma, among others (15–17). Phospholipids are the largest class of structural lipids and have been widely explored in various cancers. It has been observed that lysophosphatidylcholine (LPC) levels are increased in aggressive breast cancer cell lines (18). Individual lipid species, including palmitate-containing phosphatidylcholines (PCs), are overexpressed in malignant cancers (19). Furthermore, data obtained from lipidomics have been compared with transcriptomic data to discover specific lipid markers like cholesteryl oleate (CE) for prostate cancer and correspondingly overexpressed scavenger receptor class B type I resulting in CE accumulation (20).
Although there are several examples of lipidomics in cancer types of epithelial origin, the lipidomic profiles of cancers of mesenchymal origin such as OS have remained unexplored. In this work, the global lipidomic differences in metastatic and nonmetastatic OS cells as compared with normal fetal osteoblast cells was investigated in a high-throughput manner. Over 1,500 lipid species across various classes, including phospholipids, sphingolipids, ceramides (Cers), glycolipids, and cholesterol, were identified. Differences in lipid profiles between the classes, as well as differences between specific lipids, were observed in all the classes of lipid that were examined. Furthermore, the lipidomic signature of each class was identified using principal component analysis (PCA). Finally, a key class of lipids, diacylglycerols (DGs), was identified as being preferentially upregulated in the tumorigenic cell lines as compared with the noncancerous control cell line. The pathway of DG synthesis was blocked, and it induced apoptosis, as well as reduced migratory potential of metastatic 143B cells. Taken together, this work compares the global lipidomic profile of osteoblasts and nonmetastatic and metastatic OS cell lines. Furthermore, several differentially regulated pathways, including DGs, were identified, which might serve as biomarkers and targets for improved disease management and treatment.
MATERIALS AND METHODS
Materials
An Amplex Red kit was obtained from Thermo Scientific (catalog no. A12216). Phospholipase C (PLC) inhibitor U73122 was obtained from Enzo (catalog no. BML-ST391-0005).
Cell culture
Human cell lines FOB, HOS and 143B were cultured in DMEM (Gibco, Invitrogen, Carlsbad, CA) supplemented with 10% FBS and 1 mg·ml–1 penicillin–streptomycin (Gibco, Invitrogen) at 37°C and 5% CO2 in a humidified incubator. FOB cells were kindly provided by Dr. Avudaiappan Maran at the Mayo Clinic, Rochester, MN. All cell lines were verified via short tandem repeat analysis. The cell lines FOB, HOS, and 143B were verified at The University of Arizona Genetics Core.
Extraction of lipids
Cells were grown to 80–90% confluency. Cells were detached using scraping, and cell count was performed. The cell suspension was immediately diluted with PBS (1:1) and centrifuged for 5 min at 400 g. The cell pellet was washed three times with 1 ml of PBS, followed by centrifugation at 400 g for 5 min, and cells were resuspended in 1 ml of PBS. The cells were lysed using six freeze-thaw cycles rapidly. Total lipids from all cell lines were extracted with the Bligh and Dyer method (21). Briefly, 3.75 ml of chloroform/methanol 1:2 (v/v) was added to 1 ml of cell sample, vortexed well for 15 min, and incubated on ice for 5 min. An additional volume of 1.25 ml of chloroform and 1.25 ml of dH2O was added. Finally, following vigorous vortex for 5 min, samples were centrifuged at 150 g for 5 min at room temperature to obtain a two-phase system: aqueous top phase and organic bottom phase, from which lipids were obtained. This was dried down and submitted for MS.
Lipidomics analysis
Sample preparation.
A total of 200 µl of methanol, 100 µl of chloroform, and 30 µl of water were added into the dried sample. After centrifugation, 150 µl of aliquot from the aforementioned solution was spiked with 5 µl of 50 µg/ml internal mixture (Cer 18:1/12:0; PC 12:0/12:0; PE 14:0/14:0; phosphatidylglycerol (PG) 14:0/14:0;PS 14:0/14:0) before instrument injection.
Instrument analysis.
The samples were analyzed by using the Thermo Q-Exactive MS system (Bremen, Germany) in the Metabolomics Laboratory of the Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign. Software (Xcalibur Version 3.0.63) was used for data acquisition and analysis. The Dionex Ultimate 3000 series HPLC system (Thermo, Germering, Germany) used included a degasser, an autosampler, and a binary pump. The LC separation was performed on a Thermo Accucore C18 column (2.1 × 150 mm, 2.6 μm) with mobile phase A (60% acetonitrile: 40% H2O with 10 mM ammonium formate and 0.1% formic acid) and mobile phase B (90% isopropanol: 10% acetonitrile with 10 mM ammonium formate and 0.1% formic acid). The flow rate was 0.4 ml/min. The linear gradient was as follows: 0 min, 70% A; 4 min, 55% A; 12 min, 35% A; 18 min, 15% A; 20–25 min, 0% A; 26–33 min, 70% A. The autosampler was set to 15°C, and the column was kept at 45°C. The injection volume was 10 μl. Mass spectra were acquired under both positive (sheath gas flow rate, 50; aux gas flow rate: 13; sweep gas flow rate, 3; spray voltage, 3.5 kV; capillary temp, 263°C; Aux gas heater temp, 425°C) and negative electrospray ionization (sheath gas flow rate, 50; aux gas flow rate: 13; sweep gas flow rate, 3; spray voltage, −2.5 kV; capillary temp, 263°C; Aux gas heater temp, 425°C). The full-scan mass-spectrum resolution was set to 70,000 resolution at m/z 200 with the scan range of m/z 230~1,600, and the automatic gain control target was 1E6, with a maximum injection time of 200 ms. For MS/MS scan, the mass-spectrum resolution was set to 17,500. AGC target was 5E4 with a maximum injection time of 50 ms. Loop count was 10. Isolation window was m/z 1.0 with NCE of 25 and 30 eV.
Data analysis.
Thermo software LipidSearch (version 4.1.30) was used for lipid identification (22, 23). The lipid signal responses were normalized to total protein in the sample, and the corresponding internal standard signal response (for those lipid classes without corresponding internal standard, positive lipid ion signals were normalized with the signal of internal standard Cer 18:1/12:0 and negative-ion signals were normalized with the signal of internal standard PG 14:0/14:0). Only monoisotopic species were taken into account. Software LipidSearch uses the accurate m/z of the precursor ion and then predicts the possible fragment ions for species within the precursor ion tolerance. The LipidSearch database (not an MS/MS library) uses the known fragment ions for lipid classes and the intensity pattern based on measured spectra. Identification by the parent ion was based on the accurate mass of precursor ion with the mass shift tolerance of 5 ppm. Identification of the product ion was based on the accurate mass of precursor ion with the mass shift tolerance of 8 ppm and MS/MS spectral pattern. Additionally, isomers often coelute. Coeluting isomers are shown in the original search results as those with a different match score. The higher match score represents the mixture in the Alignment results (thus, only the higher-match-score lipid species will be presented). Further details on identification by LipidSearch software are indicated in the supplemental data.
Statistical analysis.
The high-dimensional lipidomic data of three cell lines, each in three replicates, were transformed to a lower-dimensional eigenspace using PCA in MATLAB. The lower-dimensional space is formed by three principal components that capture more than 85% of the variance in the data.
In order to identify the details of separation in lipid profiles of three cell lines, a statistical test was used to compare each lipid’s concentration in a cell line versus each of the other two, resulting in a total of three comparisons. The nonparametric Wilcoxon rank-sum test (21) implemented in MATLAB was used to calculate the p-values, which were corrected by Benjamini-Hochberg false discovery rate (FDR) control (2). Moreover, fold change was jointly used with p-values to compare lipid levels in different cell lines.
The above procedure of statistical testing used to identify regulated lipids was also executed to identify regulated lipid classes, in a global analysis of 17 lipid classes. The global lipid profile is represented by lipid levels for all the 16 lipid classes in three cell lines with three replicates for each cell line. The global lipid-level value assigned to a lipid class in a cell-line replicate is obtained by adding up the levels of all lipids belonging to that class as measured in that replicate. Finally, the global lipid profile is input to the statistical testing procedure above to identify significantly regulated lipid classes. In all lipidomic analyses, the significance is assigned if levels increase or decrease by 2-fold or greater—that is, fold change > 2 or fold change < 0.5. Moreover, as a second measure in global and individual lipid analyses, significance is assigned if FDR < 0.15.
A similar analysis was performed to characterize different lipid levels in treated versus untreated 143B cell lines. For this analysis, the more stringent criterion of FDR < 0.07 was used to assign significance, along with the same fold change threshold of two.
Identification of differentially expressed genes (DEGs) was carried by same statistical testing explained above. For gene expression analysis, significance is assigned if fold change > 2-fold or fold change < 0.5-fold and nominal p-value < 0.05 (no significantly regulated genes were found for FDR < 0.35).
Amplex Red cholesterol assay.
Lipids were extracted as outlined in the Bligh and Dyer protocol above. The lipids were suspended in reaction buffer, and the protocol was followed as outlined for Amplex Red cholesterol assay (Thermo Scientific catalog no. A12216). Briefly, 50 μl of lipid extract and 50 μl of the Amplex® Red reagent/HRP/cholesterol oxidase/cholesterol esterase working solution or working solution without esterase was added to the appropriate well and incubated in the dark at 37°C for 90 min. The plate was read using a spectramax II with excitation at 534 nm and emission at 590 nm. The samples were compared with cholesterol standard and normalized to cell number.
Cell titer blue assay.
A total of 20,000 cells were plated per well in 96-well plates and allowed to adhere overnight. In the morning, the cell media was aspirated and replaced with fresh 100 μl of cell medium and incubated with compound or DMSO vehicle control (1 μl) and allowed to incubate at 37°C and 5% CO2 in a humidified incubator for 4 h. A total of 20 μl of cell titer blue reagent was added, and the plate was incubated for 1 h before the fluorescence was read using a plate reader with excitation at 534 nm and emission at 590 nm.
Wound-healing assay.
Cells were grown to a confluent monolayer in 12-well plates, a scratch was made across the center of the plate with a pipette tip, and images were taken. The medium was replaced with fresh medium, and treatment or vehicle control was added to the wells. The cells were maintained at 37°C and 5% CO2 in a humidified incubator and images were taken every 2 h. The images were processed using ImageJ via the wound-healing plugin and normalized to 100% area at time 0.
RESULTS
Global lipidomics profile of OS cells
To evaluate the lipidomic differences in OS as compared with nontumorigenic cells, the lipidomic profiles of human osteoblast cells (FOB), HOS osteosarcoma cells, which are nonmetastatic and 143B cells, which are highly metastatic, were evaluated (24). HOS and 143B are isogenic cell lines that limit the differences in lipidomic profiles to factors that contribute to a metastatic phenotype. Total lipid was extracted from ∼90% confluent cell plates using the Bligh and Dyer method (21) for three separate experiments (n = 3). Total lipid extract was subjected to LC/MS/MS analysis by Thermo Q-Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer, and the data were searched against LIPIDSEARCHTM (Thermo Scientific) software to identify specific lipid species. The lipids were then normalized to respective internal standards and cell count from each individual sample to minimize instrument errors and differences in absolute cell densities, respectively.
Greater than 1,000 species of lipids were identified in each of the cell lines (supplemental Tables S1–S15). Mass-spectrometric methods have the advantage of high-throughput and therefore the ability to generate huge datasets in a limited time period. However, there are inherent drawbacks of mass spectrometry. For example, a precursor ion scan for the identification of phosphotidylcholine (PC) is not able to distinguish between ether versus diacyl species. For these species, an educated assumption is made to assign the molecular species. A further discussion on the mass-spectrometric identification of lipids and their nomenclature has been performed in the literature (25). Furthermore, the position of double bonds is not discernible, and therefore only the number is indicated with each species. Additionally, there are limitations in the identification of all individual species. Isobaric species (molecular species with the same nominal mass but different exact mass) are challenging to identify. For example, for PC (35:1) composition indicates that the m/z 184.0733 fragment was found but a neutral loss scan was not obtained or there was a lack of FA fragment ions, which would allow the identification of individual chains. Representative chromatograms for various species have been included in supplemental Fig. S8.
The highest number of species was identified in PCs, followed by phosphatidylethanolamines (PEs) and cardiolipin (CL). The global data were statistically analyzed using single-factor ANOVA and Tukey posthoc test. Figure 2 shows the global lipidomic changes by ANOVA. Supplemental Fig. S1 shows the log fold-change differences in global classes of lipids in HOS versus FOB, 143B versus HOS, and FOB versus 143B.
Fig. 2.
The 15 major classes of lipids detected in three cell lines: osteoblast FOB (black), nonmetastatic HOS (horizontal striped), and metastatic 143B (gray) as analyzed by ANOVA. The various lipid classes are PCs, PEs, PSs, PGs, phosphatidylinositols (PIs), CL, ChE, MGs, DGs, TGs, SMs, Cers, LPCs, and LPEs. All experiments were performed in three biological replicates (n = 3).
A similar statistical analysis of individual species is represented in Fig. 3, which shows the differentially regulated species in each class. This is different from the global changes, because specific species in each class do not differ across the three cell lines. But Fig. 3 represents what percentage of species in each cell line are different from the other cell lines. The data corresponding to Fig. 3 are given in the supplemental tables. It was noted that in classes with low numbers of individual species, the results typically appeared amplified, and therefore results for MG (seven species), Pet (7), PA (6), and LPE (4) are not as statistically robust as classes with higher total species. Supplemental Figs. S2 and S3 show the log fold-change differences in individual lipids in HOS versus FOB, 143B versus HOS, and FOB versus 143B.
Fig. 3.
The number of individual species that are significantly different among the three cell lines. Differences between FOB and HOS represents differences in lipids between normal and nonmetastatic state (yellow). Differences between 143B and HOS represents differences in lipids between nonmetastatic and metastatic state (blue). Differences between FOB and 143B represents differences in lipids between normal and metastatic state (orange).
Finally, a PCA was performed on the lipidomes of the different OS cell types (FOB, HOS, and 143B) to distinguish the normal osteoblast cells (FOB) as compared with the nonmetastatic and metastatic cells (HOS and 143B). As seen in Fig. 4A, in the PCA score plot, the OS cells (HOS and 143B) are separated in a different ellipsoid space from the normal osteoblast cells. This is also reflected in the 2D plot in Fig. 4B. The various classes and the specific species that were differentially regulated have been discussed below with respect to global changes in composition as well as variations in individual lipids within the class.
Fig. 4.
A: PCA on all three cell lines, osteoblast FOB (red), nonmetastatic (green), and metastatic 143B (blue) cells as represented in a 3D space. B: The same PCA plot with osteoblast FOB (red), nonmetastatic (green), and metastatic 143B (blue) cells as represented in a 2D space. As shown, all three cell lines occupy a different space in the plot.
PCs and LPCs.
PCs are the largest class of membrane phospholipids. LPCs are PCs that arise from partial hydrolysis of PCs. PCs and LPCs were analyzed in positive-ion mode, and 130 species of PCs and 21 species of LPCs were detected (supplemental Tables S5 and S6). Overall, similar levels of PC species were found in all three cell lines. The levels of PC in normal osteoblast FOB cells was slightly higher than HOS and 143B cells, albeit insignificantly, as analyzed by ANOVA and Wilcoxon tests.
Figure 3 represents the upregulation in specific species. As indicated in the labels, the three different colors represent the differential regulation in a class versus another. As seen in Fig. 3, 20% of species are differentially regulated in FOB versus HOS, 60% between HOS and 143B, and 50% of species are differentially regulated between FOB and 143B. PC (34:1) was the most abundant species in all three cell lines.
As PCs form a large class of structural lipids, significant alteration in PC levels is not expected. However, LPCs and subclass of PCs have been correlated with various cancers (26, 27). Herein, it was observed that the amount of LPC was significantly higher in FOB cells than in 143B cells (P = 0.015) by ANOVA. This corresponds to the nonparametric global analysis test, where LPC is downregulated in 143B with respect to both HOS and FOB cells, as shown in Fig. 2. The amount of saturated LPCs was higher in all three cell lines, as compared with the unsaturated LPC counterparts. However, both saturated and unsaturated LPCs were significantly higher in FOB cells as compared with the tumorigenic cell lines.
Among specific LPC species, higher plasma levels of LPC (18:0) have been related to lower risk of cancer (28). In this work as well, FOB cells had the highest amount of LPC (18:0) and were significantly higher than 143B cells (P = 0.014) and modestly higher than HOS cells (P = 0.099). Fig. 3 shows the comparison of individual species in all cell lines. 20% species were different between FOB and HOS, and 30% differential regulation was seen between HOS versus 143B and FOB versus 143B, respectively.
PEs and lysophosphatidylethanolamines.
PEs are positively charged phospholipids that account for 25% of all phospholipids. Lysophosphatidylethanolamines (LPEs) are derived from partial hydrolysis of PEs, similar to LPCs (from PCs). Both PEs and LPEs were measured in negative-ion mode, and 117 PE species and 4 LPE species were detected (supplemental Tables S7 and S8). Overall, no significant changes were observed in PE levels among the three cell lines by ANOVA and Wilcoxon test (Fig. 2 and supplemental Fig. S7). There was a slight gradient decrease from osteoblasts to nonmetastatic to metastatic cells, but these differences were not statistically significant. These results are in agreement with literature in breast cancer cell lines where MCF-10A nonmalignant cell lines had a higher abundance of PE than the corresponding cancerous cells (15).
The most abundant species was PE (18:0) in HOS and FOB cells. However, PE (18:1) was the most abundant in 143B metastatic cells. As observed in Fig. 3, under 30% of the PE individual species were differentially regulated in FOB versus HOS, 60% between HOS versus 143B, and 40% between FOB and 143B.
Similar to the trend observed in LPCs, the levels of LPEs were significantly higher in FOB versus 143B cells (P = 0.009). LPEs were highest in FOB cells, followed by HOS, and least in 143B cells (Fig. 2). A moderate change in LPE levels was also observed from nonmetastatic HOS to metastatic 143B cells (P = 0.072). Because of the low number of LPE species detected, robust statistical interpretation for individual species, as seen in Fig. 3 for LPE, could not be performed.
Phosphatidylserines.
Phosphatidylserines (PSs) are negatively charged membrane lipids that are typically present in the inner cell membranes (29). However, in cells undergoing apoptosis, PS asymmetrically translocates to the outer cell membrane leaflet (30). Forty-three species of PS were detected in in negative-ion mode (supplemental Table S11). The differences among the three cell lines were not significant by ANOVA and Wilcoxon tests (Fig. 2 and supplemental Fig. S7).
However, specific lipid species were differently expressed. As seen in Fig. 3, there was less than 10% difference in the individual species between FOB and HOS. However, over 50% of species were differentially regulated in HOS versus 143B and 30% of species between FOB and 143B, potentially suggesting a role for specific PS species in the metastatic process.
Sphingolipids: Cers and SMs.
SMs and Cers are lipids with a sphingosine backbone. Cers were analyzed in positive-ion mode, and a total of 124 individual Cer species were detected (supplemental Table S1). Among these, 20 CerG1, 14 CerG2, and 9 CerG3 Cers of the simple Glc series (neutral glycosphingolipids) were detected. Overall, in nonparametric global analysis, Cers were found to be significantly downregulated in HOS versus FOB cells by Wilcoxon test (supplemental Fig. S7). The tumorigenic cells exhibited lesser amounts of total Cers than their nonmalignant osteoblast counterpart. Specifically, HOS cells had the least amount of Cers followed by 143B with FOB exhibiting the highest amount (Fig. 2).
Comparing the individual species in the three cell lines, as seen in Fig. 3, there is a 45% difference between FOB and HOS, HOS and 143B, and FOB and 143B.
SMs were analyzed in positive-ion mode, and 101 SM species were detected (supplemental Table S12), and overall the differences in SM content in all three lines remained relatively consistent (Fig. 2). However, the levels of SM were less in the metastatic 143B cells, as compared with the osteoblast FOB cells and nonmetastatic HOS cells (Fig. 2).
Specifically, several SM species were significantly lower in 143B cell lines, and several species were below the threshold of detection. These were SM (d18:1/19:0), (d40:4), (d42:5), (d44:4), (d44:7), (d46:7), and (d48:2). As seen in Fig. 3, all classes showed several differentially regulated species. A 50–60% difference in individual species was observed in all three comparisons FOB versus HOS, HOS versus 143B, and FOB versus 143B.
CLs.
CL is an inner mitochondrial membrane phospholipid constituting 20% of the membrane lipid composition (31). A total of 142 CL species were detected in negative-ion mode (supplemental Table S3). As seen from Fig. 2 and supplemental Fig. S7, CL expression in HOS cells is significantly greater than in FOB and 143B cell lines.
As shown in Fig. 3, there is a 45% difference in individual species in FOB versus HOS, 65% between HOS and 143B, and 30% between FOB and 143B.
Cholesteryl esters.
Cholesteryl esters (ChEs) are FA esters of cholesterol through the hydroxyl group of cholesterol. ChEs are a biologically inert form of cholesterol storage and liberate cholesterol when required for membrane and lipoprotein formation (32). Thirty-eight distinct species of ChEs were detected in positive-ion mode (supplemental Table S2). Although the levels of ChE in FOB and HOS cells was relatively similar, the amount of ChE in 143B cells was significantly less (P = 0.045, HOS vs. 143B; and P = 0.068, FOB vs. 143B) by ANOVA (Fig. 2). This is reflected in the nonparametric global analysis in supplemental Fig. S7, where 143B was downregulated as compared with both HOS and FOB cells.
As observed in Fig. 3, HOS and FOB have only 10% species differentially regulated. However, there is a greater than 60% difference in species when 143B is compared with FOB or HOS.
Total cholesterol was not measured in our studies via high-throughput lipidomics because the ESI/MS platform is not well suited for detection of free cholesterol. However, as the interest in cholesterol and cholesterol metabolism and their correlation with cancer has increased in literature, both total cholesterol as well as ChEs in our cells through Amplex Red measurement were analyzed (33). The hydrogen peroxide generated during oxidation converts the Amplex Red to resorufin, which is a fluorescent molecule. The assay is further modified to convert cholesterol esters to free cholesterol, first by using cholesterol esterase, and thus gives a measurement of total cholesterol in the cells. In this assay, the experiment was performed with all three cell lines with and without cholesterol esterase to examine the amount of free cholesterol, ChEs, and total cholesterol in each cell line. As observed in Fig. 5A, the total cholesterol levels were greatest in FOB cells, followed by 143B cells and then HOS cells. However, none of these differences were significant. The amount of free cholesterol also followed the same pattern in levels. Finally, the difference between total cholesterol and free cholesterol was calculated to determine the amount of ChEs. As observed in Fig. 5A, the amount of ChEs was greatest in FOB followed by HOS and very low in 143B, although these differences were not statistically significant. These values indicate that the levels of esterified cholesterol are lower in 143B cells as compared with FOB and HOS cells, thereby confirming results found in the lipidomics data (Fig. 2 and supplemental Fig. S7).
Fig. 5.
A: Levels of total cholesterol (black), free cholesterol (horizontal bars), and ChE (gray) in FOB, HOS, and 143B cells, as measured by the Amplex Red detection methodology. B: Specific ChE species significantly differentially regulated in the three cell lines FOB (black), HOS (white), and 143B (gray).
Monoacylglycerols.
Monoacylglycerols (MGs) contain one glycerol moiety connected to a FA through an ester bond. MGs are catabolized by MG lipase (MAGL) to form FFA and glycerol, and the overexpression of MAGL in cancers has generated recent interest in this pathway. Seven MG species were detected in positive-ion mode with four saturated and three monounsaturated species (supplemental Table S15). A significant overexpression of MG was observed in 143B metastatic cells as compared with both nonmetastatic HOS (P = 0.0105) and osteoblast FOB (P = 0.0069) cells by ANOVA and is also reflected by the results of the nonparametric global analysis (Fig. 2). However, the difference between HOS and FOB cells was not significant. The low number of MG species does not allow for a robust statistical treatment in Fig. 3.
DGs.
DGs contain two glycerol units esterified with a FA backbone. DG levels in cancer cells has been shown to be dysregulated, signaling through the protein kinase C (PKC) pathway (34). Additionally, the levels of DG have been shown to affect the effectivity of drugs such as FA synthase inhibitors in cancer cells (35). Fifty-four DG species were detected in positive-ion mode (supplemental Table S4).
Different regulation of DG species was observed in 143B cells as compared with HOS cells (P = 0.029) and FOB cells (P = 0.07) by ANOVA (Fig. 2). Although the latter was not significant by ANOVA, both are reflected in the global Wilcoxon test as significant regulations (supplemental Fig. S7).
As seen in Fig. 3, over 60% of species were differentially regulated in 143B cells as compared with FOB and HOS cells, making this class one of the hallmark characteristics of the 143B cell line. There was only a 30% difference between FOB and HOS cells. Supplemental Fig. S4 shows the various DG species that are differentially regulated.
Phosphatidic acids.
Phosphatidic acids (PAs) are components of the membrane and generate DGs as mentioned previously. DG itself converts to PA in the presence of DG kinases that are also targets for cancer drugs (36). In this study, six PA species were detected in negative-ion mode (supplemental Table S14). Although ANOVA indicated the differences in PA levels to be not significant (Fig. 2), a global Wilcoxon test revealed that PA is downregulated in HOS cells as compared with FOB cells (supplemental Fig. S7).
Triacylglycerols.
Triacylglycerols (TGs) contain three esterified glycerols on a FA backbone. Similar to MGs and DGs, TGs are also implicated in various cancerous pathways. Particularly, increased levels of TG are associated with enhanced apoptosis (37, 38). In this study, 163 species of TGs identified in positive-ion mode (supplemental Table S13) and the levels of TGs were greatest in FOB cells, followed by HOS and least in 143B cells (Fig. 2). The difference between FOB and 143B cells was significant, with P = 0.03 by ANOVA, and was also reflected in the global Wilcoxon test (supplemental Fig. S7).
Specific species that were higher in FOB versus 143B cells were also observed in Fig. 3; there was a 20% difference in individual species levels in FOB versus HOS, 35% between HOS and 143B, and 50% between 143B and FOB.
DG inhibition results in reduced cell viability and migration
As identified in the previous section, DGs are a significantly upregulated population in the lipidome of 143B versus HOS and FOB cells. To investigate whether the inhibition of DGs will result in reduced cell viability and migration, the synthetic pathway of DG was inhibited. There are several different synthetic pathways for the formation of DGs. The chief pathway of synthesis is through the PLC-catalyzed hydrolysis of phosphatidylinositol 4,5-bisphosphate in the membrane to form DG, which remains in the membrane. The inhibition of PLC to inhibit DG synthesis and its effects on various diseased states has been investigated (39, 40). In this study, PLC inhibitor U73122 was used to inhibit DG synthesis and investigate its effects on metastatic 143B cells. First, 143B cells were incubated with 1 μM U73122 for 4 h, and lipids were extracted by the Bligh and Dyer method and analyzed by high-throughput lipidomics. As seen in Fig. 6A, the levels of DG were reduced in the treated samples as compared with the controls. Surprisingly, TG levels went up significantly after inhibitor treatment. As seen in Fig. 6B, most classes of lipids were affected. This is expected, because DG synthesis is directly linked to most phospholipid synthesis (41). Supplemental Figs. S5 and S6 show the log fold-change plots for lipids in 143B cells with U73122 treatment and those without treatment.
Fig. 6.
A: Differences in total lipidomic content of 143B cells on treatment with PLC inhibitor U73122. Solid bars represent the lipidomic composition before treatment, and striped bars represent composition after treatment with PLC inhibitor. B: Percentage changes in individual lipid species in treated versus untreated cells that are downregulated (blue) and upregulated (orange).
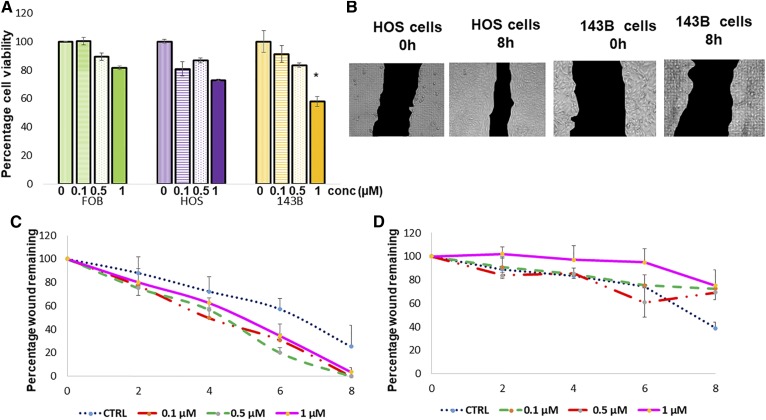
Furthermore, as seen from Fig. 7A, the cell viability was reduced in all three cell lines. The change was greatest in 143B cells, the presence of 0.1, 0.5, and 1 μM U73122, as measured by the cell titer blue assay (42). Finally, a reduction in cell migration was also observed in 143B cells in the presence of U73122, as indicated by the wound healing assay (43) in Fig. 7B–D, but the effect was not observed in HOS cells (Fig. 7D).
Fig. 7.
A: Reduction in cell viability in all three cell lines on treatment with PLC inhibitor (U73122) as measured by cell titer blue assay. B: Cell migration in 143B and HOS cells with 1 µM PLC inhibitor U73122. C: Cell migration in HOS cells at 0 to 8 h time points with different concentrations of U73122. D: Cell migration in 143B cells at 0 to 8 h time points with different concentrations of U73122. conc, concentration; CTRL, control.
Finally, we wanted to explore whether the gene expressions for the synthesis and metabolism of DGs concurred with our data. We studied published microarray gene-expression data (42) available in the Gene Expression Omnibus (GEO) database (accession number GSE66673) containing the gene expression during the early and late passages of HOS and 143B cells with three replicates (n = 3) for all cell lines (42). This dataset contained 42545 probe sets that were mapped to either a known or a model RefSeq accession number and were grouped into 20,724 unique genes. We used the data to identify DEGs in late-passage 143B versus late-passage HOS cells. Among all of the 20,724 genes, 1,415 genes passed our significance thresholds that give a DEG ratio of 0.068. In order to identify the DEGs in DG metabolism, we used the KEGG database to obtain the DG metabolic pathway. We identified three differentially identified genes in DG metabolism that were found to be upregulated in 143B versus the HOS cells—AGPAT9, PPAP2C, and MGLL, as shown in Fig. 8.
Fig. 8.
Differentially regulated genes associated with the DG synthesis and metabolism pathway in 143B (light gray) versus HOS cells (dark gray).
DISCUSSION
The lipidomic profiling of cancer cells has become an important area of research that has been facilitated by the improvement of mass-spectrometric techniques that can identify and quantify very small quantities of lipids in mixtures of hundreds of species. Although the investigation of the lipidome of breast-cancer and prostate-cancer cells has been recently explored, the investigation of lipidomic profiles of bone cancers has remained unexplored. In this work, we investigate the lipidomic profile of human OS cell lines and compare them to a human osteoblast cell line. We perform high-throughput lipidomics of osteoblasts (FOB), nonmetastatic (HOS), and metastatic (143B) cells. Various classes and hundreds of individual species in each class were identified in our results, and several of them were found to be differentially regulated.
Among the chief lipid classes, we found ChEs, CLs, MGs, DGs, and TGs, LPCs, and LPEs, as well as various individual lipid species to be significantly differentially regulated. In general, various patterns can be drawn from the global lipidomic changes represented in Fig. 2. Lipids that are differentially regulated in HOS or 143B versus FOB can be correlated with the process of tumorigenesis. Additionally, lipids that are differentially regulated in 143B versus HOS can be correlated with the metastatic process. As seen in Fig. 2, Cers are downregulated in HOS cells, and CLs are upregulated in HOS cells as compared with FOB cells. Furthermore, MGs and DGs are upregulated in 143B versus HOS cells, and LPCs and LPEs are downregulated in 143B versus HOS cells.
Cers are bioactive sphingolipids that have come to the forefront of cancer research. However, the various Cer species, depending on the FA conjugated to them, have shown various properties, including their roles in cellular proliferation or cancer cell death (44). For instance, C18-Cer treatment suppresses tumor growth in head and neck cancer cells (45); however, the C16 Cers induce head and neck squamous cell carcinoma tumor proliferation (46). Thus, a cumulative evaluation of Cer species will not be accurate in representing the correlation of lipid with OS. In our results, the levels of C18 Cers was significantly less in 143B and HOS cells, as compared with the FOB cells. C18 Cer has been recognized as a serum biomarker for monitoring chemotherapy response, as its levels increase with improvement in diseased state for head and neck squamous cell carcinoma (47).
Conversely, CLs may be implicated in tumorigenesis. Indeed, CL has been found to impact prostate cancer-cell proliferation (48). Specifically, C16-rich CL was found to stimulate proliferation, and C18-rich cells were found to reduce proliferation. As observed in supplemental Table S1, C18-rich CLs appear to have a higher expression in FOB cells as compared with 143B cells, and C16-rich CLs have a higher expression in 143B cells. For example, CL(16:0/16:0/16:0/16:0) is not detected in FOB cells, but is detected in both 143B and HOS cells, and CL(18:4/18:2/18:2/18:2) is much lower in 143B cells as compared with FOB cells.
The abundance of LPCs in nontumorigenic cell line as compared with the tumorigenic OS cell lines indicates a faster conversion of LPCs to PCs in OS cells as compared with the normal cells. Indeed, in human colorectal cancer, it has been shown that LPC acyltransferase 1 (LPCAT1), the enzyme that converts LPC to PC, is overexpressed (49), and LPCAT1 expression has been investigated as a biomarker in prostate cancer as well (50). However, the levels of LPC are reduced in 143B cells as compared with HOS cells, indicating that, although they promote tumorigenesis, they may not be completely implicated in the process of metastasis.
Cholesterol and cholesterol metabolism have been found to be highly dysregulated in cancer (51). However, the underlying mechanism of this is not very well understood. Cholesterol avidity in cancer cells is high, and it is usually stored in lipid droplets. The cholesterol concentration in cells is greatly regulated by import-export machinery through the droplets. The LDL receptors import cholesterol and are regulated by the SREBP transcriptional factors. The excess of cholesterol or of its oxidized products, oxysterols, activate liver X receptors and retinoid X receptor heterodimeric transcriptional factors. Subsequent induction of expression of ABC family transporters results in efflux of cholesterol (52). Altered expression levels and mutations of genes involved in the cholesterol homeostasis pathways have been identified in cancer cells (53). These include increases in gene copy numbers, upregulation of cholesterol synthesis gene expression, enhanced cholesterol import by LDL receptors, and decreased transport of cholesterol (53–56). However, further research is required to fully dissect the consequences of these changes and how they modulate cancer development. Cholesterol metabolites like oxysterols and steroids have also been implicated for their role in cancer (57). The accumulation of cholesterol is usually correlated with higher cancer progression and metastasis. However, in certain studies, lower levels of cholesterol have been linked to cancer (58, 59). In our studies we observed that the level of ChEs in metastatic OS cell line 143B was significantly less than the osteoblast and the nonmetastatic cell lines. On exploring this further via an Amplex Red detection of total cholesterol as well as ChEs in all three cell lines, we found that the levels of cholesterol, both free and esterified, were higher, albeit not significantly, in normal osteoblast cells than tumorigenic OS cells. This may in part be due to the lack of adipocytes in cell culture, because a significant amount of cholesterol in tumor tissue is derived from dietary cholesterol that is imported from adipocytes in the tumor microenvironment.
The other lipid-signaling pathways of interest are acylglycerols. MGs, DGs, and TGs are important lipid components of the cell, and all three were identified in our studies. Although MGs and DGs increased in metastatic cells as compared with their nonmetastatic counterparts, the levels of TGs reduced. Although all three moieties are synthesized through various pathways, they also intraconvert through various enzymatically facilitated pathways.
In our work, we show that inhibiting PLC and therefore inhibiting the DG synthesis pathway results in reduced cell viability and migration. DG is an important membrane lipid and acts as a secondary messenger. DG is known to bind directly to the PKC and protein kinase D (PKD) family, as well as to the Ras family (60, 61). The DG-related activation of PKC and PKD plays an important role in cancer, as they signal through multiple pathways and control the expression of genes controlling cell-cycle progression, tumorigenesis, and metastasis (62). Therefore, it is not surprising that the reduction of DG accumulation in cells reduces the cellular viability. The blocking of DG synthesis as a proof of concept in this work exhibits that blocking lipid synthesis is an approach toward combating tumor growth and metastasis. However, the DG pathway is an important cellular pathway, and blocking entire lipid-synthesis pathways will result in significant cellular abnormality. A more viable clinical approach would be to block specific DG species or specific species from other lipid classes. Finally, we identify genes associated with the DG pathway that are differentially regulated. AGPAT9 encodes for glycerol-3-phosphate acyltransferase 3. It catalyzes the conversion of glycerol-3-phosphate to lysophosphatidic acid, which converts to PA and subsequently to DG (63). The genetic upregulation of AGPAT9 in 143B therefore corresponds to the increased synthesis of DG and thus higher levels of DGs in 143B cells than HOS. Interestingly, AGPAT9 is also known as metastasis associated gene 1 and activates the mTOR pathway (64). PPAP2C gene encodes for lipid phosphate phosphohydrolase 2, which catalyzes the conversion of PA to DG (65), and hence increases levels of DG overall. PPAP2C has been shown to be a potential anticancer drug target by genomic screening in transformed adult mesenchymal stem cells (66). Furthermore, the knock down of PPAP2C impaired anchorage-dependent in vitro growth of cancer cell lines and impaired the in vitro growth of primary mesenchymal stem cells but not differentiated human fibroblasts by delaying entry into S phase of the cell cycle and is transcriptionally regulated by p53.
In this study, we exhibit that cancer cells have an altered lipidomic profile as compared with normal cells, and, furthermore, the lipidomic profile also changes in metastatic and nonmetastatic cells, showing that lipid composition affects the tumorigenic properties of OS. It is widely accepted that the metabolic reprogramming of cancer cells is necessary to sustain their uncontrolled proliferation. Although cells provide a homogenous system for the study of the disease, the tumor microenvironment likely affects the lipid composition further. The tumor microenvironment is unique and contains zones of hypoxia, extracellular acidosis, and necrosis. The hypoxic condition, as well as the interaction of cancer cells with the stroma and the endothelial cells, affects metabolism of various lipids, including PCs and cholesterol species (67, 68). In addition to the changes at the cellular level, including differences in unsaturation, which affects fluidity and drug resistance, changes have been investigated at the tumor-stroma interactions. Molecular interactions between cancer cells and fibroblasts in their extracellular environment, as well as their interaction with adipocytes, change the lipid composition of the cells. In fact, the FA translocation by exosomes into cells alters the lipid homeostasis and therefore disturbs various signaling pathways (69). Therefore, it is likely that the results from cellular lipidomics will not be translated unequivocally to lipidomics of tissue samples. However, we expect the trends to remain the same, favoring upregulation of lipids that promote cellular proliferation. Cellular investigations also help limit the lipid classes of interest to be investigated with scarce tissue samples. Further investigations into primary and metastatic OS tissue samples will enable targeting of specific lipid synthesis pathways for OS therapy.
CONCLUSION
This work compares the global lipidomic profile of normal osteoblast cells to nonmetastatic and metastatic OS cell lines. We show that various different lipid pathways and specific lipid species are differentially regulated in the diseased state. Furthermore, we identify that DGs are significantly upregulated in metastatic 143B cells as compared with HOS and FOB cells. We block the synthesis of DG though PLC inhibition and exhibit that this reduces cell viability as well as cell migration in 143B cells. Finally, we identify two genes involved in the DG synthesis pathway that are significantly upregulated and therefore can be potential therapeutic targets in OS.
Supplementary Material
Acknowledgments
The authors thank Dr. Zhong Li at the Roy J. Carver Biotechnology Center, University of Illinois, Urbana Champaign for assistance with lipidomics; Ms. Holly Pondenis for cell culture help; Mr. Justin Kim for editing; and Dr. Chemyong Ko and his laboratory for use of instrumentation.
Footnotes
Abbreviations:
- Cer
- ceramide
- ChE
- cholesteryl ester
- CL
- cardiolipin
- DEG
- differentially expressed gene
- DG
- diacylglycerol
- FDR
- false discovery rate
- LPC
- lysophosphatidylcholine
- LPE
- lysophosphatidylethanolamine
- MG
- monoacylglycerol
- OS
- osteosarcoma
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PCA
- principal component analysis
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PKC
- protein kinase C
- PLC
- phospholipase C
- PS
- phosphatidylserine
- TG
- triacylglycerol
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bielack S. S., Kempf-Bielack B., Delling G., Exner G. U., Flege S., Helmke K., Kotz R., Salzer-Kuntschik M., Werner M., Winkelmann W., et al. 2002. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 20: 776–790. [DOI] [PubMed] [Google Scholar]
- 2.Hughes D. P. 2009. Strategies for the targeted delivery of therapeutics for osteosarcoma. Expert Opin. Drug Deliv. 6: 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baenke F., Peck B., Miess H., and Schulze A.. 2013. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 6: 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dopico A. M., and Tigyi G. J.. 2007. A glance at the structural and functional diversity of membrane lipids. Methods Mol. Biol. 400: 1–13. [DOI] [PubMed] [Google Scholar]
- 5.Shevchenko A., and Simons K.. 2010. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 11: 593–598. [DOI] [PubMed] [Google Scholar]
- 6.Escribá P. V., Wedegaertner P. B., Goñi F. M., and Vögler O.. 2007. Lipid-protein interactions in GPCR-associated signaling. Biochim. Biophys. Acta. 1768: 836–852. [DOI] [PubMed] [Google Scholar]
- 7.Stordeur C., Puth K., Saenz J. P., and Ernst R.. 2014. Crosstalk of lipid and protein homeostasis to maintain membrane function. Biol. Chem. 395: 313–326. [DOI] [PubMed] [Google Scholar]
- 8.Corda D., and De Matteis M. A.. 2013. Lipid signalling in health and disease. FEBS J. 280: 6280. [DOI] [PubMed] [Google Scholar]
- 9.Sampaio J. L., Gerl M. J., Klose C., Ejsing C. S., Beug H., Simons K., and Shevchenko A.. 2011. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA. 108: 1903–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaidi N., Swinnen J. V., and Smans K.. 2012. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res. 72: 3709–3714. [DOI] [PubMed] [Google Scholar]
- 11.Menendez J. A., and Lupu R.. 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 7: 763–777. [DOI] [PubMed] [Google Scholar]
- 12.Guillaumond F., Bidaut G., Ouaissi M., Servais S., Gouirand V., Olivares O., Lac S., Borge L., Roques J., Gayet O., et al. 2015. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. USA. 112: 2473–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H., Volponi J. V., Oliver A. E., Parikh A. N., Simmons B. A., and Singh S.. 2011. In vivo lipidomics using single-cell Raman spectroscopy. Proc. Natl. Acad. Sci. USA. 108: 3809–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterin M., Cohen J. S., and Ringel I.. 2004. Hormone sensitivity is reflected in the phospholipid profiles of breast cancer cell lines. Breast Cancer Res. Treat. 87: 1–11. [DOI] [PubMed] [Google Scholar]
- 15.Dória M. L., Cotrim C. Z., Simões C., Macedo B., Domingues P., Domingues M. R., and Helguero L. A.. 2013. Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J. Cell. Physiol. 228: 457–468. [DOI] [PubMed] [Google Scholar]
- 16.Fhaner C. J., Liu S., Ji H., Simpson R. J., and Reid G. E.. 2012. Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal. Chem. 84: 8917–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X., Mao J., Ai J., Deng Y., Roth M. R., Pound C., Henegar J., Welti R., and Bigler S. A.. 2012. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 7: e48889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dória M. L., Cotrim Z., Macedo B., Simões C., Domingues P., Helguero L., and Domingues M. R.. 2012. Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res. Treat. 133: 635–648. [DOI] [PubMed] [Google Scholar]
- 19.Hilvo M., Denkert C., Lehtinen L., Muller B., Brockmoller S., Seppanen-Laakso T., Budczies J., Bucher E., Yetukuri L., Castillo S., et al. 2011. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 71: 3236–3245. [DOI] [PubMed] [Google Scholar]
- 20.Li J., Ren S. C., Piao H. L., Wang F. B., Yin P. Y., Xu C. L., Lu X., Ye G. Z., Shao Y. P., Yan M., et al. 2016. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Sci. Rep. 6: 20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 22.Taguchi R., and Ishikawa M.. 2010. Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J. Chromatogr. A. 1217: 4229–4239. [DOI] [PubMed] [Google Scholar]
- 23.Yamada T., Uchikata T., Sakamoto S., Yokoi Y., Fukusaki E., and Bamba T.. 2013. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J. Chromatogr. A. 1292: 211–218. [DOI] [PubMed] [Google Scholar]
- 24.Luu H. H., Kang Q., Park J. K., Si W., Luo Q., Jiang W., Yin H., Montag A. G., Simon M. A., Peabody T. D., et al. 2005. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clin. Exp. Metastasis. 22: 319–329. [DOI] [PubMed] [Google Scholar]
- 25.Liebisch G., Vizcaino J. A., Kofeler H., Trotzmuller M., Griffiths W. J., Schmitz G., Spener F., and Wakelam M. J.. 2013. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 54: 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuliszkiewicz-Janus M., Tuz M. A., and Baczynski S.. 2005. Application of 31P MRS to the analysis of phospholipid changes in plasma of patients with acute leukemia. Biochim. Biophys. Acta. 1737: 11–15. [DOI] [PubMed] [Google Scholar]
- 27.Cvetković B., Vučić V., Cvetković Z., Popović T., and Glibetić M.. 2012. Systemic alterations in concentrations and distribution of plasma phospholipids in prostate cancer patients. Med. Oncol. 29: 809–814. [DOI] [PubMed] [Google Scholar]
- 28.Kühn T., Floegel A., Sookthai D., Johnson T., Rolle-Kampczyk U., Otto W., von Bergen M., Boeing H., and Kaaks R.. 2016. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 14: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leventis P. A., and Grinstein S.. 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39: 407–427. [DOI] [PubMed] [Google Scholar]
- 30.Mariño G., and Kroemer G.. 2013. Mechanisms of apoptotic phosphatidylserine exposure. Cell Res. 23: 1247–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlame M., and Ren M.. 2009. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim. Biophys. Acta. 1788: 2080–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginsburg G. S., Atkinson D., and Small D. M.. 1984. Physical properties of cholesteryl esters. Prog. Lipid Res. 23: 135–167. [DOI] [PubMed] [Google Scholar]
- 33.Robinet P., Wang Z., Hazen S. L., and Smith J. D.. 2010. A simple and sensitive enzymatic method for cholesterol quantification in macrophages and foam cells. J. Lipid Res. 51: 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griner E. M., and Kazanietz M. G.. 2007. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer. 7: 281–294. [DOI] [PubMed] [Google Scholar]
- 35.Benjamin D. I., Li D. S., Lowe W., Heuer T., Kemble G., and Nomura D. K.. 2015. Diacylglycerol metabolism and signaling is a driving force underlying FASN inhibitor sensitivity in cancer cells. ACS Chem. Biol. 10: 1616–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mérida I., Torres-Ayuso P., Ávila-Flores A., Arranz-Nicolás J., Andrada E., Tello-Lafoz M., Liébana R., and Arcos R.. 2017. Diacylglycerol kinases in cancer. Adv. Biol. Regul. 63: 22–31. [DOI] [PubMed] [Google Scholar]
- 37.Al-Saffar N. M., Titley J. C., Robertson D., Clarke P. A., Jackson L. E., Leach M. O., and Ronen S. M.. 2002. Apoptosis is associated with triacylglycerol accumulation in Jurkat T-cells. Br. J. Cancer. 86: 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finstad H. S., Dyrendal H., Myhrstad M. C., Heimli H., and Drevon C. A.. 2000. Uptake and activation of eicosapentaenoic acid are related to accumulation of triacylglycerol in Ramos cells dying from apoptosis. J. Lipid Res. 41: 554–563. [PubMed] [Google Scholar]
- 39.Tsai F. C., Seki A., Yang H. W., Hayer A., Carrasco S., Malmersjo S., and Meyer T.. 2014. A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat. Cell Biol. 16: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stella N., Schweitzer P., and Piomelli D.. 1997. A second endogenous cannabinoid that modulates long-term potentiation. Nature. 388: 773–778. [DOI] [PubMed] [Google Scholar]
- 41.Stapleton C. M., Mashek D. G., Wang S., Nagle C. A., Cline G. W., Thuillier P., Leesnitzer L. M., Li L. O., Stimmel J. B., Shulman G. I., et al. 2011. Lysophosphatidic acid activates peroxisome proliferator activated receptor-gamma in CHO cells that over-express glycerol 3-phosphate acyltransferase-1. PLoS One. 6: e18932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed S. A., Gogal R. M. Jr., and Walsh J. E.. 1994. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J. Immunol. Methods. 170: 211–224. [DOI] [PubMed] [Google Scholar]
- 43.Liang C. C., Park A. Y., and Guan J. L.. 2007. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2: 329–333. [DOI] [PubMed] [Google Scholar]
- 44.Saddoughi S. A., and Ogretmen B.. 2013. Diverse functions of ceramide in cancer cell death and proliferation. Adv. Cancer Res. 117: 37–58. [DOI] [PubMed] [Google Scholar]
- 45.Koybasi S., Senkal C. E., Sundararaj K., Spassieva S., Bielawski J., Osta W., Day T. A., Jiang J. C., Jazwinski S. M., Hannun Y. A., et al. 2004. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem. 279: 44311–44319. [DOI] [PubMed] [Google Scholar]
- 46.Senkal C. E., Ponnusamy S., Bielawski J., Hannun Y. A., and Ogretmen B.. 2010. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 24: 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saddoughi S. A., Garrett-Mayer E., Chaudhary U., O’Brien P. E., Afrin L. B., Day T. A., Gillespie M. B., Sharma A. K., Wilhoit C. S., Bostick R., et al. 2011. Results of a phase II trial of gemcitabine plus doxorubicin in patients with recurrent head and neck cancers: serum C(1)(8)-ceramide as a novel biomarker for monitoring response. Clin. Cancer Res. 17: 6097–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sapandowski A., Stope M., Evert K., Evert M., Zimmermann U., Peter D., Page I., Burchardt M., and Schild L.. 2015. Cardiolipin composition correlates with prostate cancer cell proliferation. Mol. Cell. Biochem. 410: 175–185. [DOI] [PubMed] [Google Scholar]
- 49.Mansilla F., da Costa K. A., Wang S., Kruhoffer M., Lewin T. M., Orntoft T. F., Coleman R. A., and Birkenkamp-Demtroder K.. 2009. Lysophosphatidylcholine acyltransferase 1 (LPCAT1) overexpression in human colorectal cancer. J. Mol. Med. 87: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X., Lawrence T. J., He Z., Pound C. R., Mao J., and Bigler S. A.. 2012. The expression level of lysophosphatidylcholine acyltransferase 1 (LPCAT1) correlates to the progression of prostate cancer. Exp. Mol. Pathol. 92: 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuzu O. F., Noory M. A., and Robertson G. P.. 2016. The role of cholesterol in cancer. Cancer Res. 76: 2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorin A., Gabitova L., and Astsaturov I.. 2012. Regulation of cholesterol biosynthesis and cancer signaling. Curr. Opin. Pharmacol. 12: 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murai T. 2015. Cholesterol lowering: role in cancer prevention and treatment. Biol. Chem. 396: 1–11. [DOI] [PubMed] [Google Scholar]
- 54.Smith B., and Land H.. 2012. Anticancer activity of the cholesterol exporter ABCA1 gene. Cell Reports. 2: 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krycer J. R., and Brown A. J.. 2013. Cholesterol accumulation in prostate cancer: a classic observation from a modern perspective. Biochim. Biophys. Acta. 1835: 219–229. [DOI] [PubMed] [Google Scholar]
- 56.Llaverias G., Danilo C., Mercier I., Daumer K., Capozza F., Williams T. M., Sotgia F., Lisanti M. P., and Frank P. G.. 2011. Role of cholesterol in the development and progression of breast cancer. Am. J. Pathol. 178: 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dufour J., Viennois E., De Boussac H., Baron S., and Lobaccaro J. M.. 2012. Oxysterol receptors, AKT and prostate cancer. Curr. Opin. Pharmacol. 12: 724–728. [DOI] [PubMed] [Google Scholar]
- 58.Newman T. B., and Hulley S. B.. 1996. Carcinogenicity of lipid-lowering drugs. JAMA. 275: 55–60. [PubMed] [Google Scholar]
- 59.Ravnskov U., McCully K. S., and Rosch P. J.. 2012. The statin-low cholesterol-cancer conundrum. QJM. 105: 383–388. [DOI] [PubMed] [Google Scholar]
- 60.Carrasco S., and Merida I.. 2004. Diacylglycerol-dependent binding recruits PKCtheta and RasGRP1 C1 domains to specific subcellular localizations in living T lymphocytes. Mol. Biol. Cell. 15: 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ebinu J. O., Stang S. L., Teixeira C., Bottorff D. A., Hooton J., Blumberg P. M., Barry M., Bleakley R. C., Ostergaard H. L., and Stone J. C.. 2000. RasGRP links T-cell receptor signaling to Ras. Blood. 95: 3199–3203. [PubMed] [Google Scholar]
- 62.Garg R., Benedetti L. G., Abera M. B., Wang H., Abba M., and Kazanietz M. G.. 2014. Protein kinase C and cancer: what we know and what we do not. Oncogene. 33: 5225–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao J., Li J. L., Li D., Tobin J. F., and Gimeno R. E.. 2006. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 103: 19695–19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y., Jia H., Lin H., Tan X., Du Z., Chen H., Xu Y., Han X., Zhang J., Zhao S., et al. 2012. Metastasis-associated gene, mag-1 improves tumour microenvironmental adaptation and potentiates tumour metastasis. J. Cell. Mol. Med. 16: 3037–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leung D. W., Tompkins C. K., and White T.. 1998. Molecular cloning of two alternatively spliced forms of human phosphatidic acid phosphatase cDNAs that are differentially expressed in normal and tumor cells. DNA Cell Biol. 17: 377–385. [DOI] [PubMed] [Google Scholar]
- 66.Flanagan J. M., Funes J. M., Henderson S., Wild L., Carey N., and Boshoff C.. 2009. Genomics screen in transformed stem cells reveals RNASEH2A, PPAP2C, and ADARB1 as putative anticancer drug targets. Mol. Cancer Ther. 8: 249–260. [DOI] [PubMed] [Google Scholar]
- 67.Mori N., Natarajan K., Chacko V. P., Artemov D., and Bhujwalla Z. M.. 2003. Choline phospholipid metabolites of human vascular endothelial cells altered by cyclooxygenase inhibition, growth factor depletion, and paracrine factors secreted by cancer cells. Mol. Imaging. 2: 124–130. [DOI] [PubMed] [Google Scholar]
- 68.Ridgway N. D. 2013. The role of phosphatidylcholine and choline metabolites to cell proliferation and survival. Crit. Rev. Biochem. Mol. Biol. 48: 20–38. [DOI] [PubMed] [Google Scholar]
- 69.Beloribi-Djefaflia S., Vasseur S., and Guillaumond F.. 2016. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 5: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.