Abstract
Diabetic foot ulcers (DFUs) are a debilitating complication of diabetes in which bacterial presence, including its frequent colonizer Staphylococcus aureus, contribute to inhibition of healing. MicroRNAs (miRs) play a role in healing and host response to bacterial pathogens. However, the mechanisms by which miR response to cutaneous S. aureus contributes to DFU pathophysiology are unknown. Herein we show S. aureus inhibits wound closure and induces miR-15b-5p in acute human and porcine wound models, and in chronic DFUs. Transcriptome analyses of DFU tissue revealed induction of miR-15b-5p to be critical, regulating many cellular processes, including DNA repair and inflammatory response, by suppressing downstream targets IKBKB, WEE1, FGF2, RAD50, MSH2 and KIT. Using a human wound model we confirmed that S. aureus-triggered miR-15b-5p induction results in suppression of inflammatory- and DNA repair-related genes, IKBKB and WEE1. Inhibition of DNA repair and accumulation of DNA breaks was functionally confirmed by the presence of the pH2AX within colonized DFUs. We conclude that S. aureus induces miR-15b-5p, subsequently repressing DNA repair and inflammatory response, revealing a previously unreported mechanism of inhibition of healing in DFUs. This underscores a previously unknown role of DNA damage repair in pathophysiology of DFU colonized with S. aureus.
Introduction
The pathophysiology of diabetic foot ulcers (DFUs) is complex and poorly understood, making development of efficacious treatments challenging (Eming et al., 2014, Gregg et al., 2014). Diabetes mellitus (DM) (CDC 2014) is common and costly, with DFU costing approximately ~9-13 $billion/year (Rice et al., 2014), highlighting the unmet clinical need to better understand the mechanisms that impair healing in DFUs.
The development of DFUs is multifactorial, involving both extrinsic and intrinsic factors. External factors include wound colonization and infection, excessive pressure, and repeated trauma (Stojadinovic et al., 2012, Tsourdi et al., 2013), while intrinsic factors such as neuropathy and vascular insufficiency can alter the metabolic state/function of multiple cell types required for wound healing (Fadini et al., 2016, Hu et al., 2015, Maione et al., 2015, Tsourdi et al., 2013). A proper inflammatory response is essential to start the healing process, avoid infection, remove debris, and induce cell proliferation (Pastar et al., 2014, Yang et al., 2013), whereas dysregulated and prolonged inflammation can lead to tissue damage, inhibition of epithelialization and unresolved infection (Hu et al., 2015, Tsourdi et al., 2013, Yang et al., 2013). Patients with DFUs are highly susceptible to rapidly spreading infection that can lead to soft tissue damage, osteomyelitis and DFU-related amputations (Lavery et al., 2015, Miyajima et al., 2006). While Staphylococcus aureus is the prevalent pathogen in DFUs (Fadini et al., 2016, Gardner et al., 2013, van Asten et al., 2016), the mechanisms by which it may inhibit DFU healing remain largely unknown.
Bacterial pathogens can modulate host response through small non-coding regulatory RNAs, miRs, to establish and maintain infection (Chu et al., 2017, Eulalio et al., 2012, Kumar et al., 2016, Roy et al., 2014). However, miR response to S. aureus remains unexplored (Pastar et al., 2011, Tanaka et al., 2017). The miR-15/16 family was previously associated with bacterial infection in mice (Hsieh et al., 2012) and patients (Wang et al., 2015). Hence, we postulate that cutaneous wound colonization with S. aureus regulates the expression of miR-15 to contribute poor healing of DFUs. Indeed, we found that S. aureus induces host miR-15b-5p in both acute wounds and DFUs. S. aureus -mediated overexpression of miR-15b-5p resulted in suppression of DNA damage repair mechanisms and inflammatory response in DFUs. This effect was further confirmed by a comprehensive gene expression analysis from the DFU tissue colonized by S. aureus. Predictive analysis of upstream regulators of gene expression using Ingenuity Pathway Analysis (IPA) software identified miR-15b-5p as a master regulator in DFUs. Putative target genes of miR-15b-5p involved in DNA repair (WEE1, RAD50, MSH2 and KIT) and inflammatory response to wounding (IKBKB and FGF2) were down-regulated in DFUs. As a consequence of inhibition of DNA repair mechanisms, DNA double strand breaks (DSB) were accumulated in DFU tissue as indicated by the presence of pH2AX. Furthermore, overexpression of miR-15b-5p in keratinocytes or S. aureus infected human ex-vivo wounds down-regulated identified target genes. We found a novel mechanism of S. aureus-mediated induction of miR-15b-5p resulting in de-regulation of DNA repair mechanisms and inflammatory response, contributing to inhibition of healing in DFUs.
Results
miR-15b-5p is induced by S. aureus and is up-regulated in DFUs
Bacteria, including S. aureus, predominant in DFUs, can alter miR expression of host cells, such as the miR16/15 cluster (Cervantes-Garcia et al., 2015, Eleftheriadou et al., 2010, Hsieh et al., 2012, Jin et al., 2014). To test whether S. aureus induces miR-15b-5p expression, we used human ex-vivo and porcine in-vivo wound models (Pastar et al., 2012 , Stojadinovic and Tomic-Canic, 2013). MRSA presence in wounds resulted in inhibition of epithelialization, as observed with H&E and keratin 6 (K6) immunostaining (Figure 1a-b) (Pastar et al., 2013). Presence of S. aureus up-regulated miR-15b-5p expression in both, human ex-vivo and in porcine in-vivo acute wounds (Figure 1c). In contrast, wounding itself without presence of bacteria resulted in suppression of miR-15b-5p expression (Figure 1d). In addition, infection of in vivo porcine wounds with Pseudomonas aeruginosa (a common gram negative chronic wound colonizer) did not cause changes in miR-15b-5p expression (Figure S2a).
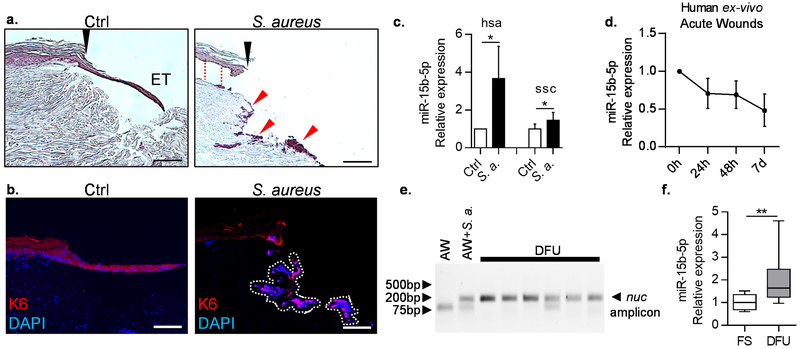
Figure 1. S. aureus infection of acute wounds induces miR-15b-5p.
a. Histology shows acute wounds 4 days post-wounding and infection of acute ex-vivo human wound model. Black arrows indicate initial site of wounding, ET = epithelial tongue; red arrows indicate bacterial aggregates. Separation of epidermis from dermis was observed in S. aureus infected wounds (red dashed line). b. Immuno-localization of K6 (red) at the wound edge and epithelial tongue upon infection with S. aureus isolate from DFU in the human skin ex-vivo model demarcates epithelialization. Nuclei are visualized with DAPI (blue). White dashed lines indicate DAPI stained bacterial aggregates. Scale bars: 50μm. c. qPCR showing up-regulation of miR-15b-5p a in S. aureus infected human (hsa, Homo sapiens) ex-vivo wounds in comparison to control wounds 4 days post infection and MRSA USA300 infected pig (ssc, Sus scrofa) wounds in vivo 2 days post infection. Paired and unpaired t-test were used respectively to determine statistical significance (n=3 independent experiments, * = p-value<0.05). d. qPCR showing suppression of miR-15b-5p in human uninfected acute wound model. Data is normalized to the 0h time point at which wounds were created (n=3 independent experiments) e. Agarose gel showing the amplification of the nuc gene in DFU samples. AW= human acute ex-vivo wound (negative control), AW + S. a. = human acute ex-vivo wound infected with S. aureus (positive control). f. qPCR showing up-regulation of miR-15b-5p in DFU (n=12 per group, FS = foot skin, Mann-Whitney U-test was used to determine statistical significance, ** = p-value<0.01).
To determine whether miR-15b-5p is induced in DFUs colonized with S. aureus, we collected full thickness tissue samples from the wound edge of DFUs, and controls of non-ulcerated human foot skin (FS) (Ramirez et al., 2015). All DFU samples had histopathological characteristics of chronic wounds (Stojadinovic et al., 2005, Stojadinovic et al., 2013) (Figure S1). We developed and successfully utilized an approach to identify S. aureus in formalin fixed-paraffin embedded (FFPE) DFU tissue. Total DNA from DFU FFPE blocks was extracted and the presence of the S. aureus-specific thermonuclease gene (nuc) (Redel et al., 2013) was analyzed by TaqMan qPCR. As expected, even without clinical signs of infection, all DFU tissue samples were positive for the nuc, which confirms colonization by S. aureus, whereas nuc gene was not amplified in control samples (Figure 1e). Furthermore, we found induction of miR-15b-5p in these DFU tissue samples (Figure 1f). We have previously shown that the epidermal cells are major contributors of the gene expression observed in full thickness biopsies (Ramirez et al., 2015). Thus, we performed laser capture microdissection of the epidermis of DFUs and generated PCR based miR arrays (data not shown) to compare it to previously published FS epidermal miR profiles (Ramirez et al., 2015), and confirmed that miR-15b-5p is up-regulated in the epidermis of DFUs in comparison to FS (Figure S2c). We conclude that S. aureus triggers miR-15b-5p induction, thus altering the host miR response to wounding.
Gene expression profiles of DFUs and functional analysis identify functional enrichment of DNA repair genes
To elucidate transcriptional alterations in DFUs and identify miR-15b-5p target genes, we used the Affymetrix Hugene 2.0 microarrays to perform a differential gene expression analysis between DFU (n=6) and FS (n=6) specimens. We identified 3900 de-regulated genes in DFUs (2-fold or more, False Discovery Rate (FDR) ≤0.05), with 75% (2829 genes) being suppressed (Figure 2a) (Gene Expression Omnibus (GEO) accession GSE80178). We then used the Ingenuity Pathway Analysis software (IPA) (Kramer et al., 2014) to find enriched cellular functions deregulated in DFUs. The most affected categories were DNA repair, cell death/survival, and regulation of gene expression, all of which were strongly suppressed in DFUs (Figure 2b). Multiple processes needed for successful wound healing such as cellular movement, cellular growth and proliferation, and the inflammatory response were also deregulated in DFU tissue (Table S1).
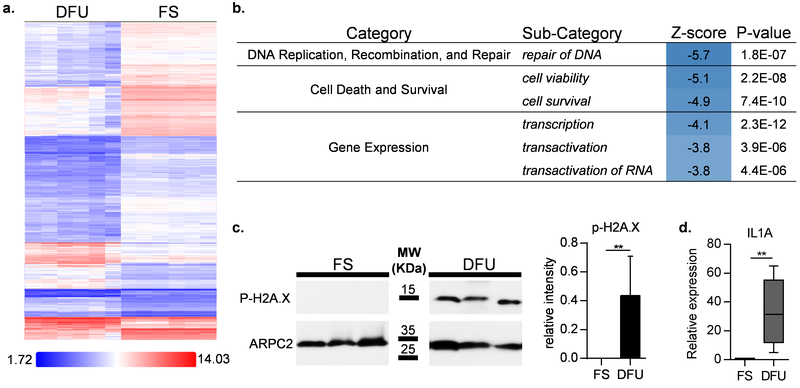
Figure 2. Microarray analysis of differentially expressed genes in DFUs vs. FS and functional enrichment analysis.
a. Heatmap of differentially expressed genes in DFUs vs. FS. The majority of DFU regulated genes were suppressed (75%). b. Top enriched functions from IPA functional enrichment analysis performed on DFU regulated genes identifies DNA repair mechanisms as the top regulated. Z-scores (activation score) represent the predicted directionality of the enriched function (Z-scores ≥ 2 are considered as induced and ≤ −2 are considered as suppressed). c. Functional assessment of DNA repair suppression was perform by quantification of pH2AX using western blot. pH2AX confirms the presence of DNA double strand breaks in DFUs, but not control FSs. Quantification of all samples shown in bar graph (n=5 per group) d. qPCR showing IL1A, a pro-inflammatory cytokine that can be induced by DNA damage, is induced in DFUs (n=5 per group, Mann-Whitney U-test was used to determine statistical significance, * = p-value<0.05, ** = p-value < 0.01).
Given the suppression of the DNA repair associated genes, we evaluated the presence of double-strand DNA breaks in DFU tissue. We detected high levels of phosphorylated-H2AX (pH2AX), a marker for DNA damage, in DFUs but not in control non-ulcerated FS, confirming the presence of double-stranded DNA breaks in DFUs (Figure 2c). Consistent with the presence of double-stranded DNA breaks in DFUs, immunoperoxidase staining of pH2AX revealed a strong nuclear staining in the epidermis of DFUs but not in FS (Figure S3).
IL1A, a direct sensor of DNA damage that acts as a triggering signal of tissue repair and inflammation upon DNA breaks (Idan et al., 2015) was found induced in these DFUs, measured by qPCR (Figure 2d), suggesting this is a downstream consequence of increased DNA damage detected in DFUs.
Network analysis and microarray validation independently confirm miR15b-5p as a master regulator in DFUs, targeting DNA repair and inflammation
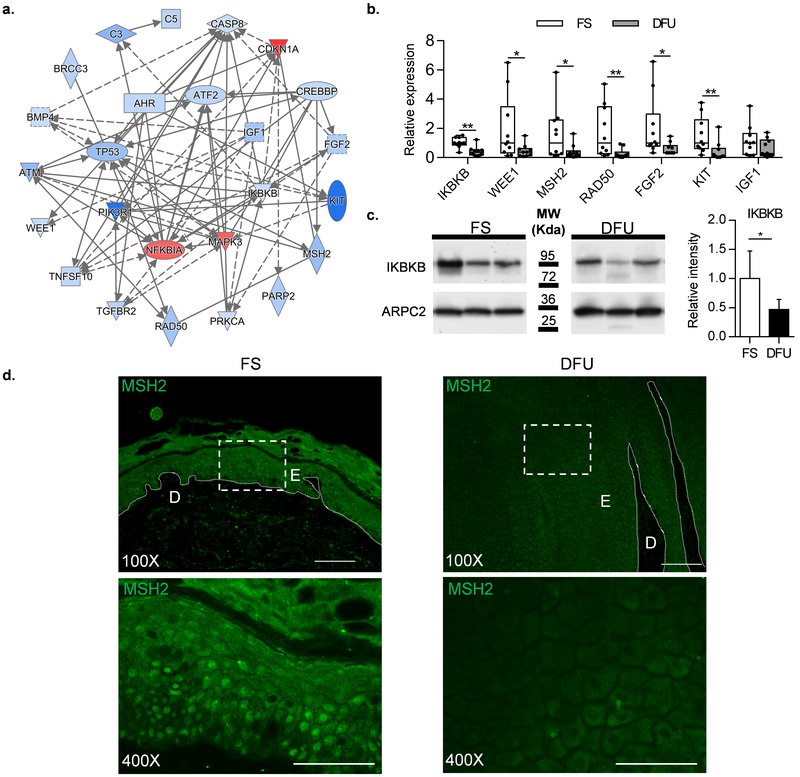
To identify the genes and their regulators driving overall gene expression suppression and down-regulation of DNA repair in DFU, we generated a regulatory network using IPA tools, comprising enriched upstream regulators and their target genes (Figure. 3a). Multiple transcription factors and genes belonging to signaling pathways were suppressed in DFUs, including inhibitor of nuclear factor kappa B kinase subunit beta (IKBKB), activating transcription factor 2 (ATF2), and genes involved in DNA repair (MutS Homolog 2-MSH2; Double Strand Break Repair Protein-RAD50; WEE1 G2 Checkpoint Kinase-WEE1; Tumor Protein P53-TP53; Proto-Oncogene Receptor Tyrosine Kinase-KIT; fibroblast growth factor 2-FGF2). The array data were confirmed by qPCR in 10 DFU tissue samples (Figure 3b). We also validated suppression of IKBKB, a key regulator of the NFKB pathway and inflammatory response, as well as MSH2, an essential DNA repair gene at the protein level using western blot and immunostaining, respectively (Figure 3c and 3d; corresponding lower magnification images and histology of these sections are shown in Figure S4). While normal skin exhibited strong nuclear staining of MSH2, overall staining was reduced in DFUs, further confirming suppression of DNA repair mechanisms in DFUs
Figure. 3. Network analysis and validation of DFU de-regulated genes.
a. Network analysis of the genes deregulated in DFUs was built using IPA focusing on genes with known regulatory functions (blue=suppression; red=induction). b. qPCR validation of microarray data. IKBKB, WEE1, MSH2, RAD50, FGF2, and KIT showed significant down-regulation in DFU compared to FS. IGF1 showed a trend of down-regulation but it did not reach statistical significance. c. Western blot confirming suppression of IKBKB in DFU compared to FS; graph represents quantification of all samples (n=5 per group). d. Immuno-fluorescence staining showing decreased expression of MSH2 in DFUs compared to FS. E=epidermis; D=Dermis; 100X Scale bars: 100 μm, 400X Scale bars: 50 μm, (n=10 per group, Mann-Whitney U test was used to determine statistical significance, * = p-value<0.05, ** = p-value < 0.01, *** = p-value < 0.001).
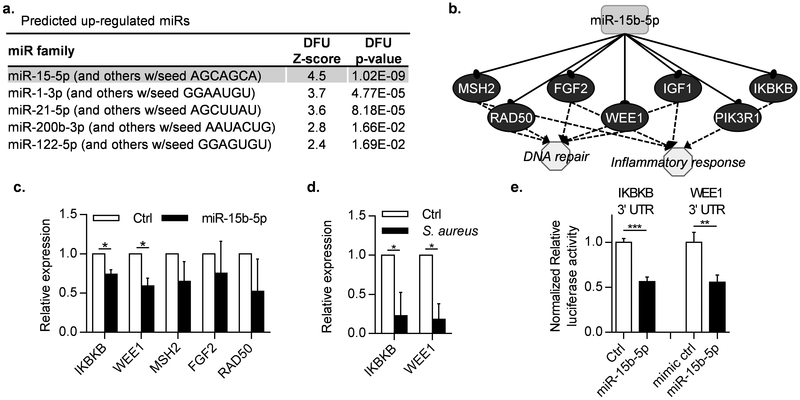
To predict potential miRs as likely upstream regulators of gene expression in DFUs we used upstream regulator analysis tools in IPA (p ≤ 0.05). This approach identified 8 miRs predicted to be up-regulated in DFUs (Figure 4a), with miR-15b-5p being the top predicted (Z-score=4.5, p=1.02*10−9). IPA predicted miR-15b-5p targeting of multiple genes within the DFU network including MSH2, RAD50, FGF2, WEE1, IGF1 and IKBKB (Figure 3b), supporting the notion that S. aureus induces miR15b-5p that in turn downregulates DNA damage repair response and dysregulates the inflammatory response.
Figure 4. miR-15b-5p, predicted to be induced in DFUs, and suppression of its targets, DNA repair and inflammatory regulatory genes.
a. IPA identified miR-15b-5p as the top enriched miR predicted to target DFU regulated genes from microarrays. b. miR-15b-5p predicted targets comprise regulators of the inflammatory response and genes involved in DNA repair. c. IKBKB and WEE1, are down-regulated in HaCaT cells transfected with mimic miR-15b-5p. MSH2 and RAD50 showed a trend of down-regulation but did not reach statistical significance. ctrl = mimic negative control #1. d. IKBKB and WEE1 are down-regulated in S. aureus infected human ex-vivo wounds in comparison to control un-infected wounds. (n=3 independent experiments, paired t-test was used to determine statistical significance, * = p-value<0.05, ** = p-value < 0.01). e. Luciferase reporter assay showing miR-15b-5p directly targets the 3’UTRs of IKBKB and WEE1. ctrl = mimic negative control #1. (n=3, unpaired t-test was used to determine statistical significance, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001).
To test whether miR-15b-5p regulates the expression of target genes dysregulated in DFUs, we transfected human keratinocytes with either the synthetic mimic miR-15b-5p or a control mimic-miR. We confirmed down-regulation of IKBKB, and WEE1 in keratinocytes overexpressing miR-15b-5p (Figure 4c). In addition to inducing miR-15b-5p in vitro, we used S. aureus to induce miR-15b-5p in human ex-vivo acute wounds and confirmed down-regulation of its targets, IKBKB and WEE1 (Figure 4d). Lastly, to validate IKBKB and WEE1 direct targeting by miR-15b, we constructed pmiRGLO luciferase reporter vectors with the 3’UTRs of IKBKB or WEE1, which were co-transfected with either miR-15b-5p or mimic-miR control. We found a significant reduction in the luciferase signal in the case of both, 3’UTR IKBKB and WEE1 constructs, unique to the cells co-transfected with miR-15b-5p, thus confirming direct targeting (Figure 4e).
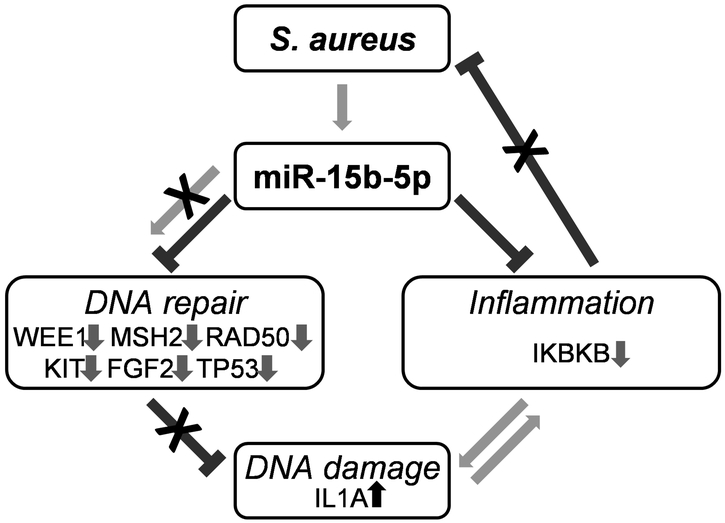
We conclude that S. aureus induces expression of miR15b-5p and corresponding suppression of its downstream target genes involved in DNA repair and inflammatory response in infected acute wounds in vivo, ex-vivo and in patients with DFUs revealing to our knowledge previously unreported mechanism that contributes to inhibition of wound closure (Figure 5).
Figure. 5. Diagram summarizing how miR-15b-5p orchestrates processes involved in pathophysiology of DFUs.
Persistent S. aureus colonization in DFUs leads to over-expression of miR-15b-5p resulting in increased DNA damage through suppression of WEE1 and chronic sub-optimal inflammation by targeting IKBKB. DNA repair mechanisms are further inhibited by down-regulation of multiple genes in DFUs including MSH2, RAD50, KIT, FGF2 and TP53. DNA damage feeds into a positive feedback loop by causing the release of IL-1A.
Discussion
Chronic wounds, including those without clinical signs of infection, are commonly colonized by both commensal and pathogenic bacteria (Bowler et al., 2001, Frank et al., 2005). S. aureus and other skin commensal bacteria can elicit cutaneous response in intact skin and also become pathogenic when there are skin barrier defects such as chronic ulcers or dermatitis (Dunyach-Remy et al., 2016, Nakatsuji et al., 2016). Here we describe, to the best of our knowledge, previously unreported mechanism by which presence of S. aureus in the wound triggers induction of miR-15b-5p, diminishing DNA repair and de-regulating inflammatory response in patients with DFUs. Although, this may not be unique to S. aureus, another common colonizer of chronic wounds, Pseudomonas aeruginosa did not show similar effect, suggesting some specificity. We based these conclusions on the data from acute human ex-vivo and porcine in-vivo wound models, and comprehensive genomic analyses of DFU tissue samples colonized with S. aureus. To the best of our knowledge, this mechanism has not been implicated in pathophysiology of DFU and aids in better understanding of DFUs, allowing new insights for more precise therapeutic approaches. It is known that DFUs are prone to infection (Mancl et al., 2013, Zhao et al., 2013) and are colonized by S. aureus (Cervantes-Garcia et al., 2015, Eleftheriadou et al., 2010, Tentolouris et al., 1999). Although mechanisms of MRSA-mediated inhibition of healing in animal models are being studied (Pastar et al., 2013), the contribution of S. aureus to DFU chronicity and inhibition of healing is not fully understood. Bacteria can enact changes in miR expression in cells (Jin et al., 2014, Naeem et al., 2012). Additionally, it has been shown that reactive oxygen species (ROS), for example triggered by bacteria in the ulcers, can induce the miR-15 family (Dhall et al., 2014, Dunnill et al., 2015, Santosa et al., 2015). Therefore, S. aureus-mediated induction of miR-15b-5p found in DFUs and infected acute wounds suggests this mechanism of action to inhibit healing. Our data further suggest that the persistent colonization of S. aureus, and consequent miR-15b-5p overexpression, may lead to more severe deep tissue infection by causing detrimental effects on DNA repair mechanisms and the inflammatory response, resulting in accumulation of DNA damage and perturbed inflammatory conditions detrimental to healing of DFUs (Figure 5).
We propose impaired DNA repair, previously unlinked to chronic wounds, as an additional culprit in the dysfunctional healing processes central to DFU formation and persistence. Taking advantage of the upstream regulator prediction built in IPA and its building tools to create a network connecting and expanding upstream regulator genes interactions that were de-regulated in DFUs, highlights miR-15b-5p as one of the ‘master’ regulators. Confirmation of down-regulation of expression of KIT, MSH2, RAD50, FGF2, WEE1, and IKBKB and particularly WEE1, RAD50 and FGF2, underscores DNA repair process in DFU pathophysiology (Cannon et al., 2013, Do et al., 2013, Guertin et al., 2013, Harfouche et al., 2010). RAD50 directly participates in the initiation and coordination of DSB repair (Cannon et al., 2013), whereas WEE1 and FGF2 are required for successful repair process. WEE1 is a DNA damage G2-M checkpoint kinase responsible for stopping DNA replication and arresting the cell cycle after DNA damage (Do et al., 2013, Guertin et al., 2013). Furthermore, FGF2 signaling pathway has been shown to be induced in keratinocyte progenitor cells upon irradiation, and its abrogation resulted in increased DNA damage (Harfouche et al., 2010). MSH2 is a part of two heterodimeric complexes MutSα (with MSH6) and MutSβ (with MSH3) that recognize and repair DNA mismatches or small insertion deletion loops, respectively. MSH2 mutations lead to deficiency in DNA repair and genomic instability (Diouf et al., 2011). miR-15b-5p induction has previously been linked to DNA breaks and increased pH2AX levels in cancer cells through targeting of WEE1 (Mei et al., 2015). Interestingly, in spite of suppression of DNA repair, accumulation of DSB often seen in cancer and previously reported activation of b-catenin and c-myc, DFUs rarely develop cancers (Park et al., 2016). Suppression of all of these miR-15b-5p target genes together with confirmed accumulation of DSB indicates accumulation of danger-associated molecular patterns that can contribute to chronic inflammation in DFU triggered by S. aureus colonization.
Other wound healing processes were also enriched in the list of DFU regulated genes, but did not exhibit clear positive or negative regulation. For example, it was expected that inflammatory response would be highly activated, and although significantly enriched (p=2.53×10−6), it was not clearly induced (Z-score=1.4) (table S1). Unresolved sub-optimal inflammatory response was also found in another type of chronic wounds, venous leg ulcers (Stone et al., 2017). Findings we present here support the notion that de-regulation of inflammatory response is what contributes to chronic presence of not very effective inflammation. We confirmed suppression of IKBKB on both mRNA and protein levels. IKBKB is a positive regulator of the inflammatory response and an important upstream modulator of NF-kB pathway. Its inhibition reduces the inflammatory response and pro-inflammatory cytokine production in response to LPS (Novoselova et al., 2014), and may contribute to diminished ability of DFUs to respond to pathogens. By suppressing the key regulator gene IKBKB, S. aureus-induced miR-15b-5p may affect multiple signaling pathways including NF-KB, IL6, IL8, IL12, PI3K/AKT, and PTEN (Ding et al., 2013, Li et al., 2010) contributing to suppression of the inflammatory phase of wound healing, which is required for a proper healing response and clearing bacterial contamination. On the other hand, unresolved DNA damage contributes to induction of IL1A in DFUs (Idan et al., 2015). One can speculate that such mixed signals, which result in the opposite regulation of inflammatory response contribute to clinically observed, perpetual and detrimental inflammation in DFUs that does not progress to proliferative phase of healing and contributes to inhibition of healing (Figure 5).
Conclusion
Induction of miR-15b-5p by S. aureus contributes to impaired DNA repair mechanisms, leading to accumulation of DSB, which subsequently serves as a facilitator for the persistent, unresolved inflammatory state found in DFUs. Taken together, S. aureus colonization can have central effect to DFU pathophysiology highlighting miR-15b-5p as one of major regulators that can serve as a potential therapeutic target and/or a biomarker.
Materials and Methods
Tissue collection
Full thickness skin/DFU samples were obtained from consenting patients receiving standard care at the University of Miami Hospital. These protocols including written informed consent were approved by the university Institutional Review Board (IRB protocols #20140473;#20090709). Patient demographics and sample characteristics are included in Supplemental Table S2. Ulcers did not have any clinical signs of infection. Biopsies were stored in RNALater (Applied Biosystems, Carlsbad, CA) for RNA isolation, snap frozen (protein isolation), or fixed in formalin (paraffin embedding).
DNA extraction from formalin fixed paraffin embedded DFU tissue (FFPE) and detection of S. aureus by PCR.
Total DNA from DFU FFPE blocks, was extracted from 3 sections (10μm thick) using the GeneRead DNA FFPE kit (QIAGEN, Valencia, CA) following manufacturer’s instructions. PCR (qPCR) reactions were performed in triplicate using 2X RT-PCR reaction mix for probes (Bio-Rad, Hercules, CA). The primers used were the following: Nuc-F-162 GTTGTAGTTTCAAGTCTAAGTAGCTC, Nuc-R-162 AACCGTATCACCATCAATC, Nuc-probe-162 ATCCAACAGTATATAGTGCAAC (Redel et al., 2013).
RNA extraction and quality control
Total RNA, including the miR fraction, was extracted using the miRNeasy kit (QIAGEN, Valencia, CA) per manufacturer’s instructions. RNA quality was assessed using the AGILENT bioanalyzer (Agilent Technologies, Palo Alto, CA) to estimate the RNA integrity number (RIN). Samples with a RIN>5 were used for mRNA profiling.
mRNA profiling of DFU tissue analysis
Transcriptome profiling was performed using Affymetrix GeneChip Human Gene 2.0 ST microarrays. CEL files alongside previously published control foot skin (FS) (GSE68186, (Ramirez et al., 2015)) were imported and processed into Expression Console™ Software (Affymetrix, Santa Clara, CA) for gene level normalization and signal summarization using default parameters. Control samples of foot skin were profiled and described previously (Ramirez et al., 2015). Very minor gene expression differences between diabetic and non-diabetic foot skin were found and both were used as controls. The output files were analyzed in Transcriptome Analysis Console (TAC) 3.0 Software to identify differentially expressed genes between DFU and control FS, and carry out clustering analysis. Genes with an FDR≤0.05 and a fold-change ≥ 2 in any direction were considered differentially regulated. The microarray profiles of DFU were deposited in GEO database, GSE80178.
IPA network and upstream regulator analysis
The list of differentially regulated genes with their fold change expression was imported into Ingenuity Pathway Analysis software (IPA, QIAGEN). A core analysis was carried out on the data set, which includes functional enrichment analysis and upstream regulator prediction. Both analyses include activation scores (Z-scores) prediction, which serves as an indication whether a function or an upstream regulator gene is activated or suppressed based on known literature incorporated into IPA database (Kramer et al., 2014). Gene networks were built using the IPA builder tools, connecting and expanding the interactions between the top predicted upstream regulators also found to be regulated in the microarray data, and discarding conflicting interactions.
Real-time qPCR gene expression and miR analysis
cDNA was made with qScript™ Synthesis kit (Quanta BioSciences Inc.,Gaithersburg, MD). ARPC2 was used as a reference gene for normalization. All real-time PCR (qPCR) reactions were performed in triplicate using PerfeCTa® SYBR® Green SuperMix (Quanta BioSciences) and quantified using the ddCT method. The primer sequences are listed in Table 1.
Table 1.
List of primers and primer sequences used for qPCR.
| Gene Symbol | Forward primer sequence (5' -> 3') | Reverse primer sequence (5' -> 3') |
|---|---|---|
| IKBKB 3’ UTR | AAAGAGCTCAGTGCTTGGAGTACGGTTTG | CCCTCTAGACACACACAATCAGCAGGAG |
| WEE1 3’ UTR | AAAGAGCTCCCTGAACACTGTGACAAGA | CCCTCTAGAAGTCAAAGACAAGTGCAAACA |
| IKBKB | CTGGCCTTTGAGTGCATCAC | CGCTAACAACAATGTCCACCT |
| ATF2 | CTGGCCTTTGAGTGCATCAC | CGCTAACAACAATGTCCACCT |
| FGF2 | AGTGTGTGCTAACCGTTACCT | ACTGCCCAGTTCGTTTCAGTG |
| IGF1 | GCTCTTCAGTTCGTGTGTGGA | GCCTCCTTAGATCACAGCTCC |
| IL1A | AGATGCCTGAGATACCCAAAACC | CCAAGCACACCCAGTAGTCT |
| MSH2 | AGTCAGAGCCCTTAACCTTTTTC | GAGAGGCTGCTTAATCCACTG |
| WEE1 | AACAAGGATCTCCAGTCCACA | GGGCAAGCGCAAAAATATCTG |
| RAD50 | TACTGGAGATTTCCCTCCTGG | AGACTGACCTTTTCACCATGC |
| TP53 | GAGGTTGGCTCTGACTGTACC | TCCGTCCCAGTAGATTACCAC |
| KIT | CGTTCTGCTCCTACTGCTTCG | CCCACGCGGACTATTAAGTCT |
| ARCP2 | TCCGGGACTACCTGCACTAC | GGTTCAGCACCTTGAGGAAG |
The qScript™ microRNA Quantification System (Quanta BioSciences, Inc.) was used for miR expression using cDNA generated by the qScript™ microRNA cDNA Synthesis Kit. 50 pg of initial RNA/PCR reaction was used on a CFX Connect™ qPCR Detection System (Bio-rad, Hercules, CA). SNORD48 was used as a reference gene to normalize miR expression.
Protein isolation and western blot
Proteins were extracted using Tissue-PE LB Kit (Geno technology, MO). Membranes were probed with anti-pH2AX Ser139 rabbit mAb (Cell Signaling, Danvers, MA) and anti-IKBKB rabbit polyclonal antibody (Sigma-Aldrich, St. Louis, MO). Membranes were incubated with their appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling) and developed using an enhanced chemoluminescence detection system (Amersham Biosciences, Arlington Heights, IL). Anti-ARPC2 antibody was used as a loading control (Santa Cruz, Dallas, TX) and quantified with Image lab software (Bio-Rad).
Immunofluorescence and immunohistochemistry
5-7 μm thick formalin-fixed, paraffin-embedded tissue sections were deparaffinized with xylene (EMD, Gibbstown, NJ), rehydrated, and H&E stained or processed for immunostaining as previously described (Ramirez et al., 2015). Anti MSH2 (H-300) rabbit polyclonal antibody (Santa-Cruz) was used at 4°C, overnight or anti K6 rabbit polyclonal antibody (gift from Dr. P. Coulombe, Johns Hopkins University) (Hutton et al., 1998). Signal was visualized using Alexa-Fluor 488 secondary antibody (Invitrogen, Carlsbad, CA) and mounted with media containing 4’,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) to visualize cell nuclei.
5-7 μm thick frozen tissue sections were fixed in cold acetone (−20°C) for 10 min washed in TBS (Tris-buffered saline) and incubated with 3% H2O2 in methanol for 10 min. Staining was performed as previously described (Ramirez et al., 2015), using anti-pH2AX Ser139 rabbit mAb (Cell Signaling, Danvers, MA) diluted 1:100 in 3% BSA in TBS with 0.3% triton X-100, Rabbit-on-Farma HRP-Polymer (Biocare Medical) and developed using Betazoid DAB chromogen kit (Biocare Medical). Nikon Eclipse E800 microscope/NIS Elements BR 3.2 software was used for collection of digital images.
Laser capture microdissection (LCM) and miR expression analysis
LCM was performed as previously described (Ramirez et al., 2015). Briefly, formalin-fixed paraffin embedded tissue blocks from DFU, were sliced into 16 to 20 sections (8-10μm thick), collected on Arcturus PEN-membrane glass slides (Life Technologies, Carlsbad, CA) and dried at 37°C for 1-2 hours. LCM was carried out on an Arcturus Veritas laser capture microdissection instrument and the epidermis was captured on CapSure® Macro LCM Caps (Life Technologies). Caps were then transferred into a tube containing 60μl of deparaffinization buffer (QIAGEN Inc., Valencia, CA) and total RNA, including the miR fraction, was extracted using the FFPE miRNeasy kit (QIAGEN Inc.) according to the manufacturer’s instructions.
The RNA concentration of the samples was measured using a NanoDrop 2000 (NanoDrop products, Wilmington, DE). MiR profiles of epidermis of 6 DFUs were generated using the miR Ready-to-Use PCR panels V2 (Exiqon) following the manufacturer’s specifications. miR-15b-5p expression was calculated using the ΔΔCT method with SNORD49 as reference, and comparing the CTs of DFUs and previously published FS (NFS and DFS) array data (Ramirez et al., 2015).
Cell culture and transfection
The human keratinocyte cell line, HaCaT, (AddexBio, San Diego, CA) was cultured in calcium-free DMEM (Gibco, Life technologies, Carlsbad, CA), supplemented FBS, 10% v/v (GE Hyclone Laboratories, Logan, UT) and penicillin-streptomycin with L-glutamine (Gibco). Cells were seeded in 24-well plates for 24h before transfection. Attractene reagent (QIAGEN) and 50nM of miR-15b-5p mimic or mimic negative control #1 (GE Dharmacon, Lafayette, CO) was used for transfection. Cells were harvested after 48h for RNA extraction.
The 3’UTRs of the WEE1 and IKBKB human genes were amplified by PCR from human genomic DNA with primers containing restriction sites for SacI and XbaI (Table 1) and cloned into the pmirGLO Dual-Luciferase miRNA Target expression vector (Promega Corporation, Madison, WI). 100ng of vector containing the UTRs and 25nM of miR-15b-5p mimic or mimic negative control #1 was used for transfections. Luciferase reporter assay was done using the Dual-Glo® Luciferase Assay system (Promega Corporation) 48h after transfection using manufacturer’s protocol.
Bacterial strains and growth conditions
Methicillin-resistant S. aureus (MRSA) USA300-0114 (Pastar et al., 2013), USA300 JE2 (Fey et al., 2013), and S. aureus isolate from the DFU were used. Todd-Hewitt broth supplemented with 0.2% yeast extract (THY) served as growth medium; oxacillin resistance screening agar base (OXOID, Carlsbad, CA) was used as selective media for MRSA colony forming units (CFU) quantification. P. aeruginosa 09-010 (Pastar et al., 2013) was grown in tryptic soy broth (TSB), while Pseudomonas Agar with CN supplement (Oxoid) was used as selective media for plate-counting.
Human ex vivo wound infection model
Healthy human skin samples were used to generate acute wounds. Wounded skin specimens were maintained at the air-liquid interface (Pastar et al., 2012, Pastar et al., 2010, Stojadinovic and Tomic-Canic, 2013). S. aureus isolate from DFU or MRSA USA300 JE2 were grown overnight in THY medium at 37 °C, then harvested by centrifugation, washed and re-suspended in DMEM. The bacterial density was adjusted to an OD600 of 0.1, corresponding to 0.8–1.2×108 colony-forming units (CFU)/ml. 10 μl of 106 CFU/mL bacterial stock solution was used for the wound inoculation. Infected and non-infected (control) skin samples were incubated at 37°C, 5% CO2 for four days and CFUs were quantified. Tissues were either used for CFU determination, fixed in 4% paraformaldehyde (Sigma-Aldrich) for H&E and histo-morphometric analyses (Pastar et al., 2012, Stojadinovic and Tomic-Canic, 2013), or preserved in RNAlater (Ambion) for RNA isolation.
Porcine wound infection model and RNA extraction from FFPE tissue samples
Wounding and infection of the animals was carried out as described previously (Pastar et al., 2013). Briefly, the back of the animals were prepped on the day of the experiment. They were anesthetized and wounded (partial thickness wounds 10 mm×7 mm) on the paravertebral area using a modified electrokeratome set at 0.5 mm deep (Pechter et al., 2012). The wounds were separated from each other by approximately 5 cm. Infection was performed by inoculating 25 μl of 106 CFU/mL of MRSA USA300 or P. aerugionsa 09-010 immediately after wounding. Wounds were covered with a polyurethane film dressing (Tegaderm; 3 M Health Care, St. Paul, MN) and secured in place by wrapping the animals with self-adherent bandages (Petflex; Andover, Salisbury, MA). Incisional biopsies from three infected and three non-infected wounds from 2 animals were obtained by creating a 3mm thick section cut through the central part of the wound, including 5mm of adjacent uninjured skin on both sides, at day 2 post-wounding. The biopsies were fixed and processed for paraffin embedding. Total RNA, including the microRNA fraction, was extracted from six 10μm sections using the FFPE miRNeasy kit (QIAGEN) according to the manufacturer’s instructions.
Statistical analyses
Data were analyzed using the software Prism (Graphpad, La Jolla, CA). Statistical significance between groups was determined using Student’s t-test or Mann-Whitney U-test. A difference between groups was considered significant at p-value ≤ 0.05.
Supplementary Material
Figure S1. Representative H&E staining of full thickness biopsies from DFU and FS. DFU samples exhibit a thicker epidermis and cornified layer than normal foot skin. E= Epidermis, D= Dermis. Scale bars: 500μm.
Figure S2. a. qPCR expression data showing miR-15b-5p is up-regulated in porcine in-vivo acute wounds by infection with S. aureus but not with P. aeruginosa. (n=6 wounds per group) b. Representative DFU section before and after LCM. E= Epidermis, D= Dermis. c. qPCR showing up-regulation of miR-15b-5p in the epidermis of DFU (n=6 per group, FS = foot skin, unpaired t-test was used to determine statistical significance, ** = p-value<0.01, n.s. = not significant).
Figure S3. Representative Immuno-peroxidase stainings of pH2AX in FS and DFU biopsies. pH2AX is expressed at low levels throughout the epidermis of FS and is induced DFUs, primarily in the epidermis of DFUs. −Ab ctrl = no primary antibody control. Scale bars: 200μm.
Figure S4. H&E staining and 40X visualization of the sections shown in Figure 3. Scale bars: 500 μm. E= Epidermis, D= Dermis. Dashed box represents the picture areas shown in Figure 3.
Figure S5. miR-15b-5p target gene expression shows corresponding suppression when compared to either controls (diabetic foot skin or non-diabetic foot skin). Each dot represents an individual patient’s sample. Distribution of microarray data for IKBKB, WEE1, MSH2, RAD50, FGF2, KIT and IGF1 expression demonstrates clustering and segregation of DFUs from diabetic foot (DFS) and normal foot skin (NFS). DFU=diabetic foot ulcer (n=6), DFS (n=3), NFS= (n=3).
Table S1. List of the functions enriched in DFU.
Table S2. Patient demographics, and sample information. AA = African-American, A= Asian, H = Hispanic, HW = Hispanic White, W = White. DFU=diabetic foot ulcer, FS= foot skin, diabetic and non-diabetic.
Acknowledgments
M.T.C., I.P., and H.R. designed research; H.R., I.P., I.J., O.S., N.O., J.G. and R.C.S. performed research; SCD, R.S.K., R.C.S., and I.P. contributed new reagents/analytic tools; M.T.C., H. R., I.P., R.C.S., I.J. O.S. and R.S.K. analyzed data; and M.T.C., I.P., and H.R. wrote the paper. We are very grateful to patients who generously participated in this research. We are also very grateful to the Wound Healing Clinical Research Team for their help and support throughout this project. We are grateful to Gregory V. Plano for the gift of S. aureus USA300 JE2, Ashley M. Rosa, Karen Garzon and Vivien Chen from the University Of Miami Miller School Of Medicine for the technical assistance, all members of MT-C laboratory for helpful criticisms and overall support, and to Barrientos Lab for technical support. M.T.C. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. This work was funded by NIH grants NR013881 (MT-C), DK086364 (MT-C), NR015649 (MT-C), University of Miami SAC Award SAC 2013-19 (MT-C) and SAC 2013-06 (IP).
Footnotes
Conflict of interest
Authors declare no conflicts of interest.
References
- Centers for Disease Control and Prevention. Centers for Disease Control and Prevention. National diabetes statistics report: estimates of diabetes and its burden in the United States, http://www.cdc.gov/diabetes/pubs/statsreport14/national-diabetes-report-web.pdf; 2014. (accessed 5 September 2017).
- Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001;14(2):244–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Kuhnlein J, Yang SH, Cheng A, Schindler D, Stark JM, et al. Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET. Proceedings of the National Academy of Sciences of the United States of America 2013;110(47):18868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Garcia E, Garcia-Gonzalez R, Resendiz-Albor A, Salazar-Schettino PM. Infections of diabetic foot ulcers with methicillin-resistant Staphylococcus aureus. Int J Low Extrem Wounds 2015;14(1):44–9. [DOI] [PubMed] [Google Scholar]
- Chu Q, Sun Y, Cui J, Xu T. Inducible microRNA-214 contributes to the suppression of NFkB mediated inflammatory response via targeting MyD88 in fish. The Journal of biological chemistry 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhall S, Do D, Garcia M, Wijesinghe DS, Brandon A, Kim J, et al. A novel model of chronic wounds: importance of redox imbalance and biofilm-forming bacteria for establishment of chronicity. PLoS One 2014;9(10):e109848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Huang S, Wang Y, Tian Q, Zha R, Shi H, et al. Genome-wide screening reveals that miR-195 targets the TNF-alpha/NF-kappaB pathway by down-regulating IkappaB kinase alpha and TAB3 in hepatocellular carcinoma. Hepatology 2013;58(2):654–66. [DOI] [PubMed] [Google Scholar]
- Diouf B, Cheng Q, Krynetskaia NF, Yang W, Cheok M, Pei D, et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat Med 2011;17(10):1298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do K, Doroshow JH, Kummar S. Wee1 kinase as a target for cancer therapy. Cell cycle 2013;12(19):3159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnill C, Patton T, Brennan J, Barrett J, Dryden M, Cooke J, et al. Reactive oxygen species (ROS) and wound healing: the functional role of ROS and emerging ROS-modulating technologies for augmentation of the healing process. Int Wound J 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunyach-Remy C, Ngba Essebe C, Sotto A, Lavigne JP. Staphylococcus aureus Toxins and Diabetic Foot Ulcers: Role in Pathogenesis and Interest in Diagnosis. Toxins (Basel) 2016;8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftheriadou I, Tentolouris N, Argiana V, Jude E, Boulton AJ. Methicillin-resistant Staphylococcus aureus in diabetic foot infections. Drugs 2010;70(14):1785–97. [DOI] [PubMed] [Google Scholar]
- Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 2014;6(265):265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Schulte L, Vogel J. The mammalian microRNA response to bacterial infections. RNA Biol 2012;9(6):742–50. [DOI] [PubMed] [Google Scholar]
- Fadini GP, Menegazzo L, Rigato M, Scattolini V, Poncina N, Bruttocao A, et al. NETosis delays diabetic wound healing in mice and humans. Diabetes 2016. [DOI] [PubMed] [Google Scholar]
- Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 2013;4(1):e00537–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C, Bayoumi I, Westendorp C. Approach to infected skin ulcers. Can Fam Physician 2005;51:1352–9. [PMC free article] [PubMed] [Google Scholar]
- Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 2013;62(3):923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg EW, Li Y, Wang J, Burrows NR, Ali MK, Rolka D, et al. Changes in diabetes-related complications in the United States, 1990-2010. The New England journal of medicine 2014;370(16):1514–23. [DOI] [PubMed] [Google Scholar]
- Guertin AD, Li J, Liu Y, Hurd MS, Schuller AG, Long B, et al. Preclinical evaluation of the WEE1 inhibitor MK-1775 as single-agent anticancer therapy. Mol Cancer Ther 2013;12(8):1442–52. [DOI] [PubMed] [Google Scholar]
- Harfouche G, Vaigot P, Rachidi W, Rigaud O, Moratille S, Marie M, et al. Fibroblast growth factor type 2 signaling is critical for DNA repair in human keratinocyte stem cells. Stem cells 2010;28(9):1639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CH, Rau CS, Jeng JC, Chen YC, Lu TH, Wu CJ, et al. Whole blood-derived microRNA signatures in mice exposed to lipopolysaccharides. J Biomed Sci 2012;19:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Jiang H, Ren H, Hu X, Wang X, Han C. AGEs and chronic subclinical inflammation in diabetes: disorders of immune system. Diabetes/metabolism research and reviews 2015;31(2):127–37. [DOI] [PubMed] [Google Scholar]
- Hutton E, Paladini RD, Yu QC, Yen M, Coulombe PA, Fuchs E. Functional differences between keratins of stratified and simple epithelia. The Journal of cell biology 1998;143(2):487–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idan C, Peleg R, Elena V, Martin T, Cicerone T, Mareike W, et al. IL-1alpha is a DNA damage sensor linking genotoxic stress signaling to sterile inflammation and innate immunity. Scientific reports 2015;5:14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Ibeagha-Awemu EM, Liang G, Beaudoin F, Zhao X, Guan le L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC genomics 2014;15:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30(4):523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Sahu SK, Kumar M, Jana K, Gupta P, Gupta UD, et al. MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cellular microbiology 2016;18(5):679–91. [DOI] [PubMed] [Google Scholar]
- Lavery LA, Davis KE, Berriman SJ, Braun L, Nichols A, Kim PJ, et al. WHS Guidelines Update: Diabetic Foot Ulcer Treatment Guidelines. Wound Repair Regen 2015. [DOI] [PubMed] [Google Scholar]
- Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 2010;11(9):799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione AG, Brudno Y, Stojadinovic O, Park LK, Smith A, Tellechea A, et al. Three-dimensional human tissue models that incorporate diabetic foot ulcer-derived fibroblasts mimic in vivo features of chronic wounds. Tissue Eng Part C Methods 2015;21(5):499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancl KA, Kirsner RS, Ajdic D. Wound biofilms: lessons learned from oral biofilms. Wound Repair Regen 2013;21(3):352–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Z, Su T, Ye J, Yang C, Zhang S, Xie C. The miR-15 family enhances the radiosensitivity of breast cancer cells by targeting G2 checkpoints. Radiat Res 2015;183(2):196–207. [DOI] [PubMed] [Google Scholar]
- Miyajima S, Shirai A, Yamamoto S, Okada N, Matsushita T. Risk factors for major limb amputations in diabetic foot gangrene patients. Diabetes research and clinical practice 2006;71(3):272–9. [DOI] [PubMed] [Google Scholar]
- Naeem A, Zhong K, Moisa SJ, Drackley JK, Moyes KM, Loor JJ. Bioinformatics analysis of microRNA and putative target genes in bovine mammary tissue infected with Streptococcus uberis. J Dairy Sci 2012;95(11):6397–408. [DOI] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression. J Invest Dermatol 2016;136(11):2192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselova EG, Khrenov MO, Glushkova OV, Lunin SM, Parfenyuk SB, Novoselova TV, et al. Anti-inflammatory effects of IKK inhibitor XII, thymulin, and fat-soluble antioxidants in LPS-treated mice. Mediators of inflammation 2014;2014:724838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Kwon HI, Kim HW, Kim JE, Ro YS, Ko JY. A Digital Squamous Cell Carcinoma Mimicking a Diabetic Foot Ulcer, With Early Inguinal Metastasis and Cancer-Related Lymphedema. Am J Dermatopathol 2016;38(2):e18–21. [DOI] [PubMed] [Google Scholar]
- Pastar I, Khan AA, Stojadinovic O, Lebrun EA, Medina MC, Brem H, et al. Induction of Specific MicroRNAs Inhibits Cutaneous Wound Healing. The Journal of biological chemistry 2012;287(35):29324–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, et al. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 2013;8(2):e56846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar I, Ramirez H, Stojadinovic O, Brem H, Kirsner RS, Tomic-Canic M. Micro-RNAs: New Regulators of Wound Healing. Surgical technology international 2011;21:51–60. [PubMed] [Google Scholar]
- Pastar I, Stojadinovic O, Krzyzanowska A, Barrientos S, Stuelten C, Zimmerman K, et al. Attenuation of the transforming growth factor beta-signaling pathway in chronic venous ulcers. Mol Med 2010;16(3-4):92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar I, Stojadinovic O, Yin NC, Ramirez H, Nusbaum AG, Sawaya A, et al. Epithelialization in Wound Healing: A Comprehensive Review. Adv Wound Care (New Rochelle) 2014;3(7):445–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechter PM, Gil J, Valdes J, Tomic-Canic M, Pastar I, Stojadinovic O, et al. Keratin dressings speed epithelialization of deep partial-thickness wounds. Wound Repair Regen 2012;20(2):236–42. [DOI] [PubMed] [Google Scholar]
- Ramirez HA, Liang L, Pastar I, Rosa AM, Stojadinovic O, Zwick TG, et al. Comparative Genomic, MicroRNA, and Tissue Analyses Reveal Subtle Differences between Non-Diabetic and Diabetic Foot Skin. PLoS One 2015;10(8):e0137133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redel H, Gao Z, Li H, Alekseyenko AV, Zhou Y, Perez-Perez GI, et al. Quantitation and composition of cutaneous microbiota in diabetic and nondiabetic men. J Infect Dis 2013;207(7):1105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JB, Desai U, Cummings AK, Birnbaum HG, Skornicki M, Parsons NB. Burden of diabetic foot ulcers for medicare and private insurers. Diabetes Care 2014;37(3):651–8. [DOI] [PubMed] [Google Scholar]
- Roy S, Elgharably H, Sinha M, Ganesh K, Chaney S, Mann E, et al. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. The Journal of pathology 2014;233(4):331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosa D, Castoldi M, Paluschinski M, Sommerfeld A, Haussinger D. Hyperosmotic stress activates the expression of members of the miR-15/107 family and induces downregulation of anti-apoptotic genes in rat liver. Scientific reports 2015;5:12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojadinovic O, Brem H, Vouthounis C, Lee B, Fallon J, Stallcup M, et al. Molecular pathogenesis of chronic wounds: the role of beta-catenin and c-myc in the inhibition of epithelialization and wound healing. Am J Pathol 2005;167(1):59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojadinovic O, Landon JN, Gordon KA, Pastar I, Escandon J, Vivas A, et al. Quality assessment of tissue specimens for studies of diabetic foot ulcers. Experimental dermatology 2013;22(3):216–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojadinovic O, Pastar I, Gordon K, Tomic-Canic M. Physiology and Pathophysiology of Wound Healing in Diabetes. In: Veves A, Giurini JM, LoGerfo FW, editors. The Diabetic Foot: Humana Press; 2012. p. pp 127–49. [Google Scholar]
- Stojadinovic O, Tomic-Canic M. Human ex vivo wound healing model. Methods Mol Biol 2013;1037:255–64. [DOI] [PubMed] [Google Scholar]
- Stone RC, Stojadinovic O, Rosa AM, Ramirez HA, Badiavas E, Blumenberg M, et al. A Bioengineered Living Cell Construct Reactivates the Acute Wound Healing Response in Venous Leg Ulcers. Science Tranlsational Medicine 2017;4;9(371):eaaf8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kim SE, Yano H, Matsumoto G, Ohuchida R, Ishikura Y, et al. MiR-142 Is Required for Staphylococcus aureus Clearance at Skin Wound Sites via Small GTPase-Mediated Regulation of the Neutrophil Actin Cytoskeleton. J Invest Dermatol 2017;137(4):931–40. [DOI] [PubMed] [Google Scholar]
- Tentolouris N, Jude EB, Smirnof I, Knowles EA, Boulton AJ. Methicillin-resistant Staphylococcus aureus: an increasing problem in a diabetic foot clinic. Diabetic medicine : a journal of the British Diabetic Association 1999;16(9):767–71. [DOI] [PubMed] [Google Scholar]
- Tsourdi E, Barthel A, Rietzsch H, Reichel A, Bornstein SR. Current aspects in the pathophysiology and treatment of chronic wounds in diabetes mellitus. BioMed research international 2013;2013:385641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Asten SA, La Fontaine J, Peters EJ, Bhavan K, Kim PJ, Lavery LA. The microbiome of diabetic foot osteomyelitis. Eur J Clin Microbiol Infect Dis 2016;35(2):293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang X, Liu X, Wang X, Xu J, Hou S, et al. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int J Clin Exp Med 2015;8(4):5683–90. [PMC free article] [PubMed] [Google Scholar]
- Yang M, Sheng L, Zhang TR, Li Q. Stem cell therapy for lower extremity diabetic ulcers: where do we stand? BioMed research international 2013;2013:462179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Usui ML, Lippman SI, James GA, Stewart PS, Fleckman P, et al. Biofilms and Inflammation in Chronic Wounds. Adv Wound Care (New Rochelle) 2013;2(7):389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Representative H&E staining of full thickness biopsies from DFU and FS. DFU samples exhibit a thicker epidermis and cornified layer than normal foot skin. E= Epidermis, D= Dermis. Scale bars: 500μm.
Figure S2. a. qPCR expression data showing miR-15b-5p is up-regulated in porcine in-vivo acute wounds by infection with S. aureus but not with P. aeruginosa. (n=6 wounds per group) b. Representative DFU section before and after LCM. E= Epidermis, D= Dermis. c. qPCR showing up-regulation of miR-15b-5p in the epidermis of DFU (n=6 per group, FS = foot skin, unpaired t-test was used to determine statistical significance, ** = p-value<0.01, n.s. = not significant).
Figure S3. Representative Immuno-peroxidase stainings of pH2AX in FS and DFU biopsies. pH2AX is expressed at low levels throughout the epidermis of FS and is induced DFUs, primarily in the epidermis of DFUs. −Ab ctrl = no primary antibody control. Scale bars: 200μm.
Figure S4. H&E staining and 40X visualization of the sections shown in Figure 3. Scale bars: 500 μm. E= Epidermis, D= Dermis. Dashed box represents the picture areas shown in Figure 3.
Figure S5. miR-15b-5p target gene expression shows corresponding suppression when compared to either controls (diabetic foot skin or non-diabetic foot skin). Each dot represents an individual patient’s sample. Distribution of microarray data for IKBKB, WEE1, MSH2, RAD50, FGF2, KIT and IGF1 expression demonstrates clustering and segregation of DFUs from diabetic foot (DFS) and normal foot skin (NFS). DFU=diabetic foot ulcer (n=6), DFS (n=3), NFS= (n=3).
Table S1. List of the functions enriched in DFU.
Table S2. Patient demographics, and sample information. AA = African-American, A= Asian, H = Hispanic, HW = Hispanic White, W = White. DFU=diabetic foot ulcer, FS= foot skin, diabetic and non-diabetic.