Abstract
Many inbred strains of mice develop spontaneous tumors as they age. Recent awareness of the impacts of mitochondrial DNA (mtDNA) on cancer and aging have inspired developing a Mitochondrial-Nuclear Exchange (MNX) mouse model in which nuclear DNA is paired with mitochondrial genomes from other strains of mouse. MNX mice exhibit mtDNA influences on tumorigenicity and metastasis upon mating with transgenic mice. However, we also wanted to investigate spontaneous tumor phenotypes as MNX mice age. Utilizing FVB/NJ, C57BL/6J, C3H/HeN, and BALB/cJ wild-type inbred strains, previously documented phenotypes were observed as expected in MNX mice with the same nuclear background. However, aging nuclear matched MNX mice exhibited decreased occurrence of mammary tumors in C3H/HeN mice containing C57BL/6J mitochondria compared to wild-type C3H/HeN mice. Although aging tumor phenotypes appear to be driven by nuclear genes, evidence suggesting that some differences are modified by the mitochondrial genome is presented.
Keywords: Cancer, mitochondria, mouse, aging
Introduction
Inbred strains of mice are an extremely valuable tool in biomedical research [1–4]. The majority of inbred strains have been selected because they develop phenotypes related to and relevant for studying human diseases. As inbred mice age they develop a variety of age-related lesions. Older mice exhibit many pathologies which may, in part, be due to organ decline [5]. Most of these pathologic lesions have been studied for the commonly used inbred strains of mice (e.g., FVB/N, C57BL/6, C3H/He, BALB/c). Each strain has unique phenotypes, including developing neoplasms [6]. Additionally, numerous genetically engineered mouse models have been generated on these inbred strain genetic backgrounds to study tumor suppressors, oncogenes, and metastasis [7–12].
The FVB/N strain resulted from inbreeding of mice homozygous for the Fv-1b allele conferring sensitivity to the B strain of Friend leukemia virus [13,14]. This strain is an excellent breeder and has been widely used to develop many transgenic models. FVB/N mice spontaneously develop lung alveolar-bronchiolar tumors by 14 months of age in both males and females [14]. Female FVB/N mice also develop pituitary adenomas, ovarian tumors, lymphomas, and histiocytic sarcomas. Male FVB/N mice develop liver hepatocellular tumors, subcutis neural crest tumors and Harderian gland adenomas.
C57BL/6 mice are also frequently used for transgenics studies in cancer, but are also widely used in cardiovascular disease, diabetes and obesity studies. C57BL/6 mice have a high susceptibility to diet-induced obesity, type-2 diabetes, atherosclerosis [15] and develop an auditory deficiency [16], but are generally resistant to developing cancers [17,18].
BALB/c mice were bred by HJ Bagg in the 1920’s [19,20]. These mice develop reticular neoplasms in both females (23%) and males (3%) [21], lung tumors in 32% of males, 30% of breeding females and 14% of virgin females, as well as renal tumors as they age [22]. BALB/c have a low incidence of mammary tumors. Mammary adenocarcinomas occur in only 5% of breeding females and 1% of virgin females [23]. Some BALB/c females develop dorso-ventral vaginal septa resulting in non-productive females [24].
C3H/He mice were developed in the 1920’s from a cross between Bagg albino and DBA strains [25]. C3H mice were chosen for their high incidence of mammary tumors [26,27]. C3H/He mice also develop hepatomas and hepatocellular carcinoma (HCC) as these mice age [28] and this strain is highly susceptible to liver carcinogenesis. In addition, some strains of C3H/He mice carry a recessive mutation for retinal degeneration [29].
Tumor phenotypes in these (and other) inbred strains are nuclear DNA driven. Additionally, genetic variation across each strain could influence these (and other) phenotypes. Some of the genetic variability underlying phenotypic differences is controlled by quantitative trait loci (QTL) that affect the phenotypes [30,31]. QTL exert measurable phenotypic variations resulting from environmental factors and/or genetic factors and can be influenced by polymorphic genes [32,30,33–35]. Historically, investigators have focused on nuclear genes, but recent interest in the mitochondrial genome has revived curiosity directed toward understanding whether mtDNA polymorphisms may be among the QTL (i.e., play a role in tumor phenotypes) [36–39].
Mitochondria are organelles most commonly associated with metabolism and apoptosis. Mutations in the mtDNA lead to many human diseases such as atherosclerosis, Alzheimer’s disease, diabetes, obesity and aging [40] as well as cancers [36].
The mouse mitochondrial genome is ~16,300 base pairs. The mitochondrial genome is a circular DNA encoding 13 proteins, 16S and 12S rRNA and 22 tRNA. All of the mtDNA-encoded proteins are part of the electron transport system, but well over 90% of mitochondrial proteins are encoded in the nucleus. Besides energy metabolism, mitochondria generate several reactive oxygen species and are integral to apoptosis and regulating cellular pH and calcium levels. Alterations in any of these metabolic processes signal changes to the epigenome [41–44].
As in humans [45,46], mtDNA sequencing of all inbred strains of mice show the presence of single nucleotide polymorphisms (SNP) that are unique among the strains [38,47–49]. These SNP allow investigators to distinguish mtDNA contributions between multiple strains of mice. Several different haplotypes were discovered across 16 strains of mice whose mtDNA was sequenced. Pertinent to this report: C57BL/6J differs from the FVB/NJ, C3H/HeN and BALB/cJ at position 9461 in the ND3 of mtDNA (C9461T). C57BL/6J and FVB/NJ differ from C3H/HeN and BALB/cJ at position 9348 in the COIII (A9348G). C3H/HeJ and C3H/HeN differ at mtDNA position 8889 in the COIII. C57BL/6J differs at 11780 in the ND4 of the mtDNA. FVB/NJ differs from C57BL/6J, C3H/HeN, and BALB/cJ at position 7778 of ATP8 of the mtDNA (T7778G). Similarities among different common inbred strains are demonstrated by diminishing numbers of SNP found between strain mtDNA [50].
Every inbred strain identifier includes the name of the source from which it was derived. For example, “J” refers to Jackson Laboratories and N refers to Envigo (formerly Harlan Laboratories). The FVB/NJ, C57BL/6J and BALB/cJ strains used in this study were purchased from Jackson Laboratories and C3H/HeN mice were obtained from Envigo. The purchasing source for inbred mice is important as mtDNA sequences differ between them (e.g., for both C3H/He and C57BL/6 strains [50,51]). In addition, even nuclear DNA-encoded genes can vary. For example, C3H/HeN mice do not contain a mutation in the toll-like receptor 4 (TLR4); whereas, C3H/HeJ mice do. Such differences are also manifest during the generation of conplastic strains, which allows comparison between strains with similar or identical nuclear genomes but differing in their mitochondrial genome, thus providing evidence that the mitochondrial genome influences phenotypes [52,53].
Point mutations and deletions are accumulated in multiple aging human and rodent tissues [54]. Since cancer is a disease closely associated with aging, it follows, then, that many regions of mtDNA also acquire mutations correlated with development of multiple human cancers [55–57]. For example, mtDNA mutations in the D-loop (transcriptional regulatory region of mtDNA) are frequently found in human hepatocellular carcinoma (HCC) [55]. There are mtDNA variants in human lung cancers found in the D-loop and in tRNAArg regions [55]. Mutations in the D-loop and NADH complex proteins were found in salivary gland adenoid cystic carcinomas [57]. A more thorough review of the mtDNA mutations can be found in this issue [36]. It has yet to be determined if any of these regions correspond to similar regions within murine mtDNA. One study has shown that there is an absence of mtDNA mutations in mouse brain tumors [51]. Maternal transmission of mtDNA mutations has been extensively studied in the mtDNA mutator mouse [58], which expresses the catalytic subunit of the DNA polymerase gamma thus leading to a proofreading deficiency. This deficiency leads to an accumulation of acquired point mutations in the mtDNA [54]. Mutations in the mutator mouse offspring show similarities to mutations observed in humans [58]. Given the relatively high mtDNA mutation rate, it is paradoxical why there are so few bona fide examples of maternally-inherited cancers.
Although several strategies have been used to assess the contributions of mtDNA to disease, strengths and limitations exist for each (reviewed in [53,36]). In order to avoid use of mutagens, off-target effects of drugs and nDNA crossover complications to studying mitochondrial genetic contributions, we developed Mitochondrial-Nuclear Exchange (MNX) mice [37,59,39,15]. MNX mice contain nuclear DNA contribution from one strain of mice and mitochondrial DNA contribution from another strain. We previously showed that mitochondrial backgrounds selectively affect tumor latency [37,39], metastasis [37,39], obesity [38], and cardiovascular disease [15]. Additionally, mitochondrial genomic backgrounds also affect nuclear DNA methylation [38], histone marks (J. McGuire and D.R Welch, in preparation) and gene expression. To date, we have developed 5 different MNX strains each with a different nuclear/mitochondrial genome [Note: since sham transferred nuclei behaved as wild-type, we purchase wild-type rather than maintaining a colony.]: FVB/NJ-mtMNX(C57BL/6J) (FC), FVB/NJ -mtMNX(BALB/cJ) (FB), C3H/HeN-mtMNX(C57BL/6J) (HC), C57BL/6J-mt MNX(C3H/HeN) (CH), C57BL/6J-mtMNX(FVB/NJ) (CF). A simplified nomenclature for the MNX strains can be found in Table 1 and will be used throughout the rest of this manuscript. Briefly, the first letter designates the nuclear genome (F=FVB; C=C57; B=BALB/c; H=C3H), while the second letter refers to the mitochondrial genome (using the same abbreviation for each strain).
Table 1.
Abbreviated nomenclature for MNX mice
| Nomenclature | nDNA | mtDNA |
|---|---|---|
| FF | FVB/NJ | FVB/NJ |
| FC | FVB/NJ | C57BL/6J |
| FB | FVB/NJ | BALB/cJ |
| CC | C57BL/6J | C57BL/6J |
| CH | C57BL/6J | C3H/HeN |
| CF | C57BL/6J | FVB/NJ |
| HH | C3H/HeN | C3H/HeN |
| HC | C3H/HeN | C57BL/6J |
| BB | BALB/cJ | BALB/cJ |
In the experiments reported here, we wanted to test whether mtDNA SNP exhibited altered tumor-associated phenotypes as MNX mice aged. While most tumor phenotypes were clearly nuclear genome- associated, some strains developed a different spectrum of tumors depending upon mtDNA SNP.
Materials and Methods
Mice
Mitochondrial Nuclear Exchange (MNX) mice were created as previously reported [15,59]. Five different mouse strains were used in this study (Table 1). Historical data regarding wild-type mice were obtained from Jackson Laboratories (FF, CC, BB) and Envigo Laboratories (HH). The MNX colonies were maintained by crossing the MNX female with males of identical nuclear background. Following weaning 15–20 MNX mice were aged until moribund and development of spontaneous tumors. All animal studies were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center.
Genotyping
To measure and to assure correct genotype, DNA was extracted from tail clips using Red Extract-N-Amp kit (Sigma, St. Louis, MO). mtDNA haplotypes were confirmed by RFLP (restriction fragment length polymorphism) analysis of PCR products to SNP that are distinguishable between the strains [15,37,39,59]. FVB/NJ and C57BL/6J mtDNA were distinguished by using the SNP located at 9461 of ND3 with a loss of the BclI restriction site. FVB/NJ and BALB/cJ were distinguished using the SNP at 9348 of COIII containing a PflFI restriction site. C3H/HeN and C57BL/6 were also distinguished using 9348 of COIII containing a PflFI restriction site.
Histology
Tissues were harvested from the mice immediately after euthanasia; fixed in 10% neutral buffered formalin solution; and, then, embedded using standard embedding procedures. Sections (5 um) were made using a microtome before dehydrating with xylenes and ethanol and prior to staining with hematoxylin and eosin. A board-certified pathologist reviewed all slides. Slides were photographed at 10X magnification on a Nikon Eclipse 80i microscope.
Results and Discussion
An emerging body of data strongly supports the existence of mtDNA QTL for many diseases, including cancer (reviewed in [36]). Preliminary data collected in our lab using MNX mice supported this connection using genetic crosses with transgenic oncogenic drivers; however, we wanted to assess whether mitochondrial polymorphisms could also alter spontaneous, autochthonous tumor development in addition to other common age-associated phenotypes. Even at the experimental design stage, we acknowledged that our cohort (n=15–20) would be miniscule compared to the massive colonies at Jackson Laboratories and Envigo. However, we had already observed that MNX mice lived longer than their wild-type counterparts and that some nuclear-mitochondrial DNA pairings altered obesity [38]. Thus, we focused on phenotypes with relatively high frequency and report the findings as they may impact outcomes for others wishing to use MNX mice for experiments.
All of the aging MNX mouse cohorts developed neoplasms with abundant mitotic fractions upon histologic examination (Table 2). At a gross level, the tissue distribution of spontaneous neoplasms corresponded highly with comparable, nDNA-matched historical cohorts. Histologic examination was performed by at least three individuals, always including a board-certified pathologist, and the consensus findings are reported below. Images of representative gross and histologic findings are presented. It is important to note that, in this study, we only measured spontaneous tumor development. No carcinogen or exogenous chemical treatments were employed. And, while a complete necropsy was performed, an exhaustive pathological exam was not. Thus, we are unable at this time to address the possibility that some lesions were metastases from another primary tumor.
Table 2.
Summary of tumor phenotypes in MNX and wild type mice. All wild type (FF, BB, CC, HH) information was obtained from historical data.
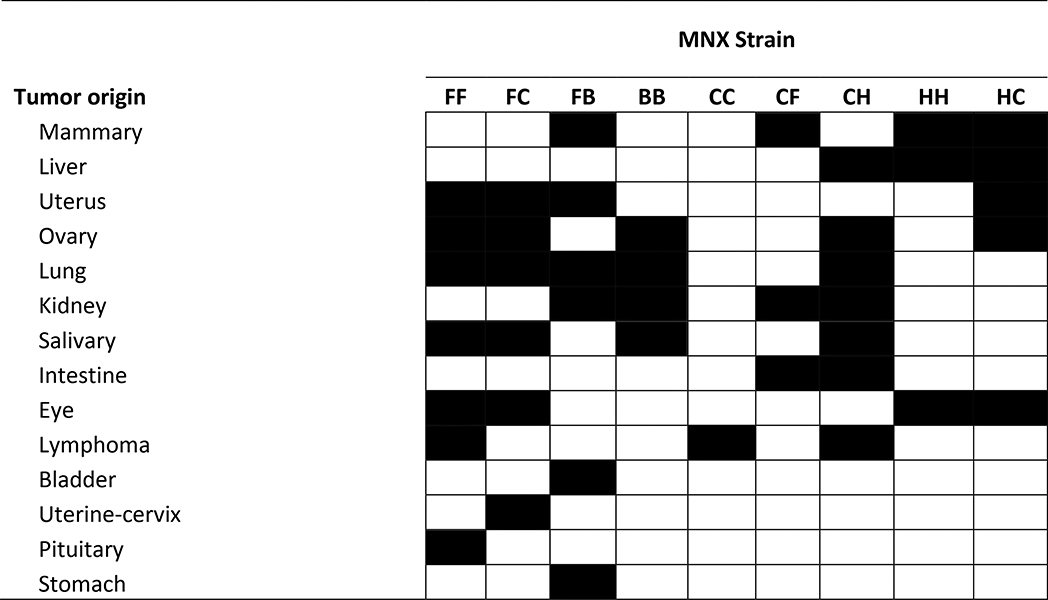 |
HH vs HC
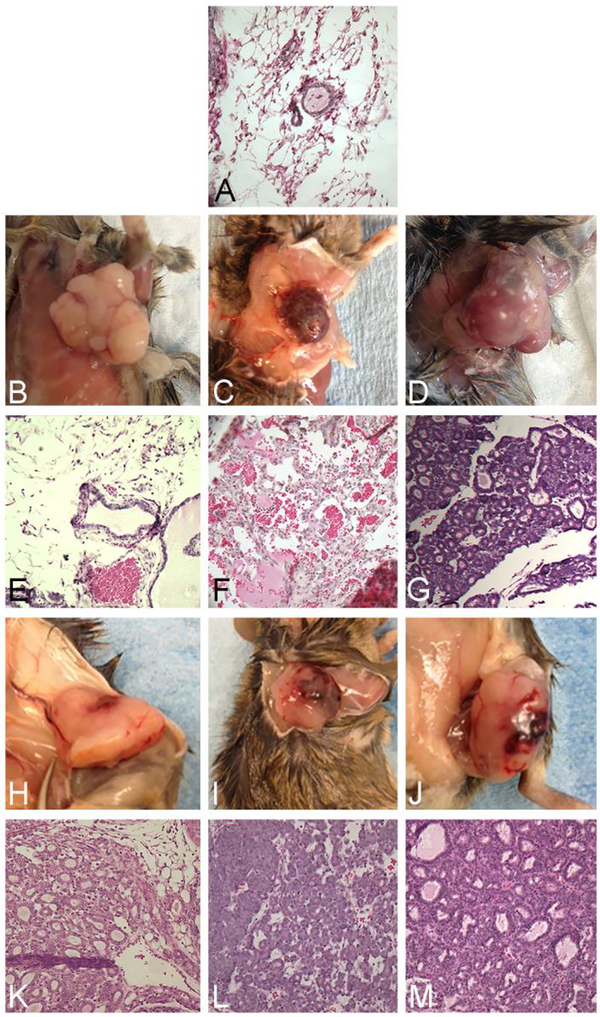
One of the most profound findings was a sharp reduction in the frequency of mammary gland neoplasms developing in MNX mice with C57BL/6 mtDNA on a C3H/HeN nuclear background. Nearly all female HH mice – 95% of parous and 88% of virgin/nonparous -- develop mammary adenocarcinomas [60]. In contrast, only 60% of breeders and 37% of virgin mice developed mammary carcinomas in HC mice at 18.8 months of age (Figure 1). Upon histologic exam, HC mammary tumors in the parous individuals were all of adenocarcinoma origin; whereas in the nonparous individuals, the spectrum of tumors was broader.
Figure 1:
Examples of spontaneous mammary adenocarcinoma development in aging female breeder and virgin HC MNX mice. Gross photographs (B-D (virgin), H-J (breeder)) and H&E stained sections (E-G, K-M) of mammary tumors are compared to a section of normal mammary gland (A).
C3H/He mice also develop hepatomas and hepatocellular carcinomas (HCC) at relatively high frequency. A small number of the HC mice developed either gross or microscopic hepatic neoplastic lesions (Figure 1). In addition to the mammary adenocarcinomas, HC mice also developed other pathologies, including: eye (cataract), enlarged heart, liver, uterine cysts, and enlarged ovaries (Figure 1).
FF vs FC
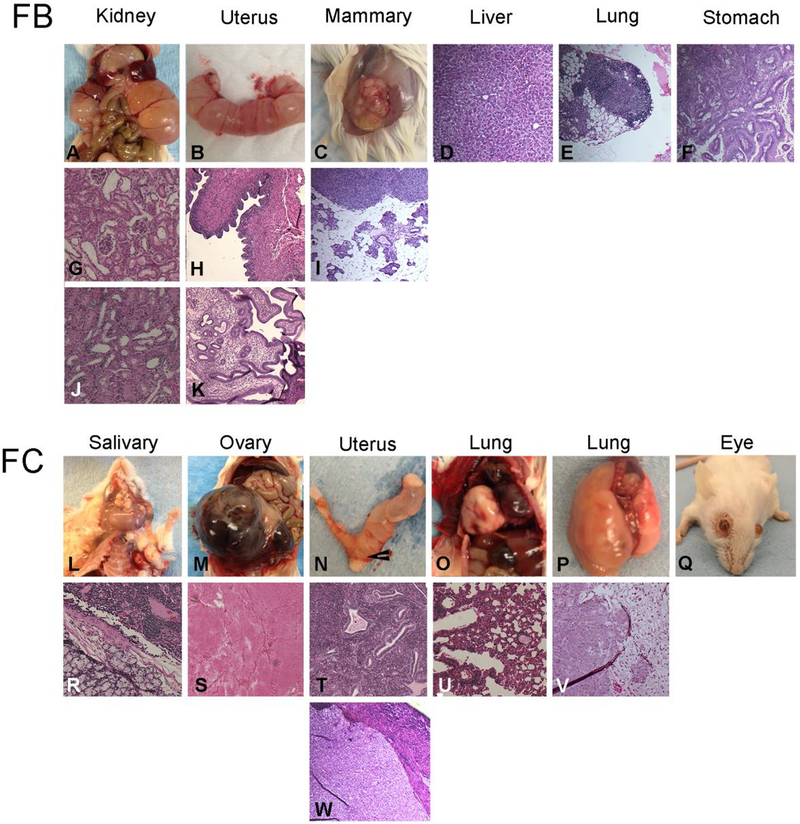
FVB/N mice develop several spontaneous lesions as they age, including lung adenocarcinomas, hepatocellular carcinoma, uterine and ovarian tumors, and salivary tumors [14]. FC mice developed a similar panel of tumors, lung adenocarcinoma and salivary tumors were observed frequently (7/20). Uterine lesions were mostly cysts. Although we did not measure blindness in our analyses, FF wild-type mice are known to become blind at around 4 months of age. We did observe enlarged eye growths around the eye sockets in the FC mice (4/20) (Figure 3)
Figure 3:
Representative aging phenotypes in FB and FC MNX mice. Panels A-K are gross and photomicrographs from FB mice. Panels L-W are from FC mice. Photomicrographs of H&E sections are aligned below gross pictures. The cystic kidneys (A) maintain some glomerular structure, but tubules are dilated (G,J). Similarly, the cystic uterus (B) is also fluid filled while maintaining relatively normal cellular architecture (H,K). A spontaneous mammary tumor (C) developed while no liver tumors were observed (D). H&E sections showing neoplasms arising in the lung (E) and in the stomach (F). Salivary gland (L,R), ovary (M,S), lung (O,P,U,V) and cervical (W) neoplasms are shown. As in FB mice, uterine cysts developed but the cellular architecture is less fluid filled than in the FB mice (T). Growths developing in the eye were observed, but not further characterized (Q) other than being always unilateral.
FF vs FB
As with FC mice, FB mice shared many phenotypes with FF wild-type mice. However, there were a few unique lesions observed. Some FB mice developed adenosquamous carcinoma at the gastroesophageal junction. We observed several cases of uterine cysts. Some of the uteri/vaginae exhibited benign simple cysts. Sporadic mammary, lung and uterus lesions were recorded (Figure 3) in addition to a single unilateral cystic kidney case showing renal tubule dilation.
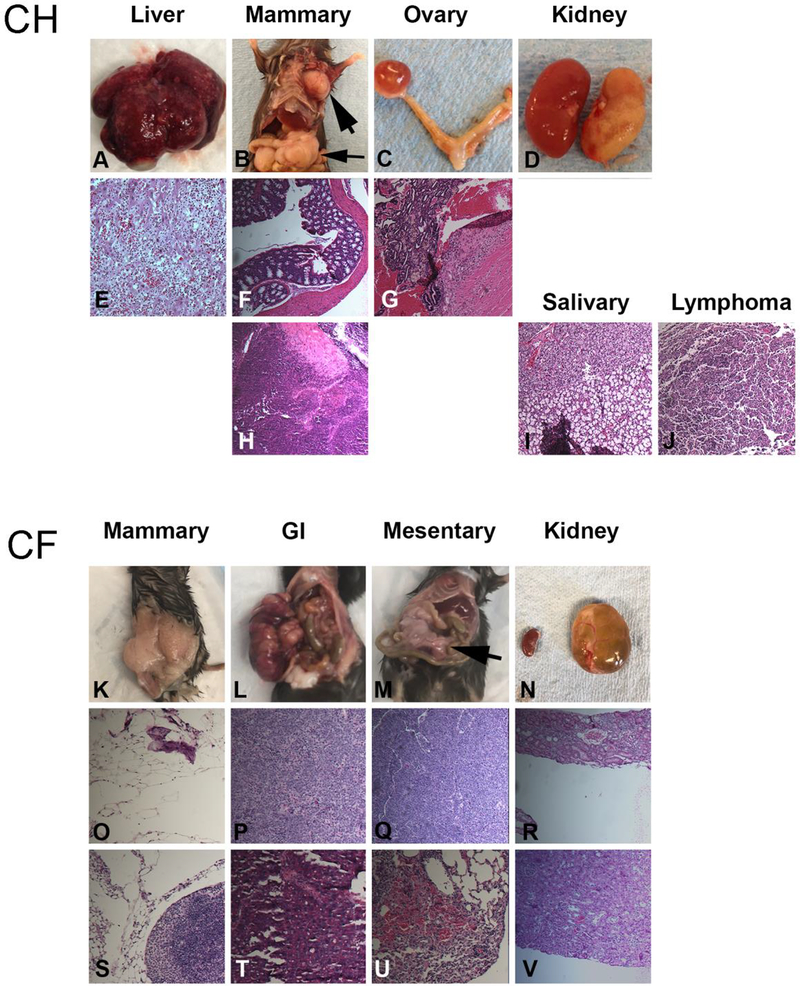
CC vs CH
C57BL/6 mice have a generally low incidence of developing cancer [17]. Some (2/15) CH mice develop inflammatory liver lesions and one mouse developed a poorly differentiated neoplasm. In addition, lymphomas and salivary tumors were also observed with similar frequency to the wild-type parental mice. Incidental tumor lesions were also observed in ovary, uterus and mammary gland (Figure 4).
Figure 4:
Representative aging phenotypes in CF and CH MNX mice. Panels A-J are gross and H&E photomicrographs from CH mice. Panels K-V are from CF mice. Liver (A,E), mammary (B,F,H), ovary (C,G), kidney (D), salivary (I) and lymphoma (J) neoplasms were observed in CH mice. Mammary (K,O,S), undifferentiated gastrointestinal tumors (L,P,T) and tumors associated with mesentery (M,Q,U) were reported in the CF mice. A cystic kidney (N, right) from one mouse is pictured with the contralateral normal kidney (N, left) for comparison. The kidney tubules were fluid filled, but some glomerular structure was maintained.
CC vs CF
Several of the CF mice develop poorly differentiated carcinomas in the gastrointestinal system. Several cases of cystic kidneys were also observed. Additional tumor phenotypes included lung, pancreas, and mammary glands. (Figure 4). CF male mice also exhibited enlarged seminal vesicles in older mice [61].
Conclusions
While susceptibility for spontaneous tumor formation appears to be largely controlled by nuclear DNA (as expected since the genome is comparatively much larger), the data presented here and in prior publications clearly demonstrate that mtDNA QTL contribute to overall susceptibility of some, but not all, neoplasms. Based upon previous findings, there is probably cooperation or antagonism between some nuclear QTL and some mitochondrial QTL that will contribute to tumor histotypes, aggressiveness, and progression. The baseline data presented in this brief report will establish a framework upon which more detailed genetic studies can be planned.
Supplementary Material
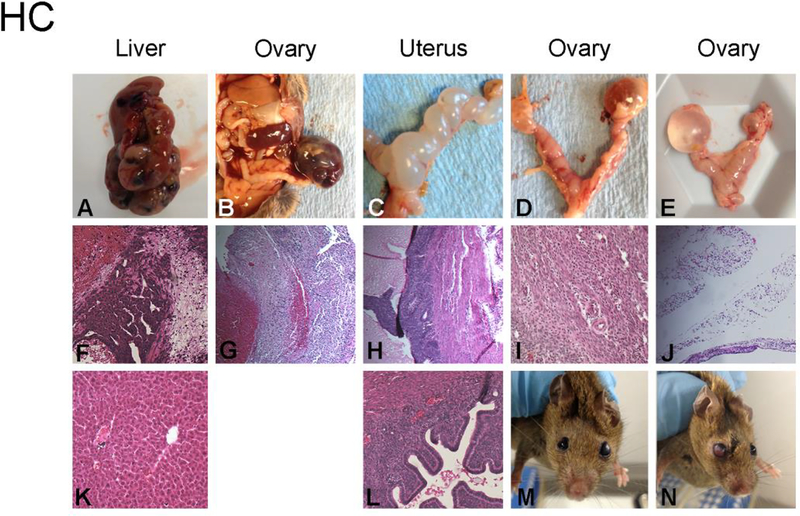
HC MNX mice have a lower incidence of HCC. A-F. Hematoxylin and eosin stained sections of normal liver. A-D. livers obtained from female mice. E-F. livers obtained from male mice. Unlike HH mice which develop hepatomas in 85% of male and female mice.
Figure 2:
Representative aging phenotypes observed in HC MNX mouse liver (A,F,K), ovary (B,G), uterus (C,D,E,H,I J,L) and eye (M,N) F is an H&E stained section of liver corresponding to the gross photograph in panel A. G is H&E stained section of the ovary photograph in panel B. Several HC mice developed lesions in the uterus and the photographs are arranged in the same column as the photograph. Panels M and N are examples of cataracts developing in the eyes of HC mice. Bilateral cataracts were not observed in our study.
Acknowledgements:
We want to thank members of the Welch lab for their helpful comments and suggestions. We also thank the Transgenic and Gene-Targeting Institutional Facility at University of Kansas Cancer Center. These studies were funded primarily by: Susan G. Komen for the Cure (SAC110037) and the National Foundation for Cancer Research with additional funding from the National Cancer Institute P30-CA168524 (RAJ; DRW).
Footnotes
Conflicts of interest: D.R. Welch is a co-holder of a patent on the MNX mice. The other authors declare no conflicts of interest.
References
- 1.Dragani TA (2003). 10 years of mouse cancer modifier loci: human relevance. Cancer Research, 63(12), 3011–3018. [PubMed] [Google Scholar]
- 2.Altman PL, & Katz DD (1979). Inbred and genetically defined strains of laboratory animals. Part 2. Hamster, guinea pig, rabbit and chicken (Vol. 1). Bethesda, MD: Federation of American Societies for Experimental Biology. [Google Scholar]
- 3.Altman PL, & Katz DD (1979). Inbred and genetically defined strains of laboratory animals. Part 1. Mouse and rat (Vol. 1). Bethesda, MD: Federation of American Societies for Experimental Biology. [Google Scholar]
- 4.Stern MC, & Conti CJ (1996). Genetic susceptibility to tumor progression in mouse skin carcinogenesis. Progress in Clinical and Biological Research, 395, 47–55. [PubMed] [Google Scholar]
- 5.Pettan-Brewer C, & Treuting PM (2011). Practical pathology of aging mice. Pathobiology of Aging and Age Related Diseases, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heston WE, & Vlahakis G (1971). Mammary tumors, plaques, and hyperplastic alveolar nodules in various combinations of mouse inbred strains and the different lines of the mammary tumor virus. International Journal of Cancer, 7(1), 141–148. [DOI] [PubMed] [Google Scholar]
- 7.Guy CT, Cardiff RD, & Muller WJ (1992). Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Molecular and Cellular Biology, 12(3), 954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardiff RD, & Muller WJ (1993). Transgenic mouse models of mammary tumorigenesis. Cancer Surveys, 16, 97–113. [PubMed] [Google Scholar]
- 9.Muller WJ, Ho J, & Siegel PM (1998). Oncogenic activation of Neu/ErbB-2 in a transgenic mouse model for breast cancer. Biochemical Society Symposia, 63, 149–157. [PubMed] [Google Scholar]
- 10.Walrath JC, Hawes JJ, Van Dyke T, & Reilly KM (2010). Genetically engineered mouse models in cancer research. Advances in Cancer Research, 106, 113–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walton JB, Farquharson M, Mason S, Port J, Kruspig B, Dowson S, et al. (2017). CRISPR/Cas9derived models of ovarian high grade serous carcinoma targeting Brca1, Pten and Nf1, and correlation with platinum sensitivity. Scientific Reports, 7(1), 16827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lampreht Tratar U, Horvat S, & Cemazar M (2018). Transgenic Mouse Models in Cancer Research. Frontiers in Oncology, 8, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taketo M, Schroeder AC, Mobraaten LE, Gunning KB, Hanten G, Fox RR, et al. (1991). FVB/N: an inbred mouse strain preferable for transgenic analyses. Proc Natl Acad Sci U S A, 88(6), 2065–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahler JF, Stokes W, Mann PC, Takaoka M, & Maronpot RR (1996). Spontaneous lesions in aging FVB/N mice. Toxicologic Pathology, 24(6), 710–716. [DOI] [PubMed] [Google Scholar]
- 15.Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, et al. (2013). Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochemical Journal, 455(2), 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng QY, Johnson KR, & Erway LC (1999). Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hearing Research, 130(1–2), 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bult CJ, Krupke DM, Begley DA, Richardson JE, Neuhauser SB, Sundberg JP, et al. (2015). Mouse Tumor Biology (MTB): a database of mouse models for human cancer. Nucleic Acids Research, 43(Database issue), D818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith CL, Blake JA, Kadin JA, Richardson JE, Bult CJ, & Mouse Genome Database G (2018). Mouse Genome Database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Research, 46(D1), D836–D842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagg HJ (1936). Functional Activity of the Mammary Gland in Relation to Extra-Chromosomal Influence in the Incidence of Mammary Tumors. Science, 83(2155), 374–375. [DOI] [PubMed] [Google Scholar]
- 20.Adair FE, & Bagg HJ (1931). Experimental and Clinical Studies on the Treatment of Cancer by Dichlorethylsulphide (Mustard Gas. Annals of Surgery, 93(1), 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebbesen P (1971). Reticulosarcoma and amyloid development in BALB/c mice inoculated with syngeneic cells from young and old donors. Journal of the National Cancer Institute, 47(6), 1241–1245. [PubMed] [Google Scholar]
- 22.Myers DD, Meier H, & Huebner RJ (1970). Prevalence of murine C-type RNA virus group specific antigen in inbred strains of mice. Life Sciences. Part 2: Biochemistry, General and Molecular Biology, 9(18), 1071–1080. [DOI] [PubMed] [Google Scholar]
- 23.Hoag WG (1963). Spontaneous Cancer in Mice. Annals of the New York Academy of Sciences, 108, 805–831. [DOI] [PubMed] [Google Scholar]
- 24.Cunliffe-Beamer TL, & Feldman DB (1976). Vaginal septa in mice: incidence, inheritance, and effect on reproductive, performance. Laboratory Animal Science, 26(6 Pt 1), 895–898. [PubMed] [Google Scholar]
- 25.Crow JF (2002). C. C. Little, cancer and inbred mice. Genetics, 161(4), 1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braunschweiger PG, Poulakos L, & Schiffer LM (1977). Cell kinetics in vivo and in vitro for C3H/He spontaneous mammary tumors. Journal of the National Cancer Institute, 59(4), 11971204. [DOI] [PubMed] [Google Scholar]
- 27.Lacour F, Delage G, & Chianale C (1975). Reduced incidence of spontaneous mammary tumors in C3H/He mice after treatment with polyadenylate-polyuridylate. Science, 187(4173), 256–257. [DOI] [PubMed] [Google Scholar]
- 28.Manenti G, Binelli G, Gariboldi M, Canzian F, De Gregorio L, Falvella FS, et al. (1994). Multiple loci affect genetic predisposition to hepatocarcinogenesis in mice. Genomics, 23(1), 118–124. [DOI] [PubMed] [Google Scholar]
- 29.Sidman RL, & Green MC (1965). Retinal Degeneration in the Mouse: Location of the Rd Locus in Linkage Group Xvii. Journal of Heredity, 56, 23–29. [DOI] [PubMed] [Google Scholar]
- 30.Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, et al. (2003). The nature and identification of quantitative trait loci: a community’s view. Nature Reviews: Genetics, 4(11), 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Little CC, & Tyzzer EE (1916). Further experimental studies on the inheritance of susceptibility to a Transplantable tumor, Carcinoma (J. W. A.) of the Japanese waltzing Mouse. Journal of Medical Research, 33(3), 393–453. [PMC free article] [PubMed] [Google Scholar]
- 32.Albert FW, & Kruglyak L (2015). The role of regulatory variation in complex traits and disease. Nature Reviews: Genetics, 16(4), 197–212. [DOI] [PubMed] [Google Scholar]
- 33.Mackay TFC (2014). Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nature Reviews Genetics, 15(1), 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drinkwater NR, & Gould MN (2012). The long path from QTL to gene. PLoS Genetics, 8(9), e1002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plomin R, Haworth CM, & Davis OS (2009). Common disorders are quantitative traits. Nature Reviews: Genetics, 10(12), 872–878. [DOI] [PubMed] [Google Scholar]
- 36.Beadnell TC, Scheid AD, Vivian CJ, & Welch DR (2018). Roles of the mitochondrial genetics in cancer metastasis: Not to be ignored any longer. Cancer and Metastasis Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brinker AE, Vivian CJ, Koestler DC, Tsue TT, Jensen RA, & Welch DR (2017). Mitochondrial Haplotype Alters Mammary Cancer Tumorigenicity and Metastasis in an Oncogenic Driver-Dependent Manner. Cancer Research, 77(24), 6941–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vivian CJ, Brinker AE, Graw S, Koestler DC, Legendre C, Gooden GC, et al. (2017). Mitochondrial Genomic Backgrounds Affect Nuclear DNA Methylation and Gene Expression. Cancer Research, 77(22), 6202–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feeley KP, Bray AW, Westbrook DG, Johnson LW, Kesterson RA, Ballinger SW, et al. (2015). Mitochondrial Genetics Regulate Breast Cancer Tumorigenicity and Metastatic Potential. Cancer Research, 75(20), 4429–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R, et al. (2016). Mitochondrial diseases. Nature Reviews Disease Primers, 2, 16080. [DOI] [PubMed] [Google Scholar]
- 41.Wallace DC, & Chalkia D (2013). Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harbor Perspectives in Biology, 5(11), a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallace DC (2005). A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual Review of Genetics, 39, 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallace DC, & Fan W (2010). Energetics, epigenetics, mitochondrial genetics. Mitochondrion, 10(1), 12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picard M, Zhang J, Hancock S, Derbeneva O, Golhar R, Golik P, et al. (2014). Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc Natl Acad Sci U S A, 111(38), E4033–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace DC (2016). Genetics: Mitochondrial DNA in evolution and disease. Nature, 535(7613), 498–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, et al. (2003). Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci U S A, 100(1), 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takibuchi G, Imanishi H, Morimoto M, Ishikawa K, Nakada K, Toyama-Sorimachi N, et al. (2013). Polymorphic mutations in mouse mitochondrial DNA regulate a tumor phenotype. Mitochondrion, 13(6), 881–887. [DOI] [PubMed] [Google Scholar]
- 48.Goios A, Gusmao L, Rocha AM, Fonseca A, Pereira L, Bogue M, et al. (2008). Identification of mouse inbred strains through mitochondrial DNA single-nucleotide extension. Electrophoresis, 29(23), 4795–4802. [DOI] [PubMed] [Google Scholar]
- 49.Sflomos G, Dormoy V, Metsalu T, Jeitziner R, Battista L, Scabia V, et al. (2016). A Preclinical Model for ERalpha-Positive Breast Cancer Points to the Epithelial Microenvironment as Determinant of Luminal Phenotype and Hormone Response. Cancer Cell, 29(3), 407–422. [DOI] [PubMed] [Google Scholar]
- 50.Goios A, Pereira L, Bogue M, Macaulay V, & Amorim A (2007). mtDNA phylogeny and evolution of laboratory mouse strains. Genome Research, 17(3), 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiebish MA, & Seyfried TN (2005). Absence of pathogenic mitochondrial DNA mutations in mouse brain tumors. BMC Cancer, 5, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roubertoux PL, Sluyter F, Carlier M, Marcet B, Maarouf-Veray F, Cherif C, et al. (2003). Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nature Genetics, 35(1), 65–69. [DOI] [PubMed] [Google Scholar]
- 53.Bussard KM, & Siracusa LD (2017). Understanding Mitochondrial Polymorphisms in Cancer. Cancer Research, 77(22), 6051–6059. [DOI] [PubMed] [Google Scholar]
- 54.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, et al. (2004). Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature, 429(6990), 417–423. [DOI] [PubMed] [Google Scholar]
- 55.Chatterjee A, Mambo E, & Sidransky D (2006). Mitochondrial DNA mutations in human cancer. Oncogene, 25(34), 4663–4674. [DOI] [PubMed] [Google Scholar]
- 56.Park CB, & Larsson NG (2011). Mitochondrial DNA mutations in disease and aging. Journal of Cell Biology, 193(5), 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, & Reddy SA (2004). The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene, 23(53), 8571–8580. [DOI] [PubMed] [Google Scholar]
- 58.Stewart JB, Freyer C, Elson JL, & Larsson NG (2008). Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nature Reviews: Genetics, 9(9), 657–662. [DOI] [PubMed] [Google Scholar]
- 59.Kesterson RA, Johnson LW, Lambert LJ, Vivian JL, Welch DR, & Ballinger SW (2016). Generation of Mitochondrial-nuclear eXchange Mice via Pronuclear Transfer. BioProtocols, 6(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCredie JA, Inch WR, & Sutherland RM (1971). Differences in growth and morphology between the spontaneous C3H mammary carcinoma in the mouse and its syngeneic transplants. Cancer, 27(3), 635–642. [DOI] [PubMed] [Google Scholar]
- 61.Pontual EV, Carvalho BE, Bezerra RS, Coelho LC, Napoleao TH, & Paiva PM (2012). Caseinolytic and milk-clotting activities from Moringa oleifera flowers. Food Chemistry, 135(3), 1848–1854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HC MNX mice have a lower incidence of HCC. A-F. Hematoxylin and eosin stained sections of normal liver. A-D. livers obtained from female mice. E-F. livers obtained from male mice. Unlike HH mice which develop hepatomas in 85% of male and female mice.