Abstract
Cardiovascular (CV)- and lifestyle-associated risk factors (RFs) are increasingly recognized as important for Alzheimer’s disease (AD) pathogenesis. Beyond the ε4 allele of apolipoprotein E (APOE), comparatively little is known about whether CV-associated genes also increase risk for AD. Using large genome-wide association studies and validated tools to quantify genetic overlap, we systematically identified single nucleotide polymorphisms (SNPs) jointly associated with AD and one or more CV-associated RFs, namely body mass index (BMI), type 2 diabetes (T2D), coronary artery disease (CAD), waist hip ratio (WHR), total cholesterol (TC), triglycerides (TG), low-density (LDL) and high-density lipoprotein (HDL). In fold enrichment plots, we observed robust genetic enrichment in AD as a function of plasma lipids (TG, TC, LDL, and HDL); we found minimal AD genetic enrichment conditional on BMI, T2D, CAD, and WHR. Beyond APOE, at conjunction FDR < 0.05 we identified 90 SNPs on 19 different chromosomes that were jointly associated with AD and CV-associated outcomes. In meta-analyses across three independent cohorts, we found four novel loci within MBLAC1 (chromosome 7, meta-p = 1.44 × 10−9), MINK1 (chromosome 17, meta-p = 1.98 × 10−7) and two chromosome 11 SNPs within the MTCH2/SPI1 region (closest gene = DDB2, meta-p = 7.01 × 10−7 and closest gene = MYBPC3, meta-p = 5.62 × 10−8). In a large ‘AD-by-proxy’ cohort from the UK Biobank, we replicated three of the four novel AD/CV pleiotropic SNPs, namely variants within MINK1, MBLAC1, and DDB2. Expression of MBLAC1, SPI1, MINK1 and DDB2 was differentially altered within postmortem AD brains. Beyond APOE, we show that the polygenic component of AD is enriched for lipid-associated RFs. We pinpoint a subset of cardiovascular-associated genes that strongly increase the risk for AD. Our collective findings support a disease model in which cardiovascular biology is integral to the development of clinical AD in a subset of individuals.
Keywords: Lipids, Polygenic enrichment, Cardiovascular, Alzheimer’s disease, Genetic pleiotropy
Introduction
There is mounting evidence that cardiovascular (CV) disease impacts Alzheimer’s disease (AD) pathogenesis. Co-occurrence of CV and AD pathology is the most common cause of dementia among the elderly [6] and imaging manifestations of vascular pathology are routinely observed in the brain on MRI scans of AD patients [41]. Observational epidemiology studies have found that cardiovascular-/lifestyle-related risk factors (RFs) are associated with dementia risk and targeting these modifiable RFs may represent a viable dementia prevention strategy [7, 32]. Recently, the National Academy of Medicine [30] and the Lancet [26] commissioned independent reports on strategies for dementia prevention. Both reports found encouraging evidence for targeting cardiovascular RFs with the Lancet commission concluding that 35% of dementia could be prevented by modifying several RFs including diabetes, hypertension, obesity, and physical inactivity.
Genetic studies have found CV-associated loci that also increase risk for late-onset AD. The ε4 allele of apolipo-protein E (APOE) is the biggest genetic risk factor for AD and encodes a lipid transport protein involved in cholesterol metabolism [29]. Genome-wide association studies (GWAS) in late-onset AD have identified single nucleotide polymorphisms (SNPs) implicated in lipid processes, such as CLU and ABCA7 [24, 37], and enrichment in cholesterol metabolism pathways [9]. Considered together, these findings suggest ‘pleiotropy’, where variations in a single gene can affect multiple, seemingly unrelated phenotypes [42].
We have previously shown that genetic enrichment in cardiovascular-/lifestyle-associated RFs and diseases (hereafter referred to as CV-associated RFs) results in improved statistical power for discovery of novel AD genes [13]. Building on this work, in the present study, we systematically evaluated shared genetic risk between AD and cardiovascular-/lifestyle-associated RFs and diseases. We focused on publicly available genetic data from cardiovascular outcomes and a combination of traits and diseases that have been epidemiologically associated with increased AD risk. Using large GWAS and validated tools to estimate pleiotropy, we sought to identify SNPs jointly associated with AD and one or more CV-associated RF, namely body mass index (BMI), type 2 diabetes (T2D), coronary artery disease (CAD), waist hip ratio (WHR), total cholesterol (TC), triglycerides (TG), low-density (LDL) and high-density lipoprotein (HDL). We additionally assessed whether the AD/CV genes showed independent replication within a large ‘AD-by-proxy’ phenotype sample that relies upon parental AD status to identify proxy cases and proxy controls [52]. Finally, we examined whether the AD/CV pleiotropic genes are differentially expressed within AD brains.
Methods
Participant samples
We evaluated complete GWAS results in the form of summary statistics (p values and odds ratios) for clinically diagnosed AD dementia [24] and eight CV-associated RFs, including BMI [47], T2D [28], CAD [31], WHR [18], and plasma lipid levels (TC, TG, LDL, and HDL [44]). We obtained publicly available AD GWAS summary statistic data from the International Genomics of Alzheimer’s Disease Project (IGAP Stages 1 and 2; for additional details, see Supplemental Information and [24]; Table 1). As our primary cohort, we used IGAP Stage 1 which consists of 17,008 AD cases (mean age = 74.7 ± 7.7 years; 59.4% female) and 37,154 controls (mean age = 76.3 ± 8.1 years; 58.6% female) drawn from four different consortia across North America and Europe with genotyped or imputed data at 7,055,881 SNPs (for a description of the AD dementia cases and controls within the IGAP Stage 1 sub-studies, please see Ref. [24]). To confirm our findings from IGAP Stage 1, we assessed the p values of pleiotropic SNPs (conjunction FDR < 0.05; see statistical analysis below) from two independent AD cohorts, namely the IGAP Stage 2 [24] sample, and a cohort of AD cases and controls drawn from the population of the United States and part of phase 2 of the Alzheimer’s Disease Genetics Consortium (ADGC2). The IGAP Stage 2 sample consisted of 8,572 AD cases (mean age = 72.5 ± 8.1 years; 61% female) and 11,312 controls (mean age = 65.5 ± 8.0 years; 43.3% female) of European ancestry with genotyped data at 11,632 SNPs (for additional details, see Ref. [24]). The ADGC2 sample consisted of 2,122 AD cases and 3,213 controls of European ancestry (for additional details, see Ref. [21]).
Table 1.
Summary data from all GWAS used in the current study
| Disease/trait | Abbr | Sample size | Cases | Controls | References | URL | Ethnicities included | SNPs | Covariate adjustments |
|---|---|---|---|---|---|---|---|---|---|
| Alzheimer’s disease | AD | 17,008 | 37,154 | [24] | http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php | EUR | 7,055,881 | Adjusted for age, sex and principal components | |
| High-density lipoprotein | HDL | 62,166 | [44] |
http://diagram-consortium.org/2015_ENGAGE_1KG/ file: HDL-C |
EUR | 9,549,055 | Adjusted for age, age2, and first 3 principal components | ||
| Low-density lipoprotein | LDL | 62,166 | [44] |
http://diagram-consortium.org/2015_ENGAGE_1KG/ file: LDL-C |
EUR | 9,545,543 | Adjusted for age, age2, and first 3 principal components | ||
| Total triglycerides | TG | 62,166 | [44] |
http://diagram-consortium.org/2015_ENGAGE_1KG/ file: TG |
EUR | 9,544,499 | Adjusted for age, age2, and first 3 principal components | ||
| Total cholesterol | TC | 62,166 | [44] |
http://diagram-consortium.org/2015_ENGAGE_1KG/ file: TC |
EUR | 9,553,380 | Adjusted for age, age2, and first 3 principal components | ||
| Body-mass index | BMI | 681,275 | [47] |
https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files file: Download Updated Meta-analysis Locke et al. + UK Biobank 2018 GZIP |
EUR | 2,336,270 | Adjusted for age, sex, recruitment center, genotyping batches, and ten principle components calculated from SNPs pre-selected by the UKB quality control team for PC analysis | ||
| Type 2 diabetes | T2D | 48,286 | 250,671 | [28] |
http://diagram-consortium.org/downloads.html file: T2D GWAS meta-analysis—European Summary Statistics |
EUR | 133,587 | Adjusted for age, sex, six axes of genetic variation, genotyping array, and body mass index | |
| Coronary artery disease | CAD | 17,283 | 137,914 | [31] |
http://www.cardiogramplusc4d.org/data-downloads/ file: UKBB.GWAS1KG.EXOME.CAD.SOFT.META.PublicRelease.300517. txt.gz |
EUR | 9,026,568 | Adjusted for array (UK Biobank versus UK BiLEVE) and the first five principal components | |
| Waist-to-hip ratio | WHR | 180,423 | [18] |
https://zenodo.org/record/l251813#.W9IlAy-ZOjQ file: whradjbmi.giant-ukbb.meta-analysis.combined.23May2018.txt.gz |
EUR | 27,375,637 | Adjusted for age, age2, sex, BMI, sex, first 5 principal components |
Abbr abbreviation, EUR European, SNPs single nucleotide polymorphisms
We further assessed the p values of our AD/CV pleiotropic SNPs in an AD-by-proxy cohort that is based on individuals of European ancestry in the UK Biobank (UKB) for whom parental AD status was available (N proxy cases = 47,793; N proxy controls = 328,320) (for additional details, see Ref. [52]). Individuals with one or two parents with AD were defined as proxy cases, while putting more weight on the proxy cases with two parents. Similarly, individuals with two parents without AD were defined as proxy controls, where older cognitively normal parents were up-weighted as proxy controls to account for the higher likelihood that younger parents may still develop AD. As the proxy phenotype is not equivalent to a clinical diagnosis of AD and may include individuals that never develop AD, we evaluated the UKB by-proxy sample separately from the IGAP and ADGC2 case control samples.
Details of the summary data and available URLs from all GWAS used in the current study are listed in Table 1. The relevant institutional review boards or ethics committees approved the research protocol of all individual GWAS used in the current analysis, and all human participants gave written informed consent.
Genetic enrichment and conjunction false discovery rates (FDR)
A brief summary of these methods follows. For details, see Supplementary methods and previous publications [2, 3, 5, 8, 12, 13, 19, 48].
We evaluated whether there is pleiotropic genetic enrichment in AD as a function of each of the eight CV-associated RFs. To do this, we compare the association with a primary trait (e.g., AD) across all SNPs and within SNP strata determined by their association with a secondary trait (e.g., BMI), and provide a visual pattern of overlap in SNP associations. For given associated phenotypes A (e.g., AD) and B (e.g., BMI), pleiotropic ‘enrichment’ of phenotype A with phenotype B exists if the proportion of SNPs or genes associated with phenotype A increases as a function of increased association with phenotype B (see Supplementary Methods). To assess for enrichment, we constructed fold-enrichment plots of nominal − log10(p) values for all AD SNPs and for subsets of SNPs determined by the significance of their association with each of the eight CV-associated RFs (e.g., − log10(p) > 1, > 2, and > 3 in CV-associated RFs). In fold-enrichment plots, the presence of enrichment is reflected as an upward deflection of the curve for phenotype A if the degree of deflection from the expected null line is dependent on the degree of association with phenotype B. More specifically, fold enrichment is computed as follows: first, we compute the empirical cumulative distribution of − log10(p) values for SNP association with a given phenotype (e.g., AD) for all SNPs, and then the cumulative − log10(p) values for each SNP stratum, which is determined by the p value of these SNPs in the conditioning phenotype (e.g., BMI). We then calculate the fold enrichment of each stratum as the ratio between the − log10(p) cumulative distribution for that stratum and the cumulative distribution for all SNPs. The x-axis shows nominal p values (− log10(p)); the y-axis shows fold enrichment. To assess for polygenic effects below the standard GWAS significance threshold, we focused the fold-enrichment plots on SNPs with nominal − log10(p) < 7.3 (corresponding to p > 5 × 10−8). The enrichment seen can be directly interpreted in terms of true discovery rate [TDR = 1 − false discovery rate (FDR)] (for additional details, see Supplemental Information).
To account for large blocks of linkage disequilibrium (LD) that may result in spurious genetic enrichment, we applied a random pruning approach, where one random SNP per LD block (defined by an r2 of 0.8) was used and averaged over 200 random pruning runs. Given prior evidence that several genetic variants within the human leukocyte antigen (HLA) region on chromosome 6 [43, 49], microtubule-associated tau protein (MAPT) region on chromosome 17 [12] and the APOE region on chromosome 19 [13] are associated with increased AD risk, one concern is that random pruning may not sufficiently account for these large LD blocks, resulting in artificially inflated genetic enrichment [8]. To better account for these large LD blocks, in our genetic enrichment analyses, we removed all SNPs in LD with r2 > 0.2 within 1 Mb of HLA, MAPT and APOE variants (based on 1000 Genomes Project LD structure).
To identify specific loci jointly involved with AD and the eight CV-associated risk factors, we computed conjunction false discovery rates (FDRs), a statistical framework that is well suited to a genetic epidemiology approach to investigate genetic pleiotropy. The standard FDR framework is based on Bayesian statistics and follows the assumption that SNPs are either associated with the phenotype (non-null) or are not associated with the phenotype (null SNPs). Within a Bayesian statistical framework, the FDR is then the probability of the SNP being null given its p value is as small as or smaller than the observed one. An extension of the standard FDR is the conjunction FDR, defined as the probability that a SNP is null for either phenotype or for both phenotypes simultaneously given its p value in both phenotypes are as small or smaller as the observed ones. The conjunction is a conservative approach requiring that loci exceed a conjunction FDR significance threshold for two traits jointly. Conjunction FDR, therefore, is more conservative and specifically pinpoints pleiotropic loci between the traits of interest. We used an overall FDR threshold of < 0.05, which means five expected false discoveries per hundred reported. Manhattan plots were constructed based on the ranking of conjunction FDR to illustrate the genomic location of the pleiotropic loci. In all analyses, we controlled for the effects of genomic inflation using intergenic SNPs (see Supplemental and previous reports for additional details [2, 5, 8, 12, 13, 19]).
For loci with conjunction FDR < 0.05, we performed a fixed-effect, inverse variance-weighted meta-analysis [46] using independent AD cohorts: IGAP Stages 1 and 2 (cases = 25,580, controls = 48,466) and ADGC2 (cases = 2122, controls = 3213). As the separate IGAP Stage 2 summary statistics are not publically available, in our meta-analysis, we used the combined IGAP Stage 1 and 2 sample which was available publically. The meta-analyses were conducted using the R package meta (http://CRAN.R-project.org/package=meta). Briefly, the fixed effects, inverse variance-weighted meta-analysis summarizes the combined statistical support across independent studies under the assumption of homogeneity of effects. Individual study estimates (log odds ratios) are averaged, weighted by the estimated standard error [23].
Functional evaluation of shared risk loci
To assess whether SNPs that are shared between AD and CV-associated RFs modify gene expression, we identified cis-expression quantitative loci (eQTLs, defined as variants within 1 Mb of a gene’s transcription start site) and regional brain expression of AD/CV SNPs in a publicly available dataset of normal control brains (UKBEC, http://braineac.org [36]). Given the evaluation of CV-associated RFs, we also evaluated eQTLs using a blood-based dataset [45].
Gene expression alterations in AD brains
To determine whether the AD/CV pleiotropic genes are differentially expressed in AD brains, we analyzed gene expression of overlapping genes in a publicly available dataset. We accessed the Mayo Clinic Brain Bank (Mayo) RNAseq study from the Accelerating Medicines Partnership-Alzheimer’s Disease (AMP-AD) portal (syn3163039; accessed April 2017). We examined gene expression in the temporal cortex of brains with neuropathologic diagnosis of AD dementia (N = 82) and elderly control brains that lacked a diagnosis of neurodegenerative disease (N = 80) [1]. Multi-variable linear regression analyses were conducted using CQN normalized gene expression measures and including age at death, gender, RNA integrity number (RIN), brain tissue source, and flow cell as biological and technical covariates.
Results
Pleiotropic enrichment in AD conditional on plasma lipid levels
For progressively stringent p value thresholds for AD SNPs [i.e., increasing values of nominal − log10(p)], we found approximately 100-fold enrichment using LDL, 75-fold enrichment using TC, 65-fold enrichment using TG, and 25-fold enrichment using HDL (Fig. 1). In comparison, we found minimal to no enrichment with BMI, T2D, CAD, and WHR. Together, these findings suggest selective genetic overlap between plasma lipids and AD. We note that these results reflect genetic enrichment in AD as a function of CV-associated RFs after the exclusion of SNPs in LD with HLA, MAPT, and APOE (see “Methods“).
Fig. 1.
Fold enrichment plots of nominal − log10 p values (corrected for inflation and excluding APOE, MAPT, and HLA regions) in Alzheimer’s disease (AD) below the standard GWAS threshold of p < 5 × 10−8 as a function of significance of association with body mass index (BMI), type 2 diabetes (T2D), coronary artery disease (CAD), waist hip ratio (WHR), total cholesterol (TC), triglycerides (TG), low-density lipoprotein (LDL), and high-density lipoprotein (HDL) at the level of p ≤ 1, p ≤ 0.1, p ≤ 0.01, respectively. Blue line indicates all SNPs
Given the long-range LD associated with the APOE/TOMM40 region [49], we focused our pleiotropy analyses on genetic variants outside chromosome 19. At a conjunction FDR< 0.05, we identified 90 SNPs, in total, across 19 chromosomes jointly associated with AD and the CV-associated RFs (Fig. 2; Table 2). After accounting for LD, we identified several AD-/CV-associated loci involved in cholesterol/lipid function including variants within ABCG5, ABCA1, and APOA4.
Fig. 2.
Conjunction Manhattan plot of conjunction − log10 (FDR) values for Alzheimer’s disease (AD) alone (black) and AD given body mass index (BMI; AD&BMI, red), type 2 diabetes (T2D; AD&T2D, blue), coronary artery disease (CAD; AD&CAD, pink), waist hip ratio (WHR; AD&WHR, magenta), total cholesterol (TC; AD&TC, green), triglycerides (TG; AD&TG, teal), low-density lipoprotein (LDL; &LDL, purple) and high-density lipoprotein (HDL, AD|HDL, maroon). SNPs with conjunction − log10 FDR > 1.3 (i.e., FDR < 0.05) are shown with large points. A black line around the large points indicates the most significant SNP in each LD block and this SNP was annotated with the closest gene, which is listed above
Table 2.
Overlapping loci between AD and CV RFs at a conjunction FDR < 0.05
| SNP | Chr | Closest gene | A1 | Reference trait | Min ConjFDR | AD p value | Reference trait p value | |
|---|---|---|---|---|---|---|---|---|
| 1 | rs61779841 | 1 | TRIT1 | A | HDL | 3.75E−02 | 5.44E−04 | 7.37E−04 |
| 2 | rs78363635 | 1 | C4BPA | C | LDL | 2.02E−02 | 8.30E−04 | 5.46E−05 |
| 3 | rsl759499 | 1 | USP24 | G | LDL | 2.50E−02 | 1.05E−03 | 1.57E−09 |
| 4 | rs6587723 | 1 | OTUD7B | C | TC | 2.89E−02 | 3.26E−04 | 1.50E−03 |
| 5 | rs1431985 | 1 | AK092251 | A | TG | 3.78E−02 | 6.63E−04 | 4.20E−04 |
| 6 | rs858952 | 2 | NRXN1 | C | BMI | 1.11E−02 | 9.45E−06 | 2.22E−04 |
| 7 | rs6733839 | 2 | BIN1 | T | HDL | 4.38E−02 | 7.11E−26 | 8.94E−04 |
| 8 | rs72796734 | 2 | ABCG5 | T | LDL | 2.02E−02 | 8.29E−04 | 2.33E−05 |
| 9 | rs55819441 | 2 | AK097952 | T | LDL | 2.30E−02 | 9.56E−04 | 1.40E−04 |
| 10 | rs7421448 | 2 | INPP5D | T | LDL | 2.58E−02 | 5.84E−04 | 1.45E−03 |
| 11 | rs12994639 | 2 | SERTAD2 | G | TC | 4.35E−02 | 1.60E−03 | 9.53E−05 |
| 12 | rs61208496 | 2 | C20RF56 | T | WHR | 3.22E−02 | 5.73E−05 | 1.88E−04 |
| 13 | rs6805910 | 3 | ARHGEF3 | C | HDL | 3.78E−02 | 6.10E−04 | 6.93E−04 |
| 14 | rs28670348 | 4 | INPP4B | G | HDL | 4.79E−02 | 1.81E−04 | 1.01E−03 |
| 15 | rsl3114818 | 4 | UBA6 | C | TC | 1.88E−02 | 6.28E−04 | 8.96E−04 |
| 16 | rs6852075 | 4 | ART3 | G | TG | 2.80E−02 | 4.02E−04 | 5.17E−04 |
| 17 | rs2074613 | 5 | HBEGF | C | BMI | 1.30E−03 | 9.29E−07 | 1.36E−05 |
| 18 | rs4912851 | 5 | SPRY4 | G | WHR | 1.99E−02 | 3.39E−05 | 2.32E−05 |
| 19 | rsl2188460 | 5 | FBXL17 | G | HDL | 4.20E−02 | 6.23E−04 | 8.49E−04 |
| 20 | rs5744712 | 5 | POLK | C | LDL | 3.15E−02 | 1.35E−03 | 1.29E−17 |
| 21 | rs6883056 | 5 | PRLR | C | LDL | 3.96E−02 | 8.48E−05 | 2.30E−03 |
| 22 | rs62383992 | 5 | FGF18 | A | TC | 3.64E−02 | 1.30E−03 | 9.12E−04 |
| 23 | rs2176298 | 5 | LOC285629 | T | TG | 2.52E−02 | 1.50E−04 | 4.56E−04 |
| 24 | rsl41129230 | 6 | HLA-B | G | HDL | 4.15E−02 | 6.73E−04 | 1.75E−04 |
| 25 | rs145749015 | 6 | HLA-DQB1 | T | HDL | 2.11E−03 | 2.71E−05 | 6.54E−06 |
| 26 | rsl15785781 | 6 | HCG18 | C | LDL | 3.17E−02 | 1.35E−03 | 1.81E−05 |
| 27 | rs9272561 | 6 | HLA-DQA1 | G | TC | 2.17E−05 | 5.37E−09 | 7.23E−07 |
| 28 | rsl15795926 | 6 | HLA-DQA2 | C | LDL | 5.84E−05 | 1.94E−06 | 1.28E−06 |
| 29 | rsl15674098 | 6 | HLA-DRA | T | LDL | 2.85E−05 | 9.28E−07 | 2.21E−08 |
| 30 | rsl16715716 | 6 | HLA-DRB1 | T | TC | 2.57E−03 | 7.87E−05 | 2.25E−05 |
| 31 | rs7774782 | 6 | PRIM2 | C | TC | 9.25E−03 | 2.93E−04 | 1.83E−04 |
| 32 | rs3103351 | 6 | SLC22A2 | G | LDL | 4.06E−02 | 1.78E−03 | 4.04E−06 |
| 33 | rsl15802139 | 6 | BTNL2 | G | T2D | 8.23E−04 | 4.39E−06 | 2.35E−07 |
| 34 | rsl14465688 | 6 | C60RF10 | G | T2D | 1.66E−02 | 9.45E−05 | 1.23E−04 |
| 35 | rs536810 | 6 | HLA-DRB5 | T | WHR | 4.51E−03 | 7.18E−06 | 4.33E−14 |
| 36 | rsl2194027 | 6 | ELOVL5 | C | TG | 1.03E−02 | 1.39E−04 | 1.53E−04 |
| 37 | rsl15813375 | 6 | HLA-C | A | TG | 3.27E−02 | 5.67E−04 | 1.05E−06 |
| 38 | rsl048365 | 7 | AP1S1 | T | BMI | 2.18E−02 | 7.84E−05 | 2.22E−04 |
| 39 | rs2597283 | 7 | BC043356 | C | BMI | 1.53E−02 | 4.20E−05 | 3.46E−04 |
| 40 | rs35991721 | 7 | MB LAC1 | T | CAD | 1.03E−02 | 5.77E−05 | 3.22E−06 |
| 41 | rs702483 | 7 | RAC1 | T | HDL | 3.82E−02 | 6.18E−04 | 3.11E−04 |
| 42 | rsl2056620 | 8 | PTK2B | T | BMI | 2.12E−02 | 7.56E−05 | 3.35E−04 |
| 43 | rs2011566 | 8 | C80RF38 | G | CAD | 4.47E−02 | 2.78E−04 | 3.83E−04 |
| 44 | rs7014168 | 8 | SOX7 | A | LDL | 1.09E−02 | 4.28E−04 | 4.01E−04 |
| 45 | rsl6895579 | 8 | TSPYL5 | A | LDL | 1.27E−03 | 8.90E−06 | 5.77E−05 |
| 46 | rsl17922969 | 8 | AK055863 | T | TC | 3.97E−02 | 1.43E−03 | 5.31E−04 |
| 47 | rsl3277568 | 8 | TRPS1 | G | TC | 3.67E−02 | 1.19E−03 | 1.17E−03 |
| 48 | rsl0991386 | 9 | ABCA1 | G | TC | 2.80E−03 | 8.54E−05 | 6.19E−07 |
| 49 | rsl2339683 | 9 | IDNK | T | LDL | 3.08E−02 | 1.31E−03 | 3.11E−04 |
| 50 | rsl1144711 | 9 | PCSK5 | G | LDL | 4.23E−02 | 5.65E−04 | 2.49E−03 |
| 51 | rs145301439 | 10 | ARMC3 | A | HDL | 1.61E−02 | 2.42E−04 | 1.57E−04 |
| 52 | rsl2784561 | 10 | CR595071 | A | LDL | 2.55E−02 | 3.80E−04 | 1.43E−03 |
| 53 | rsl2783314 | 10 | LOC219347 | A | LDL | 2.72E−02 | 2.60E−04 | 1.53E−03 |
| 54 | rsl0906257 | 10 | CCDC3 | G | TC | 1.36E−02 | 4.39E−04 | 4.72E−04 |
| 55 | rs7098392 | 10 | CHST15 | A | TC | 3.81E−02 | 1.37E−03 | 9.00E−04 |
| 56 | rs6597951 | 11 | AP2A2 | C | BMI | 1.03E−02 | 2.94E−05 | 1.38E−04 |
| 57 | rs7928842 | 11 | CELF1 | C | BMI | 2.37E−02 | 8.75E−05 | 3.19E−24 |
| 58 | rsl893306 | 11 | GUCY2EP | G | BMI | 4.26E−02 | 4.25E−05 | 1.46E−03 |
| 59 | rsl534576 | 11 | SLC39A13 | T | BMI | 1.79E−03 | 3.21E−06 | 6.62E−08 |
| 60 | rsl1039131 | 11 | DDB2 | T | TG | 6.47E−03 | 4.08E−05 | 8.55E−05 |
| 61 | rs2071305 | 11 | MYBPC3 | C | HDL | 2.58E−04 | 3.01E−06 | 2.53E−07 |
| 62 | rs3844143 | 11 | PICALM | T | LDL | 1.44E−02 | 1.94E−08 | 7.79E−04 |
| 63 | rs1263170 | 11 | APOA4 | T | TG | 3.73E−02 | 6.55E−04 | 4.33E−09 |
| 64 | rsl1039297 | 11 | PTPMT1 | A | WHR | 8.51E−03 | 1.24E−05 | 5.15E−05 |
| 65 | rs7972529 | 12 | RPL6 | G | LDL | 9.05E−03 | 3.52E−04 | 4.49E−04 |
| 66 | rs77451327 | 12 | SOAT2 | C | TC | 4.58E−02 | 9.06E−04 | 2.56E−03 |
| 67 | rsl635142 | 12 | OAS2 | A | WHR | 3.01E−02 | 5.32E−05 | 2.28E−04 |
| 68 | rs7331792 | 13 | BC038529 | A | LDL | 2.93E−02 | 1.25E−03 | 4.69E−04 |
| 69 | rs61963560 | 13 | BC035340 | A | TC | 3.61E−02 | 5.92E−04 | 1.94E−03 |
| 70 | rs7981577 | 13 | RASA3 | C | TC | 4.16E−02 | 1.37E−04 | 2.28E−03 |
| 71 | rs17125924 | 14 | FERMT2 | G | BMI | 3.65E−02 | 1.48E−05 | 1.17E−03 |
| 72 | rs650366 | 15 | FAM63B | G | TC | 1.96E−02 | 6.54E−04 | 6.86E−04 |
| 73 | rs3131575 | 15 | USP8 | G | TC | 1.42E−02 | 4.59E−04 | 4.34E−04 |
| 74 | rs16953089 | 16 | FTO | C | BMI | 3.32E−02 | 1.36E−04 | 8.62E−04 |
| 75 | rs9941245 | 16 | GPRC5B | G | BMI | 4.96E−02 | 2.29E−04 | 5.27E−16 |
| 76 | rs4985557 | 16 | MTSS1L | T | BMI | 1.02E−02 | 2.87E−05 | 1.19E−04 |
| 77 | rs9931998 | 16 | BC040927 | A | LDL | 3.45E−02 | 5.23E−04 | 1.99E−03 |
| 78 | rsl2595955 | 16 | CDH5 | G | LDL | 3.98E−02 | 1.74E−03 | 4.69E−04 |
| 79 | rs246174 | 16 | MKL2 | T | LDL | 1.93E−02 | 7.89E−04 | 5.91E−04 |
| 80 | rs79161472 | 16 | ZNF668 | A | TC | 1.78E−02 | 5.87E−04 | 6.23E−04 |
| 81 | rs4985556 | 16 | IL34 | A | T2D | 3.42E−02 | 2.11E−04 | 4.10E−04 |
| 82 | rs8062895 | 16 | DHODH | G | TC | 4.27E−02 | 1.56E−03 | 4.12E−04 |
| 83 | rs8070572 | 17 | MINK1 | C | BMI | 2.33E−02 | 4.92E−06 | 6.24E−04 |
| 84 | rs2960171 | 17 | ZNF652 | C | CAD | 2.33E−02 | 1.37E−04 | 8.72E−05 |
| 85 | rs7221196 | 17 | ITGB3 | G | LDL | 4.67E−03 | 1.78E−04 | 1.57E−07 |
| 86 | rs8071250 | 17 | PRKCA | C | LDL | 2.18E−02 | 7.56E−04 | 1.21E−03 |
| 87 | rs850520 | 17 | AK097513 | A | TG | 7.79E−03 | 1.25E−04 | 1.08E−04 |
| 88 | rs9954848 | 18 | LIPG | A | TC | 2.19E−02 | 4.58E−04 | 1.09E−03 |
| 89 | rs2298428 | 22 | YDJC | T | HDL | 6.45E−03 | 9.00E−05 | 1.58E−08 |
| 90 | rs4821116 | 22 | UBE2L3 | T | TC | 1.50E−02 | 4.02E−04 | 7.10E−04 |
Chromosome 19 SNPs are excluded
SNP single nucleotide polymorphism, Chr chromosome, Min ConjFDR minimum conjunction false discovery rate, AD Alzheimer’s disease
For the 90 pleiotropic SNPs, we conducted a meta-analysis across IGAP Stages 1 and 2 and ADGC2. We focused on SNPs found in all three cohorts and identified six variants with p < 5.0 × 10−8 (Table 3; Fig. 3a–f): (1) rs6733839 (chromosome 2, closest gene = BIN1, conditioning trait = HDL, reference allele = T, OR = 1.210, 95% CI 1.18–1.1.25, p = 1.44 × 10−45), (2) rs1534576 (chromosome 11, closest gene = SLC39A13, conditioning trait = BMI, reference allele = T, OR = 1.080, 95% CI 1.05–1.11, p = 1.49 × 10−9), (3) rs3844143 (chromosome 11, closest gene = PICALM, conditioning trait = LDL, reference allele = T, OR = 0.899, 95% CI 0.877–0.922, p = 6.52 × 10−17), (4) rs17125924 (chromosome 14, closest gene = FERMT2, conditioning trait = BMI, reference allele = G, OR = 1.130, 95% CI 1.08–1.18, p = 2.62 × 10−8), (5) rs35991721 (chromosome 7, closest gene = MBLAC1/GATS, conditioning trait = CAD, reference allele = T, OR = 0.921, 95% CI 0.896–0.947, p = 1.44 × 10−9), (6) rs536810 (chromosome 6, closest gene = HLA-DRB5, conditioning trait = WHR, reference allele = T, OR = 0.924, 95% CI 0.899–0.95, p = 1.14 × 10−8).
Table 3.
Meta-analysis using ADGC Phase 2 and IGAP stages 1 and 2 cohorts
| SNP | Chr | Closest gene | A1 | Ref trait | ADGC2 p value |
ADGC2 OR |
ADGC2 95% CI |
IGAP 1 and 2 p value |
IGAP 1 and 2 OR |
IGAP 1 and 2 95% CI |
Meta-p value | Meta-OR | Meta-95 % CI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs1431985 | 1 | AK092251 | A | TG | 6.55E−01 | 0.9779 | 0.887−1.08 | 2.40E−03 | 1.04 | 1.01−1.07 | 4.84E−03 | 1.04 | 1.01–1.06 |
| 2 | rs78363635 | 1 | C4BPA | C | LDL | 7.42E−01 | 0.9625 | 0.767−1.21 | 6.22E−03 | 1.09 | 1.02−1.16 | 1.03E−02 | 1.08 | 1.02–1.14 |
| 3 | rs55819441 | 2 | AK097952 | T | LDL | 6.35E−01 | 1.049 | 0.861−1.28 | 7.55E−02 | 0.962 | 0.922−1 | 1.01E−01 | 0.966 | 0.927–1.01 |
| 4 | rs6733839 | 2 | BIN1 | T | HDL | 8.05E−03 | 1.165 | 1.04−1.3 | 6.94E−44 | 1.22 | 1.22−1.22 | 1.44E−45 | 1.21 | 1.18–1.25 |
| 5 | rs72796734 | 2 | ABCG5 | T | LDL | 1.15E−01 | 0.8402 | 0.677−1.04 | 3.26E−02 | 0.937 | 0.883−0.995 | 1.32E−02 | 0.93 | 0.878–0.985 |
| 6 | rs7421448 | 2 | INPP5D | T | LDL | 5.75E−01 | 1.06 | 0.865−1.3 | 2.23E−04 | 0.911 | 0.867−0.957 | 5.59E−04 | 0.918 | 0.875–0.964 |
| 7 | rs858952 | 2 | NRXN1 | C | BMI | 4.67E−01 | 0.9602 | 0.861−1.07 | 3.08E−03 | 1.05 | 1.02−1.08 | 7.69E−03 | 1.04 | 1.01–1.07 |
| 8 | rsl3114818 | 4 | UBA6 | C | TC | 4.08E−01 | 0.9441 | 0.824−1.08 | 3.03E−03 | 1.05 | 1.02−1.08 | 7.50E−03 | 1.05 | 1.01–1.08 |
| 9 | rs28670348 | 4 | INPP4B | G | HDL | 9.30E−01 | 0.9918 | 0.826−1.19 | 7.76E−03 | 1.07 | 1.02−1.12 | 1.10E−02 | 1.06 | 1.01–1.12 |
| 10 | rs6852075 | 4 | ART3 | G | TG | 6.23E−01 | 0.9767 | 0.889−1.07 | 1.96E−02 | 0.969 | 0.944−0.995 | 1.77E−02 | 0.97 | 0.945–0.995 |
| 11 | rsl2188460 | 5 | FBXL17 | G | HDL | 2.50E−01 | 1.057 | 0.962−1.16 | 4.85E−03 | 1.04 | 1.01−1.07 | 2.64E−03 | 1.04 | 1.01–1.06 |
| 12 | rs2176298 | 5 | LOC285629 | T | TG | 4.52E−01 | 0.9628 | 0.872−1.06 | 1.47E−02 | 1.03 | 1.01−1.05 | 3.07E−02 | 1.03 | 1–1.06 |
| 13 | rs4912851 | 5 | SPRY4 | G | WHR | 2.32E−01 | 1.06 | 0.963−1.17 | 7.07E−03 | 1.04 | 1.01−1.07 | 3.58E−03 | 1.04 | 1.01–1.06 |
| 14 | rs536810 | 6 | HLA-DRB5 | T | WHR | 1.53E−01 | 0.9311 | 0.844−1.03 | 3.48E−08 | 0.923 | 0.897−0.95 | 1.14E−08 | 0.924 | 0.899–0.949 |
| 15 | rs2597283 | 7 | BC043356 | C | BMI | 3.53E−01 | 1.048 | 0.949−1.16 | 1.67E−07 | 0.932 | 0.908−0.957 | 1.65E−06 | 0.939 | 0.915–0.963 |
| 16 | rs35991721 | 7 | MBLAC1 | T | CAD | 7.17E−04 | 0.8255 | 0.739−0.923 | 2.62E−07 | 0.928 | 0.902−0.955 | 5.32E−09 | 0.921 | 0.896–0.947 |
| 17 | rs702483 | 7 | RAC1 | T | HDL | 5.60E−01 | 1.029 | 0.935−1.13 | 1.75E−02 | 0.97 | 0.946−0.995 | 3.17E−02 | 0.973 | 0.95–0.998 |
| 18 | rsl2056620 | 8 | PTK2B | T | BMI | 5.50E−01 | 1.029 | 0.937−1.13 | 2.66E−03 | 1.04 | 1.01−1.07 | 2.23E−03 | 1.04 | 1.01–1.07 |
| 19 | rs7014168 | 8 | SOX7 | A | LDL | 6.74E−01 | 1.024 | 0.917−1.14 | 1.39E−04 | 0.942 | 0.913−0.971 | 3.57E−04 | 0.948 | 0.92–0.976 |
| 20 | rsl0906257 | 10 | CCDC3 | G | TC | 7.75E−01 | 1.022 | 0.88−1.19 | 5.37E−03 | 1.06 | 1.02−1.1 | 5.80E−03 | 1.06 | 1.02–1.1 |
| 21 | rsl2784561 | 10 | CR595071 | A | LDL | 3.50E−01 | 0.9262 | 0.789−1.09 | 1.31E−04 | 0.918 | 0.879−0.959 | 8.51E−05 | 0.918 | 0.88–0.958 |
| 22 | rsl1039131 | 11 | DDB2 | T | TG | 2.18E−02 | 0.8803 | 0.789−0.982 | 5.18E−06 | 0.938 | 0.913−0.964 | 7.01E−07 | 0.934 | 0.91–0.96 |
| 23 | rsl263170 | 11 | APOA4 | T | TG | 8.95E−01 | 1.007 | 0.908−1.12 | 3.37E−03 | 1.05 | 1.02−1.08 | 4.22E−03 | 1.04 | 1.01–1.07 |
| 24 | rsl534576 | 11 | SLC39A13 | T | BMI | 7.03E−03 | 1.139 | 1.04−1.25 | 2.97E−08 | 1.08 | 1.05−1.11 | 1.49E−09 | 1.08 | 1.05–1.11 |
| 25 | rs1893306 | 11 | GUCY2EP | G | BMI | 6.68E−01 | 1.023 | 0.922−1.13 | 1.09E−03 | 0.953 | 0.926−0.981 | 2.35E−03 | 0.958 | 0.932–0.985 |
| 26 | rs2071305 | 11 | MYBPC3 | C | HDL | 1.12E−03 | 0.8264 | 0.737−0.927 | 1.54E−06 | 0.934 | 0.908−0.96 | 5.62E−08 | 0.928 | 0.903–0.953 |
| 27 | rs3844143 | 11 | PICALM | T | LDL | 1.62E−01 | 0.9349 | 0.851−1.03 | 1.33E−16 | 0.896 | 0.873−0.92 | 6.52E−17 | 0.899 | 0.877–0.922 |
| 28 | rsl635142 | 12 | OAS2 | A | WHR | 4.61E−01 | 1.04 | 0.937−1.15 | 1.50E−04 | 0.947 | 0.921−0.974 | 5.00E−04 | 0.953 | 0.927–0.979 |
| 29 | rs61963560 | 13 | BC035340 | A | TC | 7.43E−02 | 1.126 | 0.988−1.28 | 7.58E−04 | 0.939 | 0.905−0.974 | 5.79E−03 | 0.952 | 0.919–0.986 |
| 30 | rs7981577 | 13 | RASA3 | C | TC | 8.34E−01 | 0.9898 | 0.899−1.09 | 9.56E−02 | 0.976 | 0.949−1 | 9.76E−02 | 0.977 | 0.95–1 |
| 31 | rs17125924 | 14 | FERMT2 | G | BMI | 7.69E−01 | 1.026 | 0.864−1.22 | 1.55E−08 | 1.13 | 1.08−1.18 | 2.62E−08 | 1.13 | 1.08–1.18 |
| 32 | rs3131575 | 15 | USP8 | G | TC | 3.52E−01 | 1.055 | 0.943−1.18 | 3.74E−05 | 0.933 | 0.903−0.964 | 2.13E−04 | 0.942 | 0.913–0.972 |
| 33 | rsl6953089 | 16 | FTO | C | BMI | 1.84E−01 | 1.071 | 0.968−1.19 | 9.85E−04 | 0.954 | 0.928−0.981 | 4.63E−03 | 0.962 | 0.937–0.988 |
| 34 | rs246174 | 16 | MKL2 | T | LDL | 7.04E−01 | 0.981 | 0.889−1.08 | 4.41E−03 | 1.04 | 1.01−1.07 | 8.23E−03 | 1.04 | 1.01–1.06 |
| 35 | rs4985556 | 16 | IL34 | A | T2D | 7.48E−01 | 1.025 | 0.882−1.19 | 1.41E−06 | 1.11 | 1.06−1.16 | 2.06E−06 | 1.1 | 1.06–1.14 |
| 36 | rs4985557 | 16 | MTSS1L | T | BMI | 1.39E−01 | 1.077 | 0.976−1.19 | 9.28E−05 | 1.05 | 1.02−1.08 | 3.34E−05 | 1.06 | 1.03–1.08 |
| 37 | rs9931998 | 16 | BC040927 | A | LDL | 5.89E−01 | 1.038 | 0.907−1.19 | 4.59E−04 | 0.94 | 0.908−0.973 | 1.12E−03 | 0.946 | 0.915–0.978 |
| 38 | rs9941245 | 16 | GPRC5B | G | BMI | 9.04E−02 | 0.9022 | 0.801−1.02 | 9.42E−05 | 0.937 | 0.907−0.968 | 2.58E−05 | 0.934 | 0.905–0.964 |
| 39 | rs8070572 | 17 | MINK1 | C | BMI | 3.05E−02 | 1.195 | 1.02−1.4 | 1.49E−06 | 1.11 | 1.06−1.16 | 1.98E−07 | 1.12 | 1.07–1.17 |
| 40 | rs8071250 | 17 | PRKCA | C | LDL | 8.83E−01 | 0.9929 | 0.903−1.09 | 4.27E−04 | 0.955 | 0.931−0.98 | 5.96E−04 | 0.958 | 0.935–0.982 |
| 41 | rs9954848 | 18 | LIPG | A | TC | 4.22E−01 | 0.9605 | 0.87−1.06 | 1.61E−03 | 0.959 | 0.934−0.984 | 1.12E−03 | 0.959 | 0.935–0.983 |
| 42 | rs2298428 | 22 | YDJC | T | HDL | 6.47E−01 | 1.028 | 0.914−1.16 | 5.04E−04 | 0.942 | 0.911−0.974 | 1.34E−03 | 0.948 | 0.918–0.979 |
Bold values indicate p < 1 × 10−6
SNP single nucleotide polymorphism, Chr chromosome, Ref reference, OR odds ratio, CI confidence interval
Fig. 3.
Forest plots for a rs6733839 on chromosome 2, b rs1534576 on chromosome 11, c rs3844143 on chromosome 11, d rs17125924 on chromosome 14, e rs35991721 on chromosome 7, and f rs536810 on chromosome 6
We also identified three AD susceptibility loci at p < 1.0 × 10−6 (Table 3; Supplemental Figure 1): (1) rs11039131 (chromosome 11, closest gene = DDB2, conditioning trait = TG, reference allele = T, OR = 0.934, 95% CI 0.91–0.96, p = 7.01 × 10−7), 2) rs8070572 (chromo-some 17, closest gene = MINK1, conditioning trait = BMI, reference allele = C, OR = 1.120, 95% CI 1.07–1.17, p = 1.98 × 10−7), and (3) rs2071305 (chromosome 11, closest gene = MYBPC3, conditioning trait = HDL, reference allele = C, OR = 0.928, 95% CI 0.903–0.953, p = 5.62 × 10−8).
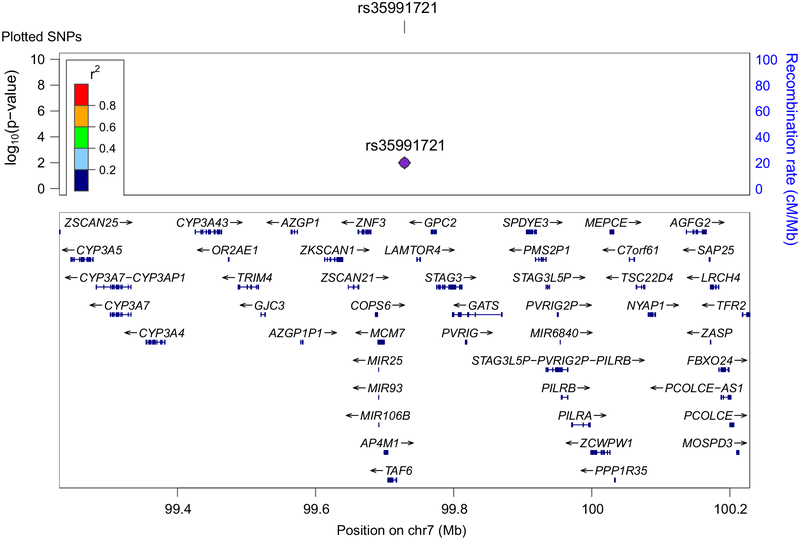
These meta-analyses point to novel AD-associated susceptibility loci. On chromosome 7, we found that the genome-wide significant rs35991721 was not in LD with the previously reported SNP rs1476679 ([24], r2 = 0.28, D′ = 0.56) and may be tagging genetic signal within GATS, STAG3 or PVRIG (Fig. 4). On chromosome 11 within the CELF1 region, we detected independent signal within rs1534576, rs11039131 and rs2071305 (Fig. 5). The genome-wide significant rs1534576 was in LD with the previously reported rs10838725 (r2 = 0.64, D′ = 0.99) indicating that these two SNPs may be tagging signal within CELF1 [24]. In contrast, rs11039131 and rs2071305 were not in LD with rs10838725 suggesting independent signal from CELF1 (Fig. 5). Of interest, rs2071305 (but not rs11039131) was in LD with rs1057233 (r2 = 0.65, D′ = 0.99), a SNP that has been associated with AD age of onset in a survival analysis [20]. Collectively, these results suggest several different AD-associated genetic variants within chromosome 11.
Fig. 4.
Regional association plots for rs35991721 on chromosome 7. Linkage disequilibrium measured in the 1000 genomes European populations
Fig. 5.
The pair-wise linkage disequilibrium patterns between rs1534576, rs11039131 rs2071305, rs10838725, and rs1057233 on chromosome 11
We also assessed whether the AD/CV pleiotropic SNPs listed in Table 2 replicated in an AD-by-proxy cohort. Of the 90 IGAP pleiotropic SNPs, 68 SNPs were available in the UKB AD-by-proxy cohort. We identified 20 significant SNPs at p < 0.05 (Table 4). The replicated variants include three of the four novel AD/CV pleiotropic SNPs, namely variants within MINK1, MBLAC1, and DDB2.
Table 4.
Replication of AD/CVD pleiotropic SNPs in a UKB AD-by-proxy cohort
| SNP | Chr | Closest Gene | BP | A1 | NMfSS | P | OR | Cfs | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rsl431985 | 1 | AK092251 | 214148246 | A | 362011 | 8.04E−01 | 1 | 1–1 |
| 2 | rs61779841 | 1 | TRIT1 | 40324666 | A | 364772 | 8.12E−01 | 1 | 1–1 |
| 3 | rs78363635 | 1 | C4BPA | 207324781 | C | 364859 | 2.05E−01 | 1.002 | 0.999–1.01 |
| 4 | rs12994639 | 2 | SERTAD2 | 64959331 | G | 364859 | 2.34E−01 | 1.002 | 0.999–1.01 |
| 5 | rs55819441 | 2 | AK097952 | 65082415 | T | 364859 | 6.27E−01 | 0.9992 | 0.996–1 |
| 6 | rs61208496 | 2 | C20RF56 | 37464230 | T | 363628 | 8.04E−03 | 1.004 | 1–1.01 |
| 7 | rs72796734 | 2 | ABCG5 | 44063731 | T | 364005 | 6.24E−01 | 1.001 | 0.997–1.01 |
| 8 | rs7421448 | 2 | fNPP5D | 233982205 | T | 364859 | 1.52E−05 | 0.9929 | 0.99–0.996 |
| 9 | rs858952 | 2 | NRXN1 | 50875879 | C | 353852 | 2.34E−01 | 1.002 | 0.999–1.01 |
| 10 | rs6805910 | 3 | ARHGEF3 | 56739923 | C | 364232 | 7.18E−01 | 0.9994 | 0.996–1 |
| 11 | rs13114818 | 4 | UBA6 | 68550295 | C | 361934 | 4.40E−01 | 0.9987 | 0.995–1 |
| 12 | rs28670348 | 4 | fNPP4B | 143625388 | G | 362077 | 9.18E−01 | 1 | 1–1 |
| 13 | rs12188460 | 5 | FBXL17 | 107172269 | G | 357888 | 4.58E−01 | 0.9988 | 0.996–1 |
| 14 | rs2074613 | 5 | HBEGF | 139714564 | C | 364859 | 1.57E−01 | 1.002 | 0.999–1 |
| 15 | rs2176298 | 5 | LOC285629 | 160388643 | T | 364192 | 1.86E−01 | 1.002 | 0.999–1 |
| 16 | rs4912851 | 5 | SPRY4 | 141815488 | G | 359562 | 9.38E−01 | 1 | 1–1 |
| 17 | rs5744712 | 5 | POLK | 74892002 | C | 364232 | 9.67E−01 | 0.9999 | 0.995–1 |
| 18 | rs62383992 | 5 | FGF18 | 170866296 | A | 356784 | 1.50E−01 | 0.9976 | 0.994–1 |
| 19 | rs6883056 | 5 | PRLR | 35080145 | C | 363845 | 4.71E−01 | 1.001 | 0.998–1 |
| 20 | rsl2194027 | 6 | ELOVL5 | 53255776 | C | 364000 | 1.47E−01 | 0.9976 | 0.994–1 |
| 21 | rs3103351 | 6 | SLC22A2 | 160716066 | G | 363577 | 4.52E−02 | 0.9967 | 0.993–1 |
| 22 | rs536810 | 6 | HLA-DRB5 | 32577497 | T | 363853 | 2.03E−04 | 0.9939 | 0.991–0.997 |
| 23 | rs7774782 | 6 | PRIM2 | 57618491 | C | 362322 | 5.06E−01 | 1.001 | 0.998–1 |
| 24 | rs1048365 | 7 | AP1S1 | 100804430 | T | 363555 | 9.45E−01 | 0.9999 | 0.997–1 |
| 25 | rs2597283 | 7 | BC043356 | 37690507 | C | 363815 | 6.48E−02 | 0.997 | 0.994–1 |
| 26 | rs35991721 | 7 | MBLAC1 | 99728790 | T | 364144 | 2.34E−04 | 0.9939 | 0.991–0.997 |
| 27 | rs702483 | 7 | RAC1 | 6426941 | T | 362242 | 1.05E−01 | 0.9973 | 0.994–1 |
| 28 | rs117922969 | 8 | AK055863 | 9257853 | T | 364639 | 5.98E−01 | 0.9991 | 0.996–1 |
| 29 | rs12056620 | 8 | PTK2B | 27291749 | T | 364405 | 1.27E−01 | 1.003 | 0.999–1.01 |
| 30 | rsl3277568 | 8 | TRPS1 | 116679547 | G | 364859 | 2.61E−01 | 1.002 | 0.999–1.01 |
| 31 | rsl6895579 | 8 | TSPYL5 | 98364076 | A | 363783 | 1.08E−01 | 1.003 | 0.999–1.01 |
| 32 | rs2011566 | 8 | C80RF38 | 95971921 | G | 363930 | 3.35E−03 | 0.9952 | 0.992–0.998 |
| 33 | rs7014168 | 8 | SOX7 | 10641965 | A | 363086 | 5.65E−02 | 0.9968 | 0.994–1 |
| 34 | rsl0991386 | 9 | ABCA1 | 107630433 | G | 348514 | 2.33E−02 | 1.004 | 1–1.01 |
| 35 | rsl1144711 | 9 | PCSK5 | 78614020 | G | 362976 | 8.55E−01 | 1 | 1–1 |
| 36 | rsl2339683 | 9 | IDNK | 86214149 | T | 358092 | 3.06E−03 | 1.005 | 1–1.01 |
| 37 | rs12784561 | 10 | CR595071 | 11712965 | A | 364859 | 1.82E−01 | 0.9978 | 0.995–1 |
| 38 | rsl45301439 | 10 | ARMC3 | 23146430 | A | 357876 | 1.27E−01 | 0.9975 | 0.994–1 |
| 39 | rsl1039131 | 11 | DDB2 | 47232038 | T | 360088 | 3.34E−02 | 0.9965 | 0.993–1 |
| 40 | rsl1039297 | 11 | PTPMT1 | 47581443 | A | 364264 | 2.80E−02 | 1.004 | 1–1.01 |
| 41 | rsl263170 | 11 | APOA4 | 116678413 | T | 359973 | 4.42E−01 | 1.001 | 0.998–1 |
| 42 | rs1534576 | 11 | SLC39A13 | 47419663 | T | 363313 | 7.46E−04 | 1.006 | 1–1.01 |
| 43 | rsl893306 | 11 | GUCY2EP | 76434820 | G | 361099 | 5.58E−01 | 0.999 | 0.996–1 |
| 44 | rs3844143 | 11 | PICALM | 85850243 | C | 364859 | 5.31E−11 | 0.9892 | 0.986–0.992 |
| 45 | rs6597951 | 11 | AP2A2 | 991530 | C | 363427 | 1.70E−01 | 0.9977 | 0.994–1 |
| 46 | rs7928842 | 11 | CELF1 | 47566352 | C | 364616 | 8.25E−01 | 0.9996 | 0.996–1 |
| 47 | rs1635142 | 12 | OAS2 | 113434518 | A | 360076 | 3.06E−01 | 0.9983 | 0.995–1 |
| 48 | rs77451327 | 12 | SOAT2 | 53524259 | C | 364824 | 7.26E−01 | 1.001 | 0.995–1.01 |
| 49 | rs61963560 | 13 | BC035340 | 113605534 | A | 359151 | 8.67E−01 | 0.9997 | 0.996–1 |
| 50 | rs7981577 | 13 | RASA3 | 114835802 | C | 364119 | 3.44E−01 | 0.9984 | 0.995–1 |
| 51 | rsl7125924 | 14 | FERMT2 | 53391680 | G | 363118 | 1.47E−03 | 1.005 | 1–1.01 |
| 52 | rs3131575 | 15 | USP8 | 50731154 | G | 364208 | 3.79E−02 | 0.9966 | 0.993–1 |
| 53 | rs650366 | 15 | FAM63B | 59061142 | G | 361213 | 4.48E−07 | 0.9917 | 0.988–0.995 |
| 54 | rsl2595955 | 16 | CDH5 | 66144173 | G | 364594 | 9.66E−01 | 0.9999 | 0.995–1 |
| 55 | rs16953089 | 16 | FTO | 54155742 | C | 353751 | 6.54E−01 | 0.9992 | 0.996–1 |
| 56 | rs246174 | 16 | MKL2 | 14379931 | T | 357267 | 8.80E−01 | 0.9997 | 0.996–1 |
| 57 | rs4985556 | 16 | fL34 | 70694000 | A | 364859 | 3.55E−03 | 1.005 | 1–1.01 |
| 58 | rs4985557 | 16 | MTSS1L | 70704974 | T | 347131 | 6.93E−01 | 1.001 | 0.996–1.01 |
| 59 | rs8062895 | 16 | DHODH | 72048632 | G | 361194 | 1.81E−01 | 0.9978 | 0.995–1 |
| 60 | rs9941245 | 16 | GPRC5B | 19916895 | G | 360821 | 8.54E−01 | 0.9997 | 0.997–1 |
| 61 | rs2960171 | 17 | ZNF652 | 47378771 | C | 364076 | 9.80E−05 | 1.006 | 1–1.01 |
| 62 | rs7221196 | 17 | TTGB3 | 45374994 | G | 359882 | 3.70E−01 | 1.001 | 0.999–1 |
| 63 | rs8070572 | 17 | MINKl | 4766937 | C | 364784 | 6.38E−03 | 1.005 | 1–1.01 |
| 64 | rs8071250 | 17 | PRKCA | 64321567 | C | 364511 | 2.11E−02 | 0.9962 | 0.993–0.999 |
| 65 | rs850520 | 17 | AK097513 | 47333067 | A | 364105 | 2.40E−04 | 1.006 | 1–1.01 |
| 66 | rs9954848 | 18 | LIPG | 47131781 | A | 364682 | 1.23E−01 | 0.9975 | 0.994–1 |
| 67 | rs2298428 | 22 | YDJC | 21982892 | T | 364859 | 1.90E−01 | 0.9978 | 0.995–1 |
| 68 | rs4821116 | 22 | UBE2L3 | 21973319 | T | 364630 | 1.10E−01 | 0.9974 | 0.994–1 |
Bold values indicate p < 0.05
SNP single nucleotide polymorphism, Chr chromosome
Shared genetic risk between CV‑associated RFs
To evaluate whether the AD susceptibility loci listed in Table 2 are associated with a single CV-associated RF or with multiple associated RFs, we constructed a matrix plot. For each of the eight CV-associated RFs, we plotted the minimum conjunction FDR for all AD/CV closest genes (Fig. 6; Supplemental Table 1). We found that some common genetic variants influencing AD risk are associated with multiple CV-associated RFs. For minimum conjunction FDR < 0.05, variants within (1) ABCA1 were associated with CAD, lipid fractions, and WHR, (2) C6ORF10 with T2D and lipid fractions and (3) SPRY4 with BMI, lipid fractions, and WHR (Fig. 6).
Fig. 6.
Matrix plot mapping minimum conjunction FDR for the non-APOE AD/CV pleiotropic genes for each CV-associated RF. Asterisk indicates the conditioning RF used to identify the most significant SNP (see Table 2 and Fig. 2)
cis‑eQTLs
We focused on the four novel genetic variants (one genome-wide significant and three suggestive SNPs, see above) and found significant cis-associations in either brain or blood tissue types (Supplemental Table 2). None of the associations replicated in both tissue types. Within blood, rs8070572 showed a significant cis-eQTLs with PLD2 (Supplemental Table 2).
Gene expression in brains from AD patients and healthy controls
To investigate whether the AD/CV pleiotropic genes are differentially expressed in AD brains, we compared gene expression in AD brains with neuropathologically normal control brains. We focused on differential expression of the closest genes from the four novel genetic variants (one genome-wide significant and three suggestive SNPs, see above) and SPI1 based on LD within chromosome 11 (see above). We used a Bonferroni-corrected p value of < 0.01 and found significant effects for differential expression of MINK1, SPI1, DDB2 and MBLAC1 (Supplemental Table 3).
Discussion
Beyond APOE, we identified 90 SNPs on 19 different chromosomes that jointly conferred increased risk for AD and cardiovascular outcomes. In meta-analyses across three independent cohorts, we found four novel genetic variants that increased risk for AD. Three of these new susceptibility loci independently replicated in an AD-by-proxy cohort. Expression of three of these AD/CV pleiotropic genes was differentially altered within AD brains. Collectively, our findings suggest that the polygenic component of AD is highly enriched for cardiovascular RFs.
In their genetic association with AD, not all cardiovascular RFs are created equal. We found minimal genetic enrichment in AD as a function of T2D, BMI, WHR, and CAD suggesting that the known comorbidity [27, 34, 40] between these CV-associated RFs and Alzheimer’s etiology are likely not genetic. In contrast, genetic enrichment in AD was predominantly localized to plasma lipids. Each of the four plasma lipid RFs resulted in a comparable level of enrichment suggesting a tight correlation between the lipid fractions. Building on our prior work leveraging statistical power from large CV GWASs for AD gene discovery [13], we found genetic variants jointly associated with AD and CV-associated RFs, many with known cholesterol/lipid function. By conditioning on plasma TC, TG, LDL, and HDL levels, we identified AD susceptibility loci within genes encoding apolipoproteins, such as APOA4, ATP-binding cassette transporters, such as ABCA1 and ABCG5, and phospholipases, such as ATP8B4 and LIPG (for a discussion on lipid genes and AD, see Ref. [14]).
Cholesterol in the brain involves metabolic pathways that work independently from those in peripheral tissue. The blood–brain barrier (BBB) prevents peripheral cholesterol from entering and leaving the brain. In the adult brain, cholesterol is synthesized predominately in astrocytes and oligodendrocytes; minimal cholesterol is synthesized in neurons. Within glial cells, cholesterol is transported by apoE and secreted into the extracellular matrix via ABCA1- and ABCG1-associated mechanisms [50]. The cholesterol then binds to the low-density receptors (LDLR) on neuronal cells. This cholesterol is critical for synapse development, synapse formation, dendrite differentiation, and synaptic transmission [50]. In the periphery, cholesterol is produced in the liver or obtained through diet. Mounting epidemiological, clinical, and animal research indicates that high plasma lipid levels (i.e., hypercholesterolemia) act as a risk factor for AD [51]. Hypercholesterolemia is thought to possibly damage the BBB, resulting in pathological cholesterol metabolism in the brain [51]. Collectively, our findings demonstrate a shared genetic basis for plasma lipids and AD. Further, we pinpoint specific genes that may be driving this genetic association.
By combining several GWASs, our results provide important insights into shared genetic risk. Conceptually similar to stepwise gatekeeper hypothesis testing [12] and a proxy phenotype approach [38], conjunction FDR identifies loci associated with two traits. These two-stage methods do not lower the statistical ‘bar’ for gene detection and maintain a constant Type I error rate. Unlike stepwise gatekeeper hypothesis testing [12] and proxy phenotype [38], which have predominantly been used in a genome-wide framework, conjunction FDR focuses on ‘hidden’ SNPs with p < 5 × 10−8, which directly translates into an effective increase in sample size [4]. Here, we used independent samples to confirm our conjunction FDR results, thereby pinpointing a subset of cardiovascular-associated genes strongly associated with AD. Our findings reinforce that specific Alzheimer’s genes, such as BIN1 and PICALM, also increase risk for cardiovascular outcomes. Importantly, using this pleiotropy informed approach, and across three independent cohorts, we found four new susceptibility loci associated with elevated Alzheimer’s risk.
In meta-analyses, we identified novel AD-associated genetic signal in one genome-wide SNP and three SNPs at p < 1 × 10−6. By conditioning on cardiovascular RFs, we detected a genetic variant within the MBLAC1/GATS/STAG3 region on chromosome 7 and with a meta-p value of 1.44 × 10−9. MBLAC1 encodes a metallo-β-lactamase domain-containing protein and shows ubiquitous expression in the brain [16]. Building on this, we found that expression of MBLAC1 was differentially altered in AD brains. We also identified a variant within MINK1 on chromosome 17. Interestingly, MINK1 expression was altered in AD brains supporting the hypothesis that phosphorylated kinases, like MINK1, are abnormal in AD [10].
On chromosome 11, our results point to AD-associated genetic signal within the MTCH2/SPI1 region that is independent of CELF1/CUGB1. We identified rs2071305 and rs11039131 that were tagging variants within MYBPC3 and DDB2, within the MTCH2 and SPI1 regions. Furthermore, rs2071305 was in LD with an AD age of onset SNP that was associated with lower expression of SPI1 in monocytes and macrophages [20, 22]. We found that SPI1 expression was altered in AD brains. SPI1 encodes a transcription factor, PU.1, that is essential for myeloid cell development and a major regulator of cellular communication in the immune system [29]. Coupled with our HLA findings, these results implicate genes expressed in microglia, astrocytes or other myeloid cell types in AD pathogenesis [39].
We identified enrichment for our novel AD/CV genetic variants within an AD-by-proxy cohort. Of the four new SNPs that strongly influenced Alzheimer’s risk, we found that MBLAC, DDB2 and MINK1 were associated with proxy AD status in the UKB sample. Importantly, five of the six IGAP/ADGC2 SNPs replicated in UKB consistent with prior work highlighting the usefulness of the by-proxy phenotype approach for AD [52]. Although a proxy phenotype is not equivalent to a clinical diagnosis of dementia, our findings illustrate that a subset of cardiovascular genes influences disease risk even in people with a genetic predisposition for developing AD.
Our pleiotropy findings suggest that complex diseases and traits have a complex genetic architecture. Although we did not evaluate causal associations using a Mendelian Randomization (MR) framework, our results have implications for the relationship between common genetic variants, CV-associated RFs and AD as an outcome. To date, MR studies have typically evaluated a single CV risk factor at a time, which is valid only if the genetic variants used for the MR influence AD exclusively via the selected CV-associated risk factor [25, 33]. For some variants, we found pleiotropy challenging the conventional MR approach for genes such as ABCA1 [17]. Instead of a single causal link [15], these results suggest two possible scenarios for genetic variants associated with multiple traits: (1) genetic variants influence cardiovascular RFs and AD independently, or (2) genetic variants influence AD through multiple cardiovascular RFs.
Several studies have explored the overall genetic relationship between CV-associated risk factors and Alzheimer’s disease. In line with our results, studies have reported significant genetic overlap between AD and plasma lipids [13, 53]. However, others have found weak casual evidence for plasma lipids and AD using MR [54] or no association between these traits using LD score regression [55]. The methods used in these studies may help explain differences from our results to some extent. As discussed above, MR analyses do not account for pleiotropic effects, which we specifically focus on in the current manuscript. Further, our pleiotropic approach allows for allelic heterogeneity and might consequently find shared genetic effects missed by the LD score regression method. Moreover, similar to our findings, others have shown minimal to no genetic overlap between CAD and T2D and AD [53]. Using MR, some have explored the causal relationship between CAD and AD risk [56] and found a lack of causal relevance of CAD for risk of late-onset AD after exclusion of APOE. Also, although CAD and AD show minimal genetic overlap, a genetic risk score for CAD has been shown to modify the association between CVD and AD [53]. Further, our understanding of the genetic relationship between BMI and AD is not well understood. We found minimal genetic overlap between BMI and AD. Others have found strong genetic overlap between BMI and AD [53], and yet others found no casual evidence between these traits [57]. These findings suggest that the genetic relationship between AD and BMI and CAD is complex and other factors may be influencing the relationship.
Our findings have clinical implications. First, given the common co-occurrence of vascular and Alzheimer’s pathology, it is highly likely that the clinically diagnosed AD individuals from our cohort have concomitant vascular brain disease, which may further contribute to their cognitive decline and dementia. As such, a plausible interpretation of our findings is that the susceptibility loci identified in this study may increase brain vulnerability to vascular and/or inflammatory insults, which in turn may exacerbate the clinical consequences of AD pathological changes. Second, no single common variant detected in this study will be clinically informative. Rather, integration of these pleiotropic variants into a cardiovascular pathway-specific, polygenic ‘hazard’ framework for predicting AD age of onset may help identify older individuals jointly at risk for cardiovascular and Alzheimer’s disease [11]. Therapeutically targeting cardiovascular RFs in these individuals may impact the Alzheimer’s disease trajectory.
This study has limitations. First, our AD patients were diagnosed largely using clinical criteria without neuropathology confirmation and this may result in misclassification of case status. However, such misclassification should reduce statistical power and bias results toward the null. Second, we focused on the closest genes as the eQTL analyses did not replicate in both brain and blood. Additional work will be required to determine the causal genes responsible for the association between these novel loci and AD. Finally, given evidence that phospholipids are proinflammatory [35], future work should evaluate whether LDL, HDL TG, or TC influence AD risk through inflammation or other mediator variables.
In summary, we show cardiovascular-associated polygenic enrichment in AD. Beyond APOE, our findings support a disease model in which lipid biology is integral to the development of clinical AD in a subset of individuals. Lastly, considerable clinical, pathological and epidemiological evidence has shown overlap between Alzheimer’s and cardiovascular risk factors. Here, we provide genetic support for this association.
Supplementary Material
Acknowledgements
We thank the Shiley-Marcos Alzheimer’s Disease Research Center at UCSD and the Memory and Aging Center at UCSF for continued support and the International Genomics of Alzheimer’s Project (IGAP) for providing summary result data for these analyses. This work was supported by Grants from the National Institutes of Health (NIH-AG046374, K01AG049152), Alzheimer’s Disease Genetics Consortium (U01 AG032984), National Alzheimer’s Coordinating Center Junior Investigator Award (RSD), RSNA Resident/Fellow Award (RSD), Foundation ASNR Alzheimer’s Imaging Grant (RSD), the Research Council of Norway (#213837, #225989, #223273, #237250/EU JPND), the South East Norway Health Authority (2013–123), Norwegian Health Association and the KG Jebsen Foundation.
Footnotes
Conflict of interest JBB served on advisory boards for Elan, Bristol-Myers Squibb, Avanir, Novartis, Genentech, and Eli Lilly and holds stock options in CorTechs Labs, Inc. and Human Longevity, Inc. AMD is a founder of and holds equity in CorTechs Labs, Inc., and serves on its Scientific Advisory Board. He is also a member of the Scientific Advisory Board of Human Longevity, Inc. (HLI), and receives research funding from General Electric Healthcare (GEHC). The terms of these arrangements have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies.
References
- 1.Allen M, Carrasquillo MM, Funk C et al. (2016) Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci Data 3:160089. 10.1038/sdata.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreassen OA, Djurovic S, Thompson WK et al. (2013) Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet 92:197–209. 10.1016/j.ajhg.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreassen OA, Thompson WK, Dale AM (2014) Boosting the power of schizophrenia genetics by leveraging new statistical tools. Schizophr Bull 40(1):13–17. 10.1093/schbul/sbt168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreassen OA, Thompson WK, Schork AJ et al. (2013) Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet 9(4):e1003455 10.1371/journal.pgen.1003455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreassen OA, Zuber V, Thompson WK, Schork AJ, Bettella F, Djurovic S et al. (2014) Shared common variants in prostate cancer and blood lipids. Int J Epidemiol 43:1205–1214. 10.1093/ije/dyu090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attems J, Jellinger KA (2014) The overlap between vascular disease and Alzheimer’s disease—lessons from pathology. BMC Med 12:206 10.1186/s12916-014-0206-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes DE, Yaffe K (2011) The projected impact of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol 10(9):819–828. 10.1016/S1474-4422(11)70072-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broce I, Karch CM, Wen N et al. (2018) Immune-related genetic enrichment in frontotemporal dementia: an analysis of genome-wide association studies. PLoS Med 15(1):e1002487 10.1371/journal.pmed.1002487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmona S, Hardy J, Guerreiro R (2018) The genetic landscape of Alzheimer disease, Chapter 26 In: Geschwind DH, Paulson HL, Klein C (eds) Handbook of clinical neurology, vol 148. Neurogenetics, Part II. Elsevier, Amsterdam, pp 395–408. 10.1016/b978-0-444-64076-5.00026-0 [DOI] [PubMed] [Google Scholar]
- 10.Dammer EB, Lee AK, Duong DM, Gearing M, Lah JJ, Levey AI et al. (2014) Quantitative phosphoproteomics of Alzheimer’s disease reveals cross-talk between kinases and small heat shock proteins. J Proteom 15:508–519. 10.1002/pmic.201400189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desikan RS, Fan CC, Wang Y et al. (2017) Genetic assessment of age-associated Alzheimer disease risk: development and validation of a polygenic hazard score. PLoS Med 14(3):e1002258 10.1371/journal.pmed.1002258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desikan RS, Schork AJ, Wang Y et al. (2015) Genetic overlap between Alzheimer’s disease and Parkinson’s disease at the MAPT locus. Mol Psychiatry 20(12):1588–1595. 10.1038/mp.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desikan RS, Schork AJ, Wang Y et al. (2015) Polygenic overlap between C-reactive protein, plasma lipids, and Alzheimer disease. Circulation 131(23):2061–2069. 10.1161/CIRCULATIONAHA.115.015489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Paolo G, Kim T-W (2011) Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci 12(5):284–296. 10.1038/nrn3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emdin CA, Khera AV, Natarajan P et al. (2017) Genetic association of waist-to-hip ratio with cardiometabolic traits, type 2 diabetes, and coronary heart disease. JAMA 317(6):626–634. 10.1001/jama.2016.21042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagerberg L, Hallström BM, Oksvold P et al. (2013) Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteom 13:397–406. 10.1074/mcp.m113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frikke-Schmidt R (2008) Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA 299:2524 10.1001/jama.299.21.2524 [DOI] [PubMed] [Google Scholar]
- 18.Pulit SL, Stoneman C, Morris AP (2018) Meta-analysis of genome-wide association studies for body fat distribution in 694,649 individuals of European ancestry. Biorxiv. 10.1101/304030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibar DP, Stein JL, Renteria ME et al. (2015) Common genetic variants influence human subcortical brain structures. Nature 520(7546):224 10.1038/nature14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang K-L, Marcora E, Pimenova AA et al. (2017) A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci 20(8):1052–1061. 10.1038/nn.4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jun G, Ibrahim-Verbaas CA, Vronskaya M et al. (2015) A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 21:108–117. 10.1038/mp.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karch CM, Ezerskiy LA, Bertelsen S, Consortium (ADGC) ADG, Goate AM (2016) Alzheimers disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 loci. PLoS One 11(2):e0148717 10.1371/journal.pone.0148717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laird NM, Mosteller F (1990) Some statistical methods for combining experimental results. Int J Technol Assess Health Care 6:5–30. 10.1017/s0266462300008916 [DOI] [PubMed] [Google Scholar]
- 24.Lambert JC, Ibrahim-Verbaas CA, Harold D et al. (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12):1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson SC, Traylor M, Malik R et al. (2017) Modifiable pathways in Alzheimer’s disease: Mendelian randomization analysis. BMJ 359:j5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livingston G, Sommerlad A, Orgeta V et al. (2017) Dementia prevention, intervention, and care. Lancet 390(10113):2673–2734. 10.1016/S0140-6736(17)31363-6 [DOI] [PubMed] [Google Scholar]
- 27.Luchsinger JA (2001) Diabetes mellitus and risk of Alzheimers disease and dementia with stroke in a multiethnic cohort. Am J Epidemiol 154:635–641. 10.1093/aje/154.7.635 [DOI] [PubMed] [Google Scholar]
- 28.Mahajan A, Wessel J, Willems SM et al. (2018) Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet 50(4):559–571. 10.1038/s41588-018-0084-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahley RW (2016) Central nervous system lipoproteins: ApoE and regulation of cholesterol metabolism. Arterioscler Thromb Vasc Biol 36(7):1305–1315. 10.1161/ATVBAHA.116.307023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Academies of Sciences, Engineering, and Medicine, Health and Medicine Division, Board on Health Sciences Policy, Committee on Preventing Dementia and Cognitive Impairment (2017) Preventing cognitive decline and dementia: a way forward. In: Downey A, Stroud C, Landis S, Leshner AI (eds) National Academies Press, Washington DC: http://www.ncbi.nlm.nih.gov/books/NBK436397/. Accessed 17 Apr 2018 [PubMed] [Google Scholar]
- 31.Nelson CP, Goel A, Butterworth AS et al. (2017) Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 49(9):1385–1391. 10.1038/ng.3913 [DOI] [PubMed] [Google Scholar]
- 32.Norton S, Matthews FE, Barnes DE, Yaffe K, Brayne C (2014) Potential for primary prevention of Alzheimer’s disease: an analysis of population-based data. Lancet Neurol 13(8):788–794. 10.1016/S1474-4422(14)70136-X [DOI] [PubMed] [Google Scholar]
- 33.Østergaard SD, Mukherjee S, Sharp SJ et al. (2015) Associations between potentially modifiable risk factors and Alzheimer disease: a Mendelian randomization study. PLoS Med 12(6):e1001841 10.1371/journal.pmed.1001841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Profenno LA, Porsteinsson AP, Faraone SV (2010) Meta-analysis of Alzheimer’s disease risk with obesity, diabetes, and related disorders. Biol Psychiatry 67(6):505–512. 10.1016/j.biopsych.2009.02.013 [DOI] [PubMed] [Google Scholar]
- 35.Que X, Hung M-Y, Yeang C et al. (2018) Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558(7709):301–306. 10.1038/s41586-018-0198-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramasamy A, Trabzuni D, Guelfi S et al. (2014) Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 17(10):1418–1428. 10.1038/nn.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reitz C (2013) Dyslipidemia and the risk of Alzheimer’s disease. Curr Atheroscler Rep 15(3):307 10.1007/s11883-012-0307-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rietveld CA, Esko T, Davies G (2014) Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. PNAS 111:13790–13794. 10.1073/pnas.1404623111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sims R, van der Lee SJ, Naj AC et al. (2017) Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet 49(9):1373–1384. 10.1038/ng.3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparks DL (2007) Cholesterol metabolism and brain amyloidosis: evidence for a role of copper in the clearance of Abeta through the liver. Curr Alzheimer Res 4(2):165–169 [DOI] [PubMed] [Google Scholar]
- 41.Staffaroni AM, Elahi FM, McDermott D et al. (2017) Neuroim-aging in dementia. Semin Neurol 37(5):510–537. 10.1055/s-0037-1608808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stearns FW (2010) One hundred years of pleiotropy: a retrospective. J Genet 186(3):767–773. 10.1534/genetics.110.122549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steele NZR, Carr JS, Bonham LW et al. (2017) Fine-mapping of the human leukocyte antigen locus as a risk factor for Alzheimer disease: a case–control study. PLoS Med 14(3):e1002272 10.1371/journal.pmed.1002272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surakka I, Horikoshi M, Mägi R (2015) The impact of low-frequency and rare variants on lipid levels. Nat Genet 47:589–597. 10.1038/ng.3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westra H-J, Peters MJ, Esko T et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45(10):1238–1243. 10.1038/ng.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willer CJ, Schmidt EM, Sengupta S et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45(11):1274–1283. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yengo L, Sidorenko J, Kemper KE et al. (2018) Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum Mol Genet. 10.1093/hmg/ddy271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokoyama JS, Wang Y, Schork AJ et al. (2016) Association between genetic traits for immune-mediated diseases and Alzheimer disease. JAMA Neurol 73(6):691–697. 10.1001/jamaneurol.2016.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu C-E, Seltman H, Peskind ER (2007) Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimers disease: patterns of linkage disequilibrium and disease/marker association. J Genom 89:655–665. 10.1016/j.ygeno.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Liu Q (2015) Cholesterol metabolism and homeostasis in the brain. Protein Cell 6:254–264. 10.1007/s13238-014-0131-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue-Shan Z, Juan P, Qi W, Zhong R, Li-Hong P, Zhi-Han T, Zhi-Sheng J, Gui-Xue W, Lu-Shan L (2016) Imbalanced cholesterol metabolism in Alzheimers disease. Clin Chim Acta 456:107–114. 10.1016/j.cca.2016.02.024 [DOI] [PubMed] [Google Scholar]
- 52.Jansen I, Savage J, Watanabe K, Bryois J, Williams D, Steinberg S, Sealock J, Karlsson I, Hagg S, Athanasiu LS (2018) Genetic meta-analysis identifies 9 novel loci and functional pathways for Alzheimers disease risk. Biorxiv. 10.1101/258533 [DOI] [Google Scholar]
- 53.Karlsson IK, Ploner A, Song C, Gatz M, Pedersen NL, Hägg S (2017) Genetic susceptibility to cardiovascular disease and risk of dementia. Transl Psychiatry. 10.1038/tp.2017.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuźma E, Hannon E, Zhou A, Lourida I, Bethel A, Levine DA, Lunnon K, Thompson-Coon J, Hyppönen E, Llewellyn DJ (2018) Which risk factors causally influence dementia? A systematic review of Mendelian randomization studies. J Alzheimers Dis 64:181–193. 10.3233/jad-180013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, Duncan L, Perry JRB, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM (2015) An atlas of genetic correlations across human diseases and traits. Nat Genet 47:1236–1241. 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grace C, Clarke R, Goel A, Farrall M, Watkins H, Hopewell JC (2018) Lack of genetic support for shared aetiology of coronary artery disease and late-onset Alzheimer’s disease. Sci Rep 1:1 10.1038/s41598-018-25460-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee S, Walter S, Kauwe JS, Saykin AJ, Bennett DA, Larson EB, Crane PK, Glymour MM (2015) Genetically predicted body mass index and Alzheimers disease–related phenotypes in three large samples: Mendelian randomization analyses. Alzheimers Dementia 11:1439–1451. 10.1016/j.jalz.2015.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.