Abstract
Objective
Sympathetic nervous system and immune cell interactions play key roles in the regulation of metabolism. For example, recent convergent studies have shown that macrophages regulate obesity through brown adipose tissue (BAT) activation and beiging of white adipose tissue (WAT) via effects upon local catecholamine availability. However, these studies have raised issues about the underlying mechanisms involved including questions regarding the production of catecholamines by macrophages, the role of macrophage polarization state and the underlying intracellular signaling pathways in macrophages that might mediate these effects.
Methods
To address such issues we generated mice lacking Irs2, which mediates the effects of insulin and interleukin 4, specifically in LyzM expressing cells (Irs2LyzM−/− mice).
Results
These animals displayed obesity resistance and preservation of glucose homeostasis on high fat diet feeding due to increased energy expenditure via enhanced BAT activity and WAT beiging. Macrophages per se did not produce catecholamines but Irs2LyzM−/− mice displayed increased sympathetic nerve density and catecholamine availability in adipose tissue. Irs2-deficient macrophages displayed an anti-inflammatory transcriptional profile and alterations in genes involved in scavenging catecholamines and supporting increased sympathetic innervation.
Conclusions
Our studies identify a critical macrophage signaling pathway involved in the regulation of adipose tissue sympathetic nerve function that, in turn, mediates key neuroimmune effects upon systemic metabolism. The insights gained may open therapeutic opportunities for the treatment of obesity.
Keywords: Macrophage, Irs2, Obesity, Inflammation, BAT, Sympathetic neurons
Abbreviations: WAT, white adipose tissue; BAT, brown adipose tissue; CAM, classically activated macrophages; AAM, alternatively activated macrophages; HFD, high fat diet; EE, energy expenditure; HOMA-IR, homeostatic model assessment-insulin resistance; BMDM, bone marrow-derived macrophages; LPS, lipopolysaccharide; ATM, adipose tissue macrophages; SAM, sympathetic-associated macrophages; CLAMS, comprehensive lab animal monitoring system; SVF, stromal vascular fraction
Highlights
-
•
Irs2 deletion in macrophages improves metabolic health and BAT activity in mice.
-
•
Irs2-deficient BMDM are protected from M1 polarisation by LPS.
-
•
BAT norepinephrine levels and TH density mirror increases in BAT activity.
-
•
Cross-talk between macrophages and BAT sympathetic neurons underpin these findings.
-
•
Macrophage Irs2 modulates energy homeostasis.
1. Introduction
Obesity and its associated co-morbidities such as type 2 diabetes are major threats to health worldwide with over 2.1 billion people currently being classified as overweight or obese [1], [2], [3]. The pathophysiology of obesity is increasingly thought to involve cross-talk between the immune and metabolic regulatory systems [4], [5], [6], [7], [8], [9]. Over-nutrition, leading to obesity, is associated with an altered adipose tissue immune profile and inflammation [4], [7], [8], [9]. This in turn leads to a systemic pro-inflammatory state causing insulin resistance, a primary factor in the pathophysiology of type 2 diabetes [4], [6], [7], [8], [9]. Macrophages sit at the core of obesity-associated inflammation [6], [9]. While now recognized as an oversimplification, two basic polarization states of macrophages have been defined: classically activated (M1) macrophages (CAM) have a pro-inflammatory phenotype while alternatively activated (M2) macrophages (AAM) have an anti-inflammatory phenotype [10], [11]. The distinct physiological roles of these macrophage phenotypes are reflected in the pathogenesis of obesity which is associated with increased adipose tissue M1 macrophage infiltration and release of inflammatory cytokines leading to antagonism of insulin signaling [7], [8], [12]. In contrast, AAM can mitigate against insulin resistance with improved insulin sensitivity associated with increased numbers of AAM in adipose tissue [7], [8], [9]. Macrophage polarization status is under complex regulation with interferon gamma, for example, promoting the generation of CAM/M1 macrophages while interleukins (IL) IL4/IL13 promote the AAM/M2 phenotype [13], [14]. IL4/IL13 may play a role in metabolic homeostasis [13]. For example, maintenance of AAMs in white adipose tissue (WAT) depends on IL4/13 signaling to STAT6 [13], [15] and this pathway also programs macrophage metabolism via PPARG, PPARD, and PGC1B away from the anaerobic metabolic profile seen in CAM [13], [16]. IL4 signaling in macrophages has also been proposed to regulate BAT thermogenesis as acute cold stress causes AAM themselves to produce catecholamines in an IL4-dependent manner, leading to BAT browning and the biogenesis of beige fat in WAT depots [17]. Consistent with these observations, IL4 administration to mice provided protection from obesity-associated insulin resistance [17]. However, these findings have been called into question by a recent study that failed to confirm that AAM synthesize catecholamines or regulate adaptive thermogenesis and did not demonstrate beneficial metabolic effects of IL4 administration to mice [18]. In contrast, it has been demonstrated that macrophages may directly regulate sympathetic nervous system function in adipose tissue including the regulation of browning [19], [20] but the potential role of IL4 in this process is unclear.
In macrophages, IL4 signals via either the type I receptor consisting of IL4RA and IL4RG subunits or the type II receptor composed of IL4RA and IL13RA1 subunits [13], [21], [22], [23]. Type I and type II receptors recruit JAK/STAT signaling with the shared IL4RA chain associating with JAK1 leading to STAT6 phosphorylation [13], [21], [22], [23]. In addition, IL4RA activates insulin receptor substrate (IRS) 2, which is also the major target of insulin and IGF1 receptors in hematopoietic cells particularly of the myeloid lineage [24], [25], [26]. When tyrosine phosphorylated, IRS2 recruits the regulatory subunits of PI3K, GRB2, and other SH2 domain containing molecules to activate a number of downstream signaling pathways [27], [28]. However, despite its potentially critical role in both IL4 and insulin/IGF1 receptor signaling, the precise in vivo function of IRS2 in macrophage biology is less clear. Defining its role is complicated by the observations that while IL4 signaling is primarily anti-inflammatory, insulin action upon macrophages may be pro-inflammatory [21], [29], [30]. Thus, IRS2 signaling may mediate divergent biological outputs but to date the precise balance of such effects in metabolic physiology is unknown. For example, myeloid-restricted deletion of the insulin receptor in mice leads to improved whole animal insulin sensitivity under conditions of high fat feeding due to reduced migration of macrophages into white adipose tissue [31]. Mice with global deletion of Irs2 develop progressive diabetes due to a combination of systemic insulin resistance, hypothalamic obesity and pancreatic beta cell failure which together have precluded detailed analysis of the specific role of macrophage IRS2 signaling in physiology [32], [33], [34]. However, a study using transplanted bone marrow from these animals demonstrated that insulin signaling via IRS2 in macrophages contributed to pro-inflammatory signals involved in the development of atherosclerosis [35]. Consistent with this concept, in vitro studies have suggested that IRS2 signaling in macrophages is required for IL4/IL4RA–mediated expression of genes characteristic of the AAM/M2 anti-inflammatory phenotype [21]. However, recent studies in Irs2-deficient macrophages also demonstrated alterations in gene expression towards an M2 phenotype with compensatory IRS1 signaling suggested to underlie this observation [29]. These findings also indicate that IRS2 signaling might act as a restraint to IL4-mediated regulation of M2 genes revealing unexpected complexities in the underlying signaling mechanisms.
Given the key potential role of IRS2 signaling as an intersection point between IL4 and insulin signaling in macrophage function and systemic metabolism we generated mice with myeloid-restricted deletion of Irs2. We found that macrophages from these animals showed a broad anti-inflammatory phenotype when exposed to inflammatory mediators but with a limited increase in the expression of markers of the AAM/M2 phenotype. Furthermore, these animals showed improved insulin sensitivity and a resistance to weight gain on a high fat diet associated with increased BAT function and increased WAT browning at ambient temperature. However, we were unable to find evidence that this phenotype was driven by intrinsic macrophage catecholamine production. In contrast, we find that macrophages associate with BAT sympathetic nerves and that deletion of Irs2 in these cells increases sympathetic innervation and local norepinephrine levels in BAT. Together these studies reveal an important role for macrophage Irs2 in systemic metabolism and reveal new details of the signaling mechanisms underlying the cross-talk between the immune and nervous system relevant to the pathogenesis of obesity and insulin resistance.
2. Methods
2.1. Animals
Experiments involving animals were designed and reported following the ARRIVE guidelines of animal experiment reporting [36]. Power calculations for number of mice for each experiment were based on reported or known effect sizes and variation, in order to maximize chances of meaningful results without the unnecessary use of experimental animals. Where possible, investigators were blinded to the genotype of both study animals and tissue/blood samples. BMDM studies were performed on 10-week old male mice.
Deletion of Irs2 in myeloid lineages was carried out using Cre-LoxP technology using mice with a previously generated floxed allele of Irs2 [37] crossed with mice expressing Cre-recombinase driven by the Lysozyme M promoter [38]. Irs2 floxed mice were backcrossed to a C57BL/6J background whereas Lysozyme M Cre mice were on a mixed C57BL/6J:129S2/Sv genetic background. Irs2 deletion and the presence or absence of the Cre recombinase were determined by PCR as described previously; the following PCR primers were used for amplification: Irs2 flox forward: 5′-ACTTGAAGGAAGCCACAGTCG-3′, Irs2 flox reverse: 5′-AGTCCACTTTCCTGACAAGC-3′, Lysozyme M Cre forward: 5′-CCCAGAAATGCCAGATTACG-3′, Lysozyme M Cre reverse: 5′- CTTGGGCTGCCAGAATTTCTC-3’ [37], [38].
Mice were maintained in a pathogen–free facility in individually ventilated cages under a controlled temperature between 21 °C and 23 °C and a regular 12 h light/dark cycle with ad libitum access to water and normal chow diet (11.5% energy from fat). The high fat diet (HFD, Research Diets, D12451) contained 45.0% energy from fat while the percentage energy from carbohydrates was less relative to the chow diet (35.0% versus 61.6% respectively). Mouse studies were performed in accordance to the United Kingdom Animals (Scientific Procedures) Act (1986) and approved by Imperial College London's Animal Welfare and Ethical Review Body.
2.2. Metabolic studies
Total fat and lean mass from live non-anesthetized mice were determined using an EchoMRI quantitative whole-body composition analyzer (Zinsser Analytic) and mice were subsequently weighed to determine percentage fat and lean mass per body weight. Energy expenditure was calculated from singly-housed mice in CLAMS cages (Columbus Instruments) at ambient temperature (22 °C) for 24 h following 24 h of acclimatization. Recordings were taken every 20 mins. During energy expenditure measurement, water and food were provided ad libitum. Assessment of food intake was performed in singly housed mice for 3 consecutive experimental days after a period of acclimatization [37].
High fat diet experiments were performed on 8-week-old control and Irs2LyzM−/− mice, which were each divided into two weight-matched groups. Littermates of mixed genotypes were caged together and each group maintained on either chow or HFD. Fasted glucose measurements, glucose tolerance tests (GTTs) and insulin tolerance tests (ITTs) were performed as previously described [37], [39], [40]. GTTs on chow diet were performed by administering 2 g/kg of glucose per body weight intraperitoneally (i.p.) at 9am after an overnight fast whereas GTTs on HFD-fed mice were performed using glucose at a lower dose (1 g/kg bodyweight). For ITTs, food was removed from mice at 8am and the animals were injected i.p. with insulin (0.5 U/kg) at 2pm. Blood glucose was measured at the times indicated in the figures using a Contour glucometer (Bayer). Insulin sensitivity on HFD was assessed in 12 h-fasted mice by the homeostatic model assessment of insulin resistance (HOMA-IR) calculated as (I0 x G0)/405 where G0 is fasting blood glucose in mg/dl and I0 is fasting insulin in μIU/ml measured simultaneously [41].
For insulin and leptin measurements, serum from fasted mice was collected from tail vein bleeds into capillary tubes (Sarstedt) and spun for 15 min at 4 °C at 2000×g. The serum supernatant was removed into a new tube and stored at −80 °C. Hormones were measured by ELISA (Millipore) according to the manufacturer's instructions. BAT catecholamines were measured by 2-CAT (A-N) ELISA (Rocky Mountain Diagnostics) according to manufacturer's instructions. Briefly, BAT tissues were homogenized in homogenization buffer (1N HCl, 0.25M EDTA, 1M Na2S2O5), and homogenates were collected after centrifugation and stored at −80 °C prior to quantification. All samples were normalized to total tissue protein concentration.
2.3. Histology
Interscapular BAT, perigonadal WAT (pWAT), and inguinal WAT (iWAT) tissues were excised and fixed in 10% neutral buffered formalin for 24 h. Tissues were then dehydrated in increasing ethanol concentrations before being embedded in paraffin and serially sectioned (5 μm). Sections were deparaffinized with xylene and rehydrated before being stained with hematoxylin and eosin [42].
For immunohistochemistry, iWAT sections were stained with polyclonal anti-UCP1 antibody (Abcam) and macrophages in pWAT were immunostained with monoclonal rat anti-MAC2 antibody (Cedarlane Laboratories) as described previously [43], [44]. Detection was performed with an HRP-conjugated secondary antibody followed by chromogenic detection using DAB substrate. Brown and white adipose tissue images were taken at 20x magnification using a light microscope (Leica DM4000 B).
For immunofluorescence staining of tyrosine hydroxylase (TH) alone or both TH and CX3CR1 staining on BAT sections, BAT tissues were fixed with 4% paraformaldehyde (PFA) and then transferred to ethanol. Sections were antigen retrieved by boiling in 10 mM citrate buffer, pH 6, blocked with serum-free blocking agent (Invitrogen) and sequentially stained with rabbit anti-TH (Millipore) and rabbit polyclonal antibody to CX3CR1 (Abcam) [45]. Secondary antibodies, goat anti-rabbit AlexaFluor-488 and goat anti-rabbit AlexaFluor-568, were from Invitrogen.
For quantitative TH density analysis, a custom ImageJ macro was written. Briefly, tissue area was identified by thresholding low intensity autofluorescence signal after application of a median filter (r = 5), with the image subsequently converted to a binary mask. Further binary operations including morphological closing and dilation and fill holes, were used to accurately identify the boundaries of the total tissue area within the image. For subsequent analysis, pixels outside the tissue area segmented were set to zero. Identification of the tissue area without the inclusion of areas occupied by lipids was achieved by using a similar method as aforementioned but without the addition of the further binary operations. Fluorescence from TH immunostaining was analyzed according to these thresholds and binary operations made accordingly. Percentage of the fluorescent signal relative to the total tissue area excluding lipids is presented as TH density.
2.4. Magnetic isolation of CD11B positive fraction from the stromal vascular fraction of perigonadal WAT
Perigonadal white adipose tissue (pWAT) was collected from HFD-fed mice and digested in a Hank's Balanced Salt Solution containing BSA and type II collagenase (Sigma–Aldrich) at 37 °C, 100 rpm for 20–30 min. The floating adipocytes were separated from the stromal vascular fraction (SVF) by centrifugation. The CD11B fraction was extracted from the SVF pellet using CD11B microbeads (Miltenyi Biotec) conjugated to monoclonal rat anti-mouse/human antibody. The mixture of SVF/antibody was passed through a LS column (Miltenyi Biotec) placed in a magnetic field. CD11B positive fraction was eluted according to the manufacturer's instructions and then processed for RNA extraction using the RNeasy mini kit (Qiagen) [46].
2.5. Macrophage culture and treatment
Bone marrow-derived macrophages (BMDMs) were cultured as previously described [47]. Briefly, femurs and tibias were cut at the proximity of each joint and bone marrow flushed out with PBS. Cells were then collected, centrifuged at 500×g, and resuspended in macrophage complete medium (1:1 DMEM/F12, 10% FBS, 2 mM l-glutamine) consisting of 25% L929 conditioned-medium until fully differentiated (7–10 days). Macrophage activation was induced in differentiated BMDMs by stimulation with bacterial lipopolysaccharide (LPS) from Escherichia coli (100 ng/ml, Sigma–Aldrich) or IL4 (10 ng/ml, R&D Systems) for 24 h [10], [21], [48]. For short-term signaling studies, BMDMs were treated with IL4 (20 ng/ml) or insulin (10 nM, Novo Nordisk) for the times indicated and homogenized in lysis buffer (50 mM Tris–HCl pH 7.5,150 mM NaCl, 1% Triton-X100, 1 mM EDTA with Complete Mini protease inhibitors and PHOSstop phosphatase inhibitors (both Roche)). Phosphorylation of AKT was analyzed by western blotting from cleared lysates normalized for total protein by BCA method (Bio-Rad) using antibodies against serine 473 phosphorylated AKT and pan AKT (both Cell Signaling, 9271 and 2920, respectively). For expression of IRS1, lysates from brain tissue and BMDM, obtained as above, were blotted using a monoclonal IRS1 antibody (Upstate, clone 4.2.2). For palmitate treatment, BMDMs were incubated with 500 μM palmitate-BSA (Agilent technologies) for the times indicated, with BSA treatment as a control. BMDMs were then processed for RT-PCR studies to measure Irs2 mRNA expression as described below.
2.6. Catecholamine measurement by LC/MS
500,000 cells were seeded in 6-well plates and treated for 24 h with either IL4 or LPS. Control (non-conditioned) and BMDM-conditioned medium was collected from each well, centrifuged at 16,000×g for 10 min at 4 °C and 50 μl of the supernatant were added to 750 μl of cold extraction solution (50% v/v methanol, 30% v/v acetonitrile, 20% v/v ddH2O). Cell culture medium extracts were kept under agitation at 4 °C for 15 min in a ThermoMixer (Eppendorf). Finally, the suspension was centrifuged for 10 min at 16,000×g at 4 °C, the supernatant transferred into autosampler vials and stored at −80 °C until further analysis. Liquid chromatography–mass spectrometry (LC–MS) analysis was performed on a Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled to a Dionex UltiMate 3000 Rapid Separation LC system. The LC system was fitted with a Raptor FluoroPhenyl column (150 mm × 2.1 mm, 2.7 μm) with the corresponding guard column (both Restek). The mobile phase was composed of 0.2% formic acid in water and methanol. The flow rate was set at 400 μl min−1, with a gradient from 2% to 40% organic, and a total run time of 8 min. The mass spectrometer was operated in full MS and positive ionization mode. Samples were randomized, to avoid bias due to machine drift, and blinded to the operator. The acquired spectra were analyzed using XCalibur Qual Browser and XCalibur Quan Browser software (Thermo Scientific) by referencing to an internal library of compounds and synthetic standards run with the same batch as the samples.
2.7. Immunocytochemistry for IRS2 in BMDM
BMDM were plated on Matrigel-coated glass coverslips (VWR). Once adhered, cells were fixed with 100% methanol for 1 min at room temperature. For IRS2 staining, cells were permeabilized (PBS with 0.1% Triton X-100 and 1% BSA) for 5 min. After fixing and permeabilization, coverslips were washed (PBS) and blocked (0.5% fish skin gelatin in PBS) overnight. Coverslips were then incubated with rabbit polyclonal anti-IRS2 primary antibody (Upstate) for 3 h at room temperature. Cells were washed and further incubated with goat anti-rabbit AlexaFluor-488 secondary antibody (Invitrogen) for 30 min in the dark. Cover slips were washed with PBS to remove unbound secondary antibody and mounted with Vectashield® mounting medium with DAPI (Vector labs) on glass slides, immobilized with clear nail polish, and visualized using a Leica SP5 confocal microscope.
2.8. mRNA expression analysis and RNA sequencing
RNA was extracted from snap-frozen tissues and BMDM using TRIzol reagent (Sigma–Aldrich) followed by chloroform treatment, and isopropanol to precipitate RNA. Following centrifugation, the pellet was washed with 70% ethanol, dried and resuspended in RNase-free H2O. RNA quality and quantity were assessed using a ND-1000 spectrophotometer Nanodrop (Thermo Fisher Scientific) for real-time PCR (RT-PCR) purposes and a Bioanalyzer 2100 (Agilent) for mRNA sequencing purposes. RNA from the cell pellet collected from CD11B positive fractions was isolated using RNeasy spin columns (Qiagen).
Total RNA was treated with DNase I (Qiagen) and 1 μg was reverse transcribed at 42 °C with SuperScript II (Invitrogen). Quantitative PCR reactions were carried out in duplicate on 384-well clear optical plates using a Taqman ABI 7900HT fast real-time PCR machine equipped with SDS 2.3 software. A list of all probes is presented in Supplemental Table 1. Experimental Ct values were normalized to specific housekeeping genes. A relative quantification using the ΔCT method or an absolute quantification using the standard curve method were used as appropriate as indicated in figure legends. Hypoxanthine phosphoribosyltransferase (Hprt) was the housekeeping gene used to normalize Ct values in macrophages. Hprt Ct values were stable across BMDM and ATM samples of both genotypes and upon IL4 and LPS treatments. To quantify mRNA in adipose tissues, the housekeeping gene Gapdh (glyceraldehyde 3 phosphate dehydrogenase) was used.
For RNA sequencing analysis, the standard Illumina library preparation protocol was carried out on 2 μg of RNA and the library prepared using the TrueSeq kit (Illumina) according to manufacturer's protocol. Each sample was divided into 8 technical replicates and sequenced on individual lanes in an Illumina flow cell. The sequencing was performed using standard Illumina protocols at the Sanger institute. Raw reads from the sequenced RNAseq libraries were mapped with Tophat splice junction aligner [49], version 2.0.8 against Ensembl mouse genome reference sequence assembly (mm9) and transcript annotations. All parameters were set to default except inner distance between mate pairs (r = 130).
Gene-based read counts were then obtained using the HTSeq count module (version 0.5.4p3) [50]. This HTSeq counts module was run using parameters m = intersection-nonempty, stranded = no. Differential expression analysis was performed on the counts data using the DESeq2 Bioconductor package (version 1.16.1) [51]. This DESeq2 package uses negative binomial model to model read counts and then performs statistical tests for differential expression of genes. Raw P values were then adjusted for multiple testing with the Benjamini-Hochberg procedure. Genes with adjusted P value of 0.05 or less were termed as differentially expressed genes.
Geneset enrichment analysis was performed to elucidate biological pathways and processes regulated by the interaction between Irs2LyzM−/− and control BMDM and with various treatments (IL4, LPS). The GSEA [52] preranked tool from GSEA version 2.2.4 was used for this purpose. Gene ontology enrichment analysis was also performed using the topGo Bioconductor package [53] separately for up and down-regulated genes in the comparisons analyzed. RNA sequencing data is available from GEO accession GSE 123180.
2.9. Statistics
Statistical analysis was carried out using Prism 7 software. Student's t test or ANOVA (or Mann–Whitney U or Kruskal–Wallis tests) were used where data was parametric or nonparametric, respectively. A P value less than 0.05 was considered significant where * represents P < 0.05, **p < 0.01 and ***p < 0.001. In all panels, data are represented as mean ± SEM unless otherwise stated. For energy expenditure (CLAMS) data on HFD, a general linear modeling (GLM) in SPSS was used with genotype as a fixed factor, body weight as covariate and energy expenditure as the dependent variable.
3. Results
3.1. Loss of Irs2 in myeloid cells improves glucose homeostasis and promotes resistance to high fat diet induced metabolic dysfunction
To investigate the role of macrophage IRS2 signaling in metabolism, we conditionally deleted Irs2 in myeloid cells in mice. To achieve this, we generated mice homozygous for a floxed allele of Irs2 [37] and heterozygous for a transgene expressing Cre recombinase from the LyzM locus (Irs2fl/flLyzM-Cretg/wt termed Irs2LyzM−/− hereafter) [38]. A combination of pure WT, Irs2fl/fl and LyzM-Cretg/wt which were phenotypically indistinguishable served as controls. We confirmed efficient deletion of Irs2 in bone-marrow derived macrophages (BMDM) by analyzing mRNA expression levels in Irs2LyzM−/− mice and control mice by RT-PCR. (Supplemental Fig. 1A). To confirm loss of IRS2 protein expression, we performed immunofluorescence staining in control BMDM and those derived from Irs2LyzM−/− mice (Irs2−/− BMDMs). This showed cytoplasmic staining for IRS2 in control BMDM and a complete absence of IRS2 in Irs2−/− BMDMs (Supplemental Fig. 1B).
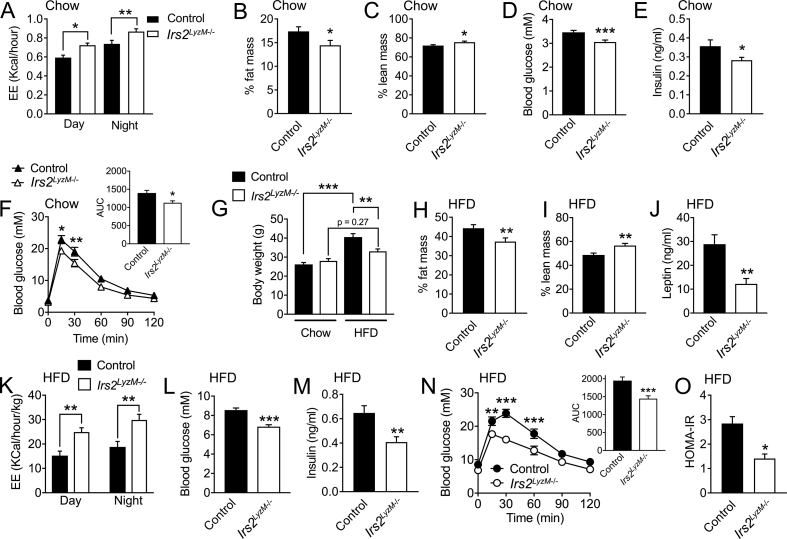
Next, we studied the body weight and metabolic phenotypes of Irs2LyzM−/− mice on a normal chow diet. At 23 weeks of age we saw no significant difference in body weight between control and Irs2LyzM−/− mice of either sex (Supplemental Fig. 1C). Assessment of percentage fat mass vs body weight by EchoMRI at earlier ages showed no differences between control and Irs2LyzM−/− mice (control: 10.5 ± 1.3% vs. 12.7 ± 1.4% in Irs2LyzM−/− mice, n = 8, P = 0.267). However, when energy expenditure was assessed by indirect calorimetry, 8-week old male Irs2LyzM−/− mice displayed significantly increased EE in both light and dark cycles (Figure 1A). Mice at this age showed a compensatory increase in food intake on a chow diet (control: 0.199 ± 0.007 g/g/day vs. 0.229 ± 0.008 g/g/day in Irs2LyzM−/− mice, n = 10, P = 0.011) similar to that seen in other mouse models with increased EE such as cold exposed mice [54] or mice with genetic interventions elevating EE (e.g. [55]). By 23 weeks of age male Irs2LyzM−/− mice showed a significant reduction in fat mass (Figure 1B) with a concomitant increase in lean body mass (Figure 1C) We next assessed glucose homeostasis in chow fed-Irs2LyzM−/− mice. At 8–10 weeks of age Irs2LyzM−/− mice had a significant reduction in both fasting glucose and insulin levels (Figure 1D, E). This was accompanied by a mild improvement in glucose clearance and insulin sensitivity (Figure 1F and ITT AUC – control: 6611 ± 311 vs. 5618 ± 225 in Irs2LyzM−/− mice, n = 8, P = 0.043). Taken together, these data suggest that deletion of Irs2 in myeloid cells leads to a mild beneficial metabolic phenotype on a chow diet associated with increased energy expenditure.
Figure 1.
Physiological parameters in Irs2LyzM−/−mice fed chow or HFD. (A) Energy expenditure (EE) in 8-week old control and Irs2LyzM−/− males during both the day and night cycles (n = 22–26). (B, C) Percentage fat mass (B) and percentage lean mass (C) in 23 weeks old control and Irs2LyzM−/− male mice on chow diet (n = 23–24). (D) Blood glucose levels in 8-week old control and Irs2LyzM−/− female mice following an overnight fast (n = 20). (E) Serum insulin levels from 8-week old control and Irs2LyzM−/− female mice following an overnight fast (n = 12–20). (F) Glucose tolerance in overnight fasted 8-week old female control and Irs2LyzM−/− mice (n = 8–9). Inset shows means of area under the curve (AUC) for both genotypes. (G) Body weight of control and Irs2LyzM−/- females fed either chow or high fat diet (HFD) from 8 weeks of age for 15 weeks (n = 8–11 for chow and n = 23 for HFD). (H, I) Percentage fat mass (H) and percentage lean mass (I) in 23-week old control and Irs2LyzM−/− female mice fed HFD for 15 weeks (n = 23). (J) Serum leptin levels following an overnight fast in control and Irs2LyzM−/− female fed HFD for 15 weeks (n = 10–12). (K) Energy expenditure (EE) values normalized to body weight in 23-week old control and Irs2LyzM−/− females fed HFD for 15 weeks (n = 18–20). (L, M) Blood glucose (L) and serum insulin levels (M) in overnight fasted 16-week old control and Irs2LyzM−/− females fed HFD for 8 weeks (n = 20–23). (N) Glucose tolerance in 6-hour fasted 16-week old control and Irs2LyzM−/− female mice fed HFD for 8 weeks (n = 21). Inset shows means of area under the curve (AUC) for both genotypes. (O) Homeostatic model assessment for insulin resistance (HOMA-IR) calculated from fasted blood glucose and serum insulin levels from mice in L and M (n = 20). In all panels, data presented are mean ± SEM with Student's t test for all panels except F, G and O, where ANOVA with repeated measure was used. (*P < 0.05, **P < 0.01 and ***P < 0.001 relative to control mice).
To assess the impact of these phenotypic changes under conditions of over-nutrition, we studied 8-week-old mice exposed to high fat diet (HFD) for 15 weeks. Control animals gained significantly more body weight on HFD compared to chow but in contrast Irs2LyzM−/− mice displayed partial resistance to weight gain (Figure 1G). Consistent with this observation Irs2LyzM−/− mice had reduced percentage fat mass, increased percentage lean mass and, as an indirect measure of body fat, reduced leptin levels (Figure 1H–J). Irs2LyzM−/− mice also displayed increased energy expenditure on HFD, when corrected for differences in body weight (Figure 1K). These increases in energy expenditure in Irs2LyzM−/− mice were also statistically significant in both the day (P = 0.007) and night (P = 0.003) cycles when using GLM (see methods 2.9) to demonstrate that genotype is a statistically significant independent contributing factor to the observed difference in EE, even after appropriately correcting for the effect of bodyweight. Assessment of glucose homeostasis in HFD-fed Irs2LyzM−/− mice showed that these animals had lower fasting glucose and insulin levels and improved glucose clearance (Figure 1L–N). Insulin sensitivity as indicated by ITT (ITT AUC on HFD – control: 10451 ± 616, n = 15 vs. 8391 ± 615, n = 8 in Irs2LyzM−/− mice, P = 0.044) and HOMA-IR was also improved compared to control mice (Figure 1O). Together these studies show that mice lacking Irs2 in myeloid cells have partial resistance to both HFD-induced weight gain and the associated disruption of glucose homeostasis.
3.2. Loss of Irs2 in myeloid cells is associated with an anti-inflammatory macrophage phenotype but does not alter macrophage numbers in WAT
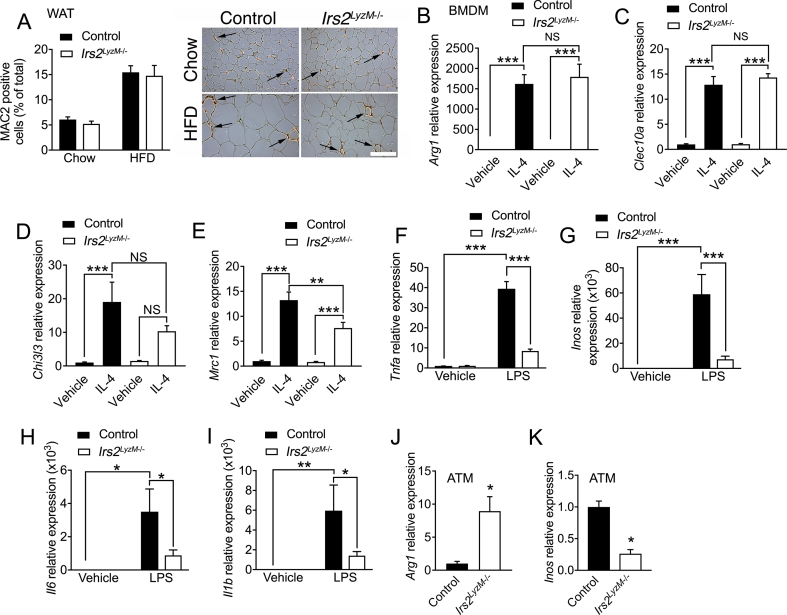
The beneficial metabolic effects seen in Irs2LyzM−/− mice on HFD prompted us to assess whether alterations in macrophage numbers in WAT might underlie these changes as previous studies have shown that myeloid deletion of the insulin receptor leads to lowered inflammation both in adipose tissue and systemically by reducing macrophage recruitment into WAT [31]. However, on both chow and HFD, WAT macrophage numbers were equivalent in control and Irs2LyzM−/− mice (Figure 2A). Polarization of macrophages away from a pro-inflammatory CAM/M1 phenotype towards an anti-inflammatory AAM/M2 phenotype also reduces adipose tissue dysfunction [56]. Therefore, we next examined the mRNA expression of candidate markers of M1/M2 polarization in BMDM isolated from control and Irs2LyzM−/− mice in response to IL4 or LPS stimulation. In response to IL4 we found equivalent expression of Arg1 and Clec10a in Irs2-deficient BMDM and mild attenuation of the induction of Chi3l3 and Mrc1 (Figure 2B–E). In contrast to the limited M2 changes we detected significant reductions in the expression of M1 markers Tnfa, Inos, Il6, and Il1b in response to LPS in Irs2-deficient macrophages, suggesting a reduction in the pro-inflammatory phenotype in these cells (Figure 2F–I). When we examined gene expression in adipose tissue macrophages (ATM) from HFD fed mice we found increased Arg1 expression and down-regulated Inos expression in Irs2LyzM−/− mice (Figure 2J, K). Taken together, these findings suggest that deletion of Irs2 in macrophages results in a markedly reduced pro-inflammatory response to LPS in macrophages rather than a broadly enhanced M2 profile when assessed by RT-PCR. Furthermore, there were no alterations in macrophage numbers in WAT upon deletion of Irs2.
Figure 2.
Characterization of bone marrow derived macrophages (BMDM) from Irs2LyzM−/−mice. (A) Quantification of MAC2 positive cells as percentage of total cell number in perigonadal white adipose tissue of control and Irs2LyzM−/− mice on chow (n = 8–9) or high fat diet (HFD) (n = 5). Representative images are shown in the right-hand panel, with arrows indicating the presence of macrophage crown-like structures visualized by MAC2 immunohistochemical staining. Scale bar = 500 μm. (B–E) Quantitative RT-PCR analysis of mRNA levels of M2 markers in control and Irs2LyzM−/− BMDM treated with vehicle or interleukin 4 (IL4, 10 ng/ml, 24 h). Hprt mRNA expression was used as an internal control and data are represented relative to control/vehicle (n = 4–7). (F–I) Analysis of mRNA levels of M1 inflammatory markers with LPS treatment (100 ng/ml, 24 h) (n = 4–6). (J, K) Arg1 and Inos mRNA levels in control and Irs2LyzM−/− adipose tissue macrophages (ATM) isolated from HFD-fed mice. Data are average of two independent RT-PCR experiments and ATMs were collected from a pool of 3–6 mice on HFD in each experiment (n = 6–12 mice per genotype total). In all panels, data presented are mean ± SEM with statistical analysis by ANOVA (A–I) or Student's t test (J, K). (*P < 0.05, **P < 0.01 and ***P < 0.001, relative to control BMDM/control BMDM with vehicle treatment).
3.3. Loss of Irs2 in myeloid cells is associated with increased browning in BAT and WAT
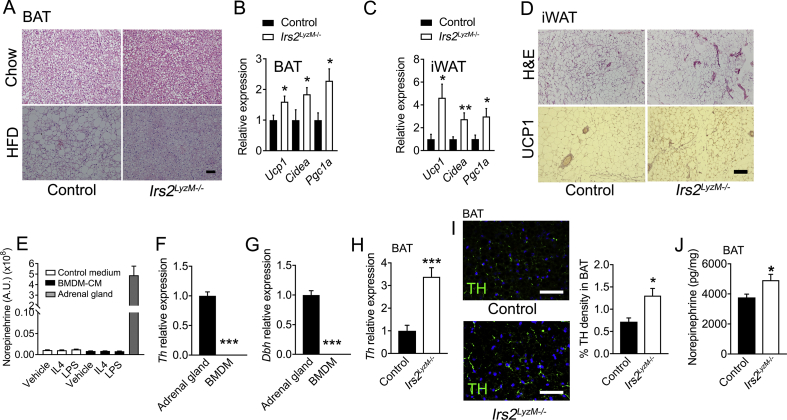
We next undertook studies to investigate the mechanisms by which loss of IRS2 signaling in myeloid cells might cause increased in energy expenditure in Irs2LyzM−/− mice. BAT thermogenesis has been suggested to be regulated by macrophage polarization status with M2 macrophages reported to release catecholamines in an IL4-dependent manner [17] although these studies have been recently challenged [18]. We found reduced lipid storage as assessed morphologically in BAT in Irs2LyzM−/− mice on both chow and HFD suggesting that there was increased activity of BAT metabolism (Figure 3A). Consistent with this histological finding, we found increased mRNA expression of Ucp1 and other markers of BAT activation including Cidea and Pgc1a in Irs2LyzM−/− mice on chow diet (Figure 3B). We also examined iWAT for indications of browning in this fat depot and found increased expression of Ucp1, Cidea and Pgc1a (Figure 3C). UCP1 protein levels as assessed by immunostaining were also increased in iWAT of Irs2LyzM−/− mice (Figure 3D). Taken together, these observations indicate that Irs2LyzM−/− mice had upregulated BAT function and browning of WAT.
Figure 3.
Assessment of brown adipose tissue function in Irs2LyzM−/−mice. (A) Representative hematoxylin and eosin (H&E) staining of interscapular brown adipose tissue (BAT) from control and Irs2LyzM−/− mice fed either chow (top panel) or high fat diet (HFD) (lower panel) (n = 4 per group). Scale bar = 500 μm. (B) Quantitative RT-PCR analysis of mRNA levels of BAT genes in Irs2LyzM−/− relative to control mice on chow diet. Gapdh mRNA expression was used as an internal control (n = 6–8). (C) Analysis of mRNA levels as in B of brown adipose genes in inguinal WAT (iWAT) of Irs2LyzM−/− relative to control mice on chow diet. Gapdh mRNA expression was used as an internal control (n = 9–11). (D) Representative H&E staining (top panel) and immunostaining of UCP1 (lower panel) in iWAT of control and Irs2LyzM−/− chow-fed mice (n = 4). Scale bar = 500 μm. (E) Norepinephrine levels in non-conditioned medium (Control medium) and BMDM-conditioned medium (BMDM-CM) with vehicle, IL4 and LPS treatment. Lysates of mouse adrenal gland were used as a positive control (n = 4 per group). (F, G) Quantitative RT-PCR analysis of Th (F) and Dbh (G) mRNA levels measured in BMDM relative to adrenal gland, both from control mice. Hprt mRNA expression was used as an internal control (n = 7). (H) Tyrosine hydroxylase (Th) mRNA levels in BAT from Irs2LyzM−/− relative to control mice. Gapdh mRNA expression was used as an internal control (n = 6–8). (I, images) TH immunostaining (green) in BAT sections from control and Irs2LyzM−/− mice. DAPI was used to visualize nuclei (blue). Scale bar = 50 μm. (I, bar chart) Quantification of percentage TH density in BAT sections (n = 3–4 mice/group and 6–12 fields of view per mouse). (J) Norepinephrine levels normalized to total protein content in BAT from control and Irs2LyzM−/− mice (n = 16). In all panels, data presented are mean ± SEM with statistical analysis by Student's t test. (*P < 0.05, **P < 0.01 and ***P < 0.001 relative to control mice).
Catecholamines play a central role in the regulation of BAT activity and WAT browning. Therefore, we next investigated whether macrophages per se might be a source of catecholamines underlying the increased browning in BAT and WAT in Irs2LyzM−/− mice. Initially, we used ELISAs to measure epinephrine and norepinephrine in conditioned medium from control and Irs2 null BMDM treated with either IL4 or LPS, but we were unable to reliably detect these catecholamines with these assays under these experimental conditions (data not shown). Therefore, we used a liquid chromatography mass spectroscopy approach to determine whether BMDM produced catecholamines. However, we were unable to detect these species at levels above those seen in media alone under the same experimental conditions despite being readily able to detect them in adrenal gland extracts (Figure 3E). To further investigate these findings, we examined the expression of the catecholamine biosynthesis enzymes Th and Dbh expression in BMDM from control mice using RT-PCR. We validated that our primers were functional using adrenal glands as a control but were unable to detect any expression of catecholamine biosynthetic enzymes in macrophages (Figure 3F, G). Taken together, these findings suggest that macrophages are not a source of norepinephrine that might underlie the upregulated BAT function and browning of WAT seen in Irs2LyzM−/− mice. Furthermore, our findings are consistent with some recent observations showing that macrophage populations do not express Th or produce catecholamines [18], [20].
3.4. Adipose tissue macrophages associate with sympathetic nerves: Irs2LyzM−/− mice display increased density of TH-positive neurons and elevated norepinephrine levels in BAT
Recent findings have suggested that CX3CR1-expressing macrophages in BAT may associate with TH-positive axons and that macrophage-specific deletion of the transcriptional repressor Mecp2, impairs BAT sympathetic innervation [45]. Furthermore, separate studies have shown both ATM and a novel macrophage sub-population (termed sympathetic-associated macrophages: SAMs) directly interact with WAT sympathetic nerves and in part regulate adipose tissue lipolysis by controling local catecholamine scavenging and catabolism [20]. In our own studies, we confirmed that CX3CR1-expressing macrophages abutted BAT TH-positive neurons (Supplemental Fig. 1D). Furthermore, we found increased Th mRNA expression and TH-nerve fibre density in the BAT of Irs2LyzM−/− mice suggesting an increase in sympathetic innervation of BAT (Figure 3H, I). Consistent with these observations, BAT norepinephrine levels were elevated in Irs2LyzM−/− mice (Figure 3J). Together these findings suggest that macrophages lacking Irs2 are able to positively influence WAT/BAT sympathetic nerve function leading to enhanced local release of norepinephrine and increase of thermogenic capacity in adipose tissues.
3.5. RNA sequencing analysis of Irs2-deficient macrophages reveals gene expression signatures of an anti-inflammatory phenotype and increased sympathetic activity in Irs2LyzM−/− mice
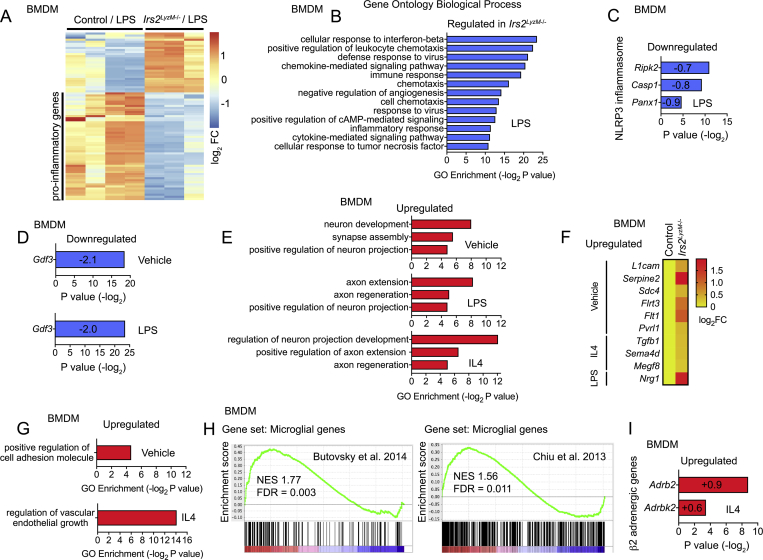
To try to gain broader insights into how deletion of Irs2 in macrophages might impart a beneficial metabolic phenotype and influence BAT function, we undertook RNA sequencing studies in BMDM from control and Irs2LyzM−/− mice. We focused our analysis on 1) macrophage inflammatory processes that may influence the ability of macrophages to scavenge and catabolize catecholamines as well as more broadly influence adipose tissue function and 2) processes by which macrophages may influence sympathetic innervation of BAT. In BMDMs treated with LPS, deletion of Irs2 was associated with a significantly reduced expression of pro-inflammatory gene categories as shown by gene ontology (GO) analysis. Down-regulated GO categories included regulation of cellular response to tumor necrosis factor, cytokine-mediated signaling pathway, inflammatory response, and cellular response to interferon beta (Figure 4A, B). Significantly down-regulated genes belonging to these categories included Inos, the Nfkb-related gene Nfkbib, Tnfa, IL-related genes (Il1a, Il19, Il7r, Il15, Il10, Il1rn, Il7r, and Il27), the Tgf beta receptor type I and a range of cytokines and chemokines (Cxcl9, Cxcl10, Ccl2, Ccl4, Ccl5, Ccl7, Clc8, Ccrl2, and Ccl12 (Figure 4A, B, and Supplemental Table 2)). The expression of genes associated with the NLRP3 inflammasome Ripk2, Casp1, and Panx1 was also reduced (Figure 4C). These data indicate that deletion of Irs2 in macrophages engenders a broad anti-inflammatory profile in these cells under the pro-inflammatory stress conditions of LPS treatment.
Figure 4.
mRNA sequencing analysis of control and Irs2LyzM−/−BMDM. (A) Heat map showing differentially regulated genes in Irs2LyzM−/− vs control BMDM following 24 h LPS treatment from RNA-seq analysis, including a large subset of downregulated pro-inflammatory genes in the Irs2LyzM−/−/LPS group (n = 4 for control, 3 for Irs2LyzM−/−). (B) Gene ontology (GO) categories of top 200 significantly regulated genes in Irs2LyzM−/− BMDM following 24 h treatment with LPS. (C) Differential expression of NLRP3 inflammasome-associated genes in LPS-treated Irs2LyzM−/− vs LPS-treated control BMDM. Log2 fold change for each gene is shown in its respective bar. (D) Differential expression of Gdf3 in both vehicle and LPS-treated Irs2LyzM−/− relative to control BMDM with respective treatments. Log2 fold change is shown in each respective bar. (E) GO categories with significant enrichment related to nerve growth and function in Irs2LyzM−/− relative to control BMDM with either vehicle, LPS or IL4 (overnight) treated conditions. (F) Heat map showing upregulated neuronal-associated genes (secreted and cell surface proteins involved in cell adhesion and cell-to-cell interactions) in Irs2LyzM−/− vs control BMDM under various conditions. (G) GO categories with significant enrichment in Irs2LyzM−/− relative to control BMDM under various conditions. (H) Gene set enrichment analysis showing increase of microglial markers in IL4-treated Irs2LyzM−/− relative to IL4-treated control BMDM. (I) Differential expression of β2 adrenergic genes Adrb2 and Adrbk2 in IL4-treated Irs2LyzM−/− relative to IL4-treated control BMDM. Log2 fold change for each gene is shown in its respective bar. In panels B–E and also G and I, P values for GO enrichment categories or individual genes are shown, calculated as described in the methods (section 2.8).
Recently it has been found that macrophages from aged mice display an inflammatory phenotype and an associated increased ability to lower WAT catecholamine levels due to a GDF3-dependent up-regulation of the catecholamine degradation machinery [19]. Consistent with these findings, relative Gdf3 expression was significantly reduced in both untreated and LPS-treated Irs2LyzM−/− BMDM compared to control BMDM. (Figure 4D). However, when we examined expression of the machinery for bioamine uptake, we did not detect Slc6a2, the noradrenaline transporter (Supplemental Table 2), which is consistent with recent findings [20] and suggests this is exclusively expressed in SAMs. In addition, we examined the expression of a number of genes involved in bioamine metabolism such as Maoa, Comt, and members of the Aldh and Akr families of enzymes involved in biogenic amine degradation. Although Maoa expression was reduced in LPS-treated Irs2LyzM−/− BMDM, in general there was no concerted change in the expression pattern of these genes that would explain the increase in WAT catecholamine levels via effects upon macrophage-clearance of catecholamines (Supplemental Table 2).
Deletion of Mecp2, a nuclear transcriptional regulator implicated in Rett syndrome, in the BAT-resident CX3CR1-expressing macrophage population recently has been shown to reduce BAT sympathetic innervation [45]. In contrast, we demonstrate increased TH-positive nerve fibre density in this tissue in Irs2LyzM−/− mice. Together these findings indicate that intrinsic macrophage signaling events might regulate pathways directly involved in nerve growth and maintenance. Consistent with this idea, in untreated Irs2LyzM−/− BMDM we found increases in the expression of genes in GO categories including positive regulation of neuron development, synapse assembly and neuron projection (Figure 4E). In Irs2LyzM−/− BMDM treated with IL4 we found increased expression in GO categories including regulation of neuron projection development and positive regulation of axon extension and axon regeneration (Figure 4E). In LPS-treated Irs2LyzM−/− BMDM, we found increased expression in GO categories including positive regulation of neuron projection and axon extension and remodeling (Figure 4E). While these categories included genes classically expressed in neurons they also included genes for secreted proteins and cell surface proteins involved in cell adhesion and cell-to-cell interaction processes through which macrophages could potentially directly be involved in the regulation of neuronal function. The genes in the GO categories involved included: L1cam, a cell adhesion molecule involved in neurite outgrowth [57]; Serpine2, which promotes neurite extension [58]; Sdc4, which plays a role in neuron adhesion processes [59]; Flrt3, which is involved in axon guidance [60]; Flt1, which has neurotrophic effects [61]; Pvrl1, an adhesion molecule with roles in glia–neuron interactions [62]; Tgfb1, which is involved in a number of elements of neuronal function [63]; Sema4d, a semaphorin that acts a soluble neurotrophic factor [64]; Megf8, a transmembrane EGF-like protein implicated in neural growth [65] and Nrg1, a nerve growth factor [66] (Figure 4F). Gene expression categories related to cell adhesion and to vascular development, which may also be involved in the control of tissue innervation [67], were also upregulated in Irs2LyzM−/− BMDM (Figure 4G). Together these findings suggest that deletion of Irs2 in macrophages potentially creates an environment in BAT that enhances sympathetic innervation.
Interestingly, we also found an increase in the expression of microglial markers in IL4 treated Irs2LyzM−/− BMDM including Cx3cr1 (Figure 4H [68], [69]), suggesting that there could be an increased association of these cells with nerves although we did not detect increased macrophage numbers in adipose tissue. Furthermore, we note that Irs2 is enriched in SAMs compared to other macrophage populations [20] again suggesting a role for this signaling molecule in these cells. We also found a significant increase in the expression of the β2 adrenergic receptor (Adrb2) and a non-significant increase in the β adrenergic receptor kinase 2 (Adrbk2) in IL4-treated Irs2LyzM−/− BMDM (Figure 4I). Together these expression profiles suggest that macrophages may play a positive role in tissue nerve function and that deletion of Irs2 increases the expression of such pathways indicating potential bidirectional interplay between macrophages and sympathetic nerves.
3.6. Irs2 expression in BMDM is increased by palmitate and deletion of Irs2 in macrophages attenuates insulin signaling but paradoxically enhances IL4 signaling
Next, we further explored potential mechanisms underlying the above gene expression profiles showing reduced inflammatory response to LPS but a paradoxically preserved response to IL4 in Irs2LyzM−/− BMDM. Insulin signaling in macrophages has been suggested to be pro-inflammatory indicating that Irs2 might mediate such events and that macrophage IRS2 may be involved in the detrimental effects of inflammatory macrophages upon insulin action in other tissues. Palmitate treatment induces an inflammatory state in BMDM and conditioned medium from these cells induces insulin resistance in muscle cells [70]. Consistent with this idea incubation of BMDM was associated with an increase in Irs2 expression (Supplemental Fig. 1E). Recent studies in BMDM from mice with global deletion of Irs2 have demonstrated enhanced IL4-dependent M2 marker expression, thought to be due to increased signaling via IRS1 [29]. As described above, we also found preservation and in some cases enhancement of IL4-stimulated gene expression in Irs2LyzM−/− BMDM. However, we found minimal expression of Irs1 and Irs3 and no Irs4 mRNA in wild type BMDM consistent with previous findings that Irs2 is the predominant Irs gene in hematopoetic cells [24–26 and Supplemental Table 2], and we saw no change in expression of Irs1 or Irs3 mRNA upon Irs2 deletion in our RNAseq data (Irs1: 0.12 log2FC; P = 0.91 and Irs3: 0.11 log2FC; P = 0.89). Indeed, we could not detect IRS1 protein expression in these cells by western blotting (Supplemental Fig. 1F). However, in Irs2LyzM−/− BMDM, we found increased AKT phosphorylation in response to IL4 treatment (Supplemental Fig. 1G). This was consistent with recent studies [29] despite attenuation of insulin-stimulated AKT phosphorylation (Supplemental Fig. 1H), indicating that normally IRS2 signaling in part plays a role in restraining IL4 signaling or that loss of IRS2 allows an alternative signaling pathway to be engaged that mediates effects upon gene expression driven by IL4.
4. Discussion
There is increasing recognition that the interplay between the immune and nervous systems plays a key role in the regulation of adipose tissue function [71]. In the current studies, we demonstrate that deletion in LyzM-expressing macrophage populations of Irs2, a major mediator of IL4 and insulin signaling, leads to resistance to diet-induced obesity via increased BAT thermogenesis through increased sympathetic innervation and increased local catecholamine availability. Underpinning these observations, we find that Irs2-deficient BMDM do not express catecholamine biosynthetic machinery but, when challenged with an inflammatory stimulus, display a transcriptional profile consistent with reduced inflammation together with alterations in the expression of genes that are involved in the regulation of tissue innervation, sympathetic nerve function, and adipose tissue lipolysis. Our studies therefore links insulin and IL4 signaling through IRS2 in macrophages to the regulation of systemic metabolism via control of adipose tissue sympathetic nerve function, adding new insights to the signaling processes involved in this neuro–immune interaction.
Previously, a series of studies has suggested that AAM produce catecholamines in an IL4-dependent manner to regulate BAT and WAT thermogenic programs and adipocyte metabolism [17]. In this work, mice with myeloid-restricted deletion of Il4ra displayed impaired M2 polarization and defective cold adaptive thermogenesis associated with a reduced production of catecholamines by macrophages [17]. However, recent evidence has challenged this model with a failure to detect either catecholamine biosynthetic apparatus in adipose tissue macrophages or beneficial effects of chronic IL4 treatment upon energy expenditure [18], [20]. While we found both increased BAT browning and an associated increase in adipose tissue noradrenaline levels in our mutant mice, we did not detect the machinery for catecholamine synthesis in BMDM or noradrenaline in BMDM conditioned medium at levels above that seen in medium alone. Recent studies have also suggested that macrophages per se are not the source of catecholamines in adipose tissue [18], [20], [72], although another recent paper has demonstrated Th expression in ATM [73]. Although our studies are consistent with the idea that macrophages in adipose tissue may not produce catecholamines, as we have performed many studies on BMDM rather than ATM our work is not definitive about this issue as resident macrophages in tissues may display distinct phenotypes from BMDM [74]. Future work will be required to address this specific question in Irs2LyzM−/−mice.
Our studies do suggest, however, that alterations in insulin and IL4 signaling may impact upon macrophage biology to engender changes in adipose tissue function via sympathetic innervation. These observations are complicated by our findings that deletion of Irs2 in macrophages causes polarization towards an anti-inflammatory phenotype with persistent rather than attenuated signaling downstream of IL4. This is consistent with recent findings in macrophages from mice with global deletion of Irs2 which also displayed a paradoxical enhancement of IL4 signaling [29] although, in our case, we did not detect upregulation of Irs1 expression in Irs2-deficient macrophages. It therefore remains to be seen what mediates the preserved IL4 signaling in Irs2-deficient macrophages in our mouse model, with other adaptor molecules downstream of IL4 receptor signaling potentially playing a role. Irs2 loss has been shown to cause enhanced rather than attenuated functional end-points, including enhanced learning and memory [75]. This in turn leads to positive effects on physiology, suggesting that in certain circumstances IRS2 may act as a brake upon specific signaling processes [75].
Taken together, our data illustrate that altered insulin and IL4 signaling in macrophages can have beneficial metabolic effects acting via alterations in sympathetic nerve function rather than release of catecholamines by macrophages per se. Recent studies have also revealed potential alternative mechanisms by which this macrophage-mediated regulation of adipose tissue sympathetic nerve function occurs. These include the scavenging and catabolism of catecholamines or the direct control of adipose tissue sympathetic innervation [19], [20], [45]. For example, it has been demonstrated that a GDF3-dependent mechanism mediates the reduction of catecholamine-induced lipolysis that occurs with ageing [19]. This study showed that age-related NLRP3 inflammasome activation increased the ability of macrophages to scavenge catecholamines in adipose tissue with elevated expression of MAOA occurring via GDF3, thereby resulting in reduced adipose tissue lipolysis [19]. Consistent with these findings we show that Irs2-deficient BMDM have reduced expression of Gdf3 which would potentially result in increased adipose tissue catecholamine availability, although it should be noted that we have not studied gene expression in ATMs per se. A parallel finding has been the identification of a specialized myeloid sub-population that is associated with sympathetic nerves [20]. These SAMS scavenge catecholamines via SLC6A2-dependent uptake and MAOA degradation pathways and abrogation of SAM function leads to increased BAT recruitment and WAT browning suggesting they may have a role in regulating adiposity [20]. While we have not isolated SAMs, we find that elements of the catecholamine scavenging machinery are expressed in BMDM (although not Slc6a2) although we do not detect concerted alterations of these genes upon Irs2 deletion in these cells that might reduce macrophage uptake and clearance of catecholamines. We also find that macrophages can be associated with BAT sympathetic nerves consistent with the findings of others and demonstrating the potential for the direct regulation of sympathetic nerve function by macrophages. Interestingly, while it appears that SAMs have a pro-inflammatory phenotype [20], macrophages lacking Irs2 have an anti-inflammatory phenotype when challenged with LPS suggesting that distinct mechanisms and myeloid populations may be involved. However, it also appears that SAMs are enriched for Irs2-expression and express LyzM [20] and so it is possible that altered SAM function contributes to the phenotype seen in our mice.
While we appear to only have partial evidence for a mechanism that might involve alterations in catecholamine scavenging by macrophages, we find increased density of sympathetic nerves in BAT and a series of transcriptional changes in BMDM that might underpin this finding. These include up-regulation of genes not only in implicated in specific components of neuronal function such as axon guidance and development but also in processes such as vascularization and cell adhesion, which may play more generic roles in tissue innervation. That macrophages can directly interact with the processes involved in regulating BAT sympathetic nerves has been illustrated recently in mice with macrophage deletion of Mecp2 [45]. The underlying mechanisms appear to involve semaphorin signaling cross-talk between macrophages and nerves and specifically macrophages expressing CX3CR1 [45]. We do not find directly equivalent gene expression alterations in semaphorin pathways in Irs2LyzM−/− BMDM, but, interestingly, these cells were enriched for microglial markers such as Cxc3r1 potentially, suggesting an increased propensity to interact with nerves.
While our work and that of others suggest that adipose tissue macrophages directly interact with sympathetic nerves to regulate catecholamine tone in this tissue, there are several lines of evidence that macrophages with an inflammatory profile may impair the ability of BAT to respond appropriately to thermogenic stimuli. For example, in vitro studies and analysis of macrophages from mice under HFD feeding shows that TNFA and IL1B directly suppress the expression of UCP1 in both BAT and WAT [76], [77]. Therefore, the markedly reduced inflammatory profile seen in Irs2LyzM−/− BMDM could contribute to the enhanced BAT activity through direct transcriptional effects on Ucp1 and other components of the thermogenic programme in addition to indirect effects via adipose tissue sympathetic nerve function. Upregulation of Irs2 by palmitate, a pro-inflammatory stimulus for BMDM that leads to secretion of insulin resistance-causing factors from macrophages, further supports the idea that deletion of Irs2 reduces macrophage inflammation and that this may be involved in the beneficial metabolic effects including improved insulin sensitivity.
It should also be noted that the LyzM-Cre mouse utilized in our studies deletes in other myeloid cell lineages. These include neutrophils which have also been implicated in the regulation of adipose tissue function and obesity-related insulin resistance [7], [8], [9], [78]. Therefore, a limitation of our study is that the in vivo phenotypes reported could potentially involve a contribution from the cell types distinct from macrophages in which LyzM-Cre is expressed.
In summary our studies indicate that the deletion of Irs2 in macrophages has beneficial metabolic effects stemming from increased activity of BAT and browning of WAT depots at ambient temperatures due to an underlying increase in adipose tissue catecholamine function. This in turn is due to pleiotropic effects upon macrophage function including a markedly reduced inflammatory profile, the expression of genes that may be involved in catecholamine scavenging and the development of an appropriate cellular environment for enhanced adipose tissue sympathetic innervation. Consistent with recent studies we show that macrophages do not produce catecholamines per se but that alteration of a component of both IL4 and insulin signaling in the LyzM population of macrophages can have profound effects on systemic metabolism. Therefore, our work adds to the growing evidence that neuro–immune interactions play a central role in adipose tissue biology and the pathophysiology of obesity and insulin resistance while identifying potential new cellular and molecular events to target for therapeutic benefit in these conditions.
Declarations of interest
None.
Author contributions
MTR, SJM, and DJW designed the research. MTR, SJM, SMAP, AIC, ASHC, DH, JG, EEI, CS, SK, and MCW performed and analysed experiments. VKY contributed reagents and tools. MTR, SJM, CS, VKY, AVP, SV, CF, and DJW drafted and/or wrote the manuscript. MTR, VKY, CF, and DJW provided funding. DJW supervised the work.
Acknowledgements
We are grateful to Chad Whilding, Naveenan Navaratnam, Alessandro Sardini, Olga Boruc, Jacques Behmoaras, and Mahrokh Nohadani for technical help with some experiments. MTR was supported by a 3 year Wellcome Trust Studentship via the Sanger Centre. VKY was supported by the Wellcome Trust (098051). Work in the CF laboratory was supported by the Medical Research Council (MRC_MC_UU_12022/6). DJW received grants from the Wellcome Trust: Project Grant (093082/Z/10/Z), a Wellcome Trust Strategic Award (098565/Z/12/Z) and funding from the Medical Research Council (MC-A654-5QB40). SV and AVP were funded by the BHF (RG/18/7/33636).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2018.11.010.
Conflict of interest
The authors declare no conflict of interest.
Supplemental Table 1.
| List of probes used in RT-PCR. (TaqMan, Applied Biosystems, all Mus Musculus). | |
|---|---|
| Interleukin 1 beta (Il1b) | Mm01336189_m1 |
| Interleukin 6 (Il6) | Mm00446190_m1 |
| Inducible nitric oxide synthase (Inos) | Mm00440502_m1 |
| Tumor necrosis factor alpha (Tnfa) | Mm00443258_m1 |
| Arginase 1 (Arg1) | Mm00475988_m1 |
| Chitinase 3-like-3 (Chi3l3) | Mm04213363_m1 |
| Mannose receptor 1 (Mrc1) | Mm00485148_m1 |
| C-type lectin domain family 10 member A (Clec10A) | Mm00546124_m1 |
| Hypoxanthine phosphoribosyltransferase (Hprt) | Mm03024075_m1 |
| Uncoupling protein 1 (Ucp1) | Mm01244861_m1 |
| Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) | Mm99999915_g1 |
| Peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (Pgc1a) | Mm01208835_m1 |
| Tyrosine hydroxylase (Th) | Mm00447557_m1 |
| Cell death-inducing DNA fragmentation factor, alpha subunit-like effector A (Cidea) | Mm00432554_m1 |
| Insulin receptor substrate 2 (Irs2) | Mm03038438_m1 |
| Dopamine beta hydroxylase (Dbh) | Mm00460472_m1 |
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
(A) Quantitative RT-PCR analysis of Irs2 mRNA expression in control and Irs2LyzM-/− BMDM. Hprt was used as an internal control (n = 5-6). (B) IRS2 immunostaining (green) in control and Irs2LyzM-/− BMDM. DAPI was used to visualize nuclei (blue). (3 mice/genotype). Scale bar = 10 μm. (C) Body weights of control and Irs2LyzM-/− male mice and female on chow diet at 23 weeks of age (n = 8). (D) TH (green) and CX3CR1 (red) immunostaining in BAT from wild type mice. DAPI was used to visualize nuclei (blue). Scale bar = 20 μm (E) Quantitative RT-PCR analysis of Irs2 mRNA expression in control BMDM treated with palmitate for 3 or 24 hours. BSA-treated BMDM were used as a control and Hprt mRNA was used as an internal gene control (n = 5-6). (F) Western blotting analysis of IRS1 expression in BMDM from control and Irs2LyzM-/−. Brain tissue lysates were used as a positive control. (G, H) Western blotting analysis of serine 473 phosphorylation of AKT in BMDM from control and Irs2LyzM-/− mice treated with IL4 (G) or insulin (H) for the times indicated. A representative blot is shown (n = 4-5). Mean values for band intensities in multiple samples quantified by densitometry are shown below the panel, relative to control samples for each time point. In all panels, data presented are mean ± SEM with Student’s t test for all panels. (*P < 0.05 and ***P < 0.001 relative to controls).
References
- 1.Collaboration N.C.D.R.F. Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19.2 million participants. Lancet. 2016;387:1377–1396. doi: 10.1016/S0140-6736(16)30054-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lauby-Secretan B., Scoccianti C., Loomis D., Grosse Y., Bianchini F., Straif K. Body fatness and cancer--viewpoint of the iarc working group. New England Journal of Medicine. 2016;375:794–798. doi: 10.1056/NEJMsr1606602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collaborators G.B.D.O., Afshin A., Forouzanfar M.H., Reitsma M.B., Sur P., Estep K. Health effects of overweight and obesity in 195 countries over 25 years. New England Journal of Medicine. 2017;377:13–27. doi: 10.1056/NEJMoa1614362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odegaard J.I., Chawla A. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nature Clinical Practice Endocrinology & Metabolism. 2008;4:619–626. doi: 10.1038/ncpendmet0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lackey D.E., Olefsky J.M. Regulation of metabolism by the innate immune system. Nature Reviews Endocrinology. 2016;12:15–28. doi: 10.1038/nrendo.2015.189. [DOI] [PubMed] [Google Scholar]
- 6.Chawla A., Nguyen K.D., Goh Y.P. Macrophage-mediated inflammation in metabolic disease. Nature Reviews Immunology. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotamisligil G.S. Foundations of immunometabolism and implications for metabolic health and disease. Immunity. 2017;47:406–420. doi: 10.1016/j.immuni.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hotamisligil G.S. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542:177–185. doi: 10.1038/nature21363. [DOI] [PubMed] [Google Scholar]
- 9.Mclaughlin T., Ackerman S.E., Shen L., Engleman E. Role of innate and adaptive immunity in obesity-associated metabolic disease. Journal of Clinical Investigation. 2017;127:5–13. doi: 10.1172/JCI88876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez F.O., Sica A., Mantovani A., Locati M. Macrophage activation and polarization. Frontiers in Bioscience. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 11.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nature Reviews Immunology. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 13.Van Dyken S.J., Locksley R.M. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annual Review of Immunology. 2013;31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wynn T.A., Chawla A., Pollard J.W. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D., Molofsky A.B., Liang H.E., Ricardo-Gonzalez R.R., Jouihan H.A., Bando J.K. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243–247. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chawla A. Control of macrophage activation and function by ppars. Circulation Research. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen K.D., Qiu Y., Cui X., Goh Y.P., Mwangi J., David T. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer K., Ruiz H.H., Jhun K., Finan B., Oberlin D.J., Van Der Heide V. Alternatively activated macrophages do not synthesize catecholamines or contribute to adipose tissue adaptive thermogenesis. Nature Medicine. 2017;23:623–630. doi: 10.1038/nm.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camell C.D., Sander J., Spadaro O., Lee A., Nguyen K.Y., Wing A. Inflammasome-driven catecholamine catabolism in macrophages blunts lipolysis during ageing. Nature. 2017;550:119–123. doi: 10.1038/nature24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pirzgalska R.M., Seixas E., Seidman J.S., Link V.M., Sanchez N.M., Mahu I. Sympathetic neuron-associated macrophages contribute to obesity by importing and metabolizing norepinephrine. Nature Medicine. 2017;23:1309–1318. doi: 10.1038/nm.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heller N.M., Qi X., Junttila I.S., Shirey K.A., Vogel S.N., Paul W.E. Type i il-4rs selectively activate irs-2 to induce target gene expression in macrophages. Science Signaling. 2008;1:ra17. doi: 10.1126/scisignal.1164795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly-Welch A., Hanson E.M., Keegan A.D. Interleukin-4 (il-4) pathway. Science's STKE. 2005 doi: 10.1126/stke.2932005cm9. 2005: cm9. [DOI] [PubMed] [Google Scholar]
- 23.Kelly-Welch A., Hanson E.M., Keegan A.D. Interleukin-13 (il-13) pathway. Science's STKE. 2005 doi: 10.1126/stke.2932005cm8. 2005: cm8. [DOI] [PubMed] [Google Scholar]
- 24.Sun X.J., Pons S., Wang L.M., Zhang Y., Yenush L., Burks D. The irs-2 gene on murine chromosome 8 encodes a unique signaling adapter for insulin and cytokine action. Molecular Endocrinology. 1997;11:251–262. doi: 10.1210/mend.11.2.9885. [DOI] [PubMed] [Google Scholar]
- 25.Sun X.J., Wang L.M., Zhang Y., Yenush L., Myers M.G., Jr., Glasheen E. Role of irs-2 in insulin and cytokine signalling. Nature. 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 26.Welham M.J., Bone H., Levings M., Learmonth L., Wang L.M., Leslie K.B. Insulin receptor substrate-2 is the major 170-kda protein phosphorylated on tyrosine in response to cytokines in murine lymphohemopoietic cells. Journal of Biological Chemistry. 1997;272:1377–1381. doi: 10.1074/jbc.272.2.1377. [DOI] [PubMed] [Google Scholar]
- 27.Withers D.J. Insulin receptor substrate proteins and neuroendocrine function. Biochemical Society Transactions. 2001;29:525–529. doi: 10.1042/bst0290525. [DOI] [PubMed] [Google Scholar]
- 28.Withers D.J., White M. Perspective: the insulin signaling system--a common link in the pathogenesis of type 2 diabetes. Endocrinology. 2000;141:1917–1921. doi: 10.1210/endo.141.6.7584. [DOI] [PubMed] [Google Scholar]
- 29.Dasgupta P., Dorsey N.J., Li J., Qi X., Smith E.P., Yamaji-Kegan K. The adaptor protein insulin receptor substrate 2 inhibits alternative macrophage activation and allergic lung inflammation. Science Signaling. 2016;9:ra63. doi: 10.1126/scisignal.aad6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keegan A.D., Zamorano J., Keselman A., Heller N.M. Il-4 and il-13 receptor signaling from 4ps to insulin receptor substrate 2: there and back again, a historical view. Frontiers in Immunology. 2018;9:1037. doi: 10.3389/fimmu.2018.01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mauer J., Chaurasia B., Plum L., Quast T., Hampel B., Bluher M. Myeloid cell-restricted insulin receptor deficiency protects against obesity-induced inflammation and systemic insulin resistance. PLoS Genetics. 2010;6 doi: 10.1371/journal.pgen.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burks D.J., Font De Mora J., Schubert M., Withers D.J., Myers M.G., Towery H.H. Irs-2 pathways integrate female reproduction and energy homeostasis. Nature. 2000;407:377–382. doi: 10.1038/35030105. [DOI] [PubMed] [Google Scholar]
- 33.Withers D.J., Burks D.J., Towery H.H., Altamuro S.L., Flint C.L., White M.F. Irs-2 coordinates igf-1 receptor-mediated beta-cell development and peripheral insulin signalling. Nature Genetics. 1999;23:32–40. doi: 10.1038/12631. [DOI] [PubMed] [Google Scholar]
- 34.Withers D.J., Gutierrez J.S., Towery H., Burks D.J., Ren J.M., Previs S. Disruption of irs-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 35.Baumgartl J., Baudler S., Scherner M., Babaev V., Makowski L., Suttles J. Myeloid lineage cell-restricted insulin resistance protects apolipoproteine-deficient mice against atherosclerosis. Cell Metabolism. 2006;3:247–256. doi: 10.1016/j.cmet.2006.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving bioscience research reporting: the arrive guidelines for reporting animal research. PLoS Biology. 2010;8 doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choudhury A.I., Heffron H., Smith M.A., Al-Qassab H., Xu A.W., Selman C. The role of insulin receptor substrate 2 in hypothalamic and beta cell function. Journal of Clinical Investigation. 2005;115:940–950. doi: 10.1172/JCI24445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clausen B.E., Burkhardt C., Reith W., Renkawitz R., Forster I. Conditional gene targeting in macrophages and granulocytes using lysmcre mice. Transgenic Research. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 39.Cantley J., Choudhury A.I., Asare-Anane H., Selman C., Lingard S., Heffron H. Pancreatic deletion of insulin receptor substrate 2 reduces beta and alpha cell mass and impairs glucose homeostasis in mice. Diabetologia. 2007;50:1248–1256. doi: 10.1007/s00125-007-0637-9. [DOI] [PubMed] [Google Scholar]
- 40.Cantley J., Selman C., Shukla D., Abramov A.Y., Forstreuter F., Esteban M.A. Deletion of the von hippel-lindau gene in pancreatic beta cells impairs glucose homeostasis in mice. Journal of Clinical Investigation. 2009;119:125–135. doi: 10.1172/JCI26934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner R.C., Holman R.R., Matthews D., Hockaday T.D., Peto J. Insulin deficiency and insulin resistance interaction in diabetes: estimation of their relative contribution by feedback analysis from basal plasma insulin and glucose concentrations. Metabolism. 1979;28:1086–1096. doi: 10.1016/0026-0495(79)90146-x. [DOI] [PubMed] [Google Scholar]
- 42.Rached M.T., Kode A., Silva B.C., Jung D.Y., Gray S., Ong H. Foxo1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. Journal of Clinical Investigation. 2010;120:357–368. doi: 10.1172/JCI39901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cinti S., Mitchell G., Barbatelli G., Murano I., Ceresi E., Faloia E. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. The Journal of Lipid Research. 2005;46:2347–2355. doi: 10.1194/jlr.M500294-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Seale P., Conroe H.M., Estall J., Kajimura S., Frontini A., Ishibashi J. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. Journal of Clinical Investigation. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf Y., Boura-Halfon S., Cortese N., Haimon Z., Sar Shalom H., Kuperman Y. Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nature Immunology. 2017;18:665–674. doi: 10.1038/ni.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prieur X., Mok C.Y., Velagapudi V.R., Nunez V., Fuentes L., Montaner D. Differential lipid partitioning between adipocytes and tissue macrophages modulates macrophage lipotoxicity and m2/m1 polarization in obese mice. Diabetes. 2011;60:797–809. doi: 10.2337/db10-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Current Protocols in Immunology Chapter. 2008;14 doi: 10.1002/0471142735.im1401s83. Unit 14 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Den Bossche J., Lamers W.H., Koehler E.S., Geuns J.M., Alhonen L., Uimari A. Pivotal advance: arginase-1-independent polyamine production stimulates the expression of il-4-induced alternatively activated macrophage markers while inhibiting lps-induced expression of inflammatory genes. Journal of Leukocyte Biology. 2012;91:685–699. doi: 10.1189/jlb.0911453. [DOI] [PubMed] [Google Scholar]
- 49.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S., Pyl P.T., Huber W. Htseq--a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexa A., Rahnenfuhrer J. R Package version 2.24.0; 2016. topGO: enrichemnt analysis for gene ontology. [Google Scholar]
- 54.Vaanholt L.M., Daan S., Schubert K.A., Visser G.H. Metabolism and aging: effects of cold exposure on metabolic rate, body composition, and longevity in mice. Physiological and Biochemical Zoology. 2009;82:314–324. doi: 10.1086/589727. [DOI] [PubMed] [Google Scholar]
- 55.Choi C.S., Fillmore J.J., Kim J.K., Liu Z., Kim S., Collier E.F. Overexpression of uncoupling protein 3 in skeletal muscle protects against fat-induced insulin resistance. Journal of Clinical Investigation. 2007;117:1995–2003. doi: 10.1172/JCI13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Odegaard J.I., Chawla A. Alternative macrophage activation and metabolism. Annual Review of Pathology: Mechanisms of Disease. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burden-Gulley S.M., Pendergast M., Lemmon V. The role of cell adhesion molecule l1 in axonal extension, growth cone motility, and signal transduction. Cell and Tissue Research. 1997;290:415–422. doi: 10.1007/s004410050948. [DOI] [PubMed] [Google Scholar]
- 58.Wagner S.L., Geddes J.W., Cotman C.W., Lau A.L., Gurwitz D., Isackson P.J. Protease nexin-1, an antithrombin with neurite outgrowth activity, is reduced in alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:8284–8288. doi: 10.1073/pnas.86.21.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin T.J., Lu K.W., Chen W.H., Cheng C.M., Lin Y.W. Roles of syndecan-4 and relative kinases in dorsal root ganglion neuron adhesion and mechanotransduction. Neuroscience Letters. 2015;592:88–93. doi: 10.1016/j.neulet.2015.02.058. [DOI] [PubMed] [Google Scholar]
- 60.Leyva-Diaz E., Del Toro D., Menal M.J., Cambray S., Susin R., Tessier-Lavigne M. Flrt3 is a robo1-interacting protein that determines netrin-1 attraction in developing axons. Current Biology. 2014;24:494–508. doi: 10.1016/j.cub.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez A., Wadhwani S., Grammas P. Multiple neurotrophic effects of vegf on cultured neurons. Neuropeptides. 2010;44:323–331. doi: 10.1016/j.npep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mori M., Rikitake Y., Mandai K., Takai Y. Roles of nectins and nectin-like molecules in the nervous system. Advances in Neurobiology. 2014;8:91–116. doi: 10.1007/978-1-4614-8090-7_5. [DOI] [PubMed] [Google Scholar]
- 63.Gomes F.C., Sousa Vde O., Romao L. Emerging roles for tgf-beta1 in nervous system development. International Journal of Developmental Neuroscience. 2005;23:413–424. doi: 10.1016/j.ijdevneu.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 64.Fujioka S., Masuda K., Toguchi M., Ohoka Y., Sakai T., Furuyama T. Neurotrophic effect of semaphorin 4d in pc12 cells. Biochemical and Biophysical Research Communications. 2003;301:304–310. doi: 10.1016/s0006-291x(02)03023-1. [DOI] [PubMed] [Google Scholar]
- 65.Engelhard C., Sarsfield S., Merte J., Wang Q., Li P., Beppu H. Megf8 is a modifier of bmp signaling in trigeminal sensory neurons. Elife. 2013;2 doi: 10.7554/eLife.01160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Esper R.M., Pankonin M.S., Loeb J.A. Neuregulins: versatile growth and differentiation factors in nervous system development and human disease. Brain Research Reviews. 2006;51:161–175. doi: 10.1016/j.brainresrev.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 67.Carmeliet P. Blood vessels and nerves: common signals, pathways and diseases. Nature Reviews Genetics. 2003;4:710–720. doi: 10.1038/nrg1158. [DOI] [PubMed] [Google Scholar]
- 68.Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G. Identification of a unique TGF-B-dependent molecular and functional signature in microglia. Nature Neuroscience. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiu I.M., Morimoto E.T.A., Goodarzi H., Liao J.T., O'Keeffe S., Phatnani H.P. A neurodegeneration-specific gene expression signature and immune profile of acutely isolated microglia from an ALS mouse model. Cell Reports. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kewalramani G., Fink L.N., Asadi F., Klip A. Palmitate-activated macrophages confer insulin resistance to muscle cells by a mechanism involving protein kinase C θ and ε. PloS One. 2011;6 doi: 10.1371/journal.pone.0026947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Villarroya F., Cereijo R., Villarroya J., Gavalda-Navarro A., Giralt M. Toward an understanding of how immune cells control brown and beige adipobiology. Cell Metabolism. 2018;27:954–961. doi: 10.1016/j.cmet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Spadaro O., Camell C.D., Bosurgi L., Nguyen K.Y., Youm Y.H., Rothlin C.V. Igf1 shapes macrophage activation in response to immunometabolic challenge. Cell Reports. 2017;19:225–234. doi: 10.1016/j.celrep.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruiz De Azua I., Mancini G., Srivastava R.K., Rey A.A., Cardinal P., Tedesco L. Adipocyte cannabinoid receptor cb1 regulates energy homeostasis and alternatively activated macrophages. Journal of Clinical Investigation. 2017;127:4148–4162. doi: 10.1172/JCI83626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Epelman S., Lavine K.J., Randolph G.J. Origin and functions of tissue macrophages. Immunity. 2014;41:21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Irvine E.E., Drinkwater L., Radwanska K., Al-Qassab H., Smith M.A., O'brien M. Insulin receptor substrate 2 is a negative regulator of memory formation. Learning & Memory. 2011;18:375–383. doi: 10.1101/lm.2111311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goto T., Naknukool S., Yoshitake R., Hanafusa Y., Tokiwa S., Li Y. Proinflammatory cytokine interleukin-1beta suppresses cold-induced thermogenesis in adipocytes. Cytokine. 2016;77:107–114. doi: 10.1016/j.cyto.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 77.Sakamoto T., Nitta T., Maruno K., Yeh Y.S., Kuwata H., Tomita K. Macrophage infiltration into obese adipose tissues suppresses the induction of ucp1 level in mice. American Journal of Physiology. Endocrinology and Metabolism. 2016;310:E676–E687. doi: 10.1152/ajpendo.00028.2015. [DOI] [PubMed] [Google Scholar]
- 78.Talukdar S., Oh D.Y., Bandyopadhyay G., Li D., Xu J., McNelis J. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nature Medicine. 2012;18:1407–1412. doi: 10.1038/nm.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.