Abstract
Objectives
Brown adipose tissue (BAT) dissipates nutritional energy as heat through uncoupling protein 1 (UCP1). The discovery of functional BAT in healthy adult humans has promoted the search for pharmacological interventions to recruit and activate brown fat as a treatment of obesity and diabetes type II. These efforts require in vivo models to compare the efficacy of novel compounds in a relevant physiological context.
Methods
We generated a knock-in mouse line expressing firefly luciferase and near-infrared red florescent protein (iRFP713) driven by the regulatory elements of the endogenous Ucp1 gene.
Results
Our detailed characterization revealed that firefly luciferase activity faithfully reports endogenous Ucp1 gene expression in response to physiological and pharmacological stimuli. The iRFP713 fluorescence signal was detected in the interscapular BAT region of cold-exposed reporter mice in an allele-dosage dependent manner. Using this reporter mouse model, we detected a higher browning capacity in female peri-ovarian white adipose tissue compared to male epididymal WAT, which we further corroborated by molecular and morphological features. In situ imaging detected a strong luciferase activity signal in a previously unappreciated adipose tissue depot adjunct to the femoral muscle, now adopted as femoral brown adipose tissue. In addition, screening cultured adipocytes by bioluminescence imaging identified the selective Salt-Inducible Kinase inhibitor, HG-9-91-01, to increase Ucp1 gene expression and mitochondrial respiration in brown and brite adipocytes.
Conclusions
In our mouse model, firefly luciferase activity serves as a bona fide reporter for dynamic regulation of Ucp1. In addition, by means of iRFP713 we are able to monitor Ucp1 expression in a non-invasive fashion.
Keywords: BAT, WAT, Firefly luciferase, iRFP713, UCP1, Thermogenesis, Browning
Highlights
-
•
A new reporter mouse model enables bona fide monitoring of endogenous Ucp1 gene expression.
-
•
The new generation near-infrared fluorescent protein iRFP713 enables noninvasive imaging of brown fat.
-
•
Reporter mice show sex-specific differences in browning propensity of gonadal WAT.
-
•
We discovered a novel brown adipose tissue depot, now adopted as femoral brown fat.
-
•
Our reporter cell assays revealed the pan-Salt-Inducible Kinase as an inhibitor of Ucp1 gene expression.
Abbreviations
- BAT
Brown adipose tissue
- cAMP
Cyclic adenosine monophosphate
- CREB
cAMP response element-binding protein
- CRTC
CREB regulated transcription coactivator
- eWAT
Epididymal WAT
- fBAT
Femoral brown adipose tissue
- FDG-PET/CT
F-18-fluorodeoxyglucose (FDG)- positron emission tomography - computed tomography
- gWAT
Gonadal WAT
- HET
Heterozygous
- iBAT
Interscapular BAT
- iWAT
Inguinal WAT
- iRFP713
Near-infrared fluorescent protein 713
- IVIS
In vivo imaging system
- KI
Knock-in
- LUC
Firefly Luciferase
- PGC-1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- poWAT
Periovarian WAT
- PPAR
Peroxisome proliferator activated receptor
- RAR
Retinoic acid receptor
- SIK
Salt-Inducible Kinase
- SVF
Stromal vascular fraction
- T2A
Thosea asigna virus 2A-like peptide
- UCP1
Uncoupling Protein 1
- WAT
White adipose tissue
- WT
Wild-type
1. Introduction
Adipose tissue depots are highly heterogeneous with respect to their morphological appearance, molecular architecture, and functional properties in mammals. White adipose tissue (WAT) stores and mobilizes triglycerides, while brown adipose tissue (BAT) is a specialized thermogenic organ that dissipates chemical energy as heat. Unlike the single large unilocular lipid droplets in white fat cells, brown adipocytes contain numerous small lipid droplets and are exceptionally rich in mitochondria equipped with uncoupling protein 1 (UCP1) [1], [2]. UCP1 catalyzes the net-transport of protons, collapses the proton motive force, and mediates the acceleration of nutrient combustion to increase heat production (reviewed in [3]).
The classical BAT can be found anterior and lateral in the interscapular, cervical, and axillary depots. In addition, inducible brown-like adipocytes are found in WAT depots, termed brite (brown-in-white) or beige adipocytes. The abundance of brite adipocytes is increased by cold exposure or treatment with β3-adrenergic receptor agonists, mimicking cold-induced norepinephrine release, or by ligands of the peroxisome proliferator-activated receptor γ (PPARγ) [4], [5], [6], [7].
The Ucp1 gene is expressed in a highly cell-specific manner restricting UCP1 synthesis to brown and brite adipocytes [8], though this view has been repeatedly challenged in the past, with reports on the detection of Ucp1 mRNA in liver [9], skeletal muscle [10], and mouse brain cortex [11]. Identification of UCP1 protein was further described in thymocytes [12], [13], neurons of torpid squirrel brain [14], and in longitudinal smooth muscle layer of uterus, epididymis, small intestine, and stomach [15]. Functionally, intestinal UCP1 may exert a role in β-adrenergically induced smooth muscle relaxation rather than thermogenesis [16]. Robust confirmation of these findings by other labs is pending. For example, expression of UCP1 in uterus smooth muscle could not be confirmed with UCP1 specific antibodies [17], and it has not been finally resolved whether Ucp1 expression in thymus is due exclusively to brown adipocytes adjacent to thymus or if there is a contribution by Ucp1 expression in thymocytes [13], [18]. Beyond the possible presence of UCP1 in non-adipose tissues, the anatomical distribution of brown adipose tissue depots in the mouse has not been finally resolved in sufficient detail. Recent analysis of a new Ucp1 reporter mouse model identified a previously unknown anatomical location of brown fat in a depot posterior to the ears [19]. Taken together, our knowledge of Ucp1 expression in cells other than adipocytes and the anatomical distribution of brown and brite adipocytes in mammals remains incomplete. Therefore, it is of vital interest to systematically profile Ucp1 gene expression in rodents expressing sensitive bona fide reporter systems.
A large body of research attests sex differences in whole-body fat distribution and adipose tissue physiology in humans [20]. Pertaining to BAT, retrospective analyses of clinical FDG-PET-CT scans suggested higher metabolic activity in women than in men [21], whereas controlled intervention studies in healthy subjects did not observe sex differences in cold-induced BAT activity [22], [23]. In rats, females have significantly higher UCP1 abundance in BAT than males, though these sex-associated divergences disappear or reverse during cold- and diet-induced thermogenesis [22], [24], [25]. Despite higher Ucp1 mRNA levels in interscapular BAT (iBAT) of female mice [26], there are no significant sex differences in UCP1 protein levels in iBAT or inguinal WAT (iWAT) [26], [27]. Regarding these conflicting data, sexual dimorphism in Ucp1 gene expression and thermogenic capacity and activity of brown and brite fat in different anatomical regions requires further investigation.
In rodents, brown and brite adipocytes have beneficial metabolic effects. Different treatment strategies for the recruitment and activation of these thermogenic adipocytes are currently under evaluation in humans to improve glucose homeostasis in type 2 diabetic subjects and to increase total energy expenditure in obese subjects. Repeated cold exposure over days leads to the recruitment of cold-inducible brown fat activity and improves glucose clearance and insulin sensitivity in pre-diabetic subjects [23], [28]. A single dose of Mirabegron, an agonist of the human β3-adrenergic receptor, activates brown fat and increases resting energy expenditure [29]. These findings encourage the search for applicable and safe behavioral and pharmacological strategies to boost energy expenditure via activation and recruitment of UCP1. Gene expression of Ucp1 is regarded an essential readout for the abundance of brown and brite adipocytes. Brown and brite adipocyte differentiation and Ucp1 expression are controlled by β3-adrenergic receptor signaling pathways (reviewed in [2]) and a set of transcriptional factors and coactivators, such as PPARs, retinoic acid receptors (RAR/RXR) [30], and PPARγ coactivator 1 alpha (PGC-1α) [31], [32]. In the search of compounds modulating transcriptional control of Ucp1 in a screen, a bona fide reporter system is needed that faithfully reflects changes in Ucp1 in cultured adipocytes and in vivo.
In the present study, we generated an Ucp1-Luciferase-T2A-iRFP713-T2A knock-in mouse model to report endogenous Ucp1 expression. Both luciferase (LUC) activity and iRFP713 faithfully reflect endogenous Ucp1 expression upon physiological and pharmacological stimulations. Taking advantage of our reporter mouse model, we revealed a sex specific difference in browning propensity, identified a new Ucp1 expressing adipose tissue depot, and, as proof of concept, tested molecules with the potential to recruit thermogenic adipocytes. Overall, our Ucp1-LUC-iRFP713 reporter mouse provides a valuable system for monitoring UCP1 both in vivo and in vitro in a rapid, simple and systematic way.
2. Material and methods
2.1. Animals
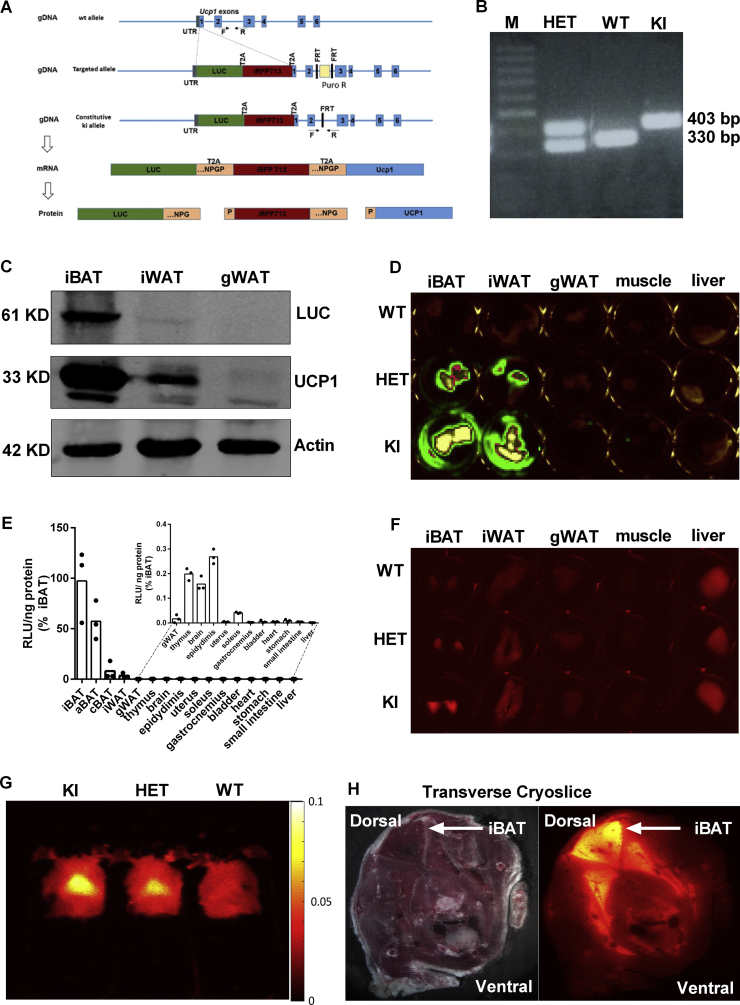
All animal experimentation was conducted in accordance with the German animal welfare law and approved by the regional government of Upper Bavaria, Germany (approval number: ROB-55.2-2532.Vet_02-16-159). The Ucp1 dual-reporter gene mouse C57BL/6NTac-Ucp1tm3588 (LUC-T2A-iRFP713-T2A-Ucp1)Arte, simultaneously expressing firefly luciferase and near-infrared fluorescent protein 713 (iRFP713; addgene 31857: piRFP, kindly provided by VIadislav Verkhusha at Albert Einstein College of Medicine, Bronx, NY, USA), was generated by a commercial provider (Taconic Biosciences GmbH) (Figure 1A). The LUC-T2A-iRFP713-T2A sequence was introduced into the 5′-untranslated region of the endogenous Ucp1 gene. In addition, the Ucp1 constitutive knock-in (KI) allele was generated after the FLPe-mediated deletion of the puromycin resistance cassette (Figure 1A). The mouse model is available on request and will be deposited in a repository.
Figure 1.
Firefly luciferase and iRFP713 reliably report tissue-specific expression characteristics of Ucp1. A. Schematic structure of the Luciferase-T2A-iRFP713-T2A knock-in construct. This transgene was integrated into the 5′ UTR of the intrinsic Ucp1 locus, encoding for the firefly luciferase protein (LUC), the near-infrared fluorescent protein (iRFP713), and the Thosea asigna virus self-cleavage 2A peptide (T2A). Genotyping primers were designed to distinguish the wildtype (330 bp) and knock-in (410) allele. B. Identification of genotyping PCR products in WT, heterozygous (HET) and KI mice. M = molecular weight marker. C. Detection of LUC, UCP1 and Actin proteins in iBAT, iWAT and gWAT of homozygous KI mice. D.Ex vivo imaging of luciferase activity in freshly isolated iBAT, iWAT, gWAT, skeletal muscle, and liver from WT, HET, and KI mice. E. Bioluminescence quantification of multiple tissues derived from KI mice, n = 3. F. Representative ex vivo scanning of iRFP713 in iBAT, iWAT, gWAT, skeletal muscle, and liver freshly isolated from WT, HET, and KI mice. G.In vivo iRFP713 imaging of cold-acclimated WT, HET, and KI mice. H. Anatomical transverse cryoslice with fluorescence imaging of a cold-exposed KI mouse indicating iRFP713 expression by iBAT (white arrow).
Unless otherwise stated, mice were maintained at 23 °C ambient temperature and 55% relative humidity, with a regular 12 h light/dark cycle in our specified-pathogen-free mouse facility. Mice had ad libitum access to a standard maintenance and breeding chow diet (SSniff Spezialdiäten GmbH, Germany) and water.
2.2. Genotyping
All mice were genotyped by PCR of genomic DNA isolated from ear tissue punches using the following primers:
Forward primer: 5′-AGACTTTCCCAAACAGCACG-3′
Reverse primer: 5′-CTTTCATTGGCCAACCGAG-3′
The amplicons obtained with these PCR primer combinations were 330 bp and 403 bp for wildtype (WT) and constitutive knock-in (KI) allele, respectively, as illustrated in Figure 1A,B.
2.3. Western blot analysis
Immunoblotting was utilized to quantify relative protein amounts as described previously [33]. Cell or tissue samples were lysed with radio immunoprecipitation assay (RIPA) buffer and 30 μg protein supernatants were resolved in an SDS-PAGE. The following primary antibodies were used: mouse anti-chicken pan-actin antibody, clone c4, MAB1501 (EMD Millipore, mouse monoclonal antibody, 42 kDa, 1:5000 dilution); rabbit anti-LUC peptide (aa 150–250), EPR17790 (Abcam, rabbit monoclonal antibody, 61 kDa, 1:10000 dilution); rabbit anti-human UCP1 peptide (aa 145–159) antibody, ab10983 (Abcam, rabbit polyclonal antibody, 33 kDa, 1:5000 dilution). Secondary antibodies were from LI-COR: IRDye 680CW donkey-anti-mouse IgG (1:20000 dilution) and IRDye 800CW goat-anti-rabbit IgG (1:20000 dilution)
2.4. Cold acclimation and CL316,243 treatment
During cold acclimation, mice were housed in groups of 2–3 individuals in type II cages (370 cm2, Tecniplast GmbH) with bedding material. Cages were transferred to a climate cabinet (HPP750life, Memmert GmbH + Co. KG) controlling ambient temperature and relative humidity. Mice were maintained in a 12 h light/dark cycle. Ambient temperature was decreased stepwise in weekly intervals from 23 °C to 18 °C, 15 °C, 10 °C and 5 °C. Cold exposure experiments started at the age of 10 weeks and lasted 4 weeks. Control mice were kept in a climate cabinet continuously maintained at 23 °C (HPP750life, Memmert GmbH + Co. KG).
The β3-adrenergic receptor agonist CL316,243 (Tocris, Bio-Techne GmbH) was injected intraperitoneally in mice at 13 weeks of age for 5 consecutive days at a daily dose of 1.0 mg/kg. Controls received vehicle injections with saline solution (0.9% NaCl, B. Braun Melsungen AG). Two days after the last CL316,243 injection, LUC bioluminescence was imaged in anesthetized mice. Afterwards mice were killed and tissues dissected for further analyses.
2.5. In vivo and ex vivo iRFP713 imaging
To detect iRFP713 near-infrared fluorescence in vivo, the mice were anesthetized with 2–3% isoflurane (Isothesia, Henry Schein Vet Pharma). Fur was removed with a commercial electrical razor (Veet Sensitive Prescision) and depilatory cream (Veet) in regions of interest. Excitation of iRFP713 was enabled by a 670 nm diode laser (300 mW, BWF1-670-300E, B&W Tek, Inc.), while fluorescence emission at 740/40 nm (ET740/40x, Chroma Technology) was detected in epi-illumination mode by iXon electron multiplying charge-coupled device (EMCCD, DV897DCS-BV, Andor Technology).
For iRFP713 imaging ex vivo, organs and tissues were dissected, placed in a 12-well cell culture plate (Sarstedt), and detected using an infrared fluorescence imaging system (Odyssey Infrared Imaging System, LI-COR Biosciences GmbH). With 700 nm laser excitation, near-infrared fluorescence emission from tissue was visualized.
2.6. In vivo, ex vivo and cellular bioluminescence imaging
In vivo bioluminescence imaging was visualized and quantified using an optical imaging device (IVIS® Lumina, Xenogen). The instrument setting was 1 min exposure, binning of small, F/Stop of 1, emission filter of open, field of view D for dorsal images, and 1 min exposure for lateral and ventral views. For in vivo imaging, mice were anesthetized with 0.5 mg/kg Medetomidinehydrochlorid (Dorbene vet, Zoetis), 5 mg/kg Midazolam (Dormicum, Roche) and 0.05 mg/kg Fentanyl (Fentadon, Albrecht) and shaved as described above. After intraperitoneal injection of 150 mg/kg VivoGlo™ Luciferin (D-Luciferin, Promega GmbH) as substrate, mice were imaged continuously in 2 min intervals. All in vivo images were quantified by the Living lmage Software (Version 2.6, Xenogen).
Ex vivo, freshly isolated tissues were directly sprayed with luciferin potassium salt solution (PJK GmbH), a specific firefly LUC substrate, and imaged with a higher performance charge-coupled-device camera (CCD camera, Hamamatsu 1394 ORCA II-ERG, sensor exposure time: 60s, binning: 4, speed index: 1).
In cell culture, adipocytes were supplied with fresh medium containing 150 μg/ml D-Luciferin just prior to imaging (IVIS® Lumina, Xenogen).
2.7. Bioluminescence quantitation
Luciferase activity was assayed in tissues (ex vivo) and in cells (in vitro) using a commercial kit system (LUC Assay System Freezer Pack E4030, Promega GmbH) with a microplate luminometer (Infinite M200 Microplate reader, Tecan). For ex vivo, 100 mg tissue was lysed in 313 μl 1 x reporter lysis buffer [34], homogenized (Miccra D-1 homogenizer, Miccra GmbH) for 30 s on ice, and centrifuged 3 min at 10,000×g and 5 °C. Clear supernatant (10 μl) was added into 96-well white-bottom microplates (Greiner Bio-one) and LUC assay substrate solution (50 μl) automatically injected for triplicate bioluminescence measurements.
For in vitro quantitation, cell cultures were lysed in 1 x reporter lysis buffer by shaking for 20 min at room temperature. Ten μl lysate was mixed with 50 μl LUC assay substrate solution, and the mixture was measured in a luminometer (Single Tube Luminometer, Titertek-Berthold GmbH). Bioluminescence readouts were normalized to protein concentration.
2.8. Gene expression analysis
Total RNA (500 ng) isolated from cells or tissues with TRIsure (Bioline) and SV Total RNA Isolation System (Promega GmbH) was reverse transcribed with SensiFast cDNA Synthesis Kit (Bioline). Real-time quantitative PCR (RT-qPCR) was performed in 12.5 μl using the SensiMix SYBR No-ROX Kit (Bioline) in the LightCycler 480 instrument II (Roche). Gene expression of target genes was normalized to the housekeeper gene general transcription factor II B (Gtf2b). Primers are listed in Supplementary Table 1.
2.9. Hematoxylin and eosin staining
Epididymal WAT, peri-ovarian WAT, and femoral BAT were dissected from 5 to 6 weeks old mice, fixed in 4% para-formaldehyde for 48 h at room temperature, and subjected to dehydration overnight as described previously [35], [36]. Paraffin embedded tissues were sectioned with a microtome (Leica) and mounted on object slides (Carl Roth). Sections were stained with hematoxylin and eosin (Carl Roth) using a multistainer (Leica).
2.10. Primary and immortalized preadipocyte culture
For primary brown and brite preadipocyte cultures, cells of the stromal vascular fraction (SVF) were isolated from iBAT and iWAT of 5-weeks-old homozygous KI male mice as described previously [33]. The SVF cells were suspended in standard high-glucose DMEM (Sigma) supplemented with 20% FBS (Biochrom), and seeded in 12-well-plates. At 90% confluency, primary preadipocyte cultures were differentiated for 2 days in induction medium (10% FBS, 850 nM insulin, 1 nM T3, 500 μM IBMX, 1 μM Dexamethasone, 125 μM Indomethacin, 1 μM Rosiglitazone), and for 7 consecutive days in differentiation medium (10% FBS, 850 nM insulin, 1 nM T3).
During differentiation, cells were chronically treated with Rosiglitazone (Biomol) at concentrations and time-points as indicated. Isoproterenol (100 nM, Sigma) and all-trans retinoid acid (1 μM, Sigma) were added overnight before harvest. The pan-salt inducible kinase (SIK) inhibitor HG-9-91-01 was continuously administered at concentrations of 100–500 nM in differentiation medium.
To immortalize brown preadipocytes, SVF cells were isolated from 5-weeks-old 129S6Sv/Ev Tac mice and transformed with the retrovirus-mediated simian virus 40 large T antigen (SV40 LT) according to a protocol supplied by Ronald Kahn using the plasmid pBABE-puro SV40 LT [37].
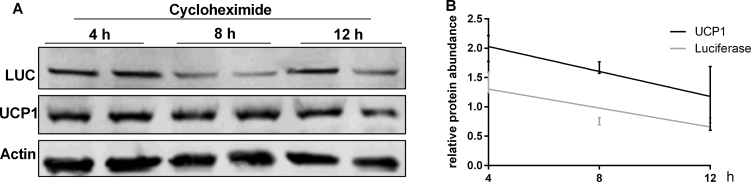
2.11. Half-life of UCP1 and LUC
Primary brown adipocytes were differentiated as described above. At day 7, cells were treated with cycloheximide (25 μg/ml) for 4 h, 8 h, and 12 h to arrest protein biosynthesis before harvest. Cells were analyzed for UCP1 and LUC levels by Western blot.
2.12. Cellular oxygen consumption rate
Cellular oxygen consumption rates (OCR, pmol/min) were measured at 37 °C by microplate respirometry (XF96 extracellular flux analyzer, Seahorse Bioscience) as described previously [33]. Briefly, after recording baseline respiration, we sequentially injected 5 μM oligomycin (Biomol), 1 μM isoproterenol (Sigma), 1 μM FCCP (Sigma) and 5 μM Antimycin A (Sigma) into each well.
2.13. Statistical analysis
Results were analyzed using a statistics software package (Graphpad prism 6). We assessed statistical significance by unpaired two-tailed t-tests, one-way ANOVA (Dunnet's Test), or two-way ANOVA (Tukey's test) for multiple comparisons. The level of significance was set to p < 0.05.
3. Results
3.1. Generation and initial characterization of the Ucp1-reporter mouse
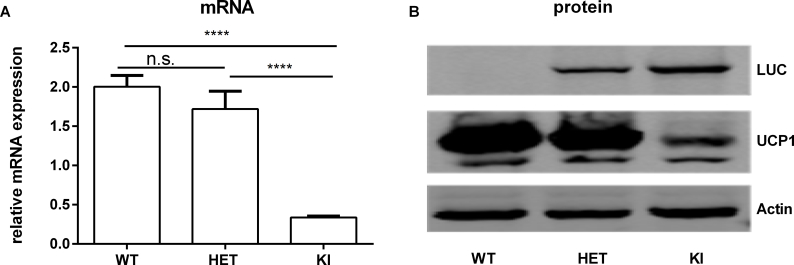
To enable monitoring of endogenous Ucp1 gene expression, we generated a reporter mouse with a Ucp1 knock-in allele (C57BL/6Ntac-Ucp1tm3588(LUC-T2A-iRFP713-T2A-Ucp1) Arte). By homologous recombination, open reading frames encoding for firefly luciferase (LUC) and near-infrared fluorescent protein (iRFP713) were positioned within the 5’ untranslated region of exon 1 of the murine Ucp1 gene (Figure 1A). Translation of all three proteins, LUC, iRFP713, and UCP1, was enabled by C-terminal T2A-tags of LUC and iRFP713, encoding for the Thosea asigna virus 2A oligopeptide [38]. We verified successful homologous recombination by PCR of genomic DNA (Figure 1B)
To evaluate functionality of the knock-in reporter allele, we first compared Ucp1 gene expression in brown fat of WT, HET, and KI mice. Notably, Ucp1 mRNA levels were comparable in WT and HET but reduced to ∼15% of WT in KI (Fig. S5A). This pattern with reduced levels of UCP1 in KI was confirmed by western blotting (see Fig. S5B). Apparently, the capacity of the KI allele to transcribe and translate Ucp1 is impaired by the T2A strategy. The WT allele, however, is dominant, as one copy of the WT allele is sufficient to maintain regular UCP1 levels in BAT of HET mice. Notably, immunodetection of LUC revealed an allele-dose dependent increase (Fig. S5B).
In KI mice, we next compared UCP1 and LUC protein levels in interscapular brown adipose tissue (iBAT), inguinal white adipose tissue (iWAT), and gonadal white adipose tissue (gWAT). UCP1 levels were most abundant in iBAT, lower in iWAT and absent in gWAT (Figure 1C), which was reflected by corresponding levels of the LUC protein (Figure 1C).
We next assessed the ex vivo activity of LUC in adipose tissue depots (iBAT, iWAT and gWAT), skeletal muscle, and in liver dissected from homozygous (KI), heterozygous (HET) and wild-type (WT) mice. Bioluminescence catalyzed by LUC was detectable in iBAT and iWAT of KI and HET mice, with much stronger light emission from fat pads of KI than HET mice (Figure 1D). These fat depot differences in LUC activity were confirmed by quantifying bioluminescence in lysates of freshly dissected adipose tissues (Fig. S1A). Subsequent comparisons of LUC activity in multiple tissues revealed detectable Ucp1 expression in epididymis, thymus, brain, soleus, and gWAT, but not in uterus or liver, nor any other tissue in our screen (Figure 1E). Notably, LUC activity in epididymis, thymus, and brain was clearly higher than in gWAT, whereas soleus was similar to gWAT (Figure 1E).
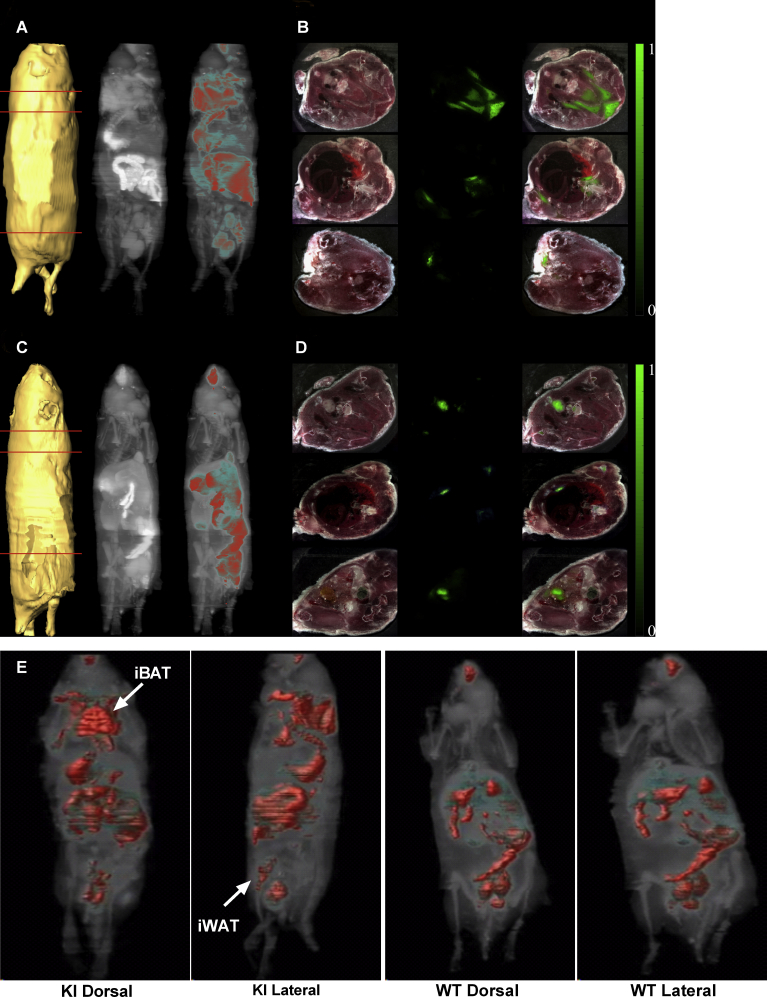
Consistent with bioluminescence, ex vivo imaging of iRFP713 revealed fluorescence signals in iBAT and iWAT of HET and KI mice (Figure 1F). Liver samples showed fluorescence in all three genotypes (Figure 1F), likely due to auto-fluorescence of hepatic bilirubin-albumin complexes [39]. In vivo imaging detected iRFP713 fluorescence signals in the iBAT region of cold-exposed transgenic mice in an allele-dosage dependent manner (Figure 1G). In addition, cryoslicing images with fluorescence microscopy (Figure 1H and S2) and the three-dimensional mode (Supplemental Movie) provided further validation of iRFP signals in iBAT of cold-exposed KI, but not in the WT mouse.
Supplementary movie related to this article can be found at https://doi.org/10.1016/j.molmet.2018.11.009.
The following are the supplementary movie related to this article:
Three-dimensional model generated by composite reconstruction of fluorescence intensities at 730/50 nm from slices acquired by the cryostat sectioning. In the KI mouse, only signals outside of the gastro-intestinal tract are due to Ucp1 gene expression. Signals within the gastrointestinal tract of WT and KI mice are due to auto-fluorescence of food in stomach and gut. Feeding of WT and KI mice prior to the iRFP713 analysis for three days with an alfa/alfa-free diet successfully reduced, but did not completely eliminate this auto/fluorescence phenomenon.
Thus, our Ucp1-LUC-iRFP713 reporter mouse can be used to image BAT and monitor Ucp1 expression in a non-invasive manner.
3.2. LUC activity faithfully reports endogenous Ucp1 expression in vivo
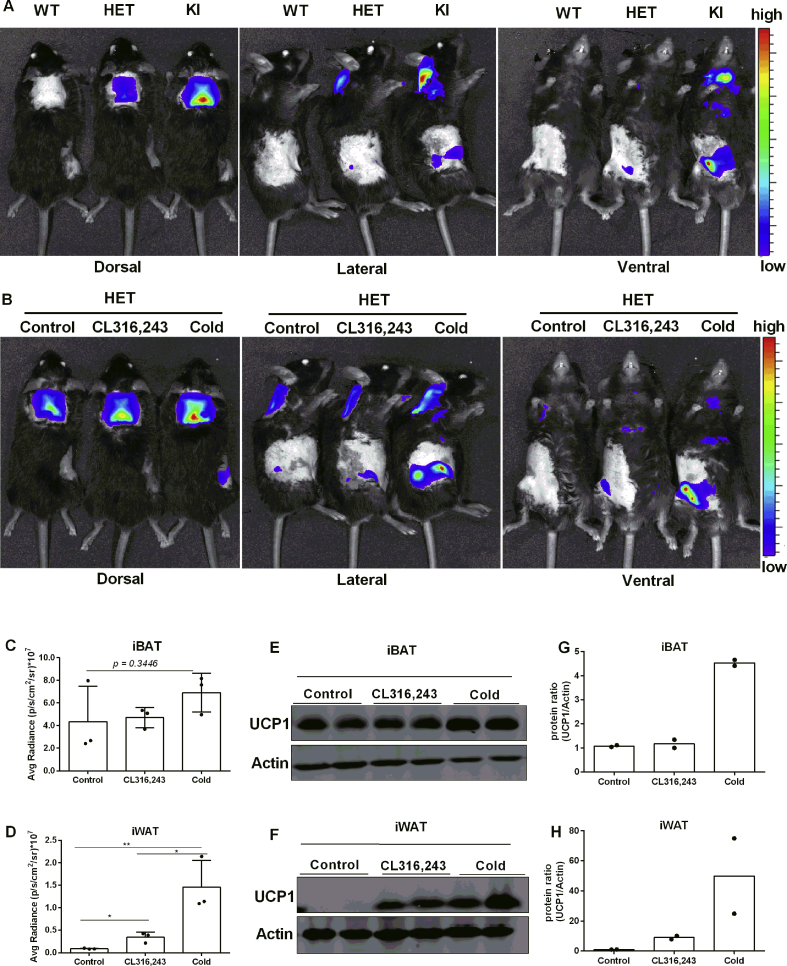
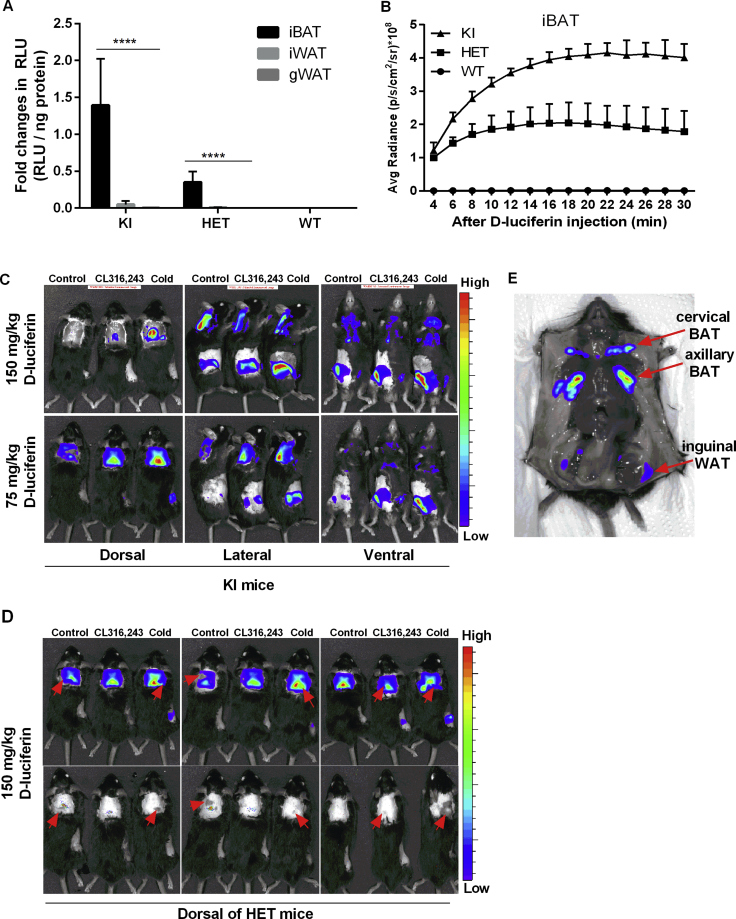
To image bioluminescence in vivo, we first analyzed the kinetics of LUC activity for the optimal detection time point. Mice were imaged continuously in 2 min intervals following D-Luciferin (150 mg/kg, i.p.) injection. In iBAT of HET and KI but not WT mice, we observed maximal bioluminescence signals approximately 20 min after injection remaining stable thereafter for 10 min (Fig. S1B). Therefore, all subsequent in vivo bioluminescence imaging was acquired 20 min after luciferin injection. First, we imaged bioluminescence in vivo in WT, HET, and KI mice housed at room temperature (23 °C) for 16 weeks. In the dorsal view, robust bioluminescence signals were detected in the interscapular region of HET and KI mice (Figure 2A). Notably, bioluminescence intensity was allele-dosage dependent, with no detectable signal in WT mice (Figure 2A). In addition, we detected bioluminescence in the anatomical position of iWAT in the lateral and ventral view, indicating the potential to monitor the browning of white fat (Figure 2A).
Figure 2.
In vivo bioluminescence imaging and quantification. A. Representative in vivo bioluminescence imaging of WT, HET, and KI mice after D-luciferin injection, n = 6. B. Representative in vivo bioluminescence imaging of HET mice under control condition (untreated), after 5 days of consecutive CL316,243 injection and after cold-acclimation. C-D. Quantification of the in vivo bioluminescence from the regions of interest, n = 3. Data were analyzed with One-way-ANOVA, *P < 0.05, **P < 0.01 (Mean ± SD). E-F. Immunoblotting of UCP1 and Actin in iBAT (15 μg protein) and iWAT (30 μg protein) isolated from HET mice. G-H. Quantification of relative UCP1 protein abundance shown in (E-F).
To test whether in vivo bioluminescence potentially reports adaptive changes in endogenous Ucp1 expression, reporter mice were either cold acclimated or repeatedly injected with the β3-adrenergic receptor agonist CL316,243. For these experiments, HET mice were used to avoid bioluminescence signal saturation observed in KI mice (Figure 2B and S1C). In iBAT, luciferase activity was not affected by CL316,243 treatment, but tended to increase in response to cold (Figure 2C), as verified by Western blotting (Figure 2E and G). As a limitation, bioluminescence measurements of iBAT were quenched by dark patches of pigmented skin in five out of nine mice, which varied in size and location (Fig. S1D). In the inguinal region no such pigmentation impaired the measurements (Fig. S1D). Both CL316,243 treated and cold acclimated mice showed increased luciferase bioluminescence, with a stronger response induced by cold (Figure 2D). Corresponding increases in UCP1 protein abundance were observed by immunoblotting (Figure 2F and H).
To eliminate saturated signal in KI mice, half dose of D-luciferin was administrated, which accordingly decreased overall light intensity, but also faithfully reflected Ucp1 variations (Fig. S1C).
In summary, our in vivo imaging results demonstrate that LUC activity is a bona fide reporter for Ucp1 gene expression in brown and brite adipose tissues.
3.3. Ucp1-imaging reveals greater browning propensity in peri-ovarian than epididymal WAT
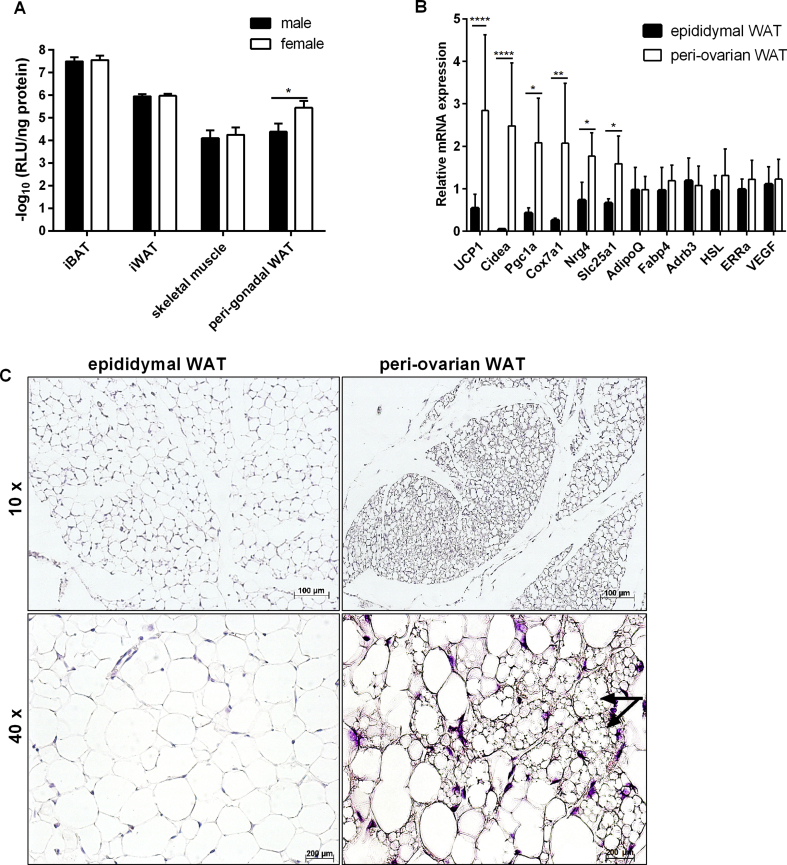
Taking advantage of our Ucp1-LUC-iRFP713 reporter model, we next investigated possible sex-specific differences in LUC activities. We systematically compared the LUC activity in tissue lysates of male and female mice. For both sexes, LUC activities were comparable in iBAT, iWAT, and skeletal muscle (Figure 3A). Comparing peri-gonadal WAT depots, we found significantly higher LUC activity in female peri-ovarian WAT (poWAT) than in male epididymal WAT (eWAT) (Figure 3A). This finding was further corroborated by analysis of gene expression (Figure 3B). Thermogenic marker genes such as Ucp1, Cell Death-Inducing DFFA-Like Effector A (Cidea), PPARγ coactivator-a (Pgc1a), cytochrome c oxidase subunit 7a isoform 1 (Cox7a1), solute carrier family 25A1 (Slc25a1), and neuregulin 4 (Nrg4) were more abundant in poWAT, while the adipocyte marker genes adiponectin (Adipoq), fatty acid binding protein 4 (Fabp4), β3-adrenergic receptor (Adrb3), hormone-sensitive lipase (Hsl), estrogen receptor alpha (ERRa), and vascular endothelial growth factor (VEGF) were expressed at similar levels in both fat pads (Figure 3B). Consistently, histological staining revealed a higher portion of smaller fat cells with multiple lipid droplets in poWAT (Figure 3C). Taken together, analysis of our reporter mouse model revealed a sex difference in gWAT with more thermogenic brite adipocytes found in females than in males.
Figure 3.
Female peri-ovarian WAT displays a higher browning propensity than male epididymal WAT. A. Bioluminescence quantification of lysates generated from iBAT, iWAT, skeletal muscle, and peri-gonadal WAT derived from 5-weeks-old male and female KI mice. Luminescence was normalized to corresponding protein concentrations, n = 6. Data were analyzed with unpaired t test, *P < 0.05 (Mean ± SD). B. Gene expression analysis of male epididymal and female peri-ovarian WAT of KI mice, n = 6. Data were analyzed with Two-way-ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001 (Mean ± SD). C. Representative hematoxylin and eosin staining of epididymal and peri-ovarian WAT; black arrows indicate the multilocular adipocytes, n = 6.
3.4. Identification of a novel brown/brite fat depot embedded in the femoral cleft
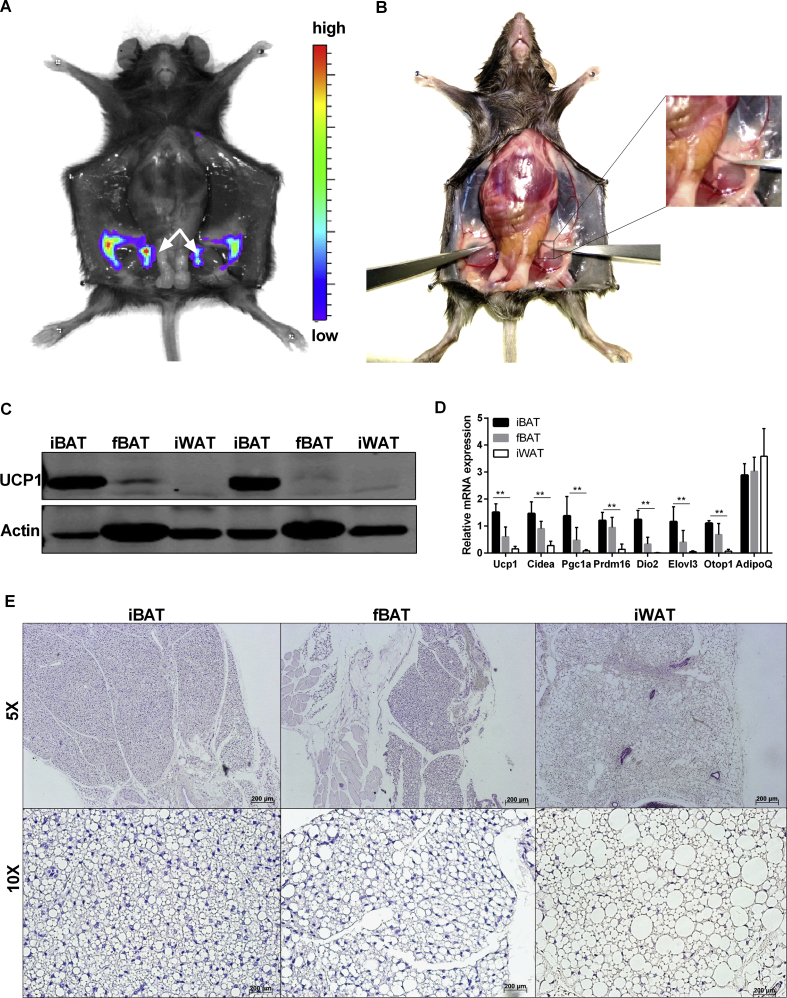
We ascertained the anatomical origin of bioluminescence emission in our reporter mice by imaging of organs and tissues in situ. Bioluminescence was detectable in adipose tissues only, with cervical BAT (cBAT), axillary BAT (aBAT), and iWAT representing the main emission sources in the ventral view (Fig. S1E). In addition, we noticed strong bioluminescence emission in a small triangular tissue embedded in the cleft of the upper femoral hind limb, adjunct to the apex of the inguinal fat depot (Figure 4A–B). Dissection of this LUC positive depot yielded about 10 mg of adipose tissue with reddish appearance. Western blot analysis of protein homogenates revealed expression of UCP1 protein in this tissue (Figure 4C) at a level intermediate to iBAT and iWAT. Thus, we named this novel, distinct tissue depot femoral BAT (fBAT) based on its anatomical location and thermogenic property. Consistently, expression of thermogenic marker genes, such as Ucp1, Cidea, Pgc1a, Dio2 and Elovl3 was intermediate to iBAT and iWAT (Figure 4D), whereas Adipoq was comparable among the three tissue depots. Finally, we observed typical adipose tissue morphology with more multilocular than unilocular adipocytes (Figure 4E). In short, fBAT represents a previously unappreciated adipose tissue depot, which shares typical morphological and molecular features with both brown and brite fat.
Figure 4.
Identification of femoral BAT as a novel UCP1-expressing tissue. A. Light emitting depots in the caudal body part of a KI mouse. White arrows point to femoral BAT (fBAT). B. Anatomical location of fBAT embedded in the superficial layer of femur muscle. C. Western blot analysis of UCP1 expression in iBAT, fBAT, and iWAT of KI mice. D. Gene expression analysis of iBAT, fBAT, and iWAT, n = 5. Data were analyzed with Two-way-ANOVA, **P < 0 .01 (Mean ± SD). E. Hematoxylin and eosin staining of iBAT, fBAT, and iWAT of KI mice.
3.5. LUC activity reliably reports UCP1 protein levels in cultured adipocytes
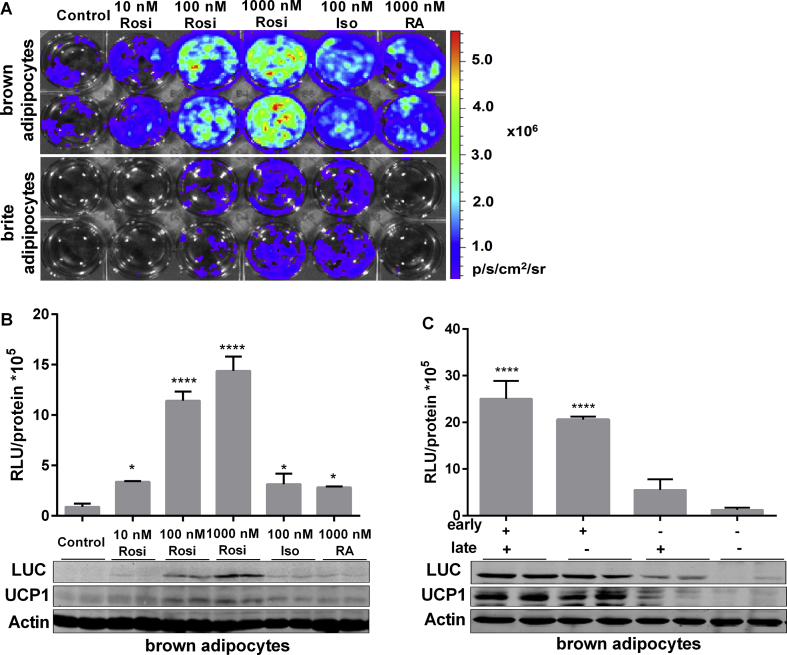
To further investigate the relationship between LUC activity and UCP1 protein in our Ucp1-LUC-iRFP713 mice, we treated primary brown and brite adipocytes in cell culture with established activators of Ucp1 transcription. Acute treatment with the β-adrenergic receptor agonist isoproterenol or the RAR/RXR ligand all-trans retinoic acid increased Ucp1-driven LUC activity. Chronic PPARγ agonist Rosiglitazone treatment boosted LUC activity in a dose-dependent manner (Figure 5A–B). Restricting Rosiglitazone treatment to the first 4 days of adipogenesis was sufficient to increase LUC activity and UCP1 protein to comparable levels as chronic treatment, whereas withdrawal of Rosiglitazone after the first 4 days sharply reduced LUC activity and UCP1 protein level (Figure 5C). Notably, as a proof of principle, imaging of brown adipocyte cultures enabled visualization of bioluminescence emission on the single cell level (Fig. S6). Therefore, Ucp1-driven LUC activity can be used to quantify changes in Ucp1 gene expression in cultured brown and brite adipocytes and reliably reflects changes in UCP1 protein levels. This cellular bioluminescence-based assay system is thus a simple and suitable alternative to conventional analysis methods to screen for putative modulators of Ucp1 gene expression.
Figure 5.
Luciferase activity reflects UCP1 expression in primary adipocytes. A.In vitro bioluminescence imaging of primary brown and beige adipocytes in response to increasing concentrations of chronic Rosiglitazone (Rosi), acute isoproterenol (Iso, 100 nM), and acute retinoic acid (RA, 1000 nM) in the differentiation medium. B. Quantification of bioluminescence intensity, and corresponding detection of Luciferase, UCP1 and Actin protein levels, n = 3. Data were analyzed with one-way-ANOVA and compared to the control group, *P < 0.05, ****P < 0.0001 (Mean ± SD). C. Luciferase activity quantification and immunoblotting analysis of Rosiglitazone time-course effects during 7-day differentiation, which was divided into an early (first 4 days) and late (last 3 days) phases, n = 3. Data were analyzed with One-way-ANOVA, compared to the negative group (without Rosi), ****P < 0.001 (Mean ± SD).
3.6. Cell based imaging reveals a novel chemical regulating Ucp1
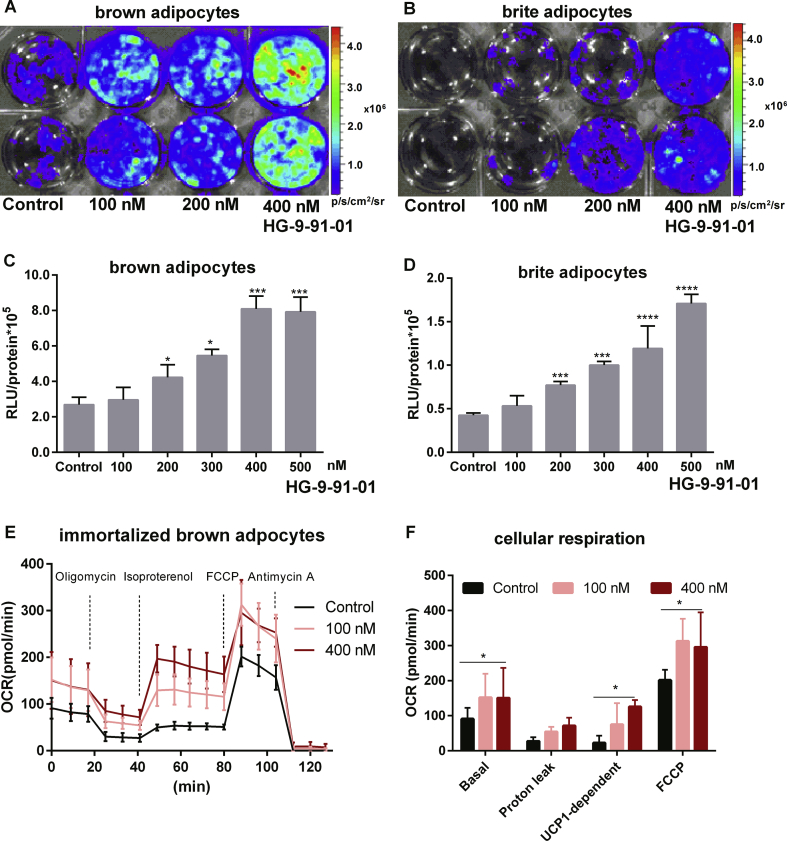
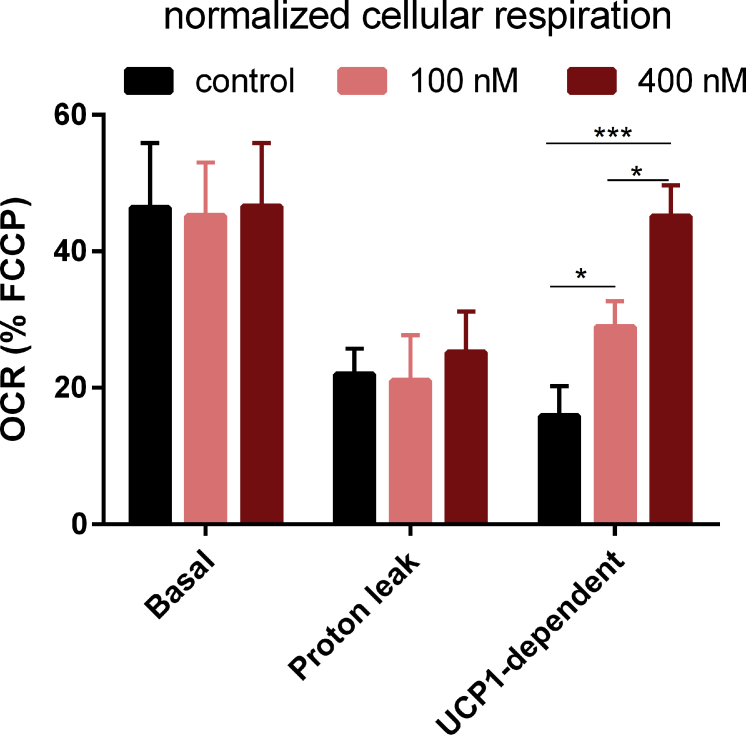
To further demonstrate the benefit of our novel reporter mouse, we utilized a cell-based imaging assay to identify novel molecules regulating Ucp1 expression. The canonical pathway driving Ucp1 expression is the cAMP/PKA/CREB signaling cascade. In hepatocytes, CREB activity is suppressed by salt-inducible kinases (SIKs), which induce sequestration of the CREB-regulated transcription co-activator and histone deacetylase 4 in the cytoplasm through a phosphorylation-dependent mechanism [40]. We therefore hypothesized that inhibition of SIK activity augments intracellular CREB activity and in turn triggers Ucp1 expression in cultured adipocytes. Utilizing primary brown and brite adipocytes generated from our reporter mice, we tested the effect of the selective SIK inhibitor HG-9-91-01 on Ucp1-driven LUC by in vitro cell imaging and LUC activity assays. In brown and brite adipocytes, chronic treatment with HG-9-91-01 during differentiation resulted in a clear dose-dependent elevation of cellular bioluminescence and LUC activity (Figure 6A–D). In HG-9-91-01-treated cells, basal respiration and Ucp1-dependent uncoupled respiration in response to isoproterenol were increased (Figure 6E–F), with the UCP1-dependent rise defined by the difference of isoproterenol-stimulated OCR minus oligomycin-insensitive OCR [33]. In addition, the maximal mitochondrial respiration rate in response to FCCP induced uncoupling was also elevated (Figure 6F). Notably, the dose-dependent increase in UCP1-dependent respiration was maintained after normalization to FCCP respiration rates (Fig. S4). Thus, we demonstrate the feasibility to identify novel Ucp1 modulators with our cell-based reporter assay system.
Figure 6.
Cell-based imaging identifies a novel regulator of Ucp1 expression. A-B. Bioluminescence imaging of primary brown and beige adipocytes from KI mice in response to different concentrations of the SIKs inhibitor HG-9-91-01. C-D. Quantification of bioluminescence of primary brown and beige adipocytes in response to increasing concentrations of the SIKs inhibitor HG-9-91-01, n = 3. *P < 0.05, ***P < 0.001, ****P < 0.0001, compared to the control cells (Mean ± SD). E. Oxygen consumption rate (OCR) of immortalized brown adipocytes from 129S6Sv/EvTac mice. Cell cultures were chronically treated with 100 nM or 400 nM HG-9-91-01 during differentiation, n = 3. F. Quantification of basal, proton leak (oligomycin-insensitive OCR), UCP1-dependent respiration [(isoproterenol-stimulated OCR) – (oligomycin-resistant OCR)] and maximal respiration induced by FCCP, N = 3, n = 16. *P < 0.05, ****P < 0.0001, Two-way-ANOVA, (Mean ± SD).
4. Discussion
The discovery of brite adipocytes and metabolically active brown adipose tissue in adult humans has broadened the interest in adipose tissue biology and sparked the search for new treatment avenues to prevent and treat obesity by recruiting and activating brown and brite adipocytes [21], [41], [42]. To accelerate progress in these fundamental and applied research activities, we now provide a novel dual-reporter gene mouse to reliably monitor Ucp1 expression and thereby the recruitment state of brown and brite adipocytes in live animals, isolated tissues and cell cultures.
Our Ucp1-LUC-iRFP713 reporter mice exhibited robust bioluminescence emission over a wide dynamic range of detection, facilitating the qualitative and quantitative analysis of Ucp1-expressing cells and tissues. In vivo, LUC activity was sufficient to monitor both the change of UCP1 content in classical brown adipose tissue and the recruitment of UCP1-positive brite adipocytes in WAT in response to cold and β-adrenergic stimulation. Bioluminescence assays of tissue lysates allowed detection of extremely low Ucp1-expression in epididymis, thymus, brain, skeletal muscle (soleus) and gonadal fat of females and males. Finally, we here prove the utility of the next generation near-infrared fluorescent protein iRFP713 to monitor Ucp1-expression in brown fat in a non-invasive manner. This reporter molecule is advantageous in terms of deeper penetration depth, lower background signals and no requirement of external substrate, since the low endogenous biliverdin levels from mammalian tissues are sufficient to trigger the engineered phytochrome-based fluorescence [43].
We wish to emphasize that all Ucp1 reporter mouse models published so far, including our new model, are suitable to monitor Ucp1 gene expression but do not report on thermogenic activity of UCP1 [44], [45], [46]. The real thermogenic activity and total capacity can be assessed with indirect calorimetry [47], [48] and new emerging technologies [49], [50], [51]. Our Ucp1-LUC-iRFP713 reporter mouse model, however, is superior to previous ones in several aspects. For instance, the ThermoMouse (MGI:5619504) carries a 98 kb BAC (bacterial artificial chromosome) transgene containing the entire Ucp1 gene with insertion of LUC and red fluorescent protein tdTomato in the first exon of Ucp1. The BAC transgene, however, randomly inserted into the Y-chromosome, therefore restricting research applications to males only [44]. Such ectopic insertion into the genome may hinder epigenetic studies and exclude identification of distant cis-elements on chromosome 8, which depend on correct genomic topology to regulate Ucp1. Similar to our reporter mouse, a recent study inserted an Ucp1-2A-LUC knock-in allele by homologous recombination into the Ucp1 locus of mice of the 129 strain thereby providing a bona fide reporter of endogenous Ucp1 gene expression [19]. This knock-in allele differed, however, by replacing the stop codon in exon 6 of Ucp1 with a 2A peptide followed by the LUC open reading frame. It remains unclear, whether this C-terminal 2A-peptide-tagging of UCP1 impairs the thermogenic function of the protein.
As a limitation of our model, the initial goal to monitor Ucp1 gene expression without altering UCP1 protein level could not be implemented. In HET mice, only a minor reduction of UCP1 mRNA and protein expression occurred in BAT (Fig. S5), most likely with no metabolic consequences. In KI mice, however, we observed a substantial reduction in UCP1 mRNA and protein, clearly demonstrating an impairment of Ucp1 gene expression by the KI allele. Further studies are needed to reveal the underlying mechanism and the metabolic consequences. For in vivo studies aiming to monitor Ucp1 gene expression, heterozygous HET mice can be used, as the WT allele is dominant and ensures regular UCP1 levels in BAT.
As a unique feature of our model, none of the transgenic reporter mouse models generated so far express a fluorescent protein suitable for UCP1-dependent in vivo imaging experiments. Under cold exposed conditions, iRFP713 was readily detectable in the brown fat of living KI mice (Figure 1G) and of its corresponding cryo-sections (Fig. S2), but not in WT controls (Fig. S2). We noticed, however, that it was technically challenging to monitor iRFP713 in mice kept at room temperature in vivo or in mature brown and brite adipocytes in vitro, due to the limited sensitivity of available optical detection systems. Altogether, our Ucp1-LUC-iRFP713 represents a valuable model to foster progress in research on brown and brite adipose tissue biology.
While Ucp1 gene expression in gonadal fat of rodents is normally low, thermogenic adipocytes with high Ucp1 expression were observed in female peri-gonadal fat of the lesser hedgehog representing a protoendothermic eutherian mammal [52]. This thermogenic capacity may have emerged early on in mammalian evolution to enable endothermia during reproduction. It is therefore of interest that LUC activity in female poWAT was increased as compared to male epididymal WAT, a finding corroborated by high expression of brown fat selective genes (Figure 3B). Abundant Ucp1 expression had been previously reported for murine poWAT, complemented by increased appearance of brown adipocyte-like mitochondria [4] and a high browning capacity in response to pharmacological stimuli [27]. The observed sex diference on Ucp1 expression and browning is depot specific as in iBAT or iWAT we detected no such effect (Figure 3A). These findings underline the possible existence of sex-dependent mechanisms in the recruitment and modulation of brite adipocytes in peri-gonadal WAT. As a limitation, the comparison of peri-ovarian and epididymal fat depots may not be feasible as they are distinct anatomical entities, which can only be found in females and males, respectively. Sex hormone replacement studies in ovariectomized female and castrated male mice may clarify whether brite adipogenesis is indeed altered by sex in the peri-gonadal fat depots.
Detection of LUC luminescence proved to be a highly sensitive reporter for Ucp1. By anatomical mapping of this signal, we identified a novel UCP1-positive fat depot, which we now adopted as ‘femoral’ brown adipose tissue (fBAT) based on complementary molecular and histological evidence (Figure 4C–E). Despite its rather small mass (∼10 mg), it is tempting to speculate that fBAT may serve in a cold environment to warm up blood vessels perfusing the hind limb muscles. Notably, because of its proximity to the apex of iWAT, fBAT may easily be mistaken as part of the subcutaneous iWAT depot, which could be expected to bias studies related to the browning of WAT. In fact, the recent establishment of an adipose tissue atlas in the mouse may have detected fBAT using PET-tracers to monitor fatty acid uptake, but subsumed the respective signals as iWAT [53]. The identification of fBAT highlights that our knowledge about the anatomical distribution of brown and brite fat in laboratory mice is yet incomplete. Notably, a small Ucp1-positive adipose tissue depot was reported posterior of the ears (eBAT) in a previously published reporter mouse model [19]. The presence of this eBAT depot, however, was neither confirmed in our Ucp1-LUC-iRFP713 reporter mice (B6N background) (Fig. S1 left), nor in the adipose tissue atlas [53]. This discrepancy may be due to the different genetic background of mouse strains used in these different studies (C57BL/6N vs. 129).
To establish a reliable in vitro imaging system, we characterized in primary cell culture the half-life time of UCP1 and LUC, respectively. Our data showed that UCP1 (t1/2 = 9 h) and LUC (t1/2 = 10 h) have comparable turnover in mature brown adipocytes (Figs. S3A and B), as reported previously [19]. Moreover, LUC activity reliably reports UCP1 protein expression when stimulated in vitro. Taken together, these data suggest that LUC activity serves as a bona fide reporter for monitoring Ucp1 expression in real-time; thus, this cellular imaging model offers a feasible platform to screen small molecules for adipocytes browning in vitro. In our first trial, chronic treatment with the SIK inhibitor HG-9-91-01 profoundly promoted the LUC activity in a dose-dependent manner and induced UCP1-mediated uncoupled respiration in brown adipocytes, which is consistent with the latest research on BAT regulation through the cAMP-SIKs axis [54]. We speculate that SIK inhibition increases CREB-mediated transactivation of the Ucp1 gene, which can now be tested in further experiments addressing the molecular mechanism. Moreover, SIK inhibition also increased the maximal respiration capacity most likely by stimulating mitochondrial biogenesis. It will be of interest to identify the molecular underpinnings of this observation.
5. Conclusion
In conclusion, in our present study, we developed a new dual-reporter gene mouse model to reliably monitor endogenous Ucp1 gene expression in live animals and in cells. This provides a valuable system to screen for new modulators of Ucp1 gene expression in cultured adipocytes and tissues in a sensitive, simple and systematic way.
Funding
This study was funded by the Else Kröner-Fresenius-Stiftung (EKFS) and the Deutsche Forschungsgemeinschaft (DFG, KL 973/11-1). HW was a fellow of the China Scholarship Council. Cryostat sectioning and imaging received funding by the Deutsche Forschungsgemeinschaft (DFG), Sonderforschungsbereich-824 (SFB-824), subproject Z3.
Contribution statement
HW and MW contributed to experimental design and data collection and drafted the manuscript. SM performed histological stainings. AK, DG, JR, and VN performed the in vivo iRFP713 imaging, analyzed the cryo data, and constructed the three-dimensional model from iRFP713 signals. YGL performed the in vitro single-cell imaging on mature brown adipocytes. TF, YGL, and MK provided counseling on brown fat physiology and data interpretation and contributed to drafting and final editing of the manuscript.
Acknowledgements
We thank Vladislav Verkhusha at Albert Einstein College of Medicine (NY, USA) for providing the iRFP plasmid, and our colleagues at TUM Siegfried Scherer and Klaus Neuhaus (Chair for Microbial Ecology) for access to the IVIS imaging instrument and Erwin Grill and Alexzander Christmann (Chair for Botany) for using the CCD camera.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2018.11.009.
Conflict of interest
On behalf of all authors, I declare that there are no any conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
Imaging and quantification of bioluminescence in Ucp1-LUC-iRFP713 reporter mice in vitro and in vivo. A. Bioluminescence quantification of iBAT, iWAT, and gWAT homogenates from WT, HET, and KI mice, n = 6. Bioluminescence was normalized to corresponding protein concentrations. Data were analyzed with two-way-ANOVA ****P < 0.0001 (Mean ± SD). B. To obtain an optimal time point for iBAT imaging, mice were injected with D-luciferin (150 mg/kg, i.p.). Afterwards, dorsal images were taken by 2-min intervals, and the Average Radiance was calculated for the region of interest superior of the iBAT location. C. The administration of 150 mg/kg D-luciferin (upper row) leads to saturated luminescence signal in KI mice (16 weeks) treated with CL316,243 during 5 consecutive days or after 4 weeks of stepwise cold acclimation. To address this issue, D-luciferin injection dose was decreased to 75 mg/kg (lower). D. Three sets of heterozygous mice were used to compare the bioluminescence intensity among control, CL316,243 and cold acclimation after administration of D-Luciferin (upper row). Pointed by arrows, pigmentation was detected above the religion of brown adipose tissue. The lower row of images was taken 1 s post D-luciferin injection demonstrating the pigmentation. E. After in vivo imaging, mice were killed and dissected to track the light-emitting tissues, among which cervical BAT, axillary BAT, and inguinal WAT were identified as main sources.
figs2.
Cryoslice imaging of Ucp1-Luc-Irfp713 reporter mice coupled with sensitive florescence camera. A. Isosurface reconstructed from slices acquired by the cryostat; the maximum projection intensity (MPI) of the fluorescence acquired in 730/50 nm in logarithmic scale; composite reconstruction of logarithmic MPI and fluorescence intensity at 730/50 nm (left to right). B. Representative cryo-sections at the locations indicated by the orange lines in KI mouse from A, color images are placed at the first column, fluorescence at the second and the composite image at the third column. Each slice has been normalized to its corresponding maximum value to demonstrate dynamic range of the acquired data. C. Equivalent to A reconstructions from a WT mouse. Signals within the gastrointestinal tract of WT and KI mice are due to auto-fluorescence of food in stomach and gut.D. Cryo-sections of the WT mouse at locations equivalent to the ones depicted in A, The normalized data show only autofluorescence and absolute absence of brown fat. E. Direct comparison between A and C demonstrates the difference between the two mice in UCP1 expression, in particular, in iBAT and iWAT.
figs3.
Half-life of UCP1 and LUC in brown adipocytes. A. Immunoblotting analysis of UCP1 and LUC protein in brown adipocytes derived from KI mice. Preadipocytes were differentiated with chronic rosiglitazone for 7 consecutive days to induce UCP1 expression. At day 7, cell cultures were treated with 25 μg/ml cycloheximide for 4 h, 8 h, and 12 h to inhibit protein biosynthesis before harvest. B. Signals of UCP1 and LUC in A were quantified and normalized to the Actin signals in the same samples. Individual protein half-life was calculated from the initial and final protein abundance.
figs4.
Normalized cellular respiration to maximal uncoupled rate. Respiration rates of basal, proton leak and UCP1-dependent were all normalized to the maximal FCCP uncoupled rate, N = 3, n = 16. *P < 0.05, ****P < 0.0001, Two-way-ANOVA, (Mean ± SD).
figs5.
Homozygous KI mice display a UCP1 deficiency phenotype in iBAT. A. Relative Ucp1 mRNA expression in interscapular BAT of WT, HET and KI mice, n = 6. Data were analyzed with One-way-ANOVA, ****P < 0.0001. B. Representative immunoblotting analysis of UCP1 and LUC proteins in interscapular BAT of WT, HET, and KI mice.
figs6.
Live single-cell imaging of luciferase bioluminescence in brown adipocytes. Living brown adipocytes were imaged with a CCD camera supplemented with D-luciferin. Blue signals correspond to bioluminescence (Scale bar = 50 μm).
References
- 1.Smorlesi A., Frontini A., Cinti S. Stem cells in aesthetic procedures: art, science, and clinical techniques. 2001. The adipose organ: morphological perspectives of adipose tissues; pp. 123–133. [Google Scholar]
- 2.Klingenspor M., Bast A., Bolze F., Li Y., Maurer S., Schweizer S. Springer; Cham: 2017. Brown adipose tissue. Adipose tissue biology; pp. 91–147. [Google Scholar]
- 3.Klingenspor M. Cold-induced recruitment of brown adipose tissue thermogenesis. Experimental Physiology. 2003;88(1):141–148. doi: 10.1113/eph8802508. [DOI] [PubMed] [Google Scholar]
- 4.Cousin B., Cinti S., Morroni M., Raimbault S., Ricquier D., Pénicaud L. Occurrence of brown adipocytes in rat white adipose tissue : molecular and morphological characterization. Journal of Cell Science. 1992;103:931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- 5.Guerra C., Koza R.A., Yamashita H., Walsh K., Kozak L.P. Emergence of brown adipocytes in white fat in mice is under genetic control effects on body weight and adiposity. The American Society for Clinical Investigation. 1998;102(2):412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue B., Rim J.-S., Hogan J.C., Coulter A.A., Koza R.A., Kozak L.P. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. Journal of Lipid Research. 2007;48(1):41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Young P., Arch J.R.S., Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Letters. 1984;167(1):10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- 8.Cannon B., Hedin A., Nedergaard J. Exclusive occurrence of thermogenin brown adipose tissue antigen in brown adipose tissue. FEBS Letters. 1982;150(1):129–132. doi: 10.1016/0014-5793(82)81319-7. [DOI] [PubMed] [Google Scholar]
- 9.Shinohara Y., Shima A., Kamida M., Terada H. Uncoupling protein is expressed in liver mitochondria and newborn rats. FEBS Letters. 1991;293:173–174. doi: 10.1016/0014-5793(91)81179-c. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida T., Umekawa T., Kumamoto K., Sakane N., Kogure A., Kondo M. β3 -Adrenergic agonist induces a functionally active uncoupling protein in fat and slow-twitch muscle fibers. American Journal of Physiology. 1998;274:469–475. doi: 10.1152/ajpendo.1998.274.3.E469. [DOI] [PubMed] [Google Scholar]
- 11.Lengacher S., Magistretti P.J., Pellerin L. Quantitative RT-PCR analysis of uncoupling protein isoforms in mouse brain cortex : methodological optimization and comparison of expression with Brown Adipose Tissue and skeletal muscle. Journal of Cerebral Blood Flow and Metabolism. 2004;24:780–788. doi: 10.1097/01.WCB.0000122743.72175.52. [DOI] [PubMed] [Google Scholar]
- 12.Carroll A.M., Haines L.R., Pearson T.W., Brennan C., Breen E.P., Porter R.K. Immunodetection of UCP1 in rat thymocytes. Biochemical Society Transactions. 2004;32:1066–1067. doi: 10.1042/BST0321066. [DOI] [PubMed] [Google Scholar]
- 13.Clarke K.J., Adams A.E., Manzke L.H., Pearson T.W., Borchers C.H., Porter R.K. A role for ubiquitinylation and the cytosolic proteasome in turnover of mitochondrial uncoupling protein 1 (UCP1) Biochimica et Biophysica Acta - Bioenergetics. 2012;1817(10):1759–1767. doi: 10.1016/j.bbabio.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 14.Laursen W.J., Mastrotto M., Pesta D., Funk O.H., Goodman J.B., Merriman D.K. Neuronal UCP1 expression suggests a mechanism for local thermogenesis during hibernation. Proceedings of the National Academy of Sciences. 2015;112(5):1607–1612. doi: 10.1073/pnas.1421419112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nibbelink M., Moulin K., Arnaud E., Duval C., Pe L., Casteilla L. Brown fat UCP1 is specifically expressed in Uterine longitudinal smooth muscle cells. The Jornal of Biological Chemistry. 2001;276(50):47291–47295. doi: 10.1074/jbc.M105658200. [DOI] [PubMed] [Google Scholar]
- 16.Shabalina I., Wiklund C., Bengtsson T., Jacobsson A., Cannon B., Nedergaard J. Uncoupling protein-1: involvement in a novel pathway for beta-adrenergic, cAMP-mediated intestinal relaxation. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2002;283(5):G1107–G1116. doi: 10.1152/ajpgi.00193.2002. [DOI] [PubMed] [Google Scholar]
- 17.Rousset S., Alves-Guerra M.-C., Ouadghiri-Bencherif S., Kozak L.P., Miroux B., Richard D. Uncoupling protein 2, but not uncoupling protein 1, is expressed in the female mouse reproductive tract. Journal of Biological Chemistry. 2003;278(46):45843–45847. doi: 10.1074/jbc.M306980200. [DOI] [PubMed] [Google Scholar]
- 18.Frontini A., Rousset S., Cassard-Doulcier A.M., Zingaretti C., Ricquier D., Cinti S. Thymus uncoupling protein 1 is exclusive to typical brown adipocytes and is not found in thymocytes. Journal of Histochemistry and Cytochemistry. 2009;55(2):183–189. doi: 10.1369/jhc.6A7013.2006. [DOI] [PubMed] [Google Scholar]
- 19.Mao L., Nie B., Nie T., Hui X., Gao X., Lin X. Visualization and quantification of browning using a Ucp1 -2A-Luciferase knock-in mouse model. Diabetes. 2017;66:407–417. doi: 10.2337/db16-0343. [DOI] [PubMed] [Google Scholar]
- 20.Nedungadi T.P., Clegg D.J. Sexual dimorphism in body fat distribution and risk for cardiovascular diseases. Journal of Cardiovascular Translational Research. 2009;2(3):321–327. doi: 10.1007/s12265-009-9101-1. [DOI] [PubMed] [Google Scholar]
- 21.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of Brown adipose tissue in adult humans. New England Journal of Medicine. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito M., Okamatsu-ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-kobayashi J. High incidence of metabolically active Brown adipose effects of cold exposure and adiposity. Diabetes. 2009;58(JULY):1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Der Lans A.A.J.J., Hoeks J., Brans B., Vijgen G.H.E.J., Visser M.G.W., Vosselman M.J. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. The Journal of Clinical Investigation. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez E., Rodríguez-Cuenca M.M.S., Amengual E.P.B., Roca P., Palou A. Sexual dimorphism in the adrenergic control of rat brown adipose tissue response to overfeeding. European Journal of Physiology. 2001;442:396–403. doi: 10.1007/s004240100556. [DOI] [PubMed] [Google Scholar]
- 25.Rodríguez A.M., Palou A. Uncoupling proteins: gender dependence and their relation to body weight control. International Journal of Obesity. 2004;28(4):500–502. doi: 10.1038/sj.ijo.0802588. [DOI] [PubMed] [Google Scholar]
- 26.Winn N.C., Grunewald Z.I., Gastecki M.L., Woodford M.L., Welly R.J., Clookey S.L. Deletion of UCP1 enhances ex vivo aortic vasomotor function in female but not male mice despite similar susceptibility to metabolic dysfunction. American Journal of Physiology. Endocrinology and Metabolism. 2018;313:402–412. doi: 10.1152/ajpendo.00096.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S.-N., Jung Y.-S., Kwon H.-J., Seong J.K., Granneman J.G., Lee Y.-H. Sex differences in sympathetic innervation and browning of white adipose tissue of mice. Biology of Sex Differences. 2016;7:67. doi: 10.1186/s13293-016-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis T.R.A. Chamber cold acclimation in man. American Physiological Society. 1961;16:1011–1015. [Google Scholar]
- 29.Cypess A.M., Weiner L.S., Roberts-Toler C., Elía E.F., Kessler S.H., Kahn P.A. Activation of human brown adipose tissue by a ??3-adrenergic receptor agonist. Cell Metabolism. 2015:33–38. doi: 10.1016/j.cmet.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rial E., Gonzälez-Barroso M., Fleury C., Iturrizaga S., Sanchis D., Jiménez-Jiménez J. Retinoids activate proton transport by the uncoupling proteins UCP1 and UCP2. The EMBO Journal. 1999;18(21):5827–5833. doi: 10.1093/emboj/18.21.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 32.Uldry M., Yang W., St-pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Fromme T., Schweizer S., Schöttl T., Klingenspor M. Taking control over intracellular fatty acid levels is essential for the analysis of thermogenic function in cultured primary brown and brite/beige adipocytes. EMBO Reports. 2014;15(10):1069–1076. doi: 10.15252/embr.201438775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manthorpe M., Cornefert-jensen F., Hartikka J., Felgner J., Rundell A.N.N., Margalith M. Gene therapy by intramuscular injection of plasmind DNA : studies on Firefly Luciferase gene expression in mice. Human Gene Therapy. 1993;4:419–431. doi: 10.1089/hum.1993.4.4-419. [DOI] [PubMed] [Google Scholar]
- 35.Madsen L., Pedersen L.M., Lillefosse H.H., Fjære E., Bronstad I., Hao Q. UCP1 induction during recruitment of brown adipocytes in white adipose tissue is dependent on cyclooxygenase activity. PloS One. 2010;5(6) doi: 10.1371/journal.pone.0011391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vegiopoulos A., Müller-decker K., Strzoda D., Schmitt I., Chichelnitskiy E., Ostertag A. Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of Brown adipocytes. Science. 2010;328:1158–1162. doi: 10.1126/science.1186034. [DOI] [PubMed] [Google Scholar]
- 37.Klein J., Fasshauer M., Klein H.H., Benito M., Ronald Kahn C. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. BioEssays. 2002;24(4):382–388. doi: 10.1002/bies.10058. [DOI] [PubMed] [Google Scholar]
- 38.Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS One. 2011;6(4):1–8. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croce A.C., Ferrigno A., Santin G., Vairetti M., Bottiroli G. Bilirubin: an autofluorescence bile biomarker for liver functionality monitoring. Journal of Biophotonics. 2014;7(10):810–817. doi: 10.1002/jbio.201300039. [DOI] [PubMed] [Google Scholar]
- 40.Patel K., Foretz M., Marion A., Campbell D.G., Gourlay R., Boudaba N. The LKB1-salt-inducible kinase pathway functions as a key gluconeogenic suppressor in the liver. Nature Communications. 2014;5:4535. doi: 10.1038/ncomms5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taittonen M., Ph D., Laine J., Savisto N. Functional brown adipose tissue in healthy adults. The New England Journal of Medicine Brief. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 42.van Marken Lichtenbelt W., Vanhommerig J., Smulders N., Drossaerts J., Kemerink G., Bouvy N. Cold-activated Brown adipose tissue in healthy men. The New England Journal of Medicine. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 43.Filonov G.S., Piatkevich K.D., Ting L.-M., Zhang J., Kim K., Verkhusha V.V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature Biotechnology. 2011;29(8):757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galmozzi A., Sonne S.B., Altshuler-Keylin S., Hasegawa Y., Shinoda K., Luijten I.H.N. ThermoMouse: an in vivo model to identify modulators of UCP1 expression in Brown adipose tissue. Cell Reports. 2014;9(5):1584–1593. doi: 10.1016/j.celrep.2014.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao L., Nie B., Nie T., Hui X., Gao X., Lin X. Visualization and quantification of browning using a Ucp1-2A-luciferase knock-in mouse model. Diabetes. 2017;66(2):407–417. doi: 10.2337/db16-0343. [DOI] [PubMed] [Google Scholar]
- 46.Kim D.I., Liao J., Emont M.P., Park M.J., Jun H., Ramakrishnan S.K. An OLTAM system for analysis of brown/beige fat thermogenic activity. International Journal of Obesity. 2018;42(4):939–945. doi: 10.1038/ijo.2017.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speakman J.R. Measuring energy metabolism in the mouse - theoretical, practical, and analytical considerations. Frontiers in Physiology. 2013;4 doi: 10.3389/fphys.2013.00034. MAR(March): 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer C.W., Willershäuser M., Jastroch M., Rourke B.C., Fromme T., Oelkrug R. Adaptive thermogenesis and thermal conductance in wild-type and UCP1-KO mice. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2010;299(5):R1396–R1406. doi: 10.1152/ajpregu.00021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khanna A., Branca R.T. Detecting brown adipose tissue activity with BOLD MRI in mice. Magnetic Resonance in Medicine. 2012;68(4):1285–1290. doi: 10.1002/mrm.24118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartelt A., Widenmaier S.B., Schlein C., Johann K., Goncalves R.L.S., Eguchi K. Brown adipose tissue thermogenic adaptation requires Nrf1- mediated proteasomal activity. Nature Medicine. 2018;24(3):292–303. doi: 10.1038/nm.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reber J., Willershäuser M., Karlas A., Paul-Yuan K., Diot G., Franz D. Non-invasive measurement of Brown fat metabolism based on optoacoustic imaging of hemoglobin gradients. Cell Metabolism. 2018;27(3) doi: 10.1016/j.cmet.2018.02.002. 689–701.e4. [DOI] [PubMed] [Google Scholar]
- 52.Oelkrug R., Goetze N., Exner C., Lee Y., Ganjam G.K., Kutschke M. Brown fat in a protoendothermic mammal fuels eutherian evolution. Nature Communications. 2013;4:2140. doi: 10.1038/ncomms3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang F., Hao G., Shao M., Nham K., An Y., Wang Q. An adipose tissue Atlas : an image-guided identification of human-like BAT and beige depots in rodents. Cell Metabolism. 2018;27:252–262. doi: 10.1016/j.cmet.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paulo E., Wu D., Wang Y., Zhang Y., Wu Y., Swaney D.L. Sympathetic inputs regulate adaptive thermogenesis in brown adipose tissue through cAMP-Salt inducible kinase axis. Scientific Reports. 2018;8(1):1–14. doi: 10.1038/s41598-018-29333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three-dimensional model generated by composite reconstruction of fluorescence intensities at 730/50 nm from slices acquired by the cryostat sectioning. In the KI mouse, only signals outside of the gastro-intestinal tract are due to Ucp1 gene expression. Signals within the gastrointestinal tract of WT and KI mice are due to auto-fluorescence of food in stomach and gut. Feeding of WT and KI mice prior to the iRFP713 analysis for three days with an alfa/alfa-free diet successfully reduced, but did not completely eliminate this auto/fluorescence phenomenon.