Abstract
Objective
The nuclear receptor PPARγ is the master regulator of adipocyte differentiation, distribution, and function. In addition, PPARγ induces terminal differentiation of several epithelial cell lineages, including colon epithelia. Loss-of-function mutations in PPARG result in familial partial lipodystrophy subtype 3 (FPDL3), a rare condition characterized by aberrant adipose tissue distribution and severe metabolic complications, including diabetes. Mutations in PPARG have also been reported in sporadic colorectal cancers, but the significance of these mutations is unclear. Studying these natural PPARG mutations provides valuable insights into structure-function relationships in the PPARγ protein. We functionally characterized a novel FPLD3-associated PPARγ L451P mutation in helix 9 of the ligand binding domain (LBD). Interestingly, substitution of the adjacent amino acid K450 was previously reported in a human colon carcinoma cell line.
Methods
We performed a detailed side-by-side functional comparison of these two PPARγ mutants.
Results
PPARγ L451P shows multiple intermolecular defects, including impaired cofactor binding and reduced RXRα heterodimerisation and subsequent DNA binding, but not in DBD-LBD interdomain communication. The K450Q mutant displays none of these functional defects. Other colon cancer-associated PPARγ mutants displayed diverse phenotypes, ranging from complete loss of activity to wildtype activity.
Conclusions
Amino acid changes in helix 9 can differently affect LBD integrity and function. In addition, FPLD3-associated PPARγ mutations consistently cause intra- and/or intermolecular defects; colon cancer-associated PPARγ mutations on the other hand may play a role in colon cancer onset and progression, but this is not due to their effects on the most well-studied functional characteristics of PPARγ.
Keywords: Lipodystrophy, FPLD3, PPARG, Nuclear receptor, Adipose tissue
Highlights
-
•
Mutations in PPARG, encoding for the nuclear receptor PPARγ, underly lipodystrophy.
-
•
A novel lipodystrophy-associated PPARγ mutant, L451P, displays impaired activity.
-
•
Mutation of the adjacent K450, detected in colon cancer, has no effect.
-
•
Other colon cancer-associated PPARγ mutants display diverse phenotypes.
-
•
Lipodystrophy but not cancer-associated PPARγ mutants consistently display defects.
1. Introduction
Unraveling the complex genetics and molecular and cellular mechanisms in adipose tissue physiology improves the understanding of adipose tissue dysfunction and its related metabolic complications. Adipose tissue dysfunction is a hallmark of pathologic conditions including obesity, type 2 diabetes mellitus (T2DM), and lipodystrophies. The nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ), encoded by the PPARG gene, is the master regulator of adipocyte differentiation, maintenance, and function [1]. Differential promoter usage and alternative splicing generates two major protein isoforms, PPARγ1 and PPARγ2 [2]. PPARγ1 is highly expressed in colon epithelial cells, adipocytes, and monocytes/macrophages. PPARγ2 contains an additional 28 amino acids at the N-terminus and is predominantly expressed in adipose tissue [1]. Both PPARγ isoforms are essential in adipogenesis [3].
PPARγ forms an obligatory heterodimer with RXRα [4]. The PPARγ/RXRα heterodimer regulates the transcription of PPARγ responsive genes by binding to PPAR-response elements (PPRE) in the promoter and/or enhancer regions. The PPARγ cistrome is gene- and cell type-specific [5]. In the absence of a ligand, PPARγ/RXRα heterodimers are able to bind to PPREs. Transcription is actively repressed via interactions with corepressors like NCoR and SMRT, which recruit histone deacetylases. Upon ligand binding, the PPARγ ligand binding domain (LBD) undergoes a conformational change that causes the release of the corepressors and binding of coactivators like SRC-1 and CBP, initiating transcription of PPARγ target genes. PPARγ can be activated by various endogenous ligands including polyunsaturated fatty acids and eicosanoids [6], [7]. PPARγ is the cognate receptor for thiazolidinediones, a class of anti-hyperglycemic agents, including rosiglitazone and pioglitazone [8]. Despite their beneficial effects in glycemic control, the clinical use of thiazolidinediones is restricted due to their side effects [9].
Loss-of-function mutations in the PPARG gene cause familial partial lipodystrophy subtype 3 (FPLD3), a rare clinical condition characterized by progressive and gradual change of subcutaneous adipose tissue distribution during peripubertal phase [10], [11], [12], [13], [14], [15]. FPLD3 patients have a marked muscular appearance due to a paucity of subcutaneous adipose tissue in the extremities and gluteal region with co-existent lipohypertrophy in face, neck, and trunk. Patients manifest with T2DM, hepatic steatosis, recurrent episodes of pancreatitis, eruptive xanthomata, and pronounced acanthosis nigricans.
The co-segregation of lipodystrophy with mutations in PPARG underscores the importance of PPARγ for adipocyte biology. In addition, PPARγ also induces the terminal differentiation in several other cell lineages, including colon, breast, prostate, and lung epithelial cells [1]. Somatic PPARG mutations have been reported in sporadic colorectal carcinomas, but functional characterization is limited and complete side-by-side comparison of their activity in a given experimental setting is currently lacking [16], [17]. In addition, high expression levels of PPARG, but no PPARG mutations, have been reported in certain cancer cell lines, including breast cancer [18] and prostate cancer [19]. Hemizygous deletions of PPARG have also been described in prostate cancers [19]. Therefore, the importance of PPARγ in tumor initiation and progression is not exactly known. However, PPARγ ligands can inhibit tumor growth [17], [18], [19], and TZDs synergistically enhance carboplantin-mediated tumor growth inhibition in a PPARγ-dependent fashion in various types of cancer [20], [21]. This synergistic effect is partly explained by a PPARγ-mediated downregulation of methallothioneins [20] or increased DNA damage and subsequent p53-mediated apoptosis [22].
Here we report the identification and functional characterization of a novel natural PPARγ L451P mutant in a family affected by FPLD3. Interestingly, substitution of the adjacent amino acid K450 was previously described in a human colon carcinoma cell line refractory to PPARγ-ligand induced growth inhibition [17]. Here we show that, in contrast to the tumor-associated K450Q mutant, the FPLD3-associated L451P mutant significantly impairs the transcriptional activity of PPARγ due to a range of molecular defects. Our findings support the view that FPLD3-associated mutations consistently cause intra- and/or intermolecular defects, while the effect of colon cancer-associated PPARγ mutations varies considerably, suggesting subtler or context-dependent effects.
2. Materials and methods
2.1. Case presentation
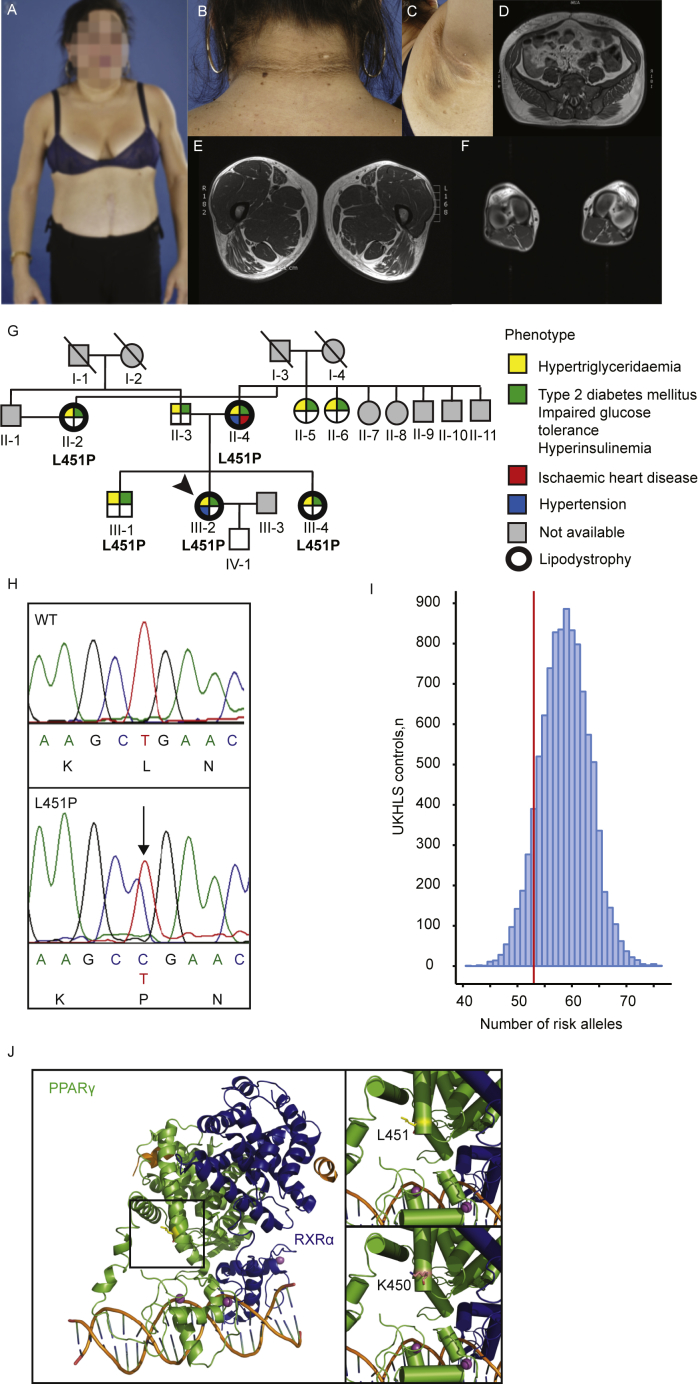
Written informed consent for study participation and publication was obtained from the patient. The index patient was referred because of a suspicion of lipodystrophy (Figure 1A–F, detailed patient description is available in the Supplemental Data). We subjected the index patient to whole exome sequencing to identify the gene responsible for the lipodystrophic phenotype. Considering the family medical history, we performed a pedigree analysis. The patient's mother was originally diagnosed with T2DM, hypertension, and hypertriglyceridemia. Furthermore, she was suffering from ischemic heart disease. The patient's sister and aunt were both initially diagnosed with T2DM and hypertriglyceridemia. The index patient's brother, who was seemingly unaffected albeit slightly overweight, appeared to have a mild T2DM and clear hypertriglyceridemia (Supplemental Data). We sequenced the candidate variant in several family members.
Figure 1.
The identification of PPARγ L451P. A-F. Clinical characteristics of the index patient. Phenotypically, she has a marked muscular habitus with trunk-sparing lipodystrophy (A, with written patient consent). In neck and axilla region patient developed acanthosis nigricans and multiple acrochordons (skin tags, B and C). Transaxial T1-weigthed MRI scans at levels of the abdomen (D) thighs (E), and calfs (F). Please note an excess of subcutaneous adipose tissue in the abdomen with a decreased amount of subcutaneous adipose tissue in the patient's lower extremities. G. Family pedigree of the index patient. Each family member is numbered for identification. The proband is indicated by the arrow. Squares and circles indicate males and females, respectively. Phenotypes are elaborated by color segments showing the presence of specific features (depicted in the figure legend). Grey symbols denote individuals that were not available for DNA analysis. Deceased individuals are indicated by a diagonal line through the symbol. No consanguinity is observed in this family pedigree. H. DNA sequence analysis showing the heterozygous L451P mutation. The chromatogram shows both alleles from the patient (lower panel) in comparison with corresponding genomic DNA from a non-affected individual (upper panel). For tracing, the nucleotide and amino acid sequence are shown. The position of the mutation in the patient is indicated by the arrow. The PPARγ nomenclature refers to PPARγ2. I. Peripheral adipose tissue expandability risk score analysis in the PPARγ L451P index patient to determine the contribution of genetic background to the FPLD3-phenotype. Genotypes for 53 previously described loci were extracted from the UKHLS GWAS dataset (EGA accession EGAD00010000890) to reconstruct a background distribution of a healthy female population. The index patient harbors 53 risk alleles (red vertical line). J. Crystal structure of an intact PPARγ–RXRα complex (PPARγ in green; RXRα in blue) bound to DNA (left panel) showing leucine 451 at the end of helix 9. The square box indicates the magnified region shown in the right panels. L451 (upper) is highlighted in yellow, K450 is highlighted in pink (bottom), both residues generated in the stick format. Protein Database entry 3DZY. The figures were generated by open source software PyMOL 099rc6 (www.pymol.org).
2.2. Materials
Rosiglitazone maleate was from Alexis. WY14643 was from Enzo Life Sciences. 15-deoxy-Δ12,14-prostaglandin J2 (abbreviated as 15d-PGJ2), was from Cayman Chemicals. Synthetic RXRα agonist 6-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)cyclopropyl]nicotinic acid (abbreviated as LG100268) was purchased from Sigma–Aldrich. Anti-PPARγ (sc-7196; RRID: AB_654710), anti-PPARα (sc-9000; RRID: AB_2165737), anti-PPARα (sc-1985; RRID: AB_2165740), anti-Gal4 DBD (sc-510; RRID: AB_627655), anti-RXRα (sc-553: RRID: AB_2184874), and anti-tubulin (Sigma Aldrich T9026: RRID: AB_477593) used for western blotting, were from Santa Cruz Biotechnology, Inc. Protein A Sepharose 4B Fast Flow beads were from Sigma–Aldrich.
2.3. DNA sequence analysis
Genomic DNA was extracted from peripheral-blood leukocytes in venous blood samples using QIAamp DNA Blood Mini Kit (Qiagen Hilden, Germany). Whole exome sequencing, mapping, and data analysis were performed as described previously [23]. The mutation identified in the index patient, corresponds to c.1352T > C and p.L451P in reference sequences NM_015869 and NP_056953.2, respectively. The identified genetic variant was validated by Sanger sequencing. The variant p.L451P was absent in the variation databases NHLBI GO Exome Sequencing Project (ESP) [24], 1000 Genomes [25], and Single Nucleotide Polymorphism Database (dbSNP) [26], and The Genome Aggregation Database (gnomAD) [27]. Amplification and Sanger sequencing of exon 6 and flanking regions were performed to genotype family members. Primers are available upon request.
2.4. Plasmids
The luciferase reporter 3xPPRE-tk-Luc was kindly provided by Dr. R.M. Evans. pGL3-mFabp4-Tk-Luc (fatty acid binding protein 4; Fabp4) was a kind gift from Dr. S. Mandrup. pGL3-mCidec-Luc (cell-death-inducing DFFA-like effector c; Cidec) was a kind gift from Dr. P.F. Marrero Gonzales. The reporter construct 5xGal4-E1BTATA-pGL3 has been described previously [28]. pCDNA3.1 vectors containing the complete coding region of hPPARγ2 wildtype (WT), a kind gift from Dr. V.K.K. Chatterjee [29], and Gal4DBD-hPPARγ-AF2 WT (amino acids 204-507) [12] were used to generate R181D/R183D, K450Q, K450P, L451G, L451P, and E455K mutants using QuikChange mutagenesis kit (Stratagene). pCDNA3.1-hPPARγ1 was used for generating Q268P, R288H, S289C, V290M, K319X, K422Q, and L423P. S414P was generated in the pCDNA3.1 vector containing PPARα. pGEX-PPARγ LBD WT was used to generate the K450Q and L451P mutants. Successful mutagenesis was verified by Sanger sequence analysis. pCDNA3.1 vectors containing RXRα, PPARγ L464R, and PPARγ L496A/E499A have been described previously [12], [29]. pGEX constructs containing RXRα, SRC1 (AA 570-780), SMRT (AA 2302-2352), and PGC1α (AA 1-338) have been described previously [12], [30], [31], [32]
2.5. Cell culture and reporter assays
For luciferase-based reporter assays the human osteosarcoma cell line U2OS and the human embryonic kidney cell line HEK293T were maintained in DMEM 4.5 g/L d-glucose supplemented with 10% fetal calf serum (Invitrogen), and 100 μg penicillin/ml and 100 μg streptomycin/ml (Invitrogen). Cells were transfected using the calcium phosphate precipitation method as described previously [12]. The results are averages of at least three independent experiments assayed in duplicate ± SEM. Student's t-tests were used. A statistically significant difference was defined as a p value of p < 0.05.
2.6. Western blot analysis
The protein expression of the transfected U2OS and HEK293T cells was determined by western blot analysis as described [12]. Anti-PPARγ, anti-PPARα, anti-Gal4 DBD, anti-RXRα, and anti-Tubulin were used for detection of the proteins. Enhanced chemiluminescence (Amersham Biosciences) was used for visualization.
2.7. NR-coregulator interaction analyses
The NR-Coregulator interaction profiling was performed as described previously [31], [33]. GST-pull down assays were performed as described previously [12].
2.8. EMSA
EMSAs were performed as described [12]. Double-stranded DNA oligomers containing a consensus PPRE and natural PPREs derived from Fabp4 (ARE6) and Cidec promoters were labeled with [γ-32P] deoxy-ATP using T4 polynucleotide kinase (Biolabs). For binding and competition analysis the following DNA oligomers were used: consensus PPRE sense probe 5′ CCGGGGACCAGGACAAAGGTCACGAAGCT 3′ (underlined characters indicate the DR1 element), mutant consensus PPRE sense probe 5′ CCGGGGACCAGCACAAAGCACACCGAAGCT 3′; mouse Fabp4 sense probe 5′ CTCTCTGGGTGAAATGTGCATTTCT 3′, and mouse Cidec sense probe 5′ CTGTGCCCTCTTGCCTAGTGC 3′.
2.9. Association of genetic variants with FPLD3 phenotype in index patient
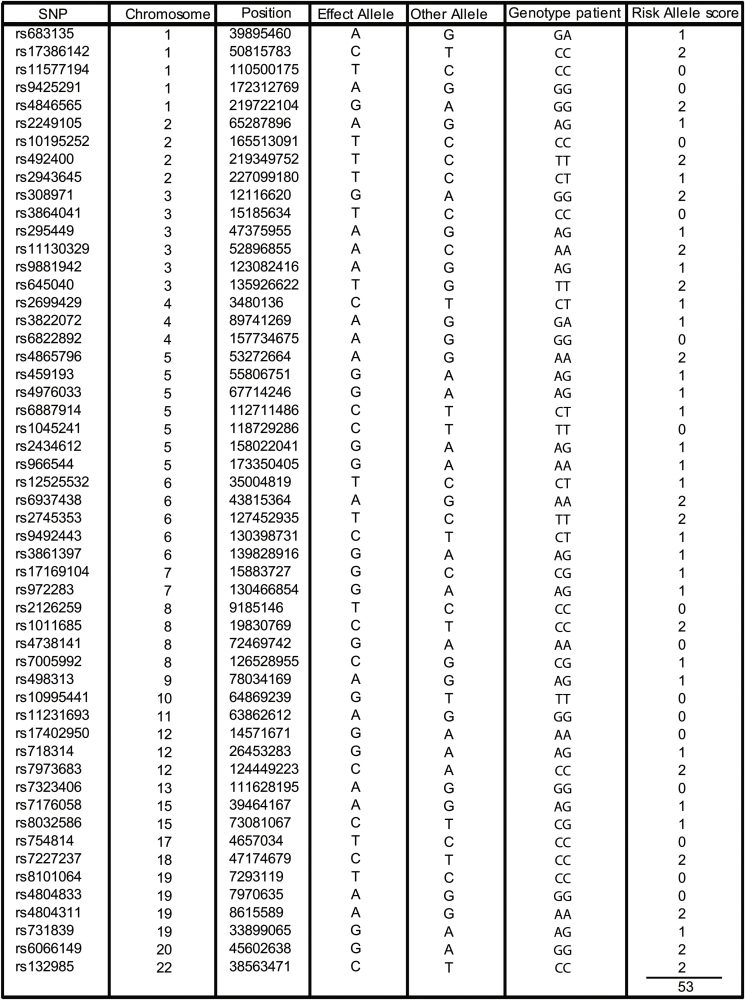
PCR was performed in the index patient to genotype the lead SNPs in 53 genomic regions that were previously identified to impair peripheral adipose tissue storage [34]. Primer sequences are available upon request. Upon download of the full dataset SNPs were extracted based on rsID from the imputed dataset (EGAS00001001232_UKHLS.UK10K+1 KG-imputed.phwe_1e-4.info_0.4.filtered). These SNPs were further processed using plink (v. 1.9b3) to extract compound genotypes and dosage. A custom R script (code and data attached in supplement) was used to further process these data and calculate the number of risk alleles in the population and calculate a Z-score.
2.10. Molecular modeling PPARγ- RXRα nuclear receptor complex
Molecular Modeling of loop 9/10 of PPARγ with and without the L451P mutation was performed using Rosetta 3.8 [35]. Briefly, modeling was based on the structure of the PPARγ–RXRα Nuclear Receptor Complex (PDB ID 3DZY) [36]. A set of 3000 models was built for the WT and L451P variant respectively, using the kinematic loop modeling protocol [37] followed by a relaxation of the structures. The structures were clustered using Calibur [38]. All models were evaluated based on energy and cluster size. Visualization and image generation was done using the PyMOL Molecular Graphics System Version 1.8 (2015) provided by SBGrid [39].
3. Results
3.1. Identification of the novel PPARγ L451P mutant in a family affected by FPLD3
Whole exome sequencing (WES) of the index patient revealed a heterozygous missense mutation in exon 6 (genomic position: chromosome 3:12475478, corresponding to c.1352T > C in reference sequences NM_015869 and NP_056953.2, respectively) in the gene PPARG, which causes the substitution of a leucine residue located at position 451 into a proline (p.L451P). This genetic variant was absent in the variation databases; the Genome Aggregation Database [27] contains the rare variant p.L451V, but whether this variant has any clinical implications, is unknown as information regarding phenotype are not available.
The clinical characteristics of the index patient closely resemble previously described cases of FPLD3, including lipodystrophy in the upper- and lower extremities with preserved truncal adipose tissue (Figure 1A, D–F), and acanthosis nigricans (Figure 1B,C). Paternal and maternal family history showed T2DM and hypertriglyceridemia (Figure 1G). This prompted us to analyze the pedigree of the index patient (III-2). The mutation was identified in her mother (II-4), sister (III-4), and an aunt (II-2). All had a clear lipodystrophic phenotype and a medical history comprising T2DM, hypertriglyceridemia, and/or cardiovascular disease. The PPARG mutation was also identified in the index patient's brother (III-1). The redistribution of adipose tissue was less obvious in the brother, he was diagnosed with T2DM during the study.
A recent population-level genetic study identified 53 genomic regions in which single nucleotide polymorphisms (SNPs) are associated with insulin resistance [34]. An increased number of these risk alleles is associated with an impaired ability to store adipose tissue in peripheral compartments [34]. While rare variants in PPARG certainly contribute to lipodystrophy, this study suggests that SNPs in regulatory regions may contribute too. Our index patient harbors 53 risk alleles (Figure 1I; Supplemental Table 1). To determine whether our patient is at an increased risk for a lipodystrophic phenotype based on her genetic background, we constructed a genetic background distribution in a healthy female population (UKHLS GWAS dataset (EGA accession EGAD00010000890); n = 5,296 female). Compared to this population distribution the index patient is not at increased risk (z-score = −1.28). Taken together, the lipodystrophic phenotype in our index patient is most likely due to the novel PPARG L451P mutation.
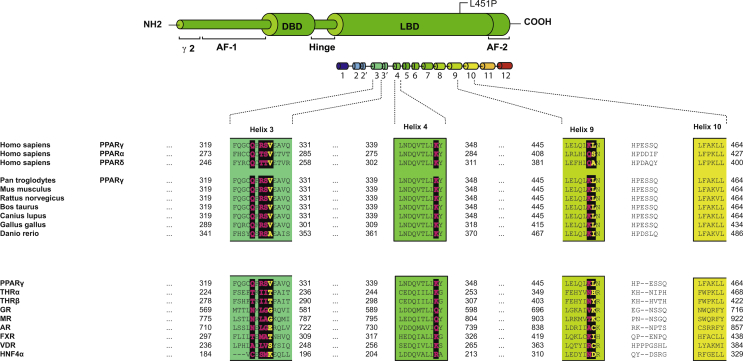
3.2. The FPLD-associated mutant L451P, but not the colon cancer-associated mutant K450Q, displays impaired transcriptional activity
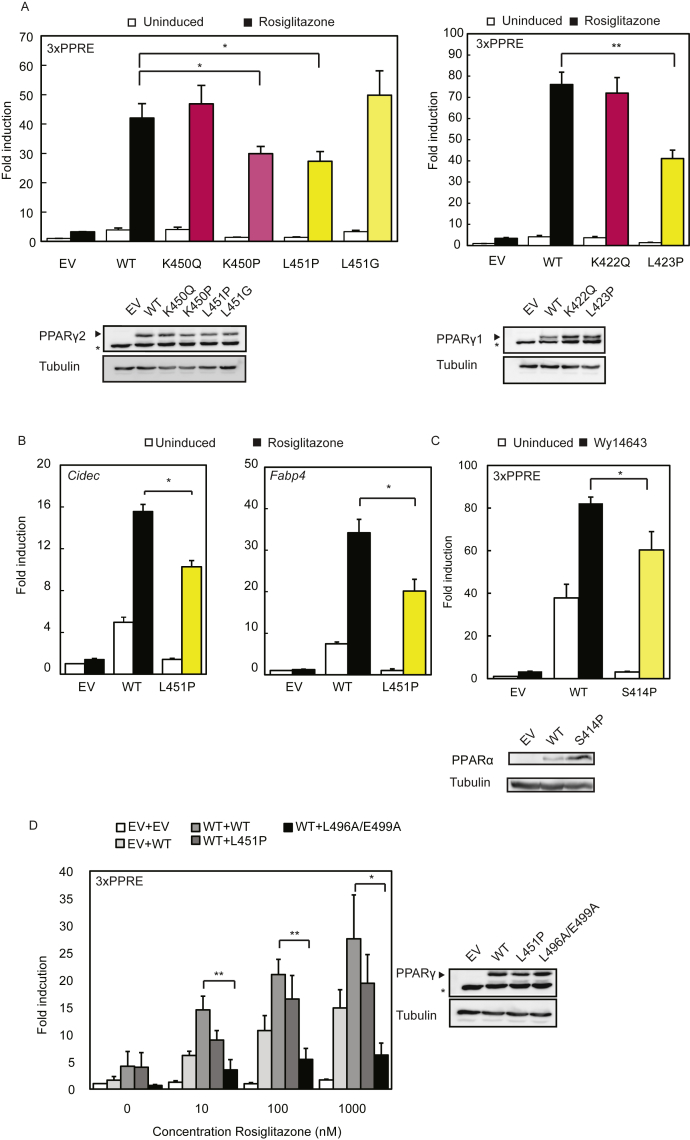
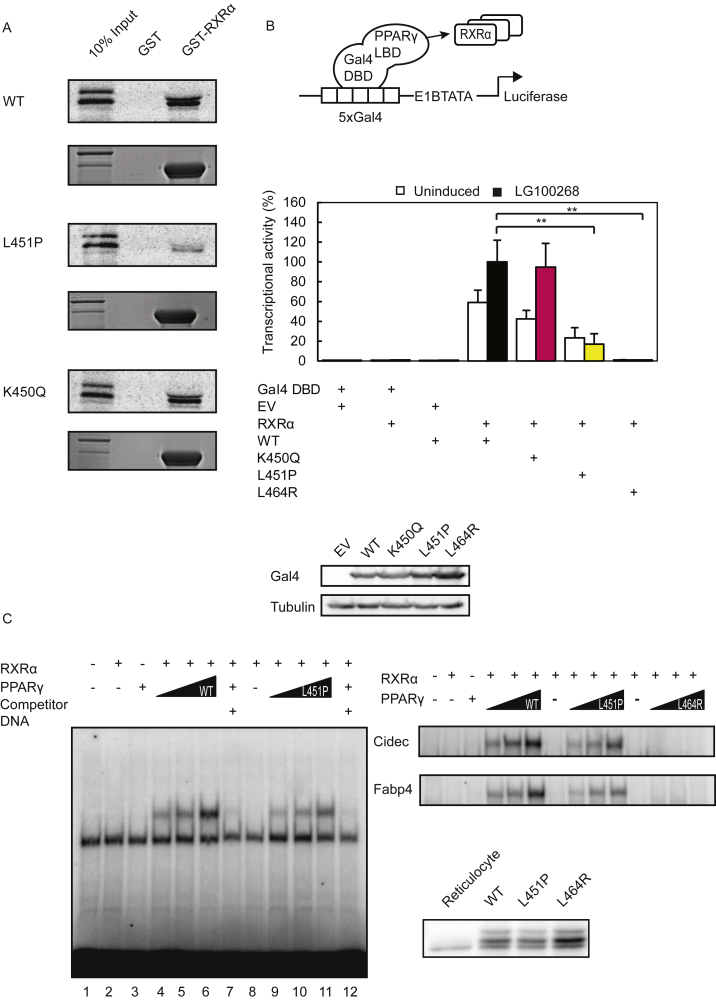
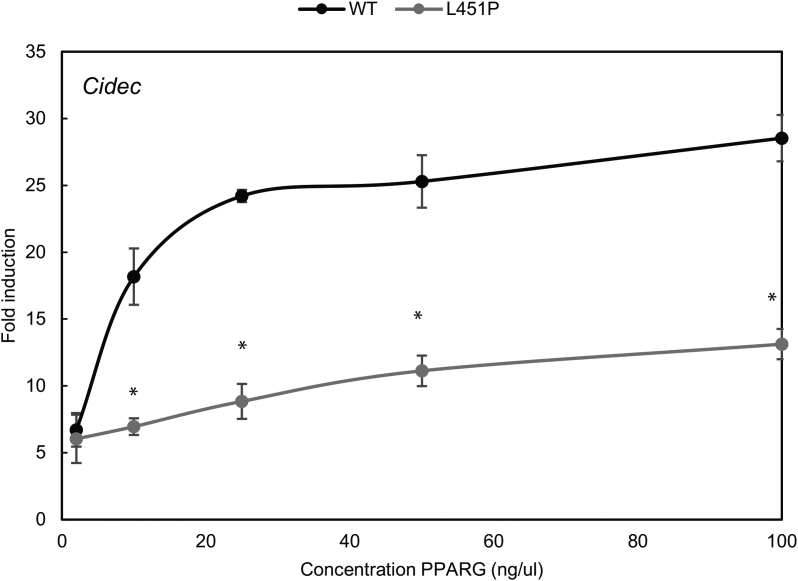
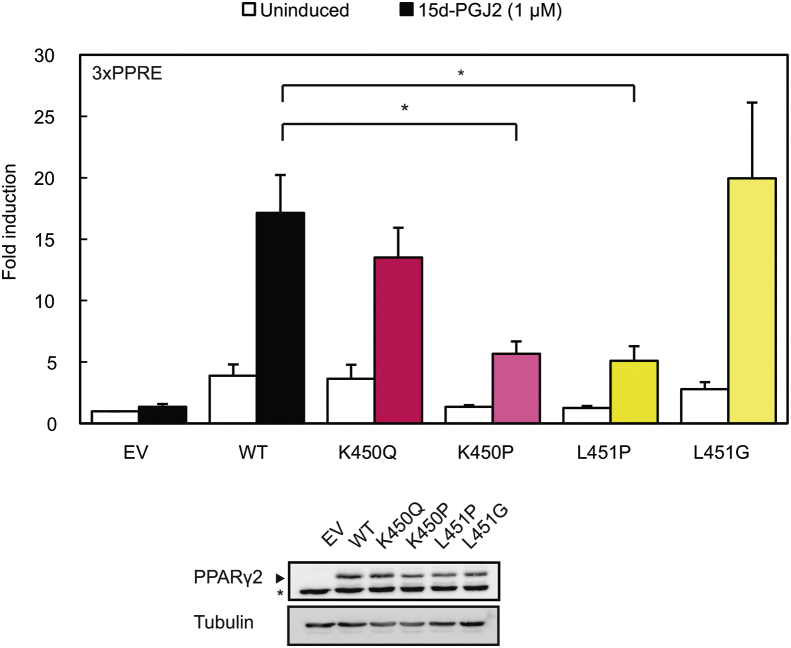
The mutation identified by WES alters a leucine residue located at position 451 at the C-terminus of helix 9 into a proline (Figure 1J). While L451P is the first FPLD3-associated PPARγ2 mutant in helix 9 of the LBD, a previous study in human colon cancer cells lines reported a K422Q mutant in PPARγ1, which corresponds to K450Q in the PPARγ2 protein (Figure 1J) [17]. As the amino acid sequence of PPARγ2 helix 9, including K450 and L451, is highly conserved among species (Supplemental Figure 1), we compared the ability of the natural L451P and K450Q mutants and some artificial mutants to activate PPARγ-mediated transcription in reporter assays. These experiments were conducted in the human osteosarcoma cell line U2OS, which express negligible levels of endogenous PPARγ but display robust activation upon PPARγ overexpression and induction [26], [40]. The transcriptional activity of the L451P mutant on a reporter construct containing three copies of a consensus PPRE (3xPPRE) was significantly impaired under both basal and induced conditions compared to the WT protein in U2OS cells (Figure 2A) and in HEK293T cells (data not shown). Increasing L451P protein concentrations led to a proportional increase in transcriptional activity, but the mutant never reached WT levels (Supplemental Figure 2). Additional mutants were analyzed to characterize the functional defect better. Mutation of L451 into a glycine residue (L451G), which may also potentially disrupt helix 9, showed no clear effect (Figure 2A). The K450Q mutant activated transcription similar to the PPARγ2 WT protein (Figure 2A and data not shown). Substitution of lysine 450 into a proline (K450P) however impaired transcriptional activity similar to the L451P mutation (Figure 2A, left panel). Similar results were obtained with 15d-PGJ2, a potential natural PPARγ ligand (Supplemental Figure 3). Increasing ligand concentrations did not compensate for the transcriptional defect observed (data not shown). The analogous mutation in the PPARγ1 isoform (L423P) displayed a similar defect (Figure 2A, right panel). In agreement with our findings for PPARγ2, the K-to-Q mutation (K422Q, corresponding to K450Q in PPARγ2) did not impair transcriptional activity. Next, the PPARγ L451P mutant was tested on reporter constructs that harbor the promoter regions of the well-characterized PPARγ target genes Cidec and Fabp4. The PPARγ2 L451P mutant also shows impaired transcriptional activity on these natural PPRE-containing promoters (Figure 2B).
Figure 2.
The FPLD-associated L451P mutant, but not the colon cancer-associated mutant K450Q, displays impaired transcriptional activity. A-D. U2OS cells were transiently cotransfected with expression vectors encoding PPARγ2 WT or mutants (A, left panel and C), PPARγ1 WT or mutants (A, right panel) or PPARα WT or S414P mutant (C). The activation of 3× peroxisome proliferator response element (PPRE)-Tk-Luc reporter (A and C), Cidec- (B), or Fabp4-Luc reporter (B), in the absence or presence of 1 μM rosiglitazone (PPARγ) or 100 μM Wy14643, is expressed as fold induction over that with empty vector (EV). D. U2OS cells were transfected with equal amounts of plasmids harboring PPARγ2 WT, L451P, and L496A/E499A in absence and presence of 1 μM rosiglitazone. Results are averages of at least three independent experiments assayed in duplicate ± SEM. *P < 0.05; **P < 0.01 cells transfected with mutant vs. WT. Expression levels of the different proteins were confirmed by western blot analysis using a PPARγ or PPARα specific antibody. The arrow indicates PPARγ and the asterisk indicates an unknown non-specific band. WT, wildtype.
The amino acid sequence and structure of PPARγ2 helix 9 is highly conserved among species (Supplemental Figure 1). The human PPARγ, PPARα, and PPARδ proteins share a high degree of homology in the sequence and structure of their LBDs, but K450 and L451 positions in PPARγ are not conserved between the 3 PPAR isotypes (Supplemental Figure 1). Substitution of serine 414 into a proline in PPARα, which corresponds to L451P in PPARγ, however, clearly inhibited the transcriptional activity of this protein (Figure 2C). The amino acid corresponding to K450 in PPARγ2 is a glutamine residue in both PPARα and PPARδ, suggesting that either a lysine or a glutamine residue is structurally tolerated at this position. This view is supported by the lack of phenotype observed with the PPARγ2 K450Q mutant (Figure 2A).
Previous studies show that FPLD3-associated PPARγ2 mutants exert their pathogenic effects by either haploinsufficiency or dominant negative activity towards the PPARγ2 wildtype protein on the level of DNA binding or cofactor binding [10]. Reporter assays using cells transfected with both PPARγ wildtype and L451P showed no decrease in transcriptional activity when compared with cells transfected with PPARγ wildtype alone, indicating that the L451P mutant exhibits no dominant-negative activity in this assay (Figure 2D). In contrast, the artificial L496A/E499A mutant displayed a strong dominant negative activity, as shown earlier [12], [29].
Taken together, these finding show that disruption of helix 9 by proline residues (PPARγ2: L451P, K450P; PPARγ1: L432P; PPARα: S414P) negatively affects PPAR function, while the natural K450Q mutation in helix 9 has no clear effect.
3.3. The L451P substitution shows that PPARγ activity does not crucially depend on PPARγ DBD-LBD interdomain communication
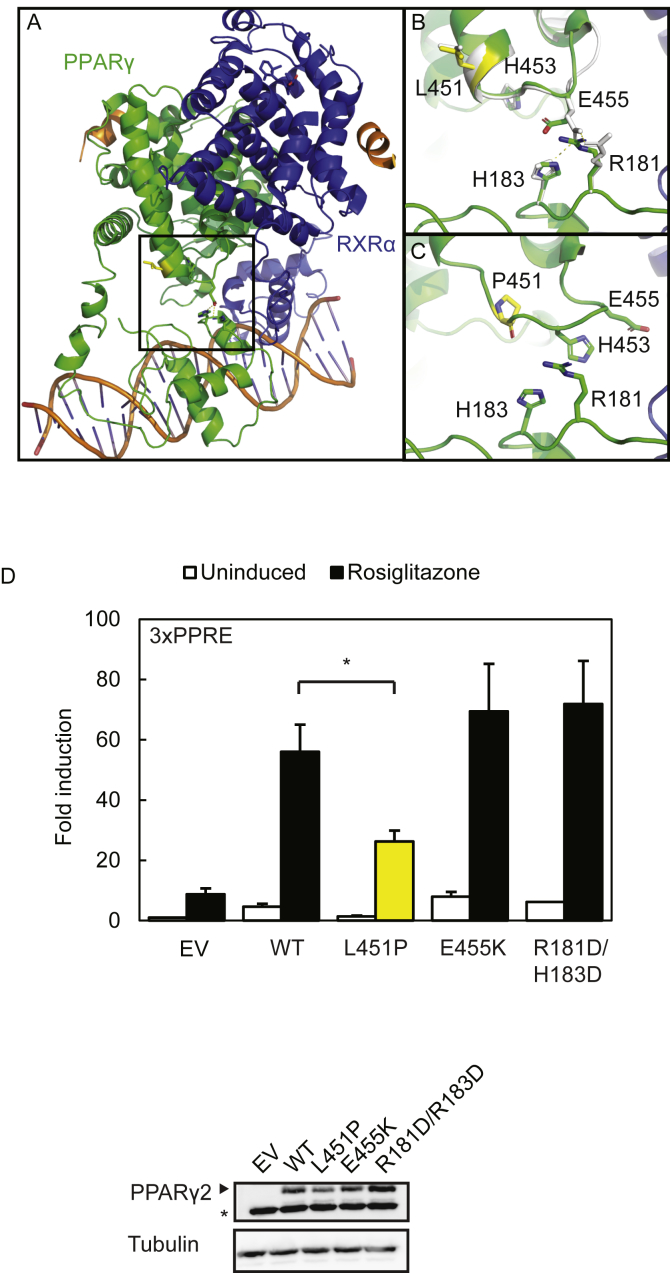
Next, we wished to identify the molecular defects of the L451P mutant (Figure 2). A recent structural and functional study on the multi-domain RARβ-RXRα heterodimer revealed that physical interactions between the RARβ DBD and LBD are implicated in allosteric signal transmission between the domains [41]. Apart from important differences in quaternary structure between the RARβ-RXRα and PPARγ–RXRα heterodimers [41], an intramolecular interaction interface - though smaller than in RARβ - is also present between the PPARγ LBD, involving the loop between helix 9 and 10, and PPARγ DBD [36], [41] (Figure 3A–C). We investigated whether the L451P substitution would interfere with the DBD-LBD interface by functionally assessing the importance of this interface for the transcriptional activity of PPARγ. Remodeling experiments on the quaternary PPARγ–RXRα heterodimer structure revealed physical interactions between a glutamic acid residue at position 455 in the loop connecting helix 9 and 10 with arginine and histidine residues at position 181 and 183 of the PPARγ DBD, respectively (Figure 3A–C). Introducing mutations at the PPARγ2 LBD-DBD interface did not compromise the transcriptional activity of PPARγ2 (Figure 3D). Our data suggest that in contrast to the larger and well-formed RARβ LBD-DBD interface [41], transcriptional activity of PPARγ does not crucially depend on the transmission of allosteric signals between the DBD-LBD intramolecular interface. Therefore, the impact of L451P on this intramolecular interdomain communication is probably limited.
Figure 3.
The L451P substitution shows that PPARγ activity does not crucially depend on PPARγ DBD-LBD interdomain communication. A. The crystal structure of an intact PPARγ–RXRα complex (PPARγ in green; RXRα in blue) bound to DNA shows that a single patch consisting of helix 9 and 10 of the PPARγ LBD is involved in the formation of PPARγ LBD-PPARγ DBD interactions. Amino acid residue 451 is highlighted in yellow generated in the stick format. The square box indicates the magnified region shown in B (PPARγ WT) and C (PPARγ L451P). B. Remodeling experiments on the crystal structures show interactions between E455 (LBD) and R181 (DBD) and H183 (DBD) in the WT situation. C. Remodeling experiments showing the disruption of the end of helix 9 by L451P and the altered configuration of amino acids at the PPARγ DBD-LBD interaction interface. Protein Database entry 3DZY. The figures were generated by PyMOL Molecular Graphics System Version 1.8 (2015) provided by SBGrid [39]. D. U2OS cells were transiently cotransfected with expression vectors encoding PPARγ WT or PPARγ mutants respectively, and 3× peroxisome proliferator response element (PPRE)-Tk-Luc reporter. Activation of the luciferase reporter, in the absence or presence of 1 μM rosiglitazone, is expressed as fold induction over that with empty vector (EV). Results are averages of three independent experiments assayed in duplicate ± SEM. *P < 0.05 cells transfected with mutant vs. WT. Expression levels of the different proteins were confirmed by western blot analysis using a PPARγ or PPARα specific antibody. The arrow indicates PPARγ and the asterisk indicates an unknown non-specific band. WT, wildtype.
3.4. L451P causes a general ligand-mediated cofactor binding defect
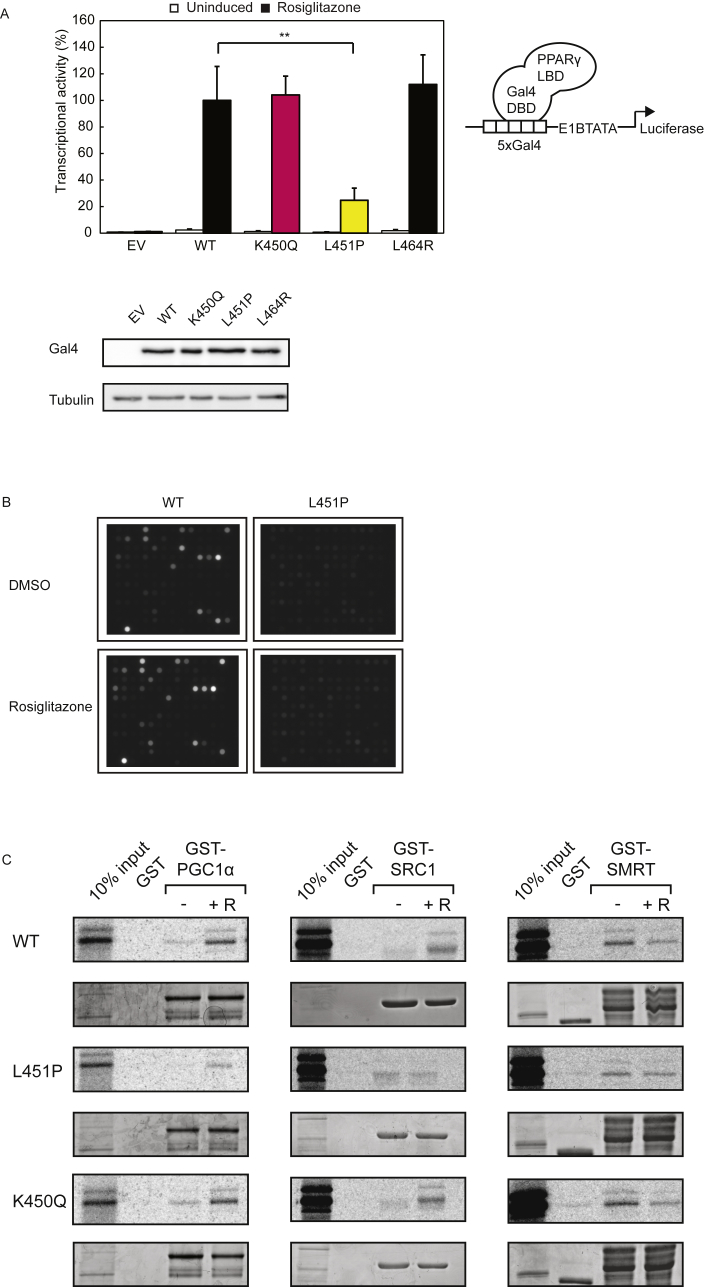
The position of L451P in the LBD suggests that it can potentially disrupt intermolecular interactions, such as ligand-mediated cofactor interactions and/or RXRα heterodimerisation. To address these options, we first performed reporter assays in which PPARγ driven transcription relies on ligand-mediated cofactor recruitment, but is independent of heterodimerisation with RXRα [12]. For this, the mutations described above were introduced into a chimeric Gal4DBD-PPARγ LBD receptor and expressed together with a Gal4 reporter gene in U2OS cells. Both the wild type Gal4-PPARγ LBD receptor and the dimerization defective L464R mutant [12] clearly activated transcription, but the L451P mutant displayed reduced activity (Figure 4A). The activity of the K450Q protein was comparable to the WT version, similar to our findings with the full-length protein (Figure 2). Even high concentrations of thiazolidinediones, including rosiglitazone and pioglitazone, or the tyrosine-based agonist GW1929 were unable to restore the transcriptional activity to WT levels (data not shown).
Figure 4.
PPARγ L451P mutant displays general ligand-mediated cofactor binding defects. A. U2OS cells were transiently transfected with chimeric Gal4 DBD-hPPARγ LBD wildtype and mutant fusion proteins and 5xGal4-E1BTATA-Luciferase. Cells were treated with or without 1 μM rosiglitazone. Shown results are average of three independent experiments assayed in duplicate ± SEM. **P < 0.01 cells transfected with mutant vs. WT. DBD, DNA binding domain; LBD, ligand binding domain. Comparable amounts of Gal4 DBD-hPPARγ LBD proteins were detected by western blot analysis using an antibody against Gal4 DBD. B. Pamgene® chips containing 154 different cofactor derived peptides (containing either LxxLL or LxxxIxxxL motifs) were incubated with recombinant GST-PPARγ-LBD or GST-PPARγ L451P-LBD and anti-GST-alexa, in the absence or presence of rosiglitazone. After 102 pump cycles a CCD camera recorded fluorescence (100 ms). Experiment was performed in triplo. Four representative images are shown. C. GST fusion proteins as indicated in the figure coupled to glutathione-Sepharose beads were incubated with [35S] methionine-labeled PPARγ (wildtype or mutant) in absence and presence of rosiglitazone (indicated by + R, 10 μM) to determine the effect of L451P on interactions with coregulators. GST alone was used as a negative control. 10% of the total lysate of the [35S] methionine-labeled PPARγ proteins used for the pull down assay was applied as control (input). Levels of GST-proteins have been confirmed by Coomassie staining.
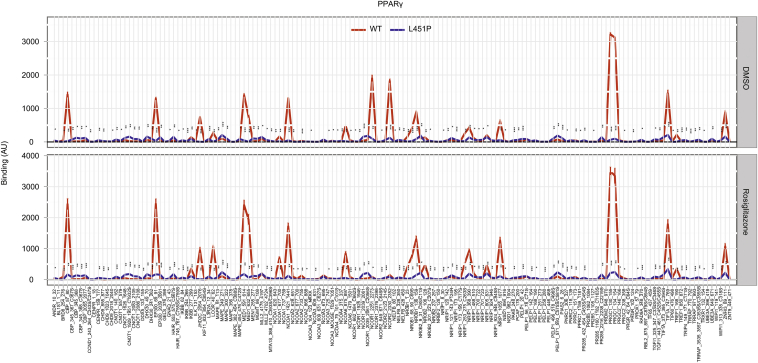
To further analyze the interaction of the L451P mutant with transcriptional cofactors, we performed MARCoNI assays, a peptide-based microarray containing 154 NR binding motifs present in different coactivators and corepressors [31], [33]. The NR-coregulator interaction profiles showed an overall impairment of binding, both in the absence and in the presence of rosiglitazone (Figure 4B and Supplementary Figure 3). In contrast, the K450Q mutant protein displayed binding profiles similar to the WT protein (data not shown). Independent GST pulldown experiments confirmed that PPARγ L451P, but not K450Q, interacts less efficiently in absence and presence of ligand with coactivators PGC1α and SRC1 and corepressor SMRT (Figure 4C). Together, these results indicate that mutation of L451 into proline, but not K450 into glutamine, causes a general ligand-mediated cofactor binding defect.
3.5. The L451P substitution disrupts PPARγ/RXRα heterodimerization and DNA binding
As leucine 451 is located relatively close to the RXRα binding interface in the LBD [36], [42] (Figure 1J), and RXRα heterodimerization is essential for subsequent DNA binding [4], we also examined the effect of the L451P substitution on these PPARγ functions. First, we performed in vitro binding assays using GST-RXRα and [35S]-methionine labeled PPARγ (wildtype and mutants). A clear reduction in RXRα heterodimerization was observed when comparing the L451P mutant protein to the WT version (Figure 5A). The K450Q mutation did not affect RXRα heterodimerization (Figure 5A), in agreement with previous data [17]. Similar findings were made when using GST-PPARγ-LBD and [35S]-methionine labeled RXRα (data not shown). To confirm and extend these findings in a cellular setting, we performed mammalian two-hybrid reporter assays in which we tethered PPARγ LBD WT and mutant versions to the DNA and coexpressed RXRα, either in the absence or presence of a synthetic RXRα ligand (LG 100268). In this setting, reporter activity depends on the interaction between the transcriptionally silent Gal4-DBD–hPPARγ-LBD protein and its ligand-activated RXRα binding partner (Figure 5B). While the activity of Gal4-DBD–hPPARγ-LBD WT and K450Q could be stimulated by RXRα and further enhanced by LG100268, the L451P protein showed only weak activity under these conditions. As a control, we used the dimerization-defective L464R mutant [12], which indeed failed to respond to the addition of RXRα protein (Figure 5B). Therefore, we conclude that substitution of L451 into proline impairs heterodimerization with RXRα; the K450Q mutant displayed no obvious dimerization defect.
Figure 5.
The L451P substitution disrupts PPARγ/RXRα heterodimerisation and DNA binding. A. GST PPARγ fusion proteins (WT or mutant) coupled to glutathione-Sepharose beads were incubated with [35S] methionine-labeled RXRα to determine the effect of L451P on heterodimerization with binding partner RXRα. GST alone was used as a negative control. 10% of the total lysate of the [35S] methionine-labeled RXRα proteins used for the pull down assay was applied as control (input). Levels of GST-proteins have been confirmed by Coomassie staining. B. HEK293T cells were transfected with chimeric Gal4 DBD-hPPARγ LBD wildtype and mutant fusion proteins and 5xGal4-E1BTATA-Luciferase, as indicated. For induction of gene expression cells were treated with 10 nM synthetic RXRα ligand (LG 100268). Shown results are average of three independent experiments assayed in duplicate. DBD, DNA binding domain; LBD, ligand binding domain. C. In vitro translated RXRα and/or PPARγ (wildtype or mutant) proteins were incubated with [32P]-labeled DNA probes in the absence or presence of 10× unlabeled wildtype and mutant probes as indicated. The formed protein-DNA complexes were separated from unbound DNA on nondenaturating SDS-polyacrylamide gels and visualized by autoradiography of dried gels. Expression levels of the different proteins were confirmed by western blot analysis using a PPARγ specific antibody. WT, wildtype.
To investigate the consequences of impaired RXRα interaction on binding of the PPARγ/RXRα heterodimer to DNA, we performed electrophoretic mobility shift assays (EMSA) using a consensus PPRE and PPREs derived from the Fabp4 and Cidec genes. Binding of the L451P mutant protein was impaired on all 3 DNA elements, which was most pronounced when lower amounts of protein were tested (Figure 5C). As a control, we again used the dimerization-defective L464R mutant [12], which indeed failed to bind DNA (Figure 5C). Taken together, these data indicate that the FPLD-associated L451P mutation results in impaired transcriptional activity due to impaired cofactor binding and reduced DNA binding, while the adjacent K450Q mutant, which was found in a human colon cancer cell line [17], displays no clear functional defects.
3.6. Colon cancer-associated PPARγ mutants display variable phenotypes
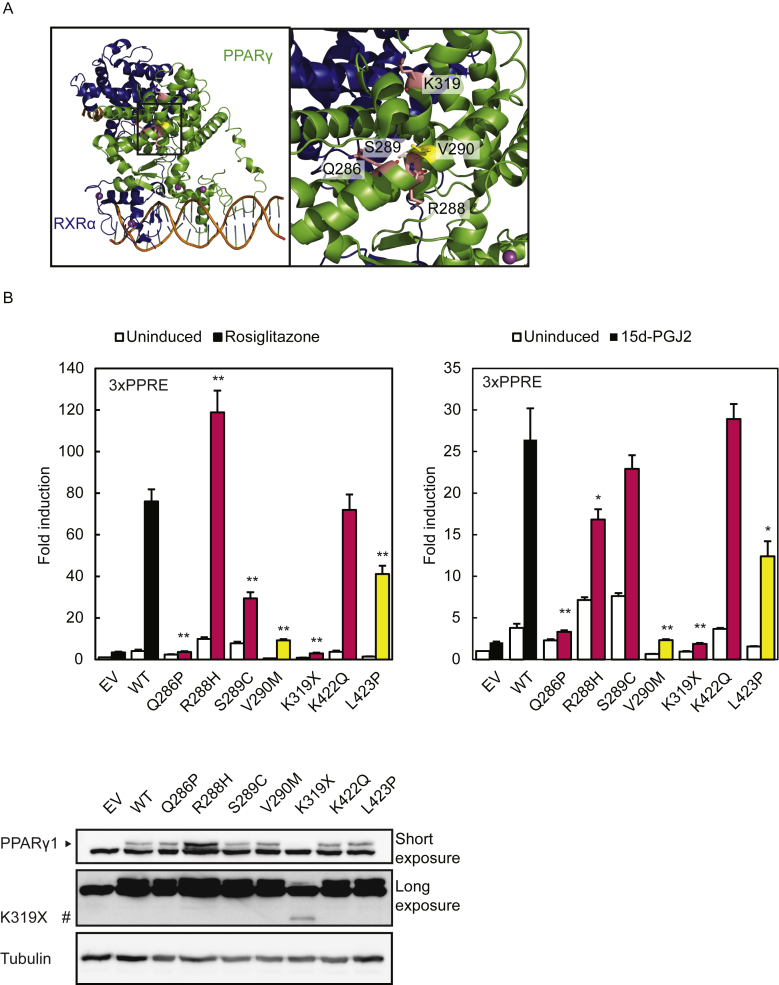
The FPLD3-associated PPARγ mutants reported so far, including the currently characterized PPARγ2 L451P, all show a consistent and profound impairment in PPARγ transcriptional activity due to different repertoires of inter- and intramolecular defects [10], [11], [12], [13], [14], [15]. Our functional characterization of the colon cancer-associated PPARγ2 K450Q (corresponding to PPARγ1 K422Q) suggests that K450Q does not display major functional defects, at least under our experimental settings. To determine whether this is a general feature of colon carcinoma-associated PPARγ1 mutants, we performed a side-by-side analysis of other previously reported colon cancer-associated PPARγ1 mutants Q286P, R288H, and K319X [16], [17]. Two additional mutants were included, S289C [43] and V290M [10]. PPARγ1 S289C is the first PPARG germline mutation that is associated with dyslipidemia and colonic polyp formation with progression to full-blown adenocarcinoma, without features of FPLD3 [43]. PPARγ1 V290M is one of the first identified FPLD3-associated mutants [10]. Whereas the activity of PPARγ1 mutants Q286P and K319X is negligible in presence of rosiglitazone and 15d-PGJ2, the transcriptional activity of PPARγ1 R288H, alike K422Q, is clearly not impaired when induced with rosiglitazone (Figure 6B). In line with a previous study [16], impaired transcriptional activity was observed when PPARγ1 R288H was treated 15d-PGJ2, a potential natural PPARγ ligand. Although PPARγ1 mutants S289C and V290M were both still able to initiate transcription, their activity was impaired (Figure 6B), in agreement with previous reports [10], [43], [44]. Interestingly, the transcriptional activity of PPARγ1 S289C reached wildtype levels upon induction with 15d-PGJ2. Collectively, these results indicate that the colon carcinoma-associated PPARγ mutants have diverse effects on transcriptional activity and suggest that colon carcinoma-associated PPARG mutations, in contrast to FPLD3-associated PPARG mutations, do not always lead to robust molecular defects. Accordingly, if the PPARG mutations are related to colon cancer onset and progression, this is not due to their effects on the most well-studied functional characteristics of PPARγ.
Figure 6.
Colon cancer-associated PPARγ mutants display variable phenotypes. A. Crystal structure of an intact PPARγ–RXRα complex (PPARγ in green; RXRα in blue) bound to DNA (left panel). The square box indicates the magnified region shown in the right panel. In pink, PPARγ Q286, R288, and S289 in helix 3 and K319. Substitution of these residues is associated with colon carcinoma. PPARγ V290, in helix 3, indicated in yellow has previously been described in FPLD3. Protein Database entry 3DZY. The figures were generated by open source software PyMOL 099rc6 (www.pymol.org). B. U2OS cells were transiently cotransfected with expression vectors encoding PPARγ1 WT or PPARγ1 mutants respectively, and 3× peroxisome proliferator response element (PPRE)-Tk-Luc reporter. Activation of the luciferase reporter, in the absence or presence of 1 μM rosiglitazone (left) or 15d-PGJ2 (right), is expressed as fold induction over that with empty vector (EV). Results are averages of at least three independent experiments assayed in duplicate ± SEM. **P < 0.01 cells transfected with mutant vs. WT. Expression levels of the different proteins were confirmed by western blot analysis using a PPARγ specific antibody. WT, wildtype. #, indicates PPARγ1 K319X.
4. Discussion
Here we show that the novel heterozygous PPARγ L451P mutation likely provides the molecular basis for the lipodystrophic phenotype in the index patient and her family. Although this is the first FPLD3-associated PPARG mutation located in helix 9, the adjacent lysine amino acid was found to be mutated (K450Q) in colorectal cancer cell lines [17]. The L451P mutant displays reduced transcriptional activity due to multiple molecular defects, including 1) general defects in ligand-mediated cofactor interactions and 2) impaired DNA binding due to reduced RXRα heterodimerization. In contrast, the K450Q mutant behaved like the wild type protein under all our experimental settings, suggesting that only dramatic amino acid changes in helix 9 will result in impaired PPARγ function. Noteworthy, our side-by-side analysis of the FPLD3-associated PPARγ L451P and V318M with the previously reported colon cancer-associated PPARγ mutants Q286P [17], R288H [17], S289C [43], K319X [17], and K422Q [16], shows that the colon cancer-associated PPARγ mutants do not always cause profound intra- and/or intermolecular defects. Interestingly, while no clear molecular defects with the K450Q mutant in PPARγ2 (present study) or the analogous K422Q mutant in PPARγ1 [17] could be identified, the PPARγ1 K422Q mutant failed to induce growth inhibition in vitro and in vivo and to induce intestinal epithelial cell differentiation when compared to the wild type protein [17]. In addition, PPARγ1 mutant S289C seems to be at the nexus of colon tumorigenesis and metabolic diseases as only the mutation carriers displayed derangements in lipid profiles without clinical features of T2DM and FPLD3 and only the mutation carriers developed dysplastic colonic polyps during the fourth-fifth decade of life, which in one patient progressed to colon cancer [43]. Together, these findings suggest that combined PPARγ transcriptome, cistrome, and interactome analyses within a specific cellular context may be required to identify specific phenotypic defects.
Although members of the nuclear receptor superfamily share a common structural architecture between their LBDs, the leucine residue at position 451 in PPARγ is not conserved (Supplemental Figure 1). As shown here, mutation of the corresponding position in PPARα reduced transcriptional activity (S414P; Figure 2A), while artificial mutants at this position in the vitamin D receptor (VDR; VDR C369G [45]) and thyroid hormone receptor β (THRβ; THRβ Y409K and neighboring residues Y406K, and R410A [46]) also show impaired LBD functions, including dimerization defects and impaired ligand- and cofactor binding. Together these findings support the view that L451 helps to maintain the integrity of helix 9, and that this helix is important for other nuclear receptors to function properly too. To the best of our knowledge, no naturally occurring mutations corresponding to the L451 position in PPARγ have been reported in other nuclear receptors so far.
In light of studying genetic PPARG variants and their effect on function, the recently developed classifier Missense InTerpretation by Experimental Response (MITER, available at http://miter.broadinstitute.org) is of particular interest [15]. MITER was generated by evaluating the effects all 9,595 possible single amino acid substitutions in PPARγ on CD36 expression using the human monocytic cell line THP-1. The MITER experimental function scores for the PPARγ2 mutants Q314P (−2,907), R316H (0.831), S317C (−0.309), V318M (−4,191), K347X (not available as nonsense mutations were not evaluated in MITER), and K450Q (−1.535) are in line with the findings of the current study. Interestingly, evaluation of the PPARγ L451P missense variant in MITER shows four-fold decreased transactivation function as compared to WT. When considered with the patient's family history and clinical presentation of lipodystrophy (corresponding to a prevalence of 20%), the MITER classifier emitted a probability of 12.7% for the L451P variant being causal for the patient's lipodystrophy. The MITER data are consistent with our own findings that the molecular effect of the L451P variant is more subtle than the canonical autosomal dominant FPLD3 variants previously reported [10], [11], [12], [13], [14], [15]. The co-segregation of lipodystrophy with PPARγ L451P strongly suggests that this PPARγ variant even without causing complete loss of function has a pathogenic effect in adipose tissue. Applying the CRISPR/CAS9 technology to adipocytes to recapitulate the pathogenicity of the heterozygous PPARG L451P would support this observation. However, this approach is technically very challenging in relevant (pre-)adipocyte cell lines, as for example the pre-adipocyte cell line 3T3L1 is polyclonal and polypoid.
The co-segregation of FPLD3 with the mutant receptor PPARγ L451P that displays reduced transcriptional activity due to multiple molecular defects, strongly indicates that this variant is causal for the condition. However, the molecular defects of the L451P mutant are subtler than previously reported FPLD3-associated PPARγ mutants, indicating that the index patient and affected family members may harbor additional co-segregating genetic variants that contribute to the lipodystrophic phenotype. Indeed, recent studies have shown that FPLD syndromes have a polygenic component. The importance of SNPs in regulatory regions in lipodystrophy has recently been depicted in a large population-level genetic study [34]. Patients with FPLD1, a subtype of FPLD with hitherto unknown genetic cause, are enriched for 53 common variants associated with limited peripheral adipose tissue storage capacity [34]. These observations not only suggest a polygenetic cause of FPLD1 [34], but also imply that genetic background affecting adipose tissue expandability can contribute to the penetrance of other types of lipodystrophy. The risk score analysis (z-score = −1.28) of the 53 loci in the index patient indicated that the index patient is not fully predisposed to a lipodystrophic phenotype based on her genetic background at these 53 SNPs. However, our catalog of genetic modifiers for FPLD is far from complete. Whereas the previously identified 53 genomic loci have been identified in subjects who are predominantly from European Caucasian origin, it remains to be defined to what extent the identified risk alleles can be applied to subjects of other genetic ancestries. The index patient in the current study is from Turkish descent and we cannot exclude that other genetic factors modify the FPLD3 phenotype. Furthermore, recent work suggests that other non-coding SNPs can modulate human metabolic disease risk by altering PPARγ binding [47]. Information from across the genome and from detailed mechanistic studies such as ours will continue to improve risk classification.
In conclusion, extensive characterization of the PPARγ L451P mutant identified here in a family affected by FPLD3 at the clinical, cellular, molecular, and genetic level revealed valuable understanding of PPARγ structure and function. We show that the newly identified FPLD3-associated L451P mutant significantly impairs the transcriptional activity of PPARγ due to a range of molecular defects that disrupt intermolecular interactions, while leaving the intramolecular LBD-DBD interactions largely intact. In contrast, the colon cancer-associated PPARγ K450Q mutation does not robustly affect PPARγ function, indicating that amino acid changes at the C-terminus of helix 9 can have differential effects on LBD integrity. Previous analyses of natural PPARγ mutants together with the data presented here support the view that FPLD3-associated mutations consistently cause intra- and/or intermolecular defects, while the effect of colon cancer-associated PPARγ mutations varies considerably, suggesting subtler or context-dependent effects.
Author contribution statement
MFB performed the experiments, data-analysis, and wrote the manuscript; MPGM and GWH performed the whole exome sequencing analysis; CD and AB performed experiments; JL performed the SNP score risk analysis; JS and AMJJB were involved in molecular modeling analysis; MGS, MNG, HM and MRS contributed to the clinical analysis. DM and RH were involved in NR-Coregulator interactions experiments and analysis. ARM advised on the SNP score risk analysis. EK designed and supervised the study. MFB and EK wrote the manuscript. All authors reviewed the manuscript.
Acknowledgements
We thank the index patient and her family for their participation in this study. We thank Dr. F. Payne (Wellcome Trust Sanger Institute, UK), Dr. I. Barroso (Wellcome Trust Sanger Institute, UK), and Prof. Dr. D. Savage (University of Cambridge, UK) for helpful discussions on the SNP analysis. Understanding Society: The UK Household Longitudinal Study is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. The survey was conducted by NatCen and the genome-wide scan data were analyzed and deposited by the Wellcome Trust Sanger Institute. Information on how to access the data can be found on the Understanding Society website https://www.understandingsociety.ac.uk/. Bioinformatics support was provided by the UMCU Bioinformatics Expertise Core (UBEC; www.ubec.nl). AMJJB and JS acknowledge financial support from the Horizon 2020 West-Life e-Infrastructure Virtual Research Environment project No. 675858. We also thank members of the Kalkhoven and Van Mil laboratories for helpful discussions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2018.12.005.
Conflict of interest
The author reports no conflicts of interest in this work.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Supplemental Figure 1.
Structural based sequence alignment of the amino acid residues surrounding PPARγ L451P between human PPARs and other members of the nuclear hormone superfamily. Human PPARγ (NP_056953.2), human PPARα (CAG30433), human PPARδ (Q03181), chimpanzee PPARγ (XP_001153669.1), mouse PPARγ (NP_035276.2), rat PPARγ (NP_037256.1), cow PPARγ (NP_851367.1), dog PPARγ (NP_001019803.1), chicken PPARγ (NP_001001460.1), and zebrafish PPARγ (NP_571542.1). THRA, thyroid receptor alpha (NP_003241.2); THRB, thyroid receptor beta (AAI06931.1); GR, glucocorticoid receptor (NP_000167.1); MR, mineralocorticoid receptor (NP_000892.2); AR, androgen receptor (P10275.2); FXR, farnesoid X receptor (NP_001193908.1); VDR, vitamin D receptor (AAH60832.1); HNFα, Hepatocyte nuclear factor 4A (CAA61133.1). Residues analogous L451 and V318 are designated in yellow and bold. Residues analogous to 314, 316, S317, K347, and K422 are indicated in pink and bold. PPARγ2 nomenclature. Dashes (−) represent the gaps included for appropriate alignment of the nuclear hormone receptors. The boundaries of the α-helices 9, and 10 are also shown.
Supplemental Figure 2.
Increasing L451P protein concentrations leads to a proportional increase in transcriptional activity, but the mutant never reaches WT levels. U2OS cells were transiently cotransfected with expression vectors encoding PPARγ WT or PPARγ L451P respectively, and a reporter construct harboring the promoter region of CIDEC. Activation of the luciferase reporter, in the absence or presence of 1 μM rosiglitazone, is expressed as fold induction over that with empty vector (EV). Results are averages of at least three independent experiments assayed in duplicate ± SEM. *P < 0.05 cells transfected with mutant vs. WT.
Supplemental Figure 3.
Proline substitutions in helix 9 impair transcriptional activity of PPARγ. U2OS cells were transiently cotransfected with expression vectors encoding PPARγ WT or PPARγ mutants respectively, and 3× peroxisome proliferator response element (PPRE)-Tk-Luc reporter. Activation of the luciferase reporter, in the absence or presence of 1 μM 15d-PGJ2, is expressed as fold induction over that with empty vector (EV). Results are averages of at least three independent experiments assayed in duplicate ± SEM. *P < 0.05 cells transfected with mutant vs. WT. Expression levels of the different proteins were confirmed by western blot analysis using a PPARγ specific antibody. The arrow indicates PPARγ and the asterisk indicates an unknown non-specific band. WT, wildtype.
Supplemental Figure 4.
Cofactor peptide binding profiles for GST-PPARγ LBD WT and L451P. GST-proteins were incubated with DMSO or 10 μM rosiglitazone. Images of all arrays were quantified and binding (AU; arbitrary units) was plotted. The experiments were performed in triple.
Supplemental Table 1.
SNP genotype of PPARγ L451P index patient. PCR was performed in the index patient to genotype the lead SNPs in 53 genomic regions that were previously identified to impair peripheral adipose tissue storage [34]. The index patients harbors 53 risk alleles.
References
- 1.Lehrke M., Lazar M.A. The many faces of PPARγ. Cell. 2005;123(6):993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Fajas L., Auboeuf D., Raspé E., Schoonjans K., Lefebvre A.-M., Saladin R. The organization, promoter analysis, and expression of the human PPARγ Gene. Journal of Biological Chemistry. 1997;272(30):18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 3.Mueller E., Drori S., Aiyer A., Yie J., Sarraf P., Chen H. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor γ isoforms. Journal of Biological Chemistry. 2002;277(44):41925–41930. doi: 10.1074/jbc.M206950200. [DOI] [PubMed] [Google Scholar]
- 4.Tontonoz P., Graves R.A., Budavari A.I., Erdjument-Bromage H., Lui M., Hu E. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPARγ and RXRa. Nucleic Acids Research. 1994;22(25):5628–5634. doi: 10.1093/nar/22.25.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefterova M.I., Haakonsson A.K., Lazar M.A., Mandrup S. PPARγ and the global map of adipogenesis and beyond. Trends in Endocrinology and Metabolism. 2014;25(6):293–302. doi: 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kliewer S.A., Lenhard J.M., Willson T.M., Patel I., Morris D.C., Lehmann J.M. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor γ and promotes adipocyte differentiation. Cell. 1995;83(5):813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 7.Forman B.M., Tontonoz P., Chen J., Brun R.P., Spiegelman B.M., Evans R.M. 15-Deoxy-Δ12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell. 1995;83(5):803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 8.Ahmadian M., Suh J.M., Hah N., Liddle C., Atkins A.R., Downes M. PPARgamma signaling and metabolism: the good, the bad and the future. Nature Medicine. 2013;19(5):557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soccio R.E., Chen E.R., Lazar M.A. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metabolism. 2014;20(4):573–591. doi: 10.1016/j.cmet.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barroso I., Gurnell M., Crowley V.E.F., Agostini M., Schwabe J.W., Soos M.A. Dominant negative mutations in human PPAR gamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402(December):880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 11.Agarwal A.K., Garg A. A novel heterozygous mutation in peroxisome proliferator-activated receptor-γ gene in a patient with familial partial lipodystrophy. The Journal of Clinical Endocrinology & Metabolism. 2002;87(1):408. doi: 10.1210/jcem.87.1.8290. [DOI] [PubMed] [Google Scholar]
- 12.Jeninga E.H., van Beekum O., van Dijk A.D.J., Hamers N., Hendriks-Stegeman B.I., Bonvin A.M.J.J. Impaired peroxisome proliferator-activated receptor gamma function through mutation of a conserved salt bridge (R425C) in familial partial lipodystrophy. Molecular Endocrinology (Baltimore, Md.) 2007;21(5):1049–1065. doi: 10.1210/me.2006-0485. [DOI] [PubMed] [Google Scholar]
- 13.Monajemi H., Zhang L., Li G., Jeninga E.H., Cao H., Maas M. Familial partial lipodystrophy phenotype resulting from a single-base mutation in deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-gamma. The Journal of Clinical Endocrinology and Metabolism. 2007;92(5):1606–1612. doi: 10.1210/jc.2006-1807. [DOI] [PubMed] [Google Scholar]
- 14.Visser M.E., Kropman E., Kranendonk M.E., Koppen a., Hamers N., Stroes E.S. Characterisation of non-obese diabetic patients with marked insulin resistance identifies a novel familial partial lipodystrophy-associated PPARγ mutation (Y151C) Diabetologia. 2011;54(7):1639–1644. doi: 10.1007/s00125-011-2142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majithia A.R., Tsuda B., Agostini M., Gnanapradeepan K., Rice R., Peloso G. Prospective functional classification of all possible missense variants in PPARG. Nature Genetics. 2016 doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarraf P., Mueller E., Smith W.M., Wright H.M., Kum J.B., Aaltonen L. a. Loss-of-function mutations in PPAR gamma associated with human colon cancer. Molecular Cell. 1999;3:799–804. doi: 10.1016/s1097-2765(01)80012-5. [DOI] [PubMed] [Google Scholar]
- 17.Gupta R.A., Sarraf P., Mueller E., Brockman J.A., Prusakiewicz J.J., Eng C. Peroxisome proliferator-activated receptor γ-mediated differentiation: a mutation in colon cancer cells reveals divergent and cell type-specific mechanisms. Journal of Biological Chemistry. 2003;278(25):22669–22677. doi: 10.1074/jbc.M300637200. [DOI] [PubMed] [Google Scholar]
- 18.Mueller E., Sarraf P., Tontonoz P., Evans R.M., Martin K.J., Zhang M. Terminal differentiation of human breast cancer through PPARγ. Molecular Cell. 1998;1(3):465–470. doi: 10.1016/S1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 19.Mueller E., Smith M., Sarraf P., Kroll T., Aiyer A., Kaufman D.S. Effects of ligand activation of peroxisome proliferator-activated receptor γ in human prostate cancer. Proceedings of the National Academy of Sciences. 2000;97(20) doi: 10.1073/pnas.180329197. 10990 LP-10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girnun G.D., Naseri E., Vafai S.B., Qu L., Szwaya J.D., Bronson R. Synergy between PPARγ ligands and platinum-based drugs in cancer. Cancer Cell. 2007;11(5):395–406. doi: 10.1016/j.ccr.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girnun G.D., Chen L., Silvaggi J., Drapkin R., Chirieac L.R., Padera R.F. Regression of drug-resistant lung cancer by the combination of rosiglitazone and carboplatin. Clinical Cancer Research. 2008;14(20) doi: 10.1158/1078-0432.CCR-08-1128. 6478 LP-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khandekar M.J., Banks A.S., Laznik-Bogoslavski D., White J.P., Choi J.H., Kazak L. Noncanonical agonist PPARγ ligands modulate the response to DNA damage and sensitize cancer cells to cytotoxic chemotherapy. Proceedings of the National Academy of Sciences. 2018;115(3) doi: 10.1073/pnas.1717776115. 561 LP-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harakalova M., van Harssel J.J.T., Terhal P.A., van Lieshout S., Duran K., Renkens I. Dominant missense mutations in ABCC9 cause Cantu syndrome. Nature Genetics. 2012;44(7):793–796. doi: 10.1038/ng.2324. [DOI] [PubMed] [Google Scholar]
- 24.Exome Variant Server., n.d. No Title. NHLBI GO Exome Sequencing Project (ESP). http://evs.gs.washington.edu/EVS/. [Accessed 1 April 2016].
- 25.Consortium, T. 1000 G.P. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalkhoven E., Teunissen H., Houweling A., Verrijzer C.P., Zantema A. The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Molecular and Cellular Biology. 2002;22(7):1961–1970. doi: 10.1128/MCB.22.7.1961-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurnell M., Wentworth J.M., Agostini M., Adams M., Collingwood T.N., Provenzano C. A dominant-negative peroxisome proliferator-activated receptor g (PPARg) mutant is a constitutive repressor and inhibits PPARg-mediated adipogenesis. Journal of Biological Chemistry. 2000;275(8):5754–5759. doi: 10.1074/jbc.275.8.5754. [DOI] [PubMed] [Google Scholar]
- 30.Chen J.D., Evans R.M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 31.Koppen A., Houtman R., Pijnenburg D., Jeninga E.H., Ruijtenbeek R., Kalkhoven E. Nuclear receptor-coregulator interaction profiling identifies TRIP3 as a novel peroxisome proliferator-activated receptor γ cofactor. Molecular & Cellular Proteomics. 2009;8(10):2212–2226. doi: 10.1074/mcp.M900209-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vega R.B., Huss J.M., Kelly D.P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Molecular and Cellular Biology. 2000;20(5) doi: 10.1128/mcb.20.5.1868-1876.2000. 1868 LP-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broekema M.F., Hollman D.A.A., Koppen A., van den Ham H.-J., Melchers D., Pijnenburg D. Profiling of 3696 nuclear receptor-coregulator interactions: a resource for biological and clinical discovery. Endocrinology. 2018 doi: 10.1210/en.2018-00149. [DOI] [PubMed] [Google Scholar]
- 34.Lotta L.A., Gulati P., Day F.R., Payne F., Ongen H., van de Bunt M. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nature Genetics. 2017;49(1):17–26. doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R. Chapter nineteen – Rosetta3: an object-oriented software suite for the simulation and design of macromolecules. In: Johnson M.L., Brand L.B.T.-M., editors. vol. 487. Academic Press; 2011. pp. 545–574. (Computer methods, part C). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T.P. Structure of the intact PPAR-gamma-RXR- alfa nuclear receptor complex on DNA. Nature. 2008;456(7220):350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein A., Kortemme T. Improvements to robotics-inspired conformational sampling in Rosetta. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0063090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S.C., Ng Y.K. Calibur: a tool for clustering large numbers of protein decoys. BMC Bioinformatics. 2010;11(1):25. doi: 10.1186/1471-2105-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morin A., Eisenbraun B., Key J., Sanschagrin P.C., Timony M.A., Ottaviano M. Collaboration gets the most out of software. ELife. 2013;2 doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tasdelen I., Berger R., Kalkhoven E. PPARγ regulates expression of carbohydrate sulfotransferase 11 (CHST11/C4ST1), a regulator of LPL cell surface binding. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0064284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandra V., Wu D., Li S., Potluri N., Kim Y., Rastinejad F. The quaternary architecture of RARβ–RXRα heterodimer facilitates domain–domain signal transmission. Nature Communications. 2017;8(1):868. doi: 10.1038/s41467-017-00981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricci C.G., Silveira R.L., Rivalta I., Batista V.S., Skaf M.S. Allosteric pathways in the PPARγ-RXRα nuclear receptor complex. Scientific Reports. 2016;6:19940. doi: 10.1038/srep19940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capaccio D., Ciccodicola a., Sabatino L., Casamassimi a., Pancione M., Fucci a. A novel germline mutation in peroxisome proliferator-activated receptor gamma gene associated with large intestine polyp formation and dyslipidemia. Biochimica et Biophysica Acta. 2010;1802(6):572–581. doi: 10.1016/j.bbadis.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Savage D.B., Tan G.D., Acerini C.L., Jebb S.A., Agostini M., Gurnell M. Human metabolic syndrome resulting from dominant-negative mutations in the nuclear receptor peroxisome proliferator-activated receptor-γ. Diabetes. 2003;52(4):910–917. doi: 10.2337/diabetes.52.4.910. [DOI] [PubMed] [Google Scholar]
- 45.Nakajima S., Hsieh J.-C., Jurutka P.W., Galligan M.A., Haussler C.A., Whitfield G.K. Examination of the potential functional role of conserved cysteine residues in the hormone binding domain of the human 1,25-dihydroxyvitamin D receptor. Journal of Biological Chemistry. 1996;271(9):5143–5149. [PubMed] [Google Scholar]
- 46.Tagami T., Yamamoto H., Moriyama K., Sawai K., Usui T., Shimatsu A. The retinoid X receptor binding to the thyroid hormone receptor: relationship with cofactor binding and transcriptional activity. Journal of Molecular Endocrinology. 2009;42(5):415–428. doi: 10.1677/JME-08-0153. [DOI] [PubMed] [Google Scholar]
- 47.Soccio R.E., Chen E.R., Rajapurkar S.R., Safabakhsh P., Marinis J.M., Dispirito J.R. Genetic variation determines PPARγ function and anti-diabetic drug response in vivo. Cell. 2015;162(1):33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.