Significance
Under starvation conditions, cells activate macroautophagy to recycle nutrients. A kinase complex that consists of Atg1 kinase, Atg13, and Atg17 is essential for macroautophagy function. This kinase complex is regulated extensively by phosphorylation events. However, the phosphatases that counteract these phosphorylations have remained elusive. Our results show that two redundant PP2C-type phosphatases, Ptc2 and Ptc3, promote autophagy by interacting with Atg1 and Atg13 and dephosphorylating them. In the absence of these phosphatases, macroautophagy, as well as selective autophagy pathways such as cytoplasm-to-vacuole targeting and genotoxin-induced targeted autophagy, are inhibited. These results suggest that PP2C-type phosphatases play a previously uncharacterized and central role in the regulation of core autophagy proteins.
Keywords: PP2C phosphatases, autophagy, ATG13, Saccharomyces cerevisiae
Abstract
Macroautophagy is orchestrated by the Atg1-Atg13 complex in budding yeast. Under nutrient-rich conditions, Atg13 is maintained in a hyperphosphorylated state by the TORC1 kinase. After nutrient starvation, Atg13 is dephosphorylated, triggering Atg1 kinase activity and macroautophagy induction. The phosphatases that dephosphorylate Atg13 remain uncharacterized. Here, we show that two redundant PP2C phosphatases, Ptc2 and Ptc3, regulate macroautophagy by dephosphorylating Atg13 and Atg1. In the absence of these phosphatases, starvation-induced macroautophagy and the cytoplasm-to-vacuole targeting pathway are inhibited, and the recruitment of the essential autophagy machinery to the phagophore assembly site is impaired. Expressing a genomic ATG13-8SA allele lacking key TORC1 phosphorylation sites partially bypasses the macroautophagy defect in ptc2Δ ptc3Δ strains. Moreover, Ptc2 and Ptc3 interact with the Atg1-Atg13 complex. Taken together, these results suggest that PP2C-type phosphatases promote macroautophagy by regulating the Atg1 complex.
Macroautophagy is a catabolic recycling pathway that is responsible for the degradation of cytoplasmic components by targeting them to the vacuole under starvation conditions (1, 2). In budding yeast, during the early steps of macroautophagy, phagophores assemble at a phagophore assembly site (PAS) adjacent to the vacuole and encapsulate cytoplasmic components (3). Then, phagophores expand to form mature autophagosomes, which eventually fuse with the vacuole (4), wherein the contents of the autophagosomes are degraded and recycled (5). In addition to the degradation of random portions of the cytoplasm to recycle nutrients under starvation conditions, specific organelles or molecules can be targeted for degradation or processing by selective autophagy (6). Selective autophagy pathways rely on the recognition of cargo via specific receptor proteins and the scaffold protein Atg11 (7, 8). One of the best-studied selective autophagy pathways is the cytoplasm-to-vacuole targeting (Cvt) pathway that targets precursor aminopeptidase I (prApe1) to the vacuole, where it is proteolytically processed by vacuolar proteases (9). The genetic requirements of the Cvt pathway and bulk macroautophagy significantly overlap; however, there are some differences. Unlike nonselective macroautophagy, the selective Cvt pathway can occur in nutrient-rich conditions. Notably, the scaffold protein Atg17 is required for nonselective macroautophagy but is dispensable for the Cvt pathway (10–12). Conversely, Atg19, the receptor protein for the Cvt pathway cargo, and the scaffold protein Atg11 are important for Cvt pathway function but are not essential for macroautophagy (12–14).
One of the sensors of nutrient availability is the target of rapamycin kinase complex 1, TORC1 (MTORC1 in mammals; hereafter we indicate mammalian homologs with a slash). When nutrients are abundant, Atg13/ATG13, an essential part of the core autophagy machinery, is kept in a hyperphosphorylated and inhibited state (15). In addition to Atg13, the central kinase in the autophagy pathway, Atg1/ULK1, is phosphorylated and inhibited by protein kinase A and TORC1-dependent phosphorylation events (16–18). When nutrients are scarce, TORC1 activity is inhibited; consequently, Atg13 is rapidly dephosphorylated. This dephosphorylation of Atg13 is required for stimulation of Atg1’s kinase activity (15). Recent work has shown that Atg1 and Atg13 binding is constitutive; however, Atg1-Atg13 interaction is enhanced upon Atg13 dephosphorylation (19–21). Atg1 and Atg13 also bind to the Atg17–Atg31–Atg29 scaffolding subcomplex to form a complete Atg1 kinase complex (22–24). The Atg1 kinase complex then localizes at the PAS and activates downstream Atg proteins to promote the formation of autophagosomes (25). Although the posttranslational regulation of the Atg1 kinase complex has been well studied, the phosphatases that are responsible for the reversal of these phosphorylation modifications remain largely uncharacterized.
Previous work from our laboratories has shown that DNA damage induces a targeted selective autophagy pathway, which we coined genotoxin-induced targeted autophagy (GTA) (26–28). GTA requires the core autophagy machinery as well as the scaffold protein Atg11. A genomewide screen to find modulators of GTA yielded Pph3, a PP4-type phosphatase, which has been demonstrated to extinguish the DNA damage checkpoint (DDC) signaling by dephosphorylation and inactivation of several targets of checkpoint kinases (29–32). Pph3 negatively regulates GTA but has no effect on rapamycin-induced autophagy (27).
Budding yeast’s PP2C-type phosphatases, Ptc2 and Ptc3, are serine/threonine-phosphatase paralogs that arose from whole genome duplication and are largely redundant (33). These phosphatases dephosphorylate a variety of targets, including those involved in the high-osmolarity glycerol pathway (34, 35), the unfolded protein response (36), the MAPK pathway (37), and general stress response (38). In addition, these phosphatases counteract DDC signaling by directly dephosphorylating the central checkpoint kinase, Rad53/CHEK2 (39). As a consequence, deletion of PTC2 and PTC3 prevents cells from extinguishing the DDC (29, 39).
Here, we asked if Ptc2 and Ptc3 phosphatases have similar functions with Pph3 for autophagy, given their similar roles in the DDC pathway. Contrary to the suppression of GTA by Pph3, we find that Ptc2 and Ptc3 promote not only GTA, but also rapamycin-induced macroautophagy and the Cvt pathway. Deletion of PTC2 and PTC3 causes the accumulation of hyperphosphorylated Atg1 and Atg13. Atg1 hyperphosphorylation in the absence of Ptc2 and Ptc3 is dependent on the kinase activity of Atg1 as well as Atg13 and Atg11. The autophagy defect of the ptc2Δ ptc3Δ mutant is suppressed by the constitutively active ATG13-8SA allele despite persistence of hyperphosphorylated Atg1. Moreover, Ptc2 and Ptc3 interact with the Atg1 kinase complex in vivo. Based on these results, we conclude that the Ptc2 and Ptc3 phosphatases are involved in the regulation of selective and nonselective autophagy pathways largely by acting on the Atg1 kinase complex.
Results
PP2C-Type Phosphatases Ptc2 and Ptc3 Promote DNA Damage-Induced and Rapamycin-Induced Autophagy.
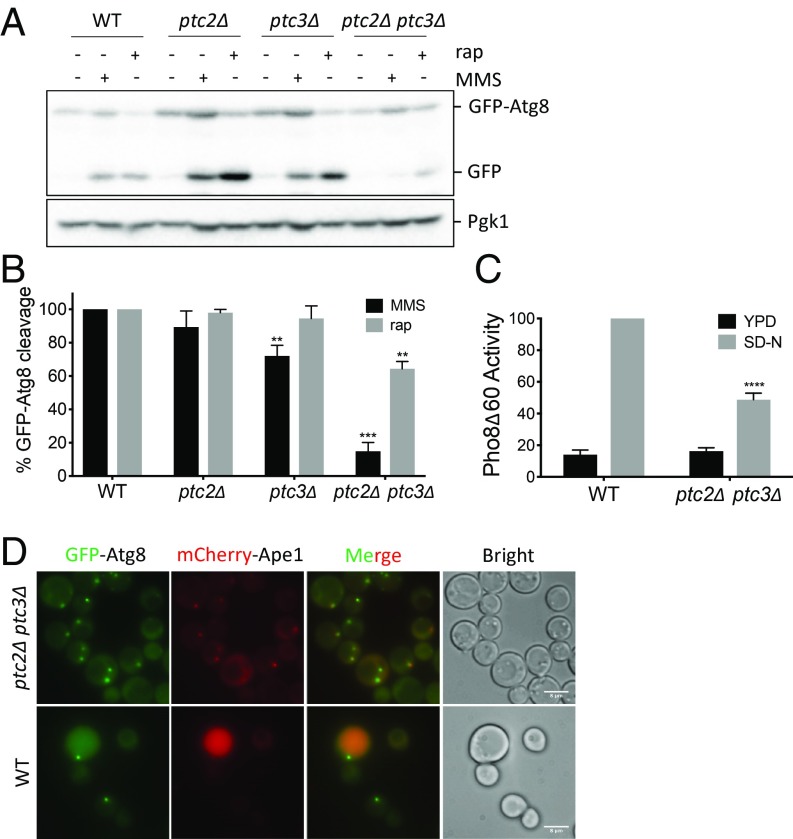
Upon autophagy induction, phosphatidylethanolamine-conjugated Atg8 localizes to the PAS. While the phagophore is expanding, a population of Atg8 is localized to both sides of the phagophore membrane. When autophagosome maturation is complete, the population of Atg8 localized on the inner membrane is delivered to the vacuole along with the autophagosome cargo. Although Atg8 is degraded, the GFP moiety is resistant to vacuolar proteases. Hence, the ratio of free GFP to GFP-Atg8 on a western blot serves as a reporter of autophagy activity (40). To investigate whether autophagy induction is altered in the phosphatase mutants, we transformed WT, ptc2Δ, ptc3Δ, and ptc2Δ ptc3Δ strains with a plasmid carrying GFP-Atg8 and treated the cells with rapamycin to induce macroautophagy or with methyl methanesulfonate (MMS) to induce DNA damage and GTA (27). The single mutants ptc2Δ and ptc3Δ showed WT levels of autophagy induction in response to rapamycin or MMS except for a small but statistically significant defect in ptc3Δ for GTA (Fig. 1 A and B). In contrast, the ptc2Δ ptc3Δ double mutant was significantly defective for rapamycin-induced autophagy and GTA (Fig. 1 A and B). In addition to the GFP-Atg8 processing assay, bulk autophagy activity can be measured by the Pho8Δ60 assay (41). This assay relies on the delivery of the truncated and cytosolic allele of the phosphatase Pho8, named Pho8Δ60, to the vacuole through bulk autophagy, where it is fully activated. Both of these assays measure complete autophagic flux, including fusion of the autophagosome with the vacuole, release of the cargo into the lumen, and subsequent cargo degradation and/or processing. In the ptc2Δ ptc3Δ double mutant, Pho8Δ60 activity was significantly reduced compared with WT after nitrogen starvation, verifying the autophagy defect (Fig. 1C).
Fig. 1.
Ptc2 and Ptc3 phosphatases redundantly promote rapamycin-induced autophagy. (A) WT and ptc2Δ ptc3Δ strains were grown in YEP-lac until they reached early exponential phase and treated with 200 ng/mL rapamycin or 0.04% MMS for 4 h. Samples were blotted for GFP to quantify GFP-Atg8 cleavage and for Pgk1 as a loading control. (B) GFP-Atg8 cleavage was calculated as the ratio of the free GFP band to the total GFP signal in the lane for three independent experiments and then normalized to WT. Error bars represent SE (**P < 0.002 and ***P < 0.0002). (C) Pho8Δ60 activity in WT cells and the phosphatase-null strain 4 h after nitrogen starvation, normalized to the WT starvation conditions for four independent experiments. Error bars represent SE (****P < 0.0001). (D) Localization of GFP-Atg8 and mCherry-Ape1 in WT and ptc2Δ ptc3Δ at 1 h after rapamycin treatment. (Scale bar: 8 μm.)
We verified the impaired autophagy activity of ptc2Δ ptc3Δ double mutant by fluorescent live-cell imaging of GFP-Atg8. Following autophagy induction, GFP-Atg8 predominantly localized to the vacuoles in WT cells as expected (42). In agreement with the GFP-Atg8 cleavage assay results, the vacuolar GFP signal was reduced in ptc2Δ ptc3Δ cells compared with WT after rapamycin treatment (Fig. 1D) or MMS treatment (SI Appendix, Fig. S1). GFP-Atg8 localization at the PAS after autophagy induction was more pronounced in ptc2Δ ptc3Δ cells compared with WT, indicating a block in GFP-Atg8 delivery to the vacuole. Additionally, in the absence of the PP2C phosphatases, we frequently observed multiple, less intense GFP-Atg8 foci that colocalized with the PAS marker mCherry-Ape1 1 h after rapamycin treatment (Fig. 1D). Overall, these results suggest that Ptc2 and Ptc3 play a partially redundant role in promoting efficient DNA damage-induced autophagy and rapamycin-induced macroautophagy.
PP2C-Type Phosphatases Ptc2 and Ptc3 Promote the Cvt Pathway.
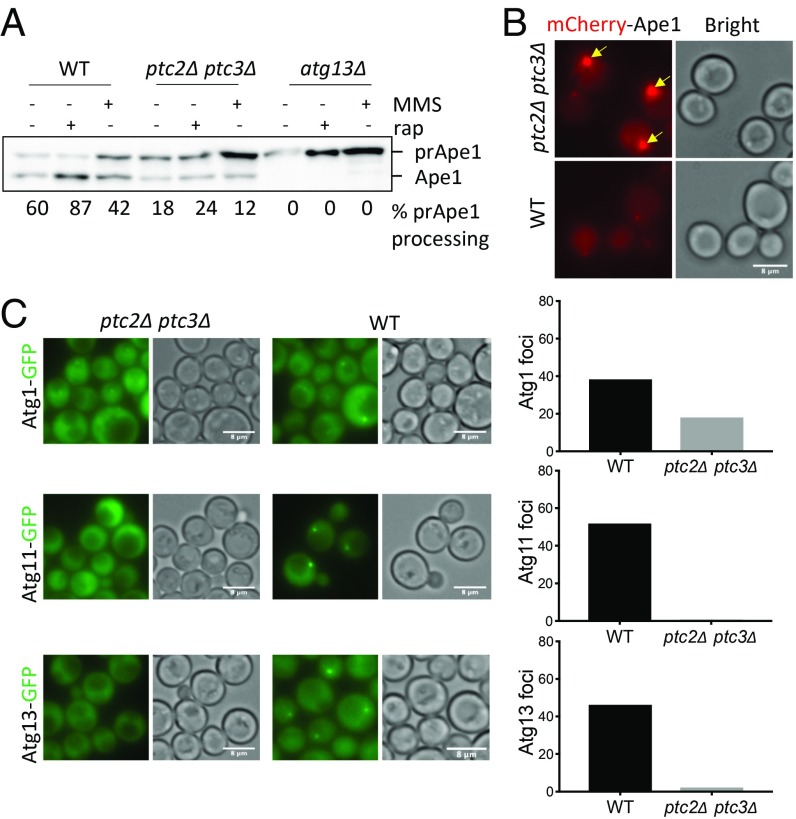
The prototypic selective autophagy pathway in yeast is the Cvt pathway (9). To assay Cvt activity quantitatively, we used the prApe1 maturation assay (43). Ape1 is synthesized in the cytosol as a precursor and then delivered through the Cvt pathway to the vacuole for its maturation by the removal of its propeptide (9). Maturation of prApe1 can be detected on a western blot with an antibody that recognizes both prApe1 and the mature form (Ape1) of the protein (44). We found that deletion of PTC2 and PTC3 impaired prApe1 maturation under basal conditions and after rapamycin or MMS treatment (Fig. 2A). However, in the ptc2Δ ptc3Δ mutant, Cvt activity was not fully abolished relative to that seen in atg13Δ cells. We also confirmed the Cvt defect of the ptc2Δ ptc3Δ strain by fluorescent live-cell imaging. We observed the accumulation of abnormally large mCherry-Ape1 foci at the PAS in ptc2Δ ptc3Δ cells 4 h after rapamycin treatment (Fig. 2B), suggesting that the Cvt pathway is significantly impaired in the absence of Ptc2 and Ptc3.
Fig. 2.
Ptc2 and Ptc3 phosphatases promote the Cvt pathway. (A) Precursor Ape1 processing in WT, ptc2Δ ptc3Δ, and atg13Δ mutants after 4-h rapamycin or MMS treatment. The ratio of mature Ape1 to total Ape1 signal for each lane is presented below every lane. (B) mCherry-Ape1 localization in WT and ptc2Δ ptc3Δ cells 4 h after rapamycin treatment. Yellow arrows mark the large perivacuolar prApe1 puncta that correspond to the PAS. (C) Localization of Atg1-GFP, Atg11-GFP, and Atg13-GFP in ptc2Δ ptc3Δ and WT cells grown in rich media. Percentages of cells with visible Atg1, Atg11, or Atg13 foci were quantified and plotted. More than 150 cells were counted for each condition. (Scale bar: B and C, 8 μm.)
The delivery of prApe1 to the vacuole is dependent on Atg1 and Atg13 (10) and the scaffold protein Atg11 (14). To see if the Cvt defect of ptc2Δ ptc3Δ is caused by the mislocalization of this necessary Atg machinery, we assayed the localization of these proteins under nutrient-replete conditions by fluorescent live-cell imaging. Atg1 puncta in the absence of these phosphatases appeared lower in signal intensity and less frequent (Fig. 2C and SI Appendix, Fig. S2). Moreover, Atg11 and Atg13 essentially did not form visible foci in the ptc2Δ ptc3Δ mutant cells (Fig. 2C and SI Appendix, Fig. S2), suggesting that Ptc2 and Ptc3 are required for the localization of Atg machinery necessary for the Cvt pathway in nutrient-replete conditions.
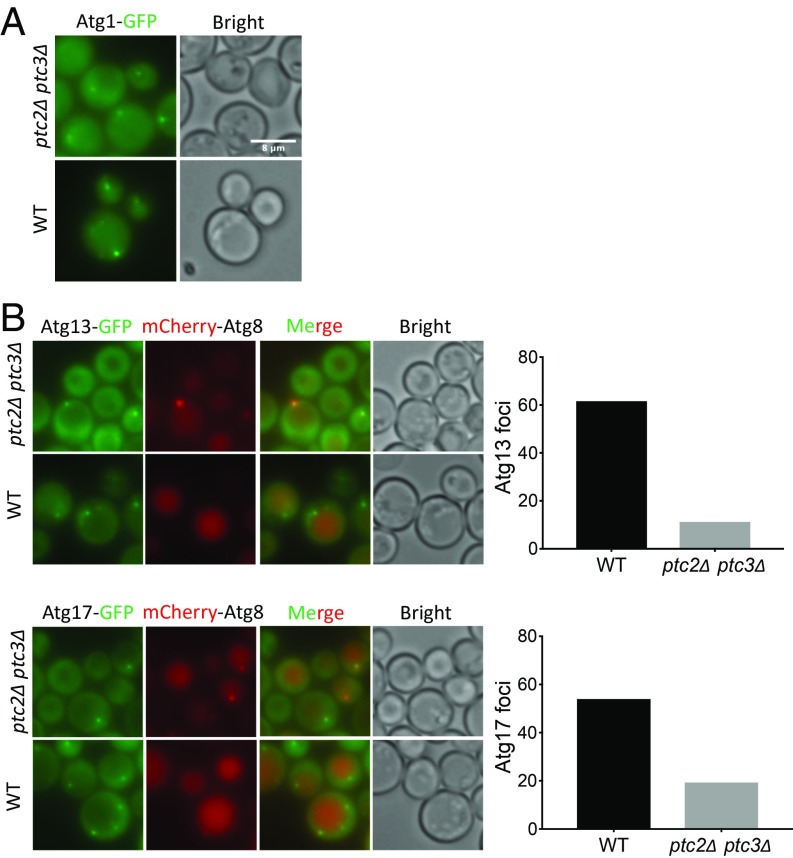
Next, we assayed the localization of Atg1, Atg11, Atg13, and Atg17 in the ptc2Δ ptc3Δ mutant by fluorescent live-cell imaging after rapamycin treatment to examine the roles of Ptc2 and Ptc3 upon autophagy induction. Similar to what we observed under nutrient-replete conditions, Atg1 puncta were weaker in the ptc2Δ ptc3Δ mutant after rapamycin treatment (Fig. 3A and SI Appendix, Fig. S3). Furthermore, the number of cells with Atg11, Atg13, and Atg17 puncta was significantly reduced in the absence of the two phosphatases (Fig. 3B and SI Appendix, Figs. S3 and S4). These results indicate that Ptc2 and Ptc3 also promote the proper recruitment of autophagy proteins to the PAS under nutrient-starvation conditions.
Fig. 3.
The essential Atg machinery is mislocalized in the absence of Ptc2 and Ptc3 phosphatases. (A) Atg1-GFP localization in ptc2Δ ptc3Δ and WT cells after 4-h rapamycin treatment. (Scale bar: 8 μm.) (B) Atg13-GFP and Atg17-GFP localization in ptc2Δ ptc3Δ and WT cells 4 h after rapamycin treatment. Percentage of cells with visible Atg13 or Atg17 foci were quantified and plotted. More than 150 cells were counted for each condition.
Atg1 Is Maintained in a Hyperphosphorylated State in the Absence of Ptc2 and Ptc3.
Next, we investigated whether Ptc2 and Ptc3 act directly on TORC1 to regulate autophagy. As a readout of TORC1 activity, we assayed the phosphorylation of the AGC kinase Sch9, a direct target of TORC1 (45). In WT and ptc2Δ ptc3Δ cells, Sch9 was phosphorylated under nutrient-replete conditions and after cycloheximide (CHX) treatment and was dephosphorylated after rapamycin treatment (SI Appendix, Fig. S5). Based on these findings, we conclude that the Ptc2 and Ptc3 phosphatases are likely to exert their effects directly on the autophagy machinery rather than by acting on TORC1 itself.
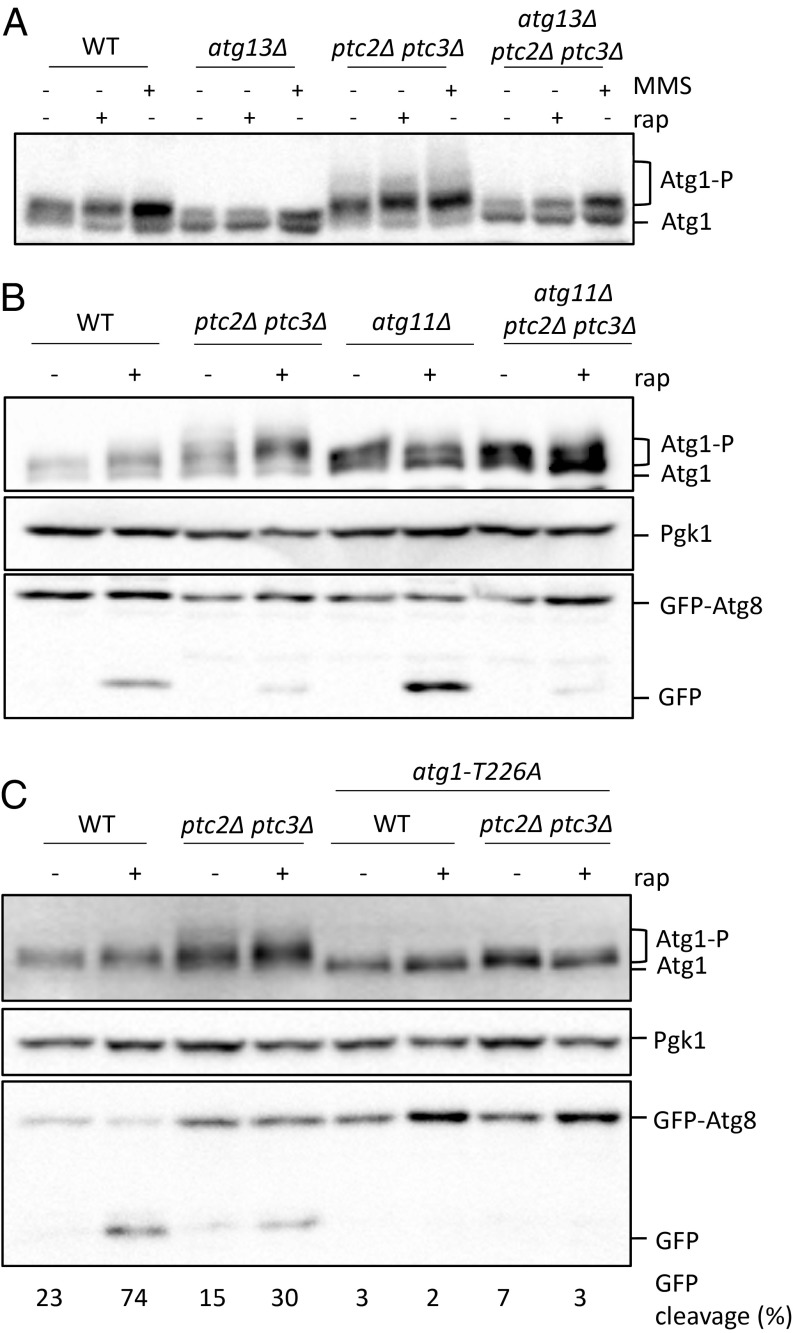
Starvation-induced inhibition of TORC1 and protein kinase A causes the activation of Atg1 through autophosphorylation (10, 18). Atg1 autophosphorylation requires the presence of Atg13, as binding to dephosphorylated Atg13 stimulates Atg1’s kinase activity after autophagy induction (46). To monitor the phosphorylation of Atg1, we used a strain carrying an N-terminal 2×FLAG-tagged Atg1 allele (7). We confirmed that Atg1 was phosphorylated after autophagy induction by rapamycin or MMS treatments in an Atg13-dependent manner (Fig. 4A) (27). In ptc2Δ and ptc3Δ single mutants, Atg1 was phosphorylated following MMS or rapamycin treatment similar to WT (SI Appendix, Fig. S6). However, in the ptc2Δ ptc3Δ double mutant, we detected an accumulation of slower-migrating Atg1 bands under nutrient-replete conditions and after the induction of autophagy (Fig. 4A). These slower-migrating bands were Atg13-dependent, as the mobility of Atg1 in the atg13Δ ptc2Δ ptc3Δ triple mutant was comparable to that of the atg13Δ single mutant (Fig. 4A).
Fig. 4.
Atg1 is hyperphosphorylated in ptc2Δ ptc3Δ strains. (A) Atg1 phosphorylation in WT, atg13Δ, ptc2Δ ptc3Δ, and atg13Δ ptc2Δ ptc3Δ strains after 4-h rapamycin or MMS treatment. Samples were blotted with anti-FLAG antibody for FLAG-Atg1 detection, Pgk1, and GFP. (B) Atg1 phosphorylation in WT, ptc2Δ ptc3Δ, atg11Δ, and atg11Δ ptc2Δ ptc3Δ strains after 4-h rapamycin treatment. (C) Atg1 phosphorylation in ptc2Δ ptc3Δ mutant strains that carry an atg1-T226A kinase-defective allele 4 h after rapamycin treatment. Numbers below lanes indicate the GFP-Atg8 cleavage for the corresponding lane.
Recent work has demonstrated that Atg1’s kinase activity is stimulated through interactions with the Atg11-bound Atg19-prApe1 Cvt cargo complex (7). We asked if the high molecular weight Atg1 bands we observe in the absence of Ptc2 and Ptc3 could result from the overstimulation of Atg1 kinase activity by Cvt cargo binding. To test this possibility, we assayed the phosphorylation of Atg1 in the ptc2Δ ptc3Δ cells lacking Atg11, Atg19, or Ape1 before or after rapamycin treatment. The appearance of slower-migrating Atg1 species was dependent on Atg11 (Fig. 4B) but not on Atg19 or Ape1 (SI Appendix, Fig. S7). Although Atg1 gel mobility was reduced in the atg11Δ ptc2Δ ptc3Δ triple mutant, this decrease did not rescue the macroautophagy defect of the ptc2Δ ptc3Δ strain as judged by the GFP-Atg8 processing assay (Fig. 4B). To see if the high-mobility Atg1 bands seen in the ptc2Δ ptc3Δ double mutant were dependent on Atg1’s kinase activity, we used CRISPR-Cas9 to create an epitope-tagged atg1-T226A point mutant, which blocks Atg1’s kinase activity and autophagy (46). As expected, replacement of ATG1 with atg1-T226A abolished the autophagy-dependent autophosphorylation of Atg1 and autophagy activity in WT cells (Fig. 4C). The atg1-T226A substitution also abolished the appearance of slower-migrating Atg1 bands in the ptc2Δ ptc3Δ double mutant (Fig. 4C). These data demonstrate that Ptc2 and Ptc3 phosphatases counteract Atg1 autophosphorylation under starvation or nutrient-replete conditions.
Next, we sought to characterize the Atg1 sites regulated by Ptc2 and Ptc3. Previous work suggests that Atg1 S34 and Atg1 S390 are regulated through phosphorylation. Atg1 S34 phosphorylation is inhibitory for Atg1’s kinase activity and autophagy, whereas Atg1 S390 phosphorylation causes the Atg1 mobility shift after autophagy induction with no obvious effect on autophagy (47). We mutated these two sites at their corresponding endogenous loci by using CRISPR-Cas9 to test if they are regulated by Ptc2 and Ptc3. As previously shown, autophagy was impaired in the atg1-S34D point mutant (4), but this substitution was not sufficient to fully eliminate the autophosphorylation of Atg1 after autophagy induction (SI Appendix, Fig. S8A). The reduction in Atg1 mobility in the atg1-S34D ptc2Δ ptc3Δ mutant could be attributed to the impaired kinase activity of atg1-S34D (47). In contrast, the atg1-S34A point mutant failed to rescue the autophagy defect in the double-phosphatase mutant. Atg1 phosphorylation was reduced compared with WT in the atg1-S34A,S390A double mutant; however, these mutations did not fully eliminate the extensive mobility shift of Atg1 when combined with ptc2Δ ptc3Δ. Moreover, neither the atg1-S34A and atg1-S390A single mutants nor the atg1-S34A,S390A double mutant were able to rescue the autophagy defect seen in ptc2Δ ptc3Δ cells (SI Appendix, Fig. S8B). Collectively, these findings indicate that the S34 and S390 sites that were previously implicated in Atg1 regulation are not directly targeted by Ptc2 and Ptc3 or that there are additional sites, yet to be identified, that play a role in Atg1 regulation.
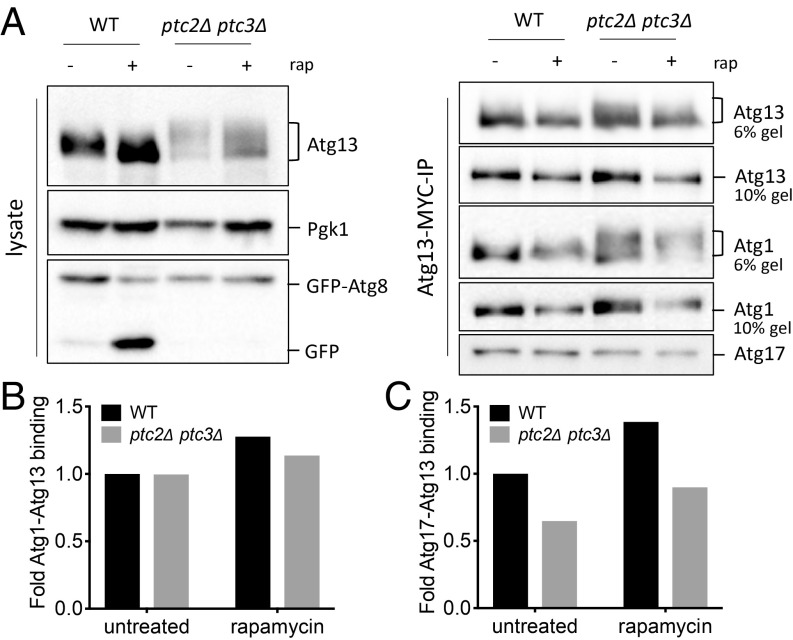
Atg13 Is Hyperphosphorylated in the ptc2Δ ptc3Δ Double Mutant.
Previous research established that Atg13 dephosphorylation is required for proper autophagy induction (15). We reasoned that one way by which Ptc2 and Ptc3 could promote autophagy is by mediating Atg13 dephosphorylation. We created an epitope-tagged Atg13 allele at the genomic locus to assay the phosphorylation of endogenous Atg13. In the ptc2Δ ptc3Δ mutant, we detected an accumulation of higher molecular weight Atg13 bands compared with WT in the absence of autophagy induction (Fig. 5A). Atg13 was only partially dephosphorylated in ptc2Δ ptc3Δ cells after autophagy induction by rapamycin, whereas phosphorylated Atg13 bands were fully diminished in the WT strain (Fig. 5A). These results suggest that the phosphatases Ptc2 and Ptc3 are involved in dephosphorylating Atg13. We also observed that Atg13 was dephosphorylated in the autophagy-defective atg1-T226A strain, suggesting that the dephosphorylation of Atg13 is independent of autophagy flux and Atg1’s kinase activity (SI Appendix, Fig. S9).
Fig. 5.
Atg1 and Atg13 interaction is unaltered in the absence of Ptc2 and Ptc3 phosphatases. (A) Atg1-Atg13-Atg17 binding in WT and ptc2Δ ptc3Δ strains before and after 4-h rapamycin treatment. Samples taken from cell lysates (Left) were blotted for anti-MYC to detect Atg13, or with anti-GFP or anti-Pgk1. Samples collected following Atg13-13×MYC immunoprecipitation (Right) were blotted for anti-FLAG to detect Atg1, anti-MYC to detect Atg13, and anti-Atg17. (B) Normalized Atg1-Atg13 binding was calculated as a ratio of FLAG-Atg1 band intensity to Atg13-MYC band intensity at 0 h for WT and ptc2Δ ptc3Δ and then normalized to WT. (C) Normalized Atg13-Atg17 binding was calculated as a ratio of Atg17 band intensity to Atg13-MYC band intensity at 0 h for WT and ptc2Δ ptc3Δ cells and then normalized to WT.
Fujioka et al. (21) have reported that Atg1-Atg13 and Atg13-Atg17 binding is enhanced upon autophagy induction and concomitant Atg13 dephosphorylation. One explanation for the autophagy defect of the ptc2Δ ptc3Δ cells would be that interactions between these essential autophagy proteins are impaired as a result of the accumulation of modified Atg1 and Atg13. We immunoprecipitated Atg13-13×MYC in the presence of phosphatase and protease inhibitors (21) to assay the binding of Atg17 and Atg1 with Atg13 (Fig. 5A). We observed that the association of Atg1-Atg13 after rapamycin treatment was modestly enriched in WT as well as in the phosphatase mutant (Fig. 5B). Conversely, the Atg13-Atg17 interaction was substantially decreased in the ptc2Δ ptc3Δ cells relative to WT before and after autophagy induction (Fig. 5C). Based on these observations, we conclude that the hyperphosphorylated Atg1 and Atg13 species observed in the phosphatase mutant still can interact, and that the autophagy defect of this mutant could be, at least in part, a result of the decreased presence of Atg17 in the Atg1 kinase complex.
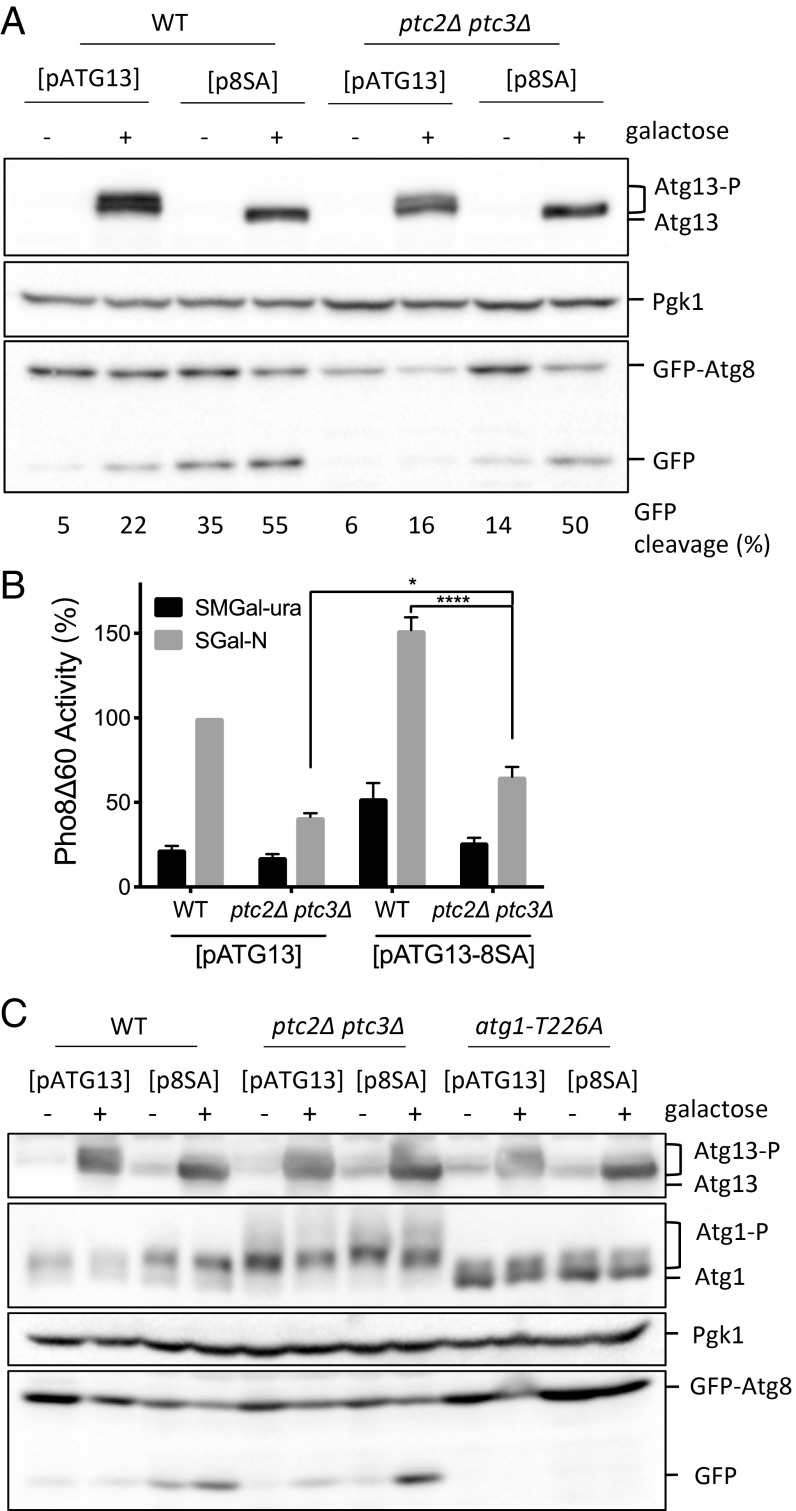
The Autophagy Defect of the ptc2Δ ptc3Δ Cells Is Partially Rescued by Expression of Constitutively Active Atg13.
Overexpression of the constitutively active ATG13-8SA is sufficient to induce autophagy that is uncoupled from TORC1 activity (15). We found that the overexpression of the constitutively active ATG13-8SA allele in a PP2C phosphatase-null strain also caused the induction of autophagy at near-WT levels as measured by the GFP-Atg8 cleavage assay (Fig. 6A). Moreover, Pho8Δ60 activity was significantly higher in a ptc2Δ ptc3Δ mutant overexpressing ATG13-8SA compared with the ptc2Δ ptc3Δ mutant alone (Fig. 6B). Overexpression of constitutively active ATG13-8SA failed to reduce the hyperphosphorylation of Atg1 in ptc2Δ ptc3Δ cells (Fig. 6C). Moreover, the overexpression ATG13-8SA in the kinase-defective atg1-T226A background did not result in autophagy induction, in agreement with the previously published results (Fig. 6C) (15). Therefore, we conclude that the impaired autophagy activity in the ptc2Δ ptc3Δ mutant strain is not the result of an inherent defect in Atg1’s kinase activity but is correlated with the failure to properly dephosphorylate Atg13.
Fig. 6.
ATG13-8SA overexpression rescues the autophagy defect of the ptc2Δ ptc3Δ mutant. (A) Overexpression of ATG13-8SA in WT and ptc2Δ ptc3Δ cells. Strains containing WT ATG13 or ATG13-8SA centromeric plasmids under galactose-inducible promoters were grown and induced with 2% galactose for 4 h. Samples were collected and blotted with anti-Atg13 antibody, anti-Pgk1 antibody, and anti-GFP antibody. The percentage of GFP-Atg8 processing for every lane is indicated below the corresponding lane. (B) Pho8Δ60 activity in WT and phosphatase-null strains carrying WT ATG13 or ATG13-8SA plasmids under galactose-inducible promoters from three independent experiments (*P < 0.05 and ***P < 0.0001). (C) Atg1 hyperphosphorylation and GFP-Atg8 processing after the overexpression of WT ATG13 or ATG13-8SA in WT, ptc2Δ ptc3Δ, and atg1-T226A strains. FLAG-Atg1 strains carrying the WT ATG13 or ATG13-8SA galactose-inducible plasmids were grown and induced as previously described.
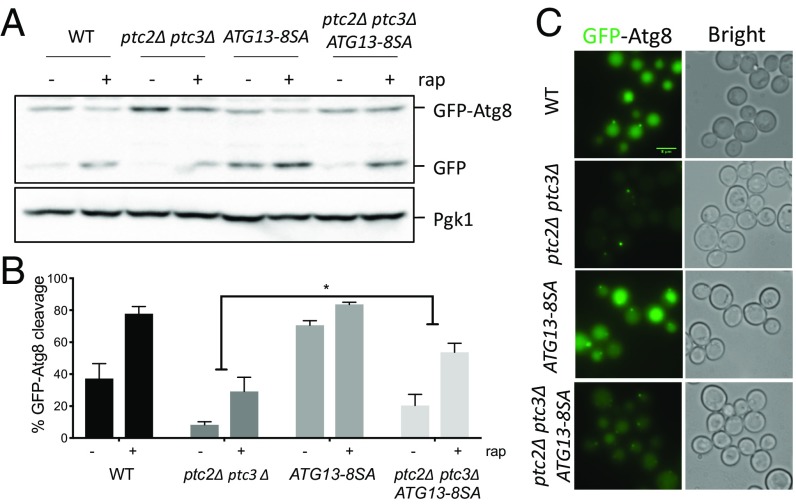
The overexpression of WT ATG13 exacerbates the activating autophosphorylation of Atg1T226A (46), indicating that Atg13 abundance itself might regulate autophagy. We also observe an increased autophagic flux in a WT strain after ATG13 overexpression (Fig. 6 A and C). To avoid overexpression, we introduced the ATG13-8SA allele at its chromosomal locus, expressed under its endogenous promoter. As expected, the ATG13-8SA strain showed increased levels of GFP-Atg8 cleavage even before rapamycin treatment (Fig. 7A). The levels of autophagy flux increased only modestly upon rapamycin treatment in this strain, again reinforcing the idea that the TORC1-mediated phosphorylation of Atg13 is the main inhibitory control over autophagy induction. GFP-Atg8 cleavage in the ptc2Δ ptc3Δ ATG13-8SA mutant was significantly higher compared with ptc2Δ ptc3Δ cells, although it did not reach WT levels (Fig. 7B). We also observed an enrichment in GFP localization to the vacuole in the ptc2Δ ptc3Δ ATG13-8SA mutant (Fig. 7C), agreeing with the GFP-Atg8 cleavage assay results. Because endogenous levels of expression of ATG13-8SA only partially rescued the autophagy defect of ptc2Δ ptc3Δ, we conclude that Ptc2 and Ptc3 could regulate other proteins in the autophagy pathway in addition to Atg13 and Atg1.
Fig. 7.
The genomic ATG13-8SA mutant partially rescues the autophagy defect of the ptc2Δ ptc3Δ mutant strain. (A) GFP-Atg8 processing in strains that carry ATG13-8SA at the endogenous locus after 4-h rapamycin treatment. Samples were collected and blotted as previously described. (B) Quantification of GFP-Atg8 processing in WT, ptc2Δ ptc3Δ, ATG13-8SA, and ptc2Δ ptc3Δ ATG13-8SA strains from three independent experiments. Error bars represent SE (P = 0.04). (C) GFP-Atg8 localization in WT, ptc2Δ ptc3Δ, ATG13-8SA, and ptc2Δ ptc3Δ ATG13-8SA strains 4 h after rapamycin treatment.
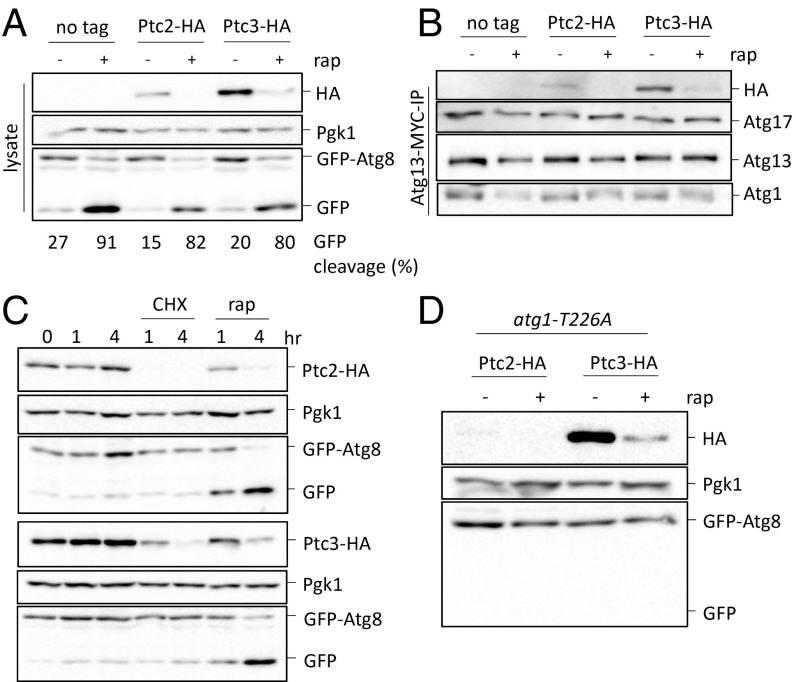
Ptc2 and Ptc3 Interact with Atg13–Atg1.
To determine if Ptc2 and Ptc3 directly interact with the Atg1-Atg13 complex, we created strains carrying Ptc2-3×HA or Ptc3-3×HA epitope-tagged alleles expressed under the proteins’ endogenous promoters in a strain that also carried epitope-tagged Atg13 and Atg1. Both of these strains had levels of GFP-Atg8 cleavage comparable to an untagged control strain (Fig. 8A). After immunoprecipitating Atg13-13×MYC, we monitored the binding of Atg13 to Atg1 and Atg17, as well as to epitope-tagged Ptc2 and Ptc3. We found that both of these phosphatases coimmunoprecipitated with Atg13, Atg1, and Atg17 (Fig. 8B). We also detected low levels of Ptc3-Atg13 binding after autophagy induction; however, because both Ptc2 and Ptc3 levels decreased upon autophagy induction (as detailed later), we were unable to detect Ptc2-Atg13 interaction under these conditions (Fig. 8 A and B). These results demonstrate that the PP2C phosphatases and the Atg1 kinase complex interact in vivo irrespective of autophagy induction.
Fig. 8.
Ptc2 and Ptc3 interact with the Atg1 kinase complex. (A) GFP-Atg8 processing in strains that carry Ptc2-HA or Ptc3-HA. An untagged strain was used as a negative control. Samples were collected 4 h after rapamycin treatment and blotted for HA for Ptc2-HA or Ptc3-HA detection, or with anti-Pgk1 or anti-GFP. Numbers below the lanes indicate the percentage of free GFP band to total GFP signal. (B) Ptc2-Ptc3 interaction with Atg1-Atg13-Atg17 after Atg13-MYC immunoprecipitation. Samples were blotted as previously described. (C) Ptc2-HA and Ptc3-HA abundance after rapamycin or 25 μg/mL CHX treatment. Samples were collected and blotted as previously described. (D) Ptc2-HA and Ptc3-HA abundance in strains that carry atg1-T226A after 4-h rapamycin treatment.
Ptc2 and Ptc3 both significantly decreased in abundance upon rapamycin treatment (Fig. 8A). This could be because these two phosphatases are targeted by autophagy upon rapamycin treatment or because of the inhibition of protein translation initiation by TORC1 deactivation (48). To differentiate between these two possibilities, we measured the abundance of Ptc2 and Ptc3 after CHX treatment. After treatment with CHX at a concentration that does not cause autophagy induction, Ptc2 was rapidly degraded and Ptc3 levels were significantly reduced (Fig. 8C). Moreover, in a strain that carries a kinase-defective atg1-T226A allele, Ptc2 and Ptc3 were still degraded following rapamycin treatment (Fig. 8D). Collectively, these results suggest that Ptc2 and Ptc3 are rapidly turned over and that their degradation is autophagy-independent.
Discussion
We have previously shown that DNA damage induces a targeted autophagy pathway, termed GTA. This pathway is selective and requires all of the core Atg machinery, the scaffold protein Atg11, as well as the essential components of the DDC signaling pathway (27). The PP2C phosphatases Ptc2 and Ptc3 were previously shown to be negative regulators of the DDC response (39, 49). For this reason, we expected an up-regulation of GTA response in the absence of these phosphatases. However, we observe a significant down-regulation of GTA and rapamycin-induced macroautophagy in the ptc2Δ ptc3Δ mutant, suggesting that Ptc2 and Ptc3 redundantly promote autophagy. Our findings also indicate that Ptc2 and Ptc3 promote the Cvt pathway. In the absence of these phosphatases, we detect an accumulation of abnormally large prApe1 structures, which might hinder the delivery of prApe1 to the vacuole. Because we ruled out the possibility of Ptc2 and Ptc3 acting directly on TORC1, we conclude that Ptc2 and Ptc3 modulate the Atg machinery downstream of TORC1.
To identify the targets of Ptc2 and Ptc3 among the Atg machinery, we turned our attention to the Atg1 kinase complex. In the absence of Ptc2 and Ptc3, we detect high-mobility Atg1 species irrespective of TORC1 status, which are dependent on Atg1’s kinase activity, as well as Atg11 and Atg13. Based on these results, it appears that the binding among Atg1, Atg13, and Atg11 stimulates the kinase activity of Atg1 (7), leading to extensive autophosphorylation of Atg1, which is counteracted by Ptc2 and Ptc3. Interestingly, the aberrant hyperphosphorylated Atg1 species in ptc2Δ ptc3Δ mutants are not dependent on the Atg19-prApe1 cargo complex, suggesting that Atg11 can stimulate Atg1’s kinase activity independently of the Atg19-prApe1 complex. Atg11 can serve as a scaffold for other selective autophagy pathways such as pexophagy, which requires Atg36 as a receptor (50). Therefore, the increase in Atg1 autophosphorylation seen in the absence of PP2C phosphatases could be caused by the stimulation of Atg1 kinase by an Atg11-cargo complex other than Atg11-Atg19-prApe1. Supporting this, recent work showed that Atg1’s kinase activity can be stimulated by Atg11 in an Atg36-dependent manner (7).
Atg13, an essential component of the core autophagy machinery, is kept in a hyperphosphorylated and inhibited state under nutrient-replete conditions, and its dephosphorylation is critical for autophagy induction. Our data show that, in the absence of Ptc2 and Ptc3, Atg13 is hyperphosphorylated irrespective of the TORC1 status, indicating that Ptc2 and Ptc3 are involved in dephosphorylation of Atg13. In agreement with this, the overexpression of the constitutively active ATG13-8SA allele bypasses the autophagy defect seen in the ptc2Δ ptc3Δ strain, whereas a single chromosomal ATG13-8SA allele partially rescues the autophagy defect of ptc2Δ ptc3Δ cells. These results illustrate that, although the autophagy defect observed in the absence of Ptc2 and Ptc3 is mainly the result of failure to dephosphorylate Atg13, additional phosphorylation sites on Atg13 or other Atg proteins are likely to be regulated by Ptc2 and Ptc3. Indeed, the phosphorylation of Atg13 at S429, which is not one of the eight TORC1 target sites, significantly reduces Atg13-Atg17 binding and impairs autophagy function (21). Therefore, it is possible that Atg13 S429 is another phosphorylation site on Atg13 that is regulated by Ptc2 and Ptc3.
PAS organization is a highly regulated process. Previous work demonstrated that the localization of the scaffold protein Atg17 to the PAS is a prerequisite for Atg13 and Atg1 recruitment (22). Although Atg1-Atg13 binding is intact despite the accumulation of aberrant Atg1 species, we show that Atg13-Atg17 binding is impaired in the absence of Ptc2 and Ptc3 phosphatases. In agreement with this finding, the localization of Atg1 and Atg13 to the PAS in the ptc2Δ ptc3Δ mutant is impaired under nutrient-deplete and nutrient-replete conditions. This defect could be caused by the regulation of Atg17 directly by Ptc2 and Ptc3 phosphatases or a defect in Atg13-Atg17 binding resulting from the accumulation of hyperphosphorylated Atg13 species observed in the ptc2Δ ptc3Δ strain as discussed earlier.
Our coimmunoprecipitation results show that Ptc2 and Ptc3 phosphatases interact with the Atg1-Atg13 complex. The rapid turnover of PP2C phosphatases suggest that the reactivation of TORC1 when the nutrient starvation is over could lead to a rapid hyperphosphorylation of Atg13 and other targets, as Ptc2 and Ptc3 are at very low levels. We note, however, that a recent study has suggested that the 3×HA epitope tag we used to study Ptc2 and Ptc3 may generally destabilize proteins (51).
Our findings suggest a previously uncharacterized role for the promiscuous Ptc2 and Ptc3 phosphatases in autophagy. Ptc2 and Ptc3 are positive regulators of macroautophagy, as well as at least two selective autophagy pathways. Ptc2 and Ptc3 are directly involved in the dephosphorylation of Atg13 and Atg1 to stimulate Atg1-Atg13-Atg17 complex formation. In the absence of Ptc2 and Ptc3, the proper PAS formation and Atg13-Atg17 binding are impaired. The autophagy defect of the phosphatase-null strain can be rescued by the overexpression of the constitutively active ATG13-8SA allele or partially rescued by expressing ATG13-8SA under its endogenous promoter, suggesting that additional Atg proteins could be involved in the modulation of autophagy by Ptc2 and Ptc3 phosphatases.
Materials and Methods
Yeast Media, Strains, and Plasmids.
Saccharomyces cerevisiae strains used in this study are derivatives of JKM179 or BY4741 (SI Appendix, Table S1). Plasmids used in the study are listed in SI Appendix, Table S2. Deletions of ORFs were created by using one-step PCR homology cassette amplification and transformed by using high-efficiency yeast transformation methods (52, 53). The primer sequences used for this study are indicated in SI Appendix, Table S3. Atg1 point mutants were created by using CRISPR-Cas9 as previously described (54). The sequences of the single-stranded oligonucleotide DNA templates used to create CRISPR-Cas9–mediated point mutants are indicated in SI Appendix, Table S3, and the CRISPR-Cas9 plasmids are indicated in SI Appendix, Table S2. The 2×FLAG-Atg1 in the Atg13-13×MYC strain was created by targeting the N terminus of Atg1 with CRISPR-Cas9 and using a PCR-amplified fragment as template. N-terminal mCherry-tagged prApe1 and N-terminal 10×MYC-tagged Atg13 were created by targeting the N terminus of the ORFs with CRISPR-Cas9. The Atg13-13×MYC, Ptc2-3×HA, and Ptc3-3×HA epitope-tagged strains were created by transforming cells with PCR-amplified cassettes that contain homology to the Atg13, Ptc2, and Ptc3 C termini, respectively (55). The GFP-Atg8, GAL-ATG13, GAL-ATG13-8SA, mCherry-Ape1, and 2×GFP plasmids were a gift from Yoshiaki Kamada (National Institute of Basic Biology, Okazaki, Japan) (15). The 2×FLAG-Atg1 VDY630 strain was a gift from Vladimir Denic (Harvard University, Cambridge, MA) (7). C-terminal 2×GFP tagging of the Atg1, Atg11, Atg13, and Atg17 was performed as previously described (56). Yeast strains were grown in 1% yeast extract, 2% peptone, 2% dextrose (YEPD) or in 1% yeast extract, 2% peptone containing 3% lactic acid (YEP-lac).
GFP-Atg8 Processing Assay.
Cells were grown in YEPD overnight, washed with YEP-lac, and inoculated into YEP-lac until they reached exponential phase. For galactose induction experiments, cells grown in YEP-lac were treated with a final concentration of 2% galactose. MMS was added at a final concentration of 0.04% to induce GTA, and rapamycin at a final concentration of 200 ng/mL was used to induce macroautophagy.
Pho8Δ60 Assay.
The Pho8Δ60 assay was performed as described previously (41). Cells were grown in YEPD and shifted to starvation conditions for 4 h. For the ATG13 overexpression assays, cells were grown in synthetic minimal medium containing dextrose (SMD)-lacking Ura to retain the ATG13 overexpression plasmids and shifted to synthetic minimal medium containing galactose (SM-GAL)-lacking Ura for 4 h.
Sample Collection and Western Blotting.
Cells were harvested from 50 mL of the early exponential-phase culture (1–10 × 106 cells per milliliter) by centrifugation. Cell pellets were washed in 1 mL of 20% TCA and quick-frozen on dry ice. TCA protein extraction was carried out as previously described (27). Affinity-isolation experiments were carried out as previously described (21) except IGEPAL-CA (Sigma-Aldrich) was used as the nonionic detergent and Halt protease inhibitor (Thermo Fisher Scientific) was used as a protease inhibitor. Cell lysis was performed by vortexing with acid-washed glass beads. The crude extract was collected and cleared by centrifugation at maximum speed twice for 15 min at 4 °C. An equal volume of the cleared lysate was mixed with protein G-agarose beads (Roche) preincubated with anti-MYC antibodies. After the overnight incubation, the beads were washed twice with 1 mL lysis buffer containing protease inhibitors, and the samples were eluted in 100 μL of 6× Laemmli buffer containing 0.9% β-mercaptoethanol by boiling at 95 °C for 5 min. Samples were run on 6% gels for the detection of hyperphosphorylated Atg1 and Atg13 species, 7% gels for Sch9 and Ape1 detection, and 10% gels for GFP-Atg8, Pgk1, and Atg13 overexpression assays. Blotting was performed by using anti-GFP antibody (ab-6556; Abcam), anti-FLAG antibody (A2220; Millipore Sigma), anti-Pgk1 antibody (ab113687; Abcam), anti-MYC antibody (9E10; Abcam), anti-HA antibody (HA.C5; Abcam), anti-Ape1 antibody (44), anti-Atg13 antibody (57), and anti-Sch9 antibody. Anti-Atg17 antibody was a gift from Yoshinori Ohsumi (Tokyo Institute of Technology, Tokyo) (11). Anti-Sch9 antibody was a gift from Robbie Loewith (University of Geneva, Geneva) (45). Membranes were developed by using Amersham ECL Prime Western Blotting Detection Reagent and visualized by using a Bio-Rad Gel Doc XR+ gel documentation system. ImageLab software (Bio-Rad) was used to crop and quantify the blots.
Fluorescence Microscopy.
Cells were grown in YEP-lac overnight and treated with MMS and rapamycin as described previously (27). Strains that contain mCherry-Atg8 plasmids were grown in leucine dropout media with 2% dextrose. Cells were harvested by centrifugation in an Eppendorf clinical centrifuge, model 5424, at 3,000 rpm for 1 min and then washed with synthetic complete media before imaging. The images were taken with a Nikon Eclipse E600 microscope and processed with Fiji software. Cells containing Atg1, Atg11, Atg13, and Atg17 foci were counted manually. For each condition, more than 150 cells were quantified.
Statistical Analysis.
Statistical analysis was performed by using Prism 7 software (GraphPad). Statistical significance was calculated by using two-way ANOVA corrected with Dunnett’s comparison test for multiple comparisons. The P values calculated are indicated in the figure legends. Error bars represent SE. For GFP-Atg8 and Pho8Δ60 assays, the data were normalized to WT.
Supplementary Material
Acknowledgments
We thank Dr. Eric Baehrecke for his insightful feedback on an earlier version of the manuscript and Dr. Vladimir Denic for useful discussions. This work was supported by National Institutes of Health Grants GM61766 and GM127029 (to J.E.H. and G.M.) and GM053396 (to D.J.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817078116/-/DCSupplemental.
References
- 1.Wen X, Klionsky DJ. An overview of macroautophagy in yeast. J Mol Biol. 2016;428:1681–1699. doi: 10.1016/j.jmb.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farré JC, Subramani S. Mechanistic insights into selective autophagy pathways: Lessons from yeast. Nat Rev Mol Cell Biol. 2016;17:537–552. doi: 10.1038/nrm.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamber RA, Shoemaker CJ, Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol Cell. 2015;59:372–381. doi: 10.1016/j.molcel.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabeya Y, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheong H, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamada Y, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 17.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and cAMP-dependent protein kinase signaling pathways coordinately control autophagy in Saccharomyces cerevisiae. Autophagy. 2010;6:294–295. doi: 10.4161/auto.6.2.11129. [DOI] [PubMed] [Google Scholar]
- 18.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft C, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh YY, Shah KH, Herman PK. An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J Biol Chem. 2011;286:28931–28939. doi: 10.1074/jbc.M111.250324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujioka Y, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21:513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 23.Kabeya Y, et al. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 24.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papinski D, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dotiwala F, et al. DNA damage checkpoint triggers autophagy to regulate the initiation of anaphase. Proc Natl Acad Sci USA. 2013;110:E41–E49. doi: 10.1073/pnas.1218065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eapen VV, et al. A pathway of targeted autophagy is induced by DNA damage in budding yeast. Proc Natl Acad Sci USA. 2017;114:E1158–E1167. doi: 10.1073/pnas.1614364114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eapen VV, Haber JE. DNA damage signaling triggers the cytoplasm-to-vacuole pathway of autophagy to regulate cell cycle progression. Autophagy. 2013;9:440–441. doi: 10.4161/auto.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JA, Hicks WM, Li J, Tay SY, Haber JE. Protein phosphatases pph3, ptc2, and ptc3 play redundant roles in DNA double-strand break repair by homologous recombination. Mol Cell Biol. 2011;31:507–516. doi: 10.1128/MCB.01168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Neill BM, et al. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc Natl Acad Sci USA. 2007;104:9290–9295. doi: 10.1073/pnas.0703252104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun LL, et al. Protein phosphatase Pph3 and its regulatory subunit Psy2 regulate Rad53 dephosphorylation and cell morphogenesis during recovery from DNA damage in Candida albicans. Eukaryot Cell. 2011;10:1565–1573. doi: 10.1128/EC.05042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keogh MC, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 33.Maeda T, Tsai AY, Saito H. Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5408–5417. doi: 10.1128/mcb.13.9.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young C, Mapes J, Hanneman J, Al-Zarban S, Ota I. Role of Ptc2 type 2C Ser/Thr phosphatase in yeast high-osmolarity glycerol pathway inactivation. Eukaryot Cell. 2002;1:1032–1040. doi: 10.1128/EC.1.6.1032-1040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warmka J, Hanneman J, Lee J, Amin D, Ota I. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol Cell Biol. 2001;21:51–60. doi: 10.1128/MCB.21.1.51-60.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welihinda AA, Tirasophon W, Green SR, Kaufman RJ. Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol Cell Biol. 1998;18:1967–1977. doi: 10.1128/mcb.18.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 38.Sharmin D, Sasano Y, Sugiyama M, Harashima S. Effects of deletion of different PP2C protein phosphatase genes on stress responses in Saccharomyces cerevisiae. Yeast. 2014;31:393–409. doi: 10.1002/yea.3032. [DOI] [PubMed] [Google Scholar]
- 39.Leroy C, et al. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol Cell. 2003;11:827–835, and correction (2003) 11:1119. doi: 10.1016/s1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 40.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 42.Kirisako T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torggler R, Papinski D, Kraft C. Assays to monitor autophagy in Saccharomyces cerevisiae. Cells. 2017;6:E23. doi: 10.3390/cells6030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urban J, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 46.Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185:871–882. doi: 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh YY, et al. The identification and analysis of phosphorylation sites on the Atg1 protein kinase. Autophagy. 2011;7:716–726. doi: 10.4161/auto.7.7.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbet NC, et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guillemain G, et al. Mechanisms of checkpoint kinase Rad53 inactivation after a double-strand break in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3378–3389. doi: 10.1128/MCB.00863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saiz-Baggetto S, Méndez E, Quilis I, Igual JC, Bañó MC. Chimeric proteins tagged with specific 3xHA cassettes may present instability and functional problems. PLoS One. 2017;12:e0183067. doi: 10.1371/journal.pone.0183067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 53.Amberg DC, Burke DJ, Strathern JN. High-efficiency transformation of yeast. CSH Protoc. 2006;2006:pdb.prot4145. doi: 10.1101/pdb.prot4145. [DOI] [PubMed] [Google Scholar]
- 54.Anand R, Memisoglu G, Haber J. 2017 Cas9-mediated gene editing in Saccharomyces cerevisiae. Protocol Exchange. Available at https://www.nature.com/protocolexchange/protocols/5791. Accessed December 1, 2018.
- 55.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 56.Li D, et al. A fluorescent tool set for yeast Atg proteins. Autophagy. 2015;11:954–960. doi: 10.1080/15548627.2015.1040971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller-Fleming L, Cheong H, Antas P, Klionsky DJ. Detection of Saccharomyces cerevisiae Atg13 by western blot. Autophagy. 2014;10:514–517. doi: 10.4161/auto.27707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.