Significance
Epidemics of community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) are of growing medical concern. To understand the emergence of virulence and antimicrobial resistance, both of which promote CA-MRSA spread, we examined an on-going disease cluster within an enclosed community by analyzing the genome sequences of CA-MRSA clones characterized by high prevalence and a profound persistence. Metabolic adaptation and a phage primed the clone for success, and then a fully optimized variant was created by selection of plasmid-mediated biocide resistance. The data provide mechanistic insight and indicate that high-risk populations are incubators for evolution of consequential phenotypes. Immediate interruption of this evolutionary pattern is essential for forestalling dissemination of resistance from high-risk communities to hospitals and the general population.
Keywords: MRSA, evolution, antimicrobial resistance, virulence
Abstract
The past two decades have witnessed an alarming expansion of staphylococcal disease caused by community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA). The factors underlying the epidemic expansion of CA-MRSA lineages such as USA300, the predominant CA-MRSA clone in the United States, are largely unknown. Previously described virulence and antimicrobial resistance genes that promote the dissemination of CA-MRSA are carried by mobile genetic elements, including phages and plasmids. Here, we used high-resolution genomics and experimental infections to characterize the evolution of a USA300 variant plaguing a patient population at increased risk of infection to understand the mechanisms underlying the emergence of genetic elements that facilitate clonal spread of the pathogen. Genetic analyses provided conclusive evidence that fitness (manifest as emergence of a dominant clone) changed coincidently with the stepwise emergence of (i) a unique prophage and mutation of the regulator of the pyrimidine nucleotide biosynthetic operon that promoted abscess formation and colonization, respectively, thereby priming the clone for success; and (ii) a unique plasmid that conferred resistance to two topical microbiocides, mupirocin and chlorhexidine, frequently used for decolonization and infection prevention. The resistance plasmid evolved through successive incorporation of DNA elements from non-S. aureus spp. into an indigenous cryptic plasmid, suggesting a mechanism for interspecies genetic exchange that promotes antimicrobial resistance. Collectively, the data suggest that clonal spread in a vulnerable population resulted from extensive clinical intervention and intense selection pressure toward a pathogen lifestyle that involved the evolution of consequential mutations and mobile genetic elements.
Community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) strains have dramatically increased the global burden of S. aureus infections. CA-MRSA consists of multiple lineages; however, specific geographic regions are usually dominated by a single subclone, with different subclones present in different regions. For example, the pandemic sequence type USA300 is dominant in the United States (1–4). Among the well-known problems caused by USA300 are outbreaks of skin abscesses in high-risk communities (e.g., jails and daycare facilities) (5). Despite considerable research, how epidemic strains of CA-MRSA become established in different settings, especially community settings, is poorly understood.
Although epidemiological risk factors, such as human activities that increase infection risk, are often the primary trigger for clonal spread of pathogens, genome-wide surveys of pathogen populations promise insight into the evolutionary processes and the genetic basis underlying the emergence of successful lineages (6–12). Indeed, genomic comparisons between successful CA-MRSA clones and distantly related strains have provided broad insight into genome evolution within S. aureus (13–17). For example, genome-wide studies have described the evolutionary history and spread of successful pandemic clones and have defined the contributions of mutation and recombination to genetic variation in the species (reviewed in ref. 18). They have also pinpointed syndrome-specific elements, such as the prophage-encoded Panton-Valentine leukocidin, that are associated with skin and soft tissue infections and that promote transmission (19). Collectively, prior work creates a framework upon which evolution studies can investigate adaptations underpinning the emergence of clones of public health concern within a background of closely related competing strains. Identification of such adaptations is key to finding new therapeutic directions, since the spectrum of adaptive changes that arises during the course of CA-MRSA spread is likely to identify genetic pathways critical for bacterial pathogenesis in vivo.
Human populations having above-average risk of infection (e.g., due to differences in exposure to infection and/or defects in preexisting immunity) are likely important for the initial spread of epidemic CA-MRSA clones. The present report describes a prolonged, ongoing spread of a USA300 clone causing infections in a high-risk community in Brooklyn, New York. Prolonged pathogen circulation represents a unique opportunity for understanding how CA-MRSA adapts to both clinical intervention and to unique aspects of bacterial pathophysiology underlying heightened infection risk. We used whole-genome sequencing of 86 isolates obtained from infected children and adults over a 2-y period, followed by comparative analyses, to reconstruct the evolutionary steps that led to the emergence of an adapted MRSA strain in a previously unrecognized high-risk population. For convenience, we call the adapted strain the USA300-Brooklyn variant (USA300-BKV). We found distinct single-nucleotide polymorphisms (SNPs) and larger structural changes that help explain the success of the USA300-BKV clone. Phenotypic analyses further showed how genetic alterations can affect virulence and antibiotic resistance. The evolution of the USA300-BKV clone supports the hypothesis that human exposures and selection pressures that occur in populations at high risk for infection result in dangerous bacterial adaptation. The study underscores the need for rapid pathogen containment in such situations.
Results
High Prevalence of MRSA Infection Among Pediatric Patients from an Orthodox Jewish Community.
Clinical reports of increasing numbers of CA-MRSA infections in our hospital among pediatric Orthodox Jewish children from Brooklyn, New York, led us to investigate the risk of MRSA infection among Orthodox Jewish patients. From May 2015 to December 2016, we identified 4,368 children aged 0 to 18 y who were admitted to our pediatric general and intensive care units (ICUs). Community members were identified on the basis of postal (zip) code, a surrogate for Orthodox-predominant neighborhoods in Brooklyn (20). Medical record review, using clinical criteria plus isolation of S. aureus from a normally sterile body site, was used to distinguish infection from colonization. The MRSA infection rate per 1,000 admissions was 10-fold higher among children from Orthodox-associated zip codes than that from other zip codes in New York City (80.2 vs. 8.1, P < 0.001) (SI Appendix, Table S1). We also determined the prevalence of MRSA colonization upon admission for the general pediatric ward and ICU. Nasal and throat swabs were collected upon admission for 451 patients. Colonization by MRSA was twofold higher in children from high-risk (Orthodox-associated) zip codes as compared to children admitted from all other zip codes, labeled “low-risk” [7% (10/127) vs. 3% (11/324), P = 0.04]. No difference was observed in methicillin-susceptible S. aureus colonization prevalence or in risk factors predisposing for MRSA acquisition. To our knowledge, correlations between the Orthodox Jewish community and risk of MRSA infection have not been previously described. The clinical characteristics of the subjects studied are described in detail elsewhere. Whether common sources serve as a reservoir for MRSA acquisition and, potentially, the spread of the USA300-BKV clone remains to be determined.
Phylogeny Indicates Spread of a Clone of CA-MRSA Strain USA300 in the Community.
We collected consecutive, single-patient isolates of MRSA from infected community members residing in Orthodox-associated zip codes during a 2-y period of medical record review (clinical characteristics and antimicrobial susceptibilities are described in Dataset S1). Ninety-two MRSA isolates from community members were obtained; 84 were from children (42 females) who ranged in age from <1 mo to 18 y, and eight isolates were from adults (four females). The mean age of children in the study was 2.9 ± 3.8 y; 71 patients presented with skin abscesses. Molecular typing results indicated that all isolates were a clone of the epidemic CA-MRSA strain USA300 [spa type t008 or a related repeat type, staphylococcal cassette chromosome mec type IV (SCCmecIV), arginine catabolic mobile genetic element (ACME), and pvl positive; these characteristics define USA300 clones (21, 22)] (Dataset S1). Thus, the USA300 clone was associated predominantly with skin infections, consistent with data indicating that the USA300 clone is currently the most frequent cause of purulent skin infection in US emergency departments (2).
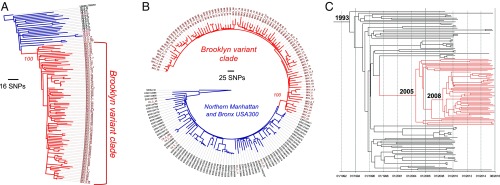
To establish whether, and to what extent, infections reflect dissemination of a specific USA300 subclone, we used whole-genome sequencing with mapping of the individual genome sequences against the reference MRSA strain, USA300_FPR3757 (NC_007793.1). Phylogenetic reconstruction was used to compare the 92 community isolates with 16 USA300 control isolates obtained from patients residing in low-risk zip codes (Dataset S1). We observed that 93% (86/92) of isolates from Orthodox community patients (USA300-BKV isolates) clustered within a unique clade, consistent with dissemination of a clone. On average, USA300-BKV isolates differed from one another by 79 SNPs, ranging from 1 to 144 SNPs. None of the control isolates was found within the clade (Fig. 1A).
Fig. 1.
Bacterial phylogeny reveals the emergence and spread of a dominant clone (USA300-BKV) in the Orthodox Jewish community. Maximum-likelihood phylogenetic trees of 92 isolates obtained from patients residing in Orthodox-associated zip codes (shown in red) compared with (A) 16 representative USA300 isolates from adults and children in the same hospital or (B) 68 USA300 strains from northern Manhattan and the Bronx. Ninety-three percent (86/92) of isolates from patients residing in Orthodox-associated zip codes clustered within a unique clade (USA300-BKV; BKV clade in red). The remaining six isolates from patients residing in high-risk zip codes clustered with contemporary isolates from our hospital and isolates from northern Manhattan and the Bronx. The trees are rooted using USA300_FPR3757 and the distantly related S. aureus isolate MRSA131 as outgroups. Bootstrapping value for the USA300-BKV clade divergence branch is indicated. (C) Bayesian phylogenetic reconstruction of BKV isolates estimated from core genome mutations. USA300-BKV clade branches are highlighted in red. Predicted dates of divergence from a most recent common ancestor are indicated.
To determine the genomic relationship between USA300-BKV isolates and other USA300 strains in New York City, we compared high-risk genomes with 68 USA300 strains that were collected in a comprehensive community-based study of MRSA transmission in northern Manhattan and the Bronx (Fig. 1B) (23, 24). Representatives of all phylogenetic subclades in the parent study were included in the analysis, capturing most of the global diversity of the USA300 lineage. Remarkably, none of these New York City USA300 strains clustered with the USA300-BKV subclade (Fig. 1B).
With the same dataset, we used a bayesian approach and the collection dates of each strain as calibration points to estimate the date of origin of the most recent common ancestor of the USA300-BKV cluster (Fig. 1C). As in the study of USA300 strains in northern Manhattan mentioned above (24), we predicted the date of divergence of the USA300 epidemic lineage from its most recent common ancestor to be around 1993. We estimated the divergence of the USA300-BKV subclade to have occurred much more recently, around 2005. Additionally, a second branching event within the clade occurred in 2008 (Fig. 1C). The nucleotide substitution rate among USA300-BKV isolates was similar to the rate of the broader USA300 lineage (24) (1.41 × 10−6 substitutions per site per year; 95% CI, 7.48 × 10−7, 2.09 × 10−6). Thus, mutation rates were not elevated in the USA300-BKV strains.
A Mutation in pyrR Enhances the Fitness of USA300-BKV Clones.
Adaptive mutations that may have contributed to the initial success of the USA300-BKV clone should appear in all isolates of the clone but be absent from non-USA300-BKV–associated isolates. USA300-BKV isolates shared 20 unique nonsynonymous SNPs and a stop-gain mutation affecting genes whose products were primarily involved in metabolism of amino acids (aroD, arcB, metK) and carbohydrates (licR, melR, glvC, setC, ddh, pyc) (SI Appendix, Table S2 and Dataset S2). Four mutated genes (licR, glvC, arcB, and fadA) have been shown to be negatively regulated through carbon catabolite repression (25–28). Carbon catabolite repression allows bacteria to preferentially utilize rapidly metabolizable carbon sources (e.g., glucose), thereby increasing fitness (29).
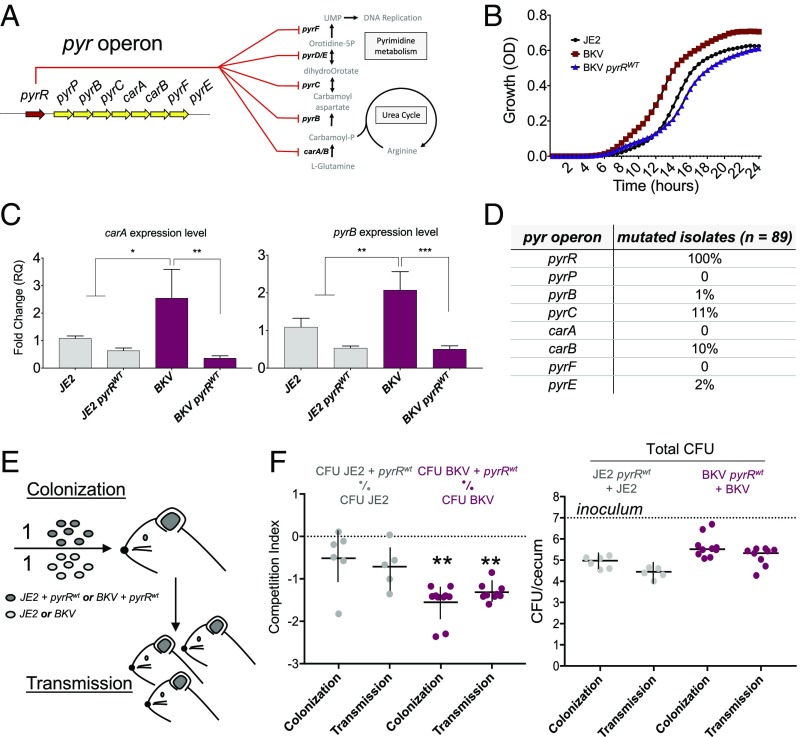
Mutation of the regulator of the pyrimidine nucleotide biosynthetic operon (pyrR) was particularly interesting. Inactivation of pyrR, a transcription repressor that responds to levels of uracil, results in up-regulation of carbamoyl phosphate synthetase (carAB). S. aureus carries only one form of carAB, which is essential for both arginine and pyrimidine biosynthesis (Fig. 2A; reviewed in ref. 30). When the sequences of pyrR orthologs were aligned, the mutation occurred near conserved residues previously identified by mutagenesis of Bacillus subtilis as inactivating pyrR, suggesting that the pyr mutation was inactivating (31, 32). Accordingly, we tested for functional effects using clinical pyr mutant strains and assayed growth in chemically defined media containing uracil (Fig. 2B). For this measurement, we employed a USA300-BKV clinical clone complemented with a chromosomally integrated wild-type (WT) pyrR. The pyrR mutation enhanced growth of the clinical isolate compared with the complemented clone, suggesting that mutation of pyrR increased fitness in energy-poor environments. Moreover, real-time quantitative PCR showed that the pyrR mutation enhanced transcription of pyr operon genes carA and pyrB (Fig. 2C), indicating inactivation of PyrR-mediated transcription repression.
Fig. 2.
A single mutation in pyrR affects S. aureus fitness in vitro and in vivo. (A) Schematic of pyrR regulation of S. aureus pyrimidine biosynthetic and urea pathways through pyr operon gene repression. (B) Growth curves of USA300-BKV isolates (BKV isolates) and pyrR-complemented BKV isolates in pyrimidine-limited chemically defined medium. (C) Real-time qPCR validation of the impact of pyrR mutation on pyr operon genes carA and pyrB. Gene expressions were normalized to pyrR gene. Data represent mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 using ANOVA with multiple t test comparisons. (D) Number of unique mutations targeting pyr genes in independent BKV-isolate genomes. (E) Schematic of the mouse model of colonization and transmission. Parental mice were orally inoculated with a 1:1 ratio of 107 cfu of USA300 strain JE2 + pyrRWT vs. JE2 (a control strain of USA300) or strain USA300-BKV + pyrRWT vs. BKV isolate. Ceca were harvested after 4 d of colonization. (F) Competitive colonization and transmission assay (competitive index, Left) and quantification of bacteria in stool (cfu, Right) were determined from the cecum of inoculated (colonization) and cohoused pups (transmission). Transmission indicates quantification of bacteria in the stool of uninoculated mice that were cohoused with inoculated mice. Median values are shown, and each symbol is the CI from one mouse (Left) or cfu from single mouse (Right). **P < 0.01 by Wilcoxon signed-rank tests.
Two additional observations suggested that the pyr mutation was biologically important. First, unique nonsynonymous mutations in other members of the pyr operon occurred in independent descendants of clones containing the USA300-BKV–associated pyrR mutation (Fig. 2D). Independent mutations in the same gene or operon evolving in parallel within individual patients (convergent evolution) suggests compensation for pyr operon overexpression through attenuating mutations. Consistent with this hypothesis, we found no evidence of mutation in pyr operon alleles among strains from control isolates from our hospital and isolates from northern Manhattan and the Bronx. Compensatory mutations may result from functional trade-offs, wherein inactivating mutations in pyrR may enhance fitness in certain vulnerable human populations, while representing a liability to clones in other environments. Second, examination of publicly available genome sequences identified a unique nonsynonymous mutation in pyrR as one of the few mutations common to all strains in an independent outbreak of CA-MRSA skin infections among 12 hospitalized children during a 6-mo outbreak in a postnatal ward (33). We found no other pyrR mutations in the public database. Collectively, these observations support the hypothesis that skin and soft tissue disease clusters exert selective pressure on pyr operon biosynthesis.
Although pyr operon-regulated arginine and pyrimidine biosynthesis is known to be critical for virulence in vivo (34–37), the pyrR mutation associated with the USA300-BKV clone did not significantly enhance virulence in a murine skin infection model in which lesion size and bacterial cfu were measured (SI Appendix, Fig. S1). These observations suggest that the virulence effects of pyrimidine biosynthesis reflect a requirement for a threshold level of expression.
The effects of pyr operon-regulated biosynthesis on commensalism are largely unknown. The majority of USA300-BKV infections in children were purulent skin and soft tissue infections [82% (64/78)] that frequently occurred in the groin/buttock area of children aged <1 y, possibly related to skin breakdown from exposure to wet or soiled diapers. At least three observations support the idea that enteric carriage is highly relevant to the spread of CA-MRSA clones. First, it is known that CA-MRSA frequently colonizes the gastrointestinal tract of infants and that broken skin can serve as a nidus for infection (38, 39). Second, in children with CA-MRSA skin infections, the rectum and perianal skin was the key site of colonization (38–41). Third, recent work involving a combination of a murine model of S. aureus gastrointestinal colonization and field studies in humans identified interactions between the global virulence regulator agr and the gut microbiota as the dominant risk for S. aureus carriage (42). To assess the in vivo impact of the pyrR mutation, we examined colonization by the USA300-BKV clone in a mouse model of gastrointestinal colonization and transmission (Fig. 2 E and F). USA300-BKV isolates showed a significant colonization and transmission advantage in the mouse intestine with respect to the same isolates carrying a WT copy of pyrR (Fig. 2F). Collectively, the data provide experimental evidence for the S. aureus pyr operon being a colonization determinant, as previously seen with Escherichia coli and Salmonella (43, 44). The data also (i) indicate that pyrR mutation confers an advantage to the bacterium during competition with other strains both in culture and in the mammalian intestine, and (ii) suggest that mutation of pyrR enhances commensal rather than pathogenic fitness.
A Mosaic Version of Phage ϕ11 in USA300-BKV Isolates Enhances Abscess Formation.
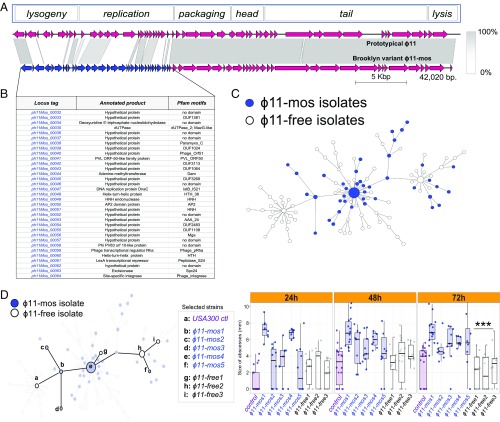
We next examined the possibility that mobile genetic elements contributed to the success of the USA300-BKV clone, beginning with an analysis of phage content. The genomes of half (41/86) of the USA300-BKV isolates contained a 42-kb prophage variant of ϕ11 (NC_004615) (Fig. 3A). Phage ϕ11 belongs to the integrase group Sa5, members of which integrate into an intergenic region in the S. aureus chromosome (45, 46). Sa5 homologs were present infrequently outside the high-risk Orthodox community in New York [16% (62/387)] (24). A mosaic block of 24 genes, which affects the lysogeny and DNA replication modules, contained sequences that are unrelated to ϕ11 or other known prophages; thus, the block might have arisen by a recombination event (Fig. 3 A and B).
Fig. 3.
USA300-BKV isolates (BKV isolates) characterized by the presence of mosaic ϕ11 (ϕ11-mos) demonstrate enhanced virulence in a murine abscess model of infection compared with phage-free control USA300 isolates. (A) Comparative genome map of the prototypical ϕ11 and the BKV isolate-associated ϕ11-mos. Arrows indicate predicted ORFs and the direction of the transcription in phage functional modules. Unique ORFs are indicated in blue. (B) Genes encoding proteins with low homology between the BKV isolate-associated ϕ11-mos and prototypical ϕ11 are indicated. (C) Distribution of ϕ11-mos among BKV isolates (blue). (D) Selected BKV isolates (n = 8) from the minimum spanning tree representing a set of phage-free (n = 3) and ϕ11-mos–containing BKV isolates (n = 5). Size of murine skin lesions [(n = 10) five mice per group with two abscesses per mouse] at 24, 48, and 72 h after s.c. infection with ∼1 × 107 cfu of the indicated strain. Statistical analyses were performed with the Kruskal–Wallis test; ***P < 0.001. The results were corrected for multiple comparisons by using the Bonferroni-corrected threshold.
Examination of the distribution of the mosaic prophage among USA300-BKV isolates showed that it clustered with the proximal portions of the phylogeny, indicating that it was a defining feature at an early stage of the lineage evolution and suggesting that, as with the pyrR mutation, it may have primed the clone for success (Fig. 3C). The absence of the phage from some of the branches of the more distal tips of the phylogeny suggests the ability to reactivate and excise, possibly owing to prophage mobilization during antimicrobial treatment. Indeed, subinhibitory concentrations of antibiotics induce bacteriophage excision and replication (47, 48); moreover, phage excision is frequent among clinical isolates associated with invasive infection (49–52). However, variants that lost the phage often represented a terminal node in phylogenic trees, suggesting that infections are likely selective for the maintenance of mosaic phage.
To determine whether the mosaic phage modulates virulence, we tested for phenotypes in several isolates representative of different phases of USA300-BKV clone evolution based on the phylogeny (Fig. 3D). A laboratory strain of USA300-LAC and a USA300 isolate from our hospital not associated with USA300-BKV were used as controls. All naturally occurring isolates from the high-risk Orthodox population demonstrated indistinguishable exoprotein profiles. Core genome-encoded toxins play an important role in CA-MRSA skin infection (53, 54), and cytotoxicity measurements can be used to determine the potential for CA-MRSA strains to cause disease (55). Cytotoxicity assays indicated that cell-free extracts from cultures of the USA300-BKV strain tended to be highly cytotoxic toward primary human neutrophils, irrespective of the presence or absence of the mosaic phage (SI Appendix, Fig. S2). Thus, in vitro analyses of cytotoxicity toward human neutrophils do not predict the relative virulence of phage-containing strains.
To determine how well in vitro data translate to the in vivo skin environment, we examined eight representative clinical USA300-BKV strains, five of which encoded the phage, in a murine skin abscess model of infection (56, 57). Each of the strains chosen was similarly cytotoxic in vitro. USA300-BKV strains that contained the mosaic phage formed larger abscesses compared with control strains and USA300-BKV strains that lacked the phage (Fig. 3D).
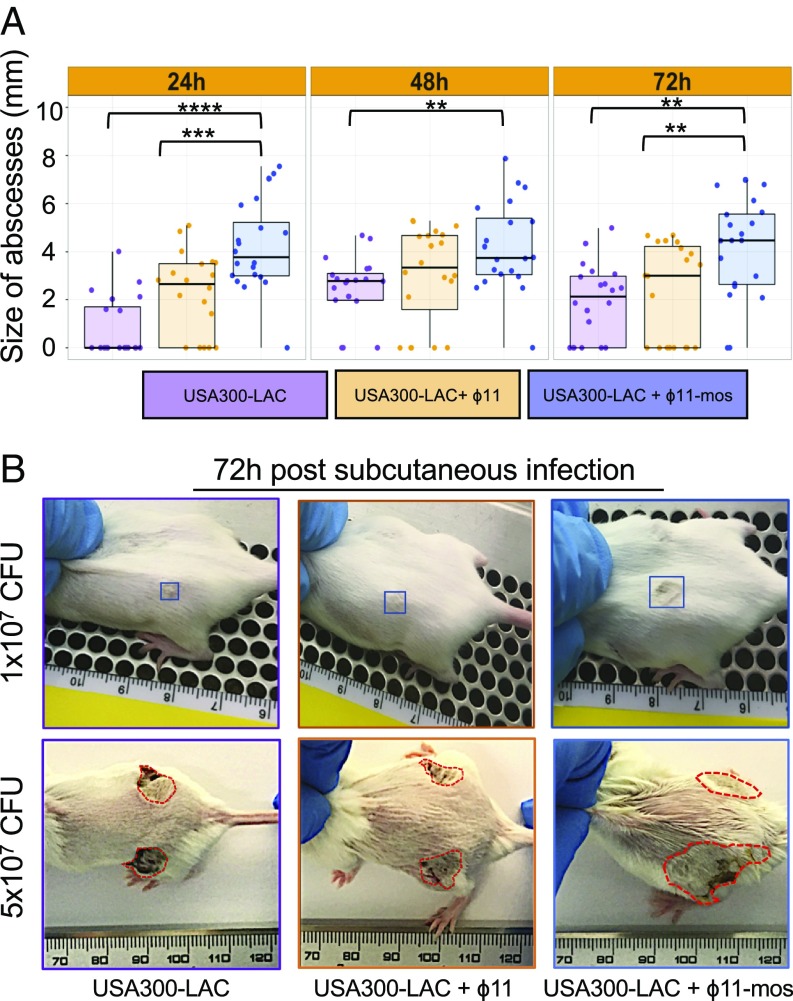
To better understand the contribution of the phage to virulence, we tested for in vivo virulence phenotypes in mosaic phage and WT ϕ11 (NC_004615, from strain RN6734) lysogens derived from strain USA300-LAC. Strikingly, a USA300-LAC strain containing mosaic ϕ11 showed a 50% increase in abscess size over USA300-LAC and USA300-LAC containing the WT ϕ11 (Fig. 4). Testing a higher bacterial inoculum revealed more prominent differences in lesion size, but not in bacterial cfu or dermonecrosis (Fig. 4 and SI Appendix, Figs. S3 and S4), perhaps owing to enhanced proinflammatory and/or cytotoxic properties of the USA300-BKV clone. Based on these data, we conclude that (i) the effect of mosaic ϕ11 was not due to cis-acting effects on genes flanking the phage attachment site (both phages integrated at the same site in the chromosome of USA300-LAC), and (ii) genes encoded within the mosaic portion of the phage enhance virulence. Additional studies are needed to determine to what extent mutation of pyrR, or other polymorphisms specific to the USA-BKV clone, modulate the effect of mosaic ϕ11. Notably, in vitro cytotoxicity failed to correlate with abscess size or phage content, suggesting that the changes that accompany phage-mediated enhanced virulence in the skin require host tissue-specific signals in vivo.
Fig. 4.
The mosaic portion of ϕ11 increases skin abscess size. USA300-LAC lysogens of mosaic ϕ11 [(ϕ11-mos) produced by induction of USA300-BKV_28] and prototypical ϕ11 (produced by induction of RN451), which differ only in the region corresponding to the mosaic block, were made and compared in a murine abscess model of infection. (A) Abscess size at 24, 48, and 72 h after s.c. infection with ∼1 × 107 cfu of the indicated strain [(n = 10) five mice per group with two abscesses per mouse]. Statistical analyses were performed with the Kruskal–Wallis test after multiple comparison correction. (B) Representative pictures of murine skin abscesses at 3 d after s.c. infection with the indicated strain at either a 1 × 107 (Top) or a 5 × 107 cfu (Bottom) inoculum.
Identification of a Staphylococcus Epidermidis-Derived Mupirocin and Chlorhexidine Resistance Plasmid Driving Expansion of USA300-BKV Clones.
DNA contigs derived from de novo DNA sequencing of all USA300-BKV strains were used to query publicly available databases [National Center for Biotechnology Information (NCBI)], resulting in identification of three plasmids. Two plasmids, present in all USA300-BKV isolates, closely matched plasmid pUSA01_ISMMS (CP007177.1), which encodes penicillinase (blaZ) and kanamycin resistance (aphA-3), and pUSA01 (NC_007790), a cryptic plasmid with no clear function. Both plasmids are widely distributed among USA300 clones (58–61).
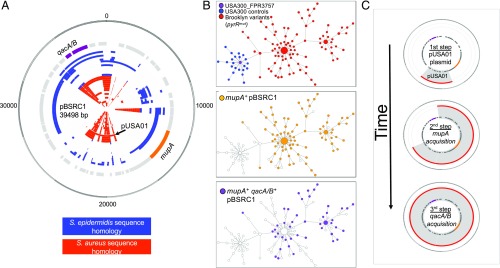
The third plasmid, designated pBSRC1, did not demonstrate homology to any known sequence. To assemble pBSRC1 sequences, two USA300-BKV isolates (BKV_A1 and BKV_A2, Dataset S1) were selected for PacBio RSII long-read sequencing. Plasmid pBSRC1, obtained from USA300-BKV isolate BKV_A2, was ∼38 kb long and contained 46 coding sequences (Fig. 5A) that included the mupA and qacA/B genes, which encode resistance to the widely used topical antimicrobials mupirocin and chlorhexidine, respectively. The pBSRC1 coding sequence matched a mixture of S. aureus and Staphylococcus epidermidis sequences (Fig. 5A).
Fig. 5.
Emergence of a dominant clone coincides with acquisition of an S. epidermidis-recombinant mupirocin resistance plasmid, pBSRC1. (A) Structure and characteristics of pBSRC1. Gene names and location are indicated in the first inner circle (gray). mupA and qacA/B are highlighted in orange and purple, respectively. Other inner circles show perfect BLAST homology results to S. epidermidis (blue) and S. aureus (red) plasmid sequences. The site of S. aureus pUSA01 integration in pBSRC1 is indicated. (B) Minimum spanning tree (MST) based on a patristic genetic comparison of 86 isolates obtained from patients residing in Orthodox-associated zip codes compared with 22 control USA300 isolates from adults and children in the same hospital (16 representative USA300 isolates from patients residing in non–high-risk zip codes, and six USA300 strains from Orthodox-associated zip-codes that were not part of the USA300-BKV clade). The first MST (Top) shows control strains (blue) and the USA300-BKV isolates (red). The second MST (Middle) highlights the distribution of mupA+ pBSRC1 containing strains (orange), whereas the third MST (Bottom) represents the distribution of qacA/B pBSRC1 containing strains (purple). Each node represents a single strain. Distance between nodes is arbitrary, whereas node size is proportional to the number of connections. (C) Working model of the stepwise assembly of pBSRC1 in USA300-BKV isolates.
Transduction of pBSRC1 to recipient strain JE2, a laboratory USA300 strain (CP020619), resulted in high-level resistance to mupirocin [minimal inhibitory concentration (MIC) >1,024 µg/mL] (Dataset S1). High-level mupirocin resistance is frequent among USA300 clades that are associated with pediatric populations in New York City (62). Since no standardized method for testing susceptibility to chlorhexidine exists, resistance is usually defined by the presence of qacA/B, which increases the risk of persistent MRSA carriage after decolonization therapy (63–66). Broth dilution assays demonstrated a twofold MIC increase (from 2 to 4 µg/mL) for chlorhexidine in transductants (Dataset S1), consistent with qacA/B–associated resistance levels reported in previous studies of USA300 strains (67, 68).
We next examined the consequences of plasmid presence in the context of the dissemination of the USA300-BKV clone. We used a network analysis approach based on patristic distances to visualize genetic relationships between each USA300-BKV isolate. Although evolution of the clone began in a linear manner, a dominant variant emerged during dissemination of the USA300-BKV clone within the community (Fig. 5B, Top).
pBSRC1 was identified in most [88% (76/86)] USA300-BKV clones. mupA was always associated with pBSRC1, the presence of which correlated with the emergence of the dominant subclone (Fig. 5B, Middle). qacA/B-containing plasmids were found in 44 isolates (49%) and were always associated with the presence of mupA (Fig. 5B, Bottom and Dataset S1). To date the acquisition of mupA and qac genes, we performed an ancestral reconstruction using a time-scaled analysis based on the maximum parsimony tree (SI Appendix, Fig. S5). The results indicated that primary acquisition of mupA was, for the most part, followed by acquisition of qacA/B. Collectively, these data suggest that the USA300-BKV variants containing pBSRC1, a dual-resistance plasmid, arose during dissemination, became dominant, and accounted for the majority of isolates during the sampling period.
pBSRC1 sequences always included the 3.1-kb cryptic pUSA01 plasmid. Thus, the probable path of evolution of pBSRC1 was sequential recombination of a mosaic plasmid containing elements from S. aureus and S. epidermidis onto an indigenous cryptic plasmid (Fig. 5C).
Discussion
We report the spread of a unique USA300 clone of CA-MRSA that was limited exclusively to an Orthodox Jewish community in Brooklyn, suggesting a previously unrecognized high-risk population. Comprehensive genomic analysis and virulence assays identified a clone-specific prophage and a metabolic change that promoted abscess formation and colonization, respectively; they appear to have primed the clonal variant for success. Key events in clonal expansion were acquisitions of resistance genes to mupirocin and chlorhexidine, agents commonly used for decolonization and prevention of infection. These adaptations in virulence and resistance, when expanded from the high-risk community to the general human population, would constitute a serious public health threat.
As expected, the USA300-BKV clone carried a number of distinct nonsynonymous mutations that were present in all isolates. These mutations primarily affected genes involved in amino acid, carbohydrate, and nucleotide biosynthesis pathways, suggesting that metabolic adaptability was critical for success of the USA300-BKV clone. Our data indicate that differences in pyrimidine metabolic pathways in the USA300-BKV strain may be advantageous in high-risk populations, both because nucleotide metabolism is linked to the expression and synthesis of virulence factors (69) and because such differences provide a priming mechanism for growth in infectious ecologies where nutrients are scarce (70).
Genomic comparisons also showed that a unique phage, present in early USA300-BKV isolates, promotes large skin abscesses that may drive CA-MRSA contagion. More detailed analysis is required to define the mosaic phage components driving the enhanced skin virulence phenotype. Intriguingly, the mosaic portion of the prophage DNA that produced virulence encodes a methyltransferase/endonuclease restriction modification (RM) system (Fig. 4B). RM systems have been associated with increased virulence and clonal spread during an outbreak of Shiga toxin-producing E. coli O104:H4 (71, 72). Regardless of whether the methylase is found to be important in staphylococcal virulence, the absence of known virulence genes encoded within the phage supports a growing body of evidence that the regulation of bacterial genes, rather than the production of a virulence factor, confers increased fitness (73–75). Interestingly, the mosaic phage was associated with virulence phenotypes in vivo, but not in vitro, suggesting that an in vivo signal is required for phage-mediated up-regulation of virulence, and potentially explaining why the involvement of lysogeny in virulence has few precedents in S. aureus research. It is also worth noting that competence for DNA uptake in staphylococci depends on the presence of bacteriophage (76–78). Thus, sequential assimilation of plasmid DNA and chlorhexidine resistance elements may have been facilitated by the recombinant ϕ11 that was initially present within USA300-BKV isolates.
Selectively advantageous changes that spread through the bacterial population will fall on distal phylogenetic branches. Thus, our data indicating that evolution of the pBSRC1 plasmid containing dual-biocide resistance coincided with the emergence of successful variants strongly suggest that this plasmid and decolonization therapy were key events in clonal spread. The pBSRC1 plasmid originated from interspecies DNA exchange of S. epidermidis and S. aureus sequences plus recombination with an indigenous cryptic plasmid, pUSA01 (Fig. 3). These data suggest that cryptic plasmids can serve as an anchor for the stepwise acquisition of elements from both species. Acquisition of the arginine catabolic mobile element of S. epidermidis by USA300 is thought to have expanded its colonization niche to the skin, increasing opportunities for abscess formation and transmission (19). Collectively, these observations point to a potential reservoir of genes in otherwise nonpathogenic staphylococcal species that primarily confer a commensal fitness advantage rather than an enhanced capacity for infection.
In conclusion, high-resolution bacterial genomics and field surveillance of clinical strains provide insight into how heightened infection risk and clinical intervention remodel pathogens with respect to virulence and antimicrobial resistance. Broadly speaking, these data lead to a two-stage framework for interpreting the phenotypes of newly emergent clones. Early on, an introduction to a vulnerable population selects for genetic changes, such as pyrR mutation and phage acquisition, that drive bacterial emergence. These data highlight the need for vigorous surveillance and early public health intervention to limit further adaptation. In later stages, clinical intervention and control measures select for rapidly spreading determinants of success, such as plasmid-borne dual resistance to chlorhexidine and mupirocin. The temporal connection between these two processes links virulence and antimicrobial resistance, potentially explaining the convergence of pvl-mediated abscess formation and methicillin resistance in CA-MRSA lineages. The result is high-consequence clones and DNA elements that threaten larger human populations, including vulnerable populations in hospitals. Unique epidemiology and pathogens make the course of dissemination unpredictable, complicating efforts to plan for the next high-risk clone. Nonetheless, we expect that a better understanding of principles and infection traits involved in the epidemiology (e.g., virulence, metabolism, and resistance) will (i) uncover new vulnerabilities that can be exploited for prevention and treatment, and (ii) facilitate integration of genomic and epidemiological analysis, allowing more effective targeting of intervention strategies.
At a more global level, the present report demonstrates how the relationship between virulence and transmissibility (79) in pathogens such as CA-MRSA, in which disease and transmission are tightly linked, can lead to a new antimicrobial-resistance threat. Historically, antimicrobial-resistant clones of S. aureus disseminate globally. To interrupt this evolutionary pattern, our results suggest that the focus of preventative strategies targeting CA-MRSA should be to influence virulence so that the infective capacity is never attained.
Methods
Study Design and Facility.
Children’s services at New York University Langone Medical Center (NYCLMC) consist of 109 beds within a larger 1,069-bed tertiary care academic medical center. The general medicine ward, pediatric ward, and ICUs manage about 24,500, 1,700, and 600 admissions a year, respectively. Ethical approval and informed consent were not required for the colonization study because all patients are routinely screened for S. aureus and MRSA nasal and throat carriage within 48 h of admission to the general pediatric ward and ICU as part of surveillance and management of health care-associated infection. Approval for medical record reviews, laboratory studies, and genome sequencing was granted by the NYULMC Institutional Review Board.
Bacterial isolates or genome sequences were obtained from three patient groups. The first group consisted of 84 consecutive single-patient isolates from pediatric patients residing in Orthodox-associated (high-risk) zip codes collected over a 2-y period from the clinical microbiology laboratory (cases, BKV_isolates, Dataset S1). Isolates were identified from every child 18 y old and younger having a high-risk zip code admitted to the general pediatric ward and ICU from May 2015 to May 2017. Eleven additional isolates were obtained from adults residing in high-risk zip codes who were admitted to the general medicine wards (BKV_A_isolates). Sites of infection and patient age are listed in Dataset S1. The second group consisted of 16 consecutive single-patient USA300 strains, including children admitted to the general pediatric ward and pediatric ICU (Ctl_isolates). Controls were collected from January 2016 to June 2016, after the clonality of isolates was established. Control zip codes represented each of the five New York City boroughs. Clonality was assigned by genotyping using a combination of methods, outlined below, as part of routine surveillance of MRSA by the NYULMC Molecular Outbreak Center. The third group consisted of 68 genome sequences of USA300 control isolates from single patients, collected and characterized in a large, community-based case-control study of CA-MRSA in northern Manhattan and the Bronx (24).
Bacterial Strains, Phage Sources, and Growth Conditions.
Clinical isolates and laboratory strains USA300-LAC (AH-LAC) and USA300-JE2 (80, 81) were used in all experiments. For pyrR complementation, SapI1 integration vector pJC1111 was used to chromosomally integrate the WT pyrR, amplified using primers pyrR-F-PstI ATATCTGCAGGATACAATTCGAAAAAGAGA and pyrR-R-BamHI ATATGGATCCGTACTGAATTAAAAGGGGTA, incorporating the endogenous promoter into the SaPI1 attC site of RN9011, as described (82). Phage 80α was used to transduce the allele into JE2 or the indicated USA300-BVK strain (83); transductants were selected on tryptic soy agar (TSA) plates containing the appropriate antimicrobial (Cd). Phage 80α was also used to transduce plasmid pBSRC1 from USA300-BKV_02 into recipient strain AH-LAC. Prototype ϕ11 and mosaic ϕ11 were obtained by mitomycin C induction of RN451 (84), which is considered to be free of prophages except for ϕ11, and clinical isolate USA300-BKV_28, respectively. USA300-AH-LAC was lysogenized as described previously (83) by plaque-purified ϕ11 and mosaic ϕ11 to create strain AH-LAC + ϕ11 and AH-LAC + mosaic ϕ11, respectively. Cells were cultured in tryptic soy broth (TSB) (Difco) or Roswell Park Memorial Institute culture medium (RPMI) 1640 (Invitrogen) supplemented with 1% casamino acids or complete defined medium [CDM; Pattee/Neveln medium (85)] with constant aeration shaking (180 rpm) or on TSA plates. Incubation was at 37 °C. Growth curves were carried out in 200-μL cultures within a 96-well plate, which was inoculated using overnight cultures of three independent colonies for each strain and then washed three times in PBS and diluted 1:1,000 in fresh medium. Optical densities at 600 nm were read at the beginning of the subculture and at the indicated time points using a Bioscreen C Analyzer.
Mouse Model of Colonization and Transmission.
We adaptated the infant mouse model to the study of CA-MRSA shedding and transmission. Intestinal carriage of S. aureus is frequent in human infants (86–88) and is thought to decrease toward adulthood. Unlike adult mice (89, 90), infant mice are highly susceptible hosts for which colonization and intralitter transmission of pathogens has been demonstrated without the use of antimicrobials (91–93). For competition experiments, WT reference strain LAC was grown separately from the pyrR-complemented strains overnight at 37 °C, diluted 100-fold, subcultured for 4 h, and mixed at a 1:1 ratio of 5 × 106 cfu. Pregnant C57BL/6J mice were obtained from The Jackson Laboratory and maintained in the Alexandria Center for Life Science West Tower animal facility. Seven-day-old pups were mouth-fed with 107 cfu of S. aureus suspended in 10 µL of sterile PBS with 20% sucrose using a blunt pipette tip. After the pup swallowed the inoculum, it was returned to its dam. In each cage, half of the pups were inoculated to study colonization, cohoused with the rest of the pups from which transmissions were detected. Cecal contents are used to evaluate colonization in infant mice, since they do not excrete stool pellets. Four days postinoculation, pups were killed by CO2 asphyxiation followed by decapitation, and the cecum was collected and resuspended in 1 mL of sterile PBS followed by homogenization using a FastPrep-24 Classic Instrument (MP Biomedicals). The cecal suspension was then performed by 10-fold serial dilutions and plated on CHROMID MRSA SMART II agar (bioMérieux). The limit of detection was 10 cfu/mL. Colonies recovered from CHROMID MRSA SMART II agar were subsequently plated on TSB with 0.25 mM CdCl2 to distinguish pyrR-complemented strains and WT. The amount of WT bacteria was calculated by subtracting the number of colonies that grew on CdCl2-containing plates from the number of colonies formed on CHROMID MRSA SMART II plates.
Genotyping, Sequencing.
All strains were genotyped by DNA sequence analysis of the protein A gene variable repeat region (spa typing) and a variety of additional DNA polymorphisms, including SCCmec, and the presence of the ACME and pvl genes as previously described (94, 95). USA300 lineage was defined by the presence of SCCmecIV, ACME, pvl, and assignment to clonal complex 8 by spa type using the Ridom SpaServer database (spa.ridom.de/mlst.shtml).
Genome Sequencing, Assembly, and Annotation.
We prepared sequencing libraries from DNA extracted from each MRSA isolate as previously described (24, 96). Whole-genome sequencing was performed using Illumina HiSeq 2000 with 100-base paired-end reads or a PacBio RSII instrument (24, 49, 96). Paired-end Illumina reads were mapped against USA300_FPR3757 reference genome using Burrows-Wheeler Aligner (BWA) (97). BWA outputs were analyzed and annotated using SAMtools (97), GATK (98), and ANNOVAR (99). SNPs in genes annotated as integrases, transposases, resolvases, maturases, or phages were removed from the analysis using custom scripts. Other mobile genetic elements, including SaPI5, phiSA2usa, phiSA3usa, SCCmecIV, and ACME, were identified using IslandPath-DIMOB and PHASTSNPs (100, 101). Raw (PacBio) long-read data were assembled using the HGAP3 version 2.2.0 pipeline (102). Subsequently, a custom postassembly pipeline (https://github.com/powerpak/pathogendb-pipeline) was used to finalize each genome. Genes were annotated using Prokka (103), and genomes were visualized using ChomoZoom (104).
Raw Illumina reads from Harris et al. (33) were downloaded from the NCBI and mapped against S. aureus HO-5096-0412 reference genome (HE681097.1). SNPs were identified, and genes sharing mutations in all high-risk strains were compared with our list of mutated genes. De novo assemblies were performed on unmapped Illumina reads selected using BWA, and contigs were generated using Velvet with the Velvet Optimizer (105). Contigs larger than 100 bp (n = 276) were subjected to BLAST to identify large sequence polymorphisms.
Phylogenetic Analyses.
Phylogenetic analysis was based on 5,326 high-confidence variable positions by specifying S. aureus MRSA131 (GCA_000187145.1) as the outgroup. Maximum-likelihood phylogenies with 1,000 bootstrap replications were obtained using PhyML (106) and the HKY model. These analyses included the two groups of control isolates from our hospital (n = 27) or the community-based case-control study in northern Manhattan (n = 68) (24). Evolutionary rates and time of emergence were determined using BEAST 1.7.5 package (107). BEAST was run for 100 million generations, sampling every 10,000 states using the HKY substitution model and strict, exponential-relaxed and lognormal-relaxed molecular clocks. Minimum spanning trees were generated using a patristic distance matrix and the Fruchterman–Reingold layout from the ape and igraph R packages (108). Tree topologies for ancestral reconstructions were generated in PhyML. Ancestral state reconstructions were performed using the phangorn R package (version 2.4.0) with the accelerated transformation function; all states and transformations were given equal weight.
Quantitative Reverse Transcriptase–PCR (qRT-PCR).
Isolation of total RNA from S. aureus cultures grown for 6 h in CDM was carried out using an RNeasy extraction kit, following the manufacturer’s instructions and the method previously described by Carroll et al. (109). qRT-PCR was performed using a one-step reaction with Reverse Transcriptase Mastermix (QuantiTect) and SYBR Green Master Mix (Qiagen) in a 7300 Real-Time PCR system (Applied Biosystems). Specific primer sets (SI Appendix, Table S3) were used to detect pyrR (this study) and carA and pyrB [Kriegeskorte et al. (69)] genes. All genes were normalized to the housekeeping gene (the 16S rRNA gene). Fold change for target genes from pyrR-complemented strains relative to the corresponding genes in WT strains was determined using the threshold cycle (2−ΔΔCT) method of analysis. Melting curve analyses were employed to verify specific single-product amplification.
Cytotoxicity Assays.
Leukopaks were obtained from deidentified donors from the New York Blood Center where written consents were obtained from all participants. Human polymorphonuclear neutrophils (hPMNs) were purified as described previously (56). Bacteriologically supernatants obtained from early logarithmic-phase growth, were used for differentiating cytotoxic activity. Briefly, overnight cultures were diluted with fresh TSB, and the diluted culture was regrown for 6 h at 37 °C. Bacteria were pelleted, and filtered culture supernatants were serially diluted and added to 2 × 105 hPMNs per well for a final volume of 100 μL per well RPMI 1640 supplemented with 10% FBS. hPMNs were intoxicated with the culture supernatant from the indicated strain for 2 h at 37 °C and 5% CO2. hPMN viability was determined using CellTiter 96 Aqueous One Solution (Promega). Cells were mixed with CellTiter and incubated at 37 °C and 5% CO2 for 1.5 h. Cell viability was measured by absorbance at 492 nm using a PerkinElmer Envision 2103 Multilabel reader (PerkinElmer).
Animal Infections.
For the skin infections, one representative strain from each group was cultivated for 3 h in TSB, washed in 1× PBS and normalized to 5 × 108 cfu. Five-week-old female ND4 Swiss Webster mice (Envigo, Inc.) were anesthetized intraperitoneally using 300 μL of avertin [2,2,2-tribromoethanol dissolved in tert-Amyl-alcohol and diluted to a final concentration of 2.5% (vol/vol) in sterile saline]. A total of 100 μL of bacteria, resulting in a 1 × 107 or a 5 × 107 cfu inoculum, as indicated, was injected s.c. into both hind flanks of shaved mice (56, 57). Subsequent lesions were measured using a digital caliper (Fisher Scientific) every 24 h, and the abscess diameter was determined. To assess bacterial burden, 8-mm punch (Integra Miltex) biopsy samples were obtained from mouse lesions at 9 d postinfection, and the tissues were homogenized, serially diluted, and enumerated on TSA (110).
Statistical Methods.
Statistically significant differences between survival curves were determined by log rank (Mantel–Cox) test. The results were corrected for multiple comparisons by using the Bonferroni-corrected threshold. Sizes of murine skin abscesses were compared using the Kruskal–Wallis test with a P value <0.05. For the proteomic data, a two-sided Student’s t test was performed, correcting for multiple testing by controlling for false discovery rate (FDR) at 5% (permutation-based FDR).
Data Availability.
All genomic data are available at the NCBI BioProject database under accession no. PRJNA497094. All other relevant data are within the paper and in Datasets S1 and S2 and SI Appendix.
Supplementary Material
Acknowledgments
This study was supported by NIH Grant R01 AI103268 and NIH National Institute of Allergy and Infectious Diseases Grant HHSN272201400019C (to R.C., V.J.T., and B.S.); NIH Research Training Grant T32 AI007180 (to W.E.S.); NIH Grants R01 AI099394 and R01 AI105129 (to V.J.T.); KiDS of NYU (J.C.F.); a generous donation by Judith and Irwin Kallman (J.C.F.); NIH Grant R01 AI119145 (to H.v.B.); NIH Grant T32 AI07647; Clinical and Translational Science Awards Program at the National Center for Advancing Translational Sciences (CTSA/NCATS) Grant KL2TR001435; the New York State Department of Health Empire Clinical Research Investigator Program (D.R.A.); and NIH Grant 1K08AI101005 (to P.J.P.). V.J.T. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All genomic data reported in this paper have been deposited in the National Center for Biotechnology Information BioProject database (accession no. PRJNA497094).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814265116/-/DCSupplemental.
References
- 1.Ammerlaan HS, et al. Secular trends in nosocomial bloodstream infections: Antibiotic-resistant bacteria increase the total burden of infection. Clin Infect Dis. 2013;56:798–805. doi: 10.1093/cid/cis1006. [DOI] [PubMed] [Google Scholar]
- 2.Moran GJ, et al. EMERGEncy ID Net Study Group Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 3.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–794. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 4.Seybold U, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42:647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 5.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson JM, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345:1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford H, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206:909–921. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl WE, et al. Ebola virus glycoprotein with increased infectivity dominated the 2013-2016 epidemic. Cell. 2016;167:1088–1098.e6. doi: 10.1016/j.cell.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepin KM, Lass S, Pulliam JR, Read AF, Lloyd-Smith JO. Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat Rev Microbiol. 2010;8:802–813. doi: 10.1038/nrmicro2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pettersson JH, et al. How did Zika virus emerge in the Pacific Islands and Latin America? MBio. 2016;7:e01239-16. doi: 10.1128/mBio.01239-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poon LL, et al. Quantifying influenza virus diversity and transmission in humans. Nat Genet. 2016;48:195–200. doi: 10.1038/ng.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urbanowicz RA, et al. Human adaptation of ebola virus during the West African outbreak. Cell. 2016;167:1079–1087.e5. doi: 10.1016/j.cell.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLeo FR, et al. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc Natl Acad Sci USA. 2011;108:18091–18096. doi: 10.1073/pnas.1111084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy AD, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansion and diversification. Proc Natl Acad Sci USA. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Planet PJ, et al. Architecture of a species: Phylogenomics of Staphylococcus aureus. Trends Microbiol. 2017;25:153–166. doi: 10.1016/j.tim.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Young BC, et al. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci USA. 2012;109:4550–4555. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald JR, Holden MT. Genomics of natural populations of Staphylococcus aureus. Annu Rev Microbiol. 2016;70:459–478. doi: 10.1146/annurev-micro-102215-095547. [DOI] [PubMed] [Google Scholar]
- 19.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher P. Identification and analysis of Orthodox Jewish enclaves in Brooklyn, New York: A GIS based approach. Middle States Geogr. 2009;42:83–89. [Google Scholar]
- 21.David MZ, et al. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for panton-valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. Medical Center. J Clin Microbiol. 2013;51:814–819. doi: 10.1128/JCM.02429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strauß L, et al. Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc Natl Acad Sci USA. 2017;114:E10596–E10604. doi: 10.1073/pnas.1702472114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafer C, Lin Y, Kornblum J, Lowy FD, Uhlemann AC. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:5845–5851. doi: 10.1128/AAC.01139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson MA, et al. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol. 2014;93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobisch S, Stülke J, Hecker M. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis. J Bacteriol. 1999;181:4995–5003. doi: 10.1128/jb.181.16.4995-5003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuxoll AS, et al. CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog. 2012;8:e1003033. doi: 10.1371/journal.ppat.1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tojo S, Satomura T, Matsuoka H, Hirooka K, Fujita Y. Catabolite repression of the Bacillus subtilis FadR regulon, which is involved in fatty acid catabolism. J Bacteriol. 2011;193:2388–2395. doi: 10.1128/JB.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto H, Serizawa M, Thompson J, Sekiguchi J. Regulation of the glv operon in Bacillus subtilis: YfiA (GlvR) is a positive regulator of the operon that is repressed through CcpA and cre. J Bacteriol. 2001;183:5110–5121. doi: 10.1128/JB.183.17.5110-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 30.Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: Repression without repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghim SY, Switzer RL. Mutations in Bacillus subtilis PyrR, the pyr regulatory protein, with defects in regulation by pyrimidines. FEMS Microbiol Lett. 1996;137:13–18. doi: 10.1111/j.1574-6968.1996.tb08075.x. [DOI] [PubMed] [Google Scholar]
- 32.Savacool HK, Switzer RL. Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J Bacteriol. 2002;184:2521–2528. doi: 10.1128/JB.184.9.2521-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris SR, et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: A descriptive study. Lancet Infect Dis. 2013;13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Date SV, et al. Global gene expression of methicillin-resistant Staphylococcus aureus USA300 during human and mouse infection. J Infect Dis. 2014;209:1542–1550. doi: 10.1093/infdis/jit668. [DOI] [PubMed] [Google Scholar]
- 35.Szafranska AK, et al. High-resolution transcriptomic analysis of the adaptive response of Staphylococcus aureus during acute and chronic phases of osteomyelitis. MBio. 2014;5:e01775-14. doi: 10.1128/mBio.01775-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valentino MD, et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. MBio. 2014;5:e01729-14. doi: 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilde AD, et al. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 2015;11:e1005341. doi: 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faden H, et al. Importance of colonization site in the current epidemic of staphylococcal skin abscesses. Pediatrics. 2010;125:e618–e624. doi: 10.1542/peds.2009-1523. [DOI] [PubMed] [Google Scholar]
- 39.McCullough AC, et al. Higher incidence of perineal community acquired MRSA infections among toddlers. BMC Pediatr. 2011;11:96. doi: 10.1186/1471-2431-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gagnaire J, et al. Epidemiology and clinical relevance of Staphylococcus aureus intestinal carriage: A systematic review and meta-analysis. Expert Rev Anti Infect Ther. 2017;15:767–785. doi: 10.1080/14787210.2017.1358611. [DOI] [PubMed] [Google Scholar]
- 41.Kumar N, David MZ, Boyle-Vavra S, Sieth J, Daum RS. High Staphylococcus aureus colonization prevalence among patients with skin and soft tissue infections and controls in an urban emergency department. J Clin Microbiol. 2015;53:810–815. doi: 10.1128/JCM.03221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piewngam P, et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature. 2018;562:532–537. doi: 10.1038/s41586-018-0616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel-Scheel J, Alpert C, Engst W, Loh G, Blaut M. Requirement of purine and pyrimidine synthesis for colonization of the mouse intestine by Escherichia coli. Appl Environ Microbiol. 2010;76:5181–5187. doi: 10.1128/AEM.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang HJ, Bogomolnaya L, McClelland M, Andrews-Polymenis H. De novo pyrimidine synthesis is necessary for intestinal colonization of Salmonella Typhimurium in chicks. PLoS One. 2017;12:e0183751. doi: 10.1371/journal.pone.0183751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iandolo JJ, et al. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene. 2002;289:109–118. doi: 10.1016/s0378-1119(02)00481-x. [DOI] [PubMed] [Google Scholar]
- 46.Xia G, Wolz C. Phages of Staphylococcus aureus and their impact on host evolution. Infect Genet Evol. 2014;21:593–601. doi: 10.1016/j.meegid.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 47.Maiques E, et al. beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol. 2006;188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ubeda C, et al. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 49.Altman DR, et al. Genome plasticity of agr-defective Staphylococcus aureus during clinical infection. Infect Immun. 2018;86:e00331-18. doi: 10.1128/IAI.00331-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goerke C, et al. Increased frequency of genomic alterations in Staphylococcus aureus during chronic infection is in part due to phage mobilization. J Infect Dis. 2004;189:724–734. doi: 10.1086/381502. [DOI] [PubMed] [Google Scholar]
- 51.Goerke C, Wirtz C, Flückiger U, Wolz C. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol Microbiol. 2006;61:1673–1685. doi: 10.1111/j.1365-2958.2006.05354.x. [DOI] [PubMed] [Google Scholar]
- 52.Salgado-Pabón W, et al. Staphylococcus aureus β-toxin production is common in strains with the β-toxin gene inactivated by bacteriophage. J Infect Dis. 2014;210:784–792. doi: 10.1093/infdis/jiu146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sampedro GR, et al. Targeting Staphylococcus aureus α-toxin as a novel approach to reduce severity of recurrent skin and soft-tissue infections. J Infect Dis. 2014;210:1012–1018. doi: 10.1093/infdis/jiu223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang R, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 55.Li M, et al. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis. 2010;202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balasubramanian D, et al. Staphylococcus aureus coordinates leukocidin expression and pathogenesis by sensing metabolic fluxes via RpiRc. MBio. 2016;7:e00818-16. doi: 10.1128/mBio.00818-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho JS, et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest. 2010;120:1762–1773. doi: 10.1172/JCI40891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glaser P, et al. Demography and intercontinental spread of the USA300 community-acquired methicillin-resistant Staphylococcus aureus lineage. MBio. 2016;7:e02183-15. doi: 10.1128/mBio.02183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kennedy AD, et al. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J Clin Microbiol. 2010;48:4504–4511. doi: 10.1128/JCM.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabat AJ, et al. Complete-genome sequencing elucidates outbreak dynamics of CA-MRSA USA300 (ST8-spa t008) in an academic hospital of Paramaribo, Republic of Suriname. Sci Rep. 2017;7:41050. doi: 10.1038/srep41050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uhlemann AC, et al. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci USA. 2014;111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Antonov NK, et al. High prevalence of mupirocin resistance in Staphylococcus aureus isolates from a pediatric population. Antimicrob Agents Chemother. 2015;59:3350–3356. doi: 10.1128/AAC.00079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Batra R, et al. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin Infect Dis. 2010;50:210–217. doi: 10.1086/648717. [DOI] [PubMed] [Google Scholar]
- 64.Johnson RC, et al. Recurrent methicillin-resistant Staphylococcus aureus cutaneous abscesses and selection of reduced chlorhexidine susceptibility during chlorhexidine use. J Clin Microbiol. 2015;53:3677–3682. doi: 10.1128/JCM.01771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee AS, et al. Impact of combined low-level mupirocin and genotypic chlorhexidine resistance on persistent methicillin-resistant Staphylococcus aureus carriage after decolonization therapy: A case-control study. Clin Infect Dis. 2011;52:1422–1430. doi: 10.1093/cid/cir233. [DOI] [PubMed] [Google Scholar]
- 66.Wassenaar TM, Ussery D, Nielsen LN, Ingmer H. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur J Microbiol Immunol (Bp) 2015;5:44–61. doi: 10.1556/EUJMI-D-14-00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mc Gann P, et al. Rapid and simultaneous detection of the chlorhexidine and mupirocin resistance genes qacA/B and mupA in clinical isolates of methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis. 2013;77:270–272. doi: 10.1016/j.diagmicrobio.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 68.Schlett CD, et al. Prevalence of chlorhexidine-resistant methicillin-resistant Staphylococcus aureus following prolonged exposure. Antimicrob Agents Chemother. 2014;58:4404–4410. doi: 10.1128/AAC.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kriegeskorte A, et al. Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. MBio. 2014;5:e01447-14. doi: 10.1128/mBio.01447-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samant S, et al. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog. 2008;4:e37. doi: 10.1371/journal.ppat.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang G, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol. 2012;30:1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forde BM, et al. Lineage-specific methyltransferases define the methylome of the globally disseminated Escherichia coli ST131 clone. MBio. 2015;6:e0160215. doi: 10.1128/mBio.01602-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y, Golding I, Sawai S, Guo L, Cox EC. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005;3:e229. doi: 10.1371/journal.pbio.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner PL, Waldor MK. Bacteriophage control of bacterial virulence. Infect Immun. 2002;70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rudkin JK, et al. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis. 2012;205:798–806. doi: 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson NE, Pattee PA. Transformation in Staphylococcus aureus: Role of bacteriophage and incidence of competence among strains. J Bacteriol. 1977;129:778–788. doi: 10.1128/jb.129.2.778-788.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson NE, Pattee PA. Genetic transformation in Staphylococcus aureus: Demonstration of a competence-conferring factor of bacteriophage origin in bacteriophage 80 alpha lysates. J Bacteriol. 1981;148:294–300. doi: 10.1128/jb.148.1.294-300.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lipsitch M, Moxon ER. Virulence and transmissibility of pathogens: What is the relationship? Trends Microbiol. 1997;5:31–37. doi: 10.1016/S0966-842X(97)81772-6. [DOI] [PubMed] [Google Scholar]
- 80.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fey PD, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen J, Yoong P, Ram G, Torres VJ, Novick RP. Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid. 2014;76:1–7. doi: 10.1016/j.plasmid.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 84.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 85.Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- 86.Björkstén B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108:516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 87.Lindberg E, et al. High rate of transfer of Staphylococcus aureus from parental skin to infant gut flora. J Clin Microbiol. 2004;42:530–534. doi: 10.1128/JCM.42.2.530-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lindberg E, Nowrouzian F, Adlerberth I, Wold AE. Long-time persistence of superantigen-producing Staphylococcus aureus strains in the intestinal microflora of healthy infants. Pediatr Res. 2000;48:741–747. doi: 10.1203/00006450-200012000-00007. [DOI] [PubMed] [Google Scholar]
- 89.Kernbauer E, Maurer K, Torres VJ, Shopsin B, Cadwell K. Gastrointestinal dissemination and transmission of Staphylococcus aureus following bacteremia. Infect Immun. 2015;83:372–378. doi: 10.1128/IAI.02272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Misawa Y, et al. Staphylococcus aureus colonization of the mouse gastrointestinal tract is modulated by wall teichoic acid, capsule, and surface proteins. PLoS Pathog. 2015;11:e1005061. doi: 10.1371/journal.ppat.1005061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fernandez MI, et al. A newborn mouse model for the study of intestinal pathogenesis of shigellosis. Cell Microbiol. 2003;5:481–491. doi: 10.1046/j.1462-5822.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 92.Klose KE. The suckling mouse model of cholera. Trends Microbiol. 2000;8:189–191. doi: 10.1016/s0966-842x(00)01721-2. [DOI] [PubMed] [Google Scholar]
- 93.Zafar MA, Kono M, Wang Y, Zangari T, Weiser JN. Infant mouse model for the study of shedding and transmission during Streptococcus pneumoniae monoinfection. Infect Immun. 2016;84:2714–2722. doi: 10.1128/IAI.00416-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Diep BA, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: Convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 95.Mendes RE, et al. Characterization of methicillin-resistant Staphylococcus aureus strains recovered from a phase IV clinical trial for linezolid versus vancomycin for treatment of nosocomial pneumonia. J Clin Microbiol. 2012;50:3694–3702. doi: 10.1128/JCM.02024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Altman DR, et al. Transmission of methicillin-resistant Staphylococcus aureus via deceased donor liver transplantation confirmed by whole genome sequencing. Am J Transplant. 2014;14:2640–2644. doi: 10.1111/ajt.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century–A clinical super-challenge. N Engl J Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 98.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gardete S, et al. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog. 2012;8:e1002505. doi: 10.1371/journal.ppat.1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Langille MG, Hsiao WW, Brinkman FS. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics. 2008;9:329. doi: 10.1186/1471-2105-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: A fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 103.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. [Google Scholar]
- 104.Pak TR, Roth FP. ChromoZoom: A flexible, fluid, web-based genome browser. Bioinformatics. 2013;29:384–386. doi: 10.1093/bioinformatics/bts695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 107.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Csardi G, Nepusz T. The igraph software package for complex network research. Inter J Complex Syst. 2006;1695:1–9. [Google Scholar]
- 109.Carroll RK, Weiss A, Shaw LN. RNA-sequencing of Staphylococcus aureus messenger RNA. Methods Mol Biol. 2016;1373:131–141. doi: 10.1007/7651_2014_192. [DOI] [PubMed] [Google Scholar]
- 110.Leech JM, Lacey KA, Mulcahy ME, Medina E, McLoughlin RM. IL-10 plays opposing roles during Staphylococcus aureus systemic and localized infections. J Immunol. 2017;198:2352–2365. doi: 10.4049/jimmunol.1601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data are available at the NCBI BioProject database under accession no. PRJNA497094. All other relevant data are within the paper and in Datasets S1 and S2 and SI Appendix.