Significance
Virulent phages can reduce populations of bacteria and help shape bacterial evolution. Here, we used three virulent phages to understand their equilibrium with V. cholerae in nutrient-limiting aquatic microcosms. It has been proposed that phages quench cholera outbreaks, but no direct evidence of phage predation in aquatic environments had been established. Here, we show that different phages possess varied abilities to infect in certain niches or stages of the host bacterial life cycle. Unveiling the phage/bacterial interactions in their natural setting is important to the understanding of cholera outbreaks and could be ultimately used to help develop a method for outbreak prediction and/or control.
Keywords: Vibrio cholerae, cholera, bacteriophage, aquatic reservoir, environment
Abstract
Vibrio cholerae, the causative agent of cholera, has reservoirs in fresh and brackish water where it interacts with virulent bacteriophages. Phages are the most abundant biological entity on earth and coevolve with bacteria. It was reported that concentrations of phage and V. cholerae inversely correlate in aquatic reservoirs and in the human small intestine, and therefore that phages may quench cholera outbreaks. Although there is strong evidence for phage predation in cholera patients, evidence is lacking for phage predation of V. cholerae in aquatic environments. Here, we used three virulent phages, ICP1, ICP2, and ICP3, commonly shed by cholera patients in Bangladesh, as models to understand the predation dynamics in microcosms simulating aquatic environments. None of the phages were capable of predation in fresh water, and only ICP1 was able to prey on V. cholerae in estuarine water due to a requirement for salt. We conclude that ICP2 and ICP3 are better adapted for predation in a nutrient rich environment. Our results point to the evolution of niche-specific predation by V. cholerae-specific virulent phages, which complicates their use in predicting or monitoring cholera outbreaks as well as their potential use in reducing aquatic reservoirs of V. cholerae in endemic areas.
The water-borne bacterium Vibrio cholerae causes cholera, an acute intestinal infection characterized by profuse secretory diarrhea that can quickly lead to severe dehydration and death if untreated. This pathogen is able to persist in aquatic environments in planktonic form or by association with phytoplankton and the chitinous carapaces of zooplankton (1) and is endemic in many countries where it forms reservoirs in fresh water and estuarine environments (2). Among the many interactions this pathogen could have in the environment, virulent phages may be of special relevance. Phages are viruses of bacteria that are present at ∼1030 in the earth’s hydrosphere (3, 4). Phages exist in dynamic equilibrium with their bacterial hosts and help drive bacterial evolution and shape ecosystems (3, 5).
In the 1920s Felix D’Herelle studied the relationship between V. cholerae and virulent phages, finding that phages were widespread in the environment a few days after the onset of an outbreak (6). Recent studies have proposed that cholera outbreaks are driven, in part, by an increase in the hyperinfectious V. cholerae population (7, 8) and quenched by an increase in virulent phages in infected humans (9, 10) and in environmental reservoirs (11, 12). However, no direct evidence of phage predation of V. cholerae in aquatic settings has been reported. It is known that phages can limit V. cholerae replication during intestinal infection (13) and are shed into the environment due to fecal contamination of waters in endemic areas. Hence, the appearance of environmental phages may simply be a consequence of phage replication in infected humans (9, 10).
Virulent phages are frequently found in cholera patient stools (14, 15). A study of a 10-y collection of such stool samples in Bangladesh revealed the presence of just three V. cholerae-specific virulent phages, ICP1, ICP2, and ICP3 (15). Of these, ICP1 was the most frequently observed. Because of the dominance of these three phages in this area, the fact that they are often shed at high titers in secretory diarrhea and the general lack of waste-water treatment facilities, it is likely that these phages frequently contact V. cholerae in the surrounding aquatic environments.
We used ICP1, ICP2, and ICP3 to study their predation abilities on V. cholerae in microcosms simulating fresh and estuarine aquatic environments. Our results show that none of the phages are able to prey on V. cholerae in fresh water, and only ICP1 is able to prey on V. cholerae in estuarine water. ICP1 can replicate in V. cholerae in fresh water that is supplemented with NaCl, hence, salinity could be a critical variable in relation to environmental phage predation.
Results
ICP1 Preys on Planktonic V. cholerae in Estuary Conditions.
Cholera epidemics occur in Bangladesh during March–May and after monsoon season in September–December. The temperature of the bodies of water in Bangladesh approaches 30 °C (16). Therefore, we first tested the ability of these phages to kill V. cholerae in a rich [Luria-Bertani (LB)] broth medium at 30 °C. When added to a multiplicity of infection (MOI) of 0.01, all three phages were able to reduce the load of V. cholerae over the first few hours. ICP1 and ICP3 reduced bacterial numbers 100-fold within 1 h, but after 4 h, numbers increased due to the outgrowth of phage-resistant (escape mutant) bacteria (SI Appendix, Fig. S1A). ICP2 infected cultures kept growing for 1 h, but by 3 h, bacterial numbers were reduced 10,000-fold. Therefore, each of the ICP phages can infect and kill V. cholerae at 30 °C. This temperature was used throughout the rest of this paper.
It has been shown that, upon dissemination of V. cholerae from cholera patient stools to fresh water, there is a major drop in osmolarity, inorganic nutrients, and carbon sources (10). An obstacle to the study of phage predation in an aquatic model is the loss of bacterial viability in this nutrient-poor environment (10, 17). Since V. cholerae shed from the host resembles the physiology of stationary growth phase bacteria (18) and because stationary phase cells exhibit better survival to osmotic shock (17), we used stationary phase cultures for adaptation to the aquatic environment before phage infection.
Phages use the bacterial cell machinery to replicate and produce progeny (19). Since shed V. cholerae resemble stationary phase bacteria, we tested if any of the phages were able to kill V. cholerae in this physiological state. Some 106 colony-forming units (CFUs) of V. cholerae from an overnight stationary phase culture were inoculated into M9 minimal media in the absence of a carbon source to avoid bacterial growth, and phages were added to a MOI of 3. ICP1 was able to reduce the V. cholerae load within 1 h by 10-fold. ICP1 killing was sustained for 3 h, but after 4 h, bacterial numbers increased due to the generation of ICP1 escape mutants (SI Appendix, Fig. S1B). ICP2 was able to kill V. cholerae by threefold within 1 h, but then, bacterial growth quickly recovered. ICP3 was not able to kill V. cholerae under these conditions.
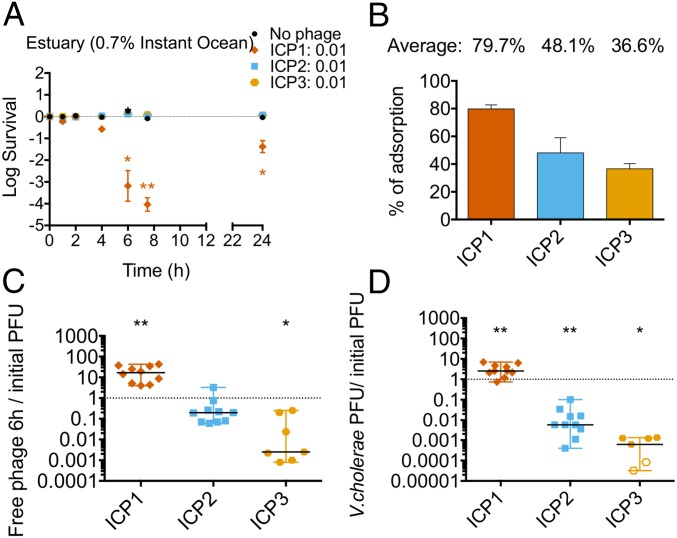
To study phage predation on planktonic V. cholerae in water in the absence of an added carbon source, 107 CFU of stationary phase V. cholerae were preadapted overnight at 30 °C with aeration in either 0.7% Instant Ocean, which we chose to mimic estuary, or fresh water obtained from a natural pond. For comparison, open ocean is equivalent to 3.5% Instant Ocean. Cultures were then infected with each phage at a MOI of 0.01, and bacterial viability was measured over time. Only ICP1 was able to replicate and kill V. cholerae in estuary conditions (Fig. 1A and SI Appendix, Fig. S2 A and B). The V. cholerae population was reduced 10-fold by 4 h and 10,000-fold by 7 h. After 24 h, viable bacterial counts rebounded by means of the growth of escape mutants. All phages were able to adsorb V. cholerae in this environment (Fig. 1B) and were stable at 30 °C with aeration in the absence of a bacterial host (SI Appendix, Fig. S3A), thus ruling out these trivial explanations for the lack of predation by ICP2 and ICP3.
Fig. 1.
ICP1 preys on V. cholerae in estuary water. Some 107 CFU of V. cholerae from an overnight culture were preadapted in 0.7% Instant Ocean overnight in the absence of an added carbon source. ICP1, ICP2, or ICP3 were added to a MOI of 0.01 to assess bacterial viability and phage replication over time. A and B show averages with the standard error. (A) Bacterial viability at 1, 2, 4, 7, and 24 h. (B) Phage adsorption after 15 min of static incubation at 24 °C. C and D show the medians and ranges of, at least, eight biological replicates. The empty symbols represent data under the limit of detection. (C) The ratio of phages in the culture supernatant at 6 h relative to the number initially added. (D) The ratio of phages associated with V. cholerae at 6 h relative to the number initially added (*P < 0.05, **P < 0.005). PFU, plaque-forming units.
The number of free and cell-associated phages after 6 h of infection was determined by infectious-center (plaque) assay. To measure cell-associated phages, the cells were washed before performing plaque assays. Removal of greater than 99% of unbound phages was confirmed by quantifying free phage before and after washing (SI Appendix, Fig. S2C). Consistent with its ability to kill V. cholerae in estuarine conditions, the titer of ICP1 in the culture supernatant increased by 15-fold (Fig. 1C). In contrast, the titers of ICP2 and ICP3 in the culture supernatant decreased 1and 3 orders of magnitude, respectively. In terms of cell-associated phages at 6 h, the titer of ICP1 increased threefold relative to the amount of phages added initially (Fig. 1D). In contrast, only 1% of ICP2 and 0.1% of ICP3 were cell associated (Fig. 1D). Thus, the majority of bound ICP2 and the overwhelming majority of ICP3 were unable to replicate in estuarylike conditions and lost their ability to subsequently form plaques.
Altogether, these results suggest that in estuarine conditions where V. cholerae is in a nongrowing planktonic state: (i) ICP1 is efficient at adsorption, predation, and replication, (ii) ICP2 is able to adsorb, but only a small percentage of these can replicate with most of the remaining losing plaque-forming ability, and (iii) ICP3 is able to adsorb but cannot replicate or maintain infectivity.
No ICP Phage Can Prey on V. cholerae in Fresh Water.
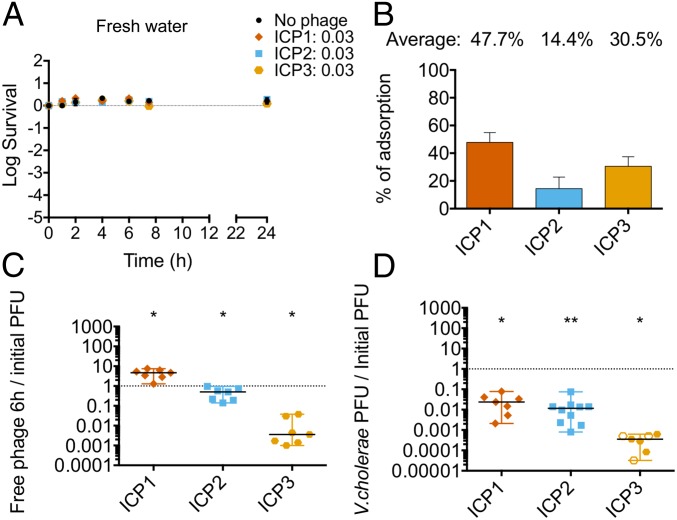
It has been suggested that fresh water is the primary aquatic reservoir for water-borne transmission of V. cholerae during epidemics (20). Therefore, understanding the phage predation dynamics in this environment is crucial. For this, we performed the same set of predation experiments as above but using fresh water obtained from a natural pond. Our results show that none of the ICP phages were able to kill V. cholerae under these conditions (Fig. 2A). Since the lack of predation did not seem to be due to either lack of adsorption (Fig. 2B) or phage stability (SI Appendix, Fig. S3B), this strongly suggests that phage predation/replication does not occur in fresh water environments. Notably, ICP1 and ICP2 had a higher adsorption rate in estuary conditions (compare Fig. 1B to Fig. 2B). In fresh water, ICP1 and ICP2 were found mostly as free phages in the supernatant after 6 h and only 1 to 2% were either associated with or within infected bacterial cells (Fig. 2 C and D). As seen previously, less than 1% of the total ICP3 added was found in the supernatant after 6 h, but less than 0.001% was able to replicate after interacting with cells.
Fig. 2.
None of the phages prey on V. cholerae in fresh water. Some 107 CFU of V. cholerae from an overnight culture were preadapted in autoclaved filter-sterilized fresh water overnight in the absence of an added carbon source. ICP1, ICP2, or ICP3 were added to a MOI of 0.01 to assess bacterial viability and phage replication over time. A and B show averages with standard errors. (A) Bacterial viability at 1, 2, 4, 7, and 24 h. (B) Phage adsorption after 15 min of static incubation at 24 °C. C and D show the medians with ranges of, at least, six biological replicates. The empty symbols represent data under the limit of detection. (C) The ratio of phages in the culture supernatant at 6 h relative to the number initially added. (D) The ratio of phages associated with V. cholerae at 6 h relative to the number initially added (*P < 0.05, **P < 0.005). PFU, plaque-forming units.
Together the results suggest that: (i) ICP1 can kill V. cholerae and replicate in a higher salinity condition than the one found in fresh water, (ii) only a small number of ICP2 phages replicate in V. cholerae in an estuary environment, and (iii) infection by ICP3 is unsuccessful in both aquatic environments.
ICP1 Predation in Estuary Water Selects for Escape Mutants.
We observed that, between 7 and 24 h post infection in estuary conditions, bacterial viability increased in the presence of ICP1 (Fig. 1A) and the likely explanation for this is the generation of escape mutants via phase variation which alters the expression of the lipopolysaccharide O1-antigen phage receptor (21). We tested this by plating surviving V. cholerae at 7 and 24 h post infection, testing the isolates for phage sensitivity by cross streaking, and then performing whole genome sequencing to identify mutations. The number of escape mutants increased over time after 7 h of infection (SI Appendix, Fig. S4A). A total of eight escape mutants after 24 h of infection were sequenced, and mutations were identified in O1-antigen biosynthetic phase variable genes wbeL and manA (21) as well as in other lipopolysaccharide biosynthetic genes (SI Appendix, Fig. S4 B and C and Table S1). At the same time, a small percentage of survivors (4%) was still sensitive to infection by ICP1 (SI Appendix, Fig. S4A, 24 h). No ICP2 or ICP3 escape mutants were detected in the estuary cultures, consistent with the lack of phage replication (Fig. 2A).
Salinity Is a Crucial Factor for ICP1 Predation.
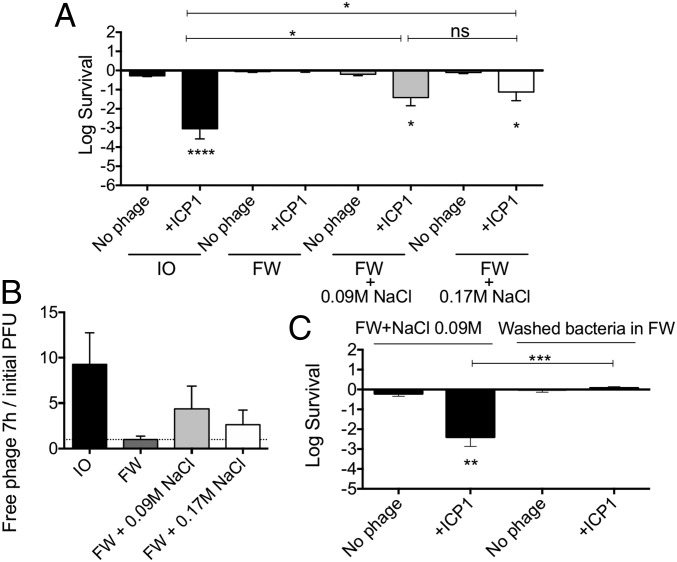
Since ICP1 was able to kill V. cholerae in estuarylike conditions and not in fresh water, we hypothesized that either the physiology of V. cholerae varies in the two environments or that a cofactor present in estuary but not in fresh water might be needed by ICP1 for predation. One major difference between estuary and fresh water is salinity. The concentration of Na+ in fresh water is about 0.3 mM (10) compared with 92.4 mM in 0.7% Instant Ocean (22). To test if ICP1 replication requires high salinity, NaCl was added to fresh water to the levels present either in our estuarine condition (0.09 M) or in LB broth (0.17 M). Cultures of V. cholerae preadapted in the two NaCl concentrations tested were infected using a MOI of 0.01. ICP1 was able to infect and kill V. cholerae in fresh water supplemented with 0.09 or 0.17 M NaCl by 7 h, albeit 100-fold less than in estuary conditions (Fig. 3A). Consistent with these levels of killing, we observed moderate levels of phage replication compared with the results from estuary water (Fig. 3B). These results suggest that NaCl alone has an impact in predation by ICP1, however, this was not sufficient to obtain the same level of predation seen in estuary conditions. Fresh water often contains growth-limiting amounts of nutrients, such as phosphate and fixed nitrogen (10), and this may prevent V. cholerae from fully supporting ICP1 infection.
Fig. 3.
A salinity threshold is needed for ICP1 predation. Some 107 CFU of V. cholerae from an overnight culture were preadapted overnight in 0.7% Instant Ocean, fresh water, or fresh water supplemented with NaCl (0.09 or 0.17 M) in the absence of an added carbon source. ICP1 was added to a MOI of 0.01 to assess bacterial viability and phage replication over time. Shown is the average with a standard error (SE) of, at least, six biological replicates. (A) Bacterial viability at 7 h. (B) The ratio of phages in the culture supernatant at 7 h relative to the number initially added. (C) Bacterial viability at 7 h (*P < 0.05, **P < 0.005, ****P < 0.0001). FW, fresh water; ns, not significant; PFU, plaque-forming units.
Because V. cholerae is a halophile, growth in high salinity may be required for the cells to become susceptible to ICP1. Alternatively, ICP1 attachment or infection may require the presence of salt. To differentiate between these, we preadapted V. cholerae in fresh water supplemented with 0.09 M NaCl, then washed and resuspended the cells in fresh water alone. If NaCl is necessary for phage attachment or infection, changing the media to fresh water alone should impair predation. Cultures were washed and infected with ICP1 at a MOI of 0.01, and viable CFU were assessed at 7 h. ICP1 exhibited impaired predation (Fig. 3C) similar to the levels seen in fresh water alone (compare Figs. 2B and 3A). These results indicate that, at the time of infection, ICP1 predation in an aquatic condition requires a minimum salinity threshold well above the salinity of fresh water.
Chitin Availability Aids Phage Predation.
It has been proposed that V. cholerae can grow in the environment using chitin as a nitrogen and carbon source (23). We first tested whether V. cholerae could utilize chitin for growth in our aquatic conditions by adding 104 CFU from a stationary phase culture into a 1% chitin suspension in either 0.7% Instant Ocean or fresh water. V. cholerae was able to grow in both, however, growth in the estuarine condition was faster and reached a higher yield compared with fresh water (SI Appendix, Fig. S5).
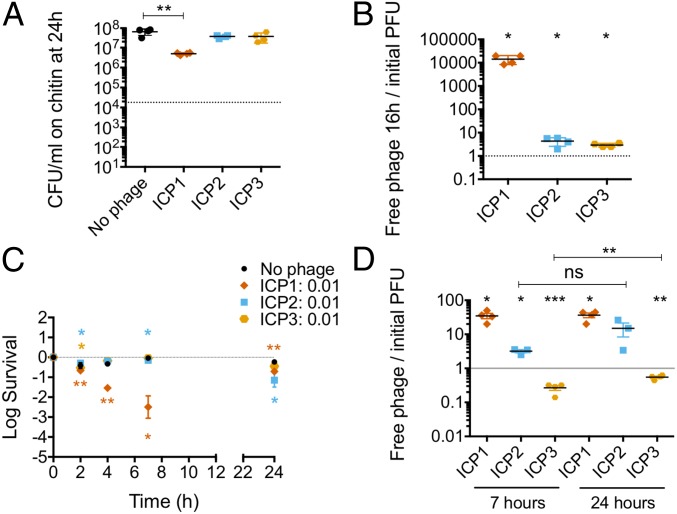
To evaluate if chitin utilization aids phage predation, we tested V. cholerae both in exponential and in stationary phases of growth. For exponential growth in the estuary condition plus chitin, preadaptation was performed for 8 h at 30 °C in static conditions (SI Appendix, Fig. S5B). Each phage was then added to a MOI of 0.01 and incubated for 16 h after which bacterial viability and phage replication were assessed. ICP1 reduced the V. cholerae population only by 10-fold (Fig. 4A) due to the generation of escape mutants during the incubation period. On the other hand, neither ICP2 nor ICP3 caused a reduction in bacterial numbers. Consistent with predation by ICP1, the phage titer increased 10,000-fold, whereas ICP2 and ICP3 titers showed no significant increase (Fig. 4B).
Fig. 4.
Vibrio cholerae is more susceptible to ICP1 and ICP2 in the presence of chitin. Some 104 CFU of V. cholerae from an overnight culture were preadapted in a 1% chitin suspension in 0.7% Instant Ocean until exponential growth. ICP1, ICP2, or ICP3 were added to a MOI of 0.01 to assess bacterial viability and phage replication over time. Shown is the average with a SE of, at least, three biological replicates. (A) Bacterial viability at 16 h. (B) Phage replication at 16 h relative to the number initially added. (C) Some 107 CFU of V. cholerae from an overnight culture were preadapted overnight for phage predation. ICP1, ICP2, or ICP3 were added to a MOI of 0.01. Bacterial viability at 1, 2, 4, 7, and 24 h in estuary conditions is shown. (D) The ratio of phages in the culture supernatant at 7 and 24 h relative to the number initially added (*P < 0.05, **P < 0.005, ****P < 0.0001). ns, not significant; PFU, plaque-forming units.
To test stationary phase cells, 107 CFU from an overnight culture were inoculated in a 1% chitin suspension in either 0.7% Instant Ocean water or fresh water, and cultures were incubated overnight at 30 °C in static conditions. Each phage was then added to a MOI of 0.01, and bacterial viability and phage replication were measured. In fresh water, none of the phages were able to kill V. cholerae (SI Appendix, Fig. S6A) or replicate (SI Appendix, Fig. S6B). In the estuary environment, we observed results similar to the estuary planktonic conditions. Only ICP1 had the ability to kill V. cholerae and to replicate (Fig. 4 C and D). Consistent with this killing, escape mutants arose; after 7 h of infection, 90% of the survivors were resistant to infection by ICP1, and this number grew to 98% within 24 h (SI Appendix, Fig. S4D). ICP2 exhibited a slightly increased ability to kill V. cholerae and to replicate compared with estuary alone; viability of V. cholerae was decreased by 10-fold at 24 h, and phage titer increased 10-fold. Consistent with this moderate level of killing, at 7 h of infection, one out of 24 isolates was resistant to ICP2 (SI Appendix, Fig. S4E). In contrast and similar to that observed in estuary alone, ICP3 was not able to reduce V. cholerae viability nor replicate (Fig. 4 C and D). Since V. cholerae was able to grow on chitin in both aquatic conditions (SI Appendix, Fig. S5A), the lack of phage predation in fresh water is not due to a lack of bacterial growth but instead appears to be impaired by the physicochemical properties of the water and/or the host’s response to growth in those conditions.
ICP2 and ICP3 Bound to V. cholerae Do Not Replicate in the Infant Mouse Small Intestine.
ICP1 replication in high salinity aquatic environments seems plausible according to our results and could explain the high prevalence of ICP1 in Bangladesh. However, it is unclear how ICP2 and ICP3 are maintained. ICP2 and ICP3 have only been observed to naturally replicate in V. cholerae that infects the intestine, and the data provided here raise doubts as to their ability to replicate on V. cholerae in nutrient-poor environmental waters. Since both phages and bacteria are shed following human infection, it is possible that ingestion of contaminated water could perpetuate phage replication. One question that arises is whether phages and bacteria are ingested as separate entities or whether they are coassociated. The former has already been demonstrated in two different animal models of intestinal infection wherein phages added before a challenge with V. cholerae or concurrently substantially decrease intestinal colonization (9, 12).
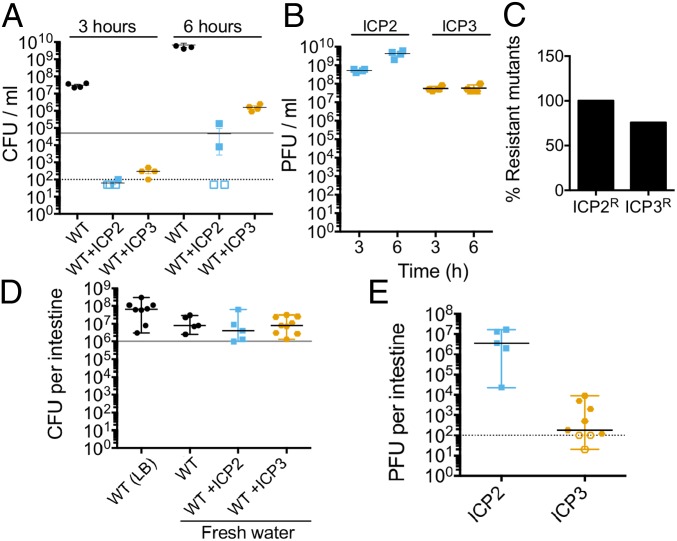
To test the latter hypothesis, we first examined the ability of cell-associated phages in fresh water to resume replication upon a shift to a nutrient rich condition. We infected 107 CFU in fresh water with each phage for 45 min at a MOI of 1 in static conditions at 24 °C. Cultures were washed to remove free phages, and the cells carrying bound phages were added to 2 mL of LB broth and grown (outgrowth) at 37 °C with aeration. After the wash step, 20% of ICP2 added and 0.01% of ICP3 added were cell associated and yielded live virions, similar to the planktonic predation results shown previously (Fig. 1D). Subsequently, both ICP2 and ICP3 reduced V. cholerae numbers after 3 h of outgrowth (Fig. 5A). This indicates that, when the small number of bacteria with live bound phages resumed growth, both ICP2 and ICP3 were able to resume replication and kill the bacteria. Within 6 h, bacterial numbers increased by means of the growth of escape mutants. Consistent with phage predation, ICP2 titer increased by 100-fold at 3 h and by 1,000-fold at 6 h (Fig. 5B); ICP3 titer increased by 5,000-fold within 3 h (Fig. 5B). Also consistent with phage predation, at 6 h of outgrowth, 100% (24/24) of the V. cholerae isolates were ICP2 escape mutants (Fig. 5C). For the ICP3 infected cultures at 6 h, 75% (22/29) of V. cholerae isolates recovered were escape mutants (Fig. 5C).
Fig. 5.
ICP2 and ICP3 replicate in a nutrient rich environment. Some 107 CFU of V. cholerae from an overnight culture were preadapted in fresh water overnight in the absence of an added carbon source. ICP2 or ICP3 was added to a MOI of 1, and adsorption was conducted for 45 min. Cultures were washed and inoculated into 2 mL of LB broth or into CD-1 mice for assessment of bacterial colonization and phage replication. The empty symbols represent data under the limit of detection. (A) Bacterial replication in LB broth at 37 °C with aeration at 3 or 6 h of outgrowth of four independent replicates. The gray line indicates bacterial concentration at time 0. (B) Phage titer from LB cultures at 3 or 6 h of outgrowth. (C) The average of percentage of resistant mutants for ICP2 and ICP3 at 6 h of outgrowth. (D) Bacterial colonization of the small intestine in CD-1 infant mice. The graph represents the median with a range of, at least, five biological replicates. The gray line indicates bacterial concentration in the inoculum. (E) Phage titer from a colonized small intestine. The graph represents the median with a range of, at least, five biological replicates. PFU, plaque-forming units.
We next examined the ability of bound phages to reduce V. cholerae colonization of the small intestine of 5-d-old mice. Some 106 CFU of stationary phase V. cholerae that were preadapted overnight in fresh water were infected with ICP2 or ICP3 for 45 min in fresh water at a MOI of 1. Cultures were washed and resuspended in fresh water for orogastric inoculation into mice. As controls, mice were also inoculated in the absence of phages with 106 CFU of V. cholerae grown overnight in LB broth or preadapted overnight in fresh water. After 24 h of infection, mice were killed, the small intestines were dissected, homogenized, and the titers of V. cholerae and phages were determined. Preadaptation overnight in fresh water did not reduce the colonization ability of V. cholerae since the load of bacteria per small intestine was not different from that of V. cholerae grown in LB (Fig. 5D). Our results show that cell-associated ICP2 and ICP3 do not limit V. cholerae infection since the V. cholerae loads present in those animals were not different from either of the control animals (Fig. 5D).
ICP3 was undetectable in a third of the animals and observed at low titer in the remaining ones (Fig. 5E). The inability of ICP3 to limit V. cholerae colonization of mice under these conditions can be explained by its inability to replicate after attachment to cells in fresh water. In contrast, ICP2 appeared to be able to replicate, at least, to some extent since all of the small intestines had a phage titer slightly higher than the number inoculated (Fig. 5D). However, this was not sufficient to reduce V. cholerae colonization of these animals. Consistent with this, no V. cholerae escape mutants to ICP2 were found among 40 isolates screened (eight from each mouse output). Thus, cell-associated ICP2 and, particularly, ICP3 fare poorly when inoculated into the infant mouse intestinal tract and, as a result, are not able to reduce the load of V. cholerae to any measurable extent.
Discussion
A better understanding of the ecology of V. cholerae and its phages in aquatic reservoirs could lead to the development of better tools for outbreak prediction and potential treatments to water supplies to reduce human infection. We used fresh and estuary water microcosms to study the interaction of three virulent phages with V. cholerae. These experiments are reductionist since in its reservoir V. cholerae has complex interactions with other organisms and it experiences changes in the physicochemical properties of the water throughout the year (24). Nevertheless, our results have relevance for understanding the impact of virulent phages on V. cholerae during cholera outbreaks during which V. cholerae and phages are shed in secretory diarrhea into bodies of water and are subsequently transmitted via consumption of contaminated fresh water.
It has been proposed that cholera outbreaks and V. cholerae populations are inversely correlated with the phage levels in the aquatic environment (11, 12), and a predation role was suggested for JSF4 (9), an ICP1-like phage (25). Our results do not support this model since ICP2 and ICP3 were shown to be incapable of preying on V. cholerae in fresh or estuary water and ICP1 was incapable of preying on V. cholerae in fresh water. However, since ICP1 is capable of predation in estuary water, it might play a role in reducing populations of V. cholerae is estuarine environments.
We also found that ICP1 is able to prey on stationary phase or nondividing V. cholerae, perhaps suggesting a role for predation in the long-term maintenance of ICP1 in nutrient-poor aquatic reservoirs. This may be a factor in the high frequency of isolation of ICP1 in patient stools and aquatic environments in Bangladesh (9, 15). ICP1 shares features with Escherichia coli T4 phage (15), and it was shown that T4 has the ability to kill stationary phase E. coli (26). However, we do not yet know how ICP1 infects stationary phase V. cholerae. It is known that genome injection by T4 requires a membrane potential (27). However, V. cholerae was highly motile, which requires a membrane potential (28), even after 48 h in fresh water in the absence of an added carbon source. Thus, the absence of a membrane potential is unlikely to explain the lack of killing by ICP1 in this setting. Our results indicate that the presence of NaCl could be an important factor for ICP1 infection. This could be due to Na+, which has been shown to be important for λ DNA injection (29).
It remains possible that ICP1 can prey on V. cholerae in fresh water due to the complex Bangladeshi climate scenario with high temperatures in summer that evaporate fresh water increasing its salinity in addition to the monsoon and flooding that lead to the mixture of aquatic reservoirs. Another possible scenario is that ICP1 in fresh water could be ingested in planktonic form or when attached to V. cholerae and replicate within the host as has been suggested (9).
ICP2 has been found sporadically in patient and environmental samples (15, 30). Our results show that ICP2 was mostly inefficient in attaching to V. cholerae and had modest predation capability in the presence of chitin in estuary conditions. ICP2 seems to thrive instead within the host and nutrient rich environments.
The last phage we studied was ICP3, a T7-like phage (15). We show that ICP3 is able to attach to V. cholerae but is incapable of replicating in fresh or estuary water and the majority becomes inactive after a short period of time. ICP3 when bound to outer membrane vesicles cannot inject its DNA, and 90% of bound ICP3 shows a full capsid (31). T7 DNA ejection needs active transport into the bacterial host that is mediated by the host RNA polymerase (28). V. cholerae remained highly motile in the nutrient-poor settings we tested, suggesting it is metabolically active, but it is possible that the host RNA polymerase is not available to aid in phage DNA transport into the cell. Despite the majority of ICP3 becoming inactive after adsorption to V. cholerae, a fraction of the phages remained intact as phages were recovered in the majority of the infected animals. This could explain why it is found sporadically in cholera patient samples and suggests that ICP3, like ICP2, has adapted to replicate within a nutrient rich microenvironment.
Because of ICP3’s fickle nature in water, it is possible that maintenance of ICP3 in Bangladesh relies primarily upon rapid transmission between people, such as occurs within households (32). Of 29 sequences reported for environmental V. cholerae phages (11, 25, 30, 33), 41% corresponded to ICP3-like phages. This result is striking since our model does not support the notion that ICP3 can replicate in nutrient-limiting aquatic environments. However, detailed information about the exact locations for sampling is lacking in these studies. It is possible that these phages were collected from surface waters near fish markets or nutrient rich microenvironments that allow V. cholerae growth (34) or near sewage where human-shed phage could be collected.
ICP2 and ICP3 also fared poorly at predation in the infant mouse small intestine after being coinoculated in cell-associated form. This contrasts with the sporadic occurrence of these phages, often at high titers, in cholera patient stools (15, 25). The inability of these phages to replicate in our animal experiments could be due to any number of reasons. For example, niches that support replication within the human small intestine may not be reproduced in infant mice. Alternatively, replication of V. cholerae in the absence of associated phages is required to occur for some period of time before free phages can successfully initiate replication. V. cholerae escape mutants have been observed in patient stools with high titers of ICP2 (35). The absence of ICP2 escape mutants in our mouse colonization experiment is consistent with a lack of sufficient phage predation needed to impose selection on V. cholerae.
Our results suggest that ICP2 and ICP3 evolved to replicate within nutrient rich environments, which might occur in certain environmental settings and within the human intestinal tract. On the other hand, our data suggest that ICP1 has evolved to be adaptable for predation on V. cholerae both in the human host and in the aquatic environments given adequate salinity. These different niche adaptations may help to maintain all three phages in the complex cholera endemic setting of Bangladesh.
To summarize, our paper highlights the evolution of niche-specific predation by three virulent phages. We examined adaptation for predation in different environments including the small intestine and aquatic environments that vary by Na+, carbon, and nitrogen source concentrations. Our results suggest that virulent phages have evolved to infect V. cholerae at different stages of this pathogen’s life cycle to ensure their survival. This paper will improve our understanding of how cholera outbreaks are impacted by the negative selection pressures of virulent phages. This will ultimately help researchers to develop more accurate models for predicting the dynamics of cholera outbreaks as well as enlighten us for the possible use of virulent phages to combat this disease.
Materials and Methods
Strains, Media, and Growth Conditions.
V. cholerae HC1037, an O1 El Tor clinical isolate (31), was grown on LB Miller broth or LB Miller agar plates at 37 °C. Fresh water was collected from Massapoag Lake. Its chemical composition is similar to pond water obtained from a cholera endemic area in Bangladesh (10, 17, 18, 36, 37). Estuary conditions were emulated by a 7 g/L (0.7%) solution of Instant Ocean Sea Salts (Spectrum Brands, Inc). Phages used in this paper were isolated from patient samples from Bangladesh and correspond to ICP1 (ICP1_2011_A; Myoviridae), ICP2 (ICP2_2004; Podoviridae), and ICP3 (ICP3_2007; Podoviridae) (15).
Phage Adsorption and Predation Assay in Aquatic Microcosms.
Stationary phase V. cholerae were diluted 1:1,000 and grown for 8 h at 37 °C with aeration. Then, 5 µL (∼108 CFU) were inoculated into 2.5 mL of autoclaved filter sterilized fresh water or 0.7% Instant Ocean and incubated overnight at 30 °C with aeration for adaptation. In some experiments, 1% shrimp chitin flakes (Sigma-Aldrich) was added, and cultures were preadapted statically at 30 °C.
After adaptation, phages were added at a MOI of 0.01. Adsorption was measured after static incubation at 24 °C for 15 min. Phage predation was assessed at 30 °C with aeration: After 1, 2, 4, 7, and 24 h, aliquots were taken to measure bacterial viability by plating, and free phages in the supernatant were measured by plaque assay as described in the SI Appendix. The survival rate of V. cholerae was calculated as follows: CFU/mL infected/CFU/mL initial and converted logarithmically.
To measure phage replication, infected cultures were centrifuged to separate bacteria from free phages in the supernatant. Some 500 µL of supernatant were filter sterilized using a 0.22 μm filter (COSTAR). The titer of the phages was measured by plaque assay.
To measure viable phages associated with V. cholerae, infected cells were washed in either fresh water or 0.7% Instant Ocean. Bacterial suspensions were serially diluted, fresh V. cholerae was added, and mixtures were allowed to incubate for 10 min at 24 °C. Each dilution was transferred to a 24-well clear plate for plaque assays.
Phage stability was measured by plaque assays after incubating each phage for 24 h in either fresh water or 0.7% Instant Ocean in the absence of a host strain. All samples were analyzed using a one-sample t test and GraphPad Prism version 6.00.
Infant Mice Colonization.
To evaluate the impact of ICP2 or ICP3 on colonization, V. cholerae preadapted overnight in fresh water were incubated with each phage at a MOI of 1 or in fresh water alone. After 45 min of incubation, bacteria were washed to remove unbound phages, and 50 µL containing ∼105 CFU were inoculated orogastrically to 5-d-old litters (both sexes) of CD1 mice (38). Twenty-four hours post infection, mice were killed, and small intestines were collected for homogenization in 1 mL of LB broth supplemented with 20% glycerol. The bacterial load was assessed by serial dilution, and plating of viable CFU on LB agar plates was supplemented with streptomycin 100 µg/mL (36, 39). Plaque-forming units were enumerated by plaque assay. Results were analyzed using a one-sample t test and GraphPad Prism version 6.00.
Animal procedures were conducted in accordance with the Tufts University School of Medicine Institutional Animal Care and Use Committee approved experimental protocol “B2016-03” and the rules of the Department of Laboratory Animal Medicine.
Supplementary Material
Acknowledgments
We thank D. Lazinski and R. Molina-Quiroz for reviewing this paper and the members of the laboratory for helpful discussions. This work was supported by the Howard Hughes Medical Institute and NIH Grant AI055058 (to A.C.). C.A.S.-V. was supported by the Pew Latin American Fellows Program in the Biomedical Sciences from Pew Charitable Trusts and a Conicyt Comisión Nacional de Investigación Científica y Tecnológica Becas Chile postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810138116/-/DCSupplemental.
References
- 1.Lutz C, Erken M, Noorian P, Sun S, McDougald D. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol. 2013;4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan MU, Shahidullah MD, Haque MS, Ahmed WU. Presence of vibrios in surface water and their relation with cholera in a community. Trop Geogr Med. 1984;36:335–340. [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses–Major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seed KD. Battling phages: How bacteria defend against viral attack. PLoS Pathog. 2015;11:e1004847. doi: 10.1371/journal.ppat.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Herelle F, Malone RH. A preliminary report of work carried out by the cholera bacteriophage enquiry. Ind Med Gaz. 1927;62:614–616. [PMC free article] [PubMed] [Google Scholar]
- 7.Butler SM, et al. Cholera stool bacteria repress chemotaxis to increase infectivity. Mol Microbiol. 2006;60:417–426. doi: 10.1111/j.1365-2958.2006.05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merrell DS, et al. Host-induced epidemic spread of the cholera bacterium. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque SM, et al. Self-limiting nature of seasonal cholera epidemics: Role of host-mediated amplification of phage. Proc Natl Acad Sci USA. 2005;102:6119–6124. doi: 10.1073/pnas.0502069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson EJ, et al. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 2008;4:e1000187. doi: 10.1371/journal.ppat.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque SM, et al. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci USA. 2005;102:1702–1707. doi: 10.1073/pnas.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen MA, Faruque SM, Mekalanos JJ, Levin BR. Modeling the role of bacteriophage in the control of cholera outbreaks. Proc Natl Acad Sci USA. 2006;103:4652–4657. doi: 10.1073/pnas.0600166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yen M, Cairns LS, Camilli A. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat Commun. 2017;8:14187. doi: 10.1038/ncomms14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Herelle F. On an invisible microbe antagonistic toward dysenteric bacilli. C R Acad Sci. 1917;165:373–375. doi: 10.1016/j.resmic.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Seed KD, et al. Evidence of a dominant lineage of Vibrio cholerae-specific lytic bacteriophages shed by cholera patients over a 10-year period in Dhaka, Bangladesh. MBio. 2011;2:e00334-10. doi: 10.1128/mBio.00334-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasan ABMSU, Rahman MZ. Change in temperature over Bangladesh associated with degrees of global warming. Asian J Appl Sci Eng. 2013;2:161–174. [Google Scholar]
- 17.Silva-Valenzuela CA, et al. Growth arrest and a persister state enable resistance to osmotic shock and facilitate dissemination of Vibrio cholerae. ISME J. 2017;11:2718–2728. doi: 10.1038/ismej.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 2013;9:e1003800. doi: 10.1371/journal.ppat.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmond GPC, Fineran PC. A century of the phage: Past, present and future. Nat Rev Microbiol. 2015;13:777–786. doi: 10.1038/nrmicro3564. [DOI] [PubMed] [Google Scholar]
- 20.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. Cholera. Lancet. 2012;379:2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seed KD, et al. Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog. 2012;8:e1002917. doi: 10.1371/journal.ppat.1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atkinson MJ, Bingman C. Elemental composition of commercial seasalts. J Aquaricult Aquat Sci. 1996;8:39–43. [Google Scholar]
- 23.Johnson CN. Fitness factors in vibrios: A mini-review. Microb Ecol. 2013;65:826–851. doi: 10.1007/s00248-012-0168-x. [DOI] [PubMed] [Google Scholar]
- 24.Vezzulli L, Colwell RR, Pruzzo C. Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb Ecol. 2013;65:817–825. doi: 10.1007/s00248-012-0163-2. [DOI] [PubMed] [Google Scholar]
- 25.Naser IB, et al. Environmental bacteriophages active on biofilms and planktonic forms of toxigenic Vibrio cholerae: Potential relevance in cholera epidemiology. PLoS One. 2017;12:e0180838. doi: 10.1371/journal.pone.0180838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryan D, El-Shibiny A, Hobbs Z, Porter J, Kutter EM. Bacteriophage T4 infection of stationary phase E. coli: Life after log from a phage perspective. Front Microbiol. 2016;7:1391. doi: 10.3389/fmicb.2016.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu B, Margolin W, Molineux IJ, Liu J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci USA. 2015;112:E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol. 2004;53:1251–1265. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- 29.Wu D, Van Valen D, Hu Q, Phillips R. Ion-dependent dynamics of DNA ejections for bacteriophage lambda. Biophys J. 2010;99:1101–1109. doi: 10.1016/j.bpj.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoque MM, et al. Quorum regulated resistance of Vibrio cholerae against environmental bacteriophages. Sci Rep. 2016;6:37956. doi: 10.1038/srep37956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes-Robles T, et al. Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J Bacteriol. 2018;200:e00792-17. doi: 10.1128/JB.00792-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris JB, et al. Susceptibility to Vibrio cholerae infection in a cohort of household contacts of patients with cholera in Bangladesh. PLoS Negl Trop Dis. 2008;2:e221–e228. doi: 10.1371/journal.pntd.0000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naser IB, et al. Analysis of the CRISPR-Cas system in bacteriophages active on epidemic strains of Vibrio cholerae in Bangladesh. Sci Rep. 2017;7:14880. doi: 10.1038/s41598-017-14839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mookerjee S, et al. Seasonal dynamics of Vibrio cholerae and its phages in riverine ecosystem of Gangetic West Bengal: Cholera paradigm. Environ Monit Assess. 2014;186:6241–6250. doi: 10.1007/s10661-014-3851-1. [DOI] [PubMed] [Google Scholar]
- 35.Seed KD, et al. Evolutionary consequences of intra-patient phage predation on microbial populations. eLife. 2014;3:e03497. doi: 10.7554/eLife.03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schild S, et al. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe. 2007;2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourassa L, Camilli A. Glycogen contributes to the environmental persistence and transmission of Vibrio cholerae. Mol Microbiol. 2009;72:124–138. doi: 10.1111/j.1365-2958.2009.06629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tischler AD, Camilli A. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun. 2005;73:5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camilli A, Beattie DT, Mekalanos JJ. Use of genetic recombination as a reporter of gene expression. Proc Natl Acad Sci USA. 1994;91:2634–2638. doi: 10.1073/pnas.91.7.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.