Significance
Facial masculinity has been considered a sexual ornament in humans, akin to peacock trains and stag antlers. Recently, studies have questioned the once-popular view that facial masculinity is a condition-dependent male ornament signaling immunocompetence (the immunocompetence handicap hypothesis). We sought to rigorously test these ideas using high-resolution phenotypic (3D facial images) and genetic data in the largest sample to date. We found no support for the immunocompetence handicap hypothesis of facial masculinity in humans. Our findings add to a growing body of evidence challenging a popular viewpoint in the field and highlight the need for a deeper understanding of the genetic and environmental factors underlying variation in facial masculinity and human sexual dimorphism more broadly.
Keywords: facial masculinity, MHC heterozygosity, sexual selection, immunocompetence handicap hypothesis, human evolution
Abstract
Recent studies have called into question the idea that facial masculinity is a condition-dependent male ornament that indicates immunocompetence in humans. We add to this growing body of research by calculating an objective measure of facial masculinity/femininity using 3D images in a large sample (n = 1,233) of people of European ancestry. We show that facial masculinity is positively correlated with adult height in both males and females. However, facial masculinity scales with growth similarly in males and females, suggesting that facial masculinity is not exclusively a male ornament, as male ornaments are typically more sensitive to growth in males compared with females. Additionally, we measured immunocompetence via heterozygosity at the major histocompatibility complex (MHC), a widely-used genetic marker of immunity. We show that, while height is positively correlated with MHC heterozygosity, facial masculinity is not. Thus, facial masculinity does not reflect immunocompetence measured by MHC heterozygosity in humans. Overall, we find no support for the idea that facial masculinity is a condition-dependent male ornament that has evolved to indicate immunocompetence.
The condition-dependent hypothesis is used to explain the evolution of male ornaments in nonhuman animals (1–5). According to this hypothesis, male ornaments (e.g., reindeer antlers and peacock trains), which grow to exaggerated proportions even though they might be detrimental to fitness (6, 7), are adaptations signaling the underlying physiological and genetic quality of the individual to females. Such traits are more sensitive to the overall growth of individuals and more variable than other traits (8–14). As growth itself is dependent on a variety of genetic and environmental factors, including immunocompetence, inbreeding, health status, and nutrient availability (15–19), slight variations in physiological and genetic quality among males are amplified to perceptible levels in sexual ornaments, making them reliable indicators of underlying health (8, 9, 13). The condition-dependent hypothesis has also been applied to humans to explain the evolution of secondary sexual characteristics, such as facial masculinity and deep voices (20–24). The apparent attraction of women to these traits, which appears to be heritable (25), is thought to be an evolutionary adaptation that helps women secure direct (e.g., investment of a healthy male) and indirect (e.g., “good genes” for their children) benefits (1, 3, 26, 27).
Among all of the factors placed under the umbrella of “condition,” immunocompetence has received considerable attention. This is, in part, because of the supposed immunosuppressive effects of androgens (28, 29), which are involved in the development of secondary sexual traits in males (22). According to the immunocompetence handicap hypothesis (ICHH), androgens mediate the allocation of resources between the competing demands of fighting infections and the development of energetically “costly” sexual ornaments (30–41). Consequently, males with more effective immune systems may withstand higher androgen levels, and the accompanying immunosuppressive burden, and can “afford” more extravagant displays. If this were true, then secondary sexual characteristics could serve as reliable (“honest”) indicators of the physiological and immunological quality of males (7, 35, 40, 42). Parts of the ICHH have found some support in humans (36, 43–45) and nonhuman animals [for review, see Roberts et al. (46)].
However, the evidence linking secondary sexual traits to the condition, immunological or otherwise, of human males is ambiguous and inconsistent across studies (44, 46–48). That androgens are immunosuppressive also does not appear to be well-supported (49). This has recently led many to question the applicability of the ICHH in humans, particularly with respect to facial masculinity (49–51). Some of the inconsistency has been attributed to methodological limitations, such as small sample size and the use of measures of perceived masculinity and attractiveness, which are influenced by sociocultural factors that are difficult to control in observational studies (50). Another limitation that has received less attention is the lack of correction for ancestry and population structure, which can lead to spurious associations. Because of these issues, a rigorous study of the link between facial masculinity and immunocompetence and/or condition is needed.
In this study, we investigated the condition-dependent hypothesis and ICHH in humans with respect to facial masculinity. Working from theory and evidence from research on condition dependence of sexual ornaments in nonhuman animals (1–5, 8), we tested three hypotheses:
Hypothesis 1: Facial masculinity is a condition-dependent male ornament in humans. If this is true, then we expect facial masculinity to be (i) more strongly correlated with overall growth in males relative to females, and (ii) more variable in males compared with females.
Hypothesis 2: Immunocompetence is associated with overall growth in humans. If immunocompetence plays a role in condition-dependent expression of secondary sexual characteristics, then it should be correlated with overall growth in humans.
Hypothesis 3: Facial masculinity reflects immunocompetence in men. Males who show greater immunocompetence should exhibit more masculine faces than males with lower immunocompetence. In contrast, facial masculinity should be less sensitive to variation in immunocompetence in females.
To test our hypotheses, we used an objective measure of facial masculinity, calculated with high resolution 3D photographs in a large sample of persons of European ancestry. We used height as a proxy for overall growth and condition as height is known to be associated with health, income, nutrition, and exposure to disease and infection (15–17). We used individual heterozygosity at the major histocompatibility locus (MHC) as a measure of immunocompetence. The MHC locus, also known in humans as the HLA complex, is located on chromosome 6 and contains around 200 genes that are involved in immune function (52). Higher genetic diversity at the MHC enables the immune system to recognize a more diverse array of foreign antigens (52–54). As a result, the MHC has experienced balancing selection in both humans and nonhumans (52, 55–58). Therefore, heterozygosity at this locus serves as a useful proxy to measure immunocompetence. Finally, we considered the effects of body size on facial masculinity (allometry) in addition to other likely confounders, such as age, weight, genome-wide heterozygosity, and population structure.
Results
Variation in Facial Masculinity.
We calculated high resolution facial masculinity (FM) for the faces of 1,233 males and females of European ancestry from 3D images using a scalar-projection approach, similar to that described in Valenzano et al. (59) (SI Appendix, Fig. S7). The 3D images were processed as described previously (60–62), allowing us to represent each face as a mesh of 7,150 points, or quasi-landmarks (QLs), each with x, y, and z coordinates. For every QL in the face, the signed difference between the coordinates of the average female and male faces represents the direction of sexual dimorphism in 3D space (SI Appendix, Fig. S7A). We defined FM for each of the 7,150 QLs (FMQL) as the degree of change in a target face (X) along these vectors. Note that this measure includes both allometric (size-dependent) and nonallometric (size-independent) components of facial masculinity (63–65). We correct for allometry, when necessary, by residualizing FM on height or by including height as a covariate, which yielded identical results (Materials and Methods and SI Appendix, Fig. S8).
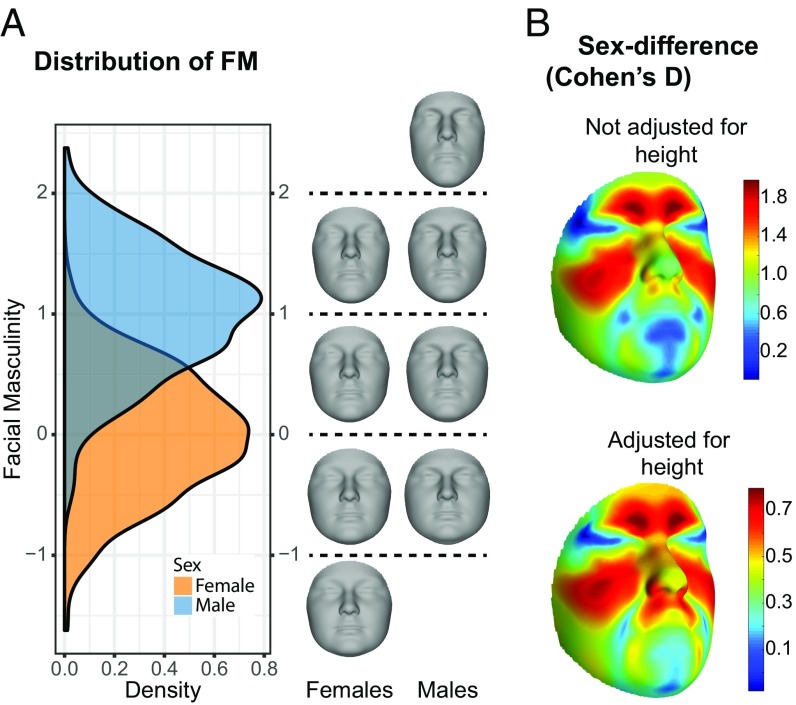
Fig. 1A shows a bimodal distribution of overall FM score (averaged across QLs—hereafter referred to as FMoverall) where values of 0 and 1 represent FMoverall of the average female and male faces, respectively. The magnitude of sex difference in FMoverall is comparable with that of height (Cohen’s D of 1.98 compared with 2.10 for height). Expectedly, the magnitude of sex difference in the nonallometric component of FMoverall was smaller (Cohen’s D = 0.76). The brow ridge, cheekbones, and nose ridge showed the greatest degree of sexual dimorphism, in agreement with previous studies (Fig. 1B) (62, 63, 66).
Fig. 1.
Variation in facial masculinity and sexual dimorphism. (A) Density plot showing a bimodal distribution of facial masculinity. (B) Heat maps showing Cohen’s D of dimorphism between average male and female faces before (Top) and after (Bottom) correction for allometry.
Facial Masculinity Is Positively Correlated with Height in both Sexes.
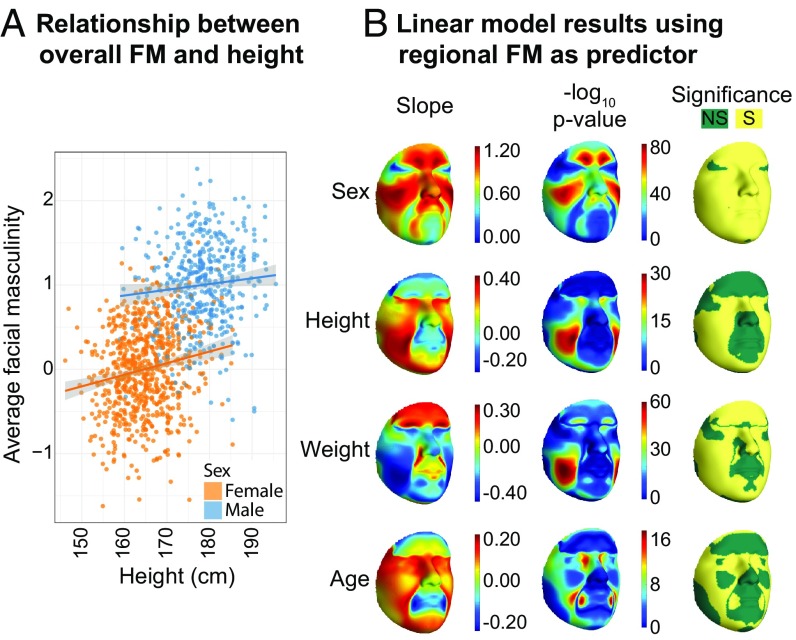
We tested the relationship between overall facial masculinity (FMoverall) and growth, using height as a predictor, with sex, age, weight, and genetic principal components (gPCs) 1 to 3 as covariates (SI Appendix, Fig. S11). FMoverall is positively correlated with height in both sexes (Fig. 2A and Table 1; T = 8.81, P = 4.17 × 10−18), suggesting that taller people have more masculine faces than shorter people. Because variation in the size of the faces was removed before calculating facial masculinity (Materials and Methods), this correlation represents allometric effects of growth on sexual dimorphism in face shape, not size (64). The effect of height on facial masculinity appears to be concentrated around the orbital region, nasal bridge, cheeks, and the chin, with masculinity in these regions increasing with height (Fig. 2B), matching previous observations on the effects of allometry on faces (63, 64). It is interesting to note that the distribution of the effect of height on masculinity across the face appears to be different from the effect of sex (Fig. 2B), which is consistent with previous observations that facial masculinity is not exclusively a result of extended overall growth in males compared with females (63, 65–68).
Fig. 2.
Facial masculinity is positively correlated with height within and between the sexes. (A) Relationship between FMoverall and height. (B) Results of linear model between FMQL and height and other covariates. Regions in yellow are significant after Bonferroni correction for 7,150 QLs (P < 0.05/7150 = 6.93 × 10−6). SI Appendix, Fig. S11 shows results for all covariates.
Table 1.
Results of linear model between FMoverall and height with covariates
| Predictor | Slope (β) | 95% CI | T statistic | P value |
| Sex | 1.240 | 1.126, 1.353 | 21.46 | 5.55 × 10−87 |
| Height | 0.260 | 0.201, 0.319 | 8.81 | 4.17 × 10−18 |
| Age | 0.109 | 0.070, 0.148 | 5.60 | 2.69 × 10−08 |
| Weight | −0.221 | −0.266, −0.176 | −9.64 | 3.07 × 10−21 |
| gPC1 | −0.083 | −0.122, −0.044 | −4.19 | 2.94 × 10−05 |
| gPC2 | −0.063 | −0.100, −0.026 | −3.25 | 1.78 × 10−03 |
| gPC3 | −0.052 | −0.089, −0.015 | −2.68 | 7.55 × 10−03 |
Slopes are standardized regression coefficients. Bonferroni cutoff for significance is 0.05/7 = 0.007. P values smaller than the Bonferroni cutoff are shown in boldface type.
The effect of height on FMoverall is not significantly different between males and females (βmale = 0.227, βfemale = 0.299, Z-scorediff = −1.18, P = 0.120), and a similar observation can be made for the regional effects of height on FMQL (SI Appendix, Fig. S12). FMoverall is also not significantly more variable in either sex (Levene’s test P = 0.37 with adjustment for height and P = 0.66 without adjustment). Greater sensitivity to growth and higher variance in males relative to females are classic signatures of male ornaments (2, 5, 8, 69), an expectation that facial masculinity does not meet. It is interesting to note that FM varies significantly along gPCs 1 and 2, suggesting that the patterns of sexual dimorphism and facial masculinity vary across populations, even within Europe (Table 1). This not only highlights the need to correct for population structure in future studies but also calls for a detailed exploration of the variation in facial shape across populations.
Height Is Positively Correlated with Immunocompetence.
We fit a linear model between height and MHC heterozygosity, while correcting for genome-wide heterozygosity, sex, age, and gPCs 1 to 3. MHC heterozygosity showed a positive correlation with height (Table 2; T = 3.18, P = 0.0015), indicating that individuals who are more heterozygous at the MHC locus tend to be taller than people who are less heterozygous. This relationship is not driven by genome-wide heterozygosity as the latter is not significantly associated with height (Table 2; T = −0.36, P = 0.72). Also note that height varies significantly along gPC1 (Table 2), which is consistent with the clinal variation in stature observed within Europe (18, 70).
Table 2.
Results of linear model between height and MHC heterozygosity
| Predictor | Slope (β) | 95% CI | T statistic | P value |
| MHC het. | 0.063 | 0.014, 0.112 | 3.18 | 1.54 × 10−03 |
| Genome-wide het. | −0.007 | −0.046, 0.032 | −0.36 | 0.72 |
| Sex | 1.452 | 1.374, 1.530 | 36.20 | 9.43 × 10−196 |
| Age | 0.022 | −0.017, 0.061 | 1.11 | 0.266 |
| gPC1 | 0.122 | 0.083, 0.161 | 6.11 | 1.36 × 10−09 |
| gPC2 | 0.051 | 0.012, 0.091 | 2.54 | 1.14 × 10−02 |
| gPC3 | 0.014 | −0.025, 0.053 | 0.70 | 0.49 |
Slopes are standardized regression coefficients. Bonferroni cutoff for significance is 0.05/7 = 0.007. P values smaller than the Bonferroni cutoff are shown in boldface type. het., heterozygosity.
Facial Masculinity Is Not Correlated with Immunocompetence.
MHC heterozygosity is not significantly correlated with overall facial masculinity regardless of whether height is included in the model (T = −0.038, P = 0.970) or not (T = 0.586, P = 0.558). Thus, neither the allometric nor the nonallometric component of facial masculinity is informative about MHC heterozygosity. MHC heterozygosity is also not significantly correlated with regional measures of facial masculinity (FMQL; SI Appendix, Fig. S13). Furthermore, there is no correlation between facial masculinity and genome-wide heterozygosity (height included as covariate: T = 0.02, P = 0.986; height not included as covariate: T = 0.65, P = 0.516). Finally, there is no difference in the effect of MHC heterozygosity on facial masculinity between males and females (βmale = 0.035, βfemale = −0.024, Z-scorediff = 0.32, P = 0.375; SI Appendix, Fig. S14).
Discussion
Condition-dependent male ornaments tend to be highly variable and more sensitive to variation in growth among males (i.e., exhibit greater allometric effects in males compared with females) (8, 10, 13, 14). Facial masculinity does not meet these expectations as neither the allometric nor the nonallometric component of facial masculinity is more variable in males compared with females, and the allometric effects of growth on facial masculinity are similar across the sexes.
Because diversity at the MHC locus is important for antigen recognition and presentation and the locus is known to be under balancing selection in many species, including humans, heterozygosity at this locus is an important marker of immunocompetence. This is supported by our finding that people who are more heterozygous at the MHC locus are taller, on average, than people who are less heterozygous, suggesting that variation at the MHC locus might be important for growth. We find that facial masculinity is not significantly associated with MHC heterozygosity, either in males or females. Our results do not support the hypothesis that facial masculinity is an indicator of immunocompetence. However, we cannot rule out the effect of other measures of immunocompetence such as non-MHC genes and antibody titers. We also find no support for the contention that FM indicates heterozygosity across the genome generally, something that has also been hypothesized previously (3, 71).
Altogether, our findings add to the growing number of studies questioning some of the evolutionary explanations behind female and male perceptions of facial masculinity (50, 51) and whether masculinity should be regarded as a condition-dependent male ornament in humans. Nevertheless, there are many questions related to facial sexual dimorphism and masculinity in humans that need to be addressed from an evolutionary standpoint. Humans show intermediate levels of allometric cranial sexual dimorphism among extant hominids (65), and we do not know whether this degree of sexual dimorphism is new to humans since their divergence from other hominins and apes. Did some aspects of facial masculinity evolve as a mechanism to intimidate rival males (72–74), or do they represent vestigial traits that have decreased over time as a result of self-domestication (75–77)? We also do not know how facial sexual dimorphism varies across human populations although we suspect that it does so considerably, both in degree and pattern, given that our results show that it varies significantly across Europe. Is genetic drift sufficient to explain these patterns? If not, can these patterns be explained by differences in perceptions of beauty and social status across populations? It is important to tackle these questions by careful comparison of the degree and pattern of sexual dimorphism within and across populations, as well as how sexual dimorphism is perceived cross-culturally.
Equally important is the need to fill gaps in our knowledge of the proximate causes underlying sexual dimorphism and facial masculinity. We know that differences in facial shape exist between male and female children as young as 3 y old (66, 68, 78) and are likely defined, in part, by the intrauterine environment during gestation (79–81). This dimorphism increases dramatically at the onset of puberty, implicating sex hormones and other endocrine processes underlying general growth during this period (66, 68, 82–85). These observations suggest that facial masculinity may arise because of extended overall growth and higher circulating androgen levels in pubertal males (66, 84, 86). However, sex differences in face shape are not merely developmental byproducts of extended overall growth in males as we and others have shown that sex has a significant effect on facial shape even after adjusting for body size (63, 65–68). Variation in facial masculinity also cannot be attributed solely to differences in circulating androgens during puberty. This is clear from the observation that, despite the fact that males exhibit higher mean and variance in androgen levels compared with females (87), they are not more variable in terms of facial masculinity. In fact, a recent report shows that the heritability of facial masculinity is similar between males and females, and the correlation between facial masculinity of same-sex siblings is similar to that of opposite-sex siblings (88). These results are indicative of a shared genetic architecture underlying facial masculinity in males and females and further serve to deemphasize the idea that facial masculinity is a male-specific ornament. It will be important to explore the effects of other hormones (e.g., estrogen and estradiol) and sex-chromosomal genes (89–91), as well as the timing of these effects. These questions are fundamental for cultivating a more mechanistic understanding of the development of sexual dimorphism, which, in turn, will lead to a better understanding of the role of sexual selection in human evolution.
Materials and Methods
Participant Recruitment.
Study participants were recruited with written informed consent in the United States through the Anthropology, DNA, and the Appearance and Perceptions of Traits (ADAPT) Study. All aspects of the study were approved by the Pennsylvania State University Institutional Review Board (no. 44929 and 45727). The 3D images were taken using the 3dMD Face system (3dMD). Height and weight were measured using an Accustat stadiometer (Genentech) and clinical scale (Tanita). Genotyping was conducted by 23andMe (23andMe) on the v4 genome-wide SNP array. After filtering out SNPs with more than 10% missing genotypes, this array comprised 567,787 SNPs.
Data Curation.
From the 2,721 participants with faces and genotype data, we removed individuals with missing covariate data, misclassified sex information, and individuals with more than 10% missing genotypes. We further restricted the analysis to unrelated individuals between 18 and 30 y of age (to reduce the effects of aging). Relatives were identified as pairs of individuals with an identity-by-state (IBS) value of at least 0.8, after which one of each pair was removed, resulting in a set of 1,921 unrelated individuals (SI Appendix, Fig. S1).
Ancestry and Population Structure.
We selected people of European ancestry as they comprised the largest sample in our dataset. To do so, we merged the genotype data from our sample (n = 1,921) with genotypes from the 1,000 Genomes Project dataset (n = 2,503) (92). Before the merge, we removed SNPs that did not intersect between the two datasets, palindromic (A/T, G/C) SNPs, and SNPs that did not meet standard quality-control criteria (SI Appendix, Fig. S1). SNPs were further pruned for linkage disequilibrium (LD) with a window size of 50 SNPs, a step size of 5 SNPs, and a variance inflation factor threshold of 2 using PLINK 1.9 (93, 94), resulting in 201,042 SNPs. Genetic ancestry was inferred using an unsupervised clustering scheme in ADMIXTURE, with K ranging from 2 to 16 (SI Appendix, Figs. S2 and S4) (95). We selected results from K = 6 as this value had a low cross-validation error (SI Appendix, Fig. S3) and showed separation based on continental ancestry (SI Appendix, Fig. S2). Then 1,249 individuals of primarily European ancestry were identified based on ADMIXTURE output by comparison with European samples from the 1,000 Genomes Project (SI Appendix, Fig. S4). We carried out principal components analysis on the genotypes of this subset and removed 16 outliers using the smartpca program in Eigensoft (SI Appendix, Fig. S5) (96, 97), leading to a sample size of 1,233 individuals. The first three genetic PCs (gPCs) were used as covariates to correct for population structure (SI Appendix, Fig. S5), which is minimal in our dataset beyond the first three gPCs (SI Appendix, Fig. S6).
Processing 3D Photographs.
High-resolution 3D images were “cleaned” to remove hair, ears, and disassociated polygons. Five positioning landmarks were placed (two on the inner corner of the eyes, two on the outer corners of the mouth, and one on the tip of the nose) to establish facial orientation. An anthropometric mask comprised of 10,000 quasi-landmarks (QLs), which was later trimmed to 7,150 QLs, was nonrigidly mapped onto all 3D surfaces such that each QL was spatially homologous across individuals (60–62). Thus, every face could be represented by a configuration of 7,150 QLs, each with three coordinates (x, y, and z). For each face, a mirror image was created by changing the sign of the x coordinates following (98), which was mapped with QLs in the same way as the original, nonreflected face. A generalized Procrustes superimposition (99) of both the original and reflected images together was performed to eliminate differences in position, orientation, and scale. The original and reflected images were then averaged to create a symmetric facial shape (62).
Calculating Facial Masculinity.
We define facial masculinity as the degree of change in the direction from an average female face to an average male face. Conversely, facial femininity is the degree of change in the opposite direction. We calculated facial masculinity (FM) per quasi-landmark (QL) for every face, using a scalar-projection approach (SI Appendix, Fig. S7) (59, 63, 100, 101). First, we generated female and male consensus faces from the sample by averaging the QL configurations across all females and all males, respectively. For every QL on the face, the signed difference between the coordinates of the male and female consensus faces was a 3D vector that represents the direction of sexual dimorphism in 3D space (SI Appendix, Fig. S7A). The goal was to calculate the degree of change in each QL of a target face X along these vectors (i.e., one for each of the 7,150 QLs), which is the FM per QL (FMQL). This could be done by computing the scalar projection of , the difference between X and the female consensus face, onto (SI Appendix, Fig. S7C):
Note that this measure represents both allometric and nonallometric components of FM. We corrected for the effects of allometry where necessary by including height as a covariate in the regression models. We show in SI Appendix that this is equivalent to other approaches, such as residualizing the original shape coordinates on height before constructing the male and female consensus faces and calculating FM (SI Appendix, Fig. S8) (63).
Genomic and MHC Heterozygosity.
We defined individual heterozygosity as the proportion of heterozygous SNPs in a region. Genome-wide heterozygosity was calculated from a total of 192,417 LD-pruned, autosomal SNPs. To measure MHC heterozygosity, we obtained a list of 195 SNPs tagging haplotype variation for the classical HLA genes in Europeans (102). We used 114 of these SNPs, the subset for which our samples were genotyped (SI Appendix, Fig. S9), to calculate MHC heterozygosity. These SNPs captured most of the HLA alleles (102) and the heterozygosity calculated using the subset of 114 SNPs was highly correlated with heterozygosity calculated using a larger subset (n = 154 SNPs) for which the sample of Europeans available in the 1000 Genomes Project dataset were genotyped (SI Appendix, Fig. S10) (92).
Data Availability.
The informed consent with which the data were collected does not allow for dissemination of identifiable data to persons not listed as researchers on the IRB protocol. Thus, the raw genotype data and 3D images cannot be made publicly available. In the interest of reproducibility, we have provided deidentified overall facial masculinity measures as well as age, sex, weight, height, ancestry, genetic PCs, and MHC and genome-wide heterozygosity, from which all results presented in this manuscript can be reproduced (Dataset S1). In addition, we provide high-density facial masculinity maps: i.e., facial masculinity calculated for every quasi-landmark (FMQL) for all 1,233 individuals used in the analyses (Dataset S2). Lastly, we provide all protocols and scripts used to process the genetic data and perform the analyses as well as a tutorial showing how FMQL can be used and visualized. This resource will allow other researchers to study variation in facial masculinity with high resolution in a large sample. All of the above are available on the following GitHub repository: https://github.com/Arslan-Zaidi/Facial_masculinity_MHC.
Supplementary Material
Acknowledgments
We thank the participants for providing the data necessary to carry out this study. We thank the members of the M.D.S. laboratory and the D.A.P. laboratory for helping with data collection; and thank Tina Lasisi and Tomás González-Zarzar for helpful discussions on the manuscript. Finally, we thank the Penn State Center for Human Evolution and Diversity (CHED), Research Fund KU Leuven (Grant BOF-C1, C14/15/081), and the Research Program of the Fund for Scientific Research–Flanders (Belgium) (Grant FWO, G078518N) for funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.M.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808659116/-/DCSupplemental.
References
- 1.Andersson M. Evolution of condition-dependent sex ornaments and mating preferences: Sexual selection based on viability differences. Evolution. 1986;40:804–816. doi: 10.1111/j.1558-5646.1986.tb00540.x. [DOI] [PubMed] [Google Scholar]
- 2.Iwasa Y, Pomiankowski A. The evolution of mate preferences for multiple sexual ornaments. Evolution. 1994;48:853–867. doi: 10.1111/j.1558-5646.1994.tb01367.x. [DOI] [PubMed] [Google Scholar]
- 3.Rowe L, Houle D. The lek paradox and the capture of genetic variance by condition dependent traits. Proc Biol Sci. 1996;263:1415–1421. [Google Scholar]
- 4.Bellamy L, Fowler K, Pomiankowski A. The use of inbreeding to assess the genetic component of condition underlying GEIs in sexual traits. In: Hunt J, Hosken D, editors. Genotype-by-Environment Interactions and Sexual Selection. John Wiley & Sons, Ltd; West Sussex, UK: 2014. pp. 213–240. [Google Scholar]
- 5.Cotton S, Fowler K, Pomiankowski A. Do sexual ornaments demonstrate heightened condition-dependent expression as predicted by the handicap hypothesis? Proc Biol Sci. 2004;271:771–783. doi: 10.1098/rspb.2004.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favored Races in the Struggle for Life. John Murray; London: 1859. [Google Scholar]
- 7.Andersson MB. Sexual Selection. Princeton Univ Press; Princeton: 1994. [Google Scholar]
- 8.Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science. 2012;337:860–864. doi: 10.1126/science.1224286. [DOI] [PubMed] [Google Scholar]
- 9.Cotton S, Fowler K, Pomiankowski A. Condition dependence of sexual ornament size and variation in the stalk-eyed fly Cyrtodiopsis dalmanni (Diptera: Diopsidae) Evolution. 2004;58:1038–1046. doi: 10.1111/j.0014-3820.2004.tb00437.x. [DOI] [PubMed] [Google Scholar]
- 10.Johns A, Gotoh H, McCullough EL, Emlen DJ, Lavine LC. Heightened condition-dependent growth of sexually selected weapons in the rhinoceros beetle, Trypoxylus dichotomus (Coleoptera: Scarabaeidae) Integr Comp Biol. 2014;54:614–621. doi: 10.1093/icb/icu041. [DOI] [PubMed] [Google Scholar]
- 11.Gotoh H, et al. Developmental link between sex and nutrition; doublesex regulates sex-specific mandible growth via juvenile hormone signaling in stag beetles. PLoS Genet. 2014;10:e1004098. doi: 10.1371/journal.pgen.1004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emlen DJ, Szafran Q, Corley LS, Dworkin I. Insulin signaling and limb-patterning: Candidate pathways for the origin and evolutionary diversification of beetle ‘horns’. Heredity (Edinb) 2006;97:179–191. doi: 10.1038/sj.hdy.6800868. [DOI] [PubMed] [Google Scholar]
- 13.Warren IA, Gotoh H, Dworkin IM, Emlen DJ, Lavine LC. A general mechanism for conditional expression of exaggerated sexually-selected traits. BioEssays. 2013;35:889–899. doi: 10.1002/bies.201300031. [DOI] [PubMed] [Google Scholar]
- 14.McCullough EL, Ledger KJ, O’Brien DM, Emlen DJ. Variation in the allometry of exaggerated rhinoceros beetle horns. Anim Behav. 2015;109:133–140. [Google Scholar]
- 15.Deaton A. Height, health, and development. Proc Natl Acad Sci USA. 2007;104:13232–13237. doi: 10.1073/pnas.0611500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkins JM, Subramanian SV, Davey Smith G, Özaltin E. Adult height, nutrition, and population health. Nutr Rev. 2016;74:149–165. doi: 10.1093/nutrit/nuv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCD Risk Factor Collaboration (NCD-RisC) A century of trends in adult human height. eLife. 2016;5:e13410. doi: 10.7554/eLife.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasgruber P, Cacek J, Kalina T, Sebera M. The role of nutrition and genetics as key determinants of the positive height trend. Econ Hum Biol. 2014;15:81–100. doi: 10.1016/j.ehb.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Bozzoli C, Deaton A, Quintana-Domeque C. Adult height and childhood disease. Demography. 2009;46:647–669. doi: 10.1353/dem.0.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little AC, Jones BC, Penton-Voak IS, Burt DM, Perrett DI. Partnership status and the temporal context of relationships influence human female preferences for sexual dimorphism in male face shape. Proc Biol Sci. 2002;269:1095–1100. doi: 10.1098/rspb.2002.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg DR, Jones BC, Little AC, Burt DM, Perrett DI. Manipulations of fundamental and formant frequencies influence the attractiveness of human male voices. Anim Behav. 2005;69:561–568. [Google Scholar]
- 22.Puts DA, Jones BC, DeBruine LM. Sexual selection on human faces and voices. J Sex Res. 2012;49:227–243. doi: 10.1080/00224499.2012.658924. [DOI] [PubMed] [Google Scholar]
- 23.Puts DA, Doll LM, Hill AK. Sexual selection on human voices. In: Weekes-Shackelford V, Shackelford T, editors. Evolutionary Perspectives on Human Sexual Psychology and Behavior, Evolutionary Psychology. Springer; New York: 2014. pp. 69–86. [Google Scholar]
- 24.Shirazi TN, Puts DA, Escasa-Dorne MJ. Filipino women’s preferences for male voice pitch: Intra-individual, life history, and hormonal predictors. Adapt Human Behav Physiol. 2018;4:188–206. [Google Scholar]
- 25.Zietsch BP, Lee AJ, Sherlock JM, Jern P. Variation in women’s preferences regarding male facial masculinity is better explained by genetic differences than by previously identified context-dependent effects. Psychol Sci. 2015;26:1440–1448. doi: 10.1177/0956797615591770. [DOI] [PubMed] [Google Scholar]
- 26.Kirkpatrick M. Good genes and direct selection in the evolution of mating preferences. Evolution. 1996;50:2125–2140. doi: 10.1111/j.1558-5646.1996.tb03603.x. [DOI] [PubMed] [Google Scholar]
- 27.Iwasa Y, Pomiankowski A. Good parent and good genes models of handicap evolution. J Theor Biol. 1999;200:97–109. doi: 10.1006/jtbi.1999.0979. [DOI] [PubMed] [Google Scholar]
- 28.Furman D, et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc Natl Acad Sci USA. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trigunaite A, Dimo J, Jørgensen TN. Suppressive effects of androgens on the immune system. Cell Immunol. 2015;294:87–94. doi: 10.1016/j.cellimm.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Grossman CJ. 1994. The role of sex steroids in immune system regulation. Bilateral Communication Between Endocrine and Immune Systems, Endocrinology and Metabolism (Progress in Research and Clinical Practice), ed Grossman CJ (Springer, New York), pp 1–11.
- 31.Chao TC, Van Alten PJ, Walter RJ. Steroid sex hormones and macrophage function: Modulation of reactive oxygen intermediates and nitrite release. Am J Reprod Immunol. 1994;32:43–52. doi: 10.1111/j.1600-0897.1994.tb00877.x. [DOI] [PubMed] [Google Scholar]
- 32.Muehlenbein MP, Bribiescas RG. Testosterone-mediated immune functions and male life histories. Am J Hum Biol. 2005;17:527–558. doi: 10.1002/ajhb.20419. [DOI] [PubMed] [Google Scholar]
- 33.Muehlenbein MP, Alger J, Cogswell F, James M, Krogstad D. The reproductive endocrine response to Plasmodium vivax infection in Hondurans. Am J Trop Med Hyg. 2005;73:178–187. [PubMed] [Google Scholar]
- 34.Muehlenbein MP, Cogswell FB, James MA, Koterski J, Ludwig GV. Testosterone correlates with Venezuelan equine encephalitis virus infection in macaques. Virol J. 2006;3:19. doi: 10.1186/1743-422X-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Folstad I, Karter AJ. Parasites, bright males, and the immunocompetence handicap. Am Nat. 1992;139:603–622. [Google Scholar]
- 36.Rantala MJ, et al. Evidence for the stress-linked immunocompetence handicap hypothesis in humans. Nat Commun. 2012;3:694. doi: 10.1038/ncomms1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rettew JA, Huet-Hudson YM, Marriott I. Testosterone reduces macrophage expression in the mouse of toll-like receptor 4, a trigger for inflammation and innate immunity. Biol Reprod. 2008;78:432–437. doi: 10.1095/biolreprod.107.063545. [DOI] [PubMed] [Google Scholar]
- 38.Lai J-J, et al. Androgen receptor influences on body defense system via modulation of innate and adaptive immune systems: Lessons from conditional AR knockout mice. Am J Pathol. 2012;181:1504–1512. doi: 10.1016/j.ajpath.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boonekamp JJ, Ros AHF, Verhulst S. Immune activation suppresses plasma testosterone level: A meta-analysis. Biol Lett. 2008;4:741–744. doi: 10.1098/rsbl.2008.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox FE. Parasites and sexual selection. Nature. 1989;341:289–290. doi: 10.1038/341289a0. [DOI] [PubMed] [Google Scholar]
- 41.Hamilton WD, Axelrod R, Tanese R. Sexual reproduction as an adaptation to resist parasites (a review) Proc Natl Acad Sci USA. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zahavi A. Mate selection-a selection for a handicap. J Theor Biol. 1975;53:205–214. doi: 10.1016/0022-5193(75)90111-3. [DOI] [PubMed] [Google Scholar]
- 43.Moore FR, et al. Evidence for the stress-linked immunocompetence handicap hypothesis in human male faces. Proc Biol Sci. 2011;278:774–780. doi: 10.1098/rspb.2010.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rantala MJ, et al. Adiposity, compared with masculinity, serves as a more valid cue to immunocompetence in human mate choice. Proc Biol Sci. 2013;280:20122495. doi: 10.1098/rspb.2012.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skrinda I, et al. Body height, immunity, facial and vocal attractiveness in young men. Naturwissenschaften. 2014;101:1017–1025. doi: 10.1007/s00114-014-1241-8. [DOI] [PubMed] [Google Scholar]
- 46.Roberts ML, Buchanan KL, Evans MR. Testing the immunocompetence handicap hypothesis: A review of the evidence. Anim Behav. 2004;68:227–239. [Google Scholar]
- 47.Thornhill R. Major histocompatibility complex genes, symmetry, and body scent attractiveness in men and women. Behav Ecol. 2003;14:668–678. [Google Scholar]
- 48.Lie HC, Rhodes G, Simmons LW. Genetic diversity revealed in human faces. Evolution. 2008;62:2473–2486. doi: 10.1111/j.1558-5646.2008.00478.x. [DOI] [PubMed] [Google Scholar]
- 49.Nowak J, Pawłowski B, Borkowska B, Augustyniak D, Drulis-Kawa Z. No evidence for the immunocompetence handicap hypothesis in male humans. Sci Rep. 2018;8:7392. doi: 10.1038/s41598-018-25694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott IML, Clark AP, Boothroyd LG, Penton-Voak IS. Do men’s faces really signal heritable immunocompetence? Behav Ecol. 2013;24:579–589. doi: 10.1093/beheco/ars092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcinkowska UM, Galbarczyk A, Jasienska G. La donna è mobile? Lack of cyclical shifts in facial symmetry, and facial and body masculinity preferences-A hormone based study. Psychoneuroendocrinology. 2018;88:47–53. doi: 10.1016/j.psyneuen.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: Expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 53.Penn DJ, Damjanovich K, Potts WK. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc Natl Acad Sci USA. 2002;99:11260–11264. doi: 10.1073/pnas.162006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janeway CA, Jr, Travers P, Walport M, Shlomchik MJ. Immunobiology: The Immune System in Health and Disease. 5th Ed. Garland Science; New York: 2001. The major histocompatibility complex and its functions; pp. 167–169. [Google Scholar]
- 55.DeGiorgio M, Lohmueller KE, Nielsen R. A model-based approach for identifying signatures of ancient balancing selection in genetic data. PLoS Genet. 2014;10:e1004561. doi: 10.1371/journal.pgen.1004561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yasukochi Y, Satta Y. Current perspectives on the intensity of natural selection of MHC loci. Immunogenetics. 2013;65:479–483. doi: 10.1007/s00251-013-0693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hedrick PW. Balancing selection and MHC. Genetica. 1998–1999;104:207–214. doi: 10.1023/a:1026494212540. [DOI] [PubMed] [Google Scholar]
- 58.Andrés AM, et al. Targets of balancing selection in the human genome. Mol Biol Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valenzano DR, Mennucci A, Tartarelli G, Cellerino A. Shape analysis of female facial attractiveness. Vision Res. 2006;46:1282–1291. doi: 10.1016/j.visres.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 60.Claes P, et al. Modeling 3D facial shape from DNA. PLoS Genet. 2014;10:e1004224. doi: 10.1371/journal.pgen.1004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claes P, Walters M, Clement J. Improved facial outcome assessment using a 3D anthropometric mask. Int J Oral Maxillofac Surg. 2012;41:324–330. doi: 10.1016/j.ijom.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 62.Claes P, et al. Sexual dimorphism in multiple aspects of 3D facial symmetry and asymmetry defined by spatially dense geometric morphometrics. J Anat. 2012;221:97–114. doi: 10.1111/j.1469-7580.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitteroecker P, Windhager S, Müller GB, Schaefer K. The morphometrics of “masculinity” in human faces. PLoS One. 2015;10:e0118374. doi: 10.1371/journal.pone.0118374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitteroecker P, Gunz P, Windhager S, Schaefer K. A brief review of shape, form, and allometry in geometric morphometrics, with applications to human facial morphology. Hystrix. 2013;24:59–66. [Google Scholar]
- 65.Schaefer K, Mitteroecker P, Gunz P, Bernhard M, Bookstein FL. Craniofacial sexual dimorphism patterns and allometry among extant hominids. Ann Anat. 2004;186:471–478. doi: 10.1016/S0940-9602(04)80086-4. [DOI] [PubMed] [Google Scholar]
- 66.Bulygina E, Mitteroecker P, Aiello L. Ontogeny of facial dimorphism and patterns of individual development within one human population. Am J Phys Anthropol. 2006;131:432–443. doi: 10.1002/ajpa.20317. [DOI] [PubMed] [Google Scholar]
- 67.Rosas A, Bastir M. Thin-plate spline analysis of allometry and sexual dimorphism in the human craniofacial complex. Am J Phys Anthropol. 2002;117:236–245. doi: 10.1002/ajpa.10023. [DOI] [PubMed] [Google Scholar]
- 68.Kesterke MJ, et al. Using the 3D facial norms database to investigate craniofacial sexual dimorphism in healthy children, adolescents, and adults. Biol Sex Differ. 2016;7:23. doi: 10.1186/s13293-016-0076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fitzpatrick S. Patterns of morphometric variation in birds’ tails: Length, shape and variability. Biol J Linn Soc Lond. 1997;62:145–162. [Google Scholar]
- 70.Robinson MR, et al. Population genetic differentiation of height and body mass index across Europe. Nat Genet. 2015;47:1357–1362. doi: 10.1038/ng.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown JL. A theory of mate choice based on heterozygosity. Behav Ecol. 1997;8:60–65. [Google Scholar]
- 72.Puts DA, et al. Sexual selection on male vocal fundamental frequency in humans and other anthropoids. Proc Biol Sci. 2016;283:20152830. doi: 10.1098/rspb.2015.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puts DA. Beauty and the beast: Mechanisms of sexual selection in humans. Evol Hum Behav. 2010;31:157–175. [Google Scholar]
- 74.Lee KS, et al. Selection on the regulation of sympathetic nervous activity in humans and chimpanzees. PLoS Genet. 2018;14:e1007311. doi: 10.1371/journal.pgen.1007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Theofanopoulou C, et al. Self-domestication in Homo sapiens: Insights from comparative genomics. PLoS One. 2017;12:e0185306. doi: 10.1371/journal.pone.0185306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilkins AS, Wrangham RW, Fitch WT. The “domestication syndrome” in mammals: A unified explanation based on neural crest cell behavior and genetics. Genetics. 2014;197:795–808. doi: 10.1534/genetics.114.165423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clark G, Henneberg M. The life history of Ardipithecus ramidus: A heterochronic model of sexual and social maturation. Anthropol Rev. 2015;78:109–132. [Google Scholar]
- 78.Matthews H, et al. Spatially dense morphometrics of craniofacial sexual dimorphism in 1-year-olds. J Anat. 2016;229:549–559. doi: 10.1111/joa.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manning J, Kilduff L, Cook C, Crewther B, Fink B. Digit ratio (2D:4D): A biomarker for prenatal sex steroids and adult sex steroids in challenge situations. Front Endocrinol (Lausanne) 2014;5:9. doi: 10.3389/fendo.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weinberg SM, Parsons TE, Raffensperger ZD, Marazita ML. Prenatal sex hormones, digit ratio, and face shape in adult males. Orthod Craniofac Res. 2015;18:21–26. doi: 10.1111/ocr.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Neave N, Laing S, Fink B, Manning JT. Second to fourth digit ratio, testosterone and perceived male dominance. Proc Biol Sci. 2003;270:2167–2172. doi: 10.1098/rspb.2003.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Verdonck A, Gaethofs M, Carels C, de Zegher F. Effect of low-dose testosterone treatment on craniofacial growth in boys with delayed puberty. Eur J Orthod. 1999;21:137–143. doi: 10.1093/ejo/21.2.137. [DOI] [PubMed] [Google Scholar]
- 83.Smith MJL, et al. Facial appearance is a cue to oestrogen levels in women. Proc Biol Sci. 2006;273:135–140. doi: 10.1098/rspb.2005.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marečková K, et al. Testosterone-mediated sex differences in the face shape during adolescence: Subjective impressions and objective features. Horm Behav. 2011;60:681–690. doi: 10.1016/j.yhbeh.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 85.Whitehouse AJO, et al. Prenatal testosterone exposure is related to sexually dimorphic facial morphology in adulthood. Proc Biol Sci. 2015;282:20151351. doi: 10.1098/rspb.2015.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koudelová J, Brůžek J, Cagáňová V, Krajíček V, Velemínská J. Development of facial sexual dimorphism in children aged between 12 and 15 years: A three-dimensional longitudinal study. Orthod Craniofac Res. 2015;18:175–184. doi: 10.1111/ocr.12096. [DOI] [PubMed] [Google Scholar]
- 87.Snyder PJ. Androgens. In: Marcus R, Feldman D, Nelson DA, Rosen CJ, editors. Osteoporosis. 3rd Ed. Academic; San Diego: 2008. pp. 1783–1792. [Google Scholar]
- 88.Lee AJ, et al. Genetic factors that increase male facial masculinity decrease facial attractiveness of female relatives. Psychol Sci. 2014;25:476–484. doi: 10.1177/0956797613510724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCarthy MM, Arnold AP. Reframing sexual differentiation of the brain. Nat Neurosci. 2011;14:677–683. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arnold AP. The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm Behav. 2009;55:570–578. doi: 10.1016/j.yhbeh.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arnold AP. A general theory of sexual differentiation. J Neurosci Res. 2017;95:291–300. doi: 10.1002/jnr.23884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 97.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klingenberg CP, Barluenga M, Meyer A. Shape analysis of symmetric structures: Quantifying variation among individuals and asymmetry. Evolution. 2002;56:1909–1920. doi: 10.1111/j.0014-3820.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 99.Rohlf FJ, Slice D. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst Zool. 1990;39:40–59. [Google Scholar]
- 100.Komori M, Kawamura S, Ishihara S. Multiple mechanisms in the perception of face gender: Effect of sex-irrelevant features. J Exp Psychol Hum Percept Perform. 2011;37:626–633. doi: 10.1037/a0020369. [DOI] [PubMed] [Google Scholar]
- 101.Holzleitner IJ, et al. Men’s facial masculinity: When (body) size matters. Perception. 2014;43:1191–1202. doi: 10.1068/p7673. [DOI] [PubMed] [Google Scholar]
- 102.de Bakker PIW, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The informed consent with which the data were collected does not allow for dissemination of identifiable data to persons not listed as researchers on the IRB protocol. Thus, the raw genotype data and 3D images cannot be made publicly available. In the interest of reproducibility, we have provided deidentified overall facial masculinity measures as well as age, sex, weight, height, ancestry, genetic PCs, and MHC and genome-wide heterozygosity, from which all results presented in this manuscript can be reproduced (Dataset S1). In addition, we provide high-density facial masculinity maps: i.e., facial masculinity calculated for every quasi-landmark (FMQL) for all 1,233 individuals used in the analyses (Dataset S2). Lastly, we provide all protocols and scripts used to process the genetic data and perform the analyses as well as a tutorial showing how FMQL can be used and visualized. This resource will allow other researchers to study variation in facial masculinity with high resolution in a large sample. All of the above are available on the following GitHub repository: https://github.com/Arslan-Zaidi/Facial_masculinity_MHC.