Significance
Whole-chromosome oligo-FISH paints using synthetic oligonucleotide libraries that can be amplified and labeled were generated for all 10 chromosomes of maize, facilitating chromosome studies with high sensitivity and specificity for genetically diverse lines. Applications include visualization of simple or complex chromosomal aberrations, establishment of chromosomal domains, illustration of mitotic and meiotic behavior, and providing insights into chromosomal relationships.
Keywords: oligo-FISH, B chromosome, karyotype, chromosomal aberrations, chromosome painting
Abstract
Whole-chromosome painting probes were developed for each of the 10 chromosomes of maize by producing amplifiable libraries of unique sequences of oligonucleotides that can generate labeled probes through transcription reactions. These paints allow identification of individual homologous chromosomes for many applications as demonstrated in somatic root tip metaphase cells, in the pachytene stage of meiosis, and in interphase nuclei. Several chromosomal aberrations were examined as proof of concept for study of various rearrangements using probes that cover the entire chromosome and that label diverse varieties. The relationship of the supernumerary B chromosome and the normal chromosomes was examined with the finding that there is no detectable homology between any of the normal A chromosomes and the B chromosome. Combined with other chromosome-labeling techniques, a complete set of whole-chromosome oligonucleotide paints lays the foundation for future studies of the structure, organization, and evolution of genomes.
Chromosome painting is typically used to determine the chromosomes involved in simple or complex rearrangements or changes in number. Chromosome painting can facilitate defining the presence, location, and identity of spontaneous or induced aberrations in chromosome structure. Whole-chromosome paints were developed for mammalian species by chromosome sorting or microdissection followed by the construction of libraries for each chromosome (1–4). In some species, collections of bacterial artificial chromosomes (BACs) spanning the length of each chromosome are used (5, 6). In these cases, it is necessary to block hybridization to the repetitive component of the genome or deplete such elements from the probe libraries.
Recently, oligonucleotide collections have been synthesized that cover portions or entire chromosomes (7–11). The current whole-chromosome oligonucleotide libraries were built on a defined set of unique sequences that have been bioinformatically chosen for chromosome specificity. These libraries have the advantage that they do not require chromosome sorting or BAC libraries for their development and can be amplified. PCR-amplified library sequences are converted into chromosome-specific probes via in vitro transcription followed by reverse transcription initiated by a fluorophore- or hapten-tagged primer.
The oligo chromosome FISH paints have numerous applications. The probes are genotype independent as an important complement to a previous method that relies on clustered repetitive sequences, which are often polymorphic among different genotypes, to identify chromosomes (12). Indeed, the oligo-FISH paints can be combined with genotype-specific probes to enhance the ascertainment of karyotype information. For example, the oligo-FISH paints could be combined with hybridization of repetitive arrays (12), individual genes (13, 14), or labeled BACs (15) to confirm physical maps, which often contain many errors. Furthermore, individual DNA transposable elements (TEs) can be detected (16) and, when combined with whole-chromosome paints, elucidate information about transposition sites on a genomic level as well as the types of chromosomal aberrations that occur with faulty transposition events (17). Combined with single transgene probes, this technique will locate the genomic position and number of transgene insertion sites in any one transformation.
Furthermore, the domain arrangement of a chromosome can be traced, which is not possible with site-specific labels. By examining the behavior of homologs as well as different chromosomes, the dynamics of chromosome action through the cell cycle including interphase can be visualized. Chromosome painting also allows the monitoring of pairing of homologous or homeologous chromosomes in prophase I of meiosis, which provides more accurate assessment of chromosome homology than pairing analysis based on metaphase I of meiosis (10, 18).
Here, oligo-FISH paints have been developed for every chromosome of a species using maize. This strategy has several advantages compared with commonly used systems; notably, it does not require chromosome sorting or dissection, has a precisely known composition of probes, does not require blocking of repetitive sequences in the genome, allows direct labeling, and can be combined with single gene/transgene/clustered repeat detection. The library of defined oligonucleotides can be used to produce regional paints or new banded paints without further bioinformatic determinations. With a proof of concept of a complete set of chromosome paints in maize, a similar system can be developed for any other model species with a sequenced genome.
Results
Chromosome FISH Paints for Each Maize Chromosome.
The strategy for the production of chromosome-specific oligonucleotide probes was to perform a k-mer analysis on a shotgun sequence library of the B73 inbred line to identify sequences that were uniquely represented in the genome. The collection of unique sequences identified for each respective chromosome was then used for the development of the 10 individual oligonucleotide libraries. Chorus (https://github.com/forrestzhang/Chorus) was used to design the oligo-based painting probes, but RepeatMasker was replaced with a k-mer method to filter out repetitive DNA sequences. Burrows–Wheeler aligner (BWA) (19) was used to replace BLAST-like alignment tool (BLAT) for mapping short sequence reads. This k-mer–based approach was more effective than RepeatMasker (www.repeatmasker.org/) to remove repeats, especially those associated with decayed TEs. A RepeatMasker-based pipeline may fail to eliminate repeats that are not identified or assembled based on a reference genome. The k-mer–based pipeline does not rely on a reference genome and can detect all potential repeats, including those that are poorly characterized.
The density of oligos is clearly lower in the centromeric and pericentromeric regions compared with the distal regions of all maize chromosomes (SI Appendix, Fig. S1). This distribution pattern of oligo density is more correlated with the density of genes than with the density of TEs and pericentric repetitive heterochromatin along maize chromosomes (20). The total number of oligos generated from the pipeline ranged from 73,443 for chromosome 9 to 178,890 for chromosome 1. Overall, 23.8%, 22.5%, and 53.7% of the probe sequences are located within exon, intron, and intergenic regions, respectively. Different numbers of oligos for each chromosome were selected to develop the painting libraries to ensure a minimum of ∼0.25 oligos/kb and to cover the entire chromosome. The numbers of selected oligos ranged from 45,967 for chromosome 9 to 91,265 for chromosome 1 (Dataset S1). The synthesis, amplification, and labeling of the libraries are described in Materials and Methods.
Using the chromosome-specific libraries, root tip metaphase chromosome material of inbred line KYS was probed with each of the 10 respective libraries (Fig. 1 and SI Appendix, Fig. S2). The chromosome oligo libraries were labeled with ATTO-550 (Materials and Methods and SI Appendix, SI Text S1). Chromosome-specific features (21) were simultaneously probed to confirm the identity of each member of the karyotype. Alternatively, the distinctive morphology of certain individual chromosomes was used.
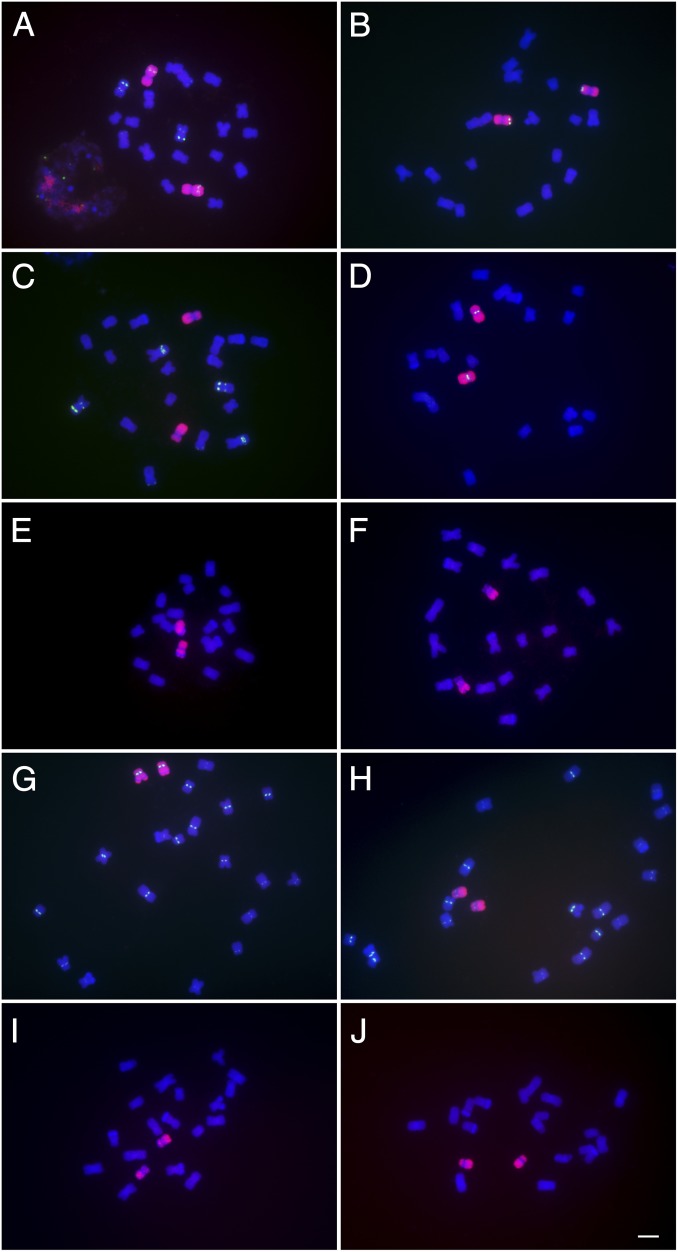
Fig. 1.
Individual oligonucleotide paints for chromosomes 1–10. Each library was labeled with ATTO-550 (red) and probed onto root tip metaphase spreads of inbred line KYS. Chromosomes were labeled with DAPI (blue), which reveals the sites of heterochromatic knobs. The confirmation of chromosome was determined by FITC or 6-FAM counterprobing in green of repetitive elements that are universally characteristic of the respective chromosome (12, 21) or previously characterized for KYS (21). (A) Chromosome 1, counterprobed with TAG microsatellite. Note the interphase nucleus at the Lower Left. (B) Chromosome 2, counterprobed with 5S ribosomal DNA. (C) Chromosome 3 lacks counterprobe of TAG microsatellite. (D) Chromosome 4, counterprobed with Cent4, a repetitive array specific to this chromosome. (E) Chromosome 5, defined by being near metacentric with a large heterochromatic knob in the long arm (gap in painting). (F) Chromosome 6, characterized by the presence of the secondary constriction (NOR). (G) Chromosome 7, counterprobed with CentC, which has a strong signal for chromosome 7 in KYS and defined by a large heterochromatic knob in the long arm. (H) Chromosome 8, counterprobed with CentC, which has a weak signal for chromosome 8 in KYS and no knob in the long arm. (I) Chromosome 9, defined by a heterochromatic knob at the tip of the short arm. (J) Chromosome 10, defined by its acrocentric morphology and absence of heterochromatic knobs. Grayscale images are presented in SI Appendix, Fig. S2. (Scale bar, 5 µm.)
In general, the intensity of FISH signals is less surrounding the primary constriction (centromere) than in distal regions as correlated with the oligo probe density noted above. Chromosomes with large visible knobs have gaps in labeling at those sites due to the highly repetitive nature of these heterochromatic positions (22). Also, the nucleolar organizing region (NOR) on chromosome 6 (Fig. 1F) has a low density of signal due to its tandemly repeated nature (23), but the small distal region beyond the NOR hybridizes to the chromosome 6 probe.
Application to Meiosis.
Meiosis is the specialized process in a eukaryotic life cycle during which the chromosome number is reduced. Synapsis and crossing over between homologs assure the faithful separation during the first division of meiosis. To examine meiotic pairing, the chromosomes are often analyzed in the pachytene stage of meiosis because they are condensed but less so than in somatic metaphase cells and exhibit distinctive morphologies among the different chromosomes (24, 25). The ability to distinguish individual chromosomes efficiently from end to end in pachynema (and other meiotic stages) would facilitate chromosomal studies of meiosis that are often precluded by the difficulty of this task.
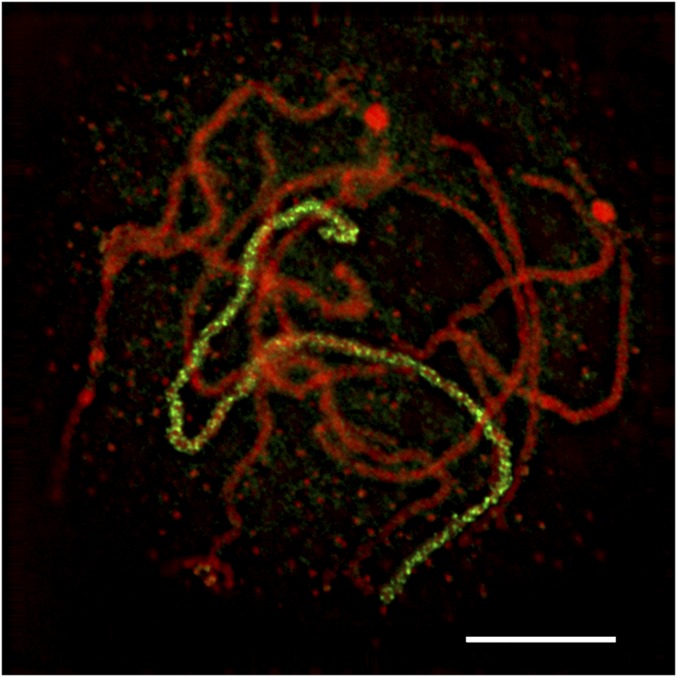
The chromosome 1-specific library was applied to meiotic pachynema (Fig. 2) (Materials and Methods). This chromosome was labeled from terminus to terminus with sufficient density to trace its path through the nucleus, which allows a better assessment of pairing extent along the entire chromosome. This example illustrates the utility of the probe collection for studies of meiotic processes beyond their applications for somatic chromosomes.
Fig. 2.
A maize meiocyte at the pachytene stage of a plant of inbred W23 was hybridized with an ATTO-550–labeled chromosome 1-specific probe. Chromosomes were stained with DAPI (shown in red), and FISH signals shown in green label the bivalent of chromosome 1. (Scale bar, 10 µm.)
Visualization of Chromosomal Domains in Interphase.
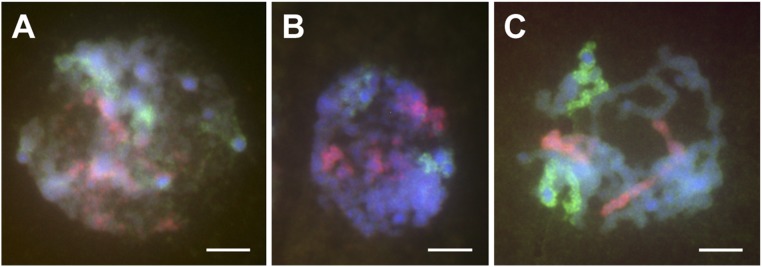
Standard methods of chromosomal identification require condensed chromosomes in metaphase or in various meiotic stages. The development of a complete set of oligo-FISH chromosomal paints permits the visualization of the chromosome territories (26) of individual chromosomes in other cell types and stages of the cell cycle. Examination of interphase or prometaphase cells in root tip preparations illustrates the visualization of homologs in the nucleus (Fig. 3). The labeling of two different chromosomes (Fig. 3) shows that each homolog resides in a separate position but within a discrete domain, albeit uncondensed and diffuse, within the nucleus as determined at this level of detection.
Fig. 3.
Chromosome paints applied to interphase nuclei. Interphase nuclei from KYS root tips were probed with labeled libraries for chromosomes 4 (ATTO-550; red) and 5 (ATTO-488; green). The chromosomes were stained with DAPI (blue). The images reveal that each chromosome occupies a discrete domain and that homologs are not associated. (A) Heterochromatic knobs are visible as blue spheres. (B) Domains becoming more discrete. (C) The image is a prometaphase cell. The heterochromatic knobs are visualized as gaps in chromosomal labeling that are blue. (Scale bar, 5 µm.)
Genotype-Independent Characterization of Chromosomal Aberrations.
Chromosomal aberrations occur spontaneously in individuals or in single cells (27). From experimental induction, they have been documented from aberrant TE transpositions (17) and from various methods of irradiation (28–30). Hundreds of translocations that serve as a valuable resource for genetic studies have been recovered in maize (31, 32), and whole-chromosome paints are useful for characterizing the nature of such aberrations.
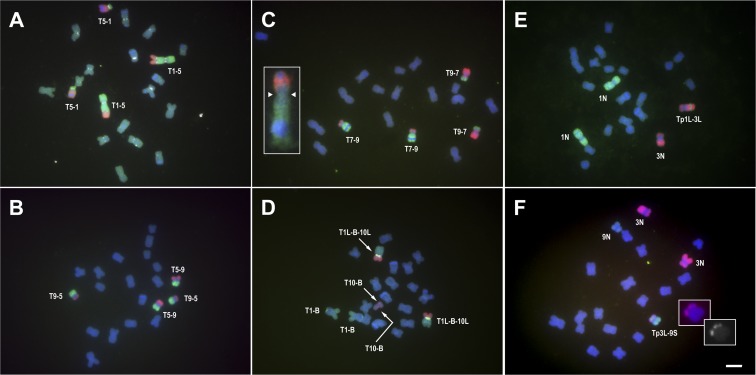
Selected chromosomal paints were applied to a variety of chromosomal aberrations to visualize the changes (Fig. 4). T1-5 (8041) is a translocation between chromosomes 1 and 5 that was recovered from maize exposed to an atomic bomb test on Eniwetok atoll on April 15, 1948 (33). It has been used in several studies in maize genetics (34–37). More recently, it was discovered that the 1-5 chromosome carries an extra set of canonical centromere sequences that are not active as a centromere (37). The painting illustrates that a centromere was apparently captured at the junction of chromosomes 1 and 5, or the translocation event fractured the centromere of chromosome 5 leaving a functional remnant at the original site and another portion at the translocation junction (Fig. 4A). The centromeric sequences at the translocation 1-5 breakpoint have apparently been in the inactive state for 70 y of propagation.
Fig. 4.
Use of whole-chromosome paints to characterize chromosomal aberrations. (A) Labeled libraries of chromosomes 1 (green) and 5 (red) were applied to homozygous material of reciprocal translocation T1-5 (8041). The centromere satellite, CentC, is labeled in white. Previous studies have demonstrated that the set of centromere sequences present at the translocation breakpoint is inactive (37). The reciprocally exchanged chromosomes are labeled T1-5 and T5-1. (B) Reciprocal translocation T5-9d was probed with the two respectively labeled libraries (5, green; 9, red). (C) Reciprocal translocation T7-9 (4363) was probed with the two respectively labeled libraries (7, green; 9, red). (Inset) Enlarged image of a T7-9 homolog from a different metaphase spread shows the breakpoint in the short arm of chromosome 7. The arrowheads at the constriction denote the position of the centromere. (D) Chromosome painting of homozygous material of T1La-B-10L18, which is a chromosome with a B centromere that has the long arm of chromosome 1 joined together with the long arm of chromosome 10, which was recovered in a test for recombination in the B chromosome (40) as described in the text. Chromosome 1 is labeled in green, and chromosome 10 in red. The T1L-B-10L chromosome is the recombinant with red and green in the opposite arms. The B chromosome centromere-specific sequence is labeled in yellow on this chromosome (Materials and Methods). The homologs of the reciprocal product of TB-1La, labeled T1-B, consist of a portion of the long arm of the B chromosome exchanged near the centromere within 1L and an intact short arm of chromosome 1. The homologs of the reciprocal product of TB-10L18, labeled T10-B, consist of the short arm of the B chromosome exchanged near the centromere in the long arm of chromosome 10 and an intact short arm. The T1-B and T10-B chromosomes have portions that are not labeled by chromosome paints because they are derived from the B chromosome. (E) Heterozygous material of transposition Tp1L-3L was labeled with paints of chromosomes 1 (green) and 3 (red). The chromosome 3 carrying a small interstitial segment of 1L is labeled Tp1L-3L. The normal chromosome 1 is labeled 1N, and normal chromosome 3 is labeled 3N. (F) Heterozygous material of Tp3L-9S was probed with labeled libraries for chromosomes 3 (red) and 9 (green). The transposition chromosome is labeled Tp3L-9S, and the normal chromosomes are labeled 9N and 3N. The Insets show a magnified region of the 3L transposition with label and in grayscale. (Scale bar, 5 µm.)
Two other reciprocal A-A translocations were examined, T5-9d (Fig. 4B) and T7-9 (4363) (Fig. 4C). Translocation 5-9d was previously reported, based on orcein staining of chromosomes, to have breakpoints at 5L 0.14 and 9L 0.10 (33). Application of individual paints to this material shows exchanges that are roughly consistent with this determination. Translocation 7-9 (4363) was originally reported to involve an exchange between the two centromeres (33). However, from observation of the labeling around the primary constriction, it is clear that the breakpoints are in the respective short arms of 7 and 9. The classical determinations of translocation breakpoints relied on orcein staining that cannot distinguish nonhomologous chromosomes and on an estimate of the cross configuration in a translocation heterozygote, which can be variable (38, 39). Thus, using the orcein method, it is particularly difficult to determine the breakpoints surrounding centromeric regions (33, 34). The contrasting chromosomal paints aid in distinguishing the nonhomologous chromosomes.
Another aberration examined was T1La-B-10L18 (40) (Fig. 4D). This chromosome was a product of a test of whether crossing over could occur in the supernumerary B chromosome. The B chromosome is nonvital and is not present in most lines of maize but is maintained by an accumulation mechanism (see below). The progenitor chromosomes were TB-1La and TB-10L18. TB-1La is a translocation between chromosome 1 and the B chromosome that has a breakpoint in the long arm of 1 (1L) and in the proximal euchromatin of the B chromosome long arm (32). TB-10L18 is an unusual B-A translocation in that it has a breakpoint on the short arm side of the B centromere that is joined with the long arm of chromosome 10 (10L) leaving the long arm of the B intact (32). To produce T1La-B-10L18, stocks bearing these two TB-A chromosomes were crossed together (40). The two translocations have a region of the B chromosome in common that, if crossing over occurred at a point along its length, the long arms of 1 and 10 would be joined in a common chromosome on opposite sides of the B chromosome centromere. The predicted recombinant chromosome was recovered (40). Material of a homozygous line was probed with libraries for chromosomes 1 and 10 together with the B-specific repeat sequence that is present at the centromeric region of the B chromosome (41–43) and at a minor site distal on the long arm of the B. The recombinant chromosome is labeled by both paints and the major site of B-specific repeat. The 1-B chromosomes from the progenitor TB-1La translocation have label from the chromosome 1 painting probe and for the small distal B repeat together with an unlabeled portion from the B long arm. The 10-B chromosome from the progenitor TB-10L18 translocation has label from the chromosome 10 library probe together with a small unlabeled region from the B short arm.
Two interstitial transposition stocks were also examined (Fig. 4 E and F). Transposition 1L-3L (Tp1L-3L) was synthesized from two reciprocal A-A translocations involving chromosomes 1 and 3: T1-3 (5267) and T1-3 (5242) (44). The previously determined breakpoints were reported as 1L 0.72, 3L 0.73 and 1L 0.90, and 3L 0.65, respectively (33). The breakpoints on the two chromosomes are oriented in proximal–distal and distal–proximal arrangements. This configuration will, upon segregation from a heterozygote of the two translocated chromosomes, produce some progeny with a viable overlap between the breakpoints (36, 45). When present in an otherwise monosomic circumstance, the overlapping regions have no competition for pairing, which will allow recombination to occur there and place the region between the breakpoints of one chromosome between a duplication of the overlap of the other chromosomal breakpoints (44). A heterozygote of this transposition was probed with labeled libraries from chromosomes 1 and 3. The region of 1L comprising an approximate 18% of the length of 1L can be visualized within the long arm of chromosome 3 (Fig. 4E). In another example, transposition 3L-9S was described by Rhoades (46). It exists in a stock carrying a deficiency of the corresponding region of chromosome arm 3L. The 3L region that spans a small segment between the genetic markers na1 and a1 now resides in 9S between the genetic markers wx1 and bz1. An individual heterozygous for the transposition was probed with labeled libraries for chromosomes 3 and 9. The transposed region of 3L can be visualized in the short arm of 9 and is missing in the normal chromosome 9 (Fig. 4F).
The examination of this variety of chromosomal aberrations illustrates the utility for different types of studies. These examples also demonstrate the ability to combine whole-chromosome paints with probes of other chromosomal features, in this case the centromere satellite, CentC, and the B chromosome-specific sequence.
The Supernumerary B Chromosome Has No Detectable Relationship to Any A Chromosome.
The B chromosome of maize is nonvital and is, in fact, missing from most lines of maize. It is maintained despite being dispensable by a drive mechanism that consists of nondisjunction at the second pollen mitosis that produces the two maize sperm (47) and then the sperm with the B chromosomes preferentially fertilizes the egg nucleus rather than the polar nuclei in the process of double fertilization (48). The evolutionary origin of the B chromosome is unknown, although presumably it was derived from a progenitor chromosome that at some point would be related to A chromosomal regions. We systematically probed the B chromosome in the background of the B73 inbred line with each of the 10 chromosome-specific libraries (SI Appendix, Fig. S3). It was not possible to detect, even with digital signal enhancement, any evidence of a relationship to any specific A chromosome (SI Appendix, Fig. S4). If the B chromosome is derived by descent from one or more A chromosomes, or their progenitors, the reason for this result is likely due to the degeneration of gene sequences from the original chromosome to the point that they are not recognized by the hybridization technique. Any homologies between the B chromosome and the single-copy A sequences in the libraries are apparently too sparse to be detected. From the standpoint of cytological analysis, there would be no confusion between the A and B chromosomes. The B chromosome possesses several specific sequences that can be used for its own analysis (41, 42).
Discussion
Chromosome oligo-FISH paints for each of the 10 chromosomes of maize were developed. They distinguish chromosomes in root tip somatic metaphase cells, paired homologs in meiosis, and even in interphase nuclei. Translocations, segmental transpositions, and complex rearrangements can be visualized and confirmed. They have been used to illustrate several issues of chromosomal structure, nuclear distribution, and chromosomal evolution. This resource will allow investigations into chromosome biology and behavior on the individual cell level.
The ability of the probe collection to label chromosomes in meiosis will facilitate studies of chromosomal behavior at this critical stage in the life cycle. Heterologous painting of a maize chromosome in an oat background by genome in situ hybridization revealed aspects of chromosome pairing in oat meiosis (49). This example illustrates the utility of painting specific chromosomes to gain insight into meiotic processes. The probe libraries reported here will permit studies within maize and allow the incorporation of the wealth of cytogenetic stocks in this species to examine the mechanics of meiosis in great depth.
The k-mer analysis developed here was successful in generating whole-chromosome paints with excellent specificity. The maize genome contains numerous duplicate genes descendant from a whole-genome duplication (50). The oligo-FISH library probes do not show any evidence of cross-hybridization to duplicate regions (Fig. 1 and SI Appendix, Fig. S2) as a result of the probe selection procedure. Nevertheless, the probe libraries are genotype independent in that they label inbred lines KYS (Fig. 1), W23 (Fig. 2), B73 (SI Appendix, Fig. S3), and various other materials of unknown heritage (Fig. 4) despite having been designed from the B73 reference sequence.
Initial labeling of the maize oligo libraries was performed using ATTO-550 and ATTO-488 because they have intrinsically bright signal output and can be visualized with standard fluorescence filter sets. At this time, characterization of complex or unknown aberrations would require potential multiple probings to the target material as performed for the B chromosome. Nevertheless, the availability of a complete set of chromosomal probes provides a comprehensive test across the whole genome, which has not been previously available in any system that does not require chromosome sorting for probe development. Future studies using mixtures of fluorescent labels (1, 4) can likely reduce the number of required probings for genome-wide determinations.
The availability of specific oligo probes for each of the 10 chromosomes (Dataset S1) could also be used to produce more specific restricted libraries. Probe collections for segmental chromosomal regions of interest could be generated from this resource. Alternating banding paints for a chromosome or even interchromosomal paints could be selected (11). The limits of such probe collections could be determined by comparing the boundaries of the desired region in the B73 reference genome to the sequentially arranged oligonucleotides (Dataset S1).
The development in maize of a complete set of chromosomal amplifiable oligo-FISH paints provides a proof of concept for any other species for which a reference genome exists. The uniqueness of the probe set derives from the k-mer analysis of a shotgun sequence and then the oligos can be assigned to chromosome from the reference genome. The paints are useful to detect chromosomal rearrangements, both extensive and minor, between nonhomologs on the single-cell level. They can visualize chromosomes, or combinations of chromosomes, at various stages of the cell cycle for a variety of investigations. Combining the paints with probes targeting specific or polymorphic features of chromosomes (12) and single-gene detection (13, 14) provides a powerful tool kit for chromosome studies.
Materials and Methods
Plant Materials.
The KYS inbred line was used to illustrate the respective chromosome labeling. Inbred line B73 with introgressed B chromosomes (>backcross 7) was used to test homology with the B chromosome. Translocation lines were originally obtained from the Maize Genetics Cooperation Stock Center at the University of Illinois at Urbana–Champaign. Other aberrations are laboratory stocks. Tp3L-9S was obtained from Erik Vollbrecht (Iowa State University, Ames, IA).
Library Sequence Selection and Synthesis.
The shotgun sequence of maize from NCBI Sequence Read Archive (SRA, SRR404240) was used for k-mer analysis. Jellyfish (51) was used to calculate the distribution of 17-mer frequency (the copy number of a given k-mer in the shotgun library) (SI Appendix, Fig. S5). For example, the k-mer frequency of “AAAAAAAATGTGGTTAA” is 37, indicating a total of 37 copies of this 17-mer in the shotgun library. The major peak of k-mer frequency distribution represents the sequence depth of the shotgun library (52). The major peak at 32 indicates that the SRR404240 library contains ∼32× of the maize genome. The k-mer count was calculated in every 17-nt window from all oligos (45 nt). The k-mer score is the total k-mer count in each 45-nt oligo and represents the relative copy number in the maize genome. The k-mer score for a single copy oligo is (45 − 17 + 1) × 32 = 928. For example, oligo “CTAAACTCTGAAGCACAATTAGCATAATTCCCACATGTAACTCTT” contains 29 17-mers so the k-mer score is 870, which is the sum of all 17-mer frequencies (SI Appendix, Fig. S6).
The maize genome sequence (AGPv3.26) was divided into 45-nt oligos in a step size of 3 nt. All oligos were mapped back to the maize genome using BWA. Oligos were first removed if they could be mapped to two or more locations with 70% homology. Oligos with dTm < 10 °C were also eliminated. The k-mer scores were calculated for the remaining oligos. Oligos with a k-mer score ranging from 348 [(45 − 17 + 1) × 12] to 1,682 [(45 − 17 + 1) × 58] were retained in the pipeline. All oligos associated with individual chromosomes were visualized along the pseudomolecules using Pandas (pandas.pydata.org/) and Matplotlib (https://matplotlib.org/) packages.
Based on the k-mer determination of unique sequences, oligonucleotides of 45 bp in length that contain a universal primer (CGTGGTCGCGTCTCA) for their amplification were synthesized at Arbor Biosciences essentially as described (10).
Library Amplification and Labeling.
The following methods are based on work by Murgha et al. (53, 54). A single PCR amplification of each library utilized only 0.07 ng of template DNA from library syntheses ranging from 300 to 800 ng. The resulting PCR product was then used as a template to transcribe RNA. Labeling of the libraries was performed via reverse transcription of the RNA using a 5′ dye-labeled version of the above defined universal primer. Fluorescent dyes ATTO-488 and ATTO-550 (ATTO-TEC) were used for these initial studies. Unincorporated primers and the RNA template were removed to yield a labeled, single-stranded cDNA probe. Detailed protocols are available in SI Appendix, SI Text S1 and are diagrammed in figure 1B of Han et al. (10) with the exception that reverse transcription was initiated with a 5′ fluorophore-tagged primer. At present, it is unknown whether repeated amplifications via PCR will generate any biases in the library composition, but the multiplicity of probes per chromosome counteracts any negative impact this might have on the ability to label a chromosome. Aliquoting early amplifications could also be used to reduce the number of amplification cycles.
Preparation of Root Tip Metaphase Chromosomes and Hybridization of Paint Probes.
Kernels of the target material were germinated and root tip metaphase chromosomes prepared as previously described (12) with modifications adjusted for whole-chromosome painting probes as described in SI Appendix, SI Text S2.
Probes for Chromosome Confirmation.
High-copy number, repetitive sequence arrays were labeled via nick translation (12) or purchased as 5′-end–labeled oligos (IDT DNA). Oligo probes used included the CentC centromere repeat (6-FAM/CCTAAAGTAGTGGATTGGGCATGTTCG) and the microsatellite TAG repeat [6-FAM/AG-(TAG)18]. The nick-translated probes included CentC (12) labeled with cyanine-5-dUTP (NEL579001EA; Perkin-Elmer), the centromere 4-specific repeat (Cent4) (14, 55) labeled with Texas Red-5-dCTP (NEL426001EA; Perkin-Elmer), and the B chromosome-specific centromere repeat (B-repeat) (GenBank; AY173950.1) labeled separately with Texas Red (above) or fluorescein-12-dUTP (NEL413001EA; Perkin-Elmer), which were hybridized together at a ratio of one part red to seven parts green to produce a yellow FISH signal.
Meiotic Analysis.
Tassels with meiotic stages were fixed in Farmer’s fixative. Chromosome spreads were prepared by the method described (56). After treating with RNase and pepsin followed by postfixation, slides were dehydrated serially in 50%, 70%, and 95% ethanol. A 20-µL hybridization mixture consisting of 50% formamide, 10% dextran sulfate, 2× SSC, 0.2% SDS, 100 ng/μL salmon sperm DNA, and 100-pmol chromosome-specific probe labeled with ATTO-550 was denatured at 85 °C for 3 min and then applied on the slide. After sealing with rubber cement, slides were denatured at 75 °C for 3 min, followed by hybridization at 37 °C overnight. Stringent washing was carried out using 20% formamide in 0.1× SSC at 42 °C for 10 min. After washing, slides were mounted with Prolong Gold mountant with DAPI (Thermo Fisher; catalog no. P36931).
Microscopy.
Images of meiotic chromosomes were acquired with a Delta Vision ELITE system (GE) operated by the SoftWorx software using an Olympus IX71 microscope. To enhance the quality of images, about 10 serial optical sections were captured with 200-nm intervals and were subjected to deconvolution with default settings by SoftWorx. The best two sectioning images with the most signal–noise ratio were projected. Finally, only brightness-contrast adjustment was performed in Photoshop.
Images of somatic nuclei and metaphase chromosomes were acquired with FISHView 7.2.7 SP7 (Applied Spectral Imaging) using an Olympus BX61 microscope fitted with a cooled CCD camera (Cool-1300QS; VDS). One or more layers of the color images were adjusted using the curves and/or brightness-contrast functions of Photoshop CC 2015. Grayscale images were generated by converting the original, unadjusted red (probe) channel first to grayscale and then digitally increasing the signal strength to check for areas of nonspecific binding. For images in SI Appendix, Fig. S2, the conversion to grayscale was performed with FISHView, and then the brightest pixel was set to white using the levels eyedropper in Photoshop. For images in SI Appendix, Fig. S4, conversion was performed in Photoshop, and a pixel with a K (black) value of 70% was set to white.
Data and Materials Availability.
Library composition of oligonucleotides is provided in Dataset S1. Chromosomal aberrations are available from the Maize Genetics Cooperation Stock Center at the University of Illinois at Urbana–Champaign (maizecoop.cropsci.uiuc.edu) or from the corresponding author.
Supplementary Material
Acknowledgments
We thank Erik Vollbrecht for sharing the transposition Tp3L-9S material and Ting Wu for discussions. Research funding is from National Science Foundation Grant IOS-01444514.
Footnotes
Conflict of interest statement: K.S. and J.-M.R. are employees of Arbor Biosciences.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813957116/-/DCSupplemental.
References
- 1.Speicher MR, Gwyn Ballard S, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet. 1996;12:368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- 2.Schröck E, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 3.Bolzer A, Craig JM, Cremer T, Speicher MR. A complete set of repeat-depleted, PCR-amplifiable, human chromosome-specific painting probes. Cytogenet Cell Genet. 1999;84:233–240. doi: 10.1159/000015266. [DOI] [PubMed] [Google Scholar]
- 4.Müller S, Neusser M, Wienberg J. Towards unlimited colors for fluorescence in-situ hybridization (FISH) Chromosome Res. 2002;10:223–232. doi: 10.1023/a:1015296122470. [DOI] [PubMed] [Google Scholar]
- 5.Lysak MA, Fransz PF, Ali HB, Schubert I. Chromosome painting in Arabidopsis thaliana. Plant J. 2001;28:689–697. doi: 10.1046/j.1365-313x.2001.01194.x. [DOI] [PubMed] [Google Scholar]
- 6.Idziak D, et al. Painting the chromosomes of Brachypodium: Current status and future prospects. Chromosoma. 2011;120:469–479. doi: 10.1007/s00412-011-0326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beliveau BJ, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci USA. 2012;109:21301–21306. doi: 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beliveau BJ, et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun. 2015;6:7147. doi: 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 2011;19:901–909. doi: 10.1007/s10577-011-9245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Y, Zhang T, Thammapichai P, Weng Y, Jiang J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics. 2015;200:771–779. doi: 10.1534/genetics.115.177642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braz GT, et al. Comparative oligo-FISH mapping: An efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics. 2018;208:513–523. doi: 10.1534/genetics.117.300344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato A, Lamb JC, Birchler JA. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA. 2004;101:13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato A, Albert PS, Vega JM, Birchler JA. Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem. 2006;81:71–78. doi: 10.1080/10520290600643677. [DOI] [PubMed] [Google Scholar]
- 14.Lamb JC, et al. Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics. 2007;175:1047–1058. doi: 10.1534/genetics.106.065573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danilova TV, Birchler JA. Integrated cytogenetic map of mitotic metaphase chromosome 9 of maize: Resolution, sensitivity, and banding paint development. Chromosoma. 2008;117:345–356. doi: 10.1007/s00412-008-0151-y. [DOI] [PubMed] [Google Scholar]
- 16.Yu W, Lamb JC, Han F, Birchler JA. Cytological visualization of DNA transposons and their transposition pattern in somatic cells of maize. Genetics. 2007;175:31–39. doi: 10.1534/genetics.106.064238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, et al. Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize. Genes Dev. 2009;23:755–765. doi: 10.1101/gad.1776909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He L, Braz GT, Torres GA, Jiang J. Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma. 2018;127:505–513. doi: 10.1007/s00412-018-0682-9. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb JC, Birchler JA. Retroelement genome painting: Cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics. 2006;173:1007–1021. doi: 10.1534/genetics.105.053165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert PS, Gao Z, Danilova TV, Birchler JA. Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet Genome Res. 2010;129:6–16. doi: 10.1159/000314342. [DOI] [PubMed] [Google Scholar]
- 22.Peacock WJ, Dennis ES, Rhoades MM, Pryor AJ. Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc Natl Acad Sci USA. 1981;78:4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips RL, Weber DF, Kleese RA, Wang SS. The nucleolus organizer region of maize (Zea mays L.): Tests for ribosomal gene compensation or magnification. Genetics. 1974;77:285–297. doi: 10.1093/genetics/77.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClintock B. A cytological demonstration of the location of an interchange between two non-homologous chromosomes of Zea mays. Proc Natl Acad Sci USA. 1930;16:791–796. doi: 10.1073/pnas.16.12.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creighton HB, McClintock B. A correlation of cytological and genetical crossing-over in Zea mays. Proc Natl Acad Sci USA. 1931;17:492–497. doi: 10.1073/pnas.17.8.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 27.Ream TS, et al. A test for ectopic exchange catalyzed by Cre recombinase in maize. Theor Appl Genet. 2005;111:378–385. doi: 10.1007/s00122-005-2031-7. [DOI] [PubMed] [Google Scholar]
- 28.Anderson EG. On the frequency and transmitted chromosome alterations and gene mutations induced by atomic bomb radiations in maize. Proc Natl Acad Sci USA. 1948;34:387–390. doi: 10.1073/pnas.34.8.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Randolph LF, Longley AE, Li CH. Cytogenetic effects in corn exposed to atomic bomb ionizing radiation at Bikini. Science. 1948;108:13–15. doi: 10.1126/science.108.2792.13. [DOI] [PubMed] [Google Scholar]
- 30.Anderson EG, Longley AE, Li CH, Retherford KL. Hereditary effects produced in maize by radiations from the Bikini atomic bomb; studies on seedlings and pollen of the exposed generation. Genetics. 1949;34:639–646. doi: 10.1093/genetics/34.6.639a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coe EH. A-A translocation: Breakpoints and stocks. In: Freeling M, Walbot V, editors. The Maize Handbook. Springer; New York: 1994. pp. 364–376. [Google Scholar]
- 32.Beckett JB. Comprehensive list of B-A translocations in maize. In: Freeling M, Walbot V, editors. The Maize Handbook. Springer; New York: 1994. pp. 336–341. [Google Scholar]
- 33.Longley AE. 1961. Breakage Points for Four Corn Translocation Series and Other Corn Chromosome Aberrations (Agriculture Research Service, US Department of Agriculture, Washington, DC) 34-16, pp 1–40. [Google Scholar]
- 34.Burnham CR, Stout JT, Weinheimer WH, Kowles RV, Phillips RL. Chromosome pairing in maize. Genetics. 1972;71:111–126. doi: 10.1093/genetics/71.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson DS. A new compound A-B translocation, TB-5S. Maize Genet Coop News Lett. 1975;49:79–81. [Google Scholar]
- 36.Birchler JA. The cytogenetic localization of the alcohol dehydrogenase-1 locus in maize. Genetics. 1980;94:687–700. doi: 10.1093/genetics/94.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Z, Fu S, Dong Q, Han F, Birchler JA. Inactivation of a centromere during the formation of a translocation in maize. Chromosome Res. 2011;19:755–761. doi: 10.1007/s10577-011-9240-5. [DOI] [PubMed] [Google Scholar]
- 38.McClintock B. The association of non-homologous parts of chromosomes in the mid-prophase of meiosis in Zea mays. Z Zellforsch Mikrosk Anat. 1933;19:191–237. [Google Scholar]
- 39.Burnham CR. Chromosomal interchanges in maize: Reduction of crossing-over and the association of non-homologous parts. Am Nat. 1934;68:81–82. [Google Scholar]
- 40.Birchler JA, Chalfoun DJ, Levin DM. Recombination in the B chromosome of maize to produce A-B-A chromosomes. Genetics. 1990;126:723–733. doi: 10.1093/genetics/126.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alfenito MR, Birchler JA. Molecular characterization of a maize B chromosome centric sequence. Genetics. 1993;135:589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamb JC, Kato A, Birchler JA. Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma. 2005;113:337–349. doi: 10.1007/s00412-004-0319-z. [DOI] [PubMed] [Google Scholar]
- 43.Jin W, et al. Molecular and functional dissection of the maize B chromosome centromere. Plant Cell. 2005;17:1412–1423. doi: 10.1105/tpc.104.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birchler JA, Levin DM. Directed synthesis of a segmental chromosomal transposition: An approach to the study of chromosomes lethal to the gametophyte generation of maize. Genetics. 1991;127:609–618. doi: 10.1093/genetics/127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gopinath DM, Burnham CR. A cytogenetic study in maize of deficiency-duplication produced by crossing interchanges involving the same chromosomes. Genetics. 1956;41:382–395. doi: 10.1093/genetics/41.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhoades MM. Studies on the cytological basis of crossing over. In: Peacock WJ, Brock RD, editors. Replication and Recombination of Genetic Material. Australian Academy of Sciences; Canberra, ACT, Australia: 1968. pp. 229–241. [Google Scholar]
- 47.Roman H. Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics. 1947;32:391–409. doi: 10.1093/genetics/32.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roman H. Directed fertilization in maize. Proc Natl Acad Sci USA. 1948;34:36–42. doi: 10.1073/pnas.34.2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bass HW, et al. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J Cell Sci. 2000;113:1033–1042. doi: 10.1242/jcs.113.6.1033. [DOI] [PubMed] [Google Scholar]
- 50.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 51.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murgha YE, Rouillard J-M, Gulari E. Methods for the preparation of large quantities of complex single-stranded oligonucleotide libraries. PLoS One. 2014;9:e94752. doi: 10.1371/journal.pone.0094752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murgha Y, et al. Combined in vitro transcription and reverse transcription to amplify and label complex synthetic oligonucleotide probe libraries. Biotechniques. 2015;58:301–307. doi: 10.2144/000114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Page BT, Wanous MK, Birchler JA. Characterization of a maize chromosome 4 centromeric sequence: Evidence for an evolutionary relationship with the B chromosome centromere. Genetics. 2001;159:291–302. doi: 10.1093/genetics/159.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang CJ, Harper L, Cande WZ. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell. 2006;18:529–544. doi: 10.1105/tpc.105.037838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.