Significance
Forest defoliation across North America by European gypsy moths (EGM), a subspecies with flightless females, has resulted in billions of dollars in economic loss. However, if established, the Asian gypsy moth subspecies, which has flight-competent females, poses a greater economic threat. Understanding gypsy moth genetics and population differences among these subspecies may aid in the design of new control strategies. Here we report the gypsy moth genome and explore the genetic features that distinguish gypsy moths from other Lepidoptera. We further examine how genetic variation among subspecies may contribute to phenotypic differences among them. Finally, we present insights into gene-expression changes of the EGM in response to virus infection, which may assist in the design of viral bioinsecticides.
Keywords: Lepidoptera, Lymantria dispar, gypsy moth, virus–host interactions
Abstract
Since its accidental introduction to Massachusetts in the late 1800s, the European gypsy moth (EGM; Lymantria dispar dispar) has become a major defoliator in North American forests. However, in part because females are flightless, the spread of the EGM across the United States and Canada has been relatively slow over the past 150 years. In contrast, females of the Asian gypsy moth (AGM; Lymantria dispar asiatica) subspecies have fully developed wings and can fly, thereby posing a serious economic threat if populations are established in North America. To explore the genetic determinants of these phenotypic differences, we sequenced and annotated a draft genome of L. dispar and used it to identify genetic variation between EGM and AGM populations. The 865-Mb gypsy moth genome is the largest Lepidoptera genome sequenced to date and encodes ∼13,300 proteins. Gene ontology analyses of EGM and AGM samples revealed divergence between these populations in genes enriched for several gene ontology categories related to muscle adaptation, chemosensory communication, detoxification of food plant foliage, and immunity. These genetic differences likely contribute to variations in flight ability, chemical sensing, and pathogen interactions among EGM and AGM populations. Finally, we use our new genomic and transcriptomic tools to provide insights into genome-wide gene-expression changes of the gypsy moth after viral infection. Characterizing the immunological response of gypsy moths to virus infection may aid in the improvement of virus-based bioinsecticides currently used to control larval populations.
The gypsy moth (Lymantria dispar) is native to Europe and Asia but was accidentally introduced into North America in Medford, Massachusetts in the 1860s (1). Since then, the gypsy moth has spread throughout much of the northeastern seaboard of the United States and Canada. Polyphagous gypsy moth caterpillars are especially devastating defoliators, feeding on well over 300 species of trees within coniferous and deciduous forests (2, 3). Defoliation of residential areas has also had significant economic impacts (2). Gypsy moth outbreaks typically occur every 5–10 y (4, 5) and result in varying degrees of defoliation, depending upon factors such as tree density and species composition in the outbreak area (2).
Three subspecies of L. dispar have been described based on morphology, female flight capability, geographic origin, and mitochondrial DNA analyses (6–8). The European gypsy moth (EGM, Lymantria dispar dispar) subspecies, which was introduced to North America, is characterized by flightless females. In contrast, the females of the two Asian subspecies: Japanese gypsy moth (Lymantria dispar japonica) and Asian gypsy moth (AGM, Lymantria dispar asiatica) have larger, more developed wings and fly (9). While L. dispar japonica is found only in Japan and is geographically restricted, the AGM inhabits most of continental Asia and eastern regions of Russia (8). The wide geographic distribution, broad diet, and female flight of the AGM make this subspecies a particular economic threat if populations are established in North America (8). The genetic determinants explaining variation in female flight ability and other phenotypic differences among these subspecies are not understood.
Strategies to control the spread of gypsy moth populations have included pheromone-baited trapping and the use of “bioinsecticides” consisting of lethal viral or bacterial pathogens of larvae (1, 10, 11). Initial testing of potential bioinsecticides has greatly benefited from the development of gypsy moth cell lines, such as the ovarian tissue-derived LD652 cell line. Since the creation of the LD652 cell line 40 y ago (12), these cells have been used to investigate the life cycle of diverse DNA viruses, including baculoviruses (12), poxviruses (13–17), and densoviruses (18). More recently, we (19, 20) and others (21) have reported RNA virus model systems in LD652 and LD652Y cells, the latter of which appears to be a derivative of LD652 cells that are persistently infected with a newly discovered iflavirus (21). These studies have been instrumental in furthering our understanding of invertebrate virus replication and gene-expression strategies. However, without an annotated L. dispar genome, virus–L. dispar interaction studies have been largely limited to the identification and characterization of viral proteins that modulate overt host cell responses, such as virus-encoded inhibitors of apoptosis (16). Moreover, transcriptome-wide studies of L. dispar responses to virus infection have not yet been conducted. Therefore, we have a limited understanding of the identity, function, and regulation of L. dispar factors that influence virus infection. Such knowledge could be applied to enhance the efficacy of virus-based bioinsecticides used to control L. dispar populations (1).
Here, we sequence and characterize the L. dispar genome. We provide a comparative analysis of genomic sequences from specimens representing major populations and subspecies worldwide. We discuss unique features of the L. dispar genome in relation to other sequenced Lepidoptera and provide insights into the genetic differences between EGM and AGM populations. Finally, we use our annotated genome in conjunction with RNA-sequencing (RNA-seq) to examine changes in L. dispar gene expression after virus infection.
Results
Genome Assembly, Quality Assessment, and Annotation.
We assembled the L. dispar dispar genome using nuclear DNA extracted from the LD652 cell line derived from North American EGM populations (12). This assembly resulted in a total size of 865 Mb, the largest among published Lepidoptera assemblies (22). Our initial assembly based on mate-pair libraries (v0) had a scaffold N50 of 0.25 Mb. However, after integration of Hi-C libraries, the N50 was further improved to 5 Mb (v1) (Table 1). This is consistent with previous observations (23, 24) that incorporation of Hi-C data can significantly enhance the continuity of genome assemblies (20× in our case). However, we were not able to obtain chromosome-level assembly.
Table 1.
Quality and composition of Lepidoptera genomes
| Feature | Ldi | Pra | Pse | Pgl | Ppo | Pxu | Dpl | Hme | Mci | Cce | Lac | Mse | Bmo | Pxy | Obr |
| Genome size, Mb | 865 | 246 | 406 | 375 | 227 | 244 | 249 | 274 | 390 | 729 | 298 | 419 | 481 | 394 | 638 |
| Heterozygosity, % | 0.3 | 1.5 | 1.2 | 2.3 | n.a. | n.a. | 0.55 | n.a. | n.a. | 1.2 | 1.5 | n.a. | n.a. | ∼2 | 0.72 |
| Scaffold N50, kb | 250/5,068* | 617 | 257 | 231 | 3672 | 6,199 | 716 | 194 | 119 | 233 | 525 | 664 | 3999 | 734 | 65.6 |
| CEGMA, % | 99.1 | 99.6 | 99.3 | 99.6 | 99.3 | 99.6 | 99.6 | 98.2 | 98.9 | 100 | 99.3 | 99.8 | 99.6 | 98.7 | 99.3 |
| CEGMA coverage by single scaffold, % | 81.7 | 88.7 | 87.4 | 86.9 | 85.8 | 88.8 | 87.4 | 86.5 | 79.2 | 85.3 | 86.8 | 86.4 | 86.8 | 84.1 | 81.4 |
| Ribosome proteins, % | 98.9 | 98.9 | 98.9 | 98.9 | 98.9 | 97.8 | 98.9 | 98.9 | 94.6 | 98.9 | 98.9 | 100 | 98.9 | 93.5 | 98.9 |
| GC content, % | 35.2 | 32.7 | 39 | 35.4 | 34 | 33.8 | 31.6 | 32.8 | 32.6 | 37.1 | 34.4 | 35.3 | 37.7 | 38.3 | 38.6 |
| Repeat, % | 36 | 22.7 | 17.2 | 22 | n.a. | n.a. | 16.3 | 24.9 | 28 | 34 | 15.5 | 24.9 | 44.1 | 34 | 53.5 |
| Exon, % | 1.8 | 7.9 | 6.2 | 5.07 | 7.49 | 8.59 | 8.4 | 6.38 | 6.36 | 3.11 | 6.96 | 5.34 | 4.03 | 6.35 | 2.9 |
| Intron, % | 17 | 33.3 | 25.5 | 25.6 | 24.8 | 45.5 | 28.1 | 25.4 | 30.7 | 24 | 31.6 | 38.3 | 15.9 | 30.7 | 17.7 |
| No. of proteins (thousands) | 13.3 | 13.2 | 16.5 | 15.7 | 15.7 | 13.1 | 15.1 | 12.8 | 16.7 | 16.5 | 17.4 | 15.6 | 14.3 | 18.1 | 16.1 |
Abbreviations: Bmo: Bombyx mori; Cce, Calycopis cecrops; Dpl, Danaus plexippus; Hme, Heliconius melpomene; Lac, Lerema accius; Ldi, Lymantria dispar; Mci, Melitaea cinxia; Mse, Manduca sexta; n.a., data not available; Obr, Operophtera brumata; Pra, Pieris rapae; Pse, Phoebis sennae; Pgl, Pterourus glaucus; Ppo, Papilio polytes; Pxu, Papilio xuthus; Pxy, Plutella xylostella.
After Hi-C.
The genome is predicted to have 13,331 protein-coding genes with exons and introns making up 1.8% and 17%, respectively, of the genomic sequence. Compared with most of the other Lepidoptera species analyzed, repetitive regions constitute a relatively large proportion (36%) of the genome (Table 1). The quality of the L. dispar genome assembly is as good as some of the best Lepidoptera genomes, and the completeness of the genome is comparable to that of other species of Lepidoptera, as based on the presence of: benchmarking universal single-copy orthologs (BUSCO) (25), core eukaryotic genes mapping approach (CEGMA) (26) genes, cytoplasmic ribosomal proteins, and independently assembled transcripts (Table 1). The heterozygosity of the LD652 cell-derived L. dispar genome was ∼0.3%, the lowest among all species compared (Table 1). This low heterozygosity may in part be attributed to the passage of these cells over time because genetic variants that confer growth advantages would likely be selected for and dominate the cell population. However, we also sequenced wild-caught L. dispar adults from around the world (Fig. 1A and Dataset S1) and found them to display relatively low heterozygosity. Among these specimens, EGM adults from North America, on average, exhibited the lowest heterozygosity (0.62%, n = 8) compared with EGM specimens from Europe (0.75%, n = 4) and AGM specimens from continental Asia (0.98%, n = 8) (Dataset S1). The reduced heterozygosity of the North American EGM specimens is consistent with these insects arising from a single founding population of EGM animals introduced into North America in the 1860s. In addition, the lower overall heterozygosity of EGM versus AGM specimens may reflect the inability of EGM females to fly, which may increase the frequency of mating between adults that are in the same local environment.
Fig. 1.
The Gypsy moth as a model organism. (A) Morphology of L. dispar populations. For each specimen, dorsal (Left) and ventral (Right) sides are shown and their voucher codes are (left to right, top to bottom): NVG-17104G03, -17104G10, -17104H08, -17105A01, -17104H01, -17105A06, -18028H06, and -18028H07. See Dataset S1 for specimen data. (B) Orthology assignment of nine insect and two mammalian genomes. Bars are subdivided to represent different types of ortholog relationships: “1:1:1” indicates universal single-copy genes present in all species; “Diptera” indicates Dipteran-specific genes, present in both D. melanogaster and Aedes aegypti; “Hymenoptera” indicates Hymenopteran-specific genes, present in N. vitripennis, Apis mellifera, and Pogonomyrmex barbatus genomes; “Insect” indicates all other insect-specific orthologs; “Mammal” indicates mammalian-specific orthologs; “N:N:N” indicates other universal genes, but absence in a single genome is tolerated; “Patchy” indicates orthologs that are present in at least one insect and one mammalian genome; “Species” indicates species-specific genes. The phylogeny on the Left is a maximum-likelihood tree of a concatenated alignment of 1,756 single-copy proteins from the 1:1:1 subgroup. The tree was rooted using mammals as the outgroup. CEGMA: these are essential genes and the presence of them in a genome is used to evaluate the quality of an assembly.
Previous studies have noted a similar ordering of genetic loci or synteny among Lepidoptera genomes (27–30). We compared our assembly to the Bombyx mori (silkworm, Bombycidae) genome by mapping exons to the B. mori assembly (31, 32). We took 1,521 L. dispar scaffolds with at least 10 exons (constituting 54% of the genome) and linked these scaffolds to a specific B. mori chromosome if at least 20% of scaffold exons mapped to the B. mori chromosome. In total, 90% of L. dispar scaffolds mapped to a single B. mori chromosome, indicating a high degree of synteny between these two assemblies. Among these scaffolds, 154 mapped to the B. mori Z chromosome and collectively encode 327 proteins, suggesting that these proteins are Z-linked in L. dispar (Dataset S2).
If L. dispar is to serve as a model to study insect or eukaryotic biology in general, it is important to identify protein-coding genes conserved in other eukaryotes. Therefore, we compared the conservation of protein-coding genes in L. dispar with select insect species representing: other Lepidoptera (B. mori), Diptera (e.g., Drosophila melanogaster), Coleoptera (e.g., Tribolium castaneum), Hymenoptera (e.g., Nasonia vitripennis), and Phthiraptera (Pediculus humanus). We also made comparisons with mouse (Mus musculus) and human genomes. All species compared shared 1,756 single-copy orthologs. Additionally, L. dispar encodes an additional 2,877 universal orthologs that are either duplicated or missing in one or two species in our comparison (Fig. 1B). Therefore, ∼35% of L. dispar protein-coding genes are represented among most of these disparate species. Importantly, we identified 4,169 orthologous protein groups shared between L. dispar and humans (Dataset S3), suggesting that the gypsy moth may be a beneficial model system for studying conserved eukaryotic mechanisms.
Comparative Analyses of the L. dispar Genome with Other Lepidoptera.
We constructed phylogenetic trees using genomic sequences from L. dispar and 18 other species of Lepidoptera with publicly available genomic sequences in Lepbase (33). Of the analyzed species, L. dispar is most related to the winter moth (Operophtera brumata, Geometridae), an invasive pest introduced to North America from Europe in the 1950s (34) (Fig. 2A).
Fig. 2.
Comparative analysis of L. dispar and other Lepidoptera species. (A) The phylogeny on the Left is a maximum-likelihood tree of a concatenated alignment of 1,756 single-copy proteins from the 1:1:1 subgroup and was rooted using Plutella xylostella as outgroup. (B) Duplication of myc genes in L. dispar. (C) Duplication of TLR genes in L. dispar. Abbreviation of the species are used and proteins from moths are colored by species. Proteins from gypsy moth are colored red. Abbreviations: atr, Amyelois transitella; aly, Achalarus lyclades; bmo, Bombyx mori; cce, Calycopis cecrops; cne, Calephelis nemesis; dpl, Danaus plexippus; hml, Heliconius melpomene; lac, Lerema accuius; ldi, Lymantria dispar; mci, Melitaea cinxia; mse, Manduca sexta; obr, Operophtera brumata; pgl, Pterourus glaucus; pra, Pieris rapae; pse, Phoebis sennae; pxu, Papilio xuthus; pxy, Plutella xylostella; tni, Trichoplusia ni.
Analysis of L. dispar protein-coding genes revealed an unusual expansion of genes encoding the conserved transcription factor Myc (Fig. 2B). Other Lepidoptera (Fig. 2B) and Drosophila encode a single myc gene (35). However, using both RNA-seq– and homology-based annotation, we found four putative Myc proteins (Fig. 2B), among which two are present in our transcriptomes. Importantly, we confirmed that all four myc genes were present in wild-caught EGM and AGM adult sequences, ruling out the possibility that the apparent expansion of Toll-like receptors (TLRs) is an artifact of genome assemblies or due to genetic anomalies resulting from passage of the LD652 cell line. Myc regulates key biological processes, such as cell growth, division, and survival (35). Therefore, expansion of Myc proteins may allow specialization of these proteins for different functions.
Another interesting gene expansion in L. dispar involves those encoding TLRs. We identified 17 putative TLR genes in L. dispar, whereas other species of Lepidoptera, such as O. brumata and Manduca sexta (tobacco hornworm, Sphingidae), only encode two to three TLRs (Fig. 2B). This is also more than D. melanogaster, which encodes nine TLRs (36). Insect TLRs are defined by their Toll/IL-1 receptor and leucine-rich repeat domains (37), and all 17 putative L. dispar TLRs encode both Toll/IL-1 receptor and leucine-rich repeat domains. Furthermore, we detected expression of five of these putative TLRs in our RNA-seq data. Mapping the reads from wild-caught EGM and AGM adults to the LD652 cell-based reference genome confirmed that these 17 copies are also present in the genomes of wild free-flying adults.
TLRs are critical components of signaling pathways involved in development and immunity in insects. In Drosophila, both developmental and immunity-related TLR pathways initiate after different upstream proteolytic cascades result in cleavage of spatzle, the TLR ligand (36). Binding of spatzle to TLRs triggers intracellular signaling pathways that result in gene-expression programs promoting dorsal-ventral patterning (developmental pathway) or antimicrobial production (immunity pathway) (36). Studies of M. sexta indicate that spatzle–TLR interaction also elicits immune responses, suggesting that this pathway is conserved in Lepidoptera (36). TLRs activate immunological responses to fungal (38), bacterial (39), and viral infections (40). Therefore, expansion of TLRs may provide L. dispar with enhanced pathogen-defense mechanisms.
Comparison of L. dispar Populations Across the Globe.
We sequenced 26 L. dispar specimens from five major geographic regions: North America, Europe, Iran, continental Asia, and Japan (Dataset S1). Mitochondrial DNA genome (mitogenome) sequences from these specimens were used to construct a phylogenetic tree to group these specimens. Our results are largely consistent with previous mitogenome studies of L. dispar populations worldwide (7, 8) (Fig. 3A). Namely, North American populations group with L. dispar dispar specimens from Europe, consistent with their origin. Furthermore, continental Asia (L. dispar asiatica) and Japanese (L. dispar japonica) populations grouped together and shared a mitochondrial gene pool. Interestingly, one specimen from the Japanese island of Hokkaido (NVG-17105A06) had a mitogenome consistent with Lymantria umbrosa, which serves as a root of the tree. L. umbrosa was initially classified as a L. dispar subspecies but mitogenome analyses led to its reclassification as a distinct species in 2007 (8). The other specimen from this island possessed a mitogenome typical of L. dispar japonica. Finally, both Iranian specimens grouped as a sister to the other L. dispar specimens.
Fig. 3.
Comparison of mitochondrial and nuclear genomes among L. dispar specimens. (A) Phylogeny of mitogenomes of 26 wild-caught specimens from continental Asia, Japan, Iran, Europe, and North America are calculated by maximum likelihood. The LD652 cell line mitogenome is also included in this analysis. Populations are presented by different colors. Iran, blue; Asian, red; European, orange; United States (North America), yellow. (B) PCA of nuclear genomes of the same 26 specimens onto first two PC axes.
Principle component analysis (PCA) of nuclear genomes from these specimens revealed intriguing differences from the mitogenome results (Fig. 3B). Three major clusters were apparent: continental Asia, Japan, and the rest. Specimens from continental Asia are well-separated from Japanese specimens and form the most genetically diverse group. The heterogeneity of this population suggests that L. dispar may have originated in Asia as a species. The specimen with the L. umbrosa mitogenome clustered with the Japanese group, suggesting that it experienced mitochondrial introgression. The third cluster includes remaining populations. Specimens from the United States are placed farther away from the rest, in agreement with a genetic bottleneck experienced during introduction. However, the Iranian specimens were not separated from European populations by their nuclear genome sequences, implying an unusual path in the evolution of their mitochondria. We suspect that these Iranian specimens belong to the European population and that they acquired mitogenomes through introgression from some other, not yet discovered population in Asia.
Genetic Variations Between European and North American L. dispar dispar.
We identified 167 proteins with significant (P < 0.05) divergence between European and North American L. dispar dispar populations (Dataset S4). Based on annotations from their best hit in UniProt, these proteins likely function in diverse cellular processes such as DNA repair [e.g., ldi371120.1, related to poly(ADP)ribose polymerase-1], metabolism (e.g., ldi479.9, related to glyceraldehyde-3-phosphate dehydrogenase 2), and translation (e.g., ldi10140.2, related to eukaryotic translation initiation factor 5B).
Interestingly, two TLR proteins (ldi40446.1 and ldi40446.2) have diverged between these populations (Dataset S4). These L. dispar TLRs are most closely related to Drosophila Toll-5 (26–28% identical) and Toll-1 (27–28% identical). Toll-5 and Toll-1 are most closely related to one another among all Drosophila TLR proteins (41). Therefore, it is perhaps not surprising that these L. dispar TLRs would be similar to both Drosophila proteins. Interestingly, Toll-5 and Toll-9 were the best enhancers of lipopolysaccharide-induced antimicrobial gene expression among Drosophila TLRs in cell culture studies, suggesting that Toll-5–related proteins may be especially important for immunity (42). Further studies will be needed to determine if these L. dispar TLRs function in a similar manner to Drosophila Toll-5, but it is possible that divergence in these TLRs may reflect pathogen-driven adaptations that enhance L. dispar survival when confronting microbes in European and North American environments.
Genetic Variations Between EGM and AGM Populations.
Compared with the EGM, AGM populations have a broader plant host range and females with larger wingspans (Fig. 1A) that are capable of flight (8, 43). To gain insights into genetic variation that may explain these phenotypic differences, we analyzed EGM and AGM genomes for regions with divergence. These two populations are very close to each other, with less than 0.07% of regions indicative of divergent position ratios of more than 0.05 (Fig. 4A). Further analyses revealed that intergenic, repetitive, and intronic regions are as much as two- to threefold more diverged than coding regions (Fig. 4B). Analysis of the upstream 100-bp region of ORFs indicates that these regions are more diverged than the coding sequences themselves, suggesting that regulatory regions between these populations are more dissimilar than coding sequences. However, we identified 278 proteins with significant divergence (P < 0.05) between EGM and AGM populations (Dataset S5). To probe the biological function of these diverged proteins, we conducted a gene ontology (GO) term analysis and found significant enrichment (P < 0.01) in 40 GO terms. These included such GO terms as “skeleton muscle adaptation” (GO:0043501) and “ionotropic glutamate receptors” (GO:0035235) among others related to gene expression and cell motility (Fig. 4C and Dataset S5).
Fig. 4.
Divergence between EGM and AGM populations. (A) Distribution of ratio of divergent positions between EGM and AGM populations in 1-kb windows. (B) The divergent positions ratio in different categories. “Adjacent” indicates a 100-bp segment upstream of genes; “Exon” indicates protein-coding regions; “Intergenic” indicates regions between genes while repetitive regions are excluded; “Intron” indicates introns excluding repetitive regions; “Repetitive” indicates repetitive regions. (C) GO-term analysis of proteins with elevated divergence between EGM and AGM populations. Related GO terms are connected by lines. The size of the GO-term circle is proportional to the number of Drosophila proteins associated with this term; the color indicates the level of significance with darker colors indicating a higher degree of significance.
Why exactly EGM females are incapable of sustained flight is unclear. However, a previous study of EGM and AGM females collected from populations around the world found that muscle strength and wing size were the best predictors of female flight ability (43). There is a positive correlation between flight endurance and flight muscle adaptation in insects (44, 45). Several diverged proteins between EGM and AGM have putative roles in muscle contraction, such as l-glutamate receptors (e.g., ldi36697.1, ldi19207.1) (46, 47) and voltage-gated calcium channel proteins (e.g., ldi3317.2, ldi3317.1) (44). Changes in intracellular calcium levels significantly regulate mechanical power output of insect flight muscles (48). Therefore, variations in voltage-gated calcium channel function in flight muscles between EGM and AGM females may result in mechanical forces that are insufficient to support female EGM flight.
The female EGM flightless phenotype may also be related to the inability of the reduced wing span of these animals to sustain flight of females (43) that have a greater body mass compared with males (Fig. 1A). Previous studies in Drosophila have identified multiple proteins that control wing size during development, such as capicua, a transcriptional regulator, and lingerer, a conserved ubiquitin-associated domain-containing protein. Capicua acts as a transcriptional repressor and its overexpression in flies results in reduced wing size (49). Lingerer also negatively regulates wing size. Flies with null alleles of lingerer abnormally up-regulate the evolutionarily conserved Hippo pathway that promotes wing development in insects, resulting in overgrowth of wing imaginal discs (50). We found the putative L. dispar capicua (ldi3634.2) and lingerer (ldi2916.2) proteins to be significantly diverged in EGM and AGM populations (Dataset S5). Therefore, if amino acid substitutions in EGM capicua and lingerer were to enhance their activity, then this could result in the smaller wings in this subspecies (43).
Previous studies suggest that Toll-1 signaling in Drosophila also regulates wing size during development by modulating mitogen-activated protein kinase-mediated wing cell death (51). Intriguingly, we found significant divergence in a TLR (ldi2259.5) between EGM and AGM populations that is most closely related to Drosophila Toll-1 (∼34% identical) and Toll-5 (28% identical). Furthermore, L. dispar homologs of snake (ldi410.4) and easter (ldi4228.6) proteases have also diverged between these subspecies (Dataset S5). During Drosophila development, TLR signaling is initiated when snake cleaves easter, which in turn cleaves spatzle, which then binds to and activates Toll-1 (36). Interestingly, capicua has recently been shown to regulate Toll-1 signaling gene targets (52). Therefore, variations in snake, easter, TLR, and capicua proteins between EGM and AGM populations may alter a TLR-centric signaling pathway that controls wing size. Functional studies will be needed to determine if ldi2259.5 functions in an analogous manner to Drosophila Toll-1 in wing-size modulation.
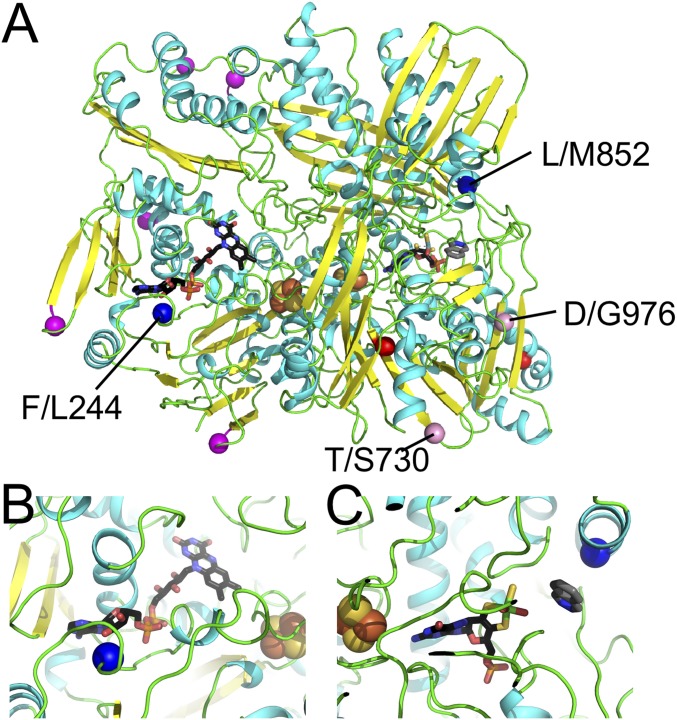
We also noted divergence between several EGM and AGM proteins with putative roles in insecticide detoxification and odorant detection. For example, we identified eight putative cytochrome p450 enzymes (ldi2653.16, ldi7704.1, ldi1892.3, ldi5170.3, ldi451.1, ldi8270.2, ldi7290.4, and ldi396463.4) that have diverged between EGM and AGM subspecies (Dataset S5). Insect p450 enzymes play key roles in the detoxification of plant toxins and insecticides, and thus their activity can significantly influence the range of plant species an insect can feed on, as well as insecticide resistance (53). Previous studies have shown that transcription of several L. dispar p450 genes is induced upon exposure to sublethal doses of insecticides, suggesting that sensing of toxins may trigger a detoxification response (54). We also found divergence of several proteins with putative chemosensory roles, such as odorant-binding proteins (e.g., ldi15505.4) and odorant receptors (e.g., ldi4228.4). The exact function of many odorant-binding proteins is unknown but they have been postulated to solubilize and transport hydrophobic pheromonal or odorant compounds to facilitate their interaction with odorant receptors in antennae (55). Adaptations to recognize specific odorants and pheromones among subspecies may enhance their ability to find mates within their respective environments. Other proteins diverged between EGM and AGM, such as aldehyde oxidases (AOX; e.g., ldi971.1), may play both chemosensory and detoxification roles. Insect AOXs catalyze the oxidation of aldehydes into carboxylic acids and inactivate odorant molecules after they bind to their receptors and transmit their signals (56). Other Lepidoptera AOXs have been shown to degrade aldehydic sex pheromones and volatile plant compounds (56–58), indicating that these enzymes may function in both communication and detoxification. Interestingly, there are 11 polymorphic sites in ldi971.1 between EGM and AGM subspecies (Dataset S5). Mapping of these sites to a human AOX structure (59) indicates that two of these positions are near active sites where the flavin adenine dinucleotide (FAD) and molybdenum cofactors are bound (Fig. 5). These polymorphisms may affect AOX ligand binding and catalytic efficiencies that could provide selective advantages to the detection of chemical stimuli or detoxification of plant volatiles found within environments inhabited by EGM and AGM populations.
Fig. 5.
An aldehyde oxidase shows elevated divergence between EGM and AGM populations. (A) L. dispar aldehyde oxidase (ldi971.1) modeled on a human aldehyde oxidase template (4UHW) is depicted. Colors indicate secondary structure: helix (cyan), strand (yellow), and loop (green) with FeS centers (orange/yellow spheres), FAD redox cofactor (black stick), molybdenum cofactor (black stick), and substrate (gray stick). Population-specific sites map to the model surface (G/S17, E/K304, C/Y366, L/Q511, C/F516, T/S730, and D/G976; Cα positions are shown with magenta/pink spheres), with two of the surface residues contributing to the dimer interface (D/G976 and T/S730; Cα positions shown with pink spheres). Two sites form the hydrophobic core of their respective subdomains (T/A725 and V/I1056; Cα positions shown with red spheres). (B and C) Zoom-in of two polymorphic residues that map near active sites (blue spheres): F/L244, which lines the FAD-binding site (black stick) (B), and L/M852, which lines the substrate-binding site (gray stick) in the molybdenum cofactor (black stick)-binding domain (C).
Gypsy Moth Cell Lines as a Model to Study Virus–Host Interactions.
A transcriptome-wide analysis of L. dispar responses to virus infection has not yet been reported. To provide initial insights into host gene-expression changes after infection, we conducted RNA-seq studies to identify differentially expressed genes (DEGs) in LD652 cells 24-h postinfection (hpi) with three different viruses. We chose Amsacta moorei entomopoxvirus (AmEPV), vaccinia virus (VACV), and vesicular stomatitis virus (VSV) for our analyses.
Both AmEPV and VACV are large double-strand DNA-encoding poxviruses that express >200 proteins and replicate exclusively in the cytoplasm of infected cells. Although originally isolated from the red hairy caterpillar (A. moorei, Arctiidae) (17, 60), AmEPV productively replicates in LD652 cells and its study in these cells has become the prototypic entomopoxvirus–insect host model system. In contrast, VACV is a vertebrate poxvirus that does not productively replicate in LD652 cells due to a failure in virion morphogenesis (61). Despite this, VACV still undergoes early gene expression, DNA replication, and postreplicative gene expression in these cells (61). Like poxviruses, VSV replicates in the cytoplasm of infected cells but encodes only five proteins in its single-strand RNA genome. We have previously shown that VSV undergoes a postentry, abortive infection in LD652 cells that can be relieved by inhibition of host transcription with actinomycin D treatment (20). Alternatively, VACV coinfection can also “rescue” VSV replication in LD652 cells (20). These observations suggest that VSV infection elicits host gene-expression changes that block infection and that VACV-encoded immunomodulatory factors may circumvent these antiviral responses. Therefore, we were interested to gain insights into potential changes in L. dispar gene expression after infection with VSV, VACV, or both viruses.
Compared with mock-infected samples, we identified 3,106 and 2,412 DEGs in AmEPV and VACV infections, respectively. Remarkably, ∼50% of AmEPV-induced DEGs were found among DEGs observed after VACV infection, suggesting that invertebrate and vertebrate poxvirus infection may invoke similar changes in the L. dispar transcriptome (Table 2 and Dataset S6). Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of up-regulated DEGs in AmEPV and VACV infections identified endocytic, ubiquitin-mediated proteolysis, phosphatidylinositol signaling, and Hippo signaling pathways as significantly enriched in both poxvirus infections (P < 0.05). Additionally, down-regulated DEGs in these infections also shared several KEGG pathway terms, including: oxidative phosphorylation, ribosome, proteasome, and RNA transport (Dataset S7). Despite these similarities, there were clear differences in the regulation of genes encoding key immunity-related factors. For example, we observed differences in regulation of the L. dispar gene encoding the NF-κB homolog relish (ldi6693.2) that acts as a transcriptional activator of antimicrobial responses (20). Relish was significantly down-regulated during AmEPV infection but not in VACV infection. In addition, a single TLR (ldi17892.4) was down-regulated in AmEPV but not in VACV infection (Dataset S6). These differences in host gene expression during AmEPV and VACV infection may reflect the degree of adaptation of these viruses to combating insect hosts.
Table 2.
Analysis of L. dispar DEGs after virus infection
| DEG | AmEPV | VACV | VSV | VSV+VACV |
| DEGs up-regulated | 1,346 | 1,264 | 11 | 1,481 |
| DEGs down-regulated | 1,760 | 1,148 | 4 | 980 |
| DEGs shared with AmEPV | NA | 1,486 | 0 | 1,515 |
| DEGs shared with VACV | 1,486 | NA | 2 | 1,990 |
| DEGs shared with VSV | 0 | 2 | NA | 1 |
| DEGs shared with VSV+VACV | 1,515 | 1,990 | 1 | NA |
| DEGs with UniProt homolog | 2,731 | 2,126 | 7 | 2,170 |
In contrast to poxvirus infection, VSV infection resulted in only 15 DEGs at 24 hpi (Table 2 and Dataset S6). One of these DEGs, ldi415371.1, was up-regulated after VSV infection but significantly down-regulated during VACV infection or VSV–VACV coinfection. This gene encodes an ortholog of the human N-α-acetyltransferase 40 protein that specifically acetylates histones H2A and H4 and thus might regulate the accessibility of chromatin to transcriptional machinery (62). The remaining 14 DEGs observed during VSV infection were not found among DEGs in VSV–VACV coinfections, suggesting that VACV infection dramatically modulates host responses to VSV infection, which may contribute to VACV-mediated relief of VSV restriction (20). The notion that VACV infection drives the majority of DEGs during coinfections with VSV is underscored by the fact that ∼81% of DEGs observed in coinfections were also found in single VACV infections (Table 2). Interestingly, L. dispar autophagy-related protein 16 (Atg16; Atg16L1 in mammals) was one of 11 DEGs up-regulated after VSV infection that was not up-regulated after VACV coinfection (Dataset S6). Autophagy is a conserved cellular process in which cytosolic components are degraded after targeting to lipid-enclosed vesicles termed “autophagosomes” (63–65). Fusion of autophagosomes with lysosomes results in the degradation of the membrane-enclosed material and this process requires Atg16 (64). Studies in Drosophila have shown that VSV infection induces autophagosome formation and that autophagy contributes to the restriction of VSV (66). Therefore, VACV coinfection may dysregulate a conserved antiviral autophagy response in L. dispar that may otherwise restrict VSV infection.
Discussion
Since the first Lepidoptera genome was reported in 2004 for B. mori (31, 32), there have been over 20 additional moth and butterfly annotated genomes published, making comparative genomics increasingly possible across Lepidoptera species (22). At 865 Mb, the L. dispar genome dwarfs the majority of sequenced Lepidoptera genomes that, on average, range from 250 to 500 Mb in size (Table 1). L. dispar is also the first member of Erebidae to be sequenced. Despite its large size, the L. dispar genome encodes a relatively small number (∼13,331) of protein-coding genes compared with other Lepidoptera with smaller genomes. This difference may be in part due to the large proportion (36%) of the genome that encodes repetitive elements. This also appears to be true for O. brumata, the closest sequenced relative of L. dispar, in which repetitive elements make up ∼50% of the 638-Mb genome (67). Given their genetic relatedness, shared history of invading North America via Europe, and significant impact on forest defoliation along the northeastern seaboard, molecular studies in L. dispar may be informative for understanding O. brumata biology and vice versa.
Comparison of genomic sequences from wild-caught specimens (Dataset S1) suggests that AGM populations are the most genetically diverse. This elevated genetic diversity suggests that L. dispar might have arisen as a species in continental Asia, although we cannot rule out the possibility that it originated elsewhere, spread to Asia, and then differentiated. The separation of L. dispar japonica from AGM was not surprising, due to the reproductive isolation of this subspecies. The lower heterozygosity of North American L. dispar dispar compared with European populations was also to be expected given the genetic bottleneck imposed on the founding population in New England and the fact that this population has remained largely confined to the northeastern seaboard and has not yet spread to other North American ecosystems that may select for different traits over time.
Analysis of protein-coding genes revealed several interesting features of L. dispar. For one, ∼35% of L. dispar proteins are conserved across other model insect species. Because L. dispar ecology is well-studied (68), its genome may be an important tool for understanding the genetic determinants of its ecology. The expansion and adaptation of protein families such as TLRs may contribute to the ability of L. dispar to inhabit environments worldwide. Recent studies of Drosophila species suggest that genetic variation of specific TLRs, such as Toll-3/4/5, may contribute to pathogen immunity in diverse environments (69, 70). Expansion and rapid evolution of TLRs has also been noted in other invertebrates, such as in Croassostrea gigas (Pacific oyster), which encodes an astounding 88 TLRs (71). Therefore, it was interesting to find significant divergence in two TLRs related to Drosophila Toll-5 and Toll-1 between North American and European L. dispar dispar populations. It is currently unclear if these L. dispar TLRs represent functional equivalents to Drosophila Toll-5 and Toll-1 but divergence in these proteins may reflect selective pressures resulting from interaction with pathogens in these different geographic locations and ecological circumstances. Previous studies have noted differences in pathogenicity among North American, European, and Asian isolates of L. dispar multicapsid nuclear polyhedrosis virus (LdMNPV) in gypsy moth larvae (72). It would be interesting to determine if TLR polymorphisms represent part of an “evolutionary arms race” between gypsy moths and natural pathogens, such as LdMNPV, that display heterogeneity in the environment.
Divergence in TLR signaling pathway components may also help to explain one of the most interesting phenotypic variations among gypsy moth populations: the inability of EGM females to fly. It was striking to find significant divergence in not only a TLR but also upstream factors (e.g., snake, easter) of this pathway given its role in regulating wing size in Diptera (51). The fact that the transcription factor capicua was also diverged and is known to regulate both wing size and TLR signaling further implies a potential role for TLR signaling in modulating wing size in the EGM. However, it is also possible that other known regulators of wing size (e.g., lingerer) or neuromuscular (e.g., voltage-gated ion channels) factors that were diverged between these populations may affect wing size and flight capabilities. Clearly, functional studies will be needed to investigate the possible role of these factors in the female EGM flightless phenotype. Why exactly EGM female flightless phenotypes are stable in European and North American environments is unclear, but female brachyptery (wing reduction) is found in many other moth species, including O. brumata (67). It has been suggested that in environments where food is abundant and distant foraging is unnecessary, female brachyptery may be a useful trade-off to increase fecundity given the energetic costs associated with flight (73, 74). Furthermore, polyphagy may allow for female brachyptery to become a stable trait in these situations. Therefore, differences in food availability in EGM and AGM habitats may contribute to selection of female brachyptery.
We also noted a large number of diverged proteins relating to chemosensory and detoxification machinery between EGM and AGM populations. Specific variations in detoxification functions between these populations may serve to define the range of food sources EGM and AGM populations can feed on and detoxify. Prominent among diverged detoxification machinery were p450 enzymes, which play critical roles in the metabolism of plant-encoded insecticidal compounds (75). Interestingly, the O. brumata genome encodes an expansion of specific p450 genes, possibly reflecting its diet preferences (67). It will be important to investigate the role of p450s in influencing differences in plant host range between EGM and AGM populations to identify environments/regions that may be particularly susceptible to these defoliators.
While divergence in coding regions may contribute to phenotypic differences between EGM and AGM populations, it is also possible that differences in noncoding regions (Fig. 4B), such as promoter sequences, may lead to variation in gene expression between these subspecies that contribute to these phenotypic differences. Future studies examining the association of genetic variation with gene-expression differences between these populations may reveal further insights into this possibility.
Despite the current use of viruses as control agents for L. dispar, we have relatively little understanding of Lepidoptera antiviral immunity compared with Diptera (76). We previously unveiled roles for L. dispar RNA interference (RNAi)-, relish-, and ubiquitin-proteasome–related immunity pathways in the restriction of RNA viruses, such as VSV (20). These studies suggested that L. dispar encodes multiple independent mechanisms to restrict virus replication. However, the gene-expression changes after RNA or DNA virus infection in L. dispar had not been investigated on a genome-wide scale. Our RNA-seq studies suggest that infection by either AmEPV or VACV results in significant changes in expression of ∼20% of L. dispar protein-coding genes, with a remarkably similar overall signature. The up-regulation of endocytic pathways by poxvirus infection (Dataset S3) may reflect a viral mechanism to enhance infection of nearby cells, as poxviruses use endocytic pathways for entry (77). These poxvirus infections also resulted in down-regulation of host ribosomal machinery, which might represent a host mechanism to shut off viral mRNA translation. Furthermore, both AmEPV and VACV induced up-regulation of ubiquitin-proteolysis machinery (Dataset S3). This may reflect an L. dispar mechanism to degrade viral proteins, as we have observed during VSV infection of LD652 cells (20). It is important to note that ∼88% of all DEGs observed in AmEPV and VACV infections encode proteins with clear homologs in UniProt databases (Table 2). Therefore, L. dispar may serve as a useful model for studying conserved virus–host interactions.
The relatively small number of DEGs observed after VSV infection may reflect the inability of this virus to productively replicate (and stimulate fulminate host responses) or the time point we analyzed may have been too late to observe altered gene-expression patterns found earlier in infection. We favor the latter scenario because inhibition of host transcription through the addition of actinomycin D to VSV cultures at 2 hpi completely restores VSV replication (20). Additional RNA-seq studies with earlier time points will be needed to investigate this further. However, it was interesting that essentially all of the VSV-induced changes in L. dispar gene expression were reversed or prevented by VACV coinfection (Table 2). This included the inhibition of VSV-induced up-regulation of Atg16, an essential component of the autophagy pathway, known to inhibit VSV in Drosophila (66). Whether autophagy inhibits RNA virus replication in Lepidoptera will require future functional studies.
Recent advancements in RNAi and genomic editing tools for Lepidoptera (78–80) have facilitated functional genomics-based studies of various aspects of Lepidoptera development (81), pheromonal communication (82), and coloration (83). Applying these tools to genome-wide functional studies of virus–host interactions in L. dispar cell lines will undoubtedly reveal key facets of antiviral immunity. Such studies may not only broaden our understanding of eukaryotic innate immunity but may also provide new strategies for compromising antiviral immunity in L. dispar, and related Lepidoptera pests, so as to enhance caterpillar susceptibility to viral bioinsecticides.
In conclusion, the sequencing and annotation of the L. dispar genome brings us closer to characterizing the genetic loci influencing key phenotypic traits, including flight capability, insecticide resistance, and pathogen susceptibility. In time, this large genome may answer many questions surrounding this tiny bug.
Materials and Methods
Sequencing Strategy.
The L. dispar dispar-derived LD652 cell line was used for DNA extraction. Two paired-end libraries with average insert sizes of 250 and 500 bp were constructed and sequenced on an Illumina Hiseq X10 instrument. Additionally, four mate-pair libraries (2, 4, 9, and 14 kb) were constructed and sequenced using an Illumina HiSEq 2500 instrument. Hi-C libraries were prepared by Novogene and sequenced on an Illumina Hiseq X10 instrument.
Genome and Transcriptome Assembly.
AdapterRemoval v2.2 (84) was used for trimming adapters and low-quality bases (quality score <20) for all libraries. For mate-pair libraries, before AdapterRemoval, the Delox script (85) was used to remove the loxP sequences and to separate true mate-pair from paired-end reads, as described previously (86). We used JELLYFISH v2.2.3 (87) to obtain k-mer frequencies in all genomic DNA libraries and QUAKE v0.3.5 was used to correct sequencing errors (88). The data processing resulted in eight libraries that were used with Platanus v1.2.4 (89) for genome assembly: 250- and 500-bp paired-end libraries; 2-, 6-, 10-, and 20-kb true mate-pair libraries; a library containing all of the paired-end reads from the mate-pair libraries; and a single-end library containing all reads whose pairs were removed in the process (86).
To remove redundant scaffolds caused by heterozygosity and repetitive regions, reads were mapped to initial assemblies and coverage of scaffolds was calculated using SAMtools (90). Scaffolds were removed if they could fully align to other regions significantly less covered in long scaffolds with high sequence identity.
For transcriptome assemblies, three strategies were used: (i) de novo assemblies by Trinity (91); (ii) reference-guided assembly by Trinity; and (iii) reference-based assembly by TopHat v2.0.10 (92) and Cufflinks v2.2.1 (93), as described previously (86). The results from all three methods were then integrated by Program to Assemble Spliced Alignment v2.0.2 (94).
Reads for the genome assembly, annotation and studies of viral infection of gypsy moth cell line LD652, and reads for population analysis of gypsy moth were deposited in NCBI as BioProject (95) ID PRJNA504524 and PRJNA505229, respectively. The assembled LD652 cell line genome was deposited in DDBJ/EMBL/GenBank (96) under the accession RJWF00000000. The version described here is RJWF01000000. Further detailed methods are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. Sean Whelan (Harvard Medical School) and Gary Luker (University of Michigan Medical School) for the provision of vesicular stomatitis virus-LUC and vaccinia virus-FL-GFP, respectively; and Drs. Richard Moyer (University of Florida) and Basil Arif (Natural Resources Canada) for the kind gifts of the Amsacta moorei entomopoxvirus-GFP (vAmΔsph/gfp) virus and LD652 cells, respectively. We are grateful to Robert K. Robbins and Brian Harris (National Museum of Natural History, Smithsonian Institution, Washington, DC) for granting access to the collection. D.B.G. was supported by funding from the University of Texas Southwestern Medical Center’s Endowed Scholars Program. N.V.G. is supported by National Institutes of Health Grants GM094575 and GM127390 and the Welch Foundation Grant I-1505.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. RJWF00000000) and NCBI Sequence Read Archive (Bioproject IDs PRJNA504524 and PRJNA505229).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818283116/-/DCSupplemental.
References
- 1.Harrison RL, Rowley DL, Keena MA. Geographic isolates of Lymantria dispar multiple nucleopolyhedrovirus: Genome sequence analysis and pathogenicity against European and Asian gypsy moth strains. J Invertebr Pathol. 2016;137:10–22. doi: 10.1016/j.jip.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Bigsby KM, Ambrose MJ, Tobin PC, Sills EO. The cost of gypsy moth sex in the city. Urban For Urban Greening. 2014;13:459–468. [Google Scholar]
- 3.Elkinton JS, Liebhold AM. Population-dynamics of gypsy-moth in North America. Annu Rev Entomol. 1990;35:571–596. [Google Scholar]
- 4.Johnson DM, Liebhold AM, Bjornstad ON, McManus ML. Circumpolar variation in periodicity and synchrony among gypsy moth populations. J Anim Ecol. 2005;74:882–892. [Google Scholar]
- 5.Haynes KJ, Liebhold AM, Johnson DM. Spatial analysis of harmonic oscillation of gypsy moth outbreak intensity. Oecologia. 2009;159:249–256. doi: 10.1007/s00442-008-1207-7. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, et al. Rapid identification of the Asian gypsy moth and its related species based on mitochondrial DNA. Ecol Evol. 2018;8:2320–2325. doi: 10.1002/ece3.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogdanowicz SM, Schaefer PW, Harrison RG. Mitochondrial DNA variation among worldwide populations of gypsy moths, Lymantria dispar. Mol Phylogenet Evol. 2000;15:487–495. doi: 10.1006/mpev.1999.0744. [DOI] [PubMed] [Google Scholar]
- 8.Djoumad A, et al. Comparative analysis of mitochondrial genomes of geographic variants of the gypsy moth, Lymantria dispar, reveals a previously undescribed genotypic entity. Sci Rep. 2017;7:14245. doi: 10.1038/s41598-017-14530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keena MA, Grinberg PS, Wallner WE. Inheritance of female flight in Lymantria dispar (Lepidoptera: Lymantriidae) Environ Entomol. 2007;36:484–494. doi: 10.1603/0046-225x(2007)36[484:ioffil]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 10.Whittle A, Lenhart S, White KA. Optimal control of gypsy moth populations. Bull Math Biol. 2008;70:398–411. doi: 10.1007/s11538-007-9260-7. [DOI] [PubMed] [Google Scholar]
- 11.Mayo JH, Straka TJ, Leonard DS. The cost of slowing the spread of the gypsy moth (Lepidoptera: Lymantriidae) J Econ Entomol. 2003;96:1448–1454. doi: 10.1603/0022-0493-96.5.1448. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin RH, Tompkins GJ, McCawley P. Gypsy moth cell lines divergent in viral susceptibility. I. Culture and identification. In Vitro. 1978;14:485–494. doi: 10.1007/BF02616088. [DOI] [PubMed] [Google Scholar]
- 13.Perera S, Krell P, Demirbag Z, Nalçacioğlu R, Arif B. Induction of apoptosis by the Amsacta moorei entomopoxvirus. J Gen Virol. 2013;94:1876–1887. doi: 10.1099/vir.0.051888-0. [DOI] [PubMed] [Google Scholar]
- 14.Muratoglu H, Nalcacioglu R, Arif BM, Demirbag Z. Genome-wide analysis of differential mRNA expression of Amsacta moorei entomopoxvirus, mediated by the gene encoding a viral protein kinase (AMV197) Virus Res. 2016;215:25–36. doi: 10.1016/j.virusres.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Liston P, Schokman N, Ho JM, Moyer RW. Amsacta moorei entomopoxvirus inhibitor of apoptosis suppresses cell death by binding Grim and Hid. J Virol. 2005;79:3684–3691. doi: 10.1128/JVI.79.6.3684-3691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Liston P, Moyer RW. Functional analysis of the inhibitor of apoptosis (iap) gene carried by the entomopoxvirus of Amsacta moorei. J Virol. 2005;79:2335–2345. doi: 10.1128/JVI.79.4.2335-2345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker MN, Greenleaf WB, Ostrov DA, Moyer RW. Amsacta moorei entomopoxvirus expresses an active superoxide dismutase. J Virol. 2004;78:10265–10275. doi: 10.1128/JVI.78.19.10265-10275.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, Graham RI, Wilson K, Wu K. Structure and transcription of the Helicoverpa armigera densovirus (HaDV2) genome and its expression strategy in LD652 cells. Virol J. 2017;14:23. doi: 10.1186/s12985-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rex EA, Seo D, Gammon DB. Arbovirus infections as screening tools for the identification of viral immunomodulators and host antiviral factors. J Vis Exp. 2018:e58244. doi: 10.3791/58244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gammon DB, et al. A single vertebrate DNA virus protein disarms invertebrate immunity to RNA virus infection. eLife. 2014;3:e02910. doi: 10.7554/eLife.02910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrillo-Tripp J, et al. Lymantria dispar iflavirus 1 (LdIV1), a new model to study iflaviral persistence in lepidopterans. J Gen Virol. 2014;95:2285–2296. doi: 10.1099/vir.0.067710-0. [DOI] [PubMed] [Google Scholar]
- 22.Triant DA, Cinel SD, Kawahara AY. Lepidoptera genomes: Current knowledge, gaps and future directions. Curr Opin Insect Sci. 2018;25:99–105. doi: 10.1016/j.cois.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Marie-Nelly H, et al. High-quality genome (re)assembly using chromosomal contact data. Nat Commun. 2014;5:5695. doi: 10.1038/ncomms6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudchenko O, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waterhouse RM, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2017;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 27.Sahara K, et al. Conserved synteny of genes between chromosome 15 of Bombyx mori and a chromosome of Manduca sexta shown by five-color BAC-FISH. Genome. 2007;50:1061–1065. doi: 10.1139/g07-082. [DOI] [PubMed] [Google Scholar]
- 28.Pringle EG, et al. Synteny and chromosome evolution in the lepidoptera: Evidence from mapping in Heliconius melpomene. Genetics. 2007;177:417–426. doi: 10.1534/genetics.107.073122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papa R, et al. Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. BMC Genomics. 2008;9:345. doi: 10.1186/1471-2164-9-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.d’Alençon E, et al. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc Natl Acad Sci USA. 2010;107:7680–7685. doi: 10.1073/pnas.0910413107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mita K, et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- 32.Xia Q, et al. Biology Analysis Group A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 33.Challis RJ, Kumar S, Dasmahapatra KK, Jiggins CD, Blaxter M. 2016. Lepbase: The lepidopteran genome database. bioRxiv, 10.1101/056994.
- 34.Burand JP, Kim W, Welch A, Elkinton JS. Identification of a nucleopolyhedrovirus in winter moth populations from Massachusetts. J Invertebr Pathol. 2011;108:217–219. doi: 10.1016/j.jip.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Grifoni D, Bellosta P. Drosophila Myc: A master regulator of cellular performance. Biochim Biophys Acta. 2015;1849:570–581. doi: 10.1016/j.bbagrm.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valanne S, Wang JH, Rämet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 37.Imler JL, Zheng L. Biology of Toll receptors: Lessons from insects and mammals. J Leukoc Biol. 2004;75:18–26. doi: 10.1189/jlb.0403160. [DOI] [PubMed] [Google Scholar]
- 38.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 39.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 40.Nakamoto M, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36:658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci USA. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ooi JY, Yagi Y, Hu X, Ip YT. The Drosophila Toll-9 activates a constitutive antimicrobial defense. EMBO Rep. 2002;3:82–87. doi: 10.1093/embo-reports/kvf004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keena MA, Côté MJ, Grinberg PS, Wallner WE. World distribution of female flight and genetic variation in Lymantria dispar (Lepidoptera: Lymantriidae) Environ Entomol. 2008;37:636–649. doi: 10.1603/0046-225x(2008)37[636:wdoffa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 44.Marden JH. Variability in the size, composition, and function of insect flight muscles. Annu Rev Physiol. 2000;62:157–178. doi: 10.1146/annurev.physiol.62.1.157. [DOI] [PubMed] [Google Scholar]
- 45.Chakravorty S, Vu H, Foelber V, Vigoreaux JO. Mutations of the Drosophila myosin regulatory light chain affect courtship song and reduce reproductive success. PLoS One. 2014;9:e90077. doi: 10.1371/journal.pone.0090077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, et al. Novel functional properties of Drosophila CNS glutamate receptors. Neuron. 2016;92:1036–1048. doi: 10.1016/j.neuron.2016.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fedorova IM, Magazanik LG, Tikhonov DB. Characterization of ionotropic glutamate receptors in insect neuro-muscular junction. Comp Biochem Physiol C Toxicol Pharmacol. 2009;149:275–280. doi: 10.1016/j.cbpc.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Gordon S, Dickinson MH. Role of calcium in the regulation of mechanical power in insect flight. Proc Natl Acad Sci USA. 2006;103:4311–4315. doi: 10.1073/pnas.0510109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L, et al. Minibrain and Wings apart control organ growth and tissue patterning through down-regulation of Capicua. Proc Natl Acad Sci USA. 2016;113:10583–10588. doi: 10.1073/pnas.1609417113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong L, et al. Growth suppressor lingerer regulates bantam microRNA to restrict organ size. J Mol Cell Biol. 2015;7:415–428. doi: 10.1093/jmcb/mjv045. [DOI] [PubMed] [Google Scholar]
- 51.Wu C, et al. Toll pathway modulates TNF-induced JNK-dependent cell death in Drosophila. Open Biol. 2015;5:140171. doi: 10.1098/rsob.140171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papagianni A, et al. Capicua controls Toll/IL-1 signaling targets independently of RTK regulation. Proc Natl Acad Sci USA. 2018;115:1807–1812. doi: 10.1073/pnas.1713930115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu N, Li M, Gong Y, Liu F, Li T. Cytochrome P450s—Their expression, regulation, and role in insecticide resistance. Pestic Biochem Physiol. 2015;120:77–81. doi: 10.1016/j.pestbp.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Sun L, Wang Z, Zou C, Cao C. Transcription profiling of 12 Asian gypsy moth (Lymantria dispar) cytochrome P450 genes in response to insecticides. Arch Insect Biochem Physiol. 2014;85:181–194. doi: 10.1002/arch.21152. [DOI] [PubMed] [Google Scholar]
- 55.Brito NF, Moreira MF, Melo AC. A look inside odorant-binding proteins in insect chemoreception. J Insect Physiol. 2016;95:51–65. doi: 10.1016/j.jinsphys.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 56.Choo YM, Pelletier J, Atungulu E, Leal WS. Identification and characterization of an antennae-specific aldehyde oxidase from the navel orangeworm. PLoS One. 2013;8:e67794. doi: 10.1371/journal.pone.0067794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He P, et al. A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genomics. 2017;18:219. doi: 10.1186/s12864-017-3592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rybczynski R, Reagan J, Lerner MR. A pheromone-degrading aldehyde oxidase in the antennae of the moth Manduca sexta. J Neurosci. 1989;9:1341–1353. doi: 10.1523/JNEUROSCI.09-04-01341.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coelho C, et al. Structural insights into xenobiotic and inhibitor binding to human aldehyde oxidase. Nat Chem Biol. 2015;11:779–783. doi: 10.1038/nchembio.1895. [DOI] [PubMed] [Google Scholar]
- 60.Granados RR, Roberts DW. Electron microscopy of a poxlike virus infecting an invertebrate host. Virology. 1970;40:230–243. doi: 10.1016/0042-6822(70)90398-3. [DOI] [PubMed] [Google Scholar]
- 61.Li Y, Yuan S, Moyer RW. The non-permissive infection of insect (gypsy moth) LD-652 cells by vaccinia virus. Virology. 1998;248:74–82. doi: 10.1006/viro.1998.9241. [DOI] [PubMed] [Google Scholar]
- 62.Hole K, et al. The human N-alpha-acetyltransferase 40 (hNaa40p/hNatD) is conserved from yeast and N-terminally acetylates histones H2A and H4. PLoS One. 2011;6:e24713. doi: 10.1371/journal.pone.0024713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamiable O, Imler JL. Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol. 2014;20:62–68. doi: 10.1016/j.mib.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagy P, et al. Drosophila Atg16 promotes enteroendocrine cell differentiation via regulation of intestinal Slit/Robo signaling. Development. 2017;144:3990–4001. doi: 10.1242/dev.147033. [DOI] [PubMed] [Google Scholar]
- 65.Zavodszky E, Vicinanza M, Rubinsztein DC. Biology and trafficking of ATG9 and ATG16L1, two proteins that regulate autophagosome formation. FEBS Lett. 2013;587:1988–1996. doi: 10.1016/j.febslet.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 66.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Derks MF, et al. The genome of winter moth (Operophtera brumata) provides a genomic perspective on sexual dimorphism and phenology. Genome Biol Evol. 2015;7:2321–2332. doi: 10.1093/gbe/evv145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Timms LL, Smith SM. Effects of gypsy moth establishment and dominance in native caterpillar communities of northern oak forests. Can Entomol. 2011;143:479–503. [Google Scholar]
- 69.Levin TC, Malik HS. Rapidly evolving Toll-3/4 genes encode male-specific toll-like receptors in Drosophila. Mol Biol Evol. 2017;34:2307–2323. doi: 10.1093/molbev/msx168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lazzaro BP, Sackton TB, Clark AG. Genetic variation in Drosophila melanogaster resistance to infection: A comparison across bacteria. Genetics. 2006;174:1539–1554. doi: 10.1534/genetics.105.054593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang L, et al. Massive expansion and functional divergence of innate immune genes in a protostome. Sci Rep. 2015;5:8693. doi: 10.1038/srep08693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shapiro M, Robertson JL, Injac MG, Katagiri K, Bell RA. Comparative infectivities of gypsy moth (Lepidoptera, Lymantriidae) nucleopolyhedrosis virus isolates from North America, Europe, and Asia. J Econ Entomol. 1984;77:153–156. [Google Scholar]
- 73.Roff DA. The evolution of wing dimorphism in insects. Evolution. 1986;40:1009–1020. doi: 10.1111/j.1558-5646.1986.tb00568.x. [DOI] [PubMed] [Google Scholar]
- 74.Barbosa P, Krischik V, Lance D. Life-history traits of forest-inhabiting flightless Lepidoptera. Am Midl Nat. 1989;122:262–274. [Google Scholar]
- 75.Feyereisen R. Insect P450 enzymes. Annu Rev Entomol. 1999;44:507–533. doi: 10.1146/annurev.ento.44.1.507. [DOI] [PubMed] [Google Scholar]
- 76.Mussabekova A, Daeffler L, Imler JL. Innate and intrinsic antiviral immunity in Drosophila. Cell Mol Life Sci. 2017;74:2039–2054. doi: 10.1007/s00018-017-2453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moss B. Membrane fusion during poxvirus entry. Semin Cell Dev Biol. 2016;60:89–96. doi: 10.1016/j.semcdb.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu H, O’Brochta DA. Advanced technologies for genetically manipulating the silkworm Bombyx mori, a model Lepidopteran insect. Proc Biol Sci. 2015;282:20150487. doi: 10.1098/rspb.2015.0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma S, et al. An integrated CRISPR Bombyx mori genome editing system with improved efficiency and expanded target sites. Insect Biochem Mol Biol. 2017;83:13–20. doi: 10.1016/j.ibmb.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 80.Mabashi-Asazuma H, Jarvis DL. CRISPR-Cas9 vectors for genome editing and host engineering in the baculovirus-insect cell system. Proc Natl Acad Sci USA. 2017;114:9068–9073. doi: 10.1073/pnas.1705836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, et al. Tissue-specific genome editing of laminA/C in the posterior silk glands of Bombyx mori. J Genet Genomics. 2017;44:451–459. doi: 10.1016/j.jgg.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 82.Garczynski SF, et al. CRISPR/Cas9 editing of the codling moth (Lepidoptera: Tortricidae) CpomOR1 gene affects egg production and viability. J Econ Entomol. 2017;110:1847–1855. doi: 10.1093/jee/tox166. [DOI] [PubMed] [Google Scholar]
- 83.Fujiwara H, Nishikawa H. Functional analysis of genes involved in color pattern formation in Lepidoptera. Curr Opin Insect Sci. 2016;17:16–23. doi: 10.1016/j.cois.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 84.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Nieuwerburgh F, et al. Illumina mate-paired DNA sequencing-library preparation using Cre-Lox recombination. Nucleic Acids Res. 2012;40:e24. doi: 10.1093/nar/gkr1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cong Q, Li W, Borek D, Otwinowski Z, Grishin NV. The bear giant-skipper genome suggests genetic adaptations to living inside yucca roots. Mol Genet Genomics. October 6, 2018 doi: 10.1007/s00438-018-1494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kelley DR, Schatz MC, Salzberg SL. Quake: Quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11:R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kajitani R, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 94.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barrett T, et al. BioProject and BioSample databases at NCBI: Facilitating capture and organization of metadata. Nucleic Acids Res. 2018;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benson DA, et al. GenBank. Nucleic Acids Res. 2018;46:D41–D47. doi: 10.1093/nar/gkx1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.