Significance
Human cytomegalovirus (HCMV) is a widespread pathogen that remains with an individual for life in a quiescent/latent state, posing little threat to an otherwise healthy person. However, when an individual’s immune system is severely compromised, HCMV can reactivate to its active/lytic state, resulting in viral spread and disease that is often fatal. The biological mechanisms underlying HCMV latency and reactivation remain poorly understood. Herein we show that the viral-encoded G protein-coupled receptor (GPCR) US28 aids in the establishment and the maintenance of viral latency. Furthermore, we find that US28 modulates host-cell proteins to suppress viral processes associated with active/lytic replication, thereby promoting latent infection. This work provides the mechanism by which HCMV modulates the host-cell environment to its advantage.
Keywords: cytomegalovirus, HCMV, latency, US28, herpesvirus
Abstract
Human cytomegalovirus (HCMV) is a ubiquitous pathogen that undergoes latency in cells of the hematopoietic compartment, although the mechanisms underlying establishment and maintenance of latency remain elusive. We previously reported that the HCMV-encoded G protein-coupled receptor (GPCR) homolog US28 is required for successful latent infection. We now show that US28 protein (pUS28) provided in trans complements the US28Δ lytic phenotype in myeloid cells, suggesting that sustained US28 expression is necessary for long-term latency. Furthermore, expression of pUS28 at the time of infection represses transcription from the major immediate early promoter (MIEP) within 24 h. However, this repression is only maintained in the presence of continual pUS28 expression provided in trans. Our data also reveal that pUS28-mediated signaling attenuates both expression and phosphorylation of cellular fos (c-fos), an AP-1 transcription factor subunit, to repress MIEP-driven transcription. AP-1 binds to the MIEP and promotes lytic replication, and in line with this we find that US28Δ infection results in an increase in AP-1 binding to the MIEP, compared with WT latent infection. Pharmacological inhibition of c-fos represses the MIEP during US28Δ infection to levels similar to those we observe during WT latent infection. Together, our data reveal that US28 is required for both establishment and long-term maintenance of HCMV latency, which is modulated, at least in part, by repressing functional AP-1 binding to the MIEP.
Human cytomegalovirus (HCMV), a β-herpesvirus, can infect a wide range of cell types, including undifferentiated myeloid cells, which represent an established, natural site of latent infection (1, 2). This latent infection underpins a lifelong, persistent infection with HCMV, present in ∼50 to 75% of the human population (3). Although healthy individuals infected with HCMV are predominantly asymptomatic, lytic reactivation from latency causes significant health complications and often mortality in immune-deficient patients (4, 5). The threat of HCMV reactivation in immunocompromised patients is most acute in transplant recipients (6, 7). While lytic HCMV infection is reduced and often controlled with antiviral therapies, these treatments target the lytic phase of infection, are problematically toxic, and are rendered less effective due to emerging drug-resistant strains (8, 9). Thus, defining novel means to prevent viral reactivation will therefore have significant clinical benefits.
Latent infection is defined as the maintenance of viral genomes in the absence of infectious virion production, coupled with the ability to reactivate lytic replication to produce infectious virus, given the proper extracellular and/or environmental cues (10). As CMVs are highly host-specific, HCMV latency studies are largely restricted to models utilizing human cells, including ex vivo primary hematopoietic cells, such as monocytes (11–13) and CD34+ hematopoietic progenitor cells (HPCs) (14–16), as well as in vitro model systems (17–23), such as Kasumi-3 cells (22) and embryonic stem cells (23). Recent advancements in humanized mouse model systems have begun to allow for in vivo HCMV studies (24–28). Defining the HCMV latent transcriptome represents a key area of continued research, and at present the exact roles of many latency-associated genes still remain unclear. However, genes such as UL135, UL138, US28, LAcmvIL-10 (latency-associated HCMV IL-10 homolog), latency unique natural antigen (LUNA), and UL144 play a role in maintaining HCMV latency and subsequent reactivation (29). HCMV also manipulates cellular factors to maintain latent infection, including cellular microRNAs (30), cell-surface protein expression (31–34), and the cellular secretome (35). Chromatinization of the major immediate early promoter (MIEP) during latency contributes to securing extended viral transcriptional silencing and establishment of latency (36), yet how this occurs is still under investigation.
The HCMV US28 gene is transcribed during both lytic and latent infection, during which the protein aids in securing latency in undifferentiated myeloid cells (37, 38). The US28-encoded protein (pUS28) is one of four G protein-coupled receptors (GPCRs) encoded by the HCMV genome (39) and is a potent signaling protein in the context of lytic infection (40). pUS28 is incorporated into the mature viral particle (37) and transcripts are expressed during both experimental (11, 15, 37, 38, 41–44) and natural latency (42, 45, 46). Although it is clear that pUS28 is necessary for HCMV latency (37, 38), whether this protein is required for the establishment and/or the maintenance of latency remains unknown. Additionally, transient expression of pUS28 in THP-1 cells, which support HCMV latency, represses the MIEP in reporter assays, a phenotype that is mediated by pUS28 signaling (38). However, the exact mechanism by which US28-directed MIEP repression is achieved in the context of viral latency remains elusive. In this current study, we report that pUS28 functions to both establish and maintain viral latency. Additionally, our data reveal that pUS28 attenuates cellular fos (c-fos), a component of the AP-1 transcription factor complex that binds to the MIEP during lytic replication. Our data reveal that pUS28-mediated attenuation of c-fos activation results in a decrease in AP-1 binding to the MIEP. Together, our findings reveal a mechanism by which pUS28 coopts the host cell to silence MIEP-driven transcription and secure viral latency.
Results
Exogenous pUS28 Expression Rescues the Lytic Growth Phenotype in US28Δ-Infected Myeloid Progenitor Cells.
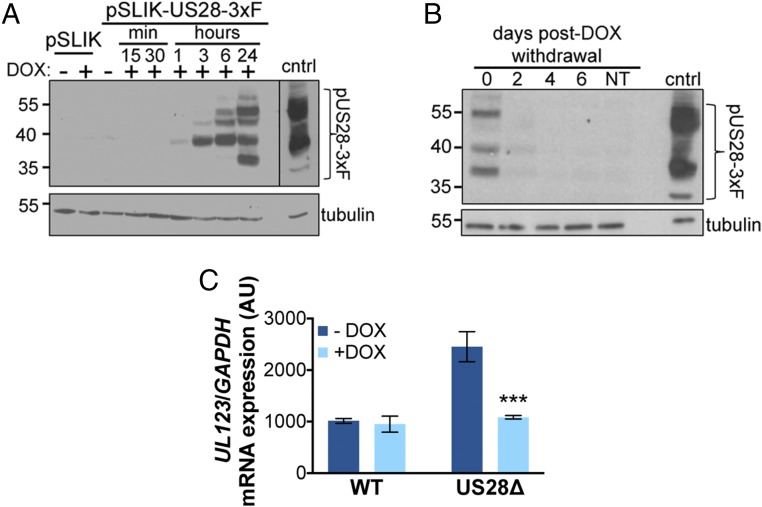
To begin to understand the role of US28 during establishment and/or maintenance of latency, we transduced THP-1 cells with a lentiviral construct (pSLIK-US28-3xF) that allows for inducible expression of pUS28 fused to a C-terminal triple-FLAG epitope tag. We first assessed the efficiency of pUS28 expression in this system by treating THP-1-pSLIK-US28-3xF or THP-1-pSLIK-hygro cells with or without doxycycline (DOX) and then harvested the cells over 24 h. We detected pUS28 expression within 1 h of induction, with the most robust expression observed 24 h posttreatment. Importantly, the expression of pUS28 is tightly regulated in this inducible system, as we did not observe pUS28 in the absence of DOX (Fig. 1A). Additionally, to understand pUS28 degradation kinetics in this system, we treated THP-1-pSLIK-US28-3xF cells with DOX for 24 h (day 0) then washed the cells to remove the DOX and harvested these cells every 2 d for 6 d to monitor pUS28 levels. Our findings indicate that pUS28 is significantly degraded 2 d after the removal of DOX and is undetectable by d 4 (Fig. 1B).
Fig. 1.
Continual pUS28 expression complements the lytic-like phenotype following US28Δ infection. (A) THP-1 cells were transduced with THP-1-pSLIK-hygro (pSLIK) or THP-1-pSLIK-US28-3xF (pSLIK-US28-3xF) and treated with (+) or without (−) DOX (1 μg/mL) to induce pUS28 expression. Cells were harvested at the indicated time points posttreatment. pUS28 expression was detected by immunoblot using the FLAG epitope tag. (B) THP-1-pSLIK-US28-3xF cells were treated with DOX for 24 h and harvested to confirm pUS28 expression (0 d). Remaining cells were washed in PBS and cultured in the absence of DOX for the duration of the experiment. Cell samples were taken at the indicated days posttreatment and all samples were then immunoblotted for pUS28 using the epitope tag. (A and B) As a control (cntrl), NuFF-1 cells were infected with US28-3xF (moi = 0.5) and cell lysates were harvested at 96 h postinfection. Tubulin is shown as a loading control. Note that due to the intensity of this control lysate in A, a shorter exposure is shown, as denoted by the black line. (C) THP-1-pSLIK-US28-3xF cells were treated without (−; dark blue) or with (+; light blue) DOX for 24 h and then infected with WT or US28Δ (moi = 1.0). DOX was replenished every 48 h and the cells were harvested at 6 dpi for UL123 expression by RT-qPCR. Samples were normalized to GAPDH, and each sample was analyzed in triplicate. Errors bars indicate SD. Statistical significance was calculated using Welch’s t test; ***P < 0.001.
We have previously shown that the deletion of the US28 ORF results in a lytic rather than latent infection of both Kasumi-3 and CD34+ HPCs (37). Thus, to determine if pUS28 provided in trans could complement this phenotype, we infected THP-1-pSLIK-US28-3xF cells in the presence or absence of DOX with either TB40/EmCherry (WT) or TB40/EmCherry-US28Δ (US28Δ). We collected the cells 7 d postinfection (dpi) and assessed viral lytic gene expression (UL123), as this serves as an indicator of MIEP activity level, and thus is a proxy for distinguishing a latent versus a lytic phenotype. Indeed, we found that DOX-induced THP-1-pSLIK-US28-3xF cells infected with US28Δ resulted in similar levels of UL123 transcript levels compared with WT infections (Fig. 1C). This finding suggests that pUS28 expression helps to maintain the suppression of viral lytic gene expression.
pUS28 Expression at the Time of Infection Fails to Maintain MIEP Repression.
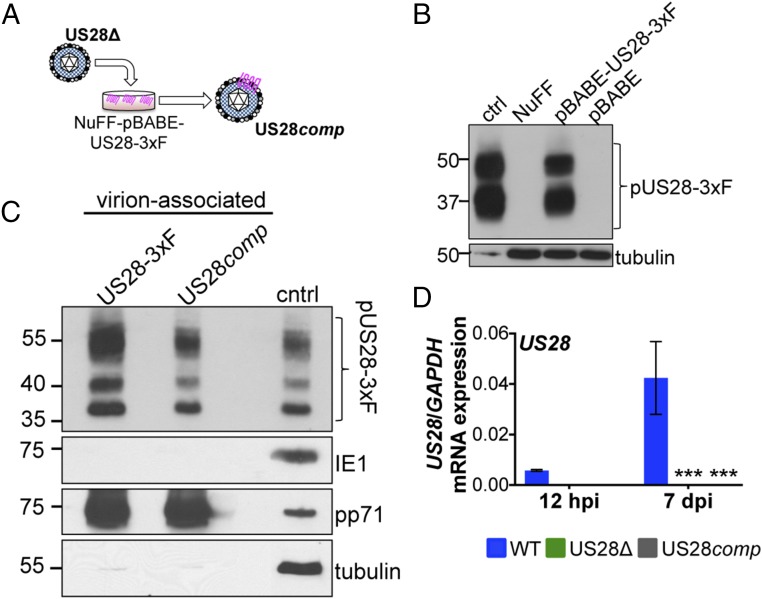
Our previous findings revealed that pUS28 is incorporated into the mature viral particle and is delivered to Kasumi-3 cells upon infection (37). Thus, we hypothesized that virion-delivered pUS28 may function early postinfection to aid in establishing a latent infection in myeloid cells. To test whether virion-delivered pUS28 could establish and/or maintain latent infections, we generated a pUS28-complemented recombinant virus, which incorporates pUS28 into the virion but lacks the US28 ORF (TB40/EmCherry-US28comp; US28comp) by growing US28Δ on a pUS28-expressing fibroblast cell line (Fig. 2 A and B). We confirmed that US28comp virus incorporates pUS28 into the virion (Fig. 2C) and that it does not express de novo US28 mRNA after infection of NuFF-1 cells (Fig. 2D). We noticed that pUS28 expression is marginally lower in US28comp virus in comparison with US28-3xF virion incorporated pUS28 (Fig. 2C), which could be due to pUS28 expression levels in the complementing cell line. Nonetheless, these data show that US28comp incorporates pUS28 into the mature viral particle but is unable to produce de novo pUS28 upon subsequent infection.
Fig. 2.
US28comp incorporates pUS28 into its virion but fails to express US28 after infection. (A) US28Δ was grown on NuFF-pBABE-US28-3xF cells. Cell-free virus was isolated and termed US28comp. (B) Lysates from NuFF-1 cells infected with TB40/E-mCherry-US28-3xF (US28-3xF; moi = 0.5, 96 h postinfection, 15 μg; ctrl), mock-infected NuFF-1 cells (30 μg; NuFF), or NuFF-1 cells stably transduced with either pBABE-US28-3xF (30 μg; NuFF-pBABE-US28-3xF) or pBABE (30 μg; NuFF-pBABE) were assessed for pUS28 expression by immunoblot using an antibody directed at the FLAG epitope tag. Tubulin is shown as a loading control. (C) US28comp virus was generated by infecting NuFF-pBABE-US28-3xF with US28Δ. As a control for virion-associated pUS28, NuFF-1 cells were infected with US28-3xF. Cell-free US28comp or US28-3xF virus was purified through a sorbitol cushion and pUS28 was detected by immunoblot for the FLAG epitope tag. Cell lysates (15 μg) from NuFF-1 cells infected with US28-3xF (moi = 0.5, 96 hpi) are shown as a control (cntrl). Samples were also probed with antibodies directed at the viral proteins IE1 and pp71, as well as cellular tubulin. (D) Kasumi-3 cells were infected with WT, US28Δ, or US28comp at a multiplicity of 1.0 TCID50 per cell and harvested 7 dpi. US28 gene expression was quantified by RT-qPCR using primers that amplify US28 (SI Appendix, Table S1). Samples were normalized to GAPDH, and each sample was analyzed in triplicate. Errors bars indicate SD. Statistical significance was calculated using two-way ANOVA and Dunnett’s post hoc analysis relative to WT at each time point; ***P < 0.001.
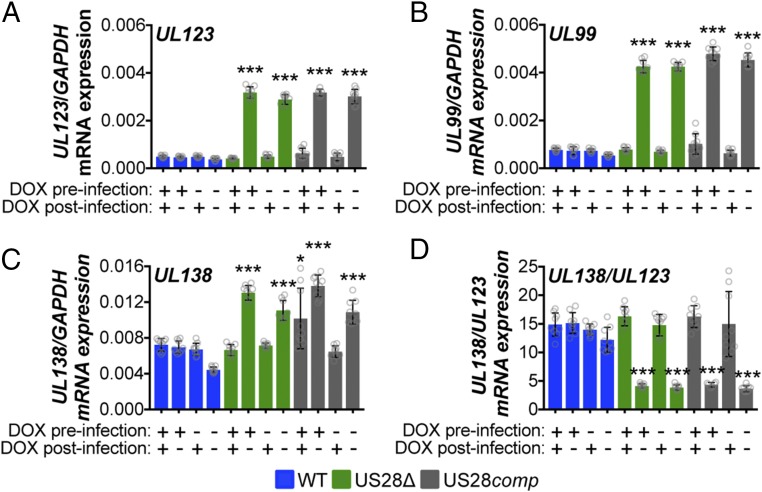
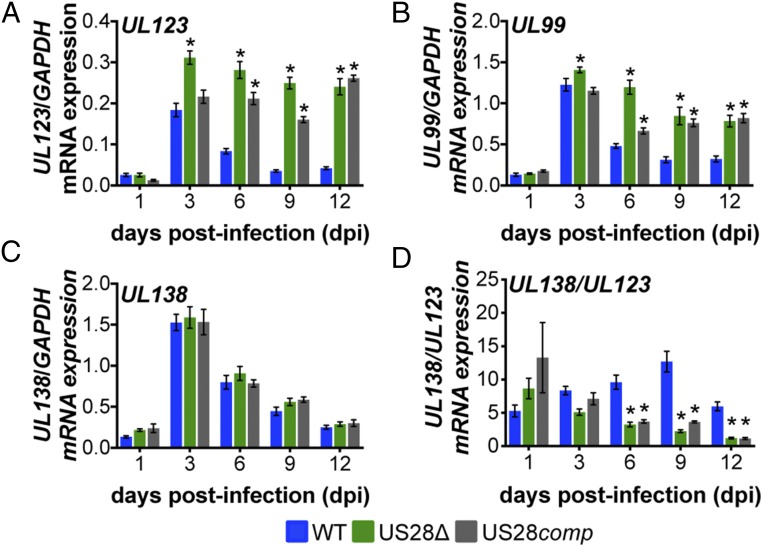
Using this complemented virus, we asked if virion-delivered pUS28 was capable of suppressing lytic gene expression in conjunction with our THP-1 cells overexpressing US28. To this end, we treated THP-pSLIK-US28-3xF with or without DOX and then infected them with WT, US28Δ, or US28comp and either maintained DOX treatment for 12 d or removed DOX treatment after infection. We found that low UL123 expression correlated with the maintenance of pUS28 expression, either by DOX treatment or by infection with WT virus. However, in cultures where pUS28 was not continually expressed for the duration of the experiment, UL123 expression was significantly higher (Fig. 3A). Using UL123 as a proxy for MIEP activity, this demonstrates that pUS28 expression must be sustained to maintain repression of MIEP-driven transcription during HCMV infection of monocytic cells. Correlating with these findings, UL99 expression, an HCMV gene associated with late lytic infection, followed a pattern similar to that of UL123 (Fig. 3B). We also assessed UL138 (Fig. 3C) and, more importantly, the ratio of UL138/UL123 mRNA expression (22, 37) to discern those cultures that favor a latent transcriptional profile (high UL138/UL123 ratio) from those that favor a lytic transcriptional profile (low UL138/UL123 ratio). The UL138/UL123 ratios for the US28Δ- and US28comp-infected cells that were cultured in the absence of exogenous pUS28 expression were significantly lower than the same cultures that received DOX after infection, as well as the WT-infected cells, regardless of treatment (Fig. 3D). Importantly, DOX treatment does not alter viral gene expression (SI Appendix, Fig. S1). Together, these findings argue that virion-delivered pUS28, or pUS28 expressed only at the time of infection, fails to maintain the suppression of lytic gene transcription.
Fig. 3.
Virion-delivered pUS28 fails to maintain suppression of lytic gene transcription. THP-1-pSLIK-US28-3xF were treated with (+) or without (−) DOX (1 μg/mL) to induce pUS28 expression (preinfection) and then infected with WT (blue), US28Δ (green), or US28comp (gray) (moi = 1.0). Infected cells were washed 2 h postinfection and cultured in the presence (+) or absence (−) of DOX, as indicated (postinfection), where cultures receiving postinfection DOX were replenished every 48 h. Cells were harvested 12 dpi and (A) UL123, (B) UL99, (C) UL138, and (D) the ratio of UL138/UL123 expression were measured by RT-qPCR. Samples were normalized to GAPDH and analyzed in triplicate. Errors bars indicate SD and statistical significance was calculated using one-way ANOVA and Tukey post hoc analysis; *P < 0.05, ***P < 0.001.
pUS28 Expression Results in Repression of MIEP-Driven Transcription.
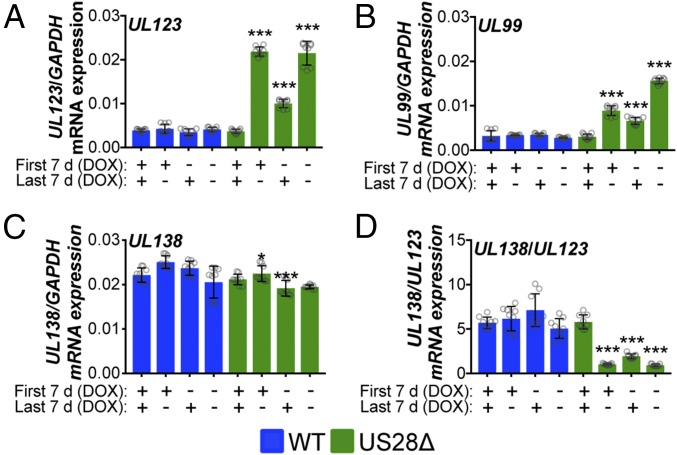
Based on our above findings, we hypothesized that pUS28 functions to repress the MIEP, both at early time points after infection to establish latency as well as throughout infection to maintain latency. To test this, we pretreated THP-1-pSLIK-US28-3xF cells with or without DOX for 48 h to induce pUS28 expression. We then infected these cells with either WT or US28Δ [multiplicity of infection (moi) = 1.0] and cultured the cells for 7 d, with or without continued DOX treatment. Cells were then washed and DOX treatment was either maintained or reversed for each culture, as indicated. We observed that continual pUS28 expression throughout infection maintained repression of the MIEP, as measured by UL123 for WT- versus US28Δ-infected cells cultured in the presence of DOX (Fig. 4A). This finding is consistent with a previous report that showed transient pUS28 expression repressed a MIEP reporter construct (38). Second, removal of DOX-induced pUS28 expression after 7 d of treatment led to a derepression of UL123 transcription by the end of the assay at day 14 (Fig. 4A), thus confirming the requirement for pUS28 in maintaining repression of MIEP-driven transcription. Finally, we observed that introduction of DOX-induced pUS28 expression for the last 7 d in US28Δ-infected cells, initially cultured in the absence of DOX, leads to an intermediate phenotype (Fig. 4A), suggesting that introduction of DOX-induced pUS28 expression later in infection can repress MIEP-driven transcription.
Fig. 4.
Sustained pUS28 expression suppresses lytic gene expression. THP-1-pSLIK-US28-3xF cells were treated with (+) or without (−) DOX (1 μg/mL) for 48 h to induce pUS28 expression and then infected with WT (blue) or US28Δ (green) (moi = 1.0). Cells were cultured for 7 d, during which cells were maintained under their original treatment conditions (First 7 d). At 7 dpi, cells were washed and treated with (+) or without (−) DOX (Last 7 d). Total RNA was harvested and (A) UL123, (B) UL99, (C) UL138, and (D) the ratio of UL138/UL123 expression were measured by RT-qPCR. Samples were normalized to GAPDH and assessed in triplicate. Errors bars indicate SD and significance calculated using one-way ANOVA and Dunnett’s post hoc analysis; *P < 0.05, ***P < 0.001.
We also assessed the expression of UL99 and found this gene is also repressed in response to pUS28 expression (Fig. 4B). Interestingly, when pUS28 expression is induced by DOX treatment for the first 7 d of infection followed by DOX withdrawal, UL99 mRNA expression increases, although it does not reach the levels we observed in the absence of DOX-induced pUS28 expression. In agreement with UL123, introduction of DOX-induced pUS28 expression only for the last 7 d of infection leads to a reduction in UL99 mRNA expression (Fig. 4B). Taken together, these data suggest that the loss of pUS28 expression results in the derepression of MIEP-driven UL123 expression, leading to induction of the lytic lifecycle, including expression of the late gene products. Furthermore, assessing UL138 (Fig. 4C) and, more importantly, the ratio of UL138/UL123 mRNA expression (22, 37) confirmed the correlation between those cultures that favor a latent transcriptional profile (high UL138/UL123 ratio) versus those that favor a lytic transcriptional profile (low UL138/UL123 ratio) (Fig. 4D). Additionally, pUS28 fails to repress lytic gene transcription in differentiated THP-1 cells in which the virus lytically replicates (47) (SI Appendix, Figs. S2 and S3), suggesting that this effect is specific to undifferentiated cells. Together, these data suggest that pUS28 suppresses lytic transcription in cells that support latency.
pUS28 Represses the MIEP Transcription at Times Consistent with the Establishment of Latent Infection in Kasumi-3 Cells.
Having observed that ectopic expression of pUS28 represses UL123 and UL99 expression in THP-1 cells, we asked whether pUS28 plays a role in suppressing lytic gene transcription during latent infection of Kasumi-3 cells, a culture system that affords us the ability to assess both functional latency and reactivation (22). We asked if virion-delivered pUS28 was capable of suppressing lytic gene expression in the context of infection and whether pUS28 contributes to the establishment of latency. We infected Kasumi-3 cells with WT, US28Δ, or US28comp (moi = 1.0) and measured expression of UL123, UL99, UL138, and UL138/UL123 over 12 d. First, we observed a burst in UL123 and UL99 mRNA expression following WT infection, which peaked 3 dpi (Fig. 5 A and B), as previously observed by others (11, 15, 48). UL123 and UL99 gene expression was lower in Kasumi-3 cells infected with WT or US28comp viruses at this time point (Fig. 5 A and B), suggesting that virion-delivered pUS28 has a repressive effect on lytic transcripts at early time points postinfection.
Fig. 5.
pUS28 represses UL123 and UL99 expression in infected Kasumi-3 cells at early times of latent infection. Kasumi-3 cells were infected with WT (blue), US28Δ (green), or US28comp (gray) (moi = 1.0) and sorted for mCherry-positive cells at 1 dpi. Cells were then harvested at the indicated dpi. (A) UL123, (B) UL99, (C) UL138, and (D) the ratio of UL138/UL123 expression were measured by RT-qPCR. Samples were normalized to GAPDH and analyzed in triplicate. Errors bars indicate SD, and statistical significance was calculated using two-way ANOVA and Dunnett’s post hoc analysis relative to WT virus at each time point; *P < 0.05.
Subsequent to this initial burst of mRNA expression, transcription of these genes decreased in the WT-infected cells compared with US28Δ- or US28comp-infected Kasumi-3 cells. Additionally, UL123 expression stabilized in US28Δ-infected cells, at an expression level that is higher than that of cells infected with WT (Fig. 5A), suggesting that pUS28 represses lytic gene transcription at early times postinfection of Kasumi-3 cells, at times consistent with the establishment of latency. Additionally, these data suggest that continued pUS28 expression represses lytic gene transcription to contribute to maintaining latency. Confirming this hypothesis, pUS28 that is delivered by US28comp virus is capable of repressing UL123 and UL99 expression at early times postinfection (Fig. 5A; WT versus US28comp at 3 dpi), but failed to repress these lytic gene products from 6 dpi onward (Fig. 5 A and B), suggesting that repression is diminished after virion-delivered pUS28 is degraded. Finally, UL138 expression (Fig. 5C), and more importantly the ratio of UL138/UL123, was significantly lower in US28Δ- or US28comp-infected cells compared with WT-infected Kasumi-3 cells from 6 dpi through the duration of the experiment, supporting the findings that these cultures display a more lytic phenotype (Fig. 5D). Overall, these data suggest that pUS28 represses MIEP-driven transcription, which helps to both establish and maintain latency.
pUS28 Repression of the MIEP Reduces Virus Production.
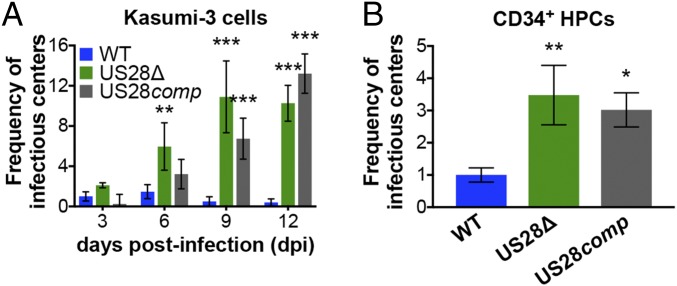
Our data so far reveal that pUS28 suppresses lytic gene transcription in cells that support viral latency. To determine if pUS28 aids in establishing and maintaining latent infection, we took advantage of the in vitro Kasumi-3 cell and ex vivo primary cord blood-derived CD34+ HPC latency models. First, we infected Kasumi-3 cells with WT, US28Δ, or US28comp (moi = 1.0) and measured the production of infectious particles by extreme limiting dilution assay (ELDA) by coculture with fibroblasts, sampling every 3 dpi for 12 d. Our results indicate that US28comp-infected Kasumi-3 cells display a phenotype similar to that of WT-infected Kasumi-3 cells until 6 dpi, after which the US28comp infections reveal an intermediate phenotype, releasing significantly more virus than WT infections, indicating a failure to maintain latency (Fig. 6A). To confirm these findings in a cell type that represents a natural site of HCMV latency, we infected cord blood-derived CD34+ HPCs with WT, US28Δ, or US28comp (moi = 2.0) and cultured the cells for 12 d under conditions that promote latency. We then harvested the cells for ELDA analysis on naïve fibroblasts, as above. We found that US28comp-infected CD34+ HPCs produce infectious virions to levels similar to those we observe in the US28Δ-infected CD34+ HPCs (Fig. 6B). Consistent with these data, viral lytic gene expression in Kasumi-3 cells and CD34+ HPCs at 12 dpi revealed increased UL123 and UL99 in US28Δ- or US28comp-infected cultures, compared with cells infected with WT (SI Appendix, Fig. S4). These data suggest that virion-delivered pUS28 contributes to establishing HCMV latency; however, it is not sufficient to maintain long-term latent infections, and therefore sustained pUS28 expression is required to suppress viral replication.
Fig. 6.
Sustained pUS28 expression is required to maintain viral latency. (A) Kasumi-3 cells (moi = 1.0) or (B) CD34+ HPCs (moi = 2.0) were infected with WT (blue), US28Δ (green), or US28comp (gray). (A and B) Cells were collected at the indicated time points and reactivation events were determined by ELDA. Data in A are presented as fold change in virus release relative to WT virus on day 3. Data in B are presented as fold change in virus release relative to WT virus. In all cases, error bars represent SDs of three biological replicates. Statistical significance was calculated relative to WT at the same time point using (A) two-way ANOVA analysis or (B) one-way ANOVA and Tukey post hoc analysis; *P < 0.05, **P < 0.01, ***P < 0.001.
pUS28 Expression Is Sufficient to Attenuate c-fos Expression and Signaling.
Our previous findings revealed that pUS28 is a potent signaling molecule during the context of infection (40). Thus, we hypothesized that pUS28’s signaling capabilities contribute to its function during latency. However, our previous work also noted that pUS28-mediated signaling is cell-type-specific (40), making it difficult to extrapolate such findings to hematopoietic cells. To begin to understand the pUS28-modulated pathways and the mechanism underlying pUS28’s contribution to establishing and maintaining latency, we performed a PCR array analysis on THP-pSLIK-US28-3xF, compared with control cells, treated with DOX for 24 h to induce pUS28 expression. This analysis revealed a subset of statistically significant up-regulated and down-regulated cellular genes (SI Appendix, Fig. S5). The most significantly up-regulated genes in the pathway analysis were CHUK (conserved helix–loop–helix ubiquitous kinase; IKKα), HRAS, JAK2 (Janus kinase 2), PIK3CB (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta), and STAT1 (signal transducer and activator of transcription 1), while c-fos, PIK3CA (PIK3C alpha), src, and STAT5A were the most significant down-regulated genes in response to pUS28 expression (SI Appendix, Fig. S5).
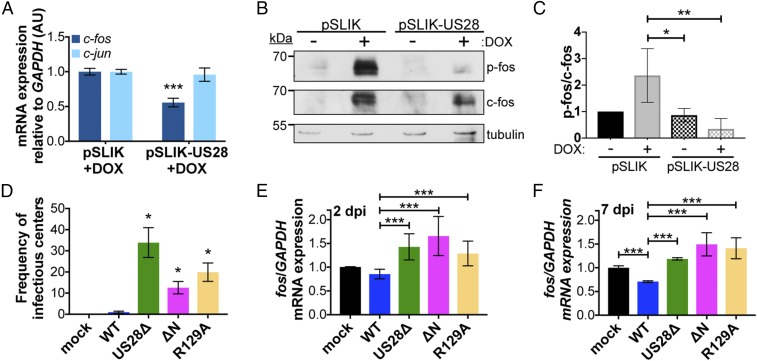
We focused on c-fos since activation of c-fos results in the heterodimerization of c-fos and c-jun to form the activator protein 1 (AP-1) complex, which binds to the MIEP, thereby aiding in its transcriptional activation (49). AP-1 complex formation is regulated by N-terminal phosphorylation of c-fos and c-jun, which results in their heterodimerization. Following complex formation, AP-1 binds to the DNA consensus sequence TGA(G/C)TCA to promote gene transcription (50, 51). Thus, we hypothesized that pUS28-mediated attenuation of c-fos transcription reduces AP-1 complex formation, thus leading to the repression of lytic transcription. We confirmed that c-fos expression is reduced in transduced THP-1 cells upon induction of pUS28 by RT-qPCR (Fig. 7A). We also assessed c-jun expression, which is significantly down-regulated during latent infection of CD34+ HPCs and Kasumi-3 cells during HCMV infection (52). Interestingly, we found pUS28 expression did not significantly impact c-jun (Fig. 7A), suggesting that pUS28 specifically targets c-fos and not both AP-1 components, and that another as-yet-unknown viral factor likely targets c-jun. We found that pUS28 also reduces c-fos protein expression and significantly decreases c-fos phosphorylation (Fig. 7 B and C). Consistent with previous findings, DOX treatment does induce both total and active forms of c-fos (53, 54), both of which are subsequently attenuated following pUS28 expression. Interestingly, pUS28 expression does not affect c-fos or c-jun expression in differentiated THP-1 cells (SI Appendix, Fig. S6), suggesting that pUS28-mediated regulation of c-fos is specific to cells that support HCMV latency.
Fig. 7.
US28 signaling attenuates both c-fos expression and activation. (A) THP-1-pSLIK-hygro (pSLIK) and THP-1-pSLIK-US28-3xF (pSLIK-US28) were treated with DOX and cells were harvested 24 h posttreatment. c-fos (dark blue) and c-jun (light blue) expression were measured relative to GAPDH. (B) Cells from A were treated with (+) or without (−) DOX and cells were harvested 24 h posttreatment. Phosphorylated fos (p-fos), total fos (c-fos), and tubulin were detected by immunoblot. Representative immunoblot shown (n = 3). (C) Quantification of the results shown in B using densitometry. Levels of p-fos were quantified relative to c-fos, shown relative to tubulin. Data were quantified from three biological replicates. Error bars indicate SD. Statistical significance was calculated using one-way ANOVA and Tukey post hoc analysis. (D–F) Kasumi-3 cells were infected with the indicated viruses (moi = 1.0) and harvested at (E) 2 or (D and F) 7 dpi. (D) The frequency of infectious virus from each latently infected culture was determined by ELDA. Data are presented as fold change in virus release relative to WT. (E and F) c-fos expression was measured relative to GAPDH. (A and D–F) Each sample was analyzed in triplicate. Error bars indicate SD. Statistical significance was calculated using Welch’s t test; *P < 0.05, **P < 0.01, ***P < 0.001.
Latent Expression of pUS28 Signals to Attenuate c-fos Expression.
To determine if pUS28-mediated signaling attenuates c-fos in the context of infection, we took advantage of two additional US28 viral recombinants that alter the ability of US28 to signal. US28-R129A has a single point mutation at amino acid position 129 in the canonical “DRY” motif (37). Mutation of this motif from DRY to DAY prevents G proteins from coupling to this domain, rendering this mutant “G protein signaling-dead” (55). The US28ΔN recombinant lacks N-terminal amino acids 2–16 (37) and is unable to interact with US28 ligands, although this mutant can still signal constitutively (55). We generated each of these signaling mutants in the TB40/EmCherry-US28-3xF background and confirmed pUS28 expression during fibroblast infection with each recombinant by immunoblot (SI Appendix, Fig. S7A). While there was a slight growth advantage with the US28-R129A mutant at 8 dpi, there was no significant difference between mutant and WT by 15 dpi (SI Appendix, Fig. S7B). Next, we infected Kasumi-3 cells under conditions that promote latency with WT, US28Δ, US28ΔN, or US28-R129A and measured the release of infectious virus by ELDA. WT-infected cells established and maintained a latent infection, while US28Δ-, US28-R129A-, and US28ΔN-infected cells all resulted in significant lytic replication (Fig. 7D), suggesting that chemokine binding and agonist-dependent signaling is required. To determine if pUS28-mediated signaling impacts c-fos expression, we latently infected Kasumi-3 cells with WT, US28Δ, US28ΔN, or US28-R129A and measured c-fos expression 2 and 7 dpi. WT virus attenuated c-fos expression as early as 2 dpi, whereas c-fos expression increased in US28Δ-, US28-R129A-, and US28ΔN-infected cells (Fig. 7E). These findings were amplified by 7 dpi, confirming that continued expression of pUS28 attenuates c-fos and pUS28-mediated signaling is important for c-fos regulation (Fig. 7F).
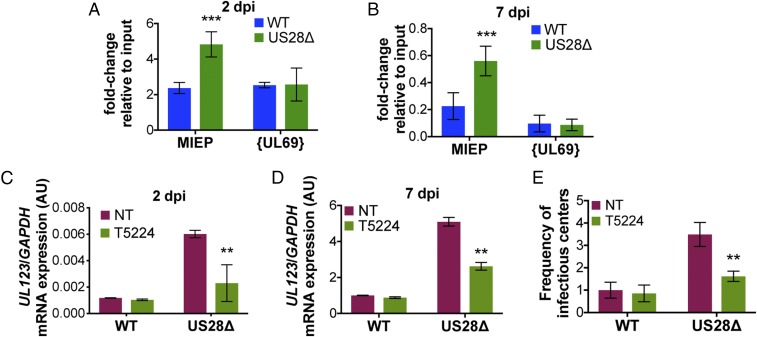
Given our observations that (i) pUS28 attenuates UL123 expression, (ii) pUS28 attenuates c-fos expression and activation, and (iii) there are AP-1 binding sites in the MIEP (56), we hypothesized that pUS28 expression in cells that support latency results in a reduction of AP-1 binding to the MIEP. To this end, we infected Kasumi-3 cells with WT or US28Δ and performed ChIP for AP-1 at the MIEP at 2 and 7 dpi using an antibody directed at c-fos. Our data reveal an increase in AP-1 bound to the MIEP in US28Δ-infected cells as early as 2 dpi (Fig. 8A), which is maintained through 7 dpi (Fig. 8B). Finally, we reasoned that if pUS28-mediated attenuation of c-fos signaling suppresses MIEP activation, then treatment of US28Δ-infected cells with a c-fos inhibitor should reduce MIEP activity. Infection of Kasumi-3 cells with either WT or US28Δ in the presence of a c-fos inhibitor (T-5224) for 2 or 7 d showed reduced UL123 expression in US28Δ-infected cultures (Fig. 8 C and D, respectively), similar to levels we observed in the presence of pUS28 exogenous expression (Fig. 1C). Additionally, this reduction in UL123 expression in the US28Δ-infected Kasumi-3 cells (Fig. 8D) results in a decrease in the production of infectious virus compared with its untreated counterpart (Fig. 8E). Together, these data show that WT virus significantly reduces AP-1–mediated activation of the MIEP by pUS28-directed attenuation of c-fos, thereby contributing to the establishment and maintenance of latency in cells that support HCMV latent infection.
Fig. 8.
pUS28 decreases c-fos binding at the MIEP, leading to transcriptional repression of IE transcripts. (A and B) Kasumi-3 cells were infected (moi = 1.0) with WT (blue) or US28Δ (green). Cells were collected (A) 2 or (B) 7 dpi and the AP-1 complex was immunoprecipitated using an anti–c-fos antibody. Coprecipitated MIEP was quantified by qPCR, and data are shown as fold change relative to input. The UL69 nonpromoter region is shown as a control. (C–E) Kasumi-3 cells were infected as in A and B in the absence (red; NT) or presence (green) of the fos inhibitor, T5224 (10 nM), and cells were harvested (C) 2 or (D and E) 7 dpi. (C and D) UL123 expression was measured and normalized to GAPDH. (E) The frequency of infectious virus from each latently infected culture was determined by ELDA. Data are presented as fold change in virus release relative to vehicle-treated WT. Each sample was analyzed in triplicate. Errors bars indicate SD and statistical significance was measured using Welch’s t test; **P < 0.01, ***P < 0.001.
Discussion
Deciphering the molecular underpinnings of the establishment and maintenance of HCMV latency is an ongoing area of intense research. Viral infection of hematopoietic cells results in what is likely a highly coordinated hijacking of the host-cell environment, whereby multifaceted processes dictate the infection outcome. The primary finding in this study is a requirement for pUS28 during both the establishment and maintenance phases of HCMV latency, dictated, at least in part, by its manipulation of host encoded c-fos. pUS28 also influences AP-1 binding to the MIEP, a cellular transcription factor that is composed of c-fos and c-jun subunits. Finally, pUS28-mediated attenuation of c-fos directly impacts MIEP-driven transcription. Our results reveal a mechanism by which pUS28 functions to suppress MIEP-driven transcription to aid in the establishment and maintenance of HCMV latency.
Viral establishment and maintenance of latency likely requires a multitude of factors, and pUS28 is a key piece of the biological puzzle that balances latency and reactivation. Suppression of the very strong lytic promoter, the MIEP, is a major determinant for a successful latent infection, and pUS28 is certainly not the only viral factor involved in this mechanism. Epigenetic silencing of the MIEP is well-established and additional viral factors, including noncoding RNAs and proteins, clearly influence cellular factors that regulate this viral promoter during infection of cells that support latency (57). While host-cell proteins like Ying Yang-1 (YY-1) and TRIM28/Kap-1 have been shown to impact MIEP transcription and viral latency, the upstream factors that lead to their regulation remain elusive, although it is attractive to speculate that viral-induced signaling, by way of pUS28, may be involved.
pUS28-mediated attenuation of the AP-1 transcription factor complex is specific to c-fos, as c-jun was unaffected. In a recent collaboration, we found that c-jun expression is significantly down-regulated during HCMV latent infection of both Kasumi-3 cells and primary CD34+ HPCs (52), although we show herein that pUS28 expression did not significantly alter c-jun expression. Together, these data suggest another viral factor regulates c-jun transcript levels during latency. HCMV has therefore devised multiple mechanisms to regulate the AP-1 complex, suggesting this transcription factor is important to regulating latency. In addition to preventing AP-1 binding at the MIEP, it will be interesting to further dissect the biological consequences of pUS28-mediated c-fos regulation. More global, unbiased approaches, such as RNA sequencing and whole-cell proteomics may reveal additional host-cell factors that are dysregulated via the pUS28:c-fos signaling axis. Indeed, these downstream events may provide an additional mechanism by which HCMV regulates the host cell during latency.
Our data also reveal that pUS28-mediated signaling is important for maintaining latency, through the regulation of c-fos in both a G protein-coupling– and ligand binding-dependent fashion. The data we present herein agree with previous findings, which showed that US28-R129A (G protein-coupling domain point mutant) expression in THP-1 cells was unable to restore lytic gene suppression after infection with HCMV Titan-US28Δ virus (38). However, the role of ligand-mediated pUS28 signaling still remains unclear. Overexpression of the US28-Y16F ligand binding mutant in THP-1 cells suppresses lytic gene expression after infection with HCMV Titan-US28Δ virus (38). It is plausible that we have yet to identify a complete list of US28 ligands, and thus perhaps the US28-Y16F mutant retains some ligand responsiveness while the US28Δ mutant is completely devoid of such interactions.
Although US28 is linked to a variety of signaling pathways during lytic infection of different cell types (55), this report directly links pUS28 to c-fos signaling. Consistent with our findings, pUS28 expression in undifferentiated THP-1 cells represses the MIEP in reporter assays and attenuates MAP kinase signaling (38), a pathway that activates c-fos (58). Conversely, pUS28 activates this same pathway in differentiated THP-1 cells (38, 59, 60), which support lytic rather than latent infection. In line with this observation, we confirmed that pUS28’s repressive effect on the MIEP, via c-fos attenuation, is specific to undifferentiated myeloid cells that support latency and does not occur in differentiated myeloid cells that support lytic infection. Whether MAP kinase signaling via pUS28 contributes to c-fos attenuation during the context of latent infection remains unknown, although it is attractive to hypothesize that pUS28’s impact on MAP kinase results in the downstream effects on c-fos expression we have observed. Furthermore, while it is possible that the alterations in the cellular environments between these different differentiation states of myeloid cells might explain the variations in pUS28 signaling activity, such conclusions remain tenuous.
How else might US28 repress the MIEP in myeloid progenitor cells? Given that pUS28 modulates a variety of signaling pathways during lytic infection (55), it is likely that future investigations will reveal additional pUS28-mediated cellular signaling pathways, some of which may aid in suppressing robust lytic gene transcription. In addition to MAP kinase signaling in THP-1s (38), US28 also activates signal transducer and activator of transcription 3-inducible nitric oxide synthase (STAT3-iNOS) in CD34+ cells (61) and PLC-β in THP-1 cells (62). How these pathways, along with others, might affect transcriptional suppression of the virus during latency in undifferentiated myeloid cells remains elusive.
Our findings described herein detail one specific mechanism by which pUS28 modulates the host-cell environment to influence HCMV latency. Further work focused on understanding additional pUS28 activities during latent infection is essential to developing therapeutic targets aimed at preventing viral reactivation.
Methods
Cells and Viruses.
Details on cells and their culture conditions, as well as details on the propagation of the TB40/E BAC-derived viruses, are included in SI Appendix, SI Materials and Methods.
Generation of Stable NuFF-1 and THP-1 Cell Lines.
For details on the generation of the pBABE-US28-3xF and pSLIK-US28-3xF constructs, as well as the US28-3xF expressing NuFF-1 cells, see SI Appendix, SI Materials and Methods.
To generate the stable THP-1-pSLIK-hygro and THP-1-pSLIK-US28-3xF lines, pSLIK-hygro or pSLIK-US28-3xF were transfected into 293T cells using Fugene 6 Transfection Reagent (Promega), as detailed above. The clarified media was then concentrated 50 times by ultracentrifugation at 82,700 × g at 4 °C for 60 min, after which the pellet was resuspended in X-VIVO15 (Lonza). The concentrated pSLIK-hygro or pSLIK-US28-3xF lentiviral particles were then used to infect 5 × 105 THP-1 cells per condition by centrifugal enhancement for 1 h at room temperature at 1,000 × g. Successfully transduced cells were selected with 450 μg/mL hygromycin, after which cell debris was removed by cushioning cells onto Ficoll Paque PLUS (GE Healthcare Life Sciences) for 35 min at room temperature at 450 × g without the brake.
RNA and Protein Analyses.
For details on RNA and protein analyses see SI Appendix, SI Materials and Methods.
Infection of THP-1, Kasumi-3, and CD34+ Cells.
THP-1 cells were infected as described elsewhere (63). In brief, 1 × 106 cells were infected at a multiplicity of 1 median tissue culture infective dose (TCID50) per cell by centrifugal enhancement at 450 × g for 35 min, without the brake, at room temperature. Infections were then returned to the culture conditions described in SI Appendix, SI Materials and Methods for an additional 75 min, after which the cells were washed with 1× PBS, replated, and incubated in X-VIVO15 media as indicated in the text. DOX induction of pUS28 expression, where stated, was performed 24–48 h before infection by adding 1 μg/mL DOX to 1 × 107 cells in RPMI. After infection, transduced THP-1 cells were washed and replated in X-VIVO15 media with 1 μg/mL DOX, where indicated. During the course of infection, transduced THP-1 cells were treated with DOX every 2 d after infection, and media was changed every 5 d, where cellular debris was removed by centrifuging cells onto Ficoll Paque PLUS (GE Healthcare Life Sciences) for 35 min at room temperature at 450 × g without the brake.
Kasumi-3 cells were infected as described previously (22, 30, 37). Briefly, cells were serum-starved in X-VIVO15 48 h before infection and then infected at a multiplicity of 1.0 TCID50 per cell by centrifugal enhancement (1,000 × g, 35 min, room temperature, with no brake) in X-VIVO15. The following day, cells were treated with trypsin to remove any virus that had not entered the cell and then cushioned onto Ficoll-Pacque (GE Healthcare Life Sciences) to remove residual virus and debris. Infected cells were washed with 1× PBS, replated in X-VIVO15, and harvested as indicated in the text.
Isolation of CD34+ HPCs is described in detail elsewhere (64). Immediately following isolation, CD34+ HPCs were infected at a multiplicity of 2.0 TCID50 per cell, as previously described (22, 30, 37, 64), in infection media consisting of IMDM (Corning) supplemented with 10% BIT9500 serum substitute (Stem Cell Technologies), 2 mM l-glutamine, 20 ng/mL low-density lipoproteins, and 50 μM 2-mercaptoethanol. The next day, cultures were washed three times in 1× PBS and replated in 0.4-μm-pore transwells over irradiated stromal cells in hLTCM, as described previously. Following 12 dpi, cells were washed three times in 1× PBS and harvested for RT-qPCR analyses and ELDA.
Assaying the Release of Infectious Virus by ELDA.
Infected Kasumi-3 cells or CD34+ HPCs were cocultured with naïve NuFF-1 cells. Briefly, infected cells were serially diluted twofold onto naïve NuFF-1 cells in X-VIVO15 for 14 d. The number of mCherry-positive wells for each dilution was counted and virus release measured using ELDA software (bioinf.wehi.edu.au/software/elda/index.html).
Chromatin Immunoprecipitation.
Cells were infected for 24 h with WT or US28Δ, fixed in 1% formaldehyde for 15 min at room temperature, and then quenched with 125 mM glycine. Cells were lysed in IP buffer (150 mM NaCl, 50 mM Tris⋅HCl, pH 7.5, 5 mM EDTA, 0.5% Igepal-CA630, and 1% Triton X-100) and debris was removed by centrifugation. DNA was sheared to 0.3- to 1-kb fragments with a MiSonix Sonicator 3000 (20% output, 0.5 s on/off, 1 min) and aliquots stored as input controls. DNA associated with AP-1 was immunoprecipitated with either normal rabbit serum (Cell Signaling) or anti-fos (ChIP grade, diluted 1:100; Cell Signaling) using protein A agarose (Millipore Sigma). DNA was eluted by boiling and was followed by proteinase K treatment. DNA from disrupted nucleosomes was precipitated and used in PCR with primers directed at the MIEP or the UL69 nonpromoter region (65) (SI Appendix, Table S1).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant AI119415 (to C.M.O.), American Heart Association Scientist Development Grant 15SDG23000029 (to C.M.O.), and University at Buffalo–SUNY and Cleveland Clinic funding (C.M.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816933116/-/DCSupplemental.
References
- 1.Mocarski ES, Courcelle CT. Cytomegaloviruses and Their Replication. 4th Ed Lippincott Williams & Wilkins; Philadelphia: 2001. [Google Scholar]
- 2.Pass RF. Cytomegaloviruses. 4th Ed Lippincott Williams & Wilkins; Philadelphia: 2001. [Google Scholar]
- 3.Khanna R, Diamond DJ. Human cytomegalovirus vaccine: Time to look for alternative options. Trends Mol Med. 2006;12:26–33. doi: 10.1016/j.molmed.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Arvin AM, Fast P, Myers M, Plotkin S, Rabinovich R. National Vaccine Advisory Committee Vaccine development to prevent cytomegalovirus disease: Report from the National Vaccine Advisory Committee. Clin Infect Dis. 2004;39:233–239. doi: 10.1086/421999. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths P, Baraniak I, Reeves M. The pathogenesis of human cytomegalovirus. J Pathol. 2015;235:288–297. doi: 10.1002/path.4437. [DOI] [PubMed] [Google Scholar]
- 6.Ljungman P, et al. Disease Definitions Working Group of the Cytomegalovirus Drug Development Forum Definitions of cytomegalovirus infection and disease in transplant patients for use in clinical trials. Clin Infect Dis. 2017;64:87–91. doi: 10.1093/cid/ciw668. [DOI] [PubMed] [Google Scholar]
- 7.Ramanan P, Razonable RR. Cytomegalovirus infections in solid organ transplantation: A review. Infect Chemother. 2013;45:260–271. doi: 10.3947/ic.2013.45.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humar A, et al. The efficacy and safety of 200 days valganciclovir cytomegalovirus prophylaxis in high-risk kidney transplant recipients. Am J Transplant. 2010;10:1228–1237. doi: 10.1111/j.1600-6143.2010.03074.x. [DOI] [PubMed] [Google Scholar]
- 9.Limaye AP, Corey L, Koelle DM, Davis CL, Boeckh M. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet. 2000;356:645–649. doi: 10.1016/S0140-6736(00)02607-6. [DOI] [PubMed] [Google Scholar]
- 10.Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol. 2006;87:1763–1779. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 11.Hargett D, Shenk TE. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci USA. 2010;107:20039–20044. doi: 10.1073/pnas.1014509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo K, Kaneshima H, Mocarski ES. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minton EJ, Tysoe C, Sinclair JH, Sissons JG. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coronel R, Takayama S, Juwono T, Hertel L. Dynamics of human cytomegalovirus infection in CD34+ hematopoietic cells and derived Langerhans-type dendritic cells. J Virol. 2015;89:5615–5632. doi: 10.1128/JVI.00305-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrum FD, Jordan CT, High K, Shenk T. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: A model for latency. Proc Natl Acad Sci USA. 2002;99:16255–16260. doi: 10.1073/pnas.252630899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maciejewski JP, et al. Infection of hematopoietic progenitor cells by human cytomegalovirus. Blood. 1992;80:170–178. [PubMed] [Google Scholar]
- 17.Dósa R, Burián K, Gönczöl E. Human cytomegalovirus latency is associated with the state of differentiation of the host cells: An in vitro model in teratocarcinoma cells. Acta Microbiol Immunol Hung. 2005;52:397–406. doi: 10.1556/AMicr.52.2005.3-4.11. [DOI] [PubMed] [Google Scholar]
- 18.Duan YL, et al. Maintenance of large numbers of virus genomes in human cytomegalovirus-infected T98G glioblastoma cells. J Virol. 2014;88:3861–3873. doi: 10.1128/JVI.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CH, et al. Factors affecting human cytomegalovirus gene expression in human monocyte cell lines. Mol Cells. 1999;9:37–44. [PubMed] [Google Scholar]
- 20.Luo MH, Fortunato EA. Long-term infection and shedding of human cytomegalovirus in T98G glioblastoma cells. J Virol. 2007;81:10424–10436. doi: 10.1128/JVI.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meier JL. Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: Role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J Virol. 2001;75:1581–1593. doi: 10.1128/JVI.75.4.1581-1593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connor CM, Murphy EA. A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J Virol. 2012;86:9854–9865. doi: 10.1128/JVI.01278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penkert RR, Kalejta RF. Human embryonic stem cell lines model experimental human cytomegalovirus latency. MBio. 2013;4:e00298-13. doi: 10.1128/mBio.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caviness K, et al. Complex interplay of the UL136 isoforms balances cytomegalovirus replication and latency. MBio. 2016;7:e01986. doi: 10.1128/mBio.01986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford LB, et al. Human cytomegalovirus induces cellular and humoral virus-specific immune responses in humanized BLT mice. Sci Rep. 2017;7:937. doi: 10.1038/s41598-017-01051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hakki M, et al. HCMV infection of humanized mice after transplantation of G-CSF-mobilized peripheral blood stem cells from HCMV-seropositive donors. Biol Blood Marrow Transplant. 2014;20:132–135. doi: 10.1016/j.bbmt.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith MS, et al. Granulocyte-colony stimulating factor reactivates human cytomegalovirus in a latently infected humanized mouse model. Cell Host Microbe. 2010;8:284–291. doi: 10.1016/j.chom.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umashankar M, et al. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog. 2011;7:e1002444. doi: 10.1371/journal.ppat.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins-McMillen D, Buehler J, Peppenelli M, Goodrum F. Molecular determinants and the regulation of human cytomegalovirus latency and reactivation. Viruses. 2018;10:E444. doi: 10.3390/v10080444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connor CM, Vanicek J, Murphy EA. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J Virol. 2014;88:5524–5532. doi: 10.1128/JVI.00481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weekes MP, et al. Proteomic plasma membrane profiling reveals an essential role for gp96 in the cell surface expression of LDLR family members, including the LDL receptor and LRP6. J Proteome Res. 2012;11:1475–1484. doi: 10.1021/pr201135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buehler J, et al. Opposing regulation of the EGF receptor: A molecular switch controlling cytomegalovirus latency and replication. PLoS Pathog. 2016;12:e1005655. doi: 10.1371/journal.ppat.1005655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JH, Collins-McMillen D, Buehler JC, Goodrum FD, Yurochko AD. Human cytomegalovirus requires epidermal growth factor receptor signaling to enter and initiate the early steps in the establishment of latency in CD34+ human progenitor cells. J Virol. 2017;91:e01206-16. doi: 10.1128/JVI.01206-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rak MA, et al. Human cytomegalovirus UL135 interacts with host adaptor proteins to regulate epidermal growth factor receptor and reactivation from latency. J Virol. 2018;92:e00919-18. doi: 10.1128/JVI.00919-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason GM, Poole E, Sissons JG, Wills MR, Sinclair JH. Human cytomegalovirus latency alters the cellular secretome, inducing cluster of differentiation (CD)4+ T-cell migration and suppression of effector function. Proc Natl Acad Sci USA. 2012;109:14538–14543. doi: 10.1073/pnas.1204836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinclair JH, Reeves MB. Human cytomegalovirus manipulation of latently infected cells. Viruses. 2013;5:2803–2824. doi: 10.3390/v5112803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humby MS, O’Connor CM. Human cytomegalovirus US28 is important for latent infection of hematopoietic progenitor cells. J Virol. 2015;90:2959–2970. doi: 10.1128/JVI.02507-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishna BA, et al. Latency-associated expression of human cytomegalovirus US28 attenuates cell signaling pathways to maintain latent infection. MBio. 2017;8:e01754-17. doi: 10.1128/mBio.01754-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chee MS, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 40.Miller WE, et al. US28 is a potent activator of phospholipase C during HCMV infection of clinically relevant target cells. PLoS One. 2012;7:e50524. doi: 10.1371/journal.pone.0050524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beisser PS, Laurent L, Virelizier JL, Michelson S. Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes. J Virol. 2001;75:5949–5957. doi: 10.1128/JVI.75.13.5949-5957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng S, et al. Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc Natl Acad Sci USA. 2017;114:E10586–E10595. doi: 10.1073/pnas.1710522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung AK, Abendroth A, Cunningham AL, Slobedman B. Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood. 2006;108:3691–3699. doi: 10.1182/blood-2005-12-026682. [DOI] [PubMed] [Google Scholar]
- 44.Goodrum F, Jordan CT, Terhune SS, High K, Shenk T. Differential outcomes of human cytomegalovirus infection in primitive hematopoietic cell subpopulations. Blood. 2004;104:687–695. doi: 10.1182/blood-2003-12-4344. [DOI] [PubMed] [Google Scholar]
- 45.Patterson BK, et al. Repertoire of chemokine receptor expression in the female genital tract: Implications for human immunodeficiency virus transmission. Am J Pathol. 1998;153:481–490. doi: 10.1016/S0002-9440(10)65591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poole E, et al. The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner. J Virol. 2013;87:4261–4271. doi: 10.1128/JVI.03497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez V, Dong JJ, Battley J, Jackson KN, Dykes BC. Human cytomegalovirus infection of THP-1 derived macrophages reveals strain-specific regulation of actin dynamics. Virology. 2012;433:64–72. doi: 10.1016/j.virol.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 48.Rossetto CC, Tarrant-Elorza M, Pari GS. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14(+) monocytes and CD34(+) cells. PLoS Pathog. 2013;9:e1003366. doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isern E, et al. The activator protein 1 binding motifs within the human cytomegalovirus major immediate-early enhancer are functionally redundant and act in a cooperative manner with the NF-kappaB sites during acute infection. J Virol. 2011;85:1732–1746. doi: 10.1128/JVI.01713-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 51.Eferl R, Wagner EF. AP-1: A double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 52.Roche KL, Nukui M, Krishna BA, O’Connor CM, Murphy EA. Selective 4-thiouracil labeling of RNA transcripts within latently infected cells after infection with human cytomegalovirus expressing functional uracil phosphoribosyltransferase. J Virol. 2018;92:e00880-18. doi: 10.1128/JVI.00880-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujioka S, et al. NF-kappaB and AP-1 connection: Mechanism of NF-kappaB-dependent regulation of AP-1 activity. Mol Cell Biol. 2004;24:7806–7819. doi: 10.1128/MCB.24.17.7806-7819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toton E, et al. The tetramethoxyflavone zapotin selectively activates protein kinase C epsilon, leading to its down-modulation accompanied by Bcl-2, c-Jun and c-Fos decrease. Eur J Pharmacol. 2012;682:21–28. doi: 10.1016/j.ejphar.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krishna BA, Miller WE, O’Connor CM. US28: HCMV’s Swiss Army knife. Viruses. 2018;10:E445. doi: 10.3390/v10080445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stinski MF, Meier JL. Immediate-early viral gene regulation and function. In: Arvin A, et al., editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge Univ Press; New York: 2007. [PubMed] [Google Scholar]
- 57.Dupont L, Reeves MB. Cytomegalovirus latency and reactivation: Recent insights into an age old problem. Rev Med Virol. 2016;26:75–89. doi: 10.1002/rmv.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okazaki K, Sagata N. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J. 1995;14:5048–5059. doi: 10.1002/j.1460-2075.1995.tb00187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boomker JM, The TH, de Leij LF, Harmsen MC. The human cytomegalovirus-encoded receptor US28 increases the activity of the major immediate-early promoter/enhancer. Virus Res. 2006;118:196–200. doi: 10.1016/j.virusres.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 60.Wen DQ, et al. Human cytomegalovirus-encoded chemokine receptor homolog US28 stimulates the major immediate early gene promoter/enhancer via the induction of CREB. J Recept Signal Transduct Res. 2009;29:266–273. doi: 10.1080/10799890903178141. [DOI] [PubMed] [Google Scholar]
- 61.Zhu D, et al. Human cytomegalovirus reprogrammes haematopoietic progenitor cells into immunosuppressive monocytes to achieve latency. Nat Microbiol. 2018;3:503–513. doi: 10.1038/s41564-018-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu SE, Miller WE. The HCMV US28 vGPCR induces potent Gαq/PLC-β signaling in monocytes leading to increased adhesion to endothelial cells. Virology. 2016;497:233–243. doi: 10.1016/j.virol.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arcangeletti MC, et al. Human cytomegalovirus reactivation from latency: Validation of a “switch” model in vitro. Virol J. 2016;13:179. doi: 10.1186/s12985-016-0634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Umashankar M, Goodrum F. Hematopoietic long-term culture (hLTC) for human cytomegalovirus latency and reactivation. Methods Mol Biol. 2014;1119:99–112. doi: 10.1007/978-1-62703-788-4_7. [DOI] [PubMed] [Google Scholar]
- 65.Cuevas-Bennett C, Shenk T. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J Virol. 2008;82:9525–9536. doi: 10.1128/JVI.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.