Significance
The intravascular processing of triglyceride-rich lipoproteins by the lipoprotein lipase (LPL)–GPIHBP1 complex is crucial for clearing triglycerides from the bloodstream and for the delivery of lipid nutrients to vital tissues. A deficiency of either LPL or GPIHBP1 impairs triglyceride processing, resulting in severe hypertriglyceridemia (chylomicronemia). Despite intensive investigation by biochemists worldwide, the structures for LPL and GPIHBP1 have remained elusive. Inspired by the recent discovery that GPIHBP1 stabilizes LPL structure and activity, we crystallized the LPL–GPIHBP1 complex and solved its structure. The structure provides insights into the ability of GPIHBP1 to preserve LPL structure and activity and also reveals how inherited defects in these proteins impair triglyceride hydrolysis and cause chylomicronemia.
Keywords: triglycerides, lipase, lipoproteins, X-ray crystallography, GPIHBP1
Abstract
Lipoprotein lipase (LPL) is responsible for the intravascular processing of triglyceride-rich lipoproteins. The LPL within capillaries is bound to GPIHBP1, an endothelial cell protein with a three-fingered LU domain and an N-terminal intrinsically disordered acidic domain. Loss-of-function mutations in LPL or GPIHBP1 cause severe hypertriglyceridemia (chylomicronemia), but structures for LPL and GPIHBP1 have remained elusive. Inspired by our recent discovery that GPIHBP1’s acidic domain preserves LPL structure and activity, we crystallized an LPL–GPIHBP1 complex and solved its structure. GPIHBP1’s LU domain binds to LPL’s C-terminal domain, largely by hydrophobic interactions. Analysis of electrostatic surfaces revealed that LPL contains a large basic patch spanning its N- and C-terminal domains. GPIHBP1’s acidic domain was not defined in the electron density map but was positioned to interact with LPL’s large basic patch, providing a likely explanation for how GPIHBP1 stabilizes LPL. The LPL–GPIHBP1 structure provides insights into mutations causing chylomicronemia.
Lipoprotein lipase (LPL), the enzyme that mediates the lipolytic processing of triglyceride-rich lipoproteins (TRLs) within capillaries, was identified more than 60 y ago (1, 2) and has been studied intensively by biochemists and physiologists worldwide. LPL is the central molecule in plasma lipid metabolism, hydrolyzing triglycerides within TRLs and releasing lipid nutrients for vital tissues (e.g., heart, skeletal muscle, and adipose tissue) (3). Genetic variation that alters the efficiency of LPL-mediated TRL processing influences both plasma triglyceride levels and the risk for coronary heart disease (4, 5). Interestingly, LPL is synthesized and secreted by parenchymal cells, primarily myocytes and adipocytes, but virtually all of the LPL in tissues is found on the surface of capillaries, where it is bound to a glycolipid-anchored protein, glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) (6, 7).
GPIHBP1, a member of the LU (Ly6/uPAR) protein family, is expressed exclusively in capillary endothelial cells (6). Based on protein homology considerations (8, 9), GPIHBP1’s LU domain (75 residues in length) is predicted to adopt a three-fingered fold, stabilized by five disulfide bonds. GPIHBP1 is unique within the LU protein family in having a highly acidic and intrinsically disordered N-terminal domain containing a sulfated tyrosine and numerous glutamates and aspartates (21 of 26 consecutive residues in human GPIHBP1 are aspartate or glutamate) (6, 10, 11). GPIHBP1 binds to the C-terminal domain of LPL (12), and both the LU domain and the acidic domain of GPIHBP1 contribute to the binding affinity (11).
From the standpoint of mammalian plasma triglyceride metabolism, GPIHBP1 can be viewed as an indispensable partner for LPL. First, GPIHBP1 is solely responsible for capturing LPL within the interstitial spaces and shuttling it across endothelial cells to its site of action in the capillary lumen (7). In the absence of GPIHBP1, LPL remains stranded in the interstitial spaces. Second, GPIHBP1-bound LPL is required for the margination of TRLs along capillaries, allowing the lipolytic processing of TRLs to proceed (13). In the absence of GPIHBP1, TRLs do not stop along capillaries and simply “flow on by” in the bloodstream. Third, the binding of GPIHBP1 stabilizes the structure of LPL and thereby preserves its enzymatic activity. In the absence of GPIHBP1, LPL’s hydrolase domain is highly susceptible to spontaneous unfolding (as judged by hydrogen–deuterium exchange/mass spectrometry studies), resulting in a rapid decline in catalytic activity (11). When LPL is complexed with GPIHBP1, the unfolding of LPL and the concomitant loss of activity is markedly inhibited. The ability of GPIHBP1 to prevent the unfolding of LPL and preserve catalytic activity depends on GPIHBP1’s disordered acidic domain (10, 11). When the acidic domain is deleted, GPIHBP1 still binds to LPL, but the protective effects of GPIHBP1 on LPL structure and activity are minimal or absent (10, 11). GPIHBP1 also inhibits the unfolding of LPL that is catalyzed by physiologic inhibitors of LPL (ANGPTL4, ANGPTL3) (14).
The central importance of both LPL and GPIHBP1 in plasma triglyceride metabolism is illustrated by the fact that missense mutations that interfere with LPL–GPIHBP1 interactions profoundly impair intravascular triglyceride processing, resulting in severe hypertriglyceridemia (chylomicronemia) (15, 16). For example, a p.C445Y mutation in LPL, first identified in the setting of chylomicronemia, has little effect on LPL catalytic activity but abolishes LPL’s ability to bind to GPIHBP1 (17). A variety of amino acid substitutions in GPIHBP1’s LU domain have been uncovered in patients with chylomicronemia, and all of those abolish GPIHBP1’s ability to bind LPL (15). The p.C445Y mutation in LPL and most disease-causing mutations in GPIHBP1 interfere with proper disulfide bond formation and protein folding (17, 18), making it difficult to draw conclusions about which amino acids participate in the LPL–GPIHBP1 binding interface.
Although LPL has been studied intensively for >60 y, its structure has never been determined, very likely because of the susceptibility of LPL’s hydrolase domain to unfolding. In the absence of a structure, the field has depended on inferences from a homology model (19) predicted from the structure of pancreatic lipase (PL), a distantly related member of the same lipase family (20). The structure of PL, solved alone or in a complex with colipase (a cofactor that facilitates lipid binding) (21, 22), suggested that LPL would have two principal domains—a conventional α/β-hydrolase domain with a catalytic triad and a C-terminal β-barrel domain that interacts with lipids (19).
Inspired by the biological importance of GPIHBP1 and the discovery that GPIHBP1 stabilizes LPL structure and activity, we sought to determine the structure of an LPL–GPIHBP1 complex, with the principal goals of clarifying interactions between the two proteins, understanding the molecular basis for mutations causing human disease, and gleaning insights into how GPIHBP1 preserves LPL structure and activity.
Results
Formation of Human LPL–GPIHBP1 Complexes.
To assess the feasibility of producing human LPL–GPIHBP1 complexes, we mixed a high concentration of human LPL (produced in CHO cells) with increasing amounts of soluble human GPIHBP1 (produced in Drosophila S2 cells) and then subjected the mixture to native polyacrylamide gel electrophoresis at pH 8.4 (Fig. 1). Because LPL is a very basic protein, only small amounts of LPL entered the gel. However, full-length soluble GPIHBP1 (GPIHBP121–151) is a highly acidic protein, allowing LPL–GPIHBP1 complexes to enter the gel and migrate as a distinct band (Fig. 1A). Our LPL/GPIHBP1 titration studies revealed that human LPL binds to human GPIHBP1 with 1:1 stoichiometry, consistent with findings reported for bovine LPL (10). Interestingly, a synthetic peptide corresponding to the disordered acidic domain of GPIHBP1 (GPIHBP121–53) also formed a complex with LPL and entered the gel. That complex also exhibited 1:1 stoichiometry (Fig. 1B).
Fig. 1.
Human GPIHBP1 binds to LPL with 1:1 stoichiometry, as judged by native polyacrylamide gel electrophoresis. In the absence of GPIHBP1, little LPL enters native polyacrylamide gels and does not migrate as a distinct band, simply because LPL is a very basic protein. GPIHBP1 is highly acidic; thus, an LPL–GPIHBP1 complex readily enters native gels and migrates as a distinct band. (A) Binding stoichiometry of LPL and GPIHBP1, determined by titrating 1.5 µM human LPL (lane 1) with increasing amounts of full-length human GPIHBP1 [0.3–3.0 µM full-length GPIHBP1 (GPIHBP121–151)] (lanes 2–11). The relative amounts of LPL–GPIHBP1 complexes (determined by scanning Coomassie blue-stained bands) are superimposed on the gels as black diamonds. These studies reveal a 1:1 binding stoichiometry. (B) A similar titration of 7.6 µM LPL (lane 1) with increasing amounts of a synthetic peptide corresponding to the N-terminal acidic domain of GPIHBP1 (GPIHBP121–53; 1–10 µM) (lanes 2–11). These experiments reveal 1:1 binding stoichiometry.
A Crystal Structure for the LPL–GPIHBP1 Complex.
The discovery that human LPL forms a stable 1:1 complex with GPIHBP1 prompted us to purify milligram amounts of the complex for protein crystallization. LPL (15 mg) was mixed with a twofold molar excess of GPIHBP1 and subjected to size-exclusion chromatography. Fractions corresponding to the LPL–GPIHBP1 complex were pooled and concentrated to 15 mg/mL. Crystals were obtained in more than 40 different conditions; however, only those grown in 200 mM magnesium acetate and 20% polyethylene glycol 3350 diffracted at a sufficiently high resolution for data collection and analysis. Crystals were harvested after 5 d, and diffraction data were collected at the European Synchrotron Radiation Facility (Grenoble, France).
The structure of the LPL–GPIHBP1 complex (2.8-Å resolution) was determined by molecular replacement methods using the coordinates for the N- and C-terminal domains of horse pancreatic lipase (PDB ID code 1HPL) and our homology model for GPIHBP1 (11). The structure was refined by manual curation (Table 1). The quality of the electron density map for defined regions within the N-terminal domain of LPL, the C-terminal domain of LPL, and GPIHBP1 is illustrated in SI Appendix, Fig. S1. The percentage of outliers in the Ramachandran plot (2.3%) was similar to that reported for other lipases [3.3% and 2.6% for human and bovine bile salt-stimulated lipase, respectively (23, 24); 2.3% for the pancreatic lipase–colipase complex (21); 4% in the triacylglycerol lipase–colipase complex (25); and 2.3% in pancreatic lipase–procolipase complex (26)]. Human LPL residues falling into disallowed regions of the Ramachandran plot were generally confined to regions where the electron density was not well defined or in regions of strained geometry (Table 1).
Table 1.
Data collection and refinement statistics
| PDB ID | 6E7K |
| Wavelength, Å | 1.000 |
| Space group | P21212 |
| Unit cell, Å | a = 101.9, b = 153.1, c = 95.7 |
| Resolution range,* Å | 95.80–2.80 (2.90–2.80) |
| Observed reflections | 122,715 |
| Unique reflections | 37,662 |
| Completeness, % | 99.4 (99.7) |
| Redundancy | 3.3 (3.2) |
| Rsym,† % | 7.4 (53.2) |
| Rpim,‡ % | 4.8 (35.7) |
| CC1/2 | 0.985 (0.728) |
| Overall <I/σ(I)> | 13.5 (1.5) |
| Wilson B factor, Å2 | 79.7 |
| Rcryst/Rfree,§ % | 19.7/23.5 |
| No. of protein, water, glycan, Ca2+ atoms | 7,873, 16, 154, 2 |
| Refined B factors for protein, water, glycan, Ca2+, Å2 | 110.7, 97.7, 148.3, 115.0 |
| Ramachandran outliers,¶ % | 2.3 |
| Bond lengths,# Å | 0.010 |
| Bond angles,# ° | 1.433 |
Values in parentheses are for the highest resolution shell.
.
, where I is the observed integrated intensity, <I> is the average integrated intensity obtained from multiple measurements, and the summation is over all observed reflections.
. Fo and Fc are the observed and calculated structure factors, respectively, and k is a scaling factor. The summation is over all measurements. Rfree is calculated as Rcryst using 5% of the reflections chosen randomly and omitted from the refinement calculations. Model stereochemistry was analyzed using MolProbity (56, 57).
Several residues falling into the disallowed area of the Ramachandran plot were located in regions of strained geometry or in regions where the electron density is not well defined. For example, D33–I37 in the N terminus of LPL had relatively poor electron density; S73 is positioned in a tight loop connecting α1 and β2; S304 is flanked by disulfide bonds; M336 precedes a proline; N359–Q360 connects β2′ and β3′; and L453–Q454 is in a tight loop connecting β11′ and β12′.
Bond lengths and angles are root-mean-square deviations from ideal values.
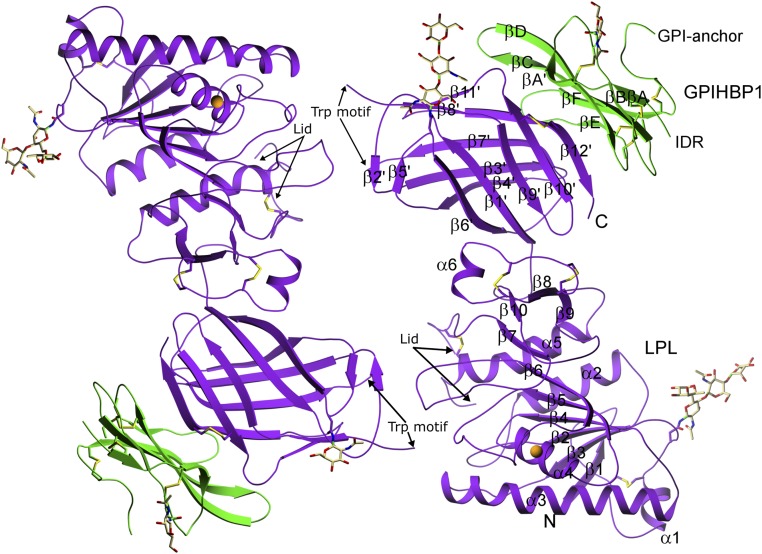
Two LPL–GPIHBP1 complexes were modeled in a head-to-tail orientation in the crystallographic asymmetric unit (Fig. 2). LPL consists of an N-terminal α/β-hydrolase domain containing 6 α-helices and 10 β-strands harboring the catalytic triad (S159, D183, and H268) and a C-terminal flattened β-barrel domain containing 12 β-strands, connected by a hinge region. Electron densities were missing for four short segments of LPL: (i) the first five amino acids of the mature protein, residues 28–32; (ii) residues 249–258 in LPL’s lid region (which controls entry of lipid substrates into the active site cleft); (iii) residues 414–420 within a hydrophobic Trp-rich motif reported to mediate TRL binding (13, 27); and (iv) the last six amino acids of the protein (residues 470–475). While 10 of the residues in LPL’s lid region (a loop extending from the C243–C266 disulfide bond) were not clearly defined in the structure, the trajectory of the amino acids adjacent to the disulfide bond revealed that the lid was in an open conformation.
Fig. 2.
Structure of the LPL–GPIHBP1 complex, as depicted by ribbon representations of the two LPL–GPIHBP1 complexes in the asymmetric crystallographic unit. LPL (purple) has five disulfide bonds (C54–C67, C243–C266, C291–C302, C305–C310, and C445–C465); a single calcium ion (orange sphere) coordinated by A194, R197, S199, D201, and D202; and two N-linked glycans (at N70 and N386). GPIHBP1 (green) has one N-linked glycan (at N78). LPL contains an N-terminal α/β-hydrolase domain (N) containing 6 α-helices and 10 β-strands and a C-terminal flattened β-barrel domain (C) containing 12 β-strands, connected by a hinge region. The numbering of the β-strands in GPIHBP1 follows the nomenclature proposed for LU domain proteins (55). IDR, intrinsically disordered region.
The LPL crystal structure revealed two N-linked glycans (N70, N386), five disulfide bonds (C54–C67, C243–C266, C291–C302, C305–C310, C445–C465), and a single calcium atom coordinated by A194, R197, S199, D201, and D202. The active site cleft is lined by the hydrophobic side chains of W82, V84, W113, Y121, Y158, L160, A185, P187, F212, I221, F239, V260, V264, and K265, which would form van der Waals interactions with (and stabilize) the hydrophobic tails of lipid substrates in the active site. Key structural features of LPL and GPIHBP1 are depicted in the primary sequences shown in SI Appendix, Fig. S2.
The two LPL molecules in the crystal structure interact reciprocally at a single site—between a C-terminal loop containing the hydrophobic Trp motif in one LPL molecule and the catalytic pocket in the N-terminal domain of the partner LPL molecule. Electron densities were observed above the catalytic pocket in both LPL–GPIHBP1 complexes (possibly corresponding to the Trp motif), but the densities differed in the two LPL molecules and could not be modeled satisfactorily, suggesting that in the absence of lipids the Trp motif can adopt several conformations. To identify potential conformations for the Trp motif (residues 414–421), we performed in silico modeling with the BioLuminate and Prime modeling suite (Schrödinger). Five models were generated by de novo loop creation for each of the two LPL molecules in the crystal structure, followed by prime energy minimization to choose the final model for each chain (28). The overall orientation of the Trp motifs in each model was similar with hydrophobic amino acids of the loop making contacts with hydrophobic amino acids lining the active site pocket and thereby burying a surface area of ∼600 Å2. In both models, W421 makes similar contacts, but the orientations of Y414, F415, W417, and W420 differ. In the model for chain A, F415 makes contacts with F212, R214, G217, S220, and I221. W417 makes contacts with F212, R219, I221, and I264. W420 interacts with W82, H120, Y121, and P122. W421 docks and makes hydrophobic interactions with the side chains of W82, Y121, P187, and I221 (SI Appendix, Fig. S3A). In the second LPL molecule, Y414 interacts with F212, F239, V263, and V264; F415 and S416 interact with L263 and V264, respectively; W420 interacts with G215, R219, and I221; and W421 contacts W82, Y121, P187, I221, and V264 (SI Appendix, Fig. S3B). In each case, the docking of the Trp motif into the catalytic pocket would explain the open lid conformation in the crystal structure. The reciprocal interaction between the two LPL molecules was not a consequence of protein crystallization because the same arrangement of molecules was observed in solution by small-angle X-ray scattering (SAXS) analyses (Fig. 3 and SI Appendix, Table S1). In the absence of lipids, we suspect that the hydrophobic Trp motif of one LPL molecule is shielded by a hydrophobic docking site (e.g., the catalytic pocket) in the second LPL molecule.
Fig. 3.
Small-angle X-ray scattering (SAXS) analysis of the LPL–GPIHBP1 complex in solution. (A) SAXS data (gray circles) and the fit of the ab initio model reconstructed using DAMMIF with P2 symmetry (green line). The Lower panel shows the error-weighted residual differences between the model fit and the experimental data. (B) The real-space distance–distribution of the SAXS data and model. The Inset shows the superposition of the crystal structure of the LPL–GPIHBP1 complex (PDB ID code 6E7K) with the filtered average ab initio model.
The absence of electron densities for GPIHBP1 residues 21–61 (which includes the entire acidic domain) was expected, given that this region is intrinsically disordered and only binds LPL transiently (10). Consistent with the presence of a classic LU fold (8, 9), GPIHBP1 has three fingers stabilized by five disulfide bonds (finger 1 containing βA–βB, amino acids 62–85; finger 2 containing βC–βD, amino acids 86–111; finger 3 containing βE–βF, amino acids 112–130) (Figs. 2 and 4).
Fig. 4.
LPL–GPIHBP1 interactions. GPIHBP1 (green sticks, showing Cα positions and key side chains) forms a concave hydrophobic surface. That surface interacts with the C-terminal domain of LPL (purple sticks, showing key LPL side chains). A semitransparent surface of LPL is colored according to electrostatic potential [red (acidic, −5 kBT), white (neutral, 0 kBT), and blue (basic, 5 kBT)]. Electrostatic potentials were calculated with DelPhi (56). All three fingers of GPIHBP1, but particularly fingers 2 and 3, interact with LPL, largely by hydrophobic contacts.
GPIHBP1–LPL Interactions.
The LPL–GPIHBP1 binding interface (∼940 Å2) involves the entire concave surface of GPIHBP1’s LU domain, involves all three fingers, and is mediated largely by hydrophobic interactions (Fig. 4). Of note, several van der Waals interactions between nonpolar stems of charged amino acids stabilize the complex (29). For example, LPL residues K464, H466, and D467 make hydrophobic interactions with S107, W109, and T111 of GPIHBP1. Finger 1 of GPIHBP1 interacts with LPL residues 443–447 and 465–466; finger 2 interacts with LPL residues 447–448 and 463–467; and finger 3 interacts with amino acids 367, 374, 369, 403–406, 447, and 464 (Fig. 4 and Dataset S1). All amino acid residues making hydrophobic contacts between LPL and GPIHBP1 are listed in Dataset S1. In addition, the side chain Nε of LPL R447 forms a hydrogen bond with the main chain carbonyl of GPIHBP1 L103. Also, the side chain of LPL E384 forms a hydrogen bond with GPIHBP1 E122, either directly or indirectly through a water molecule (the electron density map does not permit a definite conclusion).
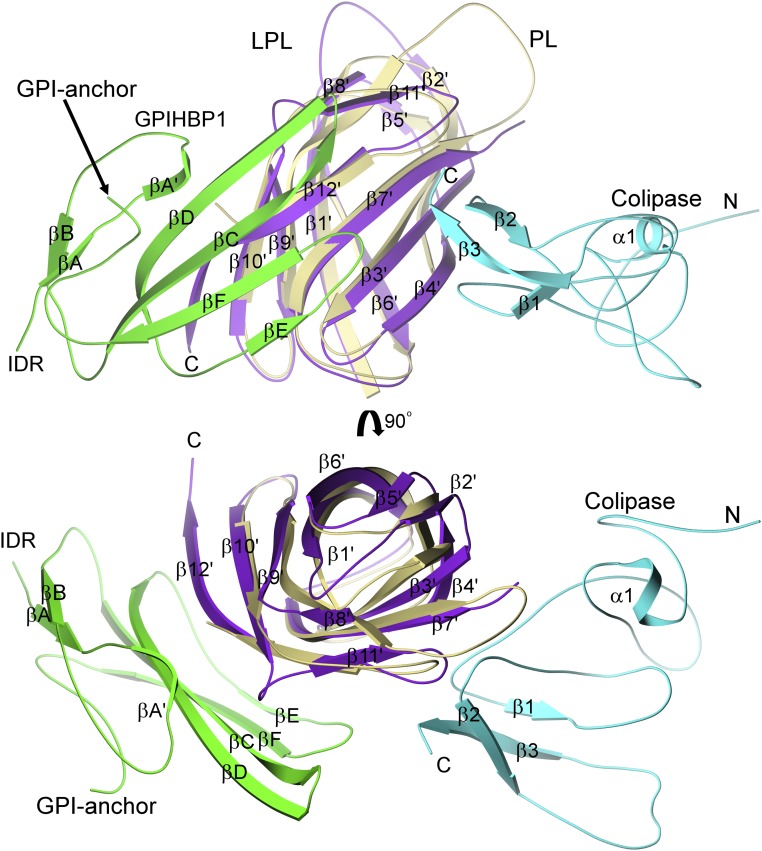
Before embarking on efforts to crystallize the LPL–GPIHBP1 complex, we suspected that the binding site for GPIHBP1 on LPL might be similar to the binding site for colipase on PL. Our suspicion was piqued by superficial similarities between GPIHBP1 and colipase: Both bind to the C-terminal domain of their lipase partner, and both are small proteins with multiple loops and five disulfide bonds. The LPL–GPIHBP1 structure revealed that GPIHBP1 and colipase bind far apart (almost on opposite sides) of the C-terminal domains of LPL and PL, respectively (Fig. 5). Unlike the situation with GPIHBP1, where interactions with LPL are largely hydrophobic and involve an extensive surface area, the binding of colipase to PL depends primarily on electrostatic interactions involving two short hairpin loops (21, 22, 30).
Fig. 5.
Distinct position for GPIHBP1 binding to LPL compared with colipase binding to pancreatic lipase. The structure of the LPL–GPIHBP1 complex (C-terminal domain of LPL in purple; GPIHBP1 in green) is superimposed on that of the pancreatic lipase (PL)–colipase complex (C-terminal domain of PL in khaki; colipase in cyan), revealing that the binding sites for GPIHBP1 and colipase on the C-terminal domains of their partner lipase (LPL and PL, respectively) are distinct. Colipase binds by polar interactions to one face of the flattened β-barrel via two hairpin loops (22), whereas GPIHBP1 interacts with both faces through a large hydrophobic interface. Both GPIHBP1 and colipase are small multifingered proteins with five disulfide bonds; however, the two proteins are evolutionarily distinct, and the arrangement of the disulfide bonds in the two proteins is different.
LPL Contains a Large Contiguous Basic Patch Spanning both N-Terminal and C-Terminal Domains.
LPL has long been recognized to have multiple positively charged heparin-binding motifs (31–34). An electrostatic surface potential map, based on our crystallographic structure, revealed that one surface of LPL has a large, flat, and contiguous basic patch (∼2,400 Å2) involving both C- and N-terminal domains and extending across LPL’s hinge region (Fig. 6). Two lysines (K472 and K473), located within the last five amino acids of LPL, were disordered and could not be assigned in the electron density map. Had those two lysines been visualized, the size of the basic patch depicted in Fig. 6 may have been even larger. The crystal structure shows that GPIHBP1’s acidic intrinsically disordered region (IDR) projects toward LPL’s basic patch. The length of the acidic IDR (>60 Å) (10) would allow it to make electrostatic interactions with LPL’s basic patch. We suspect that these electrostatic interactions account for the ability of the acidic domain to stabilize LPL structure and activity. Aside from the role of GPIHBP1’s acidic domain in stabilizing LPL structure (11), the functional relevance of LPL–GPIHBP1 electrostatic interactions is evident from the ability of GPIHBP1’s acidic domain to increase the association rate constant (ka) for LPL–GPIHBP1 complex formation by >250-fold (an effect highly dependent on the ionic strength of the buffer) (10).
Fig. 6.
Electrostatic surface of LPL showing the site for GPIHBP1 binding. GPIHBP1’s LU domain forms a concave hydrophobic surface that interacts with a hydrophobic surface on the C-terminal domain (CTD) of LPL (image on Right). LPL has a single large, contiguous basic patch (∼2,400 Å2) spanning the CTD, the hinge region, and the N-terminal catalytic domain (Middle image). The C terminus of GPIHBP1, where the GPI anchor would be attached, and the N terminus of GPIHBP1, from which GPIHBP1’s acidic intrinsically disordered region (IDR) projects, are indicated by arrows (Middle and Right images). GPIHBP1’s acidic IDR, >60 Å in length (10), is expected to project across and interact transiently with LPL’s large basic patch (Middle image). The sequence of GPIHBP1’s highly acidic IDR is shown in SI Appendix, Fig. S2. In the image on the Left, the catalytic pocket is highlighted by a yellow dotted circle. The Trp motif and lid sequences are depicted as dashed green and cyan lines, respectively. Electrostatic potentials were calculated as in Fig. 4.
Fresh Insights into LPL Mutations Causing Chylomicronemia.
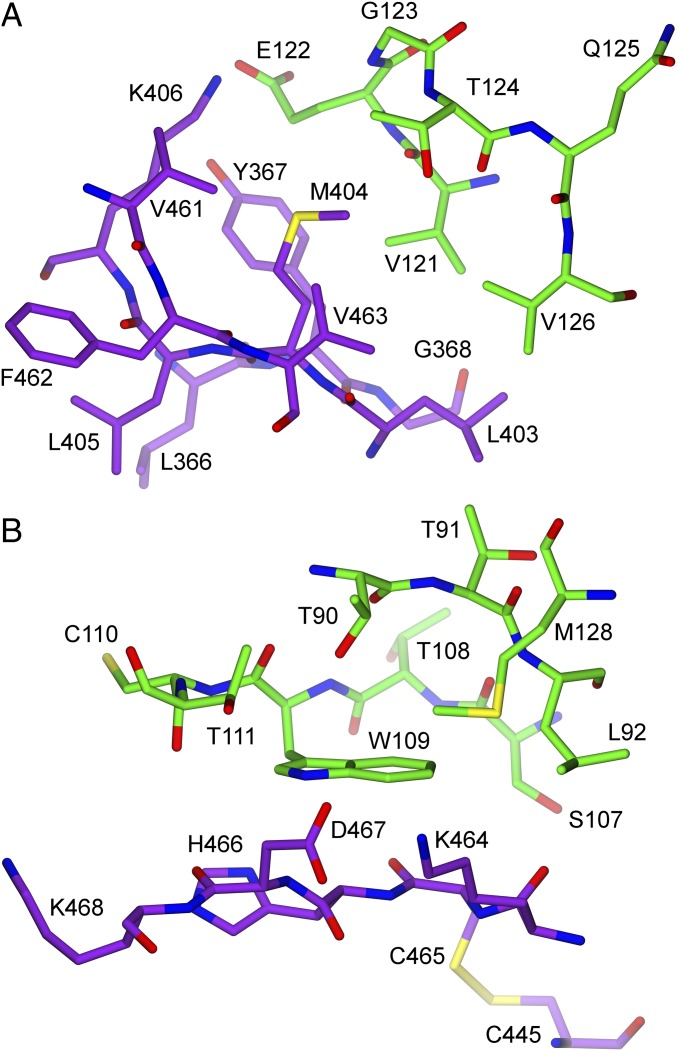
Given the very large interface between LPL’s C-terminal domain and GPIHBP1’s LU domain, we suspected that we might find missense mutations that cause chylomicronemia by interfering with LPL–GPIHBP1 interactions. We were intrigued by a report of a chylomicronemia patient with a p.M404R mutation in LPL; this mutation had been reported to abolish LPL secretion from cells (35). Our LPL–GPIHBP1 crystal structure revealed that M404 in LPL interacts with V121, E122, T124, and V126 in a hydrophobic pocket in finger 3 of GPIHBP1 (Fig. 7A and Dataset S1). It is evident from the crystal structure that this hydrophobic pocket on GPIHBP1 cannot accommodate the larger side chain of arginine. We therefore reasoned that the p.M404R mutation might actually cause disease by interfering with LPL–GPIHBP1 interactions. Indeed, the p.M404R mutation abolished binding of LPL to GPIHBP1 (Fig. 8 A and B) but had little or no effect on LPL secretion or activity (Fig. 8 B and C). As a control, we included an LPL mutant harboring a p.C445Y mutation; that mutant was shown previously to abolish LPL binding to GPIHBP1 without affecting LPL secretion, heparin binding, or catalytic activity (17). C445 in LPL is disulfide bonded to C465, and both cysteines participate in the GPIHBP1 binding interface [C445 interacts with GPIHBP1 residues K69 and S70 (Fig. 4 and Dataset S1); C465 interacts with GPIHBP1 residues K69, S70, and W109 (Fig. 4 and Dataset S1)]. As expected from earlier studies (17), LPL-C445Y did not bind to GPIHBP1 (Fig. 8 A and B). As an additional control, we tested the ability of wild-type LPL to bind to a mutant GPIHBP1 containing a p.W109S mutation. The p.W109S mutation abolishes GPIHBP1’s capacity to bind LPL and does so without interfering with proper disulfide bond formation (36). The crystal structure revealed that W109 in GPIHBP1 participates in the binding interface with LPL, interacting with LPL residues K464, C465, H466, and D467 (Fig. 7B and Dataset S1).
Fig. 7.
Schematic diagrams depicting interactions between M404 in LPL and GPIHBP1 and between W109 in GPIHBP1 and LPL. (A) Interactions of M404 in LPL (purple) with a hydrophobic pocket formed by GPIHBP1 (green) (GPIHBP1 residues V121, E122, T124, and V126; all in finger 3 of GPIHBP1’s LU domain). Replacing M404 with an arginine disrupts binding of LPL to GPIHBP1 (Fig. 8). (B) Hydrophobic interactions between W109 in GPIHBP1 and LPL. GPIHBP1-W109 is located in a hydrophobic pocket and interacts with multiple LPL residues (Dataset S1). This figure depicts the C445–C465 disulfide bond in LPL but does not show all of the hydrophobic interactions between those two cysteines and GPIHBP1. LPL-C445 interacts with GPIHBP1 residues K69 and S70; LPL-C465 interacts with multiple GPIHBP1 residues (Dataset S1). A p.C445Y mutation in LPL abolishes binding of LPL to GPIHBP1 (17), likely by disrupting the conformation of the loop established by the C445–465 disulfide bond.
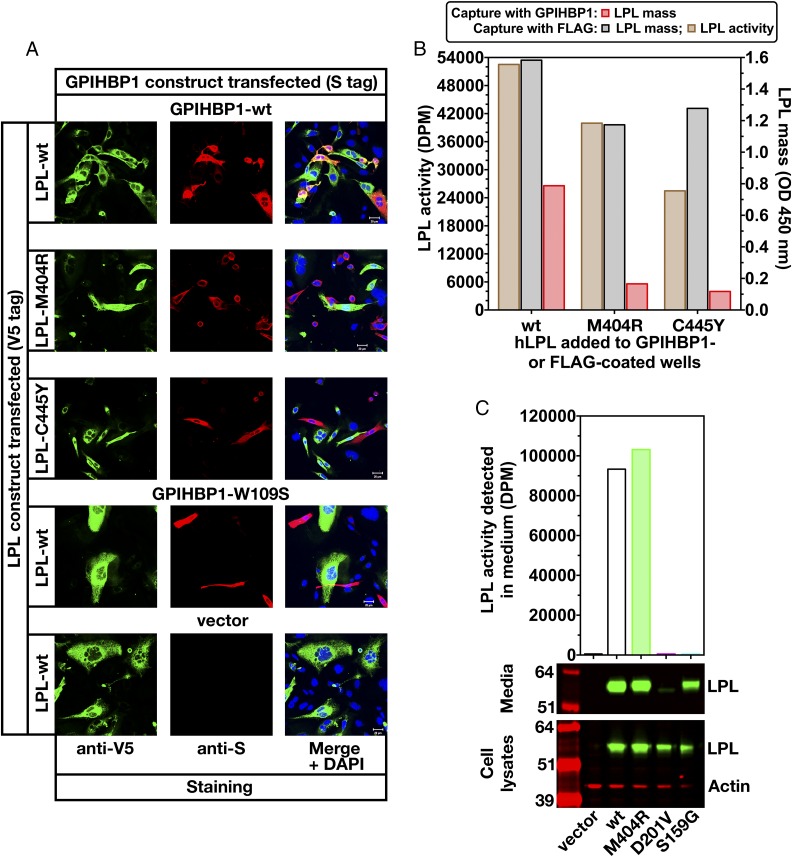
Fig. 8.
Testing the impact of an LPL missense mutation (p.M404R) on GPIHBP1 binding. (A) CHO cells that had been transfected with S-protein–tagged versions of wild-type (wt) human GPIHBP1 or GPIHBP1-W109S (or empty vector) were coplated with cells that had been transfected with V5-tagged versions of human LPL-wt, LPL-M404R, or LPL-C445Y. The p.W109S mutation in GPIHBP1 and the p.C445Y mutation in LPL interfere with LPL–GPIHBP1 interactions (17, 35). The ability of LPL to bind to GPIHBP1 on the surface of cells was assessed by immunocytochemistry. Freshly secreted LPL-wt (green) was captured by GPIHBP1 (red), resulting in colocalization of LPL and GPIHBP1 in the merged image. GPIHBP1-W109S has no capacity to bind LPL-wt; consequently, no colocalization was observed on the merged image. LPL-M404R had little or no capacity to bind to GPIHBP1 (no colocalization on the merged image). (B) LPL activity and mass assays revealed that LPL-M404R is catalytically active but binds poorly to GPIHBP1. Fresh medium from CHO cells that had been transfected with FLAG-tagged human LPL-wt, LPL-M404R, or LPL-C445Y was added to wells of a 96-well ELISA plate that had been coated with a FLAG-specific antibody (FLAG-Ab) or with human GPIHBP1. Relative amounts of LPL mass were assessed with an HRP-labeled monoclonal antibody against LPL (5D2) and plotted as optical density (OD). LPL-M404R was efficiently captured by the FLAG antibody, but little LPL-M404R was captured by GPIHBP1. LPL-M404R binding to GPIHBP1 was reduced by 79% and 85% in two independent experiments (compared with LPL-wt binding to GPIHBP1). Triglyceride hydrolase activity of LPL captured on FLAG antibody-coated wells was assessed with [3H]triolein as the substrate. LPL-wt, LPL-M404R, and LPL-C445Y were catalytically active. (C) Assessing LPL-M404R catalytic activity and secretion from cells. CHO cells were transfected with V5-tagged versions of LPL-wt, LPL-D201V, LPL-M404R, or LPL-S159G (mutation of the critical serine in LPL’s catalytic triad). The LPL in cell lysates and media was assessed by Western blotting with a V5 antibody (green). Actin (red) was used as a loading control. LPL activity in the medium was assessed with a [3H]triolein substrate. LPL-M404R was synthesized and secreted by CHO cells, and there was robust LPL-M404R catalytic activity in the cell culture medium. The enzymatic activity of LPL-M404R was 103%, 110%, 115%, and 103% of the activity of LPL-wt in four independent experiments.
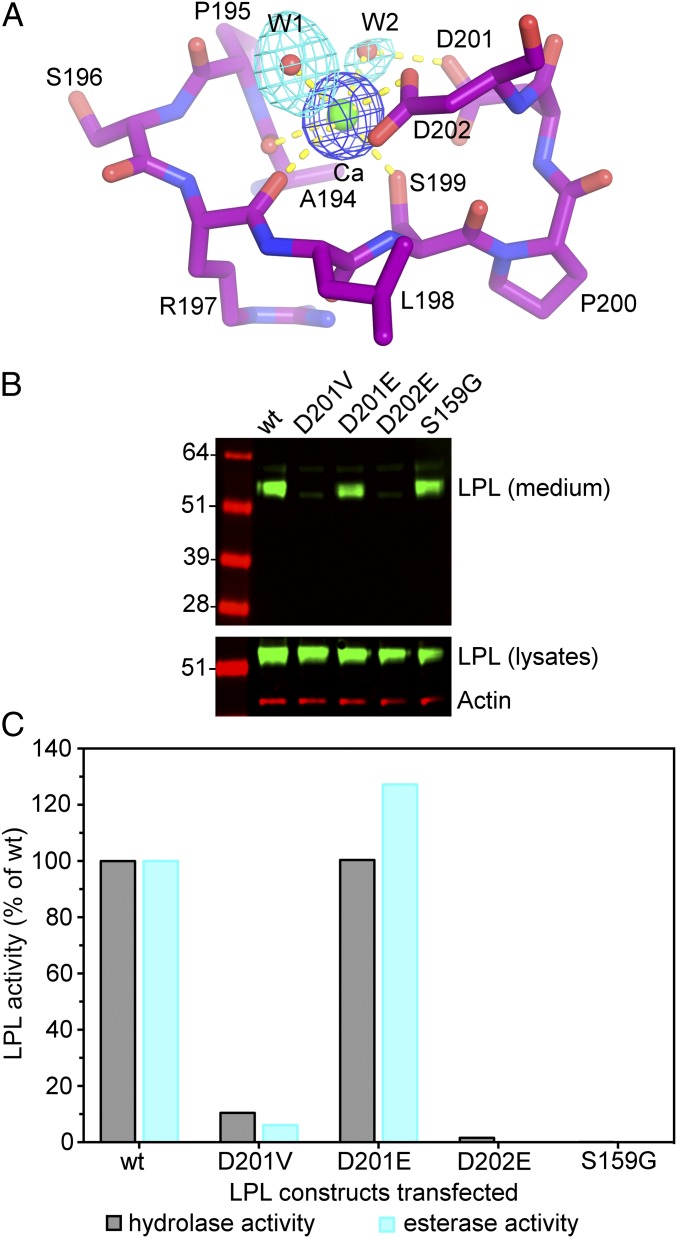
The structure of the LPL–GPIHBP1 complex also provided insights into a p.D201V mutation in LPL, first observed in two Lebanese kindred individuals with chylomicronemia (37). The mechanism by which the p.D201V mutation causes disease was never investigated. Our crystal structure revealed that the carboxylic acid side chain of D201 participates in the coordination of LPL’s calcium ion via an ordered water molecule (Fig. 9A). A valine at the same position cannot participate in calcium coordination. We predicted that defective calcium coordination in the p.D201V mutant would interfere with LPL folding and prevent LPL secretion from cells. Indeed, the p.D201V mutation eliminated LPL secretion from cells and markedly reduced LPL activity levels in the medium (Figs. 8B and 9B). Substituting glutamic acid for aspartic acid at position 201 had little effect on LPL secretion or activity because the glutamic acid side chain can displace the intervening water molecule and interact directly with the calcium ion. We predicted that a glutamate substitution at the more sterically constrained D202 (which interacts directly with the calcium ion) would not be tolerated. Indeed, LPL-D202E was not secreted from cells and LPL activity was virtually absent from the medium (Fig. 9B).
Fig. 9.
The calcium binding site in human LPL. (A) Stick representation of LPL’s Ca2+ binding site, showing main-chain and side-chain hydrogen bonding (dashed yellow lines). Difference electron density maps were contoured at 7σ (blue) and 2.8σ (cyan) for the calcium atom and water (w) molecules, respectively. (B) Assessing the impact of mutations in calcium-coordinating amino acids on LPL secretion from cells. CHO cells were transfected with FLAG-tagged versions of wild-type (wt) human LPL and several mutant LPLs (D201V, D201E, D202E, and S159G). LPL-S159G is an inactivating mutation in LPL’s catalytic triad. A Western blot with a FLAG-specific antibody (green) was used to detect LPL in cell lysates and the cell culture medium. Actin (red) was used as a loading control. (C) LPL activity in the medium of LPL-transfected cells. LPL activity was assessed with a [3H]triolein substrate (triglyceride hydrolase activity) and a DGGR substrate (esterase activity). The catalytic activities of the different LPL proteins are plotted as a percentage of those observed with LPL-wt (set at 100%). Based on the structure of LPL, we predicted that replacing D201 with a glutamate would not interfere with calcium coordination, whereas replacing D202 with a glutamate would disrupt calcium binding. D202 is buried and the extra methylene in a Glu mutation would not allow calcium coordination. D201 is located on the surface, and a Glu in that position retains the ability to coordinate calcium. LPL-D201V activity was 10.5%, 6.1%, 4.1%, and 12.9% of LPL-wt in four independent experiments; LPL-D201E activity was 100% and 127% of LPL-wt in two independent experiments; LPL-D202E activity was 1.6% and 0.0% of control in two independent experiments.
Discussion
After six decades of intensive investigation of LPL by biochemists and physiologists worldwide, we succeeded in solving, by X-ray crystallography, the atomic structure of LPL complexed to its endothelial cell partner protein, GPIHBP1. The ability to crystallize the LPL–GPIHBP1 complex depended on optimized methods for expressing and purifying the two proteins (Materials and Methods). Our suspension cultures of LPL/LMF1-transfected cells allowed us to purify >20 mg of human LPL per L of cell culture medium, and insect cell systems for expressing GPIHBP1 allowed us to purify up to 4 mg of GPIHBP1 per L of medium. While protein production capabilities were an enormous boon for the current project, the key to our success was the discovery, by Mysling et al. (11), that LPL’s hydrolase domain is highly susceptible to spontaneous unfolding and that GPIHBP1 (and in particular GPIHBP1’s disordered acidic domain) prevents the unfolding. We suspect that the propensity of LPL’s hydrolase domain to undergo unfolding largely explains our own failures, despite considerable effort, to crystallize LPL alone. After generating and purifying LPL–GPIHBP1 complexes, we quickly obtained crystals that diffracted at a resolution of 2.8 Å.
In seeking to solve the structure of a LPL–GPIHBP1 complex, two of our key goals were to define the interactions of the two proteins and to glean insights into mechanisms by which GPIHBP1 stabilizes LPL structure and activity. For years, the issue of how LPL and GPIHBP1 interact has remained elusive, whereas the mechanisms for triglyceride hydrolysis by LPL could be inferred from structural studies of other lipases (38–40). Our crystal structure was extremely helpful in defining LPL–GPIHBP1 interactions. The structure showed that human LPL and human GPIHBP1 bind with 1:1 stoichiometry, substantiating results from our titration experiments with LPL and GPIHBP1 in solution. Also, the LPL–GPIHBP1 crystal structure revealed that GPIHBP1’s LU domain interacts, largely by hydrophobic contacts, with the C-terminal domain of LPL. All three fingers of GPIHBP1 and multiple loops in LPL’s C-terminal domain participate in the binding interface. It is predominantly the concave side of the central β-sheet of GPIHBP1’s LU domain that shapes the binding interface with LPL (Figs. 4 and 5). This binding architecture resembles that for other members of the LU protein domain family. The concave surface of the extracellular LU domain of activin receptors is responsible for ligand binding (41, 42), and in the case of the urokinase receptor, the concave surfaces of its three LU domains form a large hydrophobic cavity that binds urokinase with high affinity (43, 44). The crystallographic findings represent a significant improvement in understanding LPL–GPIHBP1 interactions and aligns well with earlier hydrogen–deuterium exchange–MS analyses (11). Our earlier efforts using alanine-scanning mutagenesis studies provided limited insights, simply because the vast majority of mutations that interfered with LPL–GPIHBP1 interactions did so by preventing proper protein folding (18, 36, 45, 46).
The crystal structure revealed that one side of LPL (∼90° to the side containing the catalytic pocket) contains a large, flat, and contiguous basic patch—extending across LPL’s hinge domain and involving large portions of LPL’s N- and C-terminal domains. For years, the field has recognized that LPL contains multiple positively charged heparin-binding motifs, and the relative importance of each of these motifs was a matter of considerable discussion (31–34). Our crystal structure revealed that all of LPL’s linear heparin-binding motifs merge into one contiguous basic patch in the tertiary structure. Importantly, GPIHBP1’s N-terminal acidic IDR, although not visualized in the electron density map, was positioned to project in the direction of LPL’s basic patch. The estimated length of GPIHBP1’s extended acidic IDR (>60 Å) (10) would easily allow it to interact with the entirety of LPL’s basic patch. We suspect that the interactions of GPIHBP1’s acidic domain with LPL’s basic patch, while transient in nature, serve to stabilize LPL and inhibit the unfolding of its hydrolase domain. We speculate that the acidic domain provides a continuous but highly dynamic “chaperoning” function for LPL, stabilizing the hinge domain and limiting the propensity of LPL’s N-terminal domain to collapse and unfold.
GPIHBP1 and colipase are both small, cysteine-rich proteins, and we had initially suspected that they might bind to similar sites on their partner lipase (LPL and PL, respectively). This suspicion was proven wrong; the binding sites for GPIHBP1 and colipase for their partner lipase (LPL and PL, respectively) are located far apart—almost on opposite sides of the lipase molecules. In hindsight, the distinct binding sites are probably not too surprising. While both GPIHBP1 and colipase have multiple fingers and five disulfide bonds, they do not belong to the same protein family and the disulfide bond arrangement is different. Also, the two proteins serve different functions. Colipase functions in lipid binding (30) and interacts directly with the lid covering PL’s catalytic pocket (21, 22). GPIHBP1 is not involved in lipid binding; its functions are to escort LPL across endothelial cells and stabilize LPL structure (7, 10, 11, 14, 47). Finally, the binding interactions of colipase and GPIHBP1 differ; colipase binds to PL by electrostatic interactions (21, 22, 30), whereas GPIHBP1 binding to LPL depends largely on hydrophobic contacts.
For many years, LPL has been assumed to form a head-to-tail homodimer (19, 48–50), and recent studies have proposed that homodimer formation is required for LPL secretion from cells (51). In line with these assumptions, our crystallographic unit contained two LPL–GPIHBP1 complexes arranged in a head-to-tail orientation. The two LPL molecules interacted, in a reciprocal fashion, between the Trp motif in the C-terminal domain of one LPL and the catalytic pocket in the N-terminal region of the partner LPL. SAXS studies on relatively dilute samples in solution validated this quaternary structure. In our crystal structure, LPL’s Trp motif, particularly residues 415–421, were not resolved in the electron density map, but our in silico model suggested that the Trp motif of one LPL interacts with the catalytic pocket of the partner LPL. The head-to-tail topology of LPL–LPL interactions that we observe in our crystal structure as well as in solution by SAXS is remarkable because the presumed lipid-binding Trp motif in one LPL occludes the catalytic pocket of the partner LPL. To allow entry of triglyceride substrates into LPL’s catalytic pocket, a certain degree of flexibility of the interacting regions is likely required. The poorly defined electron densities of the Trp-rich motif would be compatible with such flexibility. One possible scenario is that the LPL–LPL interactions that we observe in the crystal structure represent a transition state that occurs physiologically in settings where TRLs are absent (e.g., LPL transiting across the interstitial spaces). At this time, our presumption regarding protein flexibility in TRL binding and entry of lipid substrates into the catalytic site remains to be clarified.
Our LPL–GPIHBP1 structure provides fresh insights into human mutations causing chylomicronemia. A p.M404R LPL missense mutation, first identified in a Swedish patient with chylomicronemia, had been reported to abolish LPL secretion and activity (35). However, based on the location of M404 in the LPL–GPIHBP1 binding interface, we suspected that it might actually cause chylomicronemia by interfering with LPL–GPIHBP1 interactions. Indeed, the p.M404R mutation abolished the ability of LPL to bind to GPIHBP1 but had little or no effect on LPL secretion or activity. The LPL–GPIHBP1 crystal structure was also instrumental in defining the mechanism for chylomicronemia in patients carrying a p.D201V LPL mutation. The carboxylic acid side chain of D201 is critical for coordinating LPL’s calcium ion, and we suspected that disrupting calcium binding would destabilize LPL folding and prevent secretion from cells. Indeed, there was no secretion of LPL-D201V from cells, nor was there secretion of LPL with a mutation in another calcium-coordinating residue (p.D202E). These studies defined a molecular mechanism for human chylomicronemia, but the fact that a mutation in a calcium-coordinating residue would elicit disease is not unprecedented. Others have documented that a mutation in a calcium-coordinating residue in desmoglein 4 causes a hair abnormality in rats (52), and missense mutations involving putative calcium-coordinating residues in fibrillin-1 have been reported to cause Marfan syndrome (53). As we look to the future, we have little doubt that insights from the LPL–GPIHBP1 structure, combined with biochemical and biophysical assays, will make it possible to define mechanisms for other “chylomicronemia mutations”—and even define mechanisms by which common LPL polymorphisms affect plasma triglyceride levels (54). Also, we suspect that the LPL–GPIHBP1 structure will ultimately prove to be helpful in designing highly stable versions of LPL for enzyme replacement therapy.
Materials and Methods
Human LPL harboring an R324A mutation was expressed in CHO cells along with lipase maturation factor 1, and LPL was purified from the medium on a butyl-Sepharose column followed by heparin-Sepharose chromatography. Human GPIHBP1 was expressed in Sf9 cells and purified to homogeneity by several chromatography procedures. The LPL was mixed with GPIHBP1, and the LPL:GPIHBP1 complex was purified by size-exclusion chromatography. Crystals were obtained in 200 mM magnesium acetate and 17–22% polyethylene glycol 3350. Diffraction data were collected at the ID30B beamline at the European Synchrotron Radiation Facility (Grenoble, France). The structure of the LPL:GPIHBP1 complex was solved by molecular replacement using coordinates for the N- and C-terminal domains from horse pancreatic lipase (PDB ID code 1HPL). SAXS data on the LPL:GPIHBP1 complex were collected on the European Molecular Biology Laboratory (EMBL) P12 beamline of the storage ring PETRA III [Deutsches Elektronen-Synchrotron (DESY), Hamburg, Germany] and used to generate low-resolution shapes. To analyze the impact of LPL mutations, CHO cells were transfected with both wild-type and mutant versions of an LPL expression vector (12, 16–18, 36). LPL and GPIHBP1 in cell lysates and cell culture medium were analyzed by Western blotting and solid-phase immunoassays. Triglyceride hydrolase activity was measured with a [3H]triolein substrate, and esterase activity was measured with a 1,2-di-O-lauryl-rac-glycero-3-glutaric acid 6′-methylresorufin ester (DGGR) substrate. A detailed description of materials and methods can be found in SI Appendix.
Supplementary Material
Acknowledgments
We are grateful to the leadership of Shire US Drug Discovery for their support; to Kevin Holmes, John Gill, and Anna Tchoudakova for a human LPL-expressing cell line; Angela Norton, Bohong Zhang, and Leigh Low for their early contributions to the production of recombinant LPL; and to Shire’s Process Development Department for providing purified LPL used in the crystallography and SAXS experiments. X-ray diffraction data were collected at the ID30B beamline at the European Synchrotron Radiation Facility (Grenoble, France), and SAXS data were collected at the EMBL P12 beamline of the storage ring PETRA III (DESY, Hamburg, Germany). We acknowledge support from Fondation Leducq Transatlantic Network Grant 12CVD04 (to S.G.Y., A.P.B., L.G.F., and M.P.); National Institutes of Health Grants HL090553 (to S.G.Y.), HL087228 (to S.G.Y.), HL125335 (to S.G.Y.), and HL139725 (to S.G.Y.); Lundbeck Foundation Grant R230-2016-2930 (to K.K.K.); and NOVO Nordisk Foundation Grants NNF17OC0026868 (to M.P.) and NNF18OC0033864 (to M.P.). Research was partly funded by Shire US Drug Discovery.
Footnotes
Conflict of interest statement: B.D., B.S.-L., C.Q.P., and M.M. are employees of Shire and hold stock and stock options in Shire. R.Z. and S.G.Y. are coordinators on a Leducq Transatlantic Network Grant. They have not collaborated directly on this project.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6E7K) and the Small-Angle Scattering Biological Data Bank, www.sasbdb.org (SASBDB ID code SASDDM9).
See Commentary on page 1480.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817984116/-/DCSupplemental.
References
- 1.Korn ED. Clearing factor, a heparin-activated lipoprotein lipase. II. Substrate specificity and activation of coconut oil. J Biol Chem. 1955;215:15–26. [PubMed] [Google Scholar]
- 2.Korn ED. Clearing factor, a heparin-activated lipoprotein lipase. I. Isolation and characterization of the enzyme from normal rat heart. J Biol Chem. 1955;215:1–14. [PubMed] [Google Scholar]
- 3.Havel RJ. Triglyceride-rich lipoproteins and plasma lipid transport. Arterioscler Thromb Vasc Biol. 2010;30:9–19. doi: 10.1161/ATVBAHA.108.178756. [DOI] [PubMed] [Google Scholar]
- 4.Khera AV, et al. Myocardial Infarction Genetics Consortium; DiscovEHR Study Group; CARDIoGRAM Exome Consortium; Global Lipids Genetics Consortium Association of rare and common variation in the lipoprotein lipase gene with coronary artery disease. JAMA. 2017;317:937–946. doi: 10.1001/jama.2017.0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stitziel NO, et al. Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med. 2016;374:1134–1144. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beigneux AP, et al. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies BSJ, et al. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010;12:42–52. doi: 10.1016/j.cmet.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, et al. Crystal structure of the urokinase receptor in a ligand-free form. J Mol Biol. 2012;416:629–641. doi: 10.1016/j.jmb.2011.12.058. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Fedarovich A, Tomlinson S, Davies C. Crystal structure of CD59: Implications for molecular recognition of the complement proteins C8 and C9 in the membrane-attack complex. Acta Crystallogr D Biol Crystallogr. 2007;63:714–721. doi: 10.1107/S0907444907015557. [DOI] [PubMed] [Google Scholar]
- 10.Kristensen KK, et al. A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proc Natl Acad Sci USA. 2018;115:E6020–E6029. doi: 10.1073/pnas.1806774115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mysling S, et al. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife. 2016;5:e12095. doi: 10.7554/eLife.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gin P, et al. Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler Thromb Vasc Biol. 2011;31:176–182. doi: 10.1161/ATVBAHA.110.214718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulbourne CN, et al. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 2014;19:849–860. doi: 10.1016/j.cmet.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mysling S, et al. The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife. 2016;5:e20958. doi: 10.7554/eLife.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young SG, Zechner R. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 2013;27:459–484. doi: 10.1101/gad.209296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong LG, et al. GPIHBP1 and plasma triglyceride metabolism. Trends Endocrinol Metab. 2016;27:455–469. doi: 10.1016/j.tem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voss CV, et al. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci USA. 2011;108:7980–7984. doi: 10.1073/pnas.1100992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plengpanich W, et al. Multimerization of glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) and familial chylomicronemia from a serine-to-cysteine substitution in GPIHBP1 Ly6 domain. J Biol Chem. 2014;289:19491–19499. doi: 10.1074/jbc.M114.558528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi Y, Nakajima T, Inoue I. Molecular modeling of the dimeric structure of human lipoprotein lipase and functional studies of the carboxyl-terminal domain. Eur J Biochem. 2002;269:4701–4710. doi: 10.1046/j.1432-1033.2002.03179.x. [DOI] [PubMed] [Google Scholar]
- 20.Winkler FK, D’Arcy A, Hunziker W. Structure of human pancreatic lipase. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 21.van Tilbeurgh H, Sarda L, Verger R, Cambillau C. Structure of the pancreatic lipase-procolipase complex. Nature. 1992;359:159–162. doi: 10.1038/359159a0. [DOI] [PubMed] [Google Scholar]
- 22.Egloff MP, Sarda L, Verger R, Cambillau C, van Tilbeurgh H. Crystallographic study of the structure of colipase and of the interaction with pancreatic lipase. Protein Sci. 1995;4:44–57. doi: 10.1002/pro.5560040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore SA, et al. The structure of truncated recombinant human bile salt-stimulated lipase reveals bile salt-independent conformational flexibility at the active-site loop and provides insights into heparin binding. J Mol Biol. 2001;312:511–523. doi: 10.1006/jmbi.2001.4979. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Wang CS, Tang J, Dyda F, Zhang XC. The crystal structure of bovine bile salt activated lipase: Insights into the bile salt activation mechanism. Structure. 1997;5:1209–1218. doi: 10.1016/s0969-2126(97)00271-2. [DOI] [PubMed] [Google Scholar]
- 25.Hermoso J, et al. Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex. J Biol Chem. 1996;271:18007–18016. doi: 10.1074/jbc.271.30.18007. [DOI] [PubMed] [Google Scholar]
- 26.van Tilbeurgh H, et al. Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography. Nature. 1993;362:814–820. doi: 10.1038/362814a0. [DOI] [PubMed] [Google Scholar]
- 27.Lookene A, Groot NB, Kastelein JJ, Olivecrona G, Bruin T. Mutation of tryptophan residues in lipoprotein lipase. Effects on stability, immunoreactivity, and catalytic properties. J Biol Chem. 1997;272:766–772. doi: 10.1074/jbc.272.2.766. [DOI] [PubMed] [Google Scholar]
- 28.Jacobson MP, et al. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 29.Dyson HJ, Wright PE, Scheraga HA. The role of hydrophobic interactions in initiation and propagation of protein folding. Proc Natl Acad Sci USA. 2006;103:13057–13061. doi: 10.1073/pnas.0605504103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe ME. Structure and function of pancreatic lipase and colipase. Annu Rev Nutr. 1997;17:141–158. doi: 10.1146/annurev.nutr.17.1.141. [DOI] [PubMed] [Google Scholar]
- 31.Hata A, et al. Binding of lipoprotein lipase to heparin. Identification of five critical residues in two distinct segments of the amino-terminal domain. J Biol Chem. 1993;268:8447–8457. [PubMed] [Google Scholar]
- 32.Hill JS, et al. Subdomain chimeras of hepatic lipase and lipoprotein lipase. Localization of heparin and cofactor binding. J Biol Chem. 1998;273:30979–30984. doi: 10.1074/jbc.273.47.30979. [DOI] [PubMed] [Google Scholar]
- 33.Sendak RA, Melford K, Kao A, Bensadoun A. Identification of the epitope of a monoclonal antibody that inhibits heparin binding of lipoprotein lipase: New evidence for a carboxyl-terminal heparin-binding domain. J Lipid Res. 1998;39:633–646. [PubMed] [Google Scholar]
- 34.Ma Y, et al. Mutagenesis in four candidate heparin binding regions (residues 279–282, 291–304, 390–393, and 439–448) and identification of residues affecting heparin binding of human lipoprotein lipase. J Lipid Res. 1994;35:2049–2059. [PubMed] [Google Scholar]
- 35.Pingitore P, et al. Identification and characterization of two novel mutations in the LPL gene causing type I hyperlipoproteinemia. J Clin Lipidol. 2016;10:816–823. doi: 10.1016/j.jacl.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Beigneux AP, et al. GPIHBP1 missense mutations often cause multimerization of GPIHBP1 and thereby prevent lipoprotein lipase binding. Circ Res. 2015;116:624–632. doi: 10.1161/CIRCRESAHA.116.305085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abifadel M, et al. Identification of the first Lebanese mutation in the LPL gene and description of a rapid detection method. Clin Genet. 2004;65:158–161. doi: 10.1111/j.0009-9163.2004.00205.x. [DOI] [PubMed] [Google Scholar]
- 38.Khan FI, et al. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front Bioeng Biotechnol. 2017;5:16. doi: 10.3389/fbioe.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brzozowski AM, et al. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature. 1991;351:491–494. doi: 10.1038/351491a0. [DOI] [PubMed] [Google Scholar]
- 40.Kim KK, Song HK, Shin DH, Hwang KY, Suh SW. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure. 1997;5:173–185. doi: 10.1016/s0969-2126(97)00177-9. [DOI] [PubMed] [Google Scholar]
- 41.Greenwald J, et al. A flexible activin explains the membrane-dependent cooperative assembly of TGF-beta family receptors. Mol Cell. 2004;15:485–489. doi: 10.1016/j.molcel.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Townson SA, et al. Specificity and structure of a high affinity activin receptor-like kinase 1 (ALK1) signaling complex. J Biol Chem. 2012;287:27313–27325. doi: 10.1074/jbc.M112.377960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin L, Gårdsvoll H, Huai Q, Huang M, Ploug M. Structure-based engineering of species selectivity in the interaction between urokinase and its receptor: Implication for preclinical cancer therapy. J Biol Chem. 2010;285:10982–10992. doi: 10.1074/jbc.M109.093492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gårdsvoll H, et al. Characterization of the functional epitope on the urokinase receptor. Complete alanine scanning mutagenesis supplemented by chemical cross-linking. J Biol Chem. 2006;281:19260–19272. doi: 10.1074/jbc.M513583200. [DOI] [PubMed] [Google Scholar]
- 45.Beigneux AP, et al. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J Biol Chem. 2009;284:30240–30247. doi: 10.1074/jbc.M109.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beigneux AP, et al. Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J Biol Chem. 2011;286:19735–19743. doi: 10.1074/jbc.M111.242024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies BS, et al. Assessing mechanisms of GPIHBP1 and lipoprotein lipase movement across endothelial cells. J Lipid Res. 2012;53:2690–2697. doi: 10.1194/jlr.M031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osborne JC, Jr, Bengtsson-Olivecrona G, Lee NS, Olivecrona T. Studies on inactivation of lipoprotein lipase: Role of the dimer to monomer dissociation. Biochemistry. 1985;24:5606–5611. doi: 10.1021/bi00341a048. [DOI] [PubMed] [Google Scholar]
- 49.Olivecrona T, Bengtsson-Olivecrona G, Osborne JC, Jr, Kempner ES. Molecular size of bovine lipoprotein lipase as determined by radiation inactivation. J Biol Chem. 1985;260:6888–6891. [PubMed] [Google Scholar]
- 50.Wong H, et al. A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc Natl Acad Sci USA. 1997;94:5594–5598. doi: 10.1073/pnas.94.11.5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doolittle MH, Ehrhardt N, Péterfy M. Lipase maturation factor 1: Structure and role in lipase folding and assembly. Curr Opin Lipidol. 2010;21:198–203. doi: 10.1097/MOL.0b013e32833854c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jahoda CA, et al. The lanceolate hair rat phenotype results from a missense mutation in a calcium coordinating site of the desmoglein 4 gene. Genomics. 2004;83:747–756. doi: 10.1016/j.ygeno.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Handford P, et al. The calcium binding properties and molecular organization of epidermal growth factor-like domains in human fibrillin-1. J Biol Chem. 1995;270:6751–6756. doi: 10.1074/jbc.270.12.6751. [DOI] [PubMed] [Google Scholar]
- 54.Sagoo GS, et al. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: A HuGE association review and meta-analysis. Am J Epidemiol. 2008;168:1233–1246. doi: 10.1093/aje/kwn235. [DOI] [PubMed] [Google Scholar]
- 55.Llinas P, et al. Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide. EMBO J. 2005;24:1655–1663. doi: 10.1038/sj.emboj.7600635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, et al. DelPhi: A comprehensive suite for DelPhi software and associated resources. BMC Biophys. 2012;5:9. doi: 10.1186/2046-1682-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.