Abstract
Rice stripe virus (RSV) is one of the most devastating viral pathogens in rice and can also cause the general chlorosis symptom in Nicotiana benthamiana plants. The chloroplast changes associated with chlorosis symptom suggest that RSV interrupts normal chloroplast functions. Although the change of proteins of the whole cell or inside the chloroplast in response to RSV infection have been revealed by proteomics, the mechanisms resulted in chloroplast-related symptoms and the crucial factors remain to be elucidated. RSV infection caused the malformation of chloroplast structure and a global reduction of chloroplast membrane protein complexes in N. benthamiana plants. Here, both the protoplast proteome and the chloroplast proteome were acquired simultaneously upon RSV infection, and the proteins in each fraction were analyzed. In the protoplasts, 1128 proteins were identified, among which 494 proteins presented significant changes during RSV; meanwhile, 659 proteins were identified from the chloroplasts, and 279 of these chloroplast proteins presented significant change. According to the label-free LC–MS/MS data, 66 nucleus-encoded chloroplast-related proteins (ChRPs), which only reduced in chloroplast but not in the whole protoplast, were identified, indicating that these nuclear-encoded ChRPswere not transported to chloroplasts during RSV infection. Gene ontology (GO) enrichment analysis confirmed that RSV infection changed the biological process of protein targeting to chloroplast, where 3 crucial ChRPs (K4CSN4, K4CR23, and K4BXN9) were involved in the regulation of protein targeting into chloroplast. In addition to these 3 proteins, 41 among the 63 candidate proteins were characterized to have chloroplast transit peptides. These results indicated that RSV infection changed the biological process of protein targeting into chloroplast and the location of ChRPs through crucial protein factors, which illuminated a new layer of RSV–host interaction that might contribute to the symptom development.
Keywords: chloroplast proteomics, rice stripe virus, nuclear/nucleus-encoded chloroplast-related protein, plant–virus interaction, chloroplast targeting
1. Introduction
1.1. Plant Virus and Chloroplast
Plant viruses are obligatory intracellular parasites and normally encode relatively fewer proteins compared to other types of pathogens. Thus, the propagation and spreading of plant viruses are exclusively dependent on host cellular machinery and metabolism [1,2,3,4]. Viral infections usually cause plant chlorosis, stunting, necrosis, or other symptoms [5,6]. Leaf chlorosis, being the most common viral symptom, implies reduced photosynthetic activity of chloroplasts [7,8]. In addition, the multiple changes associated with the symptoms of chlorosis, such as reduced chlorophyll pigmentation [9,10], changes in chloroplast structures and functions [11,12], repressed expression of nuclear-encoded chloroplast photosynthesis-related genes (ChRGs) [5,13,14], as well as metabolism accumulation of nitric oxide [15], suggest essential interactions between virus and the chloroplast [16].
Many studies indicated that virus infection always caused changes in chloroplast structure, components, and expression of ChRGs, which is assumed to be responsible for the development of viral symptoms [17,18,19,20,21,22]. For instance, there were chloroplasts with stroma vacuolization, rearrangement of the thylakoids, and cytoplasmic invagination in barley leaf cells infected with barley stripe mosaic virus (BSMV) [23]. The helper component-proteinase (HC-Pro) of potato virus Y (PVY) can interact with the chloroplast ATP synthase NtCF1β-subunit, leading to a decrease of ATP synthase complex in both the HC-Pro transgenic and the PVY-infected tobacco, suggesting that HC-Pro is involved in reducing the photosynthetic rate of PVY-infected plants [24]. The tobacco mosaic virus (TMV) coat protein (CP) can interact with the chloroplast ferredoxin I (Fd I) [25]. The chlorotic phenotype and the accumulation of the TMV CPs in the chloroplasts were enhanced by the decrease of Fd I in TMV-infected tobacco leaves, suggesting that the interaction of CP and Fd I may contribute to the occurrence of chlorosis and mosaic symptoms [26].
Nowadays, increasing studies revealed that the chloroplast, as well as its factors, can interact with or be the targets of viral components or factors to favor the replication, movement, and symptom development of viruses [16,27]. For example, the PsbP (photosystem II oxygen-evolving complex protein) interacts with alfalfa mosaic virus (AMV) CP, and its overexpression observably hindered virus replication [28]. The RuBisCO small subunit could be hijacked by the tobamovirus movement proteins to facilitate the intercellular movement of virus [29,30]. The 3′-UTR region of bamboo mosaic virus (BaMV) interacts with chloroplast phosphoglycerate kinase (CpGK). Silencing or mislocalization of the cPGK gene in N. benthamiana reduced BaMV accumulation [31]. Alternanthera mosaic virus (AltMV) RNA interference (RNAi) suppressor TGB1L88 specifically binds to the chloroplast β-ATPase (CF1β). Silencing of CF1β gene induced severe tissue necrosis in AltMV-infected N. benthamiana plants, suggesting that the interaction of CF1β and TGB1L88 weakened severe symptoms caused by AltMV infection [32]. These studies implied that the interactions of chloroplasts and plant viruses are commonly related to the formation of chlorosis symptoms during virus infection.
1.2. RSV Infection and Chloroplast Proteomics
RSV is one of the most devastating rice pathogens causing dramatic losses in rice production in East Asia [33,34]. RSV is the typical member of the genus Tenuivirus and is transmitted by the small brown plant-hopper (Laodelphax striatellus Fallén) in a persistent and circulative-propagative manner [35]. RSV-infected rice leaves initially appear white or contain yellow spots, which develop into white or yellow stripes, then the RSV-infected leaves become yellow, with necrotic areas magnified from the stripes and, finally, lead to death of the whole leaf [33,36]. It was suggested that the chlorosis symptom was caused by disruption of normal chloroplast components and functions [17,18,22]. For instance, the downregulation of ChRG genes in RSV-infected rice plants is assumed to be responsible for chlorosis of RSV symptoms [6]. Quantitative proteomics analysis of rice leaves reveal that the accumulation of 30 ChRPs was apparently reduced by RSV infection [37]. However, how the localization and function of these ChRPs were modulated during RSV infection remains obscure. So far, there is no special methodology to reveal how the localization change of the nucleus-encoded ChRP factors is affected by RSV infection at the proteomic level. In recent years, mass spectrometry-based proteomics has developed rapidly as a useful approach for the identification of all the host proteins that play roles in virus infections [17,38]. Label-free quantitative proteomics technique is growing in popularity due to faster, cleaner, and simpler mass spectrometry results with higher technical reproducibility [38,39]. In this study, we used a label-free quantitative proteomics approach to analyze protein accumulation profiles of protoplast and chloroplast from RSV-infected leaves. By combining the quantified protoplast and chloroplast proteomic data, we established a method to elucidate the localization change of nucleus-encoded ChRPs and indicate a new layer of RSV–host interaction where the targeting of nucleus-encoded ChRPs is hindered during RSV infection.
1.3. RSV Infection of Nicotiana benthamiana Plants
In nature, RSV mainly infects rice plants and many other different species of plants in the family Poaceae, including wheat (Triticum aestivum L.) and maize (Zea mays L.) [40,41]. RSV-infected rice plants exhibit common features, such as chlorotic and necrotic stripes in leaves [42]. The chlorotic and necrotic stripes appear on a few areas of the leaves at first, then the chlorotic areas enlarge and show necrosis along leaf veins, and eventually lead to plant wilting [6,43]. The development of viral symptoms suggested that the RSV infection affects the mesophyll cells unevenly in different areas in rice leaves. Therefore, rice leaves with heterogeneous symptoms of RSV infection are not suitable for explaining the general effects of the virus on chloroplast proteins, or for isolating chloroplasts and protoplasts affected by virus infection. As a susceptible host, N. benthamiana could be systemically infected by RSV and showed symptoms of unanimous chlorosis [44], which makes N. benthamiana suitable for isolating chloroplasts and protoplasts affected by virus infection and explaining the general effects on chloroplast proteins caused by RSV infection. Thus, we used N. benthamiana as the host for RSV infection to provide insights into the influence on nucleus-encoded ChRP proteins.
2. Results
2.1. The Phenotype and the Ultrastructure Change of Chloroplasts in RSV-Infected N. benthamiana Plants
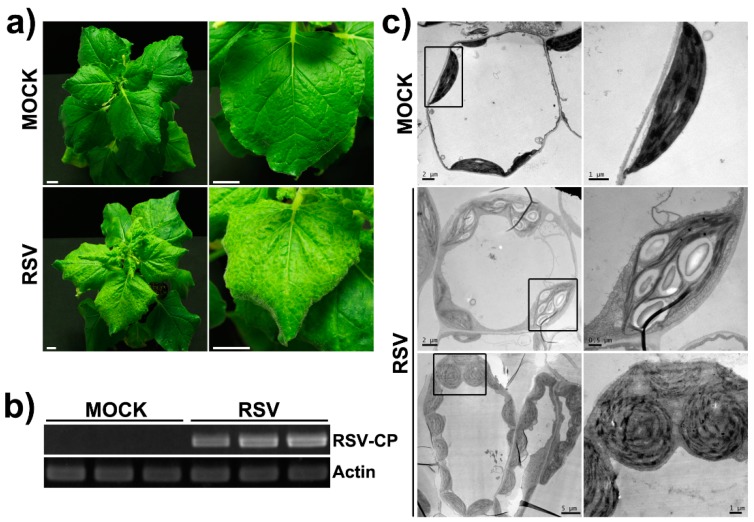
N. benthamiana plants infected with RSV began to show mild yellowing symptoms in systemic leaves at 12–15 days post-inoculation (dpi). At 20–25 dpi, RSV-infected systemic leaves exhibited distinct symptoms such as backward curling, yellow vein, and unanimous chlorosis in the whole leaf area (Figure 1a). Systemic spreading of virus was confirmed by reverse transcription PCR (RT-PCR) using specific primers to the CP gene of RSV (Figure 1b). The yellowing symptoms indicated the abnormality of the chloroplast in N. benthamiana leaf cells. By electron microscopy, the ultrastructure of chloroplasts in RSV-infected N. benthamiana plants revealed dramatic malformations. There were swollen or globular chloroplasts, enlarged and over-accumulated starch grains inside chloroplasts, and a reduction or irregular arrangement of stroma lamella (Figure 1c).
Figure 1.
Influence of rice stripe virus (RSV) infection on N. benthamiana plants and the ultrastructure of chloroplasts from N. benthamiana leaves. (a) Phenotype of RSV-infected N. benthamiana plants at 25 dpi. (b) Viral RNA accumulation in mock plants and RSV-infected plants were analyzed by RT-PCR. (c) Ultrastructure of chloroplasts from RSV-infected N. benthamiana plants. Right panels are the magnified images of the line-boxed area in the left panels, respectively. MOCK, the mock-inoculated healthy N. benthamiana plants; RSV, the RSV-infected N. benthamiana plants.
In addition, we examined the ultrastructure of chloroplast in RSV-infected Oryza sativa plants. The ultrastructure of the chloroplasts from RSV-infected rice leaves also became abnormal, such as enlarged starch grains, exaggeration of electron-dense granules, disappearance of grana stacks, altered grana with dilated thylakoids scattering into the cytoplasm, appearance of an inter-membranous sac, and broken chloroplast envelopes with disrupted membrane (Supplemental Figure S1b). RT-PCR showed that RSV was accumulated in the symptomatic leaves of the RSV-infected O. sativa plants (Supplemental Figure S1a). These results suggest that the structure of chloroplasts became abnormal in RSV-infected plant leaves.
2.2. Blue Native PAGE (BN-PAGE) of Chloroplast Membrane Complexes of RSV-Infected Plants
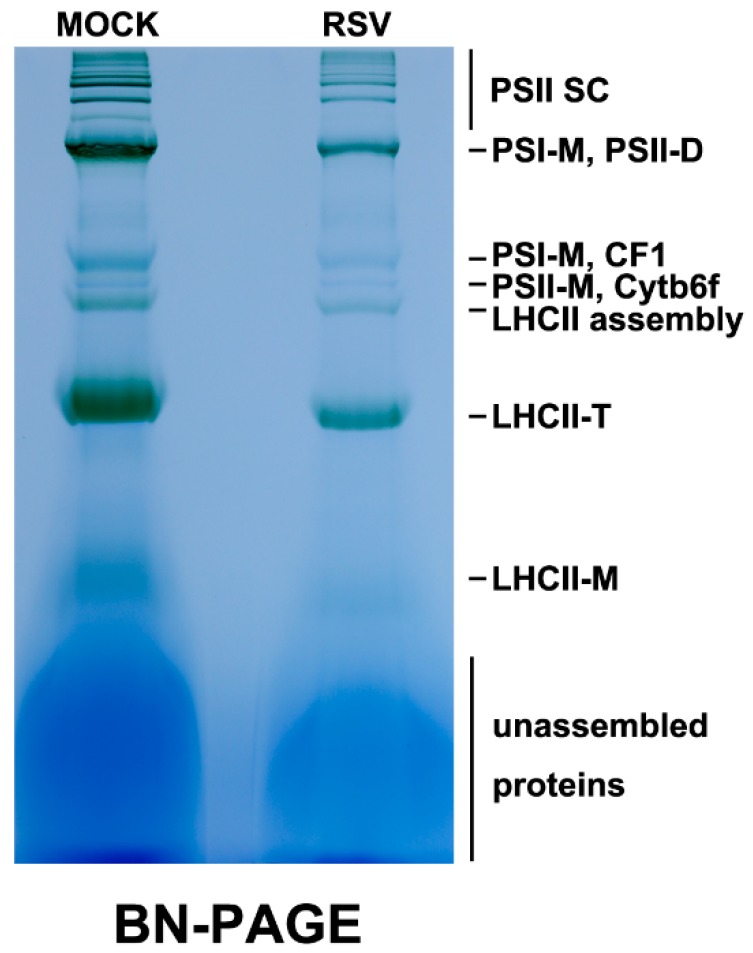
Since the structure of the chloroplasts in RSV-infected N. benthamiana leaves became abnormal, we investigated the components and the quantity of the chloroplast membrane protein complexes in RSV-infected plants. The protein complexes of chloroplast membrane isolated from healthy leaves (MOCK) and RSV-infected leaves (RSV) were analyzed by blue native PAGE (Figure 2). As illustrated in Figure 2, a global reduction of the chloroplast membrane proteins was observed after RSV infection. PSI-M and PSII-D significantly decreased the most among these complexes. These results indicated that RSV infection reduced the abundance and the constitution of the chloroplast membrane protein complexes in N. benthamiana plants, which implied that the whole constitution of the chloroplast proteins and their routine functions were highly perturbed due to RSV infection. Thus, it is assumed that RSV infection might impose influence on the transport of chloroplast proteins.
Figure 2.
Blue native PAGE of chloroplast membrane protein complexes isolated from an equal quantity of healthy leaves (MOCK) and RSV-infected leaves (RSV). MOCK, the mock-inoculated healthy N. benthamiana leaves; RSV, the RSV-infected N. benthamiana leaves. PSII-SC, PSII supercomplex; PSI-M, PSI monomer; PSII-D, PSII dimer; LHCII, PSII light-harvesting complex; LHCII-T, PSII light-harvesting complex trimer; LHCII-M, PSII light-harvesting complex monomer; Cytb6f, cytochrome b6/f complexes.
2.3. Western Blot Analysis of the Specificity of Chloroplast Protein Extracts
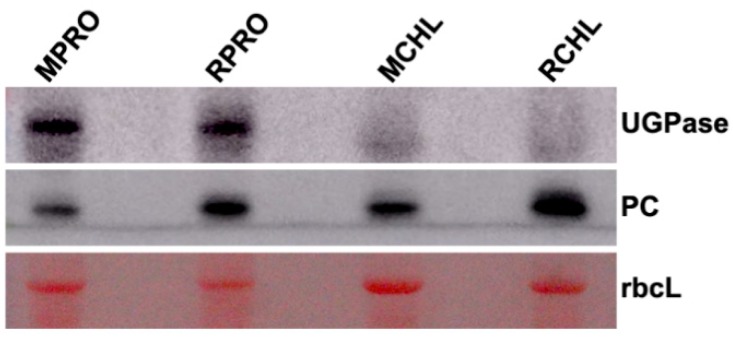
In order to investigate the nuclear-encoded ChRPs, whose process of transporting into chloroplast was influenced by RSV infection, we established a novel protoplast–chloroplast proteomic approach to screen them. The proteins of protoplast and chloroplast were extracted from mock-inoculated healthy leaves and RSV-infected N. benthamiana leaves, respectively. In order to assess the specificity of the chloroplast fraction, the marker proteins of protoplast and chloroplast were detected by WB. UDP-glucose pyrophosphorylase (UGPase) and plastocyanin (PC) were used as cytoplasmic and chloroplast markers, respectively. The WB results showed that the fraction of chloroplast did not contain UGPase, which was a specific marker in cytoplasm (Figure 3), suggesting that the chloroplast fraction did not contain cytoplasmic components. Then, the proteins extracted from the protoplasts and chloroplasts were analyzed by label-free LC–MS/MS, in hopes of finding the nucleus-encoded ChRPs, to which the process of transporting to chloroplasts was hindered by RSV infection.
Figure 3.
Analysis of the specificity of the chloroplast fraction by Western blot. MPRO, protein extracts of protoplasts from mock-inoculated healthy leaves; RPRO, protein extracts of protoplasts from RSV-infected leaves; MCHL, protein extracts of chloroplasts isolated from protoplasts from mock-inoculated healthy leaves; RCHL, protein extracts of chloroplasts isolated from protoplasts from RSV-infected leaves. UGPase, UDP-glucose pyrophosphorylase (cytoplasm marker); PC, plastocyanin (chloroplast marker); rbcL, RuBisCO large subunit.
2.4. Analysis of Protoplast and Chloroplast Proteomics by Label-Free LC–MS/MS
Total proteins were extracted from three independent biological replicates of protoplasts and chloroplasts from either the mock-inoculated healthy or the RSV-infected leaves and subjected to nano LC–MS/MS (Supplemental Figure S2). The changes of proteins in chloroplasts and protoplasts were investigated by comparative proteomic analysis combined with label-free quantification. A total of 5640 MS/MS spectra were obtained from the protoplast samples by LC–MS/MS. The acquired MS/MS spectra were subsequently analyzed by Spectronaut 10 and searched against the tomato UniProt Fasta database, the RSV proteins, and the Biognosys iRT peptide sequences, where 4668 unique peptides were assigned (Supplemental Table S1) and represented 1128 identified proteins (Supplemental Table S2). The average value of median CVs from healthy and RSV-infected protoplast samples were 19.9% and 16.2%, respectively. Totally, 1128 proteins identified from protoplast samples were taken for protein abundance comparisons between RSV-infected and the mock-inoculated control N. benthamiana leaf samples. A total of 494 proteins showed a significant difference in abundance (p < 0.05, Supplemental Table S3), and a heat map of these proteins was obtained with a fold change (FC) of at least ±0.58 (Supplemental Figure S3). A ratio of protein abundance value above 2, in RSV-infected protoplast samples compared to that of the mock-inoculated protoplast samples (RPRO/MPRO), indicated an increase in abundance, 0.5–2 indicated no abundance change, less than 0.5 indicated a decrease in abundance.
In terms of the chloroplast samples, there were 3699 MS/MS spectra and 2876 unique peptides (Supplemental Table S4), representing 659 identified proteins (Supplemental Table S5). The average value of median CVs from the mock-inoculated healthy chloroplast samples and the RSV-infected chloroplast samples were 27% and 51.5%, respectively. A total of 279 chloroplast proteins showed significant difference in abundance upon RSV infection (p < 0.05, Supplemental Table S6), and a heat map of these proteins was obtained with an FC of at least ±0.58 (Supplemental Figure S4). The ratio of the abundance in value of RCHL/MCHL above 2 indicates increased accumulation, 0.5–2 indicates little change, and less than 0.5 indicates decreased accumulation.
To further confirm the quality of the sample collection, we examined the accumulation of PC, UGPase, and fibrillarin as markers for chloroplast, protoplast, and nucleus, respectively, in the identified proteins from protoplast and chloroplast samples. It was found that PC existed in both protoplast and chloroplast samples, and UGPase and fibrillarin only existed in the protoplast. It was obvious that the chloroplast fractions were not contaminated by cytoplasm and nuclear components, which was consistent with previous WB results.
2.5. GO Enrichment Analysis of the Identified Candidate Proteins
Based on the protoplast–chloroplast proteomics, a selection filter, where the abundance of candidates only decreased in the RSV-infected chloroplast samples but not in RSV-infected protoplast samples, was used to screen the nucleus-encoded ChRPs which did not transport into chloroplasts during RSV infection. The chloroplast targeting of the candidate proteins were reconfirmed in UniProt, N. benthamiana genome, and Arabidopsis Chloroplast Target gene databases [45,46,47,48,49]. A total number of 66 nucleus-encoded ChRPs which did not transport into chloroplasts after RSV infection were obtained, as shown in Table 1.
Table 1.
The candidate proteins which the process transported to chloroplasts were blocked by RSV infection.
| UniProt AC | Protein Names | GO IDs |
|---|---|---|
| K4C8H3 | Uncharacterized protein | GO:0003842; GO:0004028; GO:0004029; GO:0005739; GO:0008270; GO:0009507; GO:0009651; GO:0010133; GO:0050897; GO:0072593 |
| Q711R1 | Allene oxide cyclase | GO:0009507; GO:0046423 |
| K4B2I9 | Uncharacterized protein | GO:0005524; GO:0006457 |
| K4BT58 | Uncharacterized protein | GO:0005509; GO:0009534; GO:0009570; GO:0009654; GO:0015979; GO:0019898; GO:0031977 |
| K4B778 | Uncharacterized protein | GO:0005741; GO:0008308 |
| A8UDS9 | Copper/zinc superoxide dismutase | GO:0006801; GO:0046872; GO:0055114 |
| A9CM21 | Voltage-dependent anion channel | GO:0005741; GO:0008308 |
| A0A075F1U0 | Constitutive plastid-lipid associated protein | |
| K4C647 | Uncharacterized protein | GO:0009570; GO:0009941; GO:0016491 |
| K4CNE8 | ATP-dependent Clp protease proteolytic subunit | GO:0000302; GO:0004252; GO:0009570; GO:0009941; GO:0010468 |
| K4B858 | Uncharacterized protein | GO:0005622 |
| Q10712 | Leucine aminopeptidase 1 | GO:0004177; GO:0008235; GO:0009507; GO:0030145 |
| K4B3J8 | Uncharacterized protein | GO:0008836; GO:0009089 |
| K4ASX2 | Uncharacterized protein | GO:0005623; GO:0045454; GO:0051920 |
| P37222 | NADP-dependent malic enzyme | GO:0004471; GO:0004473; GO:0006108; GO:0008948; GO:0009507; GO:0046872; GO:0051287 |
| A0A140G1U2 | ATP-dependent Clp protease proteolytic subunit | GO:0004252; GO:0009536 |
| K4AZT2 | Uncharacterized protein | GO:0009579; GO:0016021 |
| K4DC13 | Uncharacterized protein | GO:0005524; GO:0005759; GO:0006986; GO:0009507; GO:0046872; GO:0046914; GO:0051082; GO:0051085; GO:0051087; GO:1901671 |
| K4BXN9 | Uncharacterized protein | GO:0005524; GO:0005739; GO:0005774; GO:0006457; GO:0009408; GO:0009414; GO:0009570; GO:0009651; GO:0009704; GO:0009941; GO:0010157; GO:0045037 |
| K4D4L5 | Uncharacterized protein | GO:0005886; GO:0016655 |
| K4C841 | Uncharacterized protein | GO:0008152; GO:0016597 |
| K4BFT9 | Uncharacterized protein | GO:0016021 |
| K4B413 | Uncharacterized protein | GO:0000302; GO:0004130; GO:0005739; GO:0005774; GO:0005777; GO:0009506; GO:0009507; GO:0009735; GO:0009941; GO:0016021; GO:0020037; GO:0034599; GO:0042744 |
| K4BBZ1 | Uncharacterized protein | |
| A7U630 | Chloroplast methionine sulfoxide reductase B1 | GO:0006979; GO:0030091; GO:0033743 |
| K4CH79 | Uncharacterized protein | GO:0003735; GO:0005840; GO:0006412; GO:0009570; GO:0009941; GO:0032544 |
| Q8W3Z8 | Plastidic phosphoglucomutase | GO:0005975; GO:0016868 |
| K4CXW3 | Glutamyl-tRNA (Gln) amidotransferase subunit A | GO:0004040; GO:0005524; GO:0005739; GO:0009570; GO:0030956; GO:0032543; GO:0050567; GO:0070681 |
| K4BCV6 | Uncharacterized protein | GO:0004222; GO:0006518; GO:0009507; GO:0046872 |
| K4BCU7 | Uncharacterized protein | GO:0004747; GO:0006014 |
| K4CGD5 | Uncharacterized protein | GO:0003743; GO:0003924; GO:0005525; GO:0005622; GO:0006412; GO:0009570; GO:0009941 |
| K4BXX3 | Alpha-1,4 glucan phosphorylase | GO:0005737; GO:0005980; GO:0008184; GO:0030170 |
| K4BLP5 | Uncharacterized protein | |
| E5LBC4 | Prephenate aminotransferase | GO:0008483; GO:0009058; GO:0030170 |
| Q6T7F1 | Phosphoribosylaminoimidazole carboxylase | GO:0004638; GO:0005524; GO:0006189; GO:0046872 |
| K4C5B9 | Uroporphyrinogen decarboxylase | GO:0004853; GO:0005829; GO:0006782; GO:0006783; GO:0009570; GO:0009735; GO:0009941 |
| K4BJL2 | Uncharacterized protein | GO:0003735; GO:0005840; GO:0006412 |
| K4CPX9 | Uncharacterized protein | GO:0000027; GO:0003735; GO:0005762; GO:0006412; GO:0019843 |
| K4ASZ2 | Uncharacterized protein | GO:0003824; GO:0009058; GO:0030170 |
| K4BDV0 | Uncharacterized protein | GO:0000103; GO:0004020; GO:0004781 |
| K4ASJ9 | Uncharacterized protein | GO:0009535; GO:0009941 |
| K4BEB0 | Uncharacterized protein | GO:0000287; GO:0004614; GO:0005829; GO:0005978; GO:0006006; GO:0009570; GO:0009590; GO:0010319; GO:0019252; GO:0019388 |
| K4B075 | Uncharacterized protein | GO:0003723; GO:0004827; GO:0005524; GO:0005737; GO:0005739; GO:0006433; GO:0009553; GO:0009570; GO:0010109; GO:0017101; GO:0048316; GO:0048481 |
| K4BX77 | 3-isopropylmalate dehydrogenase | GO:0000287; GO:0003862; GO:0009098; GO:0009570; GO:0051287 |
| K4CJD3 | Uncharacterized protein | |
| A8UDS7 | ABA2 | GO:0016491; GO:0071949 |
| Q2MI78 | 30S ribosomal protein S18 | GO:0003735; GO:0006412; GO:0009507; GO:0019843; GO:0022627 |
| K4CR23 | Uncharacterized protein | GO:0003924; GO:0005525; GO:0006614; GO:0008312; GO:0009570; GO:0080085 |
| K4CNE7 | Uncharacterized protein | GO:0009535; GO:0022891; GO:0055085 |
| K4D5G2 | Uncharacterized protein | GO:0000287; GO:0005737; GO:0009570; GO:0009941; GO:0016791 |
| K4BU13 | Uncharacterized protein | GO:0000166; GO:0003729; GO:0009570 |
| K4BY59 | Uncharacterized protein | GO:0003824; GO:0009058; GO:0030170 |
| Q84RD7 | ZIP | GO:0015979; GO:0015995; GO:0046872; GO:0048529 |
| K4BPB0 | Uncharacterized protein | GO:0003735; GO:0006412; GO:0015934; GO:0022626; GO:0042254 |
| K4BXB1 | Uncharacterized protein | GO:0003735; GO:0006412; GO:0009570; GO:0009735; GO:0009941; GO:0015934 |
| K4CSN4 | Uncharacterized protein | GO:0009658; GO:0009941; GO:0016021; GO:0045037 |
| K4B2B2 | Phospho-2-dehydro-3-deoxyheptonate aldolase | GO:0003849; GO:0009073; GO:0009507 |
| A0A0H3U3I7 | Geranylgeranyl pyrophosphate synthase 5 | GO:0008299; GO:0016740 |
| K4AXX7 | Protein translocase subunit SecA | GO:0005524; GO:0006605; GO:0009658; GO:0009941; GO:0010109; GO:0015462; GO:0016020; GO:0017038 |
| K4C9K5 | Uncharacterized protein | GO:0006457; GO:0015031 |
| Q0MW94 | 1-deoxy-d-xylulose-5-phosphate reductoisomerase | GO:0008299; GO:0016853; GO:0030604; GO:0046872; GO:0070402 |
| K4BDU3 | Uncharacterized protein | GO:0009055; GO:0051536 |
| K9JIM2 | Heat shock protein 101 | GO:0005524 |
| K4BVZ1 | Uncharacterized protein | GO:0016021 |
| A0A0K1D9E3 | Granule-bound starch synthase I | |
| K4CHH4 | Uncharacterized protein | GO:0006457; GO:0015031 |
The 66 candidate proteins were subjected to BLAST against the Solanaceae genome database to gain the gene ID of the 66 proteins in Solanaceae. Gene ID of the 66 candidate proteins were submitted to gene ontology (GO) enrichment analysis using GOEAST. It was shown that 48 genes among the 66 genes have GO annotation (Supplemental Figure S5). It is worth noting that several pathways relevant to protein targeting to the chloroplast were enriched in the “biological process” category. This result indicated that RSV infection affected the pathways of protein targeting to the chloroplast. This is consistent with our proposal of the altered location of ChRPs during RSV infection, which can be validated by the protoplast–chloroplast proteomics method. Thus, this protoplast–chloroplast proteomics method successfully identifies the perturbed translocation of chloroplast proteins after RSV infection. This is the first identification of chloroplast proteins that do not transport into chloroplasts during RSV infection.
From the GO terms, three nuclear-encoded ChRPs (K4CSN4, K4CR23, and K4BXN9, Table 2) were enriched, representing the enriched chloroplast-targeting biological process (Supplemental Figure S5). This result implied that RSV infection blocked the normal localization and function of these three proteins from importing the substrate factors into the chloroplast which, in turn, lead to the abnormal location of more chloroplast proteins. These results suggest that RSV infection changed the routine biological process of protein targeting into chloroplast and, thus, resulted in the abnormal location of chloroplast proteins and may contribute to the abnormal structure and function of the chloroplasts.
Table 2.
The three representative proteins which are involved in modulating the biological process of protein targeting into chloroplast, affected by RSV infection.
| GO ID | GO Term | p-Value | UniProt Accession | Protein Name |
|---|---|---|---|---|
| GO:0045036 | protein targeting to chloroplast | 0.00242 | K4CSN4 K4CR23 K4BXN9 |
Tic110 cpSRP54 HSP90C |
2.6. Transit Peptide Analysis of the Candidate ChRPs
In addition to the above three key proteins (K4CSN4, K4CR23, and K4BXN9), the function of the other 63 candidate proteins remains to be further analyzed for their involvement in chloroplast function and relation to viral symptom development. The chloroplast transit peptide (TP) is a leader sequence that directs the chloroplast protein import into the chloroplast, making its presence crucial for the localization of the protein to the chloroplast. We analyzed and confirmed, by TargetP 1.1 (Supplemental Table S7), that these three proteins had this transit peptide. Therefore, we speculated that the three proteins might regulate the localization of chloroplast proteins by their present in the chloroplasts. In addition, according to the transit peptide analysis by TargetP 1.1 [50], the majority (65%) of the 63 candidate proteins were found to have the chloroplast transit peptide (Supplemental Table S7), which further confirmed that the candidate proteins are ChRPs with abnormal location.
GO enrichment analysis of these 41 candidate proteins with chloroplast transit peptide (Supplemental Figure S6) revealed that the associated biological pathways of protein targeting to the chloroplast, represented by the same three proteins (K4CSN4, K4CR23, and K4BXN9), were further enriched (Supplemental Figure S6), suggesting that the candidate proteins with predicted chloroplast transit peptide are more dependent on the biological process of protein targeting into the chloroplast. Thus, these three proteins play an important role in the localization of chloroplast proteins.
3. Discussions
3.1. The Coincidence of the Chloroplast Perturbation with the RSV Symptoms
Previous studies have indicated that the viral symptom of chlorosis was associated with the changes of chloroplast structure [16,20,51,52]. For instance, the severe chlorosis induced by CMV-P6 infection was basically related to the reduction in the number of the chloroplasts as well as the grana in chloroplasts [53]. Distorted thylakoid membranes may promote the shorter chlorophyll fluorescence lifetime of chlorotic leaves from plant virus-infected plant [54].
Some studies have also indicated that changes in chloroplast structure were often accompanied by changes in the chloroplast membrane protein complexes during plant infection. For instance, the TMV flavum strain infection caused a complete depletion of the monomeric and dimeric PSII complexes accompanied with chlorotic symptoms in tobacco plants [55], which is similar to our results revealed by BN-PAGE. In addition, TMV CP could embed into or form a complex with thylakoid membranes. The chloroplasts from tissues with more severe mosaic symptoms tended to accumulate more TMV CPs, up to 10–50 fold than those from symptomless leaves, thus indicating that TMV CP associated with thylakoid membranes may play a possible role in symptom development [56]. The co-localization of a tobacco protein IP-L and ToMV CP in the chloroplast thylakoid membranes suggested the interaction between the tobamovirus CPs and thylakoid membrane components, which may affect the function and stability of chloroplasts, thus leading to chlorosis [57]. These studies revealed that plant virus infection caused chlorosis-related symptoms which were coincident with the changes of chloroplast membrane complexes and chloroplast structure.
Many studies indicated that RSV infection always caused changes in chloroplast components, structure, and ChRPs in varied layers. For example, the downregulation of ChRGs in RSV-infected rice plants (Oryza sativa L.) is assumed to be responsible for the RSV chlorosis symptoms [6]. RSV disease- specific protein (SP) interacts with the ChRP 23 kDa oxygen evolving complex (OEC) PsbP, and resulted in the downregulation of PsbP in chloroplasts, which could modulate RSV symptoms through disruption of chloroplast structure and function [58]. In a recent report, iTRAQ-based quantitative proteomic analysis of rice leaves revealed that the accumulation of 30 proteins related to chloroplasts was apparently reduced by RSV infection [37]. The above studies suggested RSV infection have a tremendous influence on chloroplast structure. In this study, we found that the N. benthamiana infected by RSV exhibited tremendous changes of chloroplast ultrastructure and unanimous chlorosis lamina and yellow vein as well. It is suggested that the chloroplast ultrastructure changes and phenotypic changes were induced simultaneously in RSV-infected plants.
3.2. Targeting of ChRPs into Chloroplast is an Essential Layer of Virus–Chloroplast Interaction
The fully functioning process of ChRPs contains a series of steps. The plant virus can impose their effects on varied layers, such as transcription, post-transcription, translation, targeting into chloroplast, complex assembly, and degradation, thus contributing to virulence and related symptoms [16,55,59,60]. Plant virus protein factors, such as TMV mutant CPs which can aggregate outside of chloroplasts, may subvert chloroplast development and cause degradation of chloroplasts by interfering with the synthesis and transport of CPRPs [16,55,59,60,61,62,63]. Several research sources have reported viral proteins disturbing ChRPs from their normal transportation into chloroplasts. Tobamovirus MPs can interact with the ChRP RuBisCO small subunit (RbCS) at plasmodesma (PD), which contribute to cell-to-cell movement, as well as the systemic transport of TMV and ToMV [30]. The sugarcane mosaic virus (SCMV) HC-Pro protein specifically interacts with ChRP ferredoxin V (Fd V) via the transit peptide, and probably perturbs the importing of Fd V into chloroplasts from outside of the chloroplasts [22]. As mentioned above, RSV SP interacts with PsbP, and relocates it from the chloroplast into the cytoplasm, which is associated with the enhanced RSV symptom [58].
Here, we isolated the chloroplasts from the protoplasts and validated the specificity by WB detection of marker proteins. Then, the chloroplast proteomic and protoplast proteomic profiles were analyzed by label-free LC–MS/MS, and specific filters were set to obtain the 66 nuclear-encoded ChRPs which were not targeted into chloroplast after RSV infection. As revealed by GO enrichment analysis and chloroplast transit peptides analysis, most of these candidate proteins were associated with the biological process of protein targeting into the chloroplast. It is worth noting that several of the candidate ChRPs have also been characterized as described [37], such as P37222 (NADP-dependent malic enzyme), K4CNE8 (ATP-dependent Clp protease proteolytic subunit), and Q0MW94 (1-deoxy-d-xylulose-5-phosphate reductoisomerase). With this unique quantified chloroplast–protoplast proteomics, we demonstrated that chloroplast importation of ChRPs is an essential layer for RSV to manipulate the routine processes of plant hosts during the plant–virus interaction, which is related to virulence and symptom development.
3.3. Application of Chloroplast–Protoplast Proteomics
A growing number of studies have shown that viral proteins can associate with or transport into various organelles, and lead to malfunction or stress response of the organelles [64,65,66,67]. For instance, RSV p2 could recruit fibrillarin in the nucleus and promote virus systemic infection by manipulating nuclear functions [68]. The groundnut rosette virus (GRV) ORF3 protein enters into the nucleus by targeting and reorganizing the Cajal bodies (CBs) into multiple CB-like structures that can fuse with the nucleolus [69]. The tomato bushy stunt virus (TBSV) 33 kDa replication protein (p33) was sorted into peroxisome and endoplasmic reticulum (ER), which induced altered peroxisome rearrangements [70]. The two closely related tombusvirus carnation Italian ringspot virus (CIRV) and melon necrotic spot carmovirus (MNSV) lead to mitochondrial damage by inducing multivesicular bodies (MVBs) from the outer membrane of mitochondria [71,72,73]. Several lipid membrane-associated virus factors, such as PVX TGBp3 [74,75], rice black-streak dwarf virus (RBSDV) CP p10 [76], turnip mosaic virus (TuMV) 6K2 protein [77], and garlic virus X (GarVX) MP p11 [78], induced ER stress and the unfolded protein response (UPR), some of which could ultimately elicit a hypersensitive reaction (HR) in host plants. Plant virus MPs can modify the plasmodesmata (PDs), in favor of the virus movement, by replacing the desmotubule with expanded tubule structures [79,80], probably through interactions with plasmodesmata-located proteins (PDLPs) [80,81,82,83]. However, how the transportation of these organelle-specific localized proteins is affected by virus infection has not been studied. Thus, organelle-protoplast proteomics may be used to screen proteins with abnormal position upon virus infection. Since the chloroplast–protoplast proteomics method presented here is feasible for searching for the abnormal translocation of chloroplast proteins, the method could be applied extensively for research on nucleus-encoded proteins that can be specifically targeted into organelles other than chloroplast, which could also be influenced by pathogen infection.
3.4. Candidate for Plant Virus Resistance Improvement
GO enrichment analysis of the 66 candidate chloroplast proteins in response to RSV infection found three specific proteins which regulate the biological process of protein targeting into chloroplast. Their UniProt accessions are K4CSN4, K4CR23, and K4BXN9. In addition, we analyzed the protein levels of these three ChRPs in protoplasts after RSV infection, and found that they all showed a downregulation after RSV infection. The amino acid sequences of K4CSN4, K4CR23, and K4BXN9 were searched by BLAST, and it was shown that they had highest homology to chloroplast inner membrane translocon Tic110, chloroplast signal recognition particle 54 kDa protein (cpSRP54), and heat shock protein HSP90-5 in chloroplast (HSP90C), respectively. Many studies have pointed out that these three proteins (Tic110, cpSRP54, and HSP90C) play important roles in regulating plant growth [84,85], chloroplast structure [86], plant virus infection [8,87], and the process of protein targeting into chloroplast [88,89,90]. For instance, HSP90C is a chloroplast-localized member from a highly conserved subfamily of HSP90 molecular chaperones. It has been implicated to be involved in plant disease symptoms, photomorphogenesis, and the translocation of nuclear-encoded ChRPs to the chloroplasts [91,92]. For example, Arabidopsis HSP90C interacted with PsbO1, and functions in protein translocation into the thylakoid lumen [93]. Peach latent mosaic viroid (PLMVd) infection resulted in the silencing of HSP90C gene by an RNAi mechanism, which induced chlorosis along with growth suppression phenotypes [8,87]. HSP90C was shown to interact with import intermediates of nuclear-encoded ChRPs proteins, and the suppression of HSP90 ATPase activity can inhibit the translocations of a variety of ChRP precursors across the inner envelope membrane, indicating that Hsp90C functions in the translocation of nuclear-encoded ChRPs to the chloroplasts [92]. Thus, these three key proteins (K4CSN4, K4CR23, and K4BXN9) may play essential roles in the interaction of chloroplast and be influenced by plant viruses for infection and symptom development. It is implied that they may serve as potential antiviral targets for genetic manipulation and still need further investigation.
4. Material and methods
4.1. Plant Materials and Virus Inoculation
N. benthamiana plants were grown in pots in a 25 °C growth room under a 16 h/8 h light/dark cycle. RSV was inoculated on N. benthamiana as previously described [94,95]. In brief, the RSV inoculates were prepared from the leaves of RSV-infected adult rice plants which were collected from Zhejiang province in China. The N. benthamiana plants were mechanically inoculated with RSV at the eighth leaf growth stage (50 days after sowing). In parallel, the experimental controls were mock-inoculated with homogenized leaf tissues without RSV. The symptoms and the occurrence of disease were observed and recorded on a daily basis. N. benthamiana plants with typical systemic symptoms were examined by reverse transcription polymerase chain reaction (RT-PCR), and used in the subsequent procedures.
4.2. Isolation of Protoplast and Chloroplast
Protoplast isolation was performed as previously described [96,97,98], with w/v modifications. In brief, about 2 g of N. benthamiana leaf tissues were digested with 12.5 mL enzyme mixture (1.5% (w/v) cellulase R-10, 0.5% (w/v) macerozyme R-10, 5 mM 2-morpholinoethanesulfonic acid (MES), 0.1% (w/v) BSA, 10 mM CaCl2, and 0.4 M mannitol, pH 5.8), and incubated at 28 °C for 5 h in the dark. The cells were collected from the interface of 0.4 M mannitol-MES and 0.55 M sucrose solution after centrifugation. The purified protoplasts were diluted appropriately and counted with a hemocytometer under a microscope for protoplast yield. The viability of protoplasts was determined by the FDA staining method, as described [99].
Chloroplasts from intact protoplasts were isolated as described previously [100], and we made some modifications here. In brief, the protoplast solution was centrifuged at 300 g for 6 min, and the pellets were resuspended in protoplast lysis buffer (0.3 M sorbitol, 20 mM tricine ± KOH (pH 8.4), 5 mM EDTA, 5 mM EGTA, 10 mM NaHCO3, and 0.1% BSA), then force-filtered through two-level 15 μm nylon mesh. The released chloroplasts were immediately purified on a 40%/80% Percoll gradient by centrifugation at 3000 g for 25 min at 4 °C, then the chloroplasts at the interface of 40%/80% Percoll were collected, washed, and then resuspended in chloroplast resuspension buffer (0.3 M sorbitol, 20 mM tricine-KOH (pH 7.6), 5 mM MgCl2, 2.5 mM EDTA). The intact chloroplasts were diluted appropriately and counted with a hemocytometer under a microscope for chloroplast yield.
4.3. Blue Native PAGE Electrophoresis
A gradient BN-PAGE was performed as previously described [101,102] with some w/v modifications. In brief, the chloroplasts were lysed in hypoosmotic buffer (50 mM HEPES, 2 mM MgCl2, 1 mM EDTA, 1 mM NaF, 1 mM PMSF (pH 7.5)) for 30 min at 4 °C, and then centrifuged for 10 min at 14,000 g at 4 °C for collection of chloroplast membranes. Chloroplast membranes were washed with washing buffer (330 mM sorbitol, 50 mM BisTris–HCl (pH 7.0)), and collected by centrifugation at 300 g for 2 min at 4 °C, then resuspended in 25BTH20G buffer (20% (w/v) glycerol, 25 mM BisTris–HCl (pH 7.0)). An equal volume of resuspension buffer containing 2% (w/v) n-dodecyl-β-d-maltoside (DDM) was added with continuous mixing, and the solubilization of membrane–protein complexes was performed by incubating on ice for 30 min. Insoluble materials were removed by centrifugation at 15,000 g for 10 min. The supernatant was mixed with 0.1 volumes of 5% (w/v) Serva blue G and loaded onto a 1.0 mm-thick 4–15% (w/v) acrylamide gradient gel (101 mm × 73 mm). Electrophoresis was performed at 4 °C.
4.4. Protein Extraction
Protein extraction of protoplasts and chloroplasts was performed using the methods described by Chatterjee et al. [103]. Briefly, each sample was ground in a mortar in the presence of liquid nitrogen and extracted with 3 mL of SDS buffer (30% sucrose, 2% SDS, 0.1 M Tris-Cl, 5% β-mercaptoethanol, and 1 mM phenyl methyl sulfonyl fluoride (PMSF) (pH 8.0)). The extracts were sonicated 6 times for 15 s at 60 amps. Following sonication, 3 mL of Tris phenol buffer was added to the mixture and vortexed for 10 min at 4 °C. This mixture was centrifuged at 8000 g for 10 min at 4 °C, and phenolic phase was collected and re-extracted with 3 mL SDS buffer and shaken for 3–10 min. The mixture was centrifuged once again as before using the same settings, then the phenolic phase was collected and precipitated overnight with 4 volumes of 0.1 M ammonium acetate in methanol at −20 °C. Protein precipitates were obtained by centrifugation at 10,000 g for 30 min at 4 °C, and then washed three times with cold 0.1 M ammonium acetate, and finally with cold 80% acetone (-20 °C). The pellets were dried and used for further analyses.
4.5. Protein Digestion
The proteins were dissolved in 8 M urea, 50 mM sodium bicarbonate, then incubated in a compact Thermomixer (Thermo Scientific, Waltham, MA, USA) with 20 mM Tris(2-carboxyethyl)phosphine (TECP) for 60 min at 30 °C and 30 mM iodoacetamide at 25 °C for 40 min in darkness. The samples were then diluted with 1.6 M urea, 50 mM sodium bicarbonate, and then subjected to trypsin digestion (Sigma, St. Louis, MO, USA) at 1:50 enzyme/substrate ratio. The reaction mixture was incubated at 37 °C for 16 h, and formic acid (FA) was included to a final concentration of 1% following digestion. Prior to LC–MS/MS analysis, the resulting peptides were desalted using a Strata X 33 µm Polymeric Reversed Phase column (Phenomenex, CA, USA) and resuspended in 0.1% FA.
4.6. Mass Spectrometric Acquisition
Samples were analyzed using an Ultimate RSLC nano UHPLC system (Thermo Scientific, Waltham, MA, USA) coupled to a Q Exactive HF MS/MS (Thermo Scientific). For MS analysis, 1 μg of the samples was analyzed on an Acclaim PepMap C18 analytical column (2 μm, 75 μm id × 50 cm, Thermo Scientific) at 40 °C. The peptides were separated by a 103 min linear gradient from 4% to 16% ACN with 0.1% formic acid at 200 nL/min, followed by a linear increase to 25.6% ACN in 68 min, to 32% in 5 min, to 40% in 7 min, and to 72% ACN in 5min. For DDA acquisition, the full scan was performed between 400 and 1500 m/z. The automatic gain control target for the MS/MS scan was set to 2e5. Normalized collision energy was 27%. The HRM DIA method consisted of a survey scan at 60,000 resolution from 350 to 1200 m/z (automatic gain control target of 3e6 or 50 ms injection time). Then, 41 DIA windows were acquired at 30,000 resolution (automatic gain control target 1e6 and auto for injection time). Normalized collision energy was 30%, mass range from m/z 400 to 1000 with 41 isolation windows (29 windows of 14 m/z, followed by 5 windows of 25 m/z, 2 windows of 34.5 m/z, 5 windows of 40 m/z). The spectra were recorded in profile type.
4.7. Mass Spectrometric Raw Data Analysis
The DIA data were analyzed with Spectronaut 10, a mass spectrometer vendor-independent software from Biognosys. The default settings were used for the Spectronaut search. Retention time prediction type was set to dynamic iRT (correction factor for window 1). Interference correction on MS2 level was enabled. The false discovery rate (FDR) was set to 1% at peptide level. The DDA spectra were analyzed with the MaxQuant Version 1.5.6.5 analysis software using default settings. The minimal peptide length was set to 7. Search criteria included carbamidomethylation of cysteine as a fixed modification, oxidation of methionine and acetyl (protein N-terminus) as variable modifications. The mass tolerance for the precursor was 4.5 ppm, and for the fragment ions, was 20 ppm. The DDA files were searched against the tomato UniProt Fasta database (state 2016.06.9, 33,952 entries), the RSV proteins (state 2017.01.06, 447 entries), and the Biognosys iRT peptide sequences (11 entries). The identifications were filtered to satisfy FDR of 1% on peptide and protein level. Spectral libraries were generated using spectral library generation in Spectronaut with DDA measurements.
5. Conclusions
RSV infection caused the malformation of chloroplast structure and the whole reduction of chloroplast membrane protein complexes in N. benthamiana plants. We applied protoplast–chloroplast proteomics and established special filters to screen the nucleus-encoded ChRPs which were not transported into chloroplasts during RSV infection, and obtained 66 candidate proteins. GO enrichment analysis of these 66 candidate proteins, together with the analysis of chloroplast transit peptides, indicated that RSV infection changed the location of 66 nucleus-encoded ChRPs from chloroplast. RSV infection was indicated to regulate the relevant multiple biological process of protein targeting into chloroplast through three key proteins (K4CSN4, K4CR23, and K4BXN9), which might contribute to the abnormal chloroplast structure and function. Our results indicate the process of chloroplast transportation of nuclear-encoded ChRPs is a new layer of plant–virus interaction. Our strategy can be applied for research related to more organelles influenced by plant pathogens.
Acknowledgments
We thank Rafael A. Chavez, Carlos Garcia, Kristine Nguyen, Brian Phan at Texas A&M University AgriLife Research Center at Dallas for paper revisions.
Abbreviations
| AltMV | Alternanthera mosaic virus |
| AMV | alfalfa mosaic virus |
| BaMV | bamboo mosaic virus |
| BN-PAGE | blue native PAGE |
| BsMV | barley stripe mosaic virus |
| CB | Cajal bodies |
| CF1β | chloroplast β-ATPase |
| ChRG | photosynthesis-related genes |
| ChRP | chloroplast-related protein |
| CIRV | carnation Italian ringspot virus |
| CMV | cucumber mosaic virus |
| CP | coat protein |
| CpGK | chloroplast phosphoglycerate kinase |
| cPGK | chloroplast phosphoglycerate kinase |
| cpSRP54 | chloroplast signal recognition particle 54 kDa protein |
| dpi | days post-inoculation |
| ER | endoplasmic reticulum |
| FA | formic acid |
| FC | fold change |
| Fd I | ferredoxin I |
| FDR | false discovery rate |
| GarVX | garlic virus X |
| GO | gene ontology |
| GRV | groundnut rosette virus |
| HC-Pro | helper component-proteinase |
| HR | hypersensitive reaction |
| HSP90C | HSP90 in chloroplast |
| iTRAQ | isobaric tags for relative and absolute quantification |
| MES | 2-morpholinoethanesulfonic acid |
| MNSV | melon necrotic spot carmovirus |
| MVBs | multivesicular bodies |
| OEC | oxygen evolving complex |
| PC | plastocyanin |
| PD | plasmodesma |
| PDLP | plasmodesmata-located protein |
| PLMVd | peach latent mosaic viroid |
| PMSF | phenyl methyl sulfonyl fluoride |
| PSI | photosystem I |
| PSII | photosystem II |
| PVY | potato virus Y |
| RbCL | RuBisCO large subunit |
| RbCS | RuBisCO small subunit |
| RBSDV | rice black-streak dwarf virus |
| RNAi | RNA interference |
| RSV | rice stripe virus |
| RT-PCR | reverse transcription PCR |
| RuBisCO | ribulose-1,5-bisphosphate carboxylase/oxygenase |
| TBSV | tomato bushy stunt virus |
| TECP | Tris(2-carboxyethyl)phosphine |
| TMV | tobacco mosaic virus |
| ToMV | tomato mosaic virus |
| TP | transit peptide |
| TuMV | turnip mosaic virus |
| UGPase | UDP-glucose pyrophosphorylase |
| WB | western blot |
| MPRO | protoplasts from mock-inoculated healthy leaves |
| RPRO | protoplasts from RSV-infected leaves |
| MCHL | chloroplasts isolated from protoplasts from mock-inoculated healthy leaves |
| RCHL | chloroplasts isolated from protoplasts from RSV-infected leaves |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/20/2/253/s1.
Author Contributions
J.Z. and J.X. performed the experiments. J.Z., J.X., B.C., Z.C., W.C. and Z.D. analyzed the data. Z.Z. and X.S. provided technical support. J.Z., J.C. and F.Y. designed the program. H.Z., L.L., J.P. and Y.L. provided advices to the experiments and manuscript. All authors discussed and revised the manuscript.
Funding
This study was funded by the State Basic Research Program of China (2014CB138403), the National Nature Science Foundation of China (31500124), the International Science & Technology Cooperation Program of China (2015DFA30700), the Initial Funding of Zhejiang Academy of Agricultural Sciences and the Cultural Funding for Youth Talent of Zhejiang Academy of Agricultural Sciences (2015R21R08E03).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Yeam I. Current advances and prospectus of viral resistance in horticultural crops. Hortic. Environ. Biotechnol. 2016;57:113–122. doi: 10.1007/s13580-016-0105-x. [DOI] [Google Scholar]
- 2.Nelson R.S., Citovsky V. Plant viruses. Invaders of cells and pirates of cellular pathways. Plant Physiol. 2005;138:1809–1814. doi: 10.1104/pp.104.900167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du Y., Zhao J., Chen T., Liu Q., Zhang H., Wang Y., Hong Y., Xiao F., Zhang L., Shen Q., et al. Type I J-domain NbMIP1 proteins are required for both tobacco mosaic virus infection and plant innate immunity. PLoS Pathog. 2013;9:e1003659. doi: 10.1371/journal.ppat.1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumari R., Kumar S., Singh L., Hallan V. Movement Protein of Cucumber Mosaic Virus Associates with Apoplastic Ascorbate Oxidase. PLoS ONE. 2016;11:e0163320. doi: 10.1371/journal.pone.0163320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dardick C. Comparative expression profiling of Nicotiana benthamiana leaves systemically infected with three fruit tree viruses. Mol. Plant-Microbe Interact. 2007;20:1004–1017. doi: 10.1094/MPMI-20-8-1004. [DOI] [PubMed] [Google Scholar]
- 6.Shi B., Lin L., Wang S., Guo Q., Zhou H., Rong L., Li J., Peng J., Lu Y., Zheng H., et al. Identification and regulation of host genes related to Rice stripe virus symptom production. New Phytol. 2016;209:1106–1119. doi: 10.1111/nph.13699. [DOI] [PubMed] [Google Scholar]
- 7.Waliullah S., Mochizuki T., Sekine K.T., Atsumi G., Ali M.E., Yaeno T., Yamaoka N., Nishiguchi M., Kobayashi K. Artificial induction of a plant virus protein in transgenic tobacco provides a synchronous system for analyzing the process of leaf chlorosis. Physiol. Mol. Plant Pathol. 2014;88:43–51. doi: 10.1016/j.pmpp.2014.08.005. [DOI] [Google Scholar]
- 8.Bhor S.A., Tateda C., Mochizuki T., Sekine K.T., Yaeno T., Yamaoka N., Nishiguchi M., Kobayashi K. Inducible transgenic tobacco system to study the mechanisms underlying chlorosis mediated by the silencing of chloroplast heat shock protein 90. Virusdisease. 2017;28:1–12. doi: 10.1007/s13337-017-0361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balachandran S., Osmond C.B., Daley P.F. Diagnosis of the earliest strain-specific interactions between tobacco mosaic virus and chloroplasts of tobacco leaves in vivo by means of chlorophyll fluorescence imaging. Plant Physiol. 1994;104:1059–1065. doi: 10.1104/pp.104.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S., Cui W., Wu X., Yuan Q., Zhao J., Zheng H., Lu Y., Peng J., Lin L., Chen J., et al. Suppression of nbe-miR166h-p5 attenuates leaf yellowing symptoms of potato virus X on Nicotiana benthamiana and reduces virus accumulation. Mol. Plant Pathol. 2018;19:2384–2396. doi: 10.1111/mpp.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otulak K., Chouda M., Bujarski J., Garbaczewska G. The evidence of tobacco rattle virus impact on host plant organelles ultrastructure. Micron. 2015;70:7–20. doi: 10.1016/j.micron.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Bhat S., Folimonova S.Y., Cole A.B., Ballard K.D., Lei Z., Watson B.S., Sumner L.W., Nelson R.S. Influence of host chloroplast proteins on Tobacco mosaic virus accumulation and intercellular movement. Plant Physiol. 2013;161:134–147. doi: 10.1104/pp.112.207860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mochizuki T., Ogata Y., Hirata Y., Ohki S.T. Quantitative transcriptional changes associated with chlorosis severity in mosaic leaves of tobacco plants infected with cucumber mosaic virus. Mol. Plant Pathol. 2014;15:242–254. doi: 10.1111/mpp.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das P.P., Lin Q., Wong S.-M. Comparative proteomics of Tobacco mosaic virus-infected Nicotiana tabacum plants identified major host proteins involved in photosystems and plant defence. J. Proteom. 2018 doi: 10.1016/j.jprot.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Mwaba I., Rey M.E.C. Nitric oxide associated protein 1 is associated with chloroplast perturbation and disease symptoms in Nicotiana benthamiana infected with South African cassava mosaic virus. Virus Res. 2017;238:75–83. doi: 10.1016/j.virusres.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J., Zhang X., Hong Y., Liu Y. Chloroplast in Plant-Virus Interaction. Front. Microbiol. 2016;7:1565. doi: 10.3389/fmicb.2016.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu K., Nagy P.D. Dissecting virus-plant interactions through proteomics approaches. Curr. Proteom. 2010;7:316–327. doi: 10.2174/157016410793611792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manfre A., Glenn M., Nuñez A., Moreau R.A., Dardick C. Light quantity and photosystem function mediate host susceptibility to Turnip mosaic virus via a salicylic acid-independent mechanism. Mol. Plant-Microbe Interact. 2011;24:315–327. doi: 10.1094/MPMI-08-10-0191. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Cui H., Cui X., Wang A. The altered photosynthetic machinery during compatible virus infection. Curr. Opin. Virol. 2016;17:19–24. doi: 10.1016/j.coviro.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Revers F., García J.A. Chapter Three—Molecular Biology of Potyviruses. In: Maramorosch K., Mettenleiter T.C., editors. Advances in Virus Research. Volume 92. Academic Press, Elsevier Inc.; Amsterdam, The Netherlands: 2015. pp. 101–199. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y.-Q., Liu Z.-M., Xu J., Zhou T., Wang M., Chen Y.-T., Li H.-F., Fan Z.-F. HC-Pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin-5 in vitro and in planta. J. Gen. Virol. 2008;89:2046–2054. doi: 10.1099/vir.0.2008/001271-0. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharyya D., Gnanasekaran P., Kumar R.K., Kushwaha N.K., Sharma V.K., Yusuf M.A., Chakraborty S. A geminivirus betasatellite damages the structural and functional integrity of chloroplasts leading to symptom formation and inhibition of photosynthesis. J. Exp. Bot. 2015;66:5881–5895. doi: 10.1093/jxb/erv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarzyńska-Nowak A., Jeżewska M., Hasiów-Jaroszewska B., Zielińska L. A comparison of ultrastructural changes of barley cells infected with mild and aggressive isolates of barley stripe mosaic virus. J. Plant Dis. Prot. 2015;122:153–160. doi: 10.1007/BF03356545. [DOI] [Google Scholar]
- 24.Tu Y., Jin Y., Ma D., Li H., Zhang Z., Dong J., Wang T. Interaction between PVY HC-Pro and the NtCF1β-subunit reduces the amount of chloroplast ATP synthase in virus-infected tobacco. Sci. Rep. 2015;5:15605. doi: 10.1038/srep15605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X., Li Y., Shi M., Zhang N., Wu G., Li T., Qing L., Zhou C. In vitro binding and bimolecular fluorescence complementation assays suggest an interaction between tomato mosaic virus coat protein and tobacco chloroplast ferredoxin I. Arch. Virol. 2013;158:2611–2615. doi: 10.1007/s00705-013-1778-4. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y., Zhou T., Hong Y., Fan Z., Li H. Decreased level of ferredoxin I in tobacco mosaic virus-infected tobacco is associated with development of the mosaic symptom. Physiol. Mol. Plant Pathol. 2008;72:39–45. doi: 10.1016/j.pmpp.2008.05.004. [DOI] [Google Scholar]
- 27.Bobik K., Burch-Smith T.M. Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 2015;6:781. doi: 10.3389/fpls.2015.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balasubramaniam M., Kim B.S., Hutchenswilliams H.M., Loeschfries L.S. The photosystem II oxygen-evolving complex protein PsbP interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol. Plant-Microbe Interact. 2014;27:1107–1118. doi: 10.1094/MPMI-02-14-0035-R. [DOI] [PubMed] [Google Scholar]
- 29.Burch-Smith T.M., Zambryski P.C. Regulation of Plasmodesmal Transport and Modification of Plasmodesmata During Development and Following Infection by Viruses and Viral Proteins. In: Kleinow T., editor. Plant-Virus Interactions: Molecular Biology, Intra- and Intercellular Transport. Springer International Publishing; Cham, Switzerland: 2016. pp. 87–122. [Google Scholar]
- 30.Zhao J., Liu Q., Zhang H., Jia Q., Hong Y., Liu Y. The RubisCO small subunit is involved in Tobamovirus movement and Tm-22-mediated extreme resistance. Plant Physiol. 2013;161:374–383. doi: 10.1104/pp.112.209213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng S.-F., Huang Y.-P., Chen L.-H., Hsu Y.-H., Tsai C.-H. Chloroplast phosphoglycerate kinase is involved in the targeting of bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol. 2013;163:1598–1608. doi: 10.1104/pp.113.229666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo E.Y., Nam J., Kim H.S., Park Y.H., Hong S.M., Lakshman D., Bae H., Hammond J., Lim H.S. Selective Interaction Between Chloroplast β-ATPase and TGB1L88 Retards Severe Symptoms Caused by Alternanthera mosaic virus Infection. Plant Pathol. J. 2014;30:58–67. doi: 10.5423/PPJ.OA.09.2013.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao F., Cai Z., Hu T., Yao H., Wang L., Dong N., Wang B., Ru Z., Zhai W. Genetic Analysis and Molecular Mapping of a Novel Gene Conferring Resistance to Rice Stripe Virus. Plant Mol. Biol. Report. 2010;28:512–518. doi: 10.1007/s11105-009-0178-0. [DOI] [Google Scholar]
- 34.Cho W.K., Lian S., Kim S.M., Park S.H., Kim K.H. Current Insights into Research on Rice stripe virus. Plant Pathol. J. 2013;29:223–233. doi: 10.5423/PPJ.RW.10.2012.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toriyama S. Rice stripe virus: Prototype of a new group of viruses that replicate in plants and insects. Microbiol. Sci. 1986;3:347–351. [PubMed] [Google Scholar]
- 36.Lee S.B., Hur Y.J., Lee J.H., Kwon T., Shin D., Kim T.H., Han S.I., Cho J.H., Yoon Y.N., Kiswara G. Molecular mapping of a quantitative trait locus qSTV11Z harbouring rice stripe virus resistance gene, Stv-b. Plant Breed. 2017;136:61–66. doi: 10.1111/pbr.12419. [DOI] [Google Scholar]
- 37.Wang B., Hajano J.-U.-D., Ren Y., Lu C., Wang X. iTRAQ-based quantitative proteomics analysis of rice leaves infected by Rice stripe virus reveals several proteins involved in symptom formation. Virol. J. 2015;12:99. doi: 10.1186/s12985-015-0328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu W., Smith J.W., Huang C.M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bond N.J., Shliaha P.V., Lilley K.S., Gatto L. Improving qualitative and quantitative performance for MS(E)-based label-free proteomics. J. Proteome Res. 2013;12:2340–2353. doi: 10.1021/pr300776t. [DOI] [PubMed] [Google Scholar]
- 40.Falk B.W., Tsai J.H. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 1998;36:139–163. doi: 10.1146/annurev.phyto.36.1.139. [DOI] [PubMed] [Google Scholar]
- 41.Lian S., Jonson M.G., Cho W.K., Choi H.S., Je Y.H., Kim K.H. Generation of Antibodies Against Rice stripe virus Proteins Based on Recombinant Proteins and Synthetic Polypeptides. Plant Pathol. J. 2011;27:37–43. doi: 10.5423/PPJ.2011.27.1.037. [DOI] [Google Scholar]
- 42.Hibino H. Biology and epidemiology of rice viruses. Annu. Rev. Phytopathol. 2003;34:249–274. doi: 10.1146/annurev.phyto.34.1.249. [DOI] [PubMed] [Google Scholar]
- 43.Cho W.K., Lian S., Kim S.-M., Seo B.Y., Jung J.K., Kim K.-H. Time-Course RNA-Seq Analysis Reveals Transcriptional Changes in Rice Plants Triggered by Rice stripe virus Infection. PLoS ONE. 2015;10:e0136736. doi: 10.1371/journal.pone.0136736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong R., Wu J., Zhou Y., Zhou X. Identification of a Movement Protein of the Tenuivirus Rice Stripe Virus. J. Virol. 2008;82:12304–12311. doi: 10.1128/JVI.01696-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peltier J.-B., Emanuelsson O., Kalume D.E., Ytterberg J., Friso G., Rudella A., Liberles D.A., Söderberg L., Roepstorff P., von Heijne G., et al. Central Functions of the Lumenal and Peripheral Thylakoid Proteome of Arabidopsis Determined by Experimentation and Genome-Wide Prediction. Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koo A.J.K., Ohlrogge J.B. The Predicted Candidates of Arabidopsis Plastid Inner Envelope Membrane Proteins and Their Expression Profiles. Plant Physiol. 2002;130:823–836. doi: 10.1104/pp.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savage L.J., Imre K.M., Hall D.A., Last R.L. Analysis of Essential Arabidopsis Nuclear Genes Encoding Plastid-Targeted Proteins. PLoS ONE. 2013;8:e73291. doi: 10.1371/journal.pone.0073291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ajjawi I., Lu Y., Savage L.J., Bell S.M., Last R.L. Large-Scale Reverse Genetics in Arabidopsis: Case Studies from the Chloroplast 2010 Project. Plant Physiol. 2010;152:529–540. doi: 10.1104/pp.109.148494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myouga F., Akiyama K., Tomonaga Y., Kato A., Sato Y., Kobayashi M., Nagata N., Sakurai T., Shinozaki K. The Chloroplast Function Database II: A Comprehensive Collection of Homozygous Mutants and Their Phenotypic/Genotypic Traits for Nuclear-Encoded Chloroplast Proteins. Plant Cell Physiol. 2013;54:e2. doi: 10.1093/pcp/pcs171. [DOI] [PubMed] [Google Scholar]
- 50.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 51.Jin X., Jiang Z., Zhang K., Wang P., Cao X., Yue N., Wang X., Zhang X., Li Y., Li D., et al. Three-Dimensional Analysis of Chloroplast Structures Associated with Virus Infection. Plant Physiol. 2018;176:282–294. doi: 10.1104/pp.17.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhattacharyya D., Chakraborty S. Chloroplast: The Trojan horse in plant–virus interaction. Mol. Plant Pathol. 2017;19:504–518. doi: 10.1111/mpp.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts P.L., Wood K.R. Effects of a severe (P6) and a mild (W) strain of cucumber mosaic virus on tobacco leaf chlorophyll, starch and cell ultrastructure. Physiol. Plant Pathol. 1982;21:31IN3333–32IN3537. doi: 10.1016/0048-4059(82)90005-4. [DOI] [Google Scholar]
- 54.Lei R., Jiang H., Hu F., Yan J., Zhu S. Chlorophyll fluorescence lifetime imaging provides new insight into the chlorosis induced by plant virus infection. Plant Cell Rep. 2016 doi: 10.1007/s00299-016-2083-y. [DOI] [PubMed] [Google Scholar]
- 55.Lehto K., Tikkanen M., Hiriart J.-B., Paakkarinen V., Aro E.-M. Depletion of the photosystem II core complex in mature tobacco leaves infected by the flavum strain of tobacco mosaic virus. Mol. Plant-Microbe Interact. 2003;16:1135–1144. doi: 10.1094/MPMI.2003.16.12.1135. [DOI] [PubMed] [Google Scholar]
- 56.Reinero A., Beachy R.N. Association of TMV coat protein with chloroplast membranes in virus-infected leaves. Plant Mol. Biol. 1986;6:291–301. doi: 10.1007/BF00034936. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C., Liu Y., Sun X., Qian W., Zhang D., Qiu B. Characterization of a specific interaction between IP-L, a tobacco protein localized in the thylakoid membranes, and Tomato mosaic virus coat protein. Biochem. Biophys. Res. Commun. 2008;374:253–257. doi: 10.1016/j.bbrc.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 58.Kong L., Wu J., Lu L., Xu Y., Zhou X. Interaction between rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol. Plant. 2014;7:691–708. doi: 10.1093/mp/sst158. [DOI] [PubMed] [Google Scholar]
- 59.Pérez-Bueno M.L., Rahoutei J., Sajnani C., García-Luque I., Barón M. Proteomic analysis of the oxygen-evolving complex of photosystem II under biotec stress: Studies on Nicotiana benthamiana infected with tobamoviruses. Proteomics. 2004;4:418–425. doi: 10.1002/pmic.200300655. [DOI] [PubMed] [Google Scholar]
- 60.Seidler A. The extrinsic polypeptides of Photosystem II. Biochim Biophys Acta Bioenerg. 1996:35–60. doi: 10.1016/S0005-2728(96)00102-8. [DOI] [PubMed] [Google Scholar]
- 61.Lindbeck A.G.C., Dawson W.O., Thomson W.W. Coat protein-related polypeptides from in vitro tobacco mosaic virus coat protein mutants do not accumulate in the chloroplasts of directly inoculated leaves. Mol. Plant-Microbe Interact. 1991;4:89–94. doi: 10.1094/MPMI-4-089. [DOI] [Google Scholar]
- 62.Lindbeck A.G.C., Lewandowski D.J., Culver J.N., Thomson W.W., Dawson W.O. Mutant coat protein of tobacco mosaic virus induces acute chlorosis in expanded and developing tobacco leaves. Mol. Plant-Microbe Interact. 1992;5:235–241. doi: 10.1094/MPMI-5-235. [DOI] [Google Scholar]
- 63.Ohnishi J., Hirai K., Kanda A., Usugi T., Meshi T., Tsuda S. The coat protein of tomato mosaic virus L11Y is associated with virus-induced chlorosis on infected tobacco plants. J. Gen. Plant Pathol. 2009;75:297–306. doi: 10.1007/s10327-009-0177-x. [DOI] [Google Scholar]
- 64.Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-Dimensional Analysis of a Viral RNA Replication Complex Reveals a Virus-Induced Mini-Organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grangeon R., Agbeci M., Chen J., Grondin G., Zheng H., Laliberté J.-F. Impact on the Endoplasmic Reticulum and Golgi Apparatus of Turnip Mosaic Virus Infection. J. Virol. 2012;86:9255–9265. doi: 10.1128/JVI.01146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shaw P., Brown J. Nucleoli: Composition, Function, and Dynamics. Plant Physiol. 2012;158:44–51. doi: 10.1104/pp.111.188052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Solovyev A.G., Savenkov E.I. Factors involved in the systemic transport of plant RNA viruses: The emerging role of the nucleus. J. Exp. Bot. 2014;65:1689–1697. doi: 10.1093/jxb/ert449. [DOI] [PubMed] [Google Scholar]
- 68.Zheng L., Du Z., Lin C., Mao Q., Wu K., Wu J., Wei T., Wu Z., Xie L. Rice stripe tenuivirus p2 may recruit or manipulate nucleolar functions through an interaction with fibrillarin to promote virus systemic movement. Mol. Plant Pathol. 2015;16:921–930. doi: 10.1111/mpp.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim S.H., Ryabov E.V., Kalinina N.O., Rakitina D.V., Gillespie T., Macfarlane S., Haupt S., Brown J.W., Taliansky M. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007;26:2169–2179. doi: 10.1038/sj.emboj.7601674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCartney A.W., Greenwood J.S., Fabian M.R., White K.A., Mullen R.T. Localization of the Tomato Bushy Stunt Virus Replication Protein p33 Reveals a Peroxisome-to-Endoplasmic Reticulum Sorting Pathway. Plant Cell. 2005;17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mochizuki T., Hirai K., Kanda A., Ohnishi J., Ohki T., Tsuda S. Induction of necrosis via mitochondrial targeting of Melon necrotic spot virus replication protein p29 by its second transmembrane domain. Virology. 2009;390:239–249. doi: 10.1016/j.virol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 72.Weber-Lotfi F., Dietrich A., Russo M., Rubino L. Mitochondrial Targeting and Membrane Anchoring of a Viral Replicase in Plant and Yeast Cells. J. Virol. 2002;76:10485–10496. doi: 10.1128/JVI.76.20.10485-10496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laliberté J.-F., Sanfaçon H. Cellular Remodeling During Plant Virus Infection. Annu. Rev. Phytopathol. 2010;48:69–91. doi: 10.1146/annurev-phyto-073009-114239. [DOI] [PubMed] [Google Scholar]
- 74.Ye C.-M., Chen S., Payton M., Dickman M.B., Verchot J. TGBp3 triggers the unfolded protein response and SKP1-dependent programmed cell death. Mol. Plant Pathol. 2013;14:241–255. doi: 10.1111/mpp.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye C., Dickman M.B., Whitham S.A., Payton M., Verchot J. The Unfolded Protein Response Is Triggered by a Plant Viral Movement Protein. Plant Physiol. 2011;156:741–755. doi: 10.1104/pp.111.174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun Z., Yang D., Xie L., Sun L., Zhang S., Zhu Q., Li J., Wang X., Chen J. Rice black-streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana. Virology. 2013;447:131–139. doi: 10.1016/j.virol.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 77.Zhang L., Chen H., Brandizzi F., Verchot J., Wang A. The UPR Branch IRE1-bZIP60 in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast. PLoS Genet. 2015;11:e1005164. doi: 10.1371/journal.pgen.1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu Y., Yin M., Wang X., Chen B., Yang X., Peng J., Zheng H., Zhao J., Lin L., Yu C., et al. The unfolded protein response and programmed cell death are induced by expression of Garlic virus X p11 in Nicotiana benthamiana. J. Gen. Virol. 2016;97:1462–1468. doi: 10.1099/jgv.0.000460. [DOI] [PubMed] [Google Scholar]
- 79.Ganusova E.E., Burch-Smith T.M. Review: Plant-pathogen interactions through the plasmodesma prism. Plant Sci. 2018 doi: 10.1016/j.plantsci.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 80.Amari K., Lerich A., Schmitt-Keichinger C., Dolja V.V., Ritzenthaler C. Tubule-Guided Cell-to-Cell Movement of a Plant Virus Requires Class XI Myosin Motors. PLoS Pathog. 2011;7:e1002327. doi: 10.1371/journal.ppat.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. Specific Targeting of a Plasmodesmal Protein Affecting Cell-to-Cell Communication. PLoS Biol. 2008;6:e7. doi: 10.1371/journal.pbio.0060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.den Hollander P.W., Kieper S.N., Borst J.W., van Lent J.W.M. The role of plasmodesma-located proteins in tubule-guided virus transport is limited to the plasmodesmata. Arch. Virol. 2016;161:2431–2440. doi: 10.1007/s00705-016-2936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amari K., Boutant E., Hofmann C., Schmitt-Keichinger C., Fernandez-Calvino L., Didier P., Lerich A., Mutterer J., Thomas C.L., Heinlein M., et al. A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins. PLoS Pathog. 2010;6:e1001119. doi: 10.1371/journal.ppat.1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sandhu D., Atkinson T., Noll A., Johnson C., Espinosa K., Boelter J., Abel S., Dhatt B.K., Barta T., Singsaas E. Soybean proteins GmTic110 and GmPsbP are crucial for chloroplast development and function. Plant Sci. 2016;252:76–87. doi: 10.1016/j.plantsci.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 85.Amin P., Sy D.A., Pilgrim M.L., Parry D.H., Nussaume L., Hoffman N.E. Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 1999;121:61–70. doi: 10.1104/pp.121.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang F., Luo X., Hu B., Yong W., Xie J. YGL138 (t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice. 2013;6:1–10. doi: 10.1186/1939-8433-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Navarro B., Gisel A., Rodio M.E., Delgado S., Flores R., Di S.F. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012;70:991–1003. doi: 10.1111/j.1365-313X.2012.04940.x. [DOI] [PubMed] [Google Scholar]
- 88.Hörmann F. The Chloroplast Protein Import Machinery: A Review. Methods Mol. Biol. 2010;619:307–321. doi: 10.1007/978-1-60327-412-8_18. [DOI] [PubMed] [Google Scholar]
- 89.Erika K.B., Philipp B.J., Jürgen S., Bettina B. Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 2011;11:133. doi: 10.1186/1471-2229-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inaba A.-H., Li B.E. Arabidopsis tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell. 2005;17:1482–1496. doi: 10.1105/tpc.105.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oh S.E., Yeung C., Babaeirad R., Zhao R. Cosuppression of the chloroplast localized molecular chaperone HSP90.5 impairs plant development and chloroplast biogenesis in Arabidopsis. BMC Res. Notes. 2014;7:643. doi: 10.1186/1756-0500-7-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Inoue H., Li M., Schnell D.J. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA. 2013;110:3173–3178. doi: 10.1073/pnas.1219229110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oh S.E. Master’s Thesis. Department of Cell & Systems Biology, University of Toronto; Toronto, ON, Canada: 2012. The Role of Chloroplast Localized Molecular Chaperone HSP90 in Arabidopsis thaliana Development. [Google Scholar]
- 94.Fei Y., Zhang H.M., Adams M.J., Jian Y., Peng J.J., Antoniw J.F., Zhou Y.J., Chen J.P. Characterization of siRNAs derived from rice stripe virus in infected rice plants by deep sequencing. Arch. Virol. 2010;155:935–940. doi: 10.1007/s00705-010-0670-8. [DOI] [PubMed] [Google Scholar]
- 95.Jiang S., Lu Y., Li K., Lin L., Zheng H., Yan F., Chen J. Heat shock protein 70 is necessary for Rice stripe virus infection in plants. Mol. Plant Pathol. 2014;15:907–917. doi: 10.1111/mpp.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fitzpatrick L.M., Keegstra K. A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J. 2001;27:59–65. doi: 10.1046/j.0960-7412.2001.01061.x. [DOI] [PubMed] [Google Scholar]
- 97.Huang H., Wang Z., Cheng J., Zhao W., Li X., Wang H., Zhang Z., Sui X. An efficient cucumber (Cucumis sativus L.) protoplast isolation and transient expression system. Sci. Hortic. 2013;150:206–212. doi: 10.1016/j.scienta.2012.11.011. [DOI] [Google Scholar]
- 98.Rao A.L. Preparation and inoculation of mesophyll protoplasts from monocotyledenous and dicotyledenous hosts. Curr. Protoc. Microbiol. 2007 doi: 10.1002/9780471729259.mc16d02s4. [DOI] [PubMed] [Google Scholar]
- 99.Larkin P.J. Purification and viability determinations of plant protoplasts. Planta. 1976;128:213–216. doi: 10.1007/BF00393231. [DOI] [PubMed] [Google Scholar]
- 100.Kunst L. Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol. Biol. 1998;82:43–48. doi: 10.1385/0-89603-391-0:43. [DOI] [PubMed] [Google Scholar]
- 101.Shao J., Zhang Y., Yu J., Guo L., Ding Y. Isolation of thylakoid membrane complexes from rice by a new double-strips BN/SDS-PAGE and bioinformatics prediction of stromal ridge subunits interaction. PLoS ONE. 2011;6:e20342. doi: 10.1371/journal.pone.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qureshi M.I., D’Amici G.M., Fagioni M., Rinalducci S., Zolla L. Iron stabilizes thylakoid protein-pigment complexes in Indian mustard during Cd-phytoremediation as revealed by BN-SDS-PAGE and ESI-MS/MS. J. Plant Physiol. 2010;167:761–770. doi: 10.1016/j.jplph.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 103.Chatterjee M., Gupta S., Bhar A., Das S. Optimization of an Efficient Protein Extraction Protocol Compatible with Two-Dimensional Electrophoresis and Mass Spectrometry from Recalcitrant Phenolic Rich Roots of Chickpea (Cicer arietinum L.) Int. J. Proteom. 2012;2012:536963. doi: 10.1155/2012/536963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.