Abstract
Aquaporins (AQPs) are one diverse family of membrane channel proteins that play crucial regulatory roles in plant stress physiology. However, the heat stress responsiveness of AQP genes in soybean remains poorly understood. In this study, 75 non-redundant AQP encoding genes were identified in soybean. Multiple sequence alignments showed that all GmAQP proteins possessed the conserved regions, which contained 6 trans-membrane domains (TM1 to TM6). Different GmAQP members consisted of distinct Asn-Pro-Ala (NPA) motifs, aromatic/arginine (ar/R) selectivity filters and Froger’s positions (FPs). Phylogenetic analyses distinguished five sub-families within these GmAQPs: 24 GmPIPs, 24 GmTIPs, 17 GmNIPs, 8 GmSIPs, and 2 GmXIPs. Promoter cis-acting elements analyses revealed that distinct number and composition of heat stress and hormone responsive elements existed in different promoter regions of GmAQPs. QRT-PCR assays demonstrated that 12 candidate GmAQPs with relatively extensive expression in various tissues or high expression levels in root or leaf exhibited different expression changes under heat stress and hormone cues (abscisic acid (ABA), l-aminocyclopropane-l-carboxylic acid (ACC), salicylic acid (SA) and methyl jasmonate (MeJA)). Furthermore, the promoter activity of one previously functionally unknown AQP gene-GmTIP2;6 was investigated in transgenic Arabidopsis plants. The beta-glucuronidase (GUS) activity driven by the promoter of GmTIP2;6 was strongly induced in the heat- and ACC-treated transgenic plants and tended to be accumulated in the hypocotyls, vascular bundles, and leaf trichomes. These results will contribute to uncovering the potential functions and molecular mechanisms of soybean GmAQPs in mediating heat stress and hormone signal responses.
Keywords: soybean, aquaporin, heat stress, hormone cues, transcript expression, promoter, activated GUS, GmTIP2;6
1. Introduction
Aquaporins (AQPs), known as membrane channel proteins, transport water as well as other small solutes (AQPs). AQPs consist of six trans-membrane (TM) helical domains with two cytoplasmic termini. AQPs contain two putative Asn-Pro-Ala (NPA) motifs located in the TM helices, aromatic/arginine (ar/R) regions and Froger’s positions (FPs) [1]. AQPs belong to an ancient, abundant, and highly diversified protein super-family [2,3,4,5,6,7]. Based on the protein sequence homology and membrane localization, plant AQPs are divided into five sub-families: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), and the unrecognized X intrinsic proteins (XIPs) [8].
AQPs extensively participated in plant physiological processes under variable environmental stresses [9,10]. Transcript profiles or gene function analyses of AQPs from many plant species, such as Arabidopsis, rice, barley, sorghum, cassava, soybean, and potato, demonstrated that they were associated with drought, cold, salt, silicon, or ABA stress [11,12,13,14,15,16,17]. Recently, several publications reported that AQPs were involved in response to heat stress. In wheat, TaTIPs respond to the combined heat and drought stresses, based on the representation of expressed sequence tags (ESTs) in wheat grain-related cDNA libraries [18]. In rhododendrons, the transcripts of Rc/RpPIP2s were associated with thermonasty (leaf-curling) under freezing-rewarming cycles [19]. In strawberries, heat stress induced the gene expression of FaPIPs [20]. In Setaria viridis, heat stress activated the expression of SvPIPs [21]. In Rhazya stricta, heat stress enhanced the abundant transcripts of RsPIPs and RsTIPs [22]. In Arabidopsis, AtPIPs were highly up-regulated due to combined heat-drought stress [23]. Nevertheless, functions and mechanisms of soybean AQPs in heat stress tolerance remain obscure.
Soybean is an important economic crop and a staple food for people worldwide. Extreme heat conditions significantly reduce the productivity and weaken the global food security of soybean, especially given the growing impacts of climate changes [24,25,26,27]. In previous reports [28,29], 66 and 72 GmAQP members were identified from soybean, respectively, and the expression patterns of GmAQPs under drought or silicon stress were analyzed. However, whether GmAQP genes respond to heat stress in soybean remains poorly understood. This study focused on the investigation of correlation among expression of GmAQPs, heat stress, and different hormone signals. Gene numbers of GmAQPs were finally determined based on the recently-updated genome database Phytozome V12.1. Protein feature, sequence phylogeny, chromosomal location, and promoter elements of GmAQPs were also analyzed. Expressional patterns of 12 candidate GmAQPs with relatively extensive expression in various tissues or high expression levels in root or leaf in response to heat stress and different hormone treatments (abscisic acid (ABA), l-aminocyclopropane-l-carboxylic acid (ACC), salicylic acid (SA) and methyl jasmonate (MeJA)) were examined using quantitative real-time PCR (qRT-PCR). Additionally, the promoter activity of GmTIP2;6 was assessed using the reporter beta-glucuronidase (GUS) gene in transgenic Arabidopsis plants under both control and stressful conditions. These results will provide foundation for further elucidating the molecular mechanism of soybean GmAQPs in modulating plant thermo-tolerance.
2. Results
2.1. Identification of the Soybean AQP Family
Based on HMM, KEGG, and protein BLAST searches, a total of 75 GmAQP members were identified and annotated from the recently-updated soybean genome database Phytozome V12.1 (Table 1; Dataset S1). Among them, the length of the GmAQP CDS sequence ranged from 693 bp of GmSIP2;1 to 1092 bp of GmPIP1;9. The identified GmAQP genes encoded proteins ranging from 230 amino acids of GmSIP2;1 to 363 amino acids of GmPIP1;9. Similarly, the molecular masses of the GmAQP proteins varied from 24.08 KDa of GmTIP2;7 to 39.41 KDa of GmPIP1;9 and the pI ranged from 5.08 of GmTIP2;1 and GmTIP2;2 to 10.01 of GmTIP1;9. Compared with previous reports [28,29], we newly identified 17 GmAQPs (GmAQP9, GmAQP10, GmAQP13, GmAQP14, GmAQP18, GmAQP19, GmAQP20, GmAQP21, GmAQP22, GmAQP23, GmAQP24, GmAQP34, GmAQP54, GmAQP56, GmAQP57, GmAQP58 and GmAQP74) and 3 GmAQPs (GmAQP9, GmAQP10 and GmAQP34) not previously observed, respectively.
Table 1.
Nomenclature of aquaporin (AQP) genes in soybean.
| Gene Number | Gene Name | Gene Symbol | Chromosome Location | CDS Length (bp) | Protein Length (aa) | pI | MW (kDa) | Numbers of Phosphorylation Sites |
|---|---|---|---|---|---|---|---|---|
| GmAQP1 | GmPIP1;1 | Glyma.03G078700 | 3:18018230..18021565 | 855 | 284 | 9.10 | 30.41 | Ser: 7 Thr: 1 Tyr: 2 |
| GmAQP2 | GmPIP1;2 | Glyma.18G198300 | 18:51879812..51881980 | 864 | 287 | 9.26 | 30.89 | Ser: 8 Thr: 3 Tyr: 2 |
| GmAQP3 | GmPIP1;3 | Glyma.01G220600 | 1:54066066..54068057 | 861 | 286 | 9.13 | 30.79 | Ser: 7 Thr: 1 Tyr: 1 |
| GmAQP4 | GmPIP1;4 | Glyma.11G023200 | 11:1656129..1658174 | 861 | 286 | 8.84 | 30.74 | Ser: 6 Thr: 1 Tyr: 1 |
| GmAQP5 | GmPIP1;5 | Glyma.05G208700 | 5:41267148..41268807 | 864 | 287 | 9.00 | 30.89 | Ser: 9 Thr: 1 Tyr: 2 |
| GmAQP6 | GmPIP1;6 | Glyma.08G015300 | 8:1202356..1204135 | 870 | 289 | 8.61 | 30.89 | Ser: 9 Thr: 1 Tyr: 2 |
| GmAQP7 | GmPIP1;7 | Glyma.14G061500 | 14:4894197..4896207 | 870 | 289 | 8.60 | 30.61 | Ser: 9 Thr: 3 Tyr: 3 |
| GmAQP8 | GmPIP1;8 | Glyma.11G228000 | 11:36767510..36769078 | 870 | 289 | 7.01 | 30.77 | Ser: 11 Thr: 2 Tyr: 3 |
| GmAQP9 | GmPIP1;9 | Glyma.01G113400 | 1:1138834287..38837846 | 1092 | 363 | 9.31 | 39.41 | Ser: 14 Thr: 11 Tyr: 7 |
| GmAQP10 | GmPIP1;10 | Glyma.02G255000 | 2:44207467..44209844 | 960 | 319 | 9.38 | 34.19 | Ser: 12 Thr: 8 Tyr: 4 |
| GmAQP11 | GmPIP2;1 | Glyma.04G003200 | 4:227991..229365 | 828 | 275 | 9.45 | 29.30 | Ser: 9 Thr: 0 Tyr: 2 |
| GmAQP12 | GmPIP2;2 | Glyma.06G003200 | 6:264336..265850 | 837 | 278 | 9.35 | 29.30 | Ser: 9 Thr: 0 Tyr: 2 |
| GmAQP13 | GmPIP2;3 | Glyma.11G146500 | 11:11300751..11303007 | 861 | 286 | 6.95 | 29.55 | Ser: 8 Thr: 7 Tyr: 4 |
| GmAQP14 | GmPIP2;4 | Glyma.12G075400 | 12:5747587..5750039 | 861 | 286 | 6.19 | 30.38 | Ser: 7 Thr: 9 Tyr: 3 |
| GmAQP15 | GmPIP2;5 | Glyma.12G172500 | 12:32929324..32931027 | 864 | 287 | 8.25 | 30.67 | Ser: 9 Thr: 2 Tyr: 4 |
| GmAQP16 | GmPIP2;6 | Glyma.13G325900 | 13:40664607..40666361 | 864 | 287 | 8.26 | 30.84 | Ser: 8 Thr: 2 Tyr: 3 |
| GmAQP17 | GmPIP2;7 | Glyma.03G180900 | 3:41279731..41281496 | 861 | 286 | 8.98 | 30.79 | Ser: 12 Thr: 8 Tyr: 4 |
| GmAQP18 | GmPIP2;8 | Glyma.19G181300 | 19:44007407..44009765 | 858 | 285 | 9.15 | 30.61 | Ser: 13 Thr: 9 Tyr: 4 |
| GmAQP19 | GmPIP2;9 | Glyma.02G073600 | 2:6421649..6424849 | 858 | 285 | 8.29 | 30.56 | Ser: 10 Thr: 6 Tyr: 1 |
| GmAQP20 | GmPIP2;10 | Glyma.16G155000 | 16:31513389..31517035 | 858 | 285 | 8.29 | 30.68 | Ser: 10 Thr: 6 Tyr: 3 |
| GmAQP21 | GmPIP2;11 | Glyma.16G155100 | 16:31522994..31524889 | 858 | 285 | 8.29 | 30.41 | Ser: 10 Thr: 6 Tyr: 3 |
| GmAQP22 | GmPIP2;12 | Glyma.02G073700 | 2:6434383..6437873 | 858 | 285 | 8.59 | 30.37 | Ser: 11 Thr: 6 Tyr: 3 |
| GmAQP23 | GmPIP2;13 | Glyma.10G211000 | 10:44343751..44346957 | 891 | 296 | 7.70 | 30.44 | Ser: 9 Thr: 6 Tyr: 3 |
| GmAQP24 | GmPIP2;14 | Glyma.20G179700 | 20:41738693..41741581 | 855 | 284 | 8.29 | 31.73 | Ser: 10 Thr: 7 Tyr: 4 |
| GmAQP25 | GmTIP1;1 | Glyma.02G094700 | 2:8409966..8411440 | 759 | 252 | 5.12 | 25.96 | Ser: 3 Thr: 3 Tyr: 0 |
| GmAQP26 | GmTIP1;2 | Glyma.18G286700 | 18:60989768..60991401 | 759 | 252 | 5.49 | 26.04 | Ser: 4 Thr: 2 Tyr: 0 |
| GmAQP27 | GmTIP1;3 | Glyma.10G290600 | 10:50271428..50272965 | 759 | 252 | 6.01 | 26.02 | Ser: 3 Thr: 2 Tyr: 0 |
| GmAQP28 | GmTIP1;4 | Glyma.11G143100 | 11:10892421..10894109 | 759 | 252 | 5.37 | 25.79 | Ser: 5 Thr: 1 Tyr: 1 |
| GmAQP29 | GmTIP1;5 | Glyma.12G066200 | 12:4870480..4871652 | 738 | 245 | 6.02 | 25.03 | Ser: 5 Thr: 1 Tyr: 1 |
| GmAQP30 | GmTIP1;6 | Glyma.13G333100 | 13:41270585..41271998 | 759 | 252 | 5.16 | 26.01 | Ser: 2 Thr: 1 Tyr: 0 |
| GmAQP31 | GmTIP1;7 | Glyma.03G185900 | 3:41779243..41780564 | 753 | 250 | 6.01 | 25.45 | Ser: 0 Thr: 3 Tyr: 1 |
| GmAQP32 | GmTIP1;8 | Glyma.19G186100 | 19:44258426..44259853 | 753 | 250 | 6.01 | 25.53 | Ser: 0 Thr: 3 Tyr: 1 |
| GmAQP33 | GmTIP1;9 | Glyma.13G146300 | 13:24436182..24438466 | 753 | 250 | 10.01 | 26.54 | Ser: 8 Thr: 2 Tyr: 0 |
| GmAQP34 | GmTIP1;10 | Glyma.20G098600 | 20:34184591..34191923 | 732 | 243 | 6.17 | 25.52 | Ser: 6 Thr: 3 Tyr: 0 |
| GmAQP35 | GmTIP2;1 | Glyma.01G208200 | 1:53110677..53113455 | 750 | 249 | 5.08 | 25.28 | Ser: 4 Thr: 2 Tyr: 1 |
| GmAQP36 | GmTIP2;2 | Glyma.11G034000 | 11:2476012..2478825 | 750 | 249 | 5.08 | 25.32 | Ser: 2 Thr: 1 Tyr: 2 |
| GmAQP37 | GmTIP2;3 | Glyma.07G018000 | 7:1435523..1437651 | 747 | 248 | 5.69 | 25.23 | Ser: 6 Thr: 1 Tyr: 1 |
| GmAQP38 | GmTIP2;4 | Glyma.08G203000 | 8:16535219..16537122 | 747 | 248 | 5.69 | 25.27 | Ser: 3 Thr: 2 Tyr: 2 |
| GmAQP39 | GmTIP2;5 | Glyma.13G356000 | 13:43018922..43020336 | 744 | 247 | 5.51 | 25.07 | Ser: 2 Thr: 2 Tyr: 2 |
| GmAQP40 | GmTIP2;6 | Glyma.15G018100 | 15:1393557..1395809 | 744 | 247 | 5.50 | 25.07 | Ser: 3 Thr: 3 Tyr: 1 |
| GmAQP41 | GmTIP2;7 | Glyma.19G035400 | 19:4625496..4626575 | 714 | 237 | 5.57 | 24.08 | Ser: 4 Thr: 1 Tyr: 1 |
| GmAQP42 | GmTIP3;1 | Glyma.09G160500 | 9:35913523..35915582 | 768 | 255 | 6.54 | 27.03 | Ser: 5 Thr: 1 Tyr: 1 |
| GmAQP43 | GmTIP3;2 | Glyma.16G210000 | 16:36421819..36424304 | 768 | 255 | 6.54 | 27.11 | Ser: 5 Thr: 0 Tyr: 1 |
| GmAQP44 | GmTIP3;3 | Glyma.10G174400 | 10:40238530..40240337 | 765 | 254 | 7.13 | 27.08 | Ser: 5 Thr: 0 Tyr: 0 |
| GmAQP45 | GmTIP3;4 | Glyma.20G216100 | 20:44068541..44070258 | 765 | 254 | 7.88 | 27.07 | Ser: 5 Thr: 3 Tyr: 1 |
| GmAQP46 | GmTIP4;1 | Glyma.04G083200 | 4:7019276..7020984 | 741 | 246 | 5.71 | 25.65 | Ser: 3 Thr: 2 Tyr: 0 |
| GmAQP47 | GmTIP4;2 | Glyma.06G084600 | 6:6498818..6500103 | 741 | 246 | 5.71 | 25.63 | Ser: 5 Thr: 2 Tyr: 2 |
| GmAQP48 | GmTIP5;1 | Glyma.09G224700 | 9:41742635..41743884 | 759 | 252 | 7.82 | 26.30 | Ser: 9 Thr: 2 Tyr: 2 |
| GmAQP49 | GmNIP1;1 | Glyma.05G162600 | 5:35105884..35108185 | 813 | 270 | 9.67 | 28.68 | Ser: 8 Thr: 5 Tyr: 1 |
| GmAQP50 | GmNIP1;2 | Glyma.08G120200 | 8:9268559..9270946 | 825 | 274 | 9.48 | 29.27 | Ser: 9 Thr: 3 Tyr: 1 |
| GmAQP51 | GmNIP1;3 | Glyma.13G224900 | 13:32551102..32553703 | 822 | 273 | 7.76 | 28.93 | Ser: 6 Thr: 4 Tyr: 2 |
| GmAQP52 | GmNIP1;4 | Glyma.15G087300 | 15:6704209..6706791 | 822 | 273 | 7.74 | 28.83 | Ser: 6 Thr: 4 Tyr: 2 |
| GmAQP53 | GmNIP1;5 | Glyma.08G120100 | 8:9262302..9265834 | 816 | 271 | 6.41 | 28.91 | Ser: 9 Thr: 3 Tyr: 5 |
| GmAQP54 | GmNIP1;6 | Glyma.05G162500 | 5:35371190..35375992 | 816 | 271 | 8.87 | 28.66 | Ser: 11 Thr: 8 Tyr: 1 |
| GmAQP55 | GmNIP2;1 | Glyma.07G217700 | 7:39062920..39065820 | 789 | 262 | 8.14 | 28.13 | Ser: 4 Thr: 2 Tyr: 1 |
| GmAQP56 | GmNIP3;1 | Glyma.14G174300 | 14:43721841..43723560 | 813 | 270 | 8.23 | 28.66 | Ser: 15 Thr: 11 Tyr: 3 |
| GmAQP57 | GmNIP4;1 | Glyma.02G246700 | 2:46541265..46543675 | 786 | 261 | 7.61 | 27.61 | Ser: 23 Thr: 5 Tyr: 4 |
| GmAQP58 | GmNIP4;2 | Glyma.14G069500 | 14:5711153..5714115 | 786 | 261 | 8.25 | 27.59 | Ser: 17 Thr: 4 Tyr: 4 |
| GmAQP59 | GmNIP5;1 | Glyma.09G238200 | 9:42824943..42829709 | 882 | 293 | 8.55 | 30.44 | Ser: 28 Thr: 9 Tyr: 1 |
| GmAQP60 | GmNIP5;2 | Glyma.10G221100 | 10:44670892..44676555 | 900 | 299 | 7.68 | 31.15 | Ser: 6 Thr: 5 Tyr: 0 |
| GmAQP61 | GmNIP6;1 | Glyma.18G259500 | 18:58816436..58821548 | 888 | 295 | 6.96 | 30.55 | Ser: 19 Thr: 5 Tyr: 0 |
| GmAQP62 | GmNIP6;2 | Glyma.08G217400 | 8:17701761..17706495 | 921 | 306 | 9.13 | 31.74 | Ser: 8 Thr: 5 Tyr: 0 |
| GmAQP63 | GmNIP6;3 | Glyma.15G003900 | 15:355676..359967 | 915 | 304 | 8.25 | 31.28 | Ser: 7 Thr: 2 Tyr: 0 |
| GmAQP64 | GmNIP7;1 | Glyma.02G140500 | 2:14348789..14351092 | 891 | 296 | 8.46 | 31.42 | Ser: 10 Thr: 1 Tyr: 3 |
| GmAQP65 | GmNIP7;2 | Glyma.10G033600 | 10:2898411..2900795 | 870 | 289 | 8.69 | 30.82 | Ser: 7 Thr: 1 Tyr: 1 |
| GmAQP66 | GmSIP1;1 | Glyma.02G069800 | 2:6061309..6065568 | 921 | 306 | 9.27 | 26.64 | Ser: 0 Thr: 2 Tyr: 0 |
| GmAQP67 | GmSIP1;2 | Glyma.16G151300 | 16:30813218..30817735 | 738 | 245 | 9.27 | 26.43 | Ser: 0 Thr: 2 Tyr: 0 |
| GmAQP68 | GmSIP1;3 | Glyma.19G108400 | 19:35912781..35923174 | 747 | 248 | 9.12 | 26.56 | Ser: 1 Thr: 2 Tyr: 3 |
| GmAQP69 | GmSIP1;4 | Glyma.16G043800 | 16:4096288..4102424 | 747 | 248 | 9.10 | 26.52 | Ser: 2 Thr: 2 Tyr: 2 |
| GmAQP70 | GmSIP1;5 | Glyma.12G097800 | 12:8369034..8369846 | 720 | 239 | 9.99 | 26.02 | Ser: 4 Thr: 3 Tyr: 1 |
| GmAQP71 | GmSIP1;6 | Glyma.06G307000 | 6:48987251..48988278 | 720 | 239 | 9.91 | 25.87 | Ser: 4 Thr: 4 Tyr: 1 |
| GmAQP72 | GmSIP2;1 | Glyma.03G119300 | 3:35075328..35078322 | 693 | 230 | 9.45 | 25.24 | Ser: 6 Thr: 7 Tyr: 0 |
| GmAQP73 | GmSIP2;2 | Glyma.19G123600 | 19:37949307..37951724 | 711 | 236 | 9.45 | 25.97 | Ser: 10 Thr: 6 Tyr: 0 |
| GmAQP74 | GmXIP1;1 | Glyma.12G023600 | 12:1729006..1730580 | 939 | 312 | 7.02 | 33.75 | Ser: 6 Thr: 2 Tyr: 3 |
| GmAQP75 | GmXIP1;2 | Glyma.11G097800 | 11:7449914..7450765 | 852 | 283 | 6.50 | 30.12 | Ser: 9 Thr: 5 Tyr: 3 |
In phosphorylation site analyses, 72% of GmAQPs were found to contain all three phosphorylation sites (Ser, Thr and Tyr). Among them, Ser and Thr phosphorylation sites were found in 7 GmTIPs (GmTIP1;1, GmTIP1;2, GmTIP1;3, GmTIP1;6, GmTIP1;9, GmTIP1;10 and GmTIP4;1), 4 GmNIPs (GmNIP5;2, GmNIP6;1, GmNIP6;2 and GmNIP6;3), and 2 GmSIPs (GmSIP2;1 and GmSIP2;2). Ser and Tyr phosphorylation sites were present in GmPIP2;1, GmPIP2;2, and GmTIP3;2. Thr and Tyr phosphorylation sites were distributed in GmTIP1;7 and GmTIP1;8. Ser and Tyr phosphorylation sites existed in GmTIP3;3. Thr phosphorylation sites were located in GmSIP1;1 and GmSIP1;2.
2.2. Key Structural Features of the AQP Proteins
To understand the possible physiological role and substrate specificity of soybean AQP proteins, the TM domains, NPA motifs, ar/R selectivity filters, and FPs were investigated (Table 2; Figures S1–S6). Protein structure analyses supported that all GmAQP proteins possessed the typically conserved regions, which contained 6 TM domains (TM1 to TM6) (Figure S1). All GmPIPs and GmTIPs contained two conserved NPA motifs in LB and LE. In GmNIPs, the first NPA showed the same sequence as in PIPs and TIPs, except for GmNIP5;2, where A was replaced by S. The second NPA motif showed an A to V substitution in four NIPs (GmNIP1;6, GmNIP5;2, GmNIP6;2, and GmNIP6;3). GmSIPs showed a second NPA motif completely conserved with the other members. Instead, all of the first NPA motifs showed the replacement of A by T (GmSIP1;1, GmSIP1;2, GmSIP1;3, and GmSIP1;4), A by S (GmSIP1;5 and GmSIP1;6), and A by L (GmSIP2;1 and GmSIP1;2). In GmXIPs, A in the first NPA of GmXIP1;1 was changed to I, and N in the second NPA was changed to S. The second NPA of GmXIP1;2 was completely conserved, while the N and A residues in the first NPA were replaced by S and V.
Table 2.
Conserved amino acid residues (Asn-Pro-Ala (NPA) motifs, aromatic/arginine (ar/R) filters and Froger’s positions (FPs)) and trans-membrane (TM) domains of AQP proteins in soybean.
| Gene Name | Gene Symbol | TM Number | NPA Motifs | ar/R Selectivity Filters | FPs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LB | LE | H2 | H5 | LE1 | LE2 | P1 | P2 | P3 | P4 | P5 | |||
| GmPIP1;1 | Glyma.03G078700 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP1;2 | Glyma.18G198300 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP1;3 | Glyma.01G220600 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP1;4 | Glyma.11G023200 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP1;5 | Glyma.05G208700 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP1;6 | Glyma.08G015300 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP1;7 | Glyma.14G061500 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP1;8 | Glyma.11G228000 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP1;9 | Glyma.01G113400 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP1;10 | Glyma.02G255000 | 6 | NPA | NPA | F | H | T | R | E | S | A | F | W |
| GmPIP2;1 | Glyma.04G003200 | 6 | NPA | NPA | F | H | T | R | M | S | A | F | W |
| GmPIP2;2 | Glyma.06G003200 | 6 | NPA | NPA | F | H | T | R | M | S | A | F | W |
| GmPIP2;3 | Glyma.11G146500 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;4 | Glyma.12G075400 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;5 | Glyma.12G172500 | 6 | NPA | NPA | F | H | T | R | Q | S | A | Y | W |
| GmPIP2;6 | Glyma.13G325900 | 6 | NPA | NPA | F | H | T | R | Q | S | A | Y | W |
| GmPIP2;7 | Glyma.03G180900 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;8 | Glyma.19G181300 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;9 | Glyma.02G073600 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;10 | Glyma.16G155000 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;11 | Glyma.16G155100 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;12 | Glyma.02G073700 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;13 | Glyma.10G211000 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmPIP2;14 | Glyma.20G179700 | 6 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| GmTIP1;1 | Glyma.02G094700 | 6 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| GmTIP1;2 | Glyma.18G286700 | 6 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| GmTIP1;3 | Glyma.10G290600 | 6 | NPA | NPA | H | I | A | V | T | C | A | Y | W |
| GmTIP1;4 | Glyma.11G143100 | 6 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| GmTIP1;5 | Glyma.12G066200 | 6 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| GmTIP1;6 | Glyma.13G333100 | 6 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| GmTIP1;7 | Glyma.03G185900 | 6 | NPA | NPA | H | I | A | V | T | T | A | Y | W |
| GmTIP1;8 | Glyma.19G186100 | 6 | NPA | NPA | H | I | A | V | T | T | A | Y | W |
| GmTIP1;9 | Glyma.13G146300 | 6 | NPA | NPA | H | I | A | A | T | S | A | Y | W |
| GmTIP1;10 | Glyma.20G098600 | 6 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| GmTIP2;1 | Glyma.01G208200 | 6 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| GmTIP2;2 | Glyma.11G034000 | 6 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| GmTIP2;3 | Glyma.07G018000 | 6 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| GmTIP2;4 | Glyma.08G203000 | 6 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| GmTIP2;5 | Glyma.13G356000 | 6 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| GmTIP2;6 | Glyma.15G018100 | 6 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| GmTIP2;7 | Glyma.19G035400 | 6 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| GmTIP3;1 | Glyma.09G160500 | 6 | NPA | NPA | H | I | A | L | T | A | S | F | W |
| GmTIP3;2 | Glyma.16G210000 | 6 | NPA | NPA | H | I | A | L | T | A | S | F | W |
| GmTIP3;3 | Glyma.10G174400 | 6 | NPA | NPA | H | I | A | R | T | A | A | F | W |
| GmTIP3;4 | Glyma.20G216100 | 6 | NPA | NPA | H | I | A | R | T | A | A | F | W |
| GmTIP4;1 | Glyma.04G083200 | 6 | NPA | NPA | H | I | A | R | S | S | A | Y | W |
| GmTIP4;2 | Glyma.06G084600 | 6 | NPA | NPA | H | I | A | R | S | S | A | Y | W |
| GmTIP5;1 | Glyma.09G224700 | 6 | NPA | NPA | S | V | G | C | V | A | A | Y | W |
| GmNIP1;1 | Glyma.05G162600 | 6 | NPA | NPA | W | V | A | R | F | S | A | Y | V |
| GmNIP1;2 | Glyma.08G120200 | 6 | NPA | NPA | W | V | A | R | F | S | A | Y | V |
| GmNIP1;3 | Glyma.13G224900 | 6 | NPA | NPA | W | V | A | R | F | S | A | Y | V |
| GmNIP1;4 | Glyma.15G087300 | 6 | NPA | NPA | W | V | A | R | F | S | A | Y | V |
| GmNIP1;5 | Glyma.08G120100 | 6 | NPA | NPA | W | V | A | R | F | S | A | Y | L |
| GmNIP1;6 | Glyma.05G162500 | 6 | NPA | NPV | W | V | A | R | F | S | A | Y | L |
| GmNIP2;1 | Glyma.07G217700 | 6 | NPA | NPA | W | V | A | R | F | S | A | Y | V |
| GmNIP3;1 | Glyma.14G174300 | 6 | NPA | NPA | S | V | A | R | Y | S | A | Y | I |
| GmNIP4;1 | Glyma.02G246700 | 6 | NPA | NPA | W | V | A | R | L | S | A | Y | V |
| GmNIP4;2 | Glyma.14G069500 | 6 | NPA | NPA | W | V | A | R | L | S | A | Y | V |
| GmNIP5;1 | Glyma.09G238200 | 6 | NPA | NPA | G | S | G | R | L | T | A | Y | F |
| GmNIP5;2 | Glyma.10G221100 | 6 | NPS | NPV | A | I | G | R | Y | T | A | Y | L |
| GmNIP6;1 | Glyma.18G259500 | 6 | NPA | NPA | G | S | G | R | L | T | A | Y | F |
| GmNIP6;2 | Glyma.08G217400 | 6 | NPA | NPV | N | I | S | R | F | T | A | Y | L |
| GmNIP6;3 | Glyma.15G003900 | 6 | NPA | NPV | T | I | G | R | Y | T | A | Y | L |
| GmNIP7;1 | Glyma.02G140500 | 6 | NPA | NPA | A | V | G | R | Y | S | A | Y | M |
| GmNIP7;2 | Glyma.10G033600 | 6 | NPA | NPA | A | V | G | R | Y | S | A | Y | M |
| GmSIP1;1 | Glyma.02G069800 | 6 | NPT | NPA | I | I | P | F | M | A | A | Y | W |
| GmSIP1;2 | Glyma.16G151300 | 6 | NPT | NPA | I | I | P | F | M | A | A | Y | W |
| GmSIP1;3 | Glyma.19G108400 | 6 | NPT | NPA | V | V | P | N | M | A | A | Y | W |
| GmSIP1;4 | Glyma.16G043800 | 6 | NPT | NPA | V | V | P | N | M | A | A | Y | W |
| GmSIP1;5 | Glyma.12G097800 | 6 | NPS | NPA | N | A | P | N | L | A | A | Y | W |
| GmSIP1;6 | Glyma.06G307000 | 6 | NPS | NPA | N | A | P | N | L | A | A | Y | W |
| GmSIP2;1 | Glyma.03G119300 | 6 | NP_ | NPA | S | H | G | S | I | V | A | Y | W |
| GmSIP2;2 | Glyma.19G123600 | 6 | NP_ | NPA | S | H | G | S | I | V | A | Y | W |
| GmXIP1;1 | Glyma.12G023600 | 6 | NPI | SPA | V | V | A | R | E | C | A | F | W |
| GmXIP1;2 | Glyma.11G097800 | 6 | SPV | NPA | V | V | V | R | D | C | A | F | W |
The ar/R positions (H2, H5, LE1, and LE2) of GmAQPs showed increased sub-family specificity compared to the two NPA motifs. In GmPIPs, all selectivity filters were F-H-T-R. In GmTIPs, the ar/R positions were formed by H/S in H2, I/V in H5, A/G in LE1, and V/A/R/L/C in LE2. In GmTIPs, these selectivity filters were constituted by W/A/T/N/S/G in H2, V/S/I in H5, A/G/S in LE1, and R in LE2. GmSIPs showed I/V/N/S in H2, I/V/N/H in H5, P/G in LE1, and F/A/S in LE2. The ar/R sites in GmXIPs were quite homogeneous, with V in H2 and H5, A/V in LE1, and R in LE2. The FPs (P1, P2, P3, P4 and P5) of GmAQPs exhibited divergent combinations, such as E/Q/M-S-A-F/Y-W for GmPIPs, T/S/V-S/A/T/C-A/S-F/Y-W for GmTIPs, F/Y/L-T/S-A-Y-W/V/I/L/M/F for GmNIPs, F/N/S-M/L-A-Y-W for GmSIPs, and E/D-C-A-F-W for GmXIPs.
2.3. Chromosome Distribution of the AQP Genes
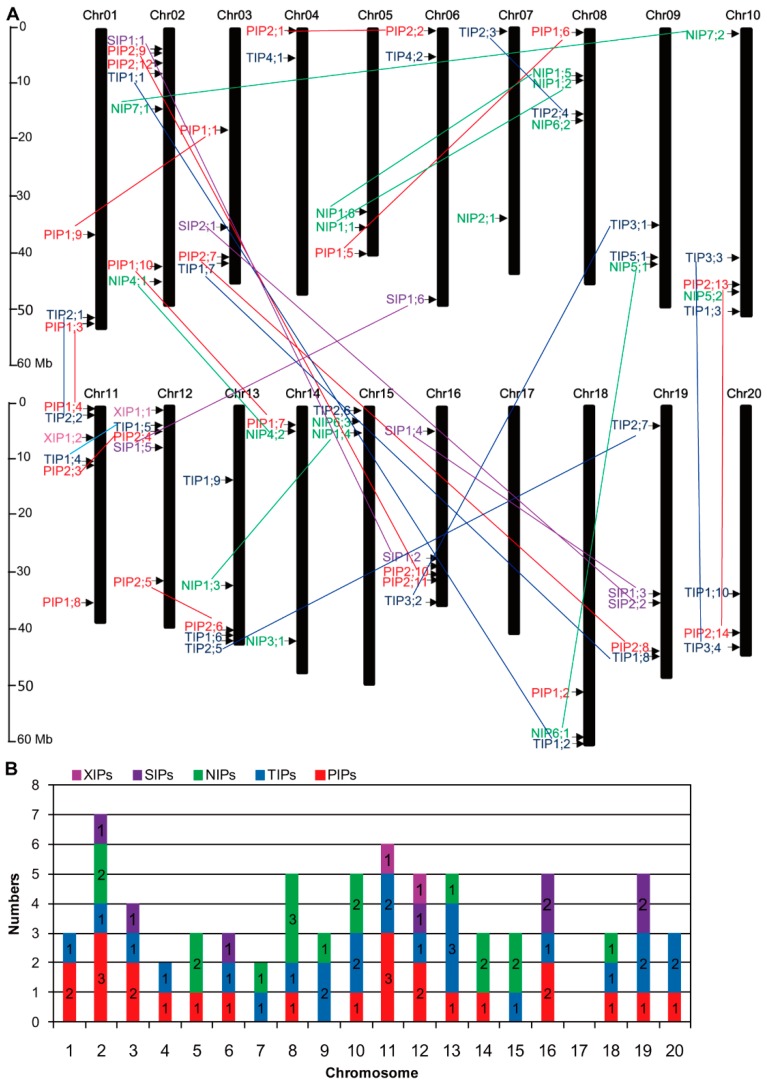
The genomic distribution of each soybean AQP was investigated, as indicated in Figure 1. Seventy-five GmAQPs were mapped on 19 chromosomes (Figure 1A). Among them, GmAQPs on chromosomes 2, 4, 6, 8, 11, 12, 14, and 15 exhibited the tendency to scatter closely to the upper end of the arm, while GmAQPs on chromosomes 1, 3, 5, 9, 10, 13, 16, 18, 19, and 20 tended to scatter closely to the lower end of the arm. Gene numbers on chromosomes 2 (7 loci each chromosome) were the maximum, whereas the gene numbers on chromosomes 4 and 7 (2 loci each chromosome) were the minimum (Figure 1B). PIPs and TIPs scattered extensively over the soybean chromosomes. While NIPs seemed to locate on chromosomes 2, 5, 7, 8, 9, 10, 13, 14, 15, and 18, SIPs were positioned on chromosomes 2, 3, 6, 12, 16 and 19. XIPs were resided on chromosomes 11 and 12 (Figure 1B).
Figure 1.
Chromosomal distribution of soybean AQP genes. (A) Graphical representation of physical locations for each AQP gene on soybean chromosomes (numbered Chr01–20). The scale on the left indicated the genomic length in megabases (Mb). PIPs, TIPs, NIPs, XIPs and SIPs were indicated with red, blue, green, purple and pink fonts, respectively. Lines represented putative gene duplications. (B) Numbers of PIPs, TIPs, NIPs, XIPs and SIPs on each soybean chromosome.
To further understand the expansion mechanism of GmAQPs, the gene duplication events were analyzed. Two duplication events (GmPIP2;9/PIP2;12 and GmPIP2;10/PIP2;11) within the same chromosome and twenty-eight duplication events (GmPIP1;1/PIP1;9, GmPIP1;3/PIP1;4, GmPIP1;5/PIP1;6, GmPIP1;7/PIP1;10, GmPIP2;1/PIP2;2, GmPIP2;3/PIP2;4, GmPIP2;5/PIP2;6, GmPIP2;7/PIP2;8, GmPIP2;9/PIP2;10, GmPIP2;13/PIP2;14, GmTIP1;1/TIP1;2, GmTIP1;4/TIP1;5, GmTIP1;7/TIP1;8, GmTIP2;1/TIP2;2, GmTIP2;3/TIP2;4, GmTIP2;5/TIP2;7, GmTIP3;1/TIP3;2, GmTIP3;3/TIP3;4, GmNIP1;1/NIP1;2, GmNIP1;3/NIP1;4, GmNIP1;5/NIP1;6, GmNIP4;1/NIP4;2, GmNIP5;1/NIP6;1, GmNIP7;1/NIP7;2, GmSIP1;1/SIP1;2, GmSIP1;3/SIP1;4, GmSIP1;5/SIP1;6 and GmSIP2;1/SIP2;2) between different chromosomes were identified, respectively.
2.4. Evolutionary Characterization of the AQP Genes
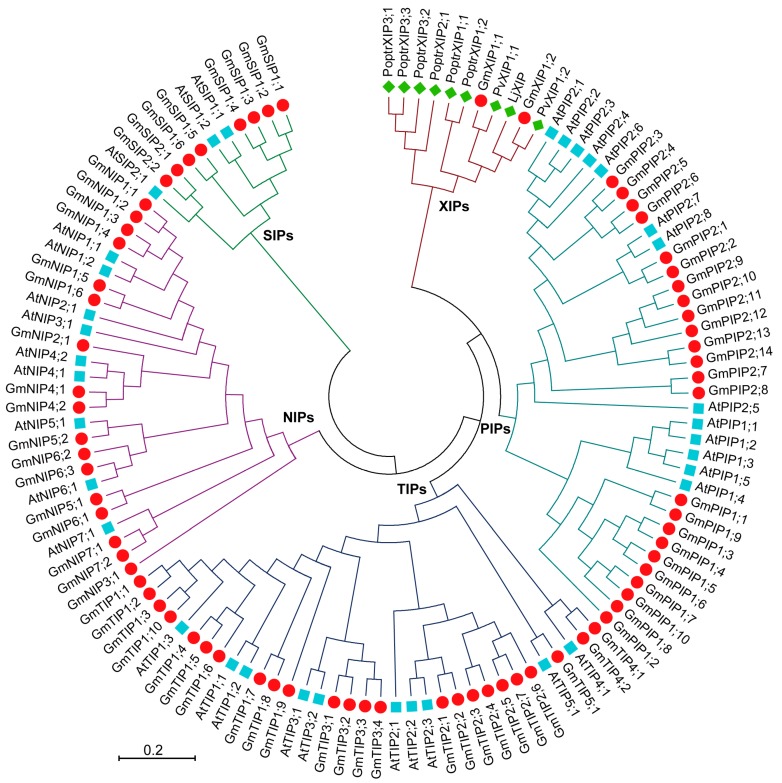
To investigate the classification and evolutionary relationship of soybean AQP proteins, the phylogenetic tree was constructed with the full-length GmAQP protein sequences from Arabidopsis, Phaseolus vulgaris, Populus trichocarpa, and Lotus japonicus (Dataset S1; Dataset S2). Soybean AQPs grouped into five sub-families (PIPs, 24; TIPs, 24; NIPs, 17; SIPs, 8; XIPs, 2) (Figure 2). Among the PIP sub-family, 24 members were divided into two groups: PIP1 with 10 members and PIP2 with 14 members. Five groups were found for the TIP sub-family (TIP1 to TIP5), with 10 members in the TIP1 group, 7 members in the TIP2 group, 4 members in the TIP3 group, 2 members in the TIP4 group, and 1 member in the TIP5 group. Seven groups belonged to the NIP sub-family (NIP1 to NIP7), with 6 members in the NIP1 group, 1 member in the NIP2 group, 1 member in the NIP3 group, 2 members in the NIP4 group, 2 members in the NIP5 group, 3 members in the NIP6 group, and 2 members in the NIP7 group. The SIP sub-family were composed of two groups (SIP1 and SIP2), with 6 members in the SIP1 group and 2 members in the SIP2 group. Only one group was identified for the XIP sub-family (XIP1), with 2 members.
Figure 2.
Phylogenetic relationships of AQP proteins from soybean, Arabidopsis, Phaseolus vulgaris, Populus trichocarpa and Lotus japonicus. The five sub-families were indicated with different colors. The red spots represent soybean AQPs. The blue squares represent Arabidopsis AQPs. The green diamonds represent Phaseolus vulgaris, Populus trichocarpa and Lotus japonicus XIPs.
Moreover, the putative orthologues of soybean GmAQPs with known Arabidopsis AtAQPs were identified. For PIP sub-family genes, GmPIP2;1 and GmPIP2;2 were the best orthology matches of Arabidopsis AtPIP2;7 and AtPIP2;8; GmPIP2;3, GmPIP2;4, GmPIP2;5, and GmPIP2;6 were the most homogeneous genes of Arabidopsis AtPIP2;1, AtPIP2;2, AtPIP2;3, AtPIP2;4, and AtPIP2;6. For TIP sub-family genes, GmTIP1;1, GmTIP1;2, GmTIP1;3, and GmTIP1;10 exhibited the closest relationship with Arabidopsis AtTIP1;3. GmTIP2;1 and GmTIP2;2 clustered closely with Arabidopsis AtTIP2;2 and AtTIP2;3. GmTIP3;1, GmTIP3;2, GmTIP3;3, and GmTIP3;4 shared a fairly close evolutionary relationship with Arabidopsis AtTIP3;1 and AtTIP3;2. GmTIP4;1 and GmTIP4;2 were phylogenetically closest to Arabidopsis AtTIP4;1. GmTIP5;1 was in the same evolutionary clade with Arabidopsis AtTIP5;1. For NIP sub-family genes, GmNIP1;1, GmNIP1;2, GmNIP1;3, and GmNIP1;4 showed the closest relationship with Arabidopsis AtNIP1;1 and AtNIP1;2. GmNIP4;1 and GmNIP4;2 gathered closely with Arabidopsis AtNIP4;1 and AtNIP4;2. GmNIP5;2 was highly homologous with Arabidopsis AtNIP5;1. GmNIP7;1 and GmNIP7;2 grouped closely with Arabidopsis AtNIP7;1. For SIP sub-family genes, GmSIP1;1, GmSIP1;2, GmSIP1;3, and GmSIP1;4 were the potential orthologs with Arabidopsis AtSIP1;1 and AtSIP1;2. GmSIP2;1 and GmSIP2;2 were highly homologous with Arabidopsis AtSIP2;1.
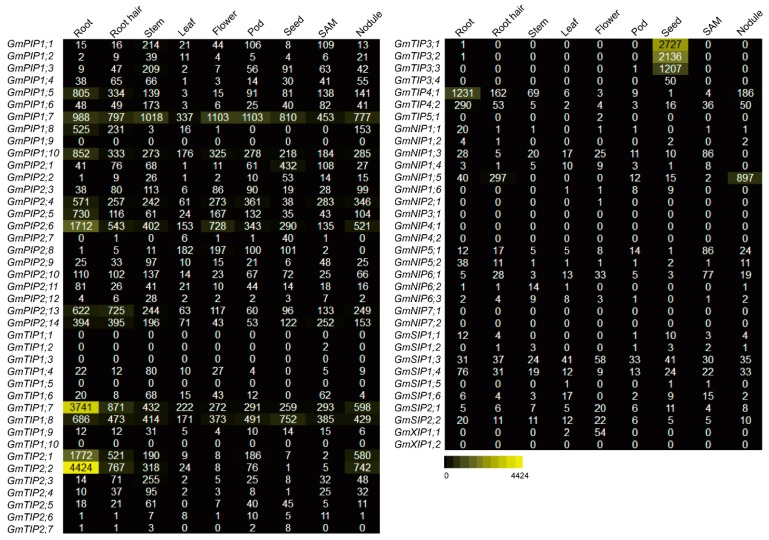
2.5. Expression Profiles of the AQP Genes in Different Tissues
To examine the tissue expression profiles of the soybean AQP genes, the RNA-seq data were retrieved from available soybean database Phytozome V12.1. For different GmAQP members, different expression profiles were represented by different colors (Figure 3). Among them, one PIP gene GmPIP1;9, four TIP genes (GmTIP1;1, GmTIP1;3, GmTIP1;5, and GmTIP1;10), five NIP genes (GmNIP3;1, GmNIP4;1, GmNIP4;2, GmNIP7;1, and GmNIP7;2), and one XIP gene GmXIP1;2 did not express in any tested tissues of soybean. In contrast, GmPIP1;7, GmPIP1;10, GmPIP2;4, GmPIP2;6, GmTIP1;7, and GmTIP1;8 were highly expressed in all the investigated tissues. Additionally, for different tissues, the expression level analyses indicated significant differentiation among different GmAQP members. GmPIP1;5, GmPIP1;8, GmPIP2;13, GmPIP2;14, GmTIP2;1, GmTIP2;2, and GmTIP4;1 were significantly expressed in roots. GmPIP2;8 was highly expressed in leaves and flowers. GmPIP2;1, GmTIP3;1, GmTIP3;2, and GmTIP3;3 were mainly expressed in seeds. GmTIP2;3 was highly expressed in stems. GmNIP1;5 was significantly expressed in root hairs and nodules.
Figure 3.
Expression profiles of soybean AQP genes in different tissues. Different expression levels in nine tissues (leaf, stem, root, flower, seed, root hair, pod, shoot apical meristem (SAM), and nodule.) were indicated in color using the RNA-seq data.
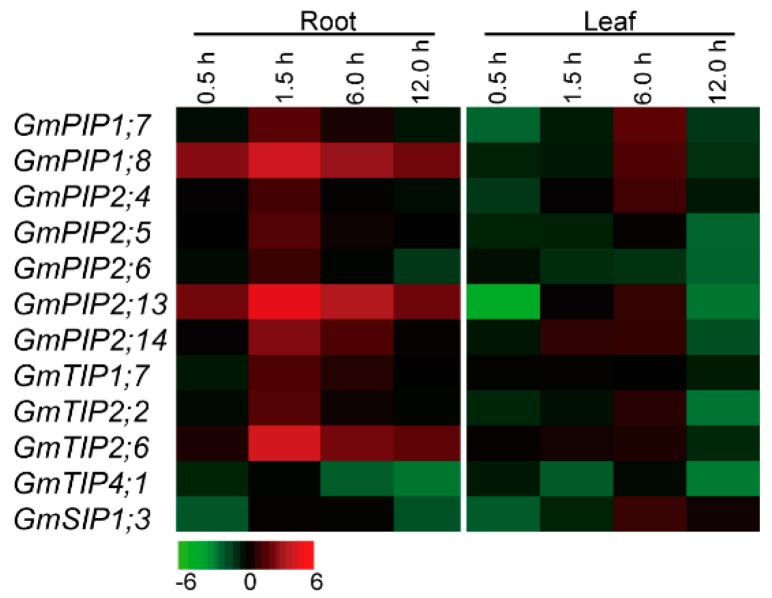
2.6. Expression Profiles of the Candidate AQP Genes in Response to Heat Stress
To explore the roles of soybean AQP genes in response to heat stress, expression profiles of 12 candidate GmAQPs were selected for investigation using qRT-PCR, due to relatively extensive expression in various tissues or high expression levels in root or leaf (Figure 3). In two different tissues, heat stress obviously up-regulated the expression of GmAQPs during the early durations (1.5 hour) in roots, whereas heat stress slightly up-regulated expression of GmAQPs during the late durations (6.0 hour) in leaves (Figure 4; Table S1). In roots, most of the analyzed members (GmPIP1;7, GmPIP1;8, GmPIP2;4, GmPIP2;5, GmPIP2;13, GmPIP2;14, GmTIP1;7, GmTIP2;2, and GmTIP2;6) were transcriptionally up-regulated, whereas GmTIP4;1 and GmSIP1;3 were extremely down-regulated after heat treatment. In leaves, the transcripts of GmPIP2;5, GmPIP2;6, GmTIP1;7, and GmTIP4;1 were inhibited by heat stress, while GmPIP1;7, GmPIP1;8, GmPIP2;4, GmPIP2;13, GmPIP2;14, GmTIP2;2, and GmSIP1;3 were firstly inhibited and then promoted. Amongst them, GmPIP1;8, GmPIP2;13, GmPIP2;14, and GmTIP2;6 were relatively dramatically induced in roots, but GmTIP4;1 was sharply repressed in both the roots and leaves under heat stress.
Figure 4.
Expression profiles of 12 candidate soybean AQP genes under heat stress treatment. 0.5, 1.5, 6.0 and 12 hour represent the treatment times. The color scales represent relative expression data.
2.7. Expression Profiles of the Candidate AQP Genes in Response to ABA, ACC, SA, and MeJA Signals
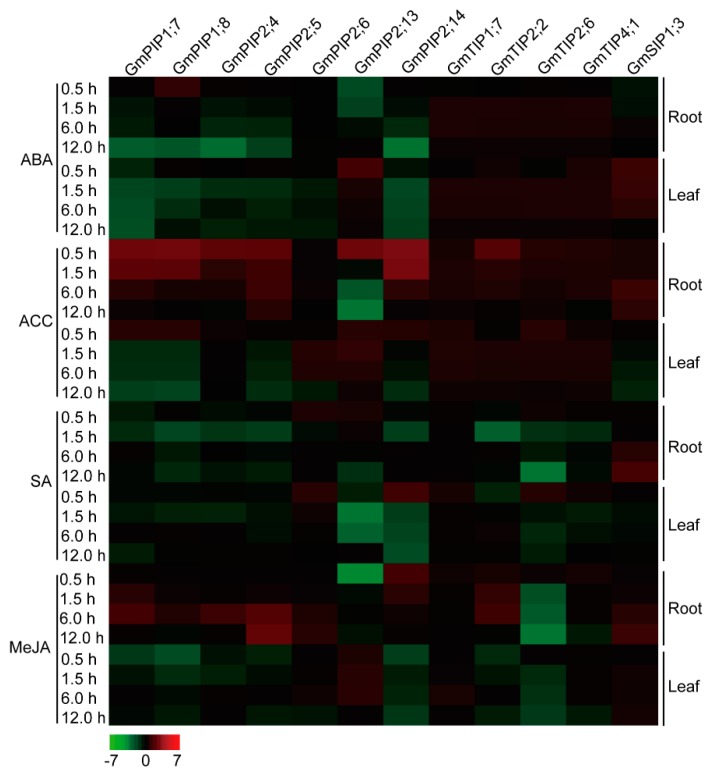
Further, these candidate soybean AQP genes were subjected to qRT-PCR analyses to evaluate their roles in response to hormone signals (Figure 5; Table S1). For ABA treatment, most transcripts of PIP genes underwent down-regulation after ABA treatment, except GmPIP1;8 and GmPIP2;13, whereas TIP genes shared up-regulation in both the roots and leaves. GmPIP1;8 was strongly up-regulated after 0.5 hour of ABA stress in roots and down-regulated in leaves. GmPIP2;13 was abundantly down-regulated after 0.5 hour of ABA stress in roots and up-regulated in leaves. For ACC treatment, most transcripts of PIP genes showed an increase in roots after 0.5 hour, 1.5 hour, and 6.0 hour of ACC stress, except GmPIP2;13. However, in leaves, a subset of PIP genes (GmPIP1;7, GmPIP1;8, GmPIP2;4, GmPIP2;5, GmPIP2;6, and GmPIP2;14) initially displayed up-regulation expression following gradual down-regulation expression. GmPIP2;13 was up-regulated only during early duration (0.5 hour) and then down-regulated in roots and continuously up-regulated in leaves. For TIP genes, all of them exhibited up-regulated expression after ACC treatment. GmSIP1;3 was up-regulated in roots and down-regulated in leaves. For SA treatment, a cluster of PIP genes (GmPIP1;7, GmPIP1;8, GmPIP2;4, and GmPIP2;5) presented down-regulation in both the roots and leaves. GmPIP2;6 was up-regulated in both the roots and leaves during early durations (0.5 and 1.5 hour) following gradual down-regulation. GmPIP2;13 was up-regulated after 0.5 hour of SA stress in roots and down-regulated in leaves. GmPIP2;14 displayed obvious down-regulation in roots and up-regulation in leaves after 0.5 hour of SA stress following down-regulation. For TIP genes, the transcriptional level of GmTIP1;7 was detected with up-regulation in both the roots and leaves. GmTIP2;2 showed sharp down-regulation after 1.5 hour of SA stress in the roots. GmTIP4;1 was up-regulated at 0.5 hour of SA stress and then down-regulated in both the roots and leaves. GmSIP1;3 displayed high transcript abundance in roots and low transcript abundance in leaves. For MeJA treatment, the expression levels of PIP genes (GmPIP1;7, GmPIP1;8, GmPIP2;4, GmPIP2;5, and GmPIP2;14) were promoted in roots and inhibited in leaves, while GmPIP2;13 was inhibited in roots and promoted in leaves. For TIP genes, GmTIP1;7 maintained up-regulation in both the roots and leaves. GmTIP2;2 showed up-regulation in roots and down-regulation in leaves. GmTIP4;1 presented up-regulation during early duration (0.5 hour) following gradual down-regulation in both the roots and leaves. The transcript of GmSIP1;3 maintained a high level in both the roots and leaves.
Figure 5.
Expression profiles of 12 candidate soybean AQP genes under abscisic acid (ABA), l-aminocyclopropane-l-carboxylic acid (ACC), salicylic acid (SA), and methyl jasmonate (MeJA) hormone treatments. 0.5, 1.5, 6.0 and 12 hour represent the treatment times. The color scales represent relative expression data.
2.8. Promoter Regulatory Elements of the Candidate AQP Genes
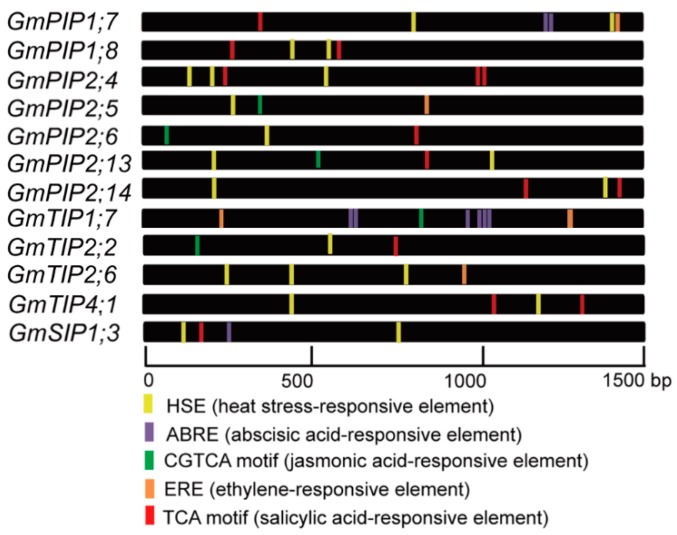
1.5 kb sequences, upstream of these 12-candidate soybean AQP coding sequences, were analyzed. The cis-acting regulatory elements were classified into two types: heat stress and hormone responsive elements (Figure 6; Table 3). Eleven GmAQPs were detected having heat stress-related elements. For instance, three HSE elements for GmPIP2;4 and GmTIP2;6; two HSE elements for GmPIP1;7, GmPIP1;8, GmTIP4;1, and GmSIP1;3 and one HSE element for GmPIP2;5 and GmTIP2;2. Moreover, 3 GmAQPs contained ABRE (response to ABA), such as six ABRE elements for GmTIP1;7; two ABRE elements for GmPIP1;7 and one ABRE element for GmSIP1;3. Five GmAQPs possessed CGTCA (response to MeJA), including one MeJA element for GmPIP2;5, GmPIP2;6, GmPIP2;13, GmTIP1;7, and GmTIP2;2. 4 GmAQPs harbored ERE (response to ethylene), such as two ERE elements for GmTIP1;7 and one ERE element for GmPIP1;7, GmPIP2;5, and GmTIP2;6. Nine GmAQPs contained TCA (response to SA), such as three TCA elements for GmPIP2;4 and two TCA elements for GmPIP1;8, GmPIP2;14, and GmTIP4;1 and two TCA elements for GmPIP1;7, GmPIP2;6, GmPIP2;13, GmTIP2;2, and GmSIP1;3. Among them, more than one hormone responsive element was observed in the promoter regions of GmPIP1;7, GmPIP2;5, GmPIP2;6, GmPIP2;13, GmTIP1;7, GmTIP2;2, and GmSIP1;3.
Figure 6.
Distribution of cis-acting elements in 12 candidate soybean AQP gene promoters. 1500 bp adjacent to the AQP coding sequence. The elements are represented by different colors. The scale bar represents 500 bp.
Table 3.
Number of cis-acting elements in 12 candidate soybean AQP gene promoters.
| Gene Name | Gene Symbol | HSE | ABRE | CGTCA | ERE | TCA |
|---|---|---|---|---|---|---|
| GmPIP1;7 | Glyma.14G061500 | 2 | 2 | 0 | 1 | 1 |
| GmPIP1;8 | Glyma.11G228000 | 2 | 0 | 0 | 0 | 2 |
| GmPIP2;4 | Glyma.12G075400 | 3 | 0 | 0 | 0 | 3 |
| GmPIP2;5 | Glyma.12G172500 | 1 | 0 | 1 | 1 | 0 |
| GmPIP2;6 | Glyma.13G325900 | 1 | 0 | 1 | 0 | 1 |
| GmPIP2;13 | Glyma.10G211000 | 2 | 0 | 1 | 0 | 1 |
| GmPIP2;14 | Glyma.20G179700 | 2 | 0 | 0 | 0 | 2 |
| GmTIP1;7 | Glyma.03G185900 | 0 | 6 | 1 | 2 | 0 |
| GmTIP2;2 | Glyma.11G034000 | 1 | 0 | 1 | 0 | 1 |
| GmTIP2;6 | Glyma.15G018100 | 3 | 0 | 0 | 1 | 0 |
| GmTIP4;1 | Glyma.04G083200 | 2 | 0 | 0 | 0 | 2 |
| GmSIP1;3 | Glyma.19G108400 | 2 | 1 | 0 | 0 | 1 |
2.9. GUS Activity of the GmTIP2;6 Promoter
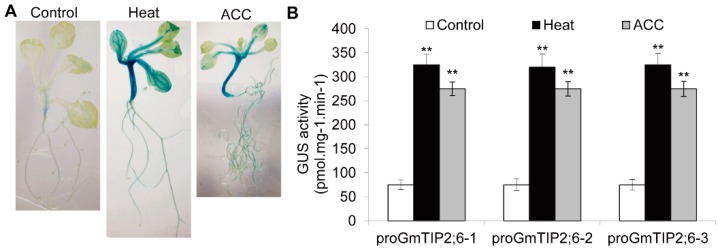
To characterize the function of AQP promoter in response to heat stress and hormone signals, the GmTIP2;6 promoter, with a relatively high number of HSE elements, was fused to the GUS reporter gene and transferred into Arabidopsis (Dataset S3). Under normal growth conditions, the expression pattern of the GUS gene driven by the GmTIP2;6 promoter was weakly detected in the hypocotyls (Figure 7A). After heat treatment, GUS activity was remarkably induced and increased in hypocotyls, roots, leaf vascular bundles, and young leaf trichomes. Similarly, after ACC treatment, the responsiveness of the GmTIP2;6 promoter was enhanced in hypocotyls, roots, leaf vascular bundles, and young leaf trichomes (Figure 7A). Further quantitative GUS activity analyses verified that the translational levels of GUS protein in the transgenic plants with heat and ACC treatments were evidently stronger than those without treatments (Figure 7B). This result confirmed that GmTIP2;6 was one heat stress and ACC hormone inducible promoter.
Figure 7.
Beta-glucuronidase (GUS) activities of GmTIP2;6 promoter under heat stress and hormone treatments. (A) Transgenic seedlings treated with heat and ACC stress. (B) Activity analyses of GUS protein in three proGmTIP2;6-GUS transgenic Arabidopsis plants under different treatments. ** indicates significant differences in comparison with the control treatment at p < 0.01 (t-test).
3. Discussion
AQPs, as representative trans-membrane transporters, have important functions in modulating plant stress tolerance [9,10]. From the recently-updated soybean genome database Phytozome V12.1, 75 putative GmAQPs were identified based on HMM profile, KEGG orthology, BlastP, and BlastN searches (Table 1). All GmAQP proteins possessed six conserved TM helices (TM1 to TM6). Divergent ar/R selectivity filters and FPs were also identified, which were essential for transport specificity of GmAQPs (Table 2). Point mutations or sequence variations of these amino acid residues could confer different substrate permeability in different GmAQP members [30,31,32]. Distinct phosphorylation sites (Ser, Thr, and Tyr) were also detected which might be involved in post-translational modifications of GmAQPs (Table 1). Various environmental conditions such as drought, salinity, or oxidative stresses could induce quantitative changes in PIP, TIP, or NIP phosphorylation at multiple sites on the N-terminal or C-terminal tail [33,34,35]. However, knowledge of the protein kinases and protein phosphatases determining aquaporin phosphorylation is still scarce. All these diverse structure characteristics may allow complex regulation modes for AQPs in response to multiple environmental and hormonal stimuli. Evolutionary analyses showed that GmAQPs were categorized into five distinct sub-families (Figure 2). Furthermore, all identified GmAQPs were phylogenetically compared to the orthologs derived from the dicotyledonous model plant Arabidopsis, which might share highly conservative functions. As AtPIP2;7 was involved in salinity-mediated transcriptional and post-translational regulation [36], it will be interesting to investigate whether their homologous genes GmPIP2;1 and GmPIP2;2 respond to salt stress. AtTIP1;3 was a pollen-specific AQP for transporting water and urea [37], and it will be interesting to test their homologous genes (GmTIP1;1, GmTIP1;2, GmTIP1;3, and GmTIP1;10) for similar permeability. AtTIP5;1 improved plant tolerance to boron toxicity [38], and its homologous gene GmTIP5;1 might also confer resistance to boron. AtNIP7;1 enhanced tolerance to arsenate toxicity [39], and its homologous genes (GmNIP7;1 and GmNIP7;2) might also contribute to arsenate stress. Compared with previous report [28], 17 GmAQP proteins were newly identified which were distributed in PIP, TIP, NIP and XIP sub-families: 11 GmPIPs (GmPIP1;9, GmPIP1;10, GmPIP2;3, GmPIP2;4, GmPIP2;8, GmPIP2;9, GmPIP2;10, GmPIP2;11, GmPIP2;12, GmPIP2;13, and GmPIP2;14), 1 GmTIP (GmTIP1;10), 4 GmNIPs (GmNIP1;6, GmNIP3;1, GmNIP4;1, and GmNIP4;2) and 1 GmXIP (GmXIP1;2). Compared with previous report [29], 2 GmPIPs (GmPIP1;9 and GmPIP1;10) and 1 GmTIP (GmTIP1;10) were newly identified. All these newly identified GmAQPs contained the typical and conserved AQP domains as shown in Figures S1–S6. They allow us to re-identify the gene numbers of soybean AQPs on the recently-updated public soybean genome database Phytozome V12.1. Our current detailed analyses will add more potentially functional AQPs to the set of soybean.
Time, location, and level of gene transcripts reflected the functions of AQPs under both favorable and stressful conditions [40,41,42]. In the present study, tissue expression patterns of GmAQPs were analyzed based on RNA-seq data from the public soybean database. Some transcripts of GmAQPs were expressed at high levels while others were expressed at low levels (Figure 3). Most GmAQPs extensively functioned in multiple tissues, and individual GmAQPs seemed to function in specific tissue, which was highly similar to AQPs in other plant species [11,12,13,14,15,16]. Some AQP gene pairs at the same evolutionary clade preferred to express in the same tissue. For instance, GmPIP2;4, GmPIP2;5, GmPIP2;6, and AtPIP2;4; GmTIP2;1, GmTIP2;2, and AtTIP2;2; and GmTIP4;1, GmTIP4;2, and AtTIP4;1 shared high transcript levels in roots. GmTIP3;1, GmTIP3;2, GmTIP3;3, GmTIP3;4, and AtTIP3;1 and AtTIP3;2 shared seed-specific expression. These results imply that soybean AQPs may function in a wide range of developmental processes.
Heat stress modulated the transcription of plant AQPs, which opened a new avenue of research for identifying soybean AQPs involved in thermo-tolerance [18,19,20,21,22,23]. However, the relationship between GmAQPs and heat resistance in soybean still remains elusive. Preliminary heat stress-related element analyses suggested that GmAQPs might be involved in modulating plant stress tolerance against heat stimuli (Figure S7). The number of heat stress-related elements in GmAQP promoters ranged from 0 to 5. Among them, 11 GmAQPs (GmPIP1;1, GmPIP1;4, GmPIP2;3, GmPIP2;4, GmPIP2;12, GmTIP1;4, GmTIP2;6, GmNIP4;1, GmNIP5;2, GmNIP7;2, and GmSIP1;6) contained more than three heat stress responsive elements. Further expression analyses also confirmed that heat stress could significantly activate or inhibit the expression of candidate GmAQPs (Figure 4). Among them, eight GmAQPs (GmPIP1;7, GmPIP1;8, GmPIP2;4, GmPIP2;5, GmPIP2;13, GmPIP2;14, GmTIP1;7, and GmTIP2;2) were favorably accumulated in roots under heat stress. Based on the public database of Arabidopsis eFP Browser [16,43], systematical microarray analyses also showed that most AtAQPs were involved in the heat stress process (Figure S8; Table S1). All these results indicate that plant AQPs serve as targets for modulating thermo-tolerance.
Plant hormones were important signal molecules that controlled plant growth and development in response to heat stimulus, including ABA, SA, and MeJA [44]. However, evidence for the molecular mechanism of AQPs’ involvement in the hormone response process remained scanty. The preliminary promoter element analyses indicated that different members of soybean GmAQPs possessed distinct hormone-related elements (Figure S7). For instance, two GmAQPs (GmPIP1;1 and GmNIP4;2), three GmAQPs (GmPIP2;9, GmTIP2;6, and GmNIP1;2), eight GmAQPs (GmPIP1;8, GmTIP2;7, GmTIP4;1, GmNIP1;1, GmNIP1;6, GmNIP6;3, GmSIP1;1, and GmSIP2;1) and three GmAQPs (GmTIP5;1, GmNIP7;1, and GmSIP1;4) contained ABA, ET, SA, or MeJA-special element, respectively. In contrast, a combination of four hormone-related elements were observed in GmPIP1;10, GmPIP2;1, and GmTIP4;2. Furthermore, gene expression analyses showed that different GmAQPs displayed distinct transcriptional changes, up-regulation or down-regulation under different hormone treatments, as evidenced by the qRT-PCR assay (Figure 5). For example, five highly up-regulated GmPIPs transcripts during ACC and MeJA hormone treatments in roots included GmPIP1;7, GmPIP1;8, GmPIP2;4, GmPIP2;5, and GmPIP2;14, and four abundantly accumulated GmTIPs during ABA and ACC hormone treatments in both the roots and leaves included GmTIP1;7, GmTIP2;2, GmTIP2;6, and GmTIP4;1. Moreover, the expression of GmSIP1;3 got enhanced and underwent significant changes during ACC, SA, and MeJA hormone treatments in roots. However, the transcript changes of four GmPIPs (GmPIP1;7, GmPIP1;8, GmPIP2;4, and GmPIP2;5) were observed with down-regulation under ABA, SA, and MeJA hormone treatments in leaves. In rice and oilseed rape, ABA highly enhanced the expression of OsTIP1;1 in the shoots and roots and BnTIP2 in the seeds [45,46]. In resurrection plant, ABA greatly decreased the expression of CpTIP in the callus [47]. In wheat, ethylene (ET) up-regulated the gene expression of wheat TaAQP8 (one PIP sub-family gene) under salt stress [48]. In rose, ET decreased the expression of RhTIP1;1 in the flower [49]. Systematical microarray analysis of AQPs under ABA, ACC, and MeJA based on the public database of Arabidopsis eFP Browser showed that most Arabidopsis AQPs were involved in the process (Figure S9; Table S1) [16,43]. Other stress-related signals such as brassinolide (BR), gibberellin (GA), and auxin (IAA) also regulated the expression of AQPs [50,51,52,53] but by, as yet, unclear mechanisms. All this evidence gives clues to the role of AQPs in complicated hormone signal transduction systems in different tissues. In this regard, it will be of interest to decipher further how AQP genes work in the interactive signaling regulation network, especially under heat stress.
Promoters, the direct indication of gene expression patterns, were extensively involved in the responses of signal molecules and environmental elicitors. It was noteworthy that we validated that soybean GmTIP2;6 promoter was a typical inducible promoter, which responded to ACC and heat stresses in hypocotyls, vascular bundles, and leaf trichomes (Figure 7). This was consistent with the qRT-PCR result that GmTIP2;6 was up-regulated by heat stress and ACC hormone (Figure 4 and Figure 5). In cotton, strong expression of the GUS gene driven by GhPIP2;7 promoter was detected in leaves of 5 to 10-day-old transgenic Arabidopsis seedlings, but GUS activity gradually became weak as the seedlings further developed. GUS activity driven by cotton GhPIP2;7 promoter was remarkably increased after mannitol treatment [54]. In Arabidopsis, AtNIP3;1 promoter-mediated GUS activity was specifically expressed in the roots [55]. The expression of the GUS gene driven by AtPIP2;7 promoter was strongly detected in cotyledons, emerging leaf primordia, and root elongation zones, and salt stress induced strong repression of AtPIP2;7 promoter activity [35]. In soybean, the GUS activity of GmTIP2;3 promoter was expressed in the root, stem, and leaf and preferentially expressed in the stele of root and stem [56]. These data indicated different promoters of AQP members played different roles in different tissues or development stages. In the continued study, it will be very meaningful to investigate the core elements of AQP promoters for quantitative and qualitative gene expression regulation.
4. Materials and Methods
4.1. Categorization of Soybean AQP Genes
The soybean genome sequences were retrieved from Phytozome V12.1 (http://phytozome.jgi.doe.gov/pz/portal.html). The keyword searches of aquaporin, Hidden Markov model profile (PF00230), and KEGG Orthology terms (PIPs, K09872; TIPs, K09873; NIPs, K09874; SIPs, K09875) were applied to identify the soybean candidate AQP members [57]. Known Arabidopsis, Zea mays, Oryza sativa, Populus trichocarapa, Phaseolus vulgaris and Lotus japonicas AQPs were also subjected to BlastP and BlastN against the soybean database with cut-off E-value of e−5 [58,59,60,61]. GmAQPs were named based on their sequence homology with known AQPs and soybean genome annotation. The decrease redundancy tool (http://web.expasy.org/decrease_redundancy/) was utilized to discard the redundant AQP sequences. Further, the resulting candidate sequences were checked for the presence of six TM domains and two NPA motifs by SMART (http://smart.embl-heidelberg.de/smart/batch.pl) and NCBI-CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) web servers. The numbers of phosphorylation sites of AQP proteins were predicted with NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/). The molecular weight (MW) and isoelectric point (pI) of AQP proteins were calculated using ExPASy (http://web.expasy.org). The chromosomal positions of GmAQPs were mapped using MapInspect software based on the starting position of all genes on each chromosome. Tandem duplications were identified manually. Adjacent genes of the same sub-group tightly linked within 20 kb of each other and the identity of the genes ≥80% are considered as tandem duplicated genes [62].
4.2. Phylogenetic Tree
The GmAQP full length protein sequences were aligned using ClustalX2 software. As soon as the ALN file was generated, MEGA6 was carried out to construct the neighbor-joining (NJ) phylogenetic tree [63]. The criteria were adopted with pairwise deletion option and Poisson correction model. Bootstrap test was performed with 1000 replicates.
4.3. Tissular Expression Profile Analyses
The RNA-seq data of GmAQP genes in different tissues, including leaf, stem, root, flower, seed, root hair, pod, SAM, and nodule, was available from Phytozome V12.1 database [64]. BAR HeatMapper Tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper.cgi) was carried out to display the expression profiles of AQP genes in heatmaps [16].
4.4. Heat Stress and Hormone Treatments
Soybean cultivar GMLN012012017, with the characteristic of heat tolerance, was used in this study. Soybean seeds were cultivated in pots in an illuminated incubator (PTC-300, Shanghai, China) adjusted to 22 °C temperature, 60% relative humidity, 16/8 h photoperiod, and25000 Lux light intensity. For high-temperature treatment, 21-day-old seedlings in pots were transferred to the illuminated incubator adjusted to 42 °C. The un-treated samples were used as the control (0 hour). For hormone treatments, the root systems of 21-day-old seedlings were washed gently with water to remove soil and then the plants were soaked into 200 mL solutions with 100 µM abscisic acid (ABA), 100 µM l-aminocyclopropane-l-carboxylic acid (ACC), 100 µM salicylic acid (SA) or 100 µM methyl jasmonate (MeJA). The samples soaked with water were used as the control (0 hour). Each single seedling was sampled at one time point (0, 0.5, 1.5, 6.0 or 12 hour). Then, whole leaves and roots of each control or treated seedling were collected and frozen in liquid nitrogen and stored at −80 °C for further analyses.
4.5. qRT-PCR
Total RNA was separately extracted from the frozen samples using a RNA Simple Total RNA Extraction Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. Then, the cDNA was synthesized using a FastQuant RT Kit (Tiangen, Beijing, China). Gene-specific primers were designed using PrimerQuest Tool (http://sg.idtdna.com/PrimerQuest/Home/) (Table S2). GmActin11 (Glyma.18G290800) was selected as the internal reference gene [65,66,67,68]. The amplification reactions were performed on Applied Biosystems StepOnePlusTM Real-Time System using KAPA SYBR®Fast qPCR Kit (Tiangen, Beijing, China) with the following parameters: initializing denaturation at 95 °C for 5 min, followed by 45 cycles of denaturation at 95 °C for 5 seconds, annealing at 58 °C for 5 seconds, and extension at 72 °C for 30 seconds. Three technical replicates were maintained for each sample. The relative expression levels were calculated as 2−ΔΔCt [69]. The heatmaps for the expression profiles of GmAQP genes were generated with BAR HeatMapper Tool [16].
4.6. Promoter Element Prediction
1.5 kb promoter regions, upstream of the AQP gene coding sequences, were extracted from Phytozome V12.1. Promoter regions were subsequently analyzed using PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to illustrate the number and composition of hormone and heat stress responsive elements.
4.7. Promoter Cloning and Arabidopsis Transformation
The promoter of GmTIP2;6 was isolated using the specific primer pairs (Table S2). To generate the proGmTIP2;6::GUS construct, the CaMV 35S promoter was replaced by the promoter of GmTIP2;6. The promoter of GmTIP2;6 was inserted into Sal I/Sma I sites and sub-cloned into the pBI121 vector whose Hind III site was replaced by three continuous sites (Hind III, Pst I, and Sal I) upstream of the CaMV 35S promoter (Figure S10). The GUS fusion construct was then introduced into Arabidopsis (Col-0) by Agrobacterium-mediated floral-dip method [70,71]. Transformed seeds were selected on MS medium with 50 mg/L kanamycin (Kan). Homozygous lines of T3 were used for the following GUS activity assays.
4.8. GUS Activity Detection
To evaluate GUS activity, the proGmTIP2;6::GUS transgenic seeds were sowed on MS medium for 5 days, and then transferred to MS medium supplemented with 100 µM ACC for 7 days. For the high-temperature treatment, 5-day-old seedlings on MS medium were transferred to the temperature-controlled chamber adjusted to 37 °C for 7 days. Seedlings on MS medium without any additions were used as controls [72]. GUS staining was conducted using standard protocols. In brief, seedlings were incubated in 10 mL tubes with 1 mg/mL 5-bromo-4-chloro-3-indolylglucuronic acid, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.03% Triton X-100, and 0.1 M sodium phosphate buffer, pH 7.0 overnight at 37 °C. Then, seedlings were immersed in 70% ethanol to remove the chlorophyll and visualized on the Leica microscope. GUS activities were measured by monitoring the cleavage of GUS substrate 4-methylumbelliferyl glucuronide as reported previously [73]. Data analyses of variance were used to compare the statistical difference based on Student’s t-test, at a significant level of p < 0.01.
Acknowledgments
This work was supported by National Key R&D Program of China (2017YFD0101500), National Natural Science Foundation of China (31601767 and 31572138), Zhejiang Provincial Important Science & Technology Specific Projects (2016C02051), Zhejiang Provincial Natural Science Foundation (LY17C150007), and Zhejiang Academy of Agricultural Sciences Program for Young Talent (2016R23R08E06).
Abbreviations
| AQP | aquaporin |
| ABA | abscisic acid |
| ACC | l-aminocyclopropane-l-carboxylic acid |
| SA | salicylic acid |
| MeJA | methyl jasmonate |
| qRT-PCR | quantitative real-time PCR |
| GUS | beta-glucuronidase |
Supplementary Materials
The following are available online at http://www.mdpi.com/1422-0067/20/2/262/s1. Dataset S1. Fasta files of 75 AQP proteins in soybean. Dataset S2. Fasta files of AQP proteins in Arabidopsis, Phaseolus vulgaris, Populus trichocarpa and Lotus japonicus. Dataset S3. Fasta file of the promoter for GmTIP2;6 in soybean. Table S1. Gene expression numbers for GmAQPs and AtAQPs in response to heat stress and hormone signals. Table S2. List of primers used in this study. Figure S1. Predicted structures of 75 AQP proteins in soybean. Figure S2. Conserved amino acid residues (NPA motifs, ar/R filter, FPs) and TM domains of PIPs in soybean. Figure S3. Conserved amino acid residues (NPA motifs, ar/R filter, FPs) and TM domains of TIPs in soybean. Figure S4. Conserved amino acid residues (NPA motifs, ar/R filter, FPs) and TM domains of NIPs in soybean. Figure S5. Conserved amino acid residues (NPA motifs, ar/R filter, FPs) and TM domains of SIPs in soybean. Figure S6. Conserved amino acid residues (NPA motifs, ar/R filter, FPs) and TM domains of XIPs in soybean. Figure S7. Distribution of cis-acting elements in 75 soybean AQP gene promoters. 1500 bp adjacent to the AQP coding sequence. The elements are represented by different colors. The scale bar represents 500 bp. Figure S8. Expression profiles of Arabidopsis AQP genes in response to heat stress. Differential sub-family AQP gene expression in response to heat stress across eight time points (0.25, 0.5, 1, 3, 4, 6, 12 and 24 hour) in two tissues (roots and shoots). Differences in gene expression changes are shown in color in the green-red scale. Figure S9. Expression profiles of Arabidopsis AQP genes in response to ABA, ACC, and MeJA signals. Differential sub-family AQP gene expression in response to ABA, ACC, and MeJA hormones across three time points (0.5, 1, and 3 hour). Differences in gene expression changes were shown in color in the green-red scale. Figure S10. Restrictive enzyme sites of pBI121 for promoter vector construction. The Hind III site upstream of the CaMV 35S promoter was replaced by three continuous sites (Hind III, Pst I, and Sal I).
Author Contributions
All authors prepared, read, and approved the final manuscript. Z.-J.F. designed the research and wrote the first draft. N.L. and G.-W.Z. performed the bioinformatic work and analyzed the experiment data. F.-G.N. and S.-C.X. managed reagents and provided analytical tools. Y.-M.G. contributed with valuable discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Fox A.R., Maistriaux L.C., Chaumont F. Toward understanding of the high number of plant aquaporin isoforms and multiple regulation mechanisms. Plant Sci. 2017;264:179–187. doi: 10.1016/j.plantsci.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 2.Chaumont F., Barrieu F., Wojcik E., Chrispeels M.J., Jung R. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 2001;125:1206–1215. doi: 10.1104/pp.125.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang J.Y., Kim D.G., Kim Y.O., Kim J.S., Kang H. An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol. Biol. 2004;54:713–725. doi: 10.1023/B:PLAN.0000040900.61345.a6. [DOI] [PubMed] [Google Scholar]
- 4.Sakurai J., Ishikawa F., Yamaguchi T., Uemura M., Maeshima M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005;46:1568–1577. doi: 10.1093/pcp/pci172. [DOI] [PubMed] [Google Scholar]
- 5.Park W., Scheffler B.E., Bauer P.J., Campbell B.T. Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.) BMC Plant Biol. 2010;10:142–158. doi: 10.1186/1471-2229-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reuscher S., Akiyama M., Mori C., Aoki K., Shibata D., Shiratake K. Genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE. 2013;8:e79052. doi: 10.1371/journal.pone.0079052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue C., Cao H., Wang L., Zhou Y., Hao X., Zeng J., Wang X., Yang Y. Molecular cloning and expression analysis of tea plant aquaporin (AQP) gene family. Plant Physiol. Biochem. 2014;83:65–76. doi: 10.1016/j.plaphy.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Abascal F., Irisarri I., Zardoya R. Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta. 2014;1840:1468–1481. doi: 10.1016/j.bbagen.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Afzal Z., Howton T.C., Sun Y.L., Shahid Mukhtar M. The roles of aquaporins in plant stress responses. J. Dev. Biol. 2016;4:9. doi: 10.3390/jdb4010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurel C., Verdoucq L., Luu D.T., Santoni V. Plant aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 2008;59:595–624. doi: 10.1146/annurev.arplant.59.032607.092734. [DOI] [PubMed] [Google Scholar]
- 11.Alexandersson E., Fraysse L., Sjövall-Larsen S., Gustavsson S., Fellert M., Karlsson M., Johanson U., Kjellbom P. Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 2005;59:469–484. doi: 10.1007/s11103-005-0352-1. [DOI] [PubMed] [Google Scholar]
- 12.Boursiac Y., Chen S., Luu D.T., Sorieul M., van den Dries N., Maurel C. Early effects of salinity on water transport in Arabidopsis roots molecular and cellular features of aquaporin expression. Plant Physiol. 2005;139:790–805. doi: 10.1104/pp.105.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Putpeerawit P., Sojikul P., Thitamadee S., Narangajavana J. Genome-wide analysis of aquaporin gene family and their responses to water-deficit stress conditions in cassava. Plant Physiol. Biochem. 2017;121:118–127. doi: 10.1016/j.plaphy.2017.10.025. [DOI] [PubMed] [Google Scholar]
- 14.Kadam S., Abril A., Dhanapal A.P., Koester R.P., Vermerris W., Jose S., Fritschi F.B. Characterization and regulation of aquaporin genes of sorghum [Sorghum bicolor (L.) Moench] in response to waterlogging stress. Front. Plant Sci. 2017;8:862. doi: 10.3389/fpls.2017.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hove R.M., Bhave M. Plant aquaporins with non-aqua functions: Deciphering the signature sequences. Plant Mol. Biol. 2011;75:413–430. doi: 10.1007/s11103-011-9737-5. [DOI] [PubMed] [Google Scholar]
- 16.Feng Z.J., Xu S.C., Liu N., Zhang G.W., Hu Q.Z., Xu Z.S., Gong Y.M. Identification of the AQP members involved in abiotic stress responses from Arabidopsis. Gene. 2018;646:64–73. doi: 10.1016/j.gene.2017.12.048. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesh J., Yu J.W., Park S.W. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol. Biochem. 2013;73:392–404. doi: 10.1016/j.plaphy.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 18.Szucs A., Jäger K., Jurca M.E., Fábián A., Bottka S., Zvara A., Barnabás B., Fehér A. Histological and microarray analysis of the direct effect of water shortage alone or combined with heat on early grain development in wheat (Triticum aestivum) Physiol. Plant. 2010;140:174–188. doi: 10.1111/j.1399-3054.2010.01394.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen K., Wang X., Fessehaie A., Yin Y., Wang X., Arora R. Is expression of aquaporins (plasma membrane intrinsic protein 2s, PIP2s) associated with thermonasty (leaf-curling) in Rhododendron. J. Plant Physiol. 2013;170:1447–1454. doi: 10.1016/j.jplph.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Christou A., Filippou P., Manganaris G.A., Fotopoulos V. Sodium hydrosulfide induces systemic thermotolerance to strawberry plants through transcriptional regulation of heat shock proteins and aquaporin. BMC Plant Biol. 2014;14:42. doi: 10.1186/1471-2229-14-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha P., Sade N., Arzani A., Wilhelmi M.D.M.R., Coe K.M., Li B., Blumwald E. Effects of abiotic stress on physiological plasticity and water use of Setaria viridis (L.) Plant Sci. 2016;251:128–138. doi: 10.1016/j.plantsci.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Obaid A.Y., Sabir J.S., Atef A., Liu X., Edris S., El-Domyati F.M., Mutwakil M.Z., Gadalla N.O., Hajrah N.H., Al-Kordy M.A., et al. Analysis of transcriptional response to heat stress in Rhazya stricta. BMC Plant Biol. 2016;16:252. doi: 10.1186/s12870-016-0938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgii E., Jin M., Zhao J., Kanawati B., Schmitt-Kopplin P., Albert A., Winkler J.B., Schäffner A.R. Relationships between drought, heat and air humidity responses revealed by transcriptome-metabolome co-analysis. BMC Plant Biol. 2017;17:120. doi: 10.1186/s12870-017-1062-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maruyama K., Ogata T., Kanamori N., Yoshiwara K., Goto S., Yamamoto Y.Y., Tokoro Y., Noda C., Takaki Y., Urawa H., et al. Design of an optimal promoter involved in the heat-induced transcriptional pathway in Arabidopsis, soybean, rice and maize. Plant J. 2017;89:671–680. doi: 10.1111/tpj.13420. [DOI] [PubMed] [Google Scholar]
- 25.Das A., Eldakak M., Paudel B., Kim D.W., Hemmati H., Basu C., Rohila J.S. Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. BioMed Res. Int. 2016;2016:6021047. doi: 10.1155/2016/6021047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdés-López O., Batek J., Gomez-Hernandez N., Nguyen C.T., Isidra-Arellano M.C., Zhang N., Joshi T., Xu D., Hixson K.K., Weitz K.K., et al. Soybean roots grown under heat stress show global changes in their transcriptional and proteomic Profiles. Front. Plant Sci. 2016;7:517. doi: 10.3389/fpls.2016.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahsan N., Donnart T., Nouri M.Z., Komatsu S. Tissue-specific defense and thermo-adaptive mechanisms of soybean seedlings under heat stress revealed by proteomic approach. J. Proteome Res. 2010;9:4189–4204. doi: 10.1021/pr100504j. [DOI] [PubMed] [Google Scholar]
- 28.Zhang D.Y., Ali Z., Wang C.B., Xu L., Yi J.X., Xu Z.L., Liu X.Q., He X.L., Huang Y.H., Khan I.A., et al. Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.) PLoS ONE. 2013;8:e56312. doi: 10.1371/journal.pone.0056312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deshmukh R.K., Vivancos J., Guérin V., Sonah H., Labbé C., Belzile F., Bélanger R.R. Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 2013;3:303–315. doi: 10.1007/s11103-013-0087-3. [DOI] [PubMed] [Google Scholar]
- 30.Beitz E., Wu B., Holm L.M., Schultz J.E., Zeuthen T. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc. Natl. Acad. Sci. USA. 2006;103:269–274. doi: 10.1073/pnas.0507225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hub J.S., de Groot B.L. Mechanism of selectivity in aquaporins and aquaglyceroporins. Proc. Natl. Acad. Sci. USA. 2008;105:1198–1203. doi: 10.1073/pnas.0707662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitani-Ueno N., Yamaji N., Zhao F.J., Ma J.F. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J. Exp. Bot. 2011;62:4391–4398. doi: 10.1093/jxb/err158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prak S., Hem S., Boudet J., Viennois G., Sommerer N., Rossignol M., Maurel C., Santoni V. Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: Role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol. Cell. Proteom. 2008;7:1019–1030. doi: 10.1074/mcp.M700566-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Daniels M.J., Yeager M. Phosphorylation of aquaporin PvTIP3; 1 defined by mass spectrometry and molecular modeling. Biochemistry. 2005;44:14443–14454. doi: 10.1021/bi050565d. [DOI] [PubMed] [Google Scholar]
- 35.Guenther J.F., Chanmanivone N., Galetovic M.P., Wallace I.S., Cobb J.A., Roberts D.M. Phosphorylation of soybean nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell. 2003;15:981–991. doi: 10.1105/tpc.009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pou A., Jeanguenin L., Milhiet T., Batoko H., Chaumont F., Hachez C. Salinity-mediated transcriptional and post-translational regulation of the Arabidopsis aquaporin PIP2;7. Plant Mol. Biol. 2016;92:731–744. doi: 10.1007/s11103-016-0542-z. [DOI] [PubMed] [Google Scholar]
- 37.Soto G., Alleva K., Mazzella M.A., Amodeo G., Muschietti J.P. AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 2008;582:4077–4082. doi: 10.1016/j.febslet.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Pang Y., Li L., Ren F., Lu P., Wei P., Cai J., Xin L., Zhang J., Chen J., Wang X. Overexpression of the tonoplast aquaporin AtTIP5;1 conferred tolerance to boron toxicity in Arabidopsis. J. Genet. Genom. 2010;37:389–397. doi: 10.1016/S1673-8527(09)60057-6. [DOI] [PubMed] [Google Scholar]
- 39.Lindsay E.R., Maathuis F.J. Arabidopsis thaliana NIP7;1 is involved in tissue arsenic distribution and tolerance in response to arsenate. FEBS Lett. 2016;590:779–786. doi: 10.1002/1873-3468.12103. [DOI] [PubMed] [Google Scholar]
- 40.Hachez C., Zelazny E.F. Chaumont modulating the expression of aquaporin genes in planta: A key to understand their physiological functions. Biochem. Biophys. Acta. 2006;1758:1142–1156. doi: 10.1016/j.bbamem.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Kapilan R., Vaziri M., Zwiazek J.J. Regulation of aquaporins in plants under stress. Biol. Res. 2018;51:4. doi: 10.1186/s40659-018-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawłowicz I., Masajada K. Aquaporins as a link between water relations and photosynthetic pathway in abiotic stress tolerance in plants. Gene. 2019;687:166–172. doi: 10.1016/j.gene.2018.11.031. [DOI] [PubMed] [Google Scholar]
- 43.Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamaoui M., Jemo M., Datla R., Bekkaoui F. Heat and drought stresses in crops and approaches for their mitigation. Front. Chem. 2018;6:26. doi: 10.3389/fchem.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Q., Umeda M., Uchimiya H. Isolation and expression analysis of two rice genes encoding the major intrinsic protein. Plant Mol. Biol. 1994;26:2003–2007. doi: 10.1007/BF00019511. [DOI] [PubMed] [Google Scholar]
- 46.Gao Y.P., Young L., Bonham-Smith P., Gusta L.V. Characterization and expression of plasma and tonoplast membrane aquaporins in primed seed of Brassica napus during germination under stress conditions. Plant Mol. Biol. 1999;40:635–644. doi: 10.1023/A:1006212216876. [DOI] [PubMed] [Google Scholar]
- 47.Mariaux J.B., Bockel C., Salamini F., Bartels D. Dessication- and abscisic acid-responsive genes encoding major intrinsic proteins (MIPs) from the resurrection plant Craterostigma plantagineum. Plant Mol. Biol. 1998;38:1089–1099. doi: 10.1023/A:1006013130681. [DOI] [PubMed] [Google Scholar]
- 48.Hu W., Yuan Q., Wang Y., Cai R., Deng X., Wang J., Zhou S., Chen M., Chen L., Huang C., et al. Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol. 2012;53:2127–2141. doi: 10.1093/pcp/pcs154. [DOI] [PubMed] [Google Scholar]
- 49.Xue J.Q., Yang F., Gao J.P. Isolation of RhTIP1;1, an aquaporin gene and its expression in rose flowers in response to ethylene and water deficit. Postharvest Biol. Tec. 2009;51:407–413. doi: 10.1016/j.postharvbio.2008.08.011. [DOI] [Google Scholar]
- 50.Phillips A.L., Huttly A.K. Cloning of two gibberellin-regulated cDNAs from Arabidopsis thaliana by subtractive hybridization: Expression of the tonoplast water channel, gamma-TIP, is increased by GA3. Plant Mol. Biol. 1994;24:603–615. doi: 10.1007/BF00023557. [DOI] [PubMed] [Google Scholar]
- 51.Morillon R., Catterou M., Sangwan R.S., Sangwan B.S., Lassalles J.P. Brassinolide may control aquaporin activities in Arabidopsis thaliana. Planta. 2001;212:199–204. doi: 10.1007/s004250000379. [DOI] [PubMed] [Google Scholar]
- 52.Gattolin S., Sorieul M., Hunter P.R., Khonsari R.H., Frigerio L. In vivo imaging of the tonoplast intrinsic protein family in Arabidopsis roots. BMC Plant Biol. 2009;9:133–141. doi: 10.1186/1471-2229-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li G., Santoni V., Maurel C. Plant aquaporins: Roles in plant physiology. Biochim. Biophys. Acta. 2014;1840:1574–1582. doi: 10.1016/j.bbagen.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J., Li D., Zou D., Luo F., Wang X., Zheng Y., Li X. A cotton gene encoding a plasma membrane aquaporin is involved in seedling development and in response to drought stress. Acta Biochim. Biophys. Sin. 2013;45:104–114. doi: 10.1093/abbs/gms096. [DOI] [PubMed] [Google Scholar]
- 55.Xu W., Dai W., Yan H., Li S., Shen H., Chen Y., Xu H., Sun Y., He Z., Ma M. Arabidopsis NIP3;1 Plays an important role in arsenic uptake and root-to-shoot translocation under arsenite stress conditions. Mol. Plant. 2015;8:722–733. doi: 10.1016/j.molp.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Zhang D., Tong J., He X., Xu Z., Xu L., Wei P., Huang Y., Brestic M., Ma H., Shao H. A novel soybean intrinsic protein gene, GmTIP2;3, involved in responding to osmotic Stress. Front. Plant Sci. 2016;6:1237. doi: 10.3389/fpls.2015.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Paula Santos Martins C., Pedrosa A.M., Du D., Gonçalves L.P., Yu Q., Gmitter F.G., Jr., Costa M.G. Genome-wide characterization and expression analysis of major intrinsic proteins during abiotic and biotic stresses in sweet orange (Citrus sinensis L. Osb.) PLoS ONE. 2015;10:e0138786. doi: 10.1371/journal.pone.0138786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johanson U., Karlsson M., Johansson I., Gustavsson S., Sjövall S., Fraysse L., Weig A.R., Kjellbom P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 2001;126:1358–1369. doi: 10.1104/pp.126.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta A.B., Sankararamakrishnan R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: Characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant. Biol. 2009;9:134. doi: 10.1186/1471-2229-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ariani A., Gepts P. Genome-wide identification and characterization of aquaporin gene family in common bean (Phaseolus vulgaris L.) Mol. Genet. Genom. 2015;290:1771–1785. doi: 10.1007/s00438-015-1038-2. [DOI] [PubMed] [Google Scholar]
- 61.Giovannetti M., Balestrini R., Volpe V., Guether M., Straub D., Costa A., Ludewig U., Bonfante P. Two putative-aquaporin genes are differentially expressed during arbuscular mycorrhizal symbiosis in Lotus japonicus. BMC Plant Biol. 2012;12:186. doi: 10.1186/1471-2229-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng Z.J., He G.H., Zheng W.J., Lu P.P., Chen M., Gong Y.M., Ma Y.Z., Xu Z.S. Foxtail millet NF-Y families: Genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Plant Sci. 2015;6:1142. doi: 10.3389/fpls.2015.01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura K., Dudley J., Nei M., Kumar S. MEGA4, molecular evolutionary genetics analysis MEGA software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 64.Libault M., Farmer A., Joshi T., Takahashi K., Langley R.J., Franklin L.D., He J., Xu D., May G., Stacey G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;63:86–99. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- 65.Zhang D., Wan Q., He X., Ning L., Huang Y., Xu Z., Liu J., Shao H. Genome-wide characterization of the ankyrin repeats gene family under salt stress in soybean. Sci. Total Environ. 2016;568:899–909. doi: 10.1016/j.scitotenv.2016.06.078. [DOI] [PubMed] [Google Scholar]
- 66.Quach T.N., Nguyen H.T., Valliyodan B., Joshi T., Xu D., Nguyen H.T. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol. Genet. Genom. 2015;290:1095–1115. doi: 10.1007/s00438-014-0978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu H., Che Z., Zeng X., Zhou X., Sitoe H.M., Wang H., Yu D. Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct. Integr. Genom. 2016;16:481–493. doi: 10.1007/s10142-016-0498-8. [DOI] [PubMed] [Google Scholar]
- 68.Zhou F., Guo Y., Qiu L.J. Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Biol. 2016;16:58–70. doi: 10.1186/s12870-016-0744-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCт method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 70.Jefferson R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 1987;5:387–405. doi: 10.1007/BF02667740. [DOI] [Google Scholar]
- 71.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhang H., Jing R., Mao X. Functional characterization of TaSnRK2.8 promoter in response to abiotic stresses by deletion analysis in transgenic Arabidopsis. Front. Plant Sci. 2017;8:1198. doi: 10.3389/fpls.2017.01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song S., Xu Y., Huang D., Miao H., Liu J., Jia C., Hu W., Valarezo A.V., Xu B., Jin Z. Identification of a novel promoter from banana aquaporin family gene (MaTIP1;2) which responses to drought and salt-stress in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2018;128:163–169. doi: 10.1016/j.plaphy.2018.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.