Abstract
Background
Diphtheria toxin (DT) is produced by toxigenic strains of the human pathogen Corynebacterium diphtheriae as well as zoonotic C. ulcerans and C. pseudotuberculosis. Toxigenic strains may cause severe respiratory diphtheria, myocarditis, neurological damage or cutaneous diphtheria. The DT encoding tox gene is located in a mobile genomic region and tox variability between C. diphtheriae and C. ulcerans has been postulated based on sequences of a few isolates. In contrast, species-specific sequence analysis of the diphtheria toxin repressor gene (dtxR), occurring both in toxigenic and non-toxigenic Corynebacterium species, has not been done yet. We used whole genome sequencing data from 91 toxigenic and 46 non-toxigenic isolates of different pathogenic Corynebacterium species of animal or human origin to elucidate differences in extracted DT, DtxR and tox-surrounding genetic elements by a phylogenetic analysis in a large sample set.
Results
Sequences of both DT and DtxR, extracted from whole genome sequencing data, could be classified in four distinct, nearly species-specific clades, corresponding to C. diphtheriae, C. pseudotuberculosis, C. ulcerans and atypical C. ulcerans from a non-toxigenic toxin gene-bearing wildlife cluster. Average amino acid similarities were above 99% for DT and DtxR within the four groups, but lower between them. For DT, subgroups below species level could be identified, correlating with different tox-comprising mobile genetic elements. In most C. diphtheriae, tox genes were located within known prophages. In contrast, in C. ulcerans diverse tox-including mobile elements could be identified: either prophages differing from C. diphtheriae prophages or an alternative pathogenicity island (PAI) described previously. One isolate showed a different, shorter tox-comprising putative PAI. Beyond the tox-overlapping elements, most isolates harbored a variety of additional prophages.
Conclusion
Our NGS data from 137 isolates indicate the existence of different genetic backgrounds of DT-mediated pathogenicity in different Corynebacterium species and evolution of once acquired pathogenicity features with the strains. Different groups of pathogenicity-related elements within C. ulcerans imply that tox transmission pathways between isolates may differ in the zoonotic species and contribute to their emerging pathogenic potential.
Electronic supplementary material
The online version of this article (10.1186/s12866-019-1402-1) contains supplementary material, which is available to authorized users.
Keywords: Bacterial infections, Diphtheria, Whole genome sequencing, Public health, Corynebacterium, Pathogenicity, Phylogeny, Diphtheria toxin, Prophages, Genomic Islands
Background
Besides several non-pathogenic species, the genus Corynebacterium comprises more than 50 medically relevant ones. Some of them have a high pathogenic potential for human and animal hosts [1, 2]. An important pathogen is Corynebacterium diphtheriae, with humans as their almost exclusive reservoir and only very rare case reports of isolates from infected animals, such as e.g. horses or pigs [3, 4]. Additional important pathogenic species include zoonotic C. ulcerans and C. pseudotuberculosis. Despite the omnipresence of vaccination programs against toxoids in most developed countries, diseases with toxigenic corynebacteria occur consistently, due to various reasons: For example vaccination gaps and/or global travel activity from and to endemic countries occur [5]. Also other factors, for example quality and availability of vaccines may be problematic, especially in low-income countries [6]. In addition to classic C. diphtheriae mediated diseases, infections with toxigenic C. ulcerans outnumber those of toxigenic C. diphtheriae since several years in central Europe. C. ulcerans is increasingly recognized as emerging pathogen with inland animal contact as the most important risk factor [7, 8] in contrast to most often abroad acquired infections by toxigenic C. diphtheriae. C. diphtheriae, as well as C. ulcerans and C. pseudotuberculosis are all able to produce various virulence- and pathogenicity-related proteins, which are often associated with cell adhesion or iron homeostasis [9]. The diphtheria toxin (DT) encoding tox gene-bearing strains may produce DT, which is one of the most potent bacterial toxins. Thus, toxigenic strains may cause severe respiratory diphtheria, myocarditis, neurological damage or severe cutaneous diphtheria. DT was one of the first bacterial toxins detected [10] and its mode of action and structure have been extensively studied [11]. It is an AB exotoxin, acting by inhibition of protein synthesis. It consists of a catalytic subunit A and an adhesion- and cell entry-mediating subunit B with a receptor-binding (R) and a translocation-mediating (T) domain [12]. The DT encoding tox gene is located in a mobile genomic region. In C. diphtheriae different tox gene-carrying prophages, introduced into the bacterial genome through lysogenic conversion, have already been described and sequenced [9, 13]. In C. ulcerans an alternative pathogenicity island (PAI) has been detected additionally [5]. Genetic tox variability within the species C. diphtheriae [14] or between single cases of C. diphtheriae and C. ulcerans has been shown [15]. However, it is not known yet if surrounding prophages or PAIs are absolutely species-specific or also transmitted between Corynebacterium species. DT is furthermore part of an iron-dependent regulation system with the diphtheria toxin repressor (DtxR) acting as main regulatory control center on DT, but also on various other virulence factors [16]. In contrast to the DT encoding tox gene, the dtxR gene occurs both in toxigenic and non-toxigenic isolates of the different Corynebacterium species and its genetic variability between them has not been analysed in detail yet.
During the last years the German Consiliary Laboratory on Diphtheria has sequenced a substantial number of genomes of Corynebacterium spp. isolates by Next Generation Sequencing (NGS), mainly in regard to various outbreak investigations. In this study we used NGS-generated Corynebacterium spp. isolate genomes, partly from previous outbreak investigations, to analyse the differences between several pathogenic Corynebacterium species regarding DT, its regulator DtxR and the tox gene-surrounding mobile elements. We aimed to investigate if differences between species can be observed on a regular basis when a larger sample set is analysed and which phylogenetic conclusions regarding the DT-mediated pathogenicity of medically relevant corynebacteria can be drawn from the results.
Results
To investigate the potential differences of DT, its iron-dependent regulator DtxR and the tox gene-surrounding, pathogenicity-related genetic elements in pathogenic corynebacteria, we conducted WGS analyses with isolates from different Corynebacterium and host species and from different geographic backgrounds (Table 1).
Table 1.
Isolates and epidemiology
| Sample name | Species | Year | Host | Host Disease | Risk factor | Toxin-PCR | Elek | Outbreak ref |
|---|---|---|---|---|---|---|---|---|
| 08–1143-CB1 | C. ulcerans | 2007 | Pig | Asymptomatic colonisation | NA | Positive | Positive | [7] |
| KL0126-cb2 | C. ulcerans | 2007 | Human | Diphtheria-like | Animal contact (pig) | Positive | Positive | [7] |
| KL0160 | C. pseudotuberculosis | 2009 | Goat | NA | NA | Negative | N.a. | – |
| KL0182 | Atypical C. ulcerans (NTTB) | 2010 | Boar | Lymphadenitis | NA | Positive | Negative | – |
| KL0183 | Atypical C. ulcerans (NTTB) | 2010 | Boar | Lymphadenitis | NA | Positive | Negative | – |
| KL0194 | C. ulcerans | 2010 | Human | NA | NA | Positive | Positive | – |
| KL0195 | C. ulcerans (NTTB) | 2010 | Human | NA | NA | Positive | Negative | – |
| KL0199 | C. ulcerans | 2010 | Human | NA | NA | Negative | N.a. | – |
| KL0246-cb3 | C. ulcerans | 2010 | Human | NA | Animal contact (cat) | Positive | Positive | [7] |
| KL0251-cb4 | C. ulcerans (NTTB) | 2010 | Cat | Asymptomatic colonisation | NA | Positive | Negative | [7] |
| KL0252-cb5 | C. ulcerans (NTTB) | 2010 | Cat | Asymptomatic colonisation | NA | Positive | Negative | [7] |
| KL0259 | Atypical C. ulcerans (NTTB) | 2011 | Boar | Lung abscess | NA | Positive | Negative | – |
| KL0260 | Atypical C. ulcerans (NTTB) | 2011 | Boar | Multiple abscesses | NA | Positive | Negative | – |
| KL0262 | C. pseudotuberculosis (NTTB) | 2011 | Buffalo | NA | NA | Positive | Negative | – |
| KL0263 | C. pseudotuberculosis (NTTB) | 2011 | Buffalo | NA | NA | Positive | Negative | – |
| KL0264 | C. pseudotuberculosis | 2011 | Human | NA | NA | Negative | Negative | – |
| KL0265 | C. pseudotuberculosis | 2011 | Human | NA | NA | Negative | Negative | – |
| KL0266 | C. pseudotuberculosis | 2011 | Human | NA | NA | Negative | Negative | – |
| KL0269 | C. pseudotuberculosis | 2011 | Goat | Abscess | NA | Negative | N.a. | – |
| KL0276 | C. diphtheriae | 2011 | Human | NA | NA | Positive | Positive | – |
| KL0315-cb6 | C. ulcerans (NTTB) | 2012 | Human | Ulcer | Animal contact (dog) | Positive | Negative | [7] |
| KL0318-cb7 | C. ulcerans (NTTB) | 2012 | Dog | Asymptomatic colonisation | NA | Positive | Negative | [7] |
| KL0330 | C. diphtheriae | 2012 | Human | Wound infection | Homeless | Negative | N.a. | [17] |
| KL0345 | C. ulcerans (NTTB) | 2012 | Cat | Rhinitis | NA | Positive | Negative | – |
| KL0349 | C. ulcerans | 2012 | Human | NA | NA | Negative | Negative | – |
| KL0350 | C. ulcerans | 2012 | Human | NA | NA | Positive | Positive | – |
| KL0355 | C. diphtheriae | 2012 | Human | Wound infection | NA | Negative | N.a. | [17] |
| KL0360 | C. diphtheriae | 2012 | Human | Wound infection | Travel (Africa) | Negative | N.a. | [17] |
| KL0371 | C. diphtheriae | 2012 | Human | Wound infection | NA | Negative | N.a. | [17] |
| KL0372 | C. diphtheriae | 2012 | Human | Wound infection | Mixed infection (S. aureus, Enterobacter spp., Pantoea spp.) | Negative | N.a. | [17] |
| KL0374 | Atypical C. ulcerans (NTTB) | 2012 | Boar | NA | NA | Positive | Negative | – |
| KL0377 | C. diphtheriae | 2012 | Human | Wound infection | Mixed infection (hemolysing Streptococcus spp., group A) | Negative | N.a. | [17] |
| KL0382 | Atypical C. ulcerans (NTTB) | 2012 | Boar | NA | NA | Positive | Negative | – |
| KL0386 | Atypical C. ulcerans (NTTB) | 2012 | Boar | NA | NA | Positive | Negative | – |
| KL0387-cb8 | C. ulcerans (NTTB) | 2012 | Human | Wound infection | Animal contact (cat) | Positive | Negative | [7] |
| KL0392-cb9 | C. ulcerans (NTTB) | 2012 | Cat | Asymptomatic colonisation | NA | Positive | Negative | [7] |
| KL0394 | Atypical C. ulcerans (NTTB) | 2013 | Boar | NA | NA | Positive | Negative | – |
| KL0395 | Atypical C. ulcerans (NTTB) | 2013 | Boar | NA | NA | Positive | Negative | – |
| KL0396 | Atypical C. ulcerans (NTTB) | 2013 | Boar | NA | NA | Positive | Negative | – |
| KL0400 | Atypical C. ulcerans (NTTB) | 2013 | Boar | Abscess | NA | Positive | Negative | – |
| KL0401 | Atypical C. ulcerans (NTTB) | 2013 | Boar | Abscess | NA | Positive | Negative | – |
| KL0433 | C. ulcerans | 2013 | Human | Wound infection | NA | Positive | Positive | – |
| KL0434 | C. diphtheriae | 2013 | Human | Wound infection (insect bite) | Travel (destination not reported) | Negative | N.a. | [17] |
| KL0438 | C. diphtheriae | 2013 | Human | Wound infection | NA | Negative | N.a. | [17] |
| KL0442 | C. ulcerans (NTTB) | 2013 | Human | Ulcer | NA | Positive | Negative | – |
| KL0451 | C. ulcerans | 2013 | Human | Sepsis, infected dialysis shunt | NA | Negative | N.a. | – |
| KL0459 | C. ulcerans | 2013 | Human | Diphtheria-like | Animal contact (dog) | Positive | Positive | – |
| KL0461 | C. diphtheriae | 2013 | Human | Wound infection | NA | Negative | N.a. | [17] |
| KL0468 | C. ulcerans (NTTB) | 2013 | Human | NA | NA | Positive | Negative | – |
| KL0472 | C. ulcerans (NTTB) | 2013 | Human | NA | NA | Positive | Negative | – |
| KL0475 | C. ulcerans | 2013 | Human | Wound infection | NA | Positive | Positive | – |
| KL0476 | C. diphtheriae | 2013 | Human | Wound infection | Travel (destination not reported) | Negative | N.a. | [17] |
| KL0479 | C. diphtheriae | 2013 | Human | Wound infection | Hepatitis C | Negative | N.a. | [17] |
| KL0483 | C. ulcerans | 2014 | Human | Wound infection | Animal contact (dog) | Positive | Positive | – |
| KL0497 | C. ulcerans | 2014 | Human | Wound infection | NA | Positive | Positive | – |
| KL0501 | C. ulcerans (NTTB) | 2014 | Human | Pus | NA | Positive | Negative | – |
| KL0507 | C. diphtheriae | 2014 | Human | Sepsis | NA | Negative | N.a. | [17] |
| KL0515 | C. ulcerans (NTTB) | 2014 | Human | Wound infection | NA | Positive | Negative | – |
| KL0522 | C. diphtheriae | 2014 | Human | Wound infection | Mixed infection (S. pyogenes), drug abuse | Negative | N.a. | [17] |
| KL0540 | C. ulcerans | 2014 | Human | Wound infection | NA | Positive | Positive | – |
| KL0541 | C. ulcerans (NTTB) | 2014 | Human | Diphtheria-like | NA | Positive | Negative | – |
| KL0547 | C. ulcerans | 2014 | Human | Wound infection | Chronic wound with C. ulcerans, Streptococcus spp. (C), S. aureus | Positive | Positive | – |
| KL0556 | C. ulcerans | 2014 | Human | Wound infection | NA | Positive | Positive | – |
| KL0557 | C. diphtheriae | 2014 | Human | Wound infection | Travel (Sri Lanka)Mixed infection (S. pyogenes) | Negative | N.a. | [17] |
| KL0565 | C. diphtheriae | 2014 | Human | Phlegmon | NA | Positive | Positive | – |
| KL0581 | Atypical C. ulcerans (NTTB) | 2014 | Boar | Abscess | NA | Positive | Negative | – |
| KL0585 | C. diphtheriae | 2015 | Human | Wound infection, deep wound | NA | Negative | N.a. | [17] |
| KL0598 | Atypical C. ulcerans (NTTB) | 2015 | Boar | Lymph node abscess | NA | Positive | Negative | – |
| KL0599 | C. diphtheriae | 2015 | Human | Wound infection, abscess | Homeless | Negative | N.a. | [17] |
| KL0603 | C. diphtheriae | 2015 | Human | Abscess | Travel (Sri Lanka) | Positive | Positive | [5] |
| KL0613 | C. diphtheriae | 2015 | Human | Ulcer | NA | Positive | Positive | [5] |
| KL0615 | Atypical C. ulcerans (NTTB) | 2015 | Boar | Abscess | NA | Positive | Negative | – |
| KL0623 | C. diphtheriae | 2015 | Human | Wound infection | Travel (asylum seeker Eritrea) | Positive | Positive | [5] |
| KL0625 | C. diphtheriae | 2015 | Human | Ulcer | Travel (asylum seeker Ethiopia), pulmonary tuberculosis | Positive | Positive | [5] |
| KL0631 | C. diphtheriae | 2015 | Human | Abscess | Travel (asylum seeker) | Positive | Positive | [5] |
| KL0633 | C. diphtheriae | 2015 | Human | Wound infection | Travel (asylum seeker Somalia) | Positive | Positive | [5] |
| KL0638 | C. diphtheriae | 2015 | Human | Tonsilitis, pharyngitis, diptheria-like | Travel (Thailand)animal contact (cat, rabbit) | Positive | Positive | [5] |
| KL0652 | C. diphtheriae | 2015 | Human | Abscess | Travel (asylum seeker) | Positive | Positive | [5] |
| KL0654 | C. diphtheriae | 2015 | Human | NA | Travel (asylum seeker Eritrea) | Positive | Positive | [5] |
| KL0655 | C. diphtheriae | 2015 | Human | NA | Travel (asylum seeker) | Positive | Positive | [5] |
| KL0663 | C. diphtheriae | 2015 | Human | Wound infection | Travel (asylum seeker Eritrea) | Positive | Positive | [5] |
| KL0670 | C. diphtheriae | 2015 | Human | NA | Travel (asylum seeker) | Positive | Positive | [5] |
| KL0675 | C. diphtheriae | 2015 | Human | Wound infection, phlegmon, sepsis | Homeless | Negative | N.a. | [17] |
| KL0676 | C. diphtheriae | 2015 | Human | Wound infection | NA | Negative | N.a. | [17] |
| KL0678 | C. diphtheriae | 2015 | Human | Wound infection | Homeless, alcohol abuse | Negative | N.a. | [17] |
| KL0691 | C. diphtheriae | 2015 | Human | Wound infection | Homeless | Negative | N.a. | [17] |
| KL0693 | C. diphtheriae | 2015 | Human | Sepsis, pneumonia | Origin: Poland | Negative | N.a. | [17] |
| KL0698 | C. diphtheriae | 2015 | Human | Wound infection | NA | Negative | N.a. | [17] |
| KL0707 | Atypical C. ulcerans (NTTB) | 2015 | Boar | NA | NA | Positive | Negative | – |
| KL0709 | Atypical C. ulcerans (NTTB) | 2015 | Boar | NA | NA | Positive | Negative | – |
| KL0713 | C. diphtheriae | 2015 | Human | Wound infection, deep wound | NA | Negative | N.a. | [17] |
| KL0747 | C. diphtheriae | 2016 | Human | Wound infection | Homeless, drug abuse | Negative | N.a. | [17] |
| KL0759 | C. diphtheriae | 2016 | Human | Olecranon bursitis | Homeless | Negative | N.a. | [17] |
| KL0762 | C. diphtheriae | 2016 | Human | Pharyngitis, tonsillitis | NA | Negative | N.a. | [17] |
| KL0768 | C. diphtheriae | 2016 | Human | Sepsis, endocarditis | Homeless | Negative | N.a. | [17] |
| KL0770 | C. diphtheriae | 2016 | Human | Wound infection, ulcer | NA | Negative | N.a. | [17] |
| KL0773 | Atypical C. ulcerans (NTTB) | 2016 | Boar | Abscess | NA | Positive | Negative | – |
| KL0774 | Atypical C. ulcerans (NTTB) | 2016 | Boar | Abscess | NA | Positive | Negative | – |
| KL0785 | C. ulcerans | 2016 | Human | Wound infection, diabetic foot | Animal contact (dog), | Positive | Positive | – |
| KL0788 | C. diphtheriae | 2016 | Human | phlegmon (post human bite) | Origin: Poland, homeless | Negative | N.a. | [17] |
| KL0796 | C. ulcerans | 2016 | Human | Wound infection | NA | Positive | Positive | – |
| KL0798 | C. diphtheriae | 2016 | Human | Wound infection, burn | Homeless | Negative | N.a. | [17] |
| KL0811 | C. diphtheriae | 2016 | Human | Wound infection | Origin: Poland, homeless, alcohol abuse | Negative | N.a. | [17] |
| KL0812 | C. diphtheriae | 2016 | Human | Wound infection, abscess | Homeless | Negative | N.a. | [17] |
| KL0818 | C. ulcerans (NTTB) | 2016 | Human | ulcer | Diabetic foot | Positive | Negative | – |
| KL0825 | C. ulcerans | 2016 | Human | Wound infection | NA | Positive | Positive | – |
| KL0832 | C. ulcerans | 2016 | Human | Wound infection | NA | Positive | Positive | – |
| KL0840 | C. ulcerans | 2016 | Human | Eczema, Erysipelas | Animal contact (cat) | Positive | Positive | – |
| KL0846 | C. ulcerans (NTTB) | 2016 | Human | Ulcer | Diabetic foot | Positive | Negative | – |
| KL0853 | C. ulcerans | 2016 | Human | Wound infection (post dog bite) | Animal contact (dog) | Negative | N.a. | – |
| KL0867 | C. ulcerans (NTTB) | 2016 | Human | Wound infection | NA | Positive | Negative | – |
| KL0870 | C. ulcerans (NTTB) | 2016 | Human | Eczema | NA | Positive | Negative | – |
| KL0876 | C. ulcerans | 2017 | Human | Wound infection | NA | Negative | N.a. | – |
| KL0880 | C. ulcerans (NTTB) | 2017 | Human | Ulcer | NA | Positive | Negative | – |
| KL0882 | Atypical C. ulcerans (NTTB) | 2017 | Boar | NA | NA | Positive | Negative | – |
| KL0883 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL0884 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL0886 | Atypical C. ulcerans (NTTB) | 2017 | Boar | NA | NA | Positive | Negative | – |
| KL0887 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL0927 | C. diphtheriae | 2017 | Human | Wound infection | Homeless | Negative | N.a. | – |
| KL0938 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL0941 | C. ulcerans | 2017 | Human | Wound infection | NA | Negative | N.a. | – |
| KL0950 | C. diphtheriae | 2017 | Human | Wound infection | Travel (Thailand)Mixed infection (S. pyogenes) | Positive | Positive | – |
| KL0956 | C. diphtheriae | 2017 | Human | Wound infection | NA | Positive | Positive | – |
| KL0957 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL0968 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL1003 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL1006 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL1007 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL1008 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL1009 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL1010 | Atypical C. ulcerans (NTTB) | 2017 | Boar | Abscess | NA | Positive | Negative | – |
| KL1015 | C. ulcerans (NTTB) | 2017 | Human | Carrier status | Animal contact (cat, dog) | Positive | Negative | – |
| KL1017 | C. ulcerans | 2017 | Cattle | Milk isolate, mastitis | NA | Negative | N.a. | – |
| KL1025 | C. ulcerans (NTTB) | 2017 | Cat | Carrier status | NA | Positive | Negative | – |
| KL1058 | C. diphtheriae | 2017 | Human | Wound infection | Travel (Angola), war victim | Negative | N.a. | – |
| KL1059 | C. diphtheriae | 2017 | Human | Wound infection | Travel (Angola), war victim | Positive | Positive | – |
NA No information available, N.a. Not analysed
Analysed isolates from C. diphtheriae were all isolated from human hosts from different geographic regions of Germany or Switzerland, partly with travel or migration history. Those of C. ulcerans were obtained from human and different animal hosts, such as cats, dogs, pigs and cattle. The isolates from a group of biochemical and genetic atypical C. ulcerans originated from a NTTB wildlife cluster of wild boar and roe deer (unpublished observations) and those of C. pseudotuberculosis from humans, buffalo or goats (Table 1). Several of the C. diphtheriae and C. ulcerans isolates belonged to different previously identified community-based or zoonotic outbreak clusters [5, 7, 17]. The NGS datasets obtained in previous outbreak analyses were bioinformatically reanalysed together with the data of currently sequenced isolates, after species identification with MALDI-TOF MS and analysis of toxin status by qPCR and by Elek test. Whole genome sequences of all 137 toxigenic and non-toxigenic isolates underwent k-mer based species confirmation. Subsequently the sequences were assembled into contigs, contigs were ordered according to corresponding reference sequences and annotated for coding sequences and other genetic elements such as rRNAs, tRNAs, repeats and additionally for known prophages.
For phylogenetic analyses of DT and its regulator DtxR, translated protein sequences of both genes were extracted from the annotated assemblies. They were aligned and finally ML phylogenies were generated and visualized in a phylogenetic tree.
In case of DT, 22 DT sequences from C. diphtheriae and 17 from C. ulcerans available on the NCBI public database were additionally included to base the initial DT comparison on more international data (Additional file 1: Table S1). These additional data originated from human hosts (C. diphtheriae and C. ulcerans) and from dogs, moles, owls (C. ulcerans) or unknown host organisms. For one of the sequenced toxigenic C. diphtheriae isolates (KL0655) the genomic region of the tox gene was poorly covered by sequencing reads, thus it was excluded from DT alignment and DT phylogeny analysis. Thereby, DT sequences of 90 of the 91 sequenced toxigenic and NTTB isolates were analysed together with 39 NCBI-derived DT sequences. With regard to DtxR, sequences of all 137 toxigenic or non-toxigenic isolates were phylogenetically analysed together.
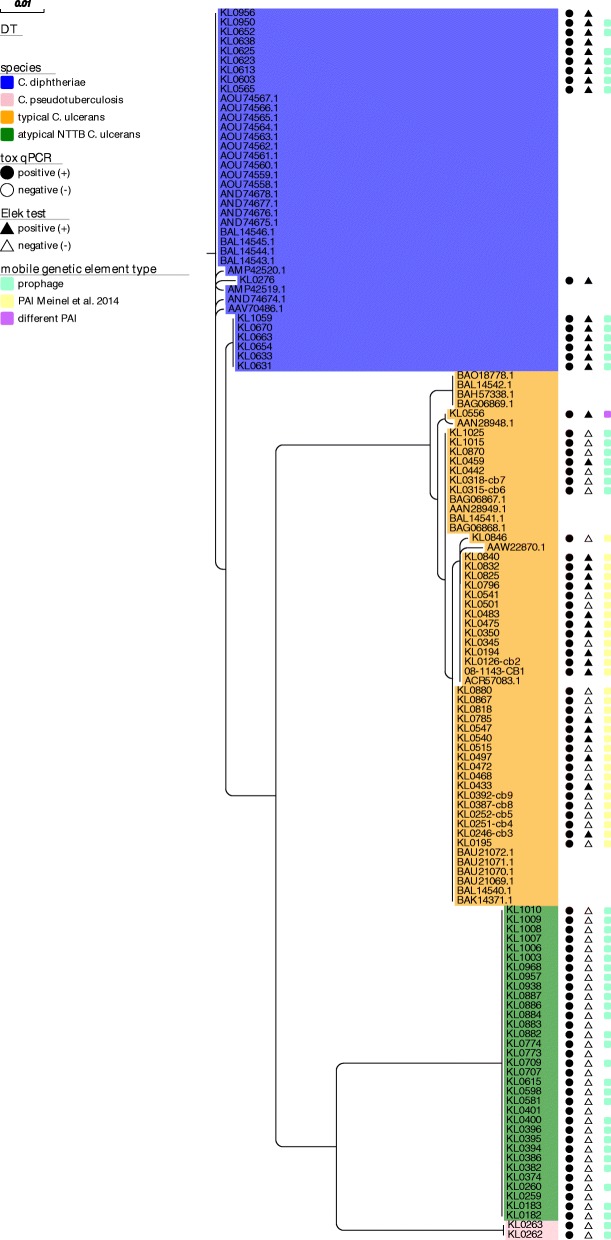
Figures 1 and 2 show that both DT and DtxR cluster in nearly species-specific distinct clades, separating the four groups of C. diphtheriae, C. ulcerans, atypical NTTB C. ulcerans and C. pseudotuberculosis.
Fig. 1.
Maximum likelihood phylogeny of translated DT sequences from different Corynebacterium species. Translated DT sequences of 90 of 91 toxigenic Corynebacterium spp. isolates were extracted, aligned together with 39 NCBI derived DT sequenced from C. diphtheriae and C. ulcerans and a maximum likelihood tree was built. Species affiliation is indicated by background coloring: C. diphtheriae = blue, C. pseudotuberculosis = pink, typical C. ulcerans = orange, atypical NTTB C. ulcerans = green. Results of tox qPCR and Elek tests are indicated by circle and triangle shaped bars, respectively: positive = black, negative = white. The type of detected tox-surrounding genetic mobile element is indicated by colored boxes on the right side of the tree: prophage = turquoise, alternative PAI described in [7] = yellow, different PAI than the one described in [7] = purple
Fig. 2.
Maximum likelihood phylogeny of translated DtxR sequences from different Corynebacterium species. Translated DtxR sequences of all 137 toxigenic and non-toxigenic Corynebacterium spp. isolates were extracted, aligned and a maximum likelihood tree was built. Species affiliation is indicated by background coloring: C. diphtheriae = blue, C. pseudotuberculosis = pink, typical C. ulcerans = orange, atypical NTTB C. ulcerans = green. Results of tox qPCR and Elek tests are indicated by circle and triangle shaped bars, respectively: positive = black, negative = white. The type of detected tox-surrounding mobile genetic regions is indicated by colored boxes on the right side of the tree: prophage = turquoise, alternative PAI described in [7] = yellow, different PAI than the one described in [7] = purple
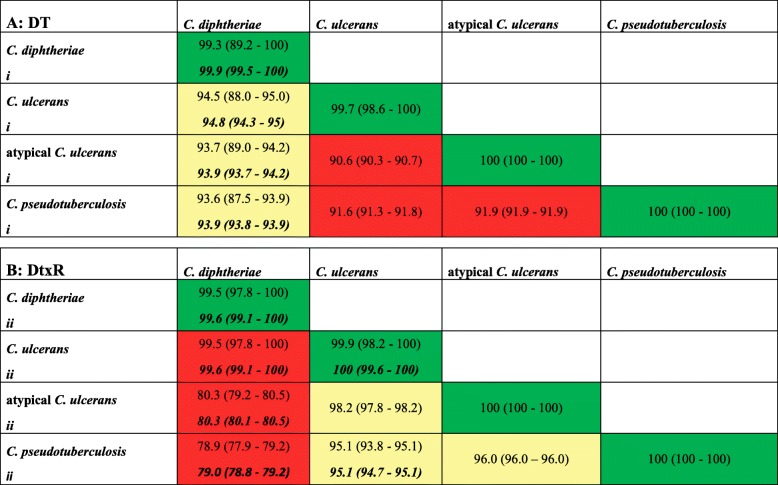
For DT (Fig. 1) the clades of C. pseudotuberculosis and atypical C. ulcerans showed identical sequences within the clades. However, it has to be noted that the toxigenic C. pseudotuberculosis clade consisted of two non-representative, related isolates only, both originating from buffalo hosts from a common submitter in 2011. In contrast, minimum sequence similarity within the clades of C. diphtheriae and C. ulcerans was at 89.2 and 98.6%, respectively (Table 2A). Interestingly, the higher DT sequence variation in C. diphtheriae was caused by only two isolates, one human isolate from a urethral swab from our sample set (KL0276) and one downloaded from NCBI (AVV70486). In both samples the higher variation was caused by significant length and sequence variations in the DT signal peptide, which is located upstream of the A-fragment of the processed DT. However, it is not clear if these variations represent factual differences or artefacts from short read sequencing or assembly. After exclusion of the two samples, minimum DT sequence similarity was at 99.5% in C. diphtheriae (Table 2A).
Table 2.
Similarity of DT (A) and DtxR (B) sequences within and between clades
Similarity percentages within and between identified DT (A) and DtxR (B) clades are shown and color-coded for high (green: > 99%), intermediate (yellow: 93 - ≤ 99%) and low (red: < 93%) similarity. Values after exclusion of samples are indicated with bold italics: i) After exclusion of C. diphtheriae isolates KL0276 and AVV70486. ii) After exclusion of C. diphtheriae belfanti isolate KL0479 and C. ulcerans isolate KL0349
Besides sporadic SNPs and the two samples with strongly differing signal peptide sequences, the SNPs found to differentiate between C. diphtheriae and C. ulcerans were mainly located in the receptor-binding (R) domain of the B fragment of DT, as already postulated on the basis of two Sanger sequenced C. ulcerans isolates and one C. diphtheriae reference sequence by [15]. Processed DT sequences from atypical NTTB C. ulcerans, however, were more similar to C. diphtheriae. Interestingly, the 33 atypical NTTB isolates showed also significant differences in the signal peptide sequence compared to the other clades, but identical sequences among them, whereas C. diphtheriae, C. pseudotuberculosis and typical C. ulcerans signal peptide sequences were very similar except for a few SNPs. Minimum DT similarity between the four clades ranged from 90.3 to 94.3% after exclusion of KL0276 and AAV70486 and mean between-clade DT similarity from 90.6 to 94.8% (Table 2A). In addition to the four main clades, several smaller branches could be identified within the two DT clades of C. diphtheriae and typical C. ulcerans based on the observed slight within-clade variations (Fig. 1).
For DtxR (Fig. 2) homogeneity within the four clades was similarly high, also in C. diphtheriae and regular C. ulcerans clades with minimum sequence similarity of 97.8 and 98.2%, respectively, although toxigenic and non-toxigenic isolates were included in the comparison of DtxR (Table 2B). The main impact on the slight within-clade variability of DtxR came from one outlier sample (KL0479) in C. diphtheriae, which was the only sample in the sample set of biotype belfanti. The same was true for within-clade variability in C. ulcerans and KL0349, a human isolate from an interlaboratory comparison. By exclusion of both samples DtxR similarities within the clades of C. diphtheriae and C. ulcerans were enhanced to ≥99.1% and ≥ 99.6%, respectively. The variability between C. diphtheriae and the other species was higher for DtxR than DT, whereas the variability between the other species (except C. diphtheriae) was higher for DT (Table 2A and B).
DT is the most important pathogenic factor within C.diphtheriae and is known to be embedded in a prophage in this species. After the discovery of nearly species-specific clades of DT and DtxR in the genus Corynebacterium we aimed to investigate if there are also species-specific differences regarding tox-surrounding genomic regions. We searched and annotated known prophage regions with the PHASTER software, which relies on a large database of viral and bacterial genomes to detect prophage regions in assembly data. Prophage sequences surrounding the tox gene could be identified for 13 of 16 WGS datasets from toxigenic C. diphtheriae, for both toxigenic C. pseudotuberculosis, for 27 of the 33 atypical NTTB C. ulcerans, but only in seven out of 39 toxigenic typical C. ulcerans isolates, respectively. Besides tox gene-surrounding prophage regions one to six different intact or incomplete prophage sequences were detected in both toxigenic and non-toxigenic isolates of the different Corynebacterium species distributed within the genomes (Additional file 2: Table S3).
Previously, we identified an additional PAI in a C. ulcerans strain reanalysed in this study (KL0251). The PAI started with a tRNA-Arg CDS with ACG codon, located approximately 6 kb upstream of the tox gene. The tRNA-Arg CDS was followed by a CDS coding for a putative integrase, another CDS carrying a Helix-turn-Helix (HTH) motif, one with a DUF955-domain suspected to be catalytically active, three coding for hypothetical proteins and two of putative integrase/transposases. The PAI was then continued with the tox gene and was terminated with a 100 bp sequence, repeating a part of the leading tRNA-Arg and downstream bases as a pseudo-tRNA repeat [7].
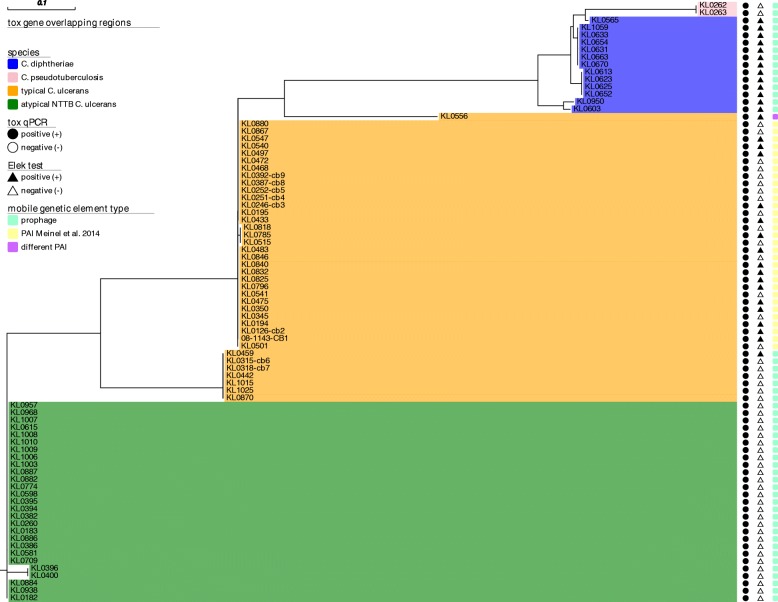
We searched for the existence of this PAI in all 137 NGS datasets. In 31 of the 32 toxigenic C. ulcerans where no prophage was detected to overlap with the tox gene, the PAI was present, but in none of the other C. ulcerans isolates or in any isolate of the other species. In each of these 31 C. ulcerans isolates, the sequence from the tRNA-Arg CDS to 200 bp downstream of the tox gene was extracted, aligned and used for a ML phylogeny together with the tox-overlapping prophage sequences detected in the other isolates (Fig. 3). The ML tree of the pathogenicity-related tox gene-surrounding nucleotide regions again classified the isolates in the same nearly species-specific groups as for DT and DtxR. However, in C. diphtheriae prophage sequences showed some extent of heterogeneity (Fig. 3, blue clade). They divided in several smaller branches. The first branch included KL0603 and KL0950. The second branch consisted of KL0613, KL0623, KL0625 and KL0652. The third branch included KL0654, KL0633, KL0631, KL0663, KL070 and KL1059 and the fourth only the single sample KL0565, located at a common node with the prophages of C. pseudotuberculosis.
Fig. 3.
Maximum likelihood phylogeny of tox gene-overlapping prophage and PAI sequences from different Corynebacterium species. Tox gene-overlapping prophage and detected alternative PAI regions of 81 toxigenic Corynebacterium spp. isolates were aligned and a maximum likelihood tree was built. Species affiliation is indicated by background coloring: C. diphtheriae = blue, C. pseudotuberculosis = pink, typical C. ulcerans = orange, atypical NTTB C. ulcerans = green. Results of tox qPCR and Elek test are indicated by circle and triangle shaped bars, respectively: positive = black, negative = white. The type of detected tox-surrounding mobile genetic regions is indicated by colored boxes on the right side of the tree: prophage = turquoise, alternative PAI described in [7] = yellow, different PAI than the one described in [7] = purple
Tox-overlapping phages of the two related C. pseudotuberculosis isolates were identical as expected (Fig. 3, pink clade). However, only in one of them an additional phage was detected, which was not located surrounding the tox gene (Additional file 2: Table S3).
In typical C. ulcerans it became obvious by substantially distinct branches that the sequences from the tox gene-surrounding prophages clearly differed from the extracted alternative PAIs (Fig. 3, orange clade) and both were independent from the underlying host species. In 28 of the 31 C. ulcerans samples the identified PAI had a length of 7.7 kb and the extracted sequences were very homogeneous. Three additional isolates (KL0515, KL0785, KL0818) of common MLST type 514 were obtained from wound infections from human patients. One of them had contact to pet dogs and two came from the same region and the same time frame in 2016. Those three showed an identical PAI, but with decreased similarity and different length (8 kb) compared to the others. The lower similarity of the PAI of these three isolates, compared to that of the other isolates was probably also due to variation in the integrase following the tRNA-Arg CDS, resulting in higher similarity to integrase accession BAM26371 than to AKN76028, as annotated in the other samples’ PAIs (Additional file 2: Table S3). Furthermore, the PAI of those three samples showed one SNP in the pseudo-tRNA repeat. KL0556 was an outlier of the C. ulcerans clade and was located on an own branch of the tree (Fig. 3). The extracted sequence between tRNA-Arg and 200 bp downstream the tox gene was shorter (4.96 kb) compared to the other PAIs (7.7–8 kb) and the annotations detected in the other isolates were not present here. The region included four hypothetical proteins between the tRNA and the tox gene, with one of them being also a putative integrase, and additionally three SNPs in the pseudo-tRNA repeat.
Prophages in atypical NTTB C. ulcerans built a separate branch (Fig. 3, green clade) as it was also the case for DT (Fig. 1) and DtxR (Fig. 2). In all except one isolate their tox-embedding prophages were classified with questionable completeness status by the annotation software. This was mainly due to lacking overlap of the found CDS with the used databases of known prophage sequences, although a substantial extent of CDS was classified as phage sequences with specific functions. Most of the regions had a length of 31–33 kb or 43 kb (KL0396, KL0400). Only in KL0884 a prophage with a length of 51 kb was classified as intact by PHASTER. Despite its different length, the region overlapping with the shorter prophages of the other atypical NTTB C. ulcerans showed high similarity to them.
Generally, tox-overlapping prophages and PAIs showed different GC contents compared to the average GC content of the whole genome assemblies, as expected (Additional file 2: Table S3). In C. diphtheriae GC content was reduced by 1.2–1.5% and in atypical NTTB C. ulcerans by 1.5–3.8%. However, in phage regions of C. ulcerans a reduction by only 0.3–0.6% occurred, while in GC content reduction in PAI regions was substantially higher (5.6–5.8%). GC content of the shorter, different PAI in KL0556 was even reduced by 6.4%.
Discussion
With the WGS data from different Corynebacterium species infecting different host organisms, we demonstrated pathogen species-specific clades of DT and tox gene-surrounding mobile, pathogenicity related genetic elements in toxigenic and NTTB isolates, as well as of DtxR in toxigenic, NTTB and non-toxigenic isolates (Table 1). For the tox-surrounding phage or PAI regions we found various branches, matching with those found in DT and DtxR. Surprisingly, a notable extent of heterogeneity of the tox-surrounding phage regions was found within C. diphtheriae for which research is done for decades. For example Chen and co-workers similarly found in 2008 that the lytic corynephage P1201 showed only 9% shared gene content with the only previously sequenced prophage BFK20 and that the mosaic-like genome of P1201 indicates extensive horizontal gene transfer among P1201, Gordonia terrae phage GTE5, mycobacteriophages and several regions of Corynebacterium spp. Genomes [13]. This finding may be true for further corynephages and at least partly explain the variations observed in our data. However, why other Corynebacterium species seem to be less prone to this variation than C. diphtheriae is not clear, but it may be associated with host species adaption and interaction.
Datasets of several C. diphtheriae isolates reanalysed in this study were part of a previous outbreak investigation of isolates from African refugees and could be classified in two outbreak clusters of epidemiological related samples by whole genome SNP profiles in the previous study. A third cluster detected back then included non-toxigenic samples that were not included in the current study [5]. Interestingly, in the current study the separation of the two included outbreak clusters could be reconstructed even on the level of DT and of the respective surrounding prophage, but not as clearly on DtxR level. Outbreak cluster 1 with isolates KL0633, KL0654, KL0663, KL0670 was affiliated to the above mentioned branch 3 of the C. diphtheriae clade and outbreak cluster 2 with isolates KL0623, KL0265, KL0652 to branch 2. Horizontal gene transfer in C. diphtheriae is an important factor for propagation of pathogenicity. However, this higher similarity of mobile pathogenicity-related elements in closely related strains indicates that important virulence features like DT and underlying prophages may at least be partly retained in bacterial populations and diverge along the evolutionary path with the bacterial core genome, potentially due to their advantages in host interaction or infectivity.
Additionally to the known tox-surrounding prophages, we found an alternative tox-surrounding PAI described in [7] in a majority of C. ulcerans isolates, which was not present in any isolate of the other analysed Corynebacterium species. Having a closer look at the CDS annotations of this PAI, which comprised several sequences with putative integrase activity, it could be assumed that this PAI also developed from a former not yet described prophage sequence. In general, the different pathogenicity-related features exist in zoonotic C. ulcerans across different host species. Interestingly, isolate KL0556 built an exception to all other C. ulcerans isolates, regarding its tox-surrounding PAI. The extracted region between tRNA-Arg and 200 bp downstream of tox was much shorter than the PAI of the other samples and showed different CDS annotations. The differences to the PAIs of the other C. ulcerans isolates with the same start and end annotations might be due to assembly problems, although the differences in annotated CDS sequences render this explanation of being a technical artefact very unlikely. The reason for it might also be evolution from previously more similar sequences. Another probably even more likely reason would be, especially regarding the differences of the integrase CDS, the pseudo tRNA-repeat and the lower GC content, that this isolate harbours another not yet described PAI, which might indicate the existence of a higher number of yet unknown corynephages.
Generally, besides tox-surrounding prophages or PAIs, we found a varying number of incomplete prophages in all our analysed strains. Similar observations have been made by Subedi et al. [18] in three different C. ulcerans strains, belonging to two different lineages, independent of the infected host organism. They linked the incompleteness of the prophages to the draft version of their NGS assembly data, but found that those prophages lead to significant diversity within C. ulcerans, consistent with earlier publications. Those assumptions concluded from the data of the three strains in [18] could very well be reconstructed in our larger dataset with more than 40 C. ulcerans isolates, and based on our data be expanded to at least C. diphtheriae and eventually also be postulated for C. pseudotuberculosis.
Additionally to the existence of various prophages within the strains the question of their propagation arises. Trost et al. described that C. diphtheriae comprises a stable, but open core genome of approx. 1600 genes (approximately 70% of the genome) being complemented by more variable gene content of 30%. Especially the variable content is to some extent subject to horizontal gene transfer. Furthermore, a recent study from Mansfield et al. [14] even discovered a DT-like protein family in bacterial lineages outside Corynebacterium genus, including Austwickia and Streptomyces. These genes show a certain degree of similarity to tox, but are surrounded by different loci. In all except one genus they did not discover any dtxR-like genes. The authors concluded that tox-like genes have not simply been exchanged between genera via corynephages in recent times, but either may have been acquired by independent mechanisms or the genomic evidence of the acquisition was lost over time.
Based on this knowledge and our results, which clearly demarcate clades between the different Corynebacterium species and show a certain degree of diversity within those clades, we conclude besides the fact that horizontal gene transfer via prophages is indeed likely to occur within species of the genus Corynebacterium, sequences may be based on different genetic backgrounds. Furthermore, once acquired elements also seem to evolve along with other virulence and pathogenicity related features within the different species. The alternative PAI exclusively present in C. ulcerans furthermore implies that DT transmission pathways between isolates may be different between human-pathogenic C. diphtheriae and the zoonotic species and contribute to the emerging pathogenic potential of the latter ones.
However, it has to be noted that despite the considerable evidence, our conclusions are based on results from short read assemblies with varying missing parts. Finishing of representative genomes which can be achieved by third generation sequencing technologies in future studies can strengthen the significance of our conclusions.
Conclusion
With our phylogenetic analyses of DT, DtxR and tox gene-embedding mobile elements from WGS data of 91 toxigenic and 46 non-toxigenic isolates of different pathogenic Corynebacterium species, we were able to detect four nearly species-specific clades and verify previous postulations of species-specific DT differences based on a meaningful sample set. The clear clade-like separation of tox gene and surrounding genetic elements and the slight intra-clade variations even allow grouping of some related sample clusters. Furthermore a PAI, previously detected in a single isolate only could be classified as a general feature in a substantial number of zoonotic C. ulcerans across different hosts. These observations imply that the pathogenicity related features have been acquired a considerable time ago, by different and maybe independent events. They may furthermore correlate with host and environmental niche adaption in the different Corynebacterium species. Furthermore the features have probably been retained through evolution of strains due to their striking advantages in host interaction and infectivity. This conclusion on the important and potent pathogenicity factor DT and surrounding mobile elements gives a good example of how pathogenicity in Corynebacterium spp. could generally be established and propagated. This can also be of considerable value for research on related pathogenic bacteria. Furthermore, the findings are of special value for public health as the identified differences will be reflected in pathogen-host-interaction and may at least partly explain differences in disease manifestation, progression or treatment success. For example reduced immunity against strains with different corynephages and toxin types after vaccination with standard toxoid may occur. Furthermore, C. ulcerans is increasingly identified in human infections. These and especially the still rare but consistently occurring cases of pharyngeal diphtheria, caused by toxigenic C. ulcerans which are more often found in wound infections, may be associated with the differences in pathogenicity factors and an expanded host reservoir. Therefore, these topics may also be of reasonable interest for public health related applied research in the future.
Methods
Aim and design of the study
We aimed to investigate if differences in DT, its regulator DtxR and the tox gene-surrounding pathogenicity-related elements between different Corynebacterium species can be observed on a regular basis when a larger sample set is analysed and which phylogenetic conclusions regarding the DT-mediated pathogenicity of medically relevant corynebacteria can be drawn from the results. Therefore, we performed phylogenetic analyses with translated versions of the WGS-generated respective genes and genetic regions.
Isolates selection
For phylogenetic comparison of DT, tox gene-surrounding pathogenicity related elements and DtxR, isolates from different Corynebacterium species or their previously generated whole genome sequencing (WGS) data were selected from the culture collection of the German Consiliary Laboratory on Diphtheria. Toxigenic isolates from different years, geographic regions and hosts were selected and complemented with non-toxigenic isolates from the same bacterial and host species for comparison (C. diphtheriae: 17 toxigenic, 34 non-toxigenic; typical C. ulcerans: 18 toxigenic, 21 non-toxigenic tox gene-bearing (NTTB), 7 non-toxigenic; atypical C. ulcerans from a wildlife cluster: 33 NTTB; C. pseudotuberculosis: 2 toxigenic, 5 non-toxigenic). For DT alignment 39 published DT sequences from C. diphtheriae (N = 22) and C. ulcerans (N = 17), originating from humans (C. diphtheria and C. ulcerans) or from dogs, moles and owls (C. ulcerans), were downloaded from NCBI and included (Additional file 1: Table S1).
Bacteriological analyses
Bacteriological species identification was performed using MALDI-TOF MS analysis (microflex LT Mass Spectrometer, MALDI Biotyper™; Bruker Daltonics, MBT 7311 MSP library, Bremen, Germany) as described in [19]. The presence of tox gene was investigated by real-time PCR [20] and positive PCR results were phenotypically confirmed by a modified Elek test [21].
Next generation sequencing and data analyses
Whole genome sequencing (WGS) of the isolates was performed on an Illumina MiSeq with 2 × 250 bp paired-end reads (Illumina, San Diego, CA, USA), after DNA isolation on the Promega Maxwell system and library preparation by Nextera XT, as reported previously [17].
Bioinformatics analyses started with quality control per sequencing run and sample using Illumina SAV software [22] and fastQC [23], respectively. Species identity and absence of contaminations from other species was checked with kraken [24]. Reads were trimmed for quality and from short, truncated reads with trimmomatic [25]. De novo assemblies were done with SPAdes assembler [26]. Assembly quality was checked with QUAST [27]. Contigs were ordered with the mauve ContigOrderer tool [28], along genbank-formatted reference genomes of toxigenic C. diphtheria NCTC 13129 (accession BX248353), C. ulcerans FRC58 (accession CP0011913.1) and C. pseudotuberculosis C231 (accession NC_017301). Reordered contigs were concatenated to continuous input files for subsequent annotation steps. Annotation of the draft genomes was done with prokka [29] using the default database and an additional Corynebacterium genus database, built with the prokka_database_maker script [30], based on NCBI-deposited sequence data from 305 tax IDs as database input (Additional file 3). Prophage detection, annotation and phage sequence extraction was performed with PHASTER [31] using the PHASTER url API.
Translated protein sequences of DT and DtxR were extracted from the annotated genbank-formatted draft genomes with open source python scripts [32] with the respective locus tag as identifier. For subsequent steps 39 NCBI-derived DT sequences were included (accession numbers in Additional file 1: Tables S1).
Extracted DT, DtxR and phage sequences were aligned using muscle [33] and maximum likelihood (ML) trees were built with FastTree, using the default Shimodaira-Hasegawa test on the three alternate topologies and 1000 resamples [34]. Similarity percentages of DT and DtxR alignments were calculated with CLC genomics workbench (Qiagen).
NGS raw datasets analysed during the current study are available in the NCBI sequence read archive (SRA) at https://www.ncbi.nlm.nih.gov/sra under BioProjects PRJNA416260 and PRJNA490531 (accession numbers in Additional file 1: Table S2).
Additional files
NCBI accession numbers of downloaded DT sequences (Table S1) and SRA accession numbers of analysed WGS data (Table S2). (DOCX 28 kb)
Detected tox, dtxR, prophage and PAI coordinates, annotations and GC content (Table S3): For each of the 137 isolates, coordinates of tox and dtxR genes are given along with their species affiliation and their tox status analysed by qPCR and modified Elek test. All detected prophage sequences and PAIs are shown one line per region. For prophages and PAIs, information about region type and their overlapping or non-overlapping behavior in regard to the tox gene is indicated. For detected prophages, results from the prophage annotation software PHASTER [31] are given. For the alternative PAI, genomic start coordinates of the individual CDS sequences and structural annotations are indicated as well. Average GC content of the draft genome and the specific prophage/PAI regions is given. (XLSX 88 kb)
Tax IDs used for the generation of the Corynebacterium spp. annotation database in prokka [29]. (DOCX 21 kb)
Acknowledgements
We thank Sabine Wolf, Jasmin Fräßdorf, Katja Meindl and Marion Lindermayer for their excellent laboratory work on DNA isolation, library preparation and sequencing, as well as Wolfgang Schmidt for the participation in cultivation and microbiological characterization of the corynebacteria. The work was done as part of our involvement in the European Study Group of Public Health Microbiology (ESGPHM) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID).
Funding
The study was partly financially supported by the Bavarian State Ministry of Health and Care as well as by the German Federal Ministry of Health via the Robert Koch Institute and its National Reference Laboratories Network (09-49, FKZ 1369-359).
Availability of data and materials
The datasets generated and analysed during the current study are available in the NCBI sequence read archive (SRA) at https://www.ncbi.nlm.nih.gov/sra under BioProjects PRJNA416260 and PRJNA490531 (detailed accession numbers in Additional file 1: Table S2) and at the NCBI protein and nucleotide databases at https://www.ncbi.nlm.nih.gov (Additional file 1: Table S1).
Abbreviations
- CDS
Coding sequence
- DT
Diphtheria toxin
- DtxR
Diphtheria toxin regulator
- dtxR
Diphtheria toxin regulator gene
- DUF
Domain of unknown function
- HTH
Helix-turn-helix (motif)
- ML
Maximum likelihood
- MLST
Multi locus sequence typing
- NGS
Next generation sequencing
- PAI
Pathogenicity island
- SNP(s)
Single Nucleotide Polymorphism(s)
- tox
Diphtheria toxin gene
- WGS
Whole genome sequencing
Authors’ contributions
AD, AB, RK and AS conceived experiments. RK and AB performed micro- and molecular biological analysis. AD performed NGS data acquisition and analysis. AD, AB and AS wrote the manuscript. All authors contributed, read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alexandra Dangel, Email: alexandra.dangel@lgl.bayern.de.
Anja Berger, Email: anja.berger@lgl.bayern.de.
Regina Konrad, Email: regina.konrad@lgl.bayern.de.
Andreas Sing, Email: andreas.sing@lgl.bayern.de.
References
- 1.Bernard K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol. 2012;50(10):3152–3158. doi: 10.1128/JCM.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Von Graevenitz A, Bernard K. The genus Corynebacterium - medical. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes: volume 3: archaea Bacteria: Firmicutes, Actinomycetes. New York: Springer New York; 2006. pp. 819–842. [Google Scholar]
- 3.Leggett BA, De Zoysa A, Abbott YE, Leonard N, Markey B, Efstratiou A. Toxigenic Corynebacterium diphtheriae isolated from a wound in a horse. Vet Rec. 2010;166(21):656–657. doi: 10.1136/vr.b4846. [DOI] [PubMed] [Google Scholar]
- 4.Poor AP, Moreno LZ, Matajira CEC, Parra BM, Gomes VTM, Silva APS, et al. Characterization of Corynebacterium diphtheriae, C. confusum and C. amycolatum isolated from sows with genitourinary infection. Vet Microbiol. 2017;207:149–152. doi: 10.1016/j.vetmic.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Meinel DM, Kuehl R, Zbinden R, Boskova V, Garzoni C, Fadini D, et al. Outbreak investigation for toxigenic Corynebacterium diphtheriae wound infections in refugees from Northeast Africa and Syria in Switzerland and Germany by whole genome sequencing. Clin Microbiol Infect. 2016;22(12):1003 e1–1003 e8. doi: 10.1016/j.cmi.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 6.UNICEF Supply Division. Diphtheria Tetanus and Pertussis Vaccine Supply Update October 2016. 2016. Available online at: https://www.unicef.org/supply/index_67674.html.
- 7.Meinel DM, Margos G, Konrad R, Krebs S, Blum H, Sing A. Next generation sequencing analysis of nine Corynebacterium ulcerans isolates reveals zoonotic transmission and a novel putative diphtheria toxin-encoding pathogenicity island. Genome Med. 2014;6(11):113. doi: 10.1186/s13073-014-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hacker E, Antunes CA, Mattos-Guaraldi AL, Burkovski A, Tauch A. Corynebacterium ulcerans, an emerging human pathogen. Future Microbiol. 2016;11:1191–1208. doi: 10.2217/fmb-2016-0085. [DOI] [PubMed] [Google Scholar]
- 9.Trost E, Blom J, Soares Sde C, Huang IH, Al-Dilaimi A, Schroder J, et al. Pangenomic study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J Bacteriol. 2012;194(12):3199–3215. doi: 10.1128/JB.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiRita VJ. Classic spotlight: phage bring punch to the party. J Bacteriol. 2016;198(2):202. doi: 10.1128/JB.00644-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collier RJ. Diphtheria toxin: mode of action and structure. Bacteriol Rev. 1975;39(1):54–85. doi: 10.1128/br.39.1.54-85.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes RK. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J Infect Dis. 2000;181(Suppl 1):S156–S167. doi: 10.1086/315554. [DOI] [PubMed] [Google Scholar]
- 13.Chen CL, Pan TY, Kan SC, Kuan YC, Hong LY, Chiu KR, et al. Genome sequence of the lytic bacteriophage P1201 from Corynebacterium glutamicum NCHU 87078: evolutionary relationships to phages from Corynebacterineae. Virology. 2008;378(2):226–232. doi: 10.1016/j.virol.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Mansfield MJ, Sugiman-Marangos S, Melnyk RA, Doxey AC. Identification of a diphtheria toxin-like gene family beyond the Corynebacterium genus. FEBS Lett. 2018. Epub 31 July 2018. 10.1002/1873-3468.13208. [DOI] [PubMed]
- 15.Sing A, Hogardt M, Bierschenk S, Heesemann J. Detection of differences in the nucleotide and amino acid sequences of diphtheria toxin from Corynebacterium diphtheriae and Corynebacterium ulcerans causing extrapharyngeal infections. J Clin Microbiol. 2003;41(10):4848–4851. doi: 10.1128/JCM.41.10.4848-4851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittchen M, Busche T, Gaspar AH, Lee JH, Ton-That H, Kalinowski J, et al. Transcriptome sequencing of the human pathogen Corynebacterium diphtheriae NCTC 13129 provides detailed insights into its transcriptional landscape and into DtxR-mediated transcriptional regulation. BMC Genomics. 2018;19(1):82. doi: 10.1186/s12864-018-4481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dangel A, Berger A, Konrad R, Bischoff H, Sing A. Geographically diverse clusters of nontoxigenic Corynebacterium diphtheriae infection, Germany, 2016-2017. Emerg Infect Dis. 2018;24(7):1239–1245. doi: 10.3201/eid2407.172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subedi R, Kolodkina V, Sutcliffe IC, Simpson-Louredo L, Hirata R, Jr, Titov L, et al. Genomic analyses reveal two distinct lineages of Corynebacterium ulcerans strains. New Microbes New infect. 2018;25:7–13. doi: 10.1016/j.nmni.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konrad R, Berger A, Huber I, Boschert V, Hormansdorfer S, Busch U, et al. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry as a tool for rapid diagnosis of potentially toxigenic Corynebacterium species in the laboratory management of diphtheria-associated bacteria. Euro Surveill. 2010;15(43) Epub 23 Nov 2010. [DOI] [PubMed]
- 20.Schuhegger R, Lindermayer M, Kugler R, Heesemann J, Busch U, Sing A. Detection of toxigenic Corynebacterium diphtheriae and Corynebacterium ulcerans strains by a novel real-time PCR. J Clin Microbiol. 2008;46(8):2822–2823. doi: 10.1128/JCM.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engler KH, Glushkevich T, Mazurova IK, George RC, Efstratiou A. A modified Elek test for detection of toxigenic corynebacteria in the diagnostic laboratory. J Clin Microbiol. 1997;35(2):495–498. doi: 10.1128/jcm.35.2.495-498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Illumina Inc. Sequencing Analysis Viewer Software. Available from: http://emea.support.illumina.com/sequencing/sequencing_software/sequencing_analysis_viewer_sav.html.
- 23.Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. [Google Scholar]
- 24.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 30.Ehrlich R. Prokka database maker script. 2016. [Google Scholar]
- 31.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leimbach A. Collection of scripts for bacterial genomics. Zenodo. 10.5281/zenodo.215824.2016 [12 Nov 2018]; Available online at: https://github.com/aleimba/bac-genomics-scripts/blob/master/cds_extractor/cds_extractor.pl.
- 33.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NCBI accession numbers of downloaded DT sequences (Table S1) and SRA accession numbers of analysed WGS data (Table S2). (DOCX 28 kb)
Detected tox, dtxR, prophage and PAI coordinates, annotations and GC content (Table S3): For each of the 137 isolates, coordinates of tox and dtxR genes are given along with their species affiliation and their tox status analysed by qPCR and modified Elek test. All detected prophage sequences and PAIs are shown one line per region. For prophages and PAIs, information about region type and their overlapping or non-overlapping behavior in regard to the tox gene is indicated. For detected prophages, results from the prophage annotation software PHASTER [31] are given. For the alternative PAI, genomic start coordinates of the individual CDS sequences and structural annotations are indicated as well. Average GC content of the draft genome and the specific prophage/PAI regions is given. (XLSX 88 kb)
Tax IDs used for the generation of the Corynebacterium spp. annotation database in prokka [29]. (DOCX 21 kb)
Data Availability Statement
The datasets generated and analysed during the current study are available in the NCBI sequence read archive (SRA) at https://www.ncbi.nlm.nih.gov/sra under BioProjects PRJNA416260 and PRJNA490531 (detailed accession numbers in Additional file 1: Table S2) and at the NCBI protein and nucleotide databases at https://www.ncbi.nlm.nih.gov (Additional file 1: Table S1).