Abstract
Neurofibromatosis type 1 (NF1), a genetic disorder linked to inactivating mutations or a homozygous deletion of the Nf1 gene, is characterized by tumorigenesis, cognitive dysfunction, seizures, migraine, and pain. Omic studies on human NF1 tissues identified an increase in the expression of collapsin response mediator protein 2 (CRMP2), a cytosolic protein reported to regulate the trafficking and activity of presynaptic N-type voltage-gated calcium (Cav2.2) channels. Because neurofibromin, the protein product of the Nf1 gene, binds to and inhibits CRMP2, the neurofibromin–CRMP2 signaling cascade will likely affect Ca2+ channel activity and regulate nociceptive neurotransmission and in vivo responses to noxious stimulation. Here, we investigated the function of neurofibromin–CRMP2 interaction on Cav2.2. Mapping of >275 peptides between neurofibromin and CRMP2 identified a 15-amino acid CRMP2–derived peptide that, when fused to the tat transduction domain of HIV-1, inhibited Ca2+ influx in dorsal root ganglion neurons. This peptide mimics the negative regulation of CRMP2 activity by neurofibromin. Neurons treated with tat-CRMP2/neurofibromin regulating peptide 1 (t-CNRP1) exhibited a decreased Cav2.2 membrane localization, and uncoupling of neurofibromin–CRMP2 and CRMP2–Cav2.2 interactions. Proteomic analysis of a nanodisc-solubilized membrane protein library identified syntaxin 1A as a novel CRMP2-binding protein whose interaction with CRMP2 was strengthened in neurofibromin-depleted cells and reduced by t-CNRP1. Stimulus-evoked release of calcitonin gene–related peptide from lumbar spinal cord slices was inhibited by t-CNRP1. Intrathecal administration of t-CNRP1 was antinociceptive in experimental models of inflammatory, postsurgical, and neuropathic pain. Our results demonstrate the utility of t-CNRP1 to inhibit CRMP2 protein–protein interactions for the potential treatment of pain.
Keywords: Neurofibromatosis type 1, N-type voltage-gated calcium channel, CRMP2, Syntaxin 1A, Protein–protein interaction, Proteomics, CGRP, Hyperalgesia, Allodynia, Inflammatory pain, Postsurgical pain, gp120-induced peripheral neuropathy
1. Introduction
Neurofibromatosis type 1 (NF1) is a relatively common genetic disease primarily linked to nervous system tumors that can be associated with significant chronic pain.9,13,15,24 Although, NF1-related pain conditions are not well described, there is clearly pain that is not directly related to tumor burden.13 The mechanisms involved in the enhanced pain sensitivity in patients with NF1 remain unresolved. It has been postulated that the increased pain perception in patients with NF may involve the sensitization of small-diameter nociceptive sensory neurons that are known to mediate the transmission of the reported neurological complications of pain and itch. Small-diameter capsaicin-sensitive sensory neurons isolated from mice with a heterozygous mutation of the Nf1 gene (coding for the protein neurofibromin) were shown to have augmented excitability compared with wild-type neurons.79 Furthermore, Nicol et al. reported that peak current densities for both tetrodotoxin (TTX)-sensitive and TTX-resistant sodium currents78 were significantly larger in the Nf1+/− sensory neurons. Stimulus-evoked release of the neuropeptides–substance P and calcitonin gene–related peptide (CGRP)—was also significantly higher from sensory neurons isolated from Nf1+/− mice.31 Nf1+/− mice (male), however, did not exhibit increased sensitivity to acute, inflammatory, or neuropathic pain.61 By contrast, female Nf1+/− mice were reported to have increased hyperalgesia.46 These findings suggested that Nf1 heterozygosity alone was insufficient to increase pain in mice, highlighting the possible existence of additional mechanisms that may underlie reports of increased pain in patients with NF1.
We, and others, reported that sensory neurons from Nf1+/− mice exhibit increased N-type voltage-gated Ca2+ (Cav2.2) currents, and this likely accounts for the increased release of neuropeptides that occurs in Nf1+/− sensory neurons.16,77 Notably, the levels of mRNA for the α subunits of Cav2.2 were not different between the genotypes,16 invoking the possibility of neurofibromin-dependent pathways in the upregulation of Cav2.2 activity. Our previous work identified the axonal growth and specification collapsin response mediator protein 2 (CRMP2) as a novel regulator of Cav2.2 activity: direct binding between Cav2.2 and CRMP2 leads to a CRMP2-mediated increase in Ca2+ current density and increased transmitter release in sensory neurons.11 In addition, CRMP2 was reported to bind to neurofibromin.41,65 Loss of neurofibromin increased CRMP2 phosphorylation,65 which in turn increased its association with Cav2.2.7 A further unresolved question was to examine how the CRMP2-neurofibromin complex controlled the activity of Cav2.2, a channel with clear links to heightened sensitivity resulting from inflammation and/or neuropathic pain.8,49 In this study, we identify a 15-amino acid peptide derived from the C-terminus of neurofibromin that disrupted the interaction between CRMP2 and neurofibromin; reduced calcium influx in a heterogeneous population of sensory neurons; and blunted evoked CGRP release. This peptide reproduced the inhibition of CRMP2 by neurofibromin by blocking CRMP2 protein–protein interactions (PPIs). Here, targeting CRMP2/neurofibromin interaction disrupted CRMP2’s interaction with syntaxin 1A, a protein involved in synaptic vesicle docking and neurotransmitter release.70,73,86 Of importance, intrathecal administration of the peptide reversed mechanical allodynia and/or thermal hyperalgesia in experimental models of inflammatory, postsurgical, and neuropathic pain.
2. Methods
2.1. Animals
Pathogen-free, adult male and female Sprague-Dawley rats (225–250 g; Harlan Laboratories) were housed in temperature-controlled (23 ± 3˚C) and light-controlled (12-h light/12-h dark cycle; lights on 07:00–19:00) rooms with standard rodent chow and water available ad libitum. The Institutional Animal Care and Use Committee of the College of Medicine at the University of Arizona approved all experiments. All procedures were conducted in accordance with the Guide for Care and Use of Laboratory Animals published by the National Institutes of Health and the ethical guidelines of the International Association for the Study of Pain. Animals were randomly assigned to treatment or control groups for the behavioral experiments. Animals were initially housed 3 per cage but individually housed after the intrathecal cannulation on a 12-hour light-dark cycle with food and water ad libitum. All behavioral experiments were performed by experimenters who were blinded to the experimental groups and treatments.
2.2. Materials
All chemicals, unless noted were purchased from Sigma (St. Louis, MO). Validated antibodies were purchased as follows: anti-CRMP2 polyclonal antibody (Sigma-Aldrich Cat# C2993 Research Resource Identifier (RRID):AB_1078573), CRMP2 pSer522 (ECM Biosciences Cat# CP2191, RRID:AB_2094486), anti-Cav2.2 polyclonal antibody (Cat# TA308673; Origene Technologies, Inc, Rockville, MD), antineurofibromin C-terminal (Abcam Cat# ab17963, RRID:AB_444142), βIII-Tubulin (Cat# G712A; Promega, Madison, WI), syntaxin 1A (Millipore Cat# AB5820, RRID: AB_2216165), and Actin (Sigma-Aldrich Cat# A2228 RRID:AB_476697). Peptides (Table 1) were synthesized and HPLC-purified to >95% purity by Genscript Inc, (Piscataway, NJ).
Table 1.
CRMP2- and neurofibromin-derived peptides used in this study.
| Peptide* | Sequence (N → C termini) | Molecular weight (g/mol) |
|---|---|---|
| t-CNRP1 | HVTEGSGRYIPRKPF | 3285.79 |
| t-CNRP2 | PDFVYKRIKARSRLA | 3361.97 |
| t-CNRP3 | AQVHAENGDIIAEEQ | 3165.50 |
| t-NCRP1 | KGYRHPSPAIVARTV | 3193.74 |
| t-NCRP2 | YLQSFGFNGLWRFAG | 3289.91 |
| t-NCRP3 | RVSHGQIKQIIRILS | 3304.79 |
t is the trans-acting activator of transcription (TAT) protein transduction domain of HIV type 1.
YGRKKRRQRRR is the TAT sequence fused to the N-terminal region of the peptides used in this study to make them cell-penetrant. CNRP, CRMP2-neurofibromin regulating peptide; NCRP, neurofibromin-CRMP2 regulating peptide.
2.3. Peptide spot arrays and far-Western assay
Far-Western protein–binding affinity assays were performed as previously described.5 Peptide spot arrays (15-mers with an overlap of 6 or 5 residues) spanning full-length CRMP2 (residues 1–572 of the rat sequence) and the proximal C-terminus residues 2260 to 2818 of rat neurofibromin were constructed using the SPOTS-synthesis method. Standard 9-fluorenylmethoxy carbonyl (Fmoc) chemistry was used to synthesize the peptides and spot them onto nitrocellulose membranes prederivatized with a polyethylene glycerol spacer (Intavis AG). Fmoc-protected and Fmoc-activated amino acids were spotted in 20 by 30 arrays on 150 mm by 100-mm membranes using an Intavis MultiPep robot. The nitrocellulose membrane containing the immobilized peptides was soaked in N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer (10 mM CAPS, pH 11.0 with 20% vol/vol methanol) for 30 minutes, washed once with Tris-buffered 0.1% Tween 20 (TBST), and then blocked for 1 hour at room temperature (RT) with gentle shaking in TBST containing 5% (mass/vol) nonfat milk and then incubated with rat spinal cord protein (see below) for 1 hour at RT with gentle shaking. Next, the membranes were incubated in primary antibody for CRMP2, neurofibromin, or syntaxin 1A for 2 hours at RT with gentle shaking, followed by washing with TBST. Finally, the membranes were incubated in secondary antibody (goat anti-rabbit DyLight 800, Cat# 355571, Thermo Fisher) for 45 minutes, washed for 30 minutes in TBST and visualized by infrared fluorescence (Li-Cor Odyssey Clx Imaging System; LI-COR Biosciences, Lincoln, NE). Four independent peptide spot arrays were used in this study.
2.4. Generation of spinal cord lysates
Lumbar segment of spinal cord lysates prepared from adult Sprague-Dawley rats were generated by homogenization and sonication in lysis buffer (50 mM Tris-HCl, pH 7.4, 50 mM NaCl, 2 mM MgCl2, 1% [vol/vol] NP40, 0.5% [mass/vol] sodium deoxycholate, 0.1% [mass/vol] sodium dodecyl sulfate [SDS]) as previously described.11 The lysis buffer included freshly added protease inhibitors (Cat# B14002; Biotools, Houston, TX), phosphatase inhibitors (Cat# B15002, Biotools), and Benzonase (Cat# 71206; Millipore, Billerica, MA). Protein concentrations were determined using the bicinchoninic acid protein assay (Cat# PI23225, Thermo scientific).
2.5. CRMP2 protein purification
GST-CRMP2 protein was purified as previously described.6,76 Briefly, after a 19-hour long induction at 16˚C with 0.5 mM IPTG, bacterial cells expressing recombinant CRMP2-GST were resuspended in 50 mM NaH2PO4, pH 7.5, 500 mM NaCl, 10% glycerol, 0.5 mM TCEP, supplemented with complete EDTA-free protease inhibitors (Roche, Basel, Switzerland). Disruption of the bacteria was performed by 2 rounds of high-pressure homogenization at 10,000 PSI with an LM10 microfluidizer (Microfluidics, Westwood), and the lysate was centrifuged 45 minutes at 4,500g at 4˚C. The supernatant was loaded on a GST-Trap HP column (GE Healthcare, Uppsala, Sweden) equilibrated with 50 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol, and 0.5 mM TCEP. After a washing step with 50 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol, and 0.5 mM TCEP, CRMP2-GST was eluted with a glutathione gradient. The fractions of interest for CRMP2-GST were loaded on a HiLoad Superdex size exclusion column (GE Healthcare) and eluted with 50 mM NaH2PO4, pH 7.5 10% glycerol, 0.5 mM TCEP. The eluted proteins were concentrated with Amicon Ultra 15 centrifugal filters (Regenerated cellulose 10,000 NMWL; Merck Millipore, Darmstadt, Germany) and flash frozen on dry ice and stored at −80˚C until use. Protein concentration was determined by a Pierce assay using bovine serum albumin (BSA) as a standard. The purity of the protein was verified with SDS-PAGE and structure/activity by checking its ability to bind tubulin (data not shown).
2.6. GST pull-down and Western blotting
Glutathione magnetic beads (Cat# B23702, Biotools), preincubated with purified CRMP2-GST (~0.4 µM), were incubated overnight with 300 µg of total protein from spinal cord lysates at 4˚C in the absence or presence of the indicated peptides with gentle rotation. Beads were washed 3 times with lysis buffer before resuspension in Laemmli buffer and denaturation (5 minutes at 95˚C) and immunoblotting as described previously.4,36,83 Immunoblots were revealed by enhanced luminescence (WBKLS0500, Millipore) before exposure to photographic film. Films were scanned, digitized, and quantified using Un-Scan-It gel version 6.1 scanning software (Silk Scientific Inc, Orem, UT).
2.7. Preparation of acutely dissociated dorsal root ganglion neurons
Dorsal root ganglia from all levels were acutely dissociated using methods as described previously.23 Rat DRG neurons were isolated from 150 to 174 g Sprague-Dawley rats using previously developed procedures.54 In brief, removing dorsal skin and muscle and cutting the vertebral bone processes parallel to the dissection stage-exposed DRG. Dorsal root ganglia were then collected, trimmed at their roots, and digested in 3 mL bicarbonate-free, serum-free, sterile DMEM (Cat# 11965, Thermo Fisher Scientific) solution containing neutral protease (3.125 mg.ml-1, Cat#LS02104; Worthington, Lakewood, NJ) and collagenase type I (5 mg/mL, Cat# LS004194, Worthington, Lakewood, NJ) and incubated for 60 minutes at 37˚C under gentile agitation. Dissociated DRG neurons (~1.5 × 106) were then gently centrifuged to collect cells and washed with DRG media DMEM containing 1% penicillin/streptomycin sulfate from 10,000 µg/mL stock, 30 ng/mL nerve growth factor, and 10% fetal bovine serum (Hyclone) before plating onto poly-D-lysine– and laminin-coated glass 12-or 15-mm coverslips. Small-diameter neurons were selected to target Aδ- and c-fiber nociceptive neurons. For rat DRG cultures, small cells were considered to be ~ < 30 µm. All cultures were used within 48 hours.
2.8. Calcium imaging in acutely dissociated dorsal root ganglion neurons
Dorsal root ganglion neurons were loaded for 30 minutes at 37˚C with 3 µM Fura-2AM (Cat# F1221, Thermo Fisher, stock solution prepared at 1 mM in DMSO, 0.02% pluronic acid, Cat#P-3000 MP, Life technologies) to follow changes in intracellular calcium ([Ca2+]c) in a standard bath solution containing 139 mM NaCl, 3 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 10 mM Na HEPES, pH 7.4, 5 mM glucose exactly as previously described.5 In some experiments, incubation with ɷ-conotoxin-GVIA (500 nM, N-type)21 was used to determine the fraction of N-type channels inhibited by t-CNRP1. Fluorescence imaging was performed with an inverted microscope, Nikon Eclipse TE2000-U, using objective Nikon Super Fluor ×20 0.75 NA and a Photometrics cooled CCD camera Cool-SNAPHQ (Roper Scientific, Tucson, AZ) controlled by MetaFluor 6.3 software (Molecular Devices, Downingtown, PA). The excitation light was delivered by a Lambda-LS system (Sutter Instruments, Novato, CA). The excitation filters (340 ± 5 and 380 ± 7) were controlled by a Lambda 10 to 2 optical filter change (Sutter Instruments). Fluorescence was recorded through a 505-nm dichroic mirror at 535 ± 25 nm. To minimize photobleaching and phototoxicity, the images were taken every ~5 seconds during the time course of the experiment using the minimal exposure time that provided acceptable image quality. The changes in [Ca2+]c were monitored by following a ratio of F340/F380, calculated after subtracting the background from both channels.
2.8.1. Constellation pharmacology
These experiments were performed as described previously,54,75 but with the following modifications. Dorsal root ganglia neurons were loaded at 37˚C with 3 µM Fura-2AM for 30 minutes in Tyrode solution (at ~310 mOsm) containing 119 mM NaCl, 2.5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 25 mM HEPES, pH 7.4, and 30 Mm glucose. After a 1-minute baseline measurement, Ca2+ influx was stimulated by the addition of the following receptor agonists: 400 nM menthol, 50 µM histamine, 10 µM adenosine triphosphate (ATP), 200 µM allyl isothiocyanate (AITC), 1 mM acetylcholine (Ach), and 100 nM capsaicin diluted in Tyrode solution. At the end of the constellation pharmacology protocol, cell viability was assessed by depolarization-induced Ca2+ influx using and an excitatory KCl solution comprising 32 mM NaCl, 90 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 25 mM HEPES, pH 7.4, and 30 mM glucose. After the 1-minute baseline measurement, each trigger was applied for 15 seconds in the order indicated above in 6-minute intervals. After each trigger, bath solution was continuously perfused over the cells to wash off excess of the trigger. This process was automated using software WinTask x64 (Version 5.1; WinTask) that controlled the perfusion of the standard bath solution and triggers through Valvelink 8.2 software (Automate Scientific). For the t-CNRP1 condition, 10 µM of the peptide was added to the Tyrode solution during the loading with Fura-2AM. Fluorescence imaging was performed under the same conditions noted above for calcium imaging. A cell was defined as a “responder” if its fluorescence ratio of 340 nm/380 nm was greater than 10% of the baseline value calculated using the average fluorescence in the 30 seconds preceding application of the trigger.
2.9. Immunocytochemistry, confocal microscopy, and colocalization analysis
Immunocytochemistry was performed on DRG incubated with vehicle (control) or peptide (10 µM) for 24 hours.6,11,23 Briefly, cells were fixed using 4% paraformaldehyde and 4% sucrose for 20 minutes at RT. Permeabilization was achieved by a 30-minute incubation in phosphate-buffered saline (PBS), 0.1% Triton X-100, after which nonspecific–binding sites were saturated by PBS containing 3% BSA for 30 minutes at RT. Cell staining was performed with primary antibodies (Cav2.2 and CRMP2) in PBS with 3% BSA overnight at 4˚C. The cells were then washed thrice in PBS, and incubated with PBS containing 3% BSA and secondary antibodies (Alexa 488 goat antimouse and Alexa 594 goat antirabbit secondary antibodies [Life Technologies]) for 1 hour at RT. Coverslips were mounted and stored at 4˚C until analysis. Immunofluorescent micrographs were acquired on a Nikon C1si scanning confocal microscope using CFI Plan APO VC ×60 oil immersion objective with 1.4 numerical aperture. For all quantitative comparisons among cells under differing experimental conditions, camera gain and other relevant settings were kept constant. Nikon NIS-Elements software was used for quantifying cellular membrane fluorescence. Membrane immunoreactivity was calculated by measuring the signal intensity in the area contiguous to the boundary of the cell. Annular regions of interest (ROIs) were assigned to individual neurons. A perimeter of a neuron was manually traced, and then an inner curve was obtained at a constant distance from the perimeter. A region enclosed by the 2 curves represented the annular membrane ROI. All images were corrected for background by subtracting the average background fluorescence (areas within the field of view not containing cells) from the ROI. Finally, colocalization analysis, including calculation of the Pearson correlation coefficient, was performed using NIS-Elements software.
2.10. Whole-cell patch recordings of Ca2+ currents in acutely dissociated dorsal root ganglion neurons
Recordings were obtained from acutely dissociated DRG neurons as described previously.35,57 To isolate calcium currents, Na+ and K+ currents were blocked with 500 nM TTX (Alomone Laboratories) and 30 mM tetraethylammonium chloride (Sigma). Extracellular recording solution (at ~310 mOsm) consisted of the following (in mM): 110 N-methyl-D-glucamine (NMDG), 10 BaCl2, 30 tetraethylammonium chloride, 10 HEPES, 10 glucose, pH at 7.4, 0.001 TTX, 0.01 nifedipine. The intracellular recording solution (at ~310 mOsm) consisted of the following (in mM): 150 CsCl2, 10 HEPES, 5 Mg-ATP, 5 BAPTA, and pH at 7.4. To isolate the contributions of N-type (Cav2.2) channel subtype, we used the following subunit-selective blockers (all purchased from Alomone Labs, Jerusalem): Nifedipine (10 µM, L-type); v-agatoxin GIVA (200 nM, P/Q-type)52; SNX-482 (200 nM, R-type)59; and 3,5-dichloro-N-[1-(2,2-dimethyl-tetrahydro-pyran-4-ylmethyl)-4-fluoro-piperidin-4-ylmethyl]-benzamide (TTA-P2, 1 µM, T-type).12 Fire-polished recording pipettes, 2 to 5 MΩ resistance, were used for all recordings. Whole-cell recordings were obtained with an HEKA EPC-10 USB (HEKA Instruments Inc, Bellmore, NY); data were acquired with a Patchmaster (HEKA) and analyzed with a Fitmaster (HEKA). Capacitive artifacts were fully compensated, and series resistance was compensated by ~70%. Recordings made from cells with greater than a 5 mV shift in series resistance compensation error were excluded from analysis. All experiments were performed at RT (~23˚C).
The Boltzmann relation was used to determine the voltage dependence for activation of ICa, wherein the conductance-voltage curve was fit by the equation G/Gmax = 1/[1 + exp (V0.5 − Vm)/k], where G is the conductance G = I/(Vm − ECa), Gmax is the maximal conductance obtained from the Boltzmann fit under control conditions, V0.5 is the voltage for half-maximal activation, Vm is the membrane potential, and k is a slope factor. ECa is the reversal potential for ICa and was determined for each individual neuron. The values of ICa around the reversal potential were fit with a linear regression line to establish the voltage at which the current was zero. The Boltzmann parameters were determined for each individual neuron and then used to calculate the mean ± SEM.
2.11. Proximity ligation assay
Proximity ligation assay (PLA) was performed as described previously57 to visualize PPIs by microscopy and is based on paired complementary oligonucleotide-labeled secondary antibodies that can hybridize and amplify a red fluorescent signal only when bound to 2 corresponding primary antibodies whose targets are in close proximity. Dorsal root ganglion neurons were incubated overnight with 10 µM of t-CNRP1 or 0.1% DMSO as a control before fixation using 4% paraformaldehyde for 20 minutes at RT. Blocking and permeabilization was performed by incubating the cells with PBS, 0.1% Triton X-100 with 3% BSA for 30 minutes at RT. Primary antibodies were incubated for 1 hour at RT in PBS, 0.1% Triton X-100, 3% BSA before 3 washes in PBS, 0.1% Triton for 5 minutes at RT. The proximity ligation reaction and amplification of signal were performed according to the manufacturer’s protocol using the Duolink Detection Kit with PLA PLUS and MINUS probes for mouse and rabbit antibodies (Duolink, Sigma). DAPI (4′,6-Diamidino-2-phenylindole, dihydrochloride) stain was used to detect cell nuclei. Immunofluorescent micrographs were acquired on a Nikon Eclipse Ti/U microscope with a photometrics cooled CCD camera CoolSNAP ES2 (Roper Scientific, Planegg, Germany) controlled by NIS-Elements software (version 4.30; Nikon instruments), using a 60X plan Apo 1.4 numerical aperture objective. ImageJ was used to count the number of PLA dots per cell and which was normalized to the area analyzed.
2.12. Synaptosome preparation from spinal cord
Spinals cords were dissected from Sprague-Dawley rats and the lumbar region isolated. Only the dorsal horn of the spinal cord was used as this structure contains the synapses arising from the DRG. Synaptosomes isolation was performed according to Ref. 62. Fresh tissues were homogenized in ice-cold sucrose 0.32M, HEPES 10 mM, and pH 7.4 buffer. The homogenates were centrifuged at 1000g for 10 minutes at 4˚C to pellet the insoluble material. The supernatant was harvested and centrifuged at 12,000g for 20 minutes at 4˚C to pellet a crude membrane fraction. The pellet was then resuspended in a hypotonic buffer (4 mM HEPES, 1 mM EDTA, and pH 7.4) and the resulting synaptosomes pelleted by centrifugation at 12,000g for 20 minutes at 4˚C.
2.13. Nanodisc-solubilized membrane protein library preparation
Nanodisc-solubilized membrane protein libraries were created as previously described.48,82 Briefly, MSP1E3D1, a His-tagged membrane scaffold protein (MSP),14 was purified as previously described67 to a concentration of around 200 µM. Exogenous 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, Avanti Polar Lipids) was solubilized in chloroform, and the concentration was determined by phosphate analysis. POPC was then dried with nitrogen and stored under vacuum overnight. Dried POPC was solubilized in 0.1 M sodium cholate to 50 mM. Membrane scaffold protein and cholate-solubilized POPC were mixed using a molar ratio of 110 POPC:MSP. Supplemental cholate was added to achieve a final cholate concentration of 25 mM. Synaptosomal proteins were solubilized in dodecyl maltoside detergent and mixed with MSP, POPC, and cholate in a ratio of 10 µg membrane proteins/nmol MSP. The reconstitution mixture was incubated for 2 hours at 4˚C. Amberlite XAD-2 hydrophobic beads (Sigma Aldrich, St. Louis, MO) were added in 0.5 to 0.8 g/mL of solution to initiate self-assembly of Nanodiscs. After overnight incubation on a shaker at 4˚C, the reconstitution mixture was separated from the Amberlite beads. Nanodiscs were purified first by immobilized metal affinity chromatography and then size exclusion chromatography using a HisTrap HP and a Superdex 200 Increase 10/300 GL column (Amersham-Pharmacia Biotech, Piscataway, NJ) respectively. Nanodisc fractions from 2 assembly reactions were pooled.
2.14. Immunoprecipitation of CRMP2 from nanodiscsolubilized synaptosomal protein libraries and preparation for proteomics identification
The nanodisc-laden synaptosomal membrane proteins were incubated with vehicle (0.01% DMSO) or 10 µM t-CNRP1 for 15 minutes on ice and then subjected to immunoprecipitation with isotype-specific IgG or 5 µg CRMP2 antibody, overnight at 4˚C, as described previously.56 After elution in 25 mM glycine (pH 2.0), the sample was brought up to 1.0% SDS and boiled at 95˚C for 5 minutes to disassemble the Nanodiscs. To remove the large excess of MSP, we then incubated the eluates with Nickel magnetic beads (Cat# B23601; Bimake, Houston, TX) for 1 hour at RT. The MSP remained bound on the Nickel magnetic beads, whereas the proteins contained in the supernatant were precipitated using 100% ice-cold acetone and centrifuged at 15,000g at 4˚C for 10 minutes. The pellets, containing solubilized proteins, were resuspended in Laemmli buffer containing 100 mM DTT, boiled at 95˚C to ensure complete solubilization of the pelleted proteins and then resolved by SDS-PAGE on 4% to 20% Novex gels (Cat# EC60285BOX; Thermo Fisher Scientific, Waltham, MA). Gels were stained with Coomassie brilliant blue (Cat# 1610436; Biorad, Hercules, CA). Gel slices were excised, digested with trypsin, and their protein content analyzed by mass spectrometry at the Arizona Proteomics Consortium.
2.14.1. Database searching
Tandem mass spectra were extracted. Charge state deconvolution and deisotoping were not performed. All tandem mass spectrometry (MS/MS) samples were analyzed using Sequest (Thermo Fisher Scientific, San Jose, CA, version 1.3.0.339). Sequest was set up to search RattusNorvegicus_UniprotKB_2016_0406_cont.fasta assuming the digestion enzyme trypsin. Sequest was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 10.0 PPM. Oxidation of methionine and carbamidomethyl of cysteine was specified in Sequest as variable modifications.
2.14.2. Criteria for protein identification
Scaffold (version 4.5.1; Proteome Software Inc, Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 20.0% probability to achieve a false discovery rate less than 0.1% by the Scaffold Local false discovery rate algorithm. Protein identifications were accepted if they could be established at greater than 95.0% probability. Protein probabilities were assigned by the Protein Prophet algorithm.58 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
2.15. Calcitonin gene–related peptide release from lumbar slices
Rats were anesthetized with 5% isofluorane and then decapitated. Two vertebral incisions (cervical and lumbar) were made to expose the spinal cord. Pressure was applied to a saline-filled syringe inserted into the lumbar vertebral foramen, and the spinal cord was extracted. Only the lumbar region of the spinal cord was used for the CGRP release assay. Baseline treatments (#1 and #2) involved bathing the spinal cord in the aforementioned standard Tyrode solution. The aforementioned excitatory solution consisting of 90 mM KCl was paired with the treatment for fraction #4. These fractions (5 minutes, 700 µL each) were collected for measurement of CGRP release. Samples were immediately stored in a −20˚C freezer. t-CNRP1 (10 µM) or vehicle (0.9% saline) was added to the pretreatment and cotreatment fractions (#3 and 4). The concentration of CGRP released into the buffer was measured by enzyme-linked immunosorbent assay (Cat# 589001; Cayman Chemical, Ann Arbor, MI).
2.16. Implantation of intrathecal catheter
For intrathecal drug administration, rats were chronically implanted with catheters as described by Yaksh and Rudy.87 Rats were anesthetized with halothane and placed in a stereotactic head holder. The occipital muscles were separated from their occipital insertion and retracted caudally to expose the cisternal membrane at the base of the skull. Polyethylene tubing was passed caudally from the cisterna magna to the level of the lumbar enlargement. Animals were allowed to recover and were examined for evidence of neurologic injury. Animals with evidence of neuromuscular deficits were excluded.
2.17. Measurement of thermal withdrawal latency
The method of Hargreaves et al.26 was used. Rats were acclimated within Plexiglas enclosures on a clear glass plate maintained at 30˚C. A radiant heat source (high-intensity projector lamp) was focused onto the plantar surface of the hind paw. When the paw was withdrawn, a motion detector halted the stimulus and a timer. A maximal cutoff of 33.5 seconds was used to prevent tissue damage.
2.18. Testing of allodynia
The assessment of tactile allodynia (ie, a decreased threshold to paw withdrawal after probing with normally innocuous mechanical stimuli) consisted of testing the withdrawal threshold of the paw in response to probing with a series of calibrated fine (von Frey) filaments. Each filament was applied perpendicularly to the plantar surface of the paw of rats held in suspended wire mesh cages. Withdrawal threshold was determined by sequentially increasing and decreasing the stimulus strength (the “up and down” method), and data were analyzed with the nonparametric method of Dixon, as described by Chaplan et al.10 and expressed as the mean withdrawal threshold.
2.19. Carrageenan-induced acute inflammatory pain and paw edema
Acute inflammatory conditions were induced by injection of 50 µL 2% carrageenan in the dorsal surface of the hind paw. To test the antinociceptive activity of t-CNRP1 under acute inflammatory conditions, thermal paw withdrawal latency (PWL) was measured as described in the thermal withdrawal latency section. In a separate experiment, hind paw edema was assessed by measuring the paw thickness. Measurements of paw thickness were made before carrageenan administration and 3 hours after carrageenan administration. Peptide (t-CNRP1, 10 µM) or vehicle was then administered and measurements were again made 3 hours after drug administration. Animals were randomly assigned to treatment conditions. All behavioral testing procedures were performed with the experimenter blinded to the treatment conditions.
2.20. Paw incision model of postoperative pain
An animal model of surgical pain was generated by plantar incision as previously described.3 Male Sprague-Dawley rats were anesthetized with isoflurane vaporized through a nose cone. The plantar aspect of the left hind paw was scrubbed with betadine and 70% alcohol 3 times. A 1-cm long incision, starting 0.5 cm from the heel and extending toward the toes, was made with a number 11 blade, through the skin and fascia of the plantar aspect of the left hind paw including the underlying muscle. The plantaris muscle was then elevated and longitudinally incised, leaving the muscle origin and insertion intact. After hemostasis with gentle pressure, the skin was closed with 2 mattress sutures of 5–0 nylon on a curved needle. Rats received an injection of gentamicin (1 mL/kg of 8 mg/mL solution, s.c.) and were allowed to recover from the anesthesia before returning to their home cage. Sham animals were anesthetized and the left hind paw scrubbed with betadine 3 times, then 70% ethanol, but no incision was made. Animals were allowed to recover for 24 hours, and then paw withdrawal thresholds were measured at 24 hours after surgery.
2.21. HIV sensory neuropathy
Mechanical allodynia is produced by intrathecal administration of the HIV-1 envelope glycoprotein, gp120.51 Seven days after implantation of an intrathecal catheter, baseline behavioral measurements were obtained and then rats were randomly assigned to 2 groups. On days 10, 12, and 14, rats were injected i.t. with 300 ng of gp120 (Cat#4961, HIV-1 BaL gp120 recombinant protein, NIH-AIDS Reagent program) in a final volume of 20 µL in 0.9% saline and 0.1% BSA.
2.22. Statistical analyses
All values represent the mean ± SEM, unless noted otherwise. All data were first tested for a Gaussian distribution using a Shapiro–Wilk test (GraphPad Prism 7 Software). The statistical significance of differences between means was determined by Student t test, parametric analysis of variance (ANOVA) followed by post hoc comparisons (Tukey) using GraphPad Prism 7 Software. All behavioral data were analyzed by nonparametric 2-way ANOVA (post hoc: Student–Newman–Keuls) in FlashCalc (Dr Michael H. Ossipov, University of Arizona, Tucson, AZ). Differences were considered to be significant if P ≤ 0.05. All data were plotted in GraphPad Prism 7. No outlier data were removed.
3. Results
3.1. Reciprocal mapping of interaction domains between CRMP and neurofibromin
A strong convergence of signaling pathways for neurofibromin and CRMP2 was suggested by findings that neurofibromin directly binds to CRMP2 and modulates CRMP2-dependent neurite outgrowth.65 In separate studies, it was reported that CRMP2 binds to voltage-gated calcium channels, particularly N-type (Cav2.2), and regulates calcium influx through these channels.5,6,11 That Cav2.2 currents were increased in sensory16 and hippocampal77 neurons from mice with a haploinsufficiency of Nf1 suggests that the CRMP2–neurofibromin protein complex could be a determinant of Cav2.2 currents.
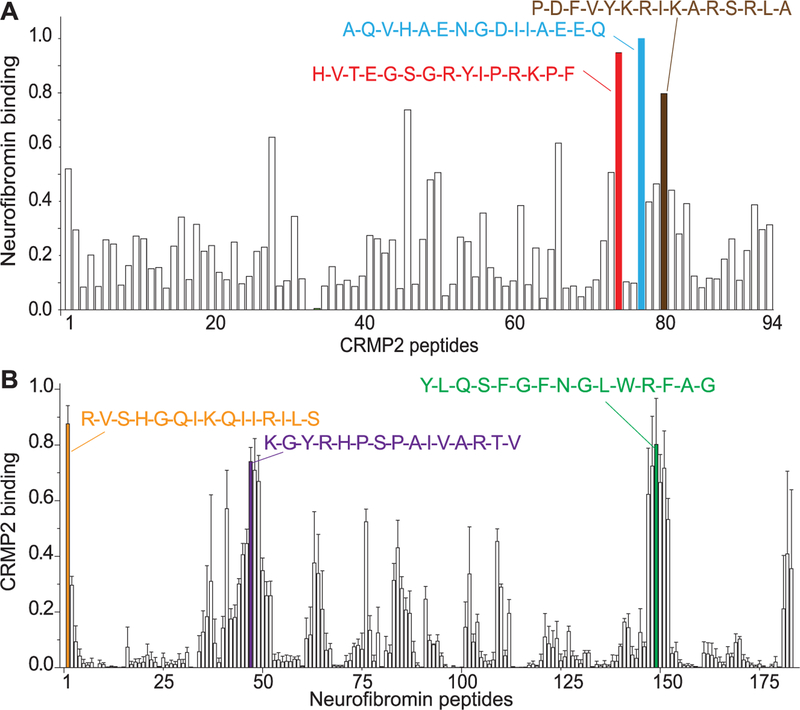
To test this hypothesis, we began by developing tools to map the putative interaction(s). A peptide array representing the entire protein sequence of CRMP2 was tiled as 15-mer peptides, with an increment of 6 amino acids, and then incubated with purified synaptosomes and probed for neurofibromin. Using this far-Western approach, we found 3 peptides originating from CRMP2 that bound highly to neurofibromin (Fig. 1A); these peptides were named CRMP2-neurofibromin regulating peptides (CNRPs 1–3) (Table 1). As a positive control, strong signal for neurofibromin was observed on spots, wherein amino acids 1357 to 1473 of the neurofibromin interacting adaptor protein syndecan-233 were tiled. Conversely, no binding of neurofibromin was observed to an array of spots with amino acids 1470 to 1564 of syndecan-2, which does not bind neurofibromin,33 thus serving as a negative control for our far-Western experiments. Next, we tiled the reported CRMP2-interacting C-terminus domain of neurofibromin65 as 15-mer peptides with an increment of 6 amino acids and performed a far-Western with CRMP2 antibodies. Here, we probed for total CRMP2, p522 CRMP2 (phosphorylation by Cdk5 at serine 522) and p555 CRMP2 (phosphorylation by ROCK at threonine 555). The rationale for probing for the phosphorylated forms of CRMP2 was guided by previous reports of the CRMP2–neurofibromin interaction regulating CRMP2’s phosphorylation status.65 This resulted in the identification of 3 neurofibromin peptides that highly bound CRMP2 or phosphorylated CRMP2 (Fig. 1B and Supplementary Figure 1, available online at http://links.lww.com/PAIN/A465); these were designated as neurofibromin–CRMP2 regulating peptides (NCRPs 1–3) (Table 1).
Figure 1.
Mapping reciprocal binding between CRMP2 and the C-terminus of neurofibromin. Far-Western assay of full-length CRMP2 (572 amino acids) (A) or the C-terminus region of neurofibromin (B) spanning amino acids 2260 to 2818, a region reported to bind CRMP2,65 in overlapping 6 amino acid steps. The blots were blocked, then overlaid with adult rat brain lysate in solubilization buffer, washed briefly, and then incubated with a neurofibromin antibody (Santa Cruz) (A) or polyclonal CRMP2 (Sigma) or monoclonal CRMP2 + polyclonal CRMP2 phospho-Ser522 and Ser555 antibodies (Millipore) (B). The blots were scanned and the spot intensities normalized to the maximum intensity in each series. The normalized data were then averaged and the mean ± SEM is shown in the graphs above (n = 2 for A and n = 3 for B). The single amino acid sequences of selected top binders are illustrated above the bars. For this and other figures in this article, unless otherwise stated, error bars represent mean ± SEM.
3.2. CRMP2–neurofibromin regulating peptide 1 controls depolarization-evoked Ca2+ influx
To investigate the function of the CRMP2/neurofibromin interaction, we designed fusions of all 6 peptides by appending to them the trans-acting activator of transcription domain of the HIV-1 protein69 to facilitate their penetration into cells. Our goal was to identify the component(s) of the CRMP2–neurofibromin interaction that regulate Cav2.2 activity. Thus, we screened the t-CNRP and t-NCRP peptides (t-refers to 11 amino acid cell-penetrant region of the transcription protein) for their ability to regulate depolarization-evoked Ca2+ influx. Ca2+ imaging was performed on rat sensory neurons preincubated overnight with 10 µM of the indicated peptides. Ca2+ influx was evoked by the application of a 90 mM KCl solution, a concentration known to recruit mostly Cav2 Ca2+ channels.80 Here, only 2 peptides, both originating from CRMP2, were able to inhibit Ca2+ influx (Fig. 2A). A concentration–response analysis indicated that the peptides inhibited Ca2+ influx in a concentration-dependent manner with IC50 values of 5.3 ± 0.4 µM (n 5 4) for t-CNRP1 and 58.8 ± 0.9 µM for t-CNRP3 (n = 4) (Fig. 2B). The concentration–response curves for inhibition of Ca2+ influx were statistically different between the 2 peptides (P < 0.05, 1-way ANOVA with Tukey post hoc test), indicating that t-CNRP1 was both more potent and efficacious compared with t-CNRP3 (Fig. 2B). An acute application of t-CNRP1 was sufficient to inhibit calcium influx (Supplementary Figure 2, available online at http://links.lww.com/PAIN/A466). We also observed a slight increase of Ca2+ influx with t-NCRP1 and t-NCRP2 peptides (Fig. 2A), both originating from neurofibromin. These results identify the existence of a regulatory node, between CRMP2 and neurofibromin, which can be targeted, with the CRMP2-derived peptide t-CNRP1, to control calcium influx.
Figure 2.
Inhibition of calcium influx by a CRMP2–neurofibromin regulating peptide (CNRP). (A) Peak calcium responses of DRG neurons incubated for 30 minutes with 30 µM t-CNRPs, 30 µM t-NRCPs, or vehicle (n = 24–35 cells per condition). Asterisks indicate statistical significance compared with untreated cells (P < 0.05, 1-way ANOVA with Dunnett post hoc test). (B) Concentration-dependent effect of t-CNRP1 and t-CNRP3 on depolarization-evoked Ca2+ influx in sensory neurons. Percent inhibition of peak Ca2+ influx normalized to the control is plotted for various concentrations of t-CNRP1 or t-CNRP3. The data were fitted in GraphPad Prism with a log (inhibitor) vs normalized response with variable slope curve which yielded an IC50 = 1.25 µM for t-CNRP1 and 58.8 µM for t-CNRP3. (C) A single subunit of CRMP2 (PDB ID 5UQC19) with a zoomed-in view of the CNRP1 peptide is shown (CNRP1 side chains drawn as sticks). Inset, the tetrameric structure with the CNRP1 peptide drawn as a red cartoon, highlighting the surface location of the peptide. ANOVA, analysis of variance; DRG, dorsal root ganglion.
The t-CNRP3 peptide contains a previously identified interaction domain (the calcium channel–binding domain 3 or CBD3) between CRMP2 and Cav2.2,5,57 which inhibits Ca2+ influx,5,66,83 and thus the inhibition observed with t-CNRP3 is likely a result of this overlapping peptide sequence and unrelated to disruption of a CRMP2-neurofibromin interaction. By contrast, the t-CNRP1 peptide is located at the surface of CRMP2 and is readily accessible for PPIs in the tetrameric form (Fig. 2C). For these reasons, we selected the t-CNRP1 peptide for further characterization of its possible regulation of Cav2.2 activity.
3.3. t-CNRP1 inhibits Ca2+ currents in sensory neurons
We next interrogated the effects of t-CNRP1 on Ca2+ currents in sensory neurons. As control, we also tested the t-NCRP1 peptide, which did not inhibit depolarization-evoked Ca2+ influx (Fig. 2A). Sensory neurons were treated overnight with the indicated peptides (10 µM), and Ca2+ currents were recorded (Fig. 3A). No change was observed for Ca2+ current densities and biophysical properties in neurons treated with t-NCRP1 compared with the control (Vehicle: 0.1% DMSO) (Fig. 3). Consistent with the Ca2+ imaging data, t-CNRP1–treated cells showed an ~56% inhibition of total Ca2+ current compared with control (Fig. 3). The biophysical properties of the Ca2+ currents were not changed in any of the conditions tested (Fig. 3E and Table 2). At least ~40% of the calcium current through Cav2.2 was blocked by t-CNRP1 (Supplementary Figure 3, available online at http://links.lww.com/PAIN/A467). The observed decrease in the peak current density suggests a lesser availability of the Ca2+ channels at the surface of the sensory neurons.
Figure 3.
t-CNRP1 decreases Ca2+ currents in sensory neurons. (A) Voltage protocol: currents were evoked by 200 millisecond prepulses between −60 mV and +50 mV from a holding potential of −90 mV. (B) Representative family of current traces are illustrated from sensory neurons treated with DMSO (0.1%; vehicle control) or treated with 10 µM of the indicated peptides. (C) Summary of the total calcium current vs voltage relationship (in pA/pF) from sensory neurons treated with DMSO (0.1%; vehicle control) or treated with 10 µM of the indicated peptides. (D) Peak current density, at +10 mV, for the indicated conditions. n = 8 to 9 cells per condition (*P < 0.05, 1-way analysis of variance with Tukey post hoc analysis). (E) The normalized conductance G/Gmax-voltage relations for the voltage dependence of activation for the indicated conditions. The continuous lines through the data points represent the Boltzmann fits.
Table 2.
Comparison of biophysical properties of calcium channel currents after treatment with peptides in DRG cells.
| Voltage dependence of activation | Voltage dependence of inactivation | |||
|---|---|---|---|---|
| V1/2 (mV) | Slope (mV/e–fold) | V1/2 (mV) | Slope (mV/e–fold) | |
| Vehicle (0.1% DMSO) | 4.4 ± 2.1 (9) | 8.5 ± 0.8 (9) | –23.0 ± 1.8 (7) | 9.6 ± 1.1 (7) |
| t-CNRP1 | 3.5 ± 2.6 (9) | 6.9 ± 1.0 (9) | –19.0 ± 2.1 (6) | 8.9 ± 1.5 (6) |
| t-NCRP1 | 2.7 ± 2.2 (8) | 6.6 ± 1.0 (8) | –18.9 ± 1.9 (6) | 9.8 ± 1.7 (6) |
n value provided in parentheses. Comparative calcium current statistics from DRG cells treated overnight with 10 µM of the indicated peptides. Comparative current densities and Boltzmann properties of half-maximal effect (V1/2) and slope (k) for activation and fast inactivation are presented. Data show mean ± SEM; no significant difference was found, Kruskal–Wallis test with pairwise comparisons.
DRG, dorsal root ganglion.
3.4. t-CNRP1 controls the trafficking of the N-type Ca2+ channel by inhibiting its interaction with CRMP2
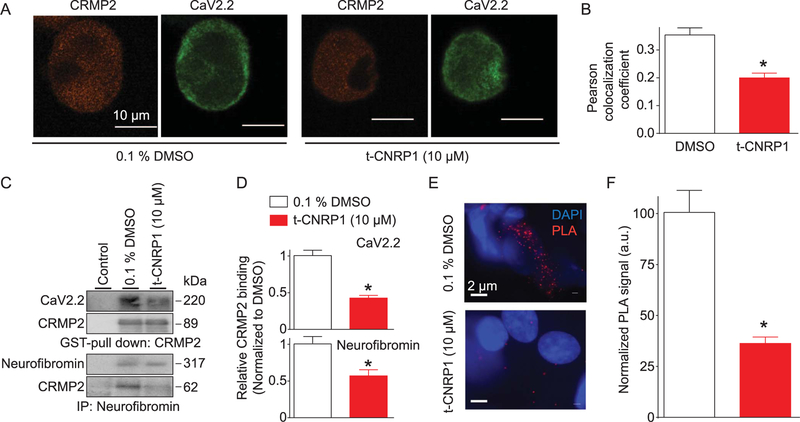
The Ca2+ peak current density is decreased by t-CNRP1, a peptide arising from CRMP2. Decreased peak current density may be due to a decreased amount of the N-type Ca2+ channel at the plasma membrane, an event previously reported to be dependent on CRMP2.23 To test this possibility, we analyzed Cav2.2 membrane localization using confocal microscopy (Fig. 4A). By costaining with Cav2.2 and CRMP2, we observed decreased membrane localization of Cav2.2 in sensory neurons treated overnight with 10 µM t-CNRP1 compared with those treated with vehicle (DMSO) (Fig. 4A). The Pearson correlation coefficient (PCC) was used to quantify the degree of colocalization between the fluorophores for the 2 proteins; the PCC relies on the deviation from the mean. A high PCC value verified the membrane colocalization, and this was decreased by t-CNRP1 (Fig. 4B).
Figure 4.
t-CNRP1 controls Cav2.2 trafficking and CRMP2/Cav2.2 interaction. (A) Representative micrographs of DRG neurons immunostained with Cav2.2 and CRMP2 after treatment with 10 µM t-CNRP1. Scale bar is 10 µm. (B) Bar graphs of Pearson colocalization coefficient values for surface colocalization of CRMP2 and Cav2.2 in DRG neurons treated with vehicle (0.1% DMSO) or 10 µM of t-CNRP1 (n = 10–12 cells). (C) Pull down with CRMP2-GST (top blots) or immunoprecipitation (IP) with neurofibromin antibody (bottom blots) from spinal cord lysates in the absence or presence of t-CNRP1. (D) Relative CRMP2 binding to Cav2.2 (top) or neurofibromin (bottom) (n = 3). (E) Representative micrographs showing proximity ligation assay (PLA) signal for CRMP2/Cav2.2 interaction in sensory neurons treated with (0.1% DMSO) or 10 µM of t-CNRP1. DAPI (blue) signal shows the nucleus of the cell. Scale bar is 2 µm. (I) Summary of the number of PLA dots, normalized to the area of the cell analyzed (n = 14–16 cells each). *P < 0.05 vs control, Student t test. DRG, dorsal root ganglion.
Diminished colocalization of CRMP2/Cav2.2 is likely a result of the dissociation of the previously reported interaction between these 2 proteins.6,11 Therefore, we next tested whether t-CNRP1 peptide could interfere with the CRMP2/Cav2.2 interaction. Rat spinal cord lysates were incubated with glutathione beads preadsorbed with purified CRMP2-GST in the presence of DMSO (0.3%, control) or 10 µM of t-CNRP1, and then CRMP2-bound proteins were recovered by boiling in Laemmli buffer in reducing conditions, followed by immunoblotting. Probing of the CRMP2-enriched fraction with a Cav2.2 antibody demonstrated a robust interaction between Cav2.2 and CRMP2 (Fig. 4C; top blot, lane 2). This interaction was inhibited by t-CNRP1 (Fig. 4D).
Because t-CNRP1 was designed based on a CRMP2 interaction domain with neurofibromin, we also investigated whether the peptide could disrupt the CRMP2/neurofibromin interaction. Coimmunoprecipitation of neurofibromin with CRMP2 (Fig. 4C; third blot, lane 2) was decreased in the presence of t-CNRP1 (Fig. 4D).
Finally, to confirm the disruption of CRMP2 interaction with Cav2.2 in sensory neurons, we used a PLA. This assay allows for the detection and quantification of protein complexes in situ. When proteins are within 30 nm of each other, a PLA signal (fluorescent amplification product) is generated (Fig. 4E). Direct protein interactions between CRMP2 and Cav2.2 in DRG sensory neurons were evident with PLA (Fig. 4E). Treatment of DRG neurons with t-CNRP1 decreased the PLA signal by ~66% compared with vehicle-treated neurons (Fig. 4F). Collectively, these results demonstrate that t-CNRP1 decreases Cav2.2-dependent Ca2+ influx, currents, and surface localization through disruption of CRMP2’s binding to Cav2.2 and neurofibromin.
3.5. Identification of a novel CRMP2 interacting partner, syntaxin 1A
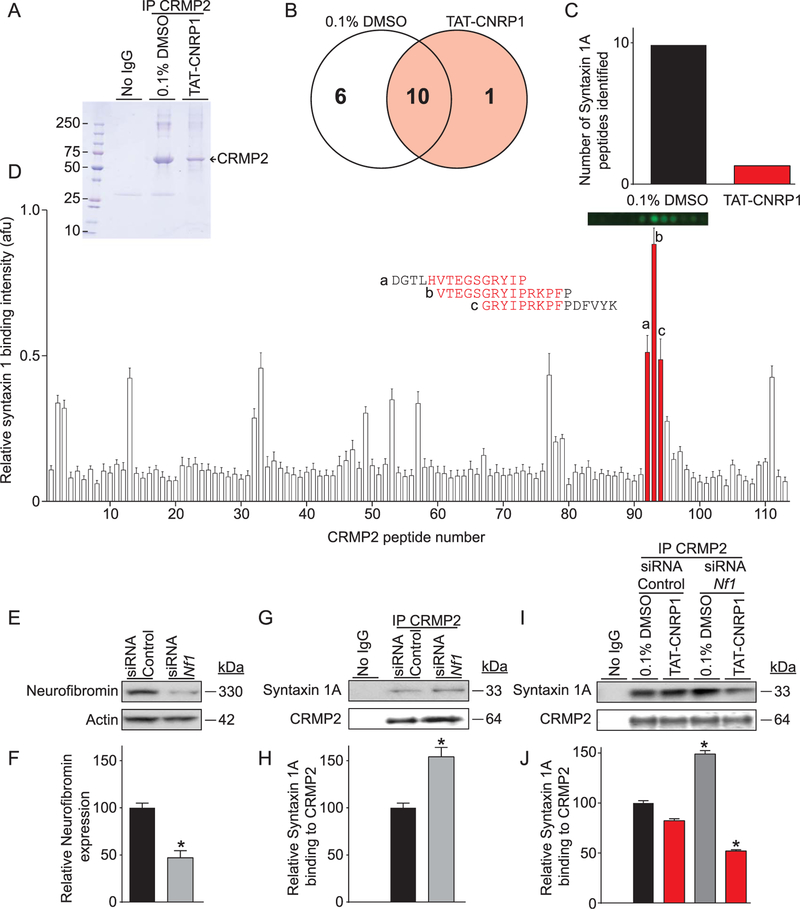
Thus far, our findings demonstrate that uncoupling the CRMP2/ neurofibromin interaction, with t-CNRP1, leads to decreases in surface Cav2.2 and Ca2+ influx. And although it was previously reported that CRMP2 binds to neurofibromin,65 the functional consequence of a connection between a CRMP2/neurofibromin interaction and a CRMP2/Cav2.2 interaction is an open question, despite reports that Cav2.2 mRNA and currents are increased in sensory neurons with reduced neurofibromin levels.16,77 We hypothesized that the CRMP2/neurofibromin interaction could be preventing CRMP2 from interacting with protein partners involved in Cav2.2 trafficking and that t-CNRP1 could inhibit these interactions. To identify these proteins, we constructed a synaptic membrane library embedded in nanodiscs (self-assembling nanoscale phospholipid bilayers stabilized by engineered membrane scaffold; Supplementary Figure 4, available online at http://links.lww.com/PAIN/A469). Notably, this approach allows us to assess CRMP2 binding to membrane proteins in the absence of detergents, a component usually essential for solubilizing membrane proteins. In other words, the nanodiscs preserve the integrity of membrane proteins by serving as a virtually native environment while simultaneously increasing their accessibility to biochemical assays like mass spectrometry. We coimmunoprecipitated CRMP2-bound proteins from the nanodics library in the presence of t-CNRP1 or 0.1% DMSO (Fig. 5A). Mass spectrometry analysis of the samples revealed 17 proteins identified with >95% confidence (Fig. 5B). Among these proteins, we identified syntaxin 1A, a well-established regulator of Cav2.2,73 whose binding to CRMP2 was disrupted by t-CNRP1 (10 peptides identified in 0.1% DMSO CRMP2 immunoprecipitates vs 1 peptide found in the presence of t-CNRP1) (Fig. 5C).
Figure 5.
Identification of a CRMP2/syntaxin 1A interaction. (A) Coomassie brilliant blue staining of an eluate from CRMP2 immunoprecipitation (or isotype-specific control IgG) in the presence of 10 µM t-CNRP1 or 0.1% DMSO as a control. The band corresponding to CRMP2 is indicated. (B) Venn diagram of the proteins identified in the indicated immunoprecipitations. (C) Bar graph showing the number of peptides detected for syntaxin 1A in CRMP2 immunoprecipitates with 10 µM t-CNRP1 or 0.1% DMSO. (D) Far-Western assay on full-length CRMP2 tiled as 15-mer peptides with an increment of 5 amino acids. Syntaxin 1A binding to each peptide is shown as mean ± SEM (n = 4). Strong syntaxin 1A binding was found for peptides #92 to 95 from CRMP2 whose sequence is indicated next to the bar graph. Representative dot blot fluorescence is shown for these peptides. (E) Representative immunoblot showing neurofibromin knockdown after Nf1 siRNA transfection in CAD cells. (F) Bar graph showing neurofibromin expression level relative to control siRNA transfection, mean ± SEM (n = 3). (G) Representative immunoblot showing syntaxin 1A binding to CRMP2 after neurofibromin knockdown in CAD cells. (H) Bar graph showing increased syntaxin 1A binding to CRMP2 after neurofibromin knockdown, mean ± SEM (n = 3). (I) Representative immunoblot showing inhibition of syntaxin 1A binding to CRMP2 by 10 µM t-CNRP1 after neurofibromin knockdown, compared with 0.1% DMSO as a control. (J) Bar graph showing decreased syntaxin 1A binding to CRMP2 by 10 µM t-CNRP1 compared with 0.1% DMSO, mean ± SEM (n = 3). *P < 0.05, Kruskal–Wallis or Mann–Whitney test. CAD, catecholamine A–differentiated cells.
To further understand the syntaxin 1A/CRMP2 interaction, we next sought to define the precise sites of the interaction using the CRMP2 peptide array. Overlaying CRMP2 15-mer peptide arrays with a 5 amino acid overlapping interval with purified synaptosomes and probing with syntaxin 1A revealed several peptides with high binding (Fig. 5D). The highest binding peptide contained residues between amino acids aspartic acid 456 and lysine 480 of CRMP2. Remarkably, this region, which consists of 3 contiguous 15-mer peptides constituting the binding domain between CRMP2 and syntaxin 1A, contains the entire t-CNRP1 sequence (Fig. 5D). These results identify a singular “Janus”/2-faced domain on CRMP2 that coordinates both the CRMP2/neurofibromin and CRMP2/Syntaxin 1A interactions.
3.6. Neurofibromin inhibits CRMP2 interactions with syntaxin 1A
To reconcile how t-CNRP1 could participate in both sets of interactions, we hypothesized that the CRMP2/neurofibromin interaction would compete with CRMP2/syntaxin 1A interaction as suggested by our proteomics data (Fig. 5C). To test if neurofibromin’s interaction with CRMP2 could regulate CRMP2 interaction with syntaxin, a short interfering RNA (siRNA) approach was used to knockdown neurofibromin (Fig. 5E). Transfection of an siRNA against Nf1 designed to knockdown neurofibromin65 in catecholamine A–differentiated cells, a model neuronal cell line with constitutive CRMP2 and neurofibromin expression, resulted in a 50% reduction in neurofibromin levels (Fig. 5F) similar to those reported in mice with a heterozygous loss of Nf1. We then tested CRMP2 binding to syntaxin 1A by coimmunoprecipitation (Fig. 5G). We found an increased association between CRMP2 and syntaxin 1A when neurofibromin levels were reduced in catecholamine A–differentiated cells (Figs. 5G and H). This increased association was blocked by t-CNRP1 (Figs. 5I and J). These results show that the previously unknown interaction between CRMP2 and syntaxin 1A is regulated by neurofibromin and that it can be inhibited using t-CNRP1.
3.7. Constellation pharmacology to “fingerprint” sensory neuronal responses to t-CNRP1
The results obtained thus far demonstrate the potential of t-CNRP1 to inhibit Ca2+ influx through control of Cav2.2 membrane localization. However, these studies do not the address the type/class of neuron targeted by t-CNRP1. To test this, we used the recently described phenotypic screening method termed constellation pharmacology,74,75 which uses subtype-selective pharmacological agents to elucidate cell-specific combinations (constellations) of key signaling proteins that define specific cell types. The constellation pharmacology protocol consists of sequential challenges, applied 6 minutes apart, to test activity of Ca2+ permeable ligand-gated ion channels, metabotropic receptors and voltage-gated Ca2+ channels. At the end of each experiment, a membrane-depolarizing agent (ie KCl) is used to ensure neuronal viability; neurons that did not respond to this trigger were not analyzed. In each case, the readout is a change in Ca2+ fluorescence. Examples of typical Ca2+ traces of DRG incubated with vehicle (0.1% DMSO) (Fig. 6A) or t-CNRP1 (10 µM) (Fig. 6B) demonstrate the heterogeneity in responses. Of importance, consistent with our earlier data (Figs. 2 and 3), t-CNRP1 reproducibly decreased the K+-evoked Ca2+ influx, despite being applied after several challenges of the constellation pharmacology protocol.
Figure 6.
Functional “fingerprinting” of DRG neuronal subclasses after treatment with t-CNRP1. (A) Ca2+-imaging traces of DRG responding to constellation pharmacology agonist or membrane-potential triggers. Each trace represents the response of a different neuron. In a typical experimental trial, the responses of >300 individual neurons were monitored simultaneously. Selected traces are shown. Colored columns indicate the ~15-s application of challenge compounds or high [K+]o; after each application, the free compound or high [K+]o was continuously washed out of the well with room temperature bath solution. The y-axis shows the Fura-2AM fluorescence ratio (F340/F380) for each trace. Raw, unfiltered traces are presented. Triggers used were menthol (400 nM), histamine (50 µM), ATP (10 µM), AITC (200 µM), acetylcholine (1 mM), capsaicin (100 nM), and KCl (90 mM). Dorsal root ganglion neurons were treated with either vehicle (0.01% DMSO) (A) or t-CNRP1 (10 µM) (B) and the neuronal populations analyzed by the constellation pharmacology protocol. All cells were selected based on their response to the depolarizing pulse of KCl. Size of each cell is indicated by the surface area value in each trace. (C) Bar graph showing the overall number of functional DRG neuron classes identified. t-CNRP1 increased the number of functional DRG neuron classes detected. (D) The response of DRG neurons to one or more constellation pharmacology triggers was analyzed. The polar plot indicates the percentage of cells that responded to the indicated number of triggers independent of which compound they responded to. The number 0 corresponds to the proportion of cells that responded to KCl only and no other trigger. After t-CNRP1 treatment, less cells responded to only KCl and increased number of cells responded to one or more constellation pharmacology triggers. (E) Polar plot showing the percent of cells responding to major classes of triggers. Data are from 4 independent experiments with a total n = 1541 for control and n = 1642 for 10 µM t-CNRP1. Bar graphs showing the average peak (F) or area under the curve (AUC) (G) calcium response in the major functional neuronal populations identified in panel (E). Area under the curve was calculated with Prism software using the trapezoid rule. t-CNRP1 did not decrease the peak calcium influx in response to any trigger except Cap for peak or Ach and Cap for AUC (*P < 0.05; Student t test). An AUC for KCl was not calculated in these experiments because KCl was the last trigger (see traces in A and B). (H) Bar graphs showing the average peak response to depolarization for cells showing a response to the indicated trigger independent of which other compound they responded to. T-CNRP1 decreased the peak KCl response for all subclasses except Menth responding DRG neurons. (*P < 0.05; Student t test). Ach, acetylcholine; AITC, allyl isothiocyanate; ATP, adenosine triphosphate; DRG, dorsal root ganglion; Hist, histamine; Menth, menthol; Cap, capsaicin; KCl, potassium chloride.
We applied the constellation pharmacology protocol to DRG neurons treated overnight with 10 µM t-CNRP1 (n 5 1622) or 0.1% DMSO (vehicle, n = 1541). Data from 3 independent experiments were collected, and the responses of each neuron to each constellation pharmacology trigger were analyzed. Only DRG with responses >10% over the baseline fluorescence were considered in our analyses. We first asked if t-CNRP1 treatment altered the overall competence of the neurons by assessing if the DRG respond to the same number of agonist challenges, independent of which compound they responded to (Fig. 6C). t-CNRP1 increased the number of neurons responding to receptor agonist challenge compared with vehicle-treated DRG (Fig. 6C). Consistent with this, an increased number of different functional neuronal subclasses were found in sensory neurons treated with t-CNRP1. We next asked if t-CNRP1 could change the sensitivity of sensory neurons to the different agonist challenges. We analyzed the percent of cells responding to a defined trigger, independent of any other trigger the cells may have responded to. In t-CNRP1–treated DRG, the percent responders to the various triggers were increased compared with vehicle-treated neurons (Fig. 6D). These results demonstrate that t-CNRP1 increases the functional competence of sensory neurons.
The above analyses do not take into account the extent of Ca2+ influx following each challenge. Therefore, here we analyzed peak Ca2+ responses of responders to the various triggers. t-CNRP1 did not change the peak response to acetylcholine, allyl isothiocyanate (AITC), ATP, histamine, and menthol but increased the Ca2+ influx elicited by capsaicin. This suggests a sensitization of TRPV1 channels (Fig. 6F). The area under the curve (AUC) was quantified for each trigger to assess if the extent of the Ca2+ response could be altered by t-CNRP1. This revealed a decreased AUC in t-CNRP1–treated DRG neurons for acetylcholine and for capsaicin (Fig. 6G).
We next asked if the inhibition of K+-evoked Ca2+ influx by t-CNRP1 could be recapitulated in certain functional classes identified above. We investigated the average peak Ca2+ response to KCl stimulation of neurons that responded to a trigger, independent of any other compounds they responded to. In t-CNRP1–treated DRG, the average peak Ca2+ response to KCl was lower in acetylcholine, AITC, ATP, histamine, and capsaicin responding but not in menthol-sensitive DRG neurons compared with control (Fig. 6H). This shows that t-CNRP1–mediated inhibition of Cav2.2 is not efficient in the menthol-sensitive subpopulation of DRG neurons. Menthol responsiveness has been involved in analgesic mechanisms.42,64 Thus, the preservation of depolarization-induced Ca2+ influx in menthol-responding DRG neurons might indicate a beneficial effect of t-CNRP1 in inducing analgesia.
Dorsal root ganglion neurons are heterogeneous in size.28 Although cell size approximates the functional class of DRG neurons, it is not an absolute predictor of functional type of neuron. Having identified classes of neurons that respond differently to the triggers, we next asked if some of these changes could be due to the size heterogeneity of the neurons. There were no differences in the numbers of cells across different cell sizes between vehicle- or t-CNRP1–treated DRG neurons (Supplementary Figure 5, available online at http://links.lww.com/PAIN/A470). These results highlight that t-CNRP1 targets all neuronal size classes except for menthol-responding DRG neurons. The constellation pharmacology experiments identify that distinct neuron classes altered after t-CNRP1 treatment and may contribute to a better understanding of how this peptide may achieve antinociceptive efficacy.
3.8. t-CNRP1 reduces K+-evoked transmitter release from spinal cord ex vivo
The release of the neuropeptide CGRP by presynaptic terminals from small-diameter sensory neurons is known to be a major mediator of pain signaling.88 Activity of the N-type Ca2+ channel has been directly linked to CGRP release in nerve terminals,40,44 and controlling this channel’s interaction with CRMP2 has proven efficient in inhibiting CGRP release from sensory neurons.5,20 Furthermore, Cav2.2-dependent neurotransmitter release relies on syntaxin 1A recruitment.27,37 Our data suggest that t-CNRP1 inhibits Cav2.2 by uncoupling of CRMP2 and syntaxin 1A interaction. We, therefore, hypothesized that t-CNRP1 could inhibit the release of the pro-nociceptive neurotransmitter CGRP. We used an ex vivo approach by dissecting the lumbar region of rat spinal cord to perform a CGRP release assay, using high potassium as the trigger. After 3 washes, samples were collected every 10 minutes and CGRP content measured by an enzyme-linked immunosorbent assay. Basal CGRP release was ~8.5 ± 1.9 pg·ml−1·mg−1 (Fig. 7, fractions #1 & 2). t-CNRP1 (10 µM) or vehicle (0.1% DMSO) was added (Fig. 7, fraction #3) 10 minutes before stimulation with 90 mM KCl (Fig. 7, fraction #4). Under basal conditions, t-CNRP1 treatment did not elicit any CGRP release from the spinal cords (Fig. 7, fraction #3). Depolarization resulted in increased CGRP release (vehicle, ~127.8 ± 17.3 pg·ml−1·mg−1), which was significantly inhibited (−58%) by t-CNRP1 (~58.9 ± 3.1) (Fig. 7, fraction #4). These results show that t-CNRP1 inhibition of Cav2.2 results in decreased evoked CGRP release in the spinal cord.
Figure 7.
CGRP release from spinal cord is inhibited by t-CNRP1. Spinal cords isolated from naive adult rats were used to assess depolarization (90 mM KCl)- induced CGRP release. Depolarization-triggered CGRP release from the spinal cords was significantly attenuated by pre- and co-incubation of t-CNRP1 (10 µM) with 90 mM KCl (10 min/fraction). t-CNRP1 alone did not have any effect on CGRP release (fraction #3) and reduced evoked CGRP release by 58% compared with control (fraction #4, *P < 0.05 vs control; 2-way ANOVA post hoc Sidak test). Y-axis shows immunoreactive CGRP levels in the bath solution and normalized to the weight of each spinal cord. ANOVA, analysis of variance; CGRP, calcitonin gene–related peptide.
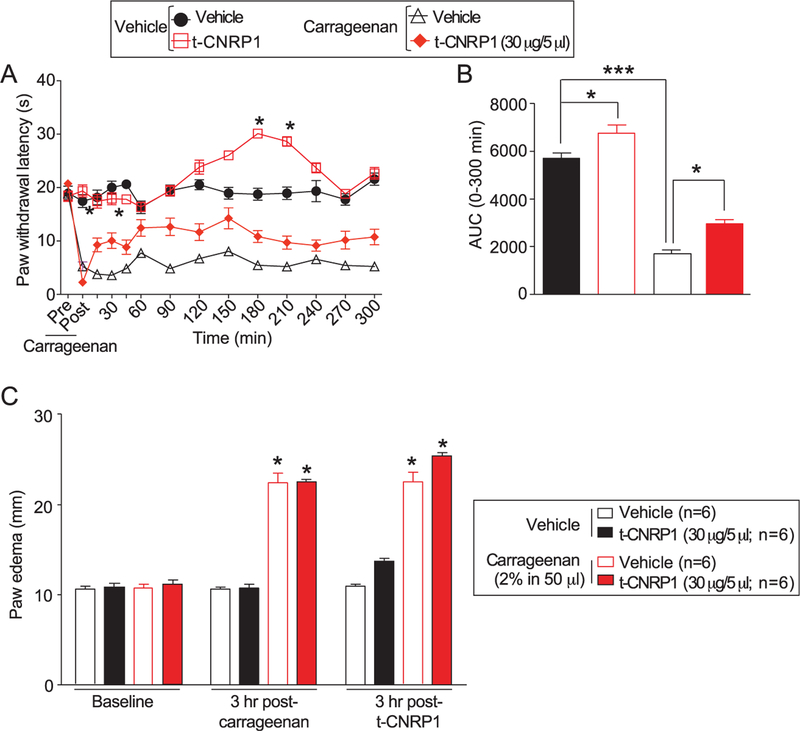
3.9. t-CNRP1 reverses carrageenan-induced inflammatory pain
The above data showing inhibition of the pro-nociceptive neuropeptide CGRP by t-CNRP1 imply an antinociceptive activity of the peptide. Therefore, we tested the efficacy of t-CNRP1 in reversing inflammatory pain. Administration of carrageenan (50 µL of 2%) produced a significant decrease in PWLs 3 hours postcarrageenan injection (Figs. 8A–C); PWLs were decreased by ~80% 3 hours after carrageenan injection. The intrathecal administration of t-CNRP1 (30 µg/5 µL i.th.) significantly attenuated carrageenan-induced thermal hypersensitivity after 60 minutes and lasted 3 hours compared with vehicle group (Fig. 8A). In sham animals, saline administration in the paw, intrathecal administration of t-CNRP1 (20 µg/5 µL i.th) produced analgesia at 180 minutes and 210 minutes after injection. This was evidenced by quantifying the AUC which showed significant overall analgesia (sham animals) or antinociception (carrageenan animals) elicited by t-CNRP1 (Fig. 8B). Administration of carrageenan (50 µL of 2%) produced a significant increase in paw thickness 3 hours postcarrageenan injection (Fig. 8C). The carrageenan-induced edema was not blunted by t-CNRP1 (Fig. 8C).
Figure 8.
t-CNRP1 reduces carrageenan-induced inflammation. Spinal administration of (A) t-CNRP1 (20 µg/5 µL i.t.) significantly attenuated carrageenan (50 µL, 2%, intrapaw)-induced thermal hypersensitivities in rats. Paw withdrawal latencies were significantly decreased 3 hours after carrageenan injection. Administration of t-CNRP1 (20 µg/5 µL i.t.) significantly attenuated carrageenan-induced thermal hypersensitivity compared with vehicle group (n = 6, *P < 0.05). Data analyzed using 2-way analysis of variance followed by Bonferroni post hoc tests where time was treated as “within-subjects” factor, whereas treatment was treated as “between” subjects factor. (B) Summary of the data in panel A plotted as area under the curve (AUC) between 0 and 300 minutes. (C) Intrapaw administration of carrageenan (50 µL 2%) produced significant paw edema in rats 3 hours after carrageenan treatment. Edema was not blocked by t-CNRP1 administration (20 µg/5 µL i.t.). Data are expressed as mean ± SEM. *P < 0.05, ***P < 0.001 when compared with precarrageenan values.
3.10. t-CNRP1 reverses postsurgical pain
After showing the beneficial effect of t-CNRP1 in a model of inflammatory pain, we tested the antinociceptive potential of this peptide on thermal hyperalgesia and tactile allodynia induced by an incision of the plantaris muscle of the rat hind paw, a model of postoperative surgical pain.3 Gabapentin has been shown to be antinociceptive in this model, implicating involvement of Cav2.2.22 An incision of the rat plantaris muscle led to an induction of thermal and tactile hypersensitivity (Figs. 9A–B). Both nociceptive responses were maintained during the 6-hour experimental period in vehicle-treated animals. Spinal administration of t-CNRP1 (30 µg/5 µL i.th.) blocked the tactile allodynia at 30 minutes after injection and then at 210 minutes and 300 minutes (Fig. 9A). Thermal hyperalgesia was reversed for at least 6 hours (Fig. 9B). In sham-injured animals, t-CNRP1 demonstrated an analgesic activity, for PWL at 150 and 180 minutes after administration (Fig. 9B). The vehicle (saline) did not affect the nociceptive responses. The quantification of the AUC showed significant antinociception elicited by t-CNRP1 (Figs. 9C–D).
Figure 9.
t-CNRP1 reduces plantar incision-induced thermal hyperalgesia and mechanical allodynia. Rats received a plantar incision on the left hind paw. Paw withdrawal thresholds (PWTs) were significantly decreased 24 hours after incision. t-CNRP1 (20 µg/5 µL) or vehicle (saline) were injected into the intrathecal space and PWTs measured. Paw withdrawal thresholds were significantly reversed at the indicated times after injection of t-CNRP1 (A) (n = 6; *P < 0.05, **P < 0.01, ***P < 0.001; 2-way analysis of variance [ANOVA] with a Bonferroni post hoc test) where time was treated as “within-subjects” factor, whereas treatment was treated as “between” subjects factor. Likewise, paw withdrawal latencies (PWLs) were significantly decreased 24 hours after incision. Injection of t-CNRP1 (B) significantly reversed PWLs at the indicated times (n = 6; *P < 0.05, **P < 0.01, ***P < 0.001; 2-way analysis of variance [ANOVA] with a Bonferroni post hoc test). Area under the curve (AUC), using the trapezoid method, for PWT (C; summary for data shown in A) and PWL (D; summary for data shown in B) are shown.
3.11. t-CNRP1 reverses HIV-induced sensory neuropathy
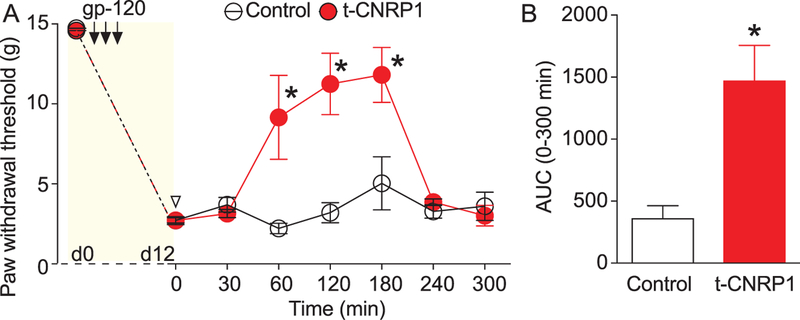
HIV-induced sensory neuropathy (HIV-SN) is a frequently occurring neurological complication of HIV infection.45 Cav2.2 has been found to contribute to pain in patients with HIV-SN.60 Therefore, we tested the antinociceptive potential of t-CNRP1 on mechanical allodynia induced by intrathecal injections of the HIV-1 envelope glycoprotein (gp120), which results in robust mechanical allodynia.51,89 Intrathecal injections of gp120 led to the gradual lowering of paw withdrawal thresholds compared with baseline (ie, pre-gp120) (Fig. 10A); this mechanical allodynia was reversed by an intrathecal administration of t-CNRP1 (30 µg/5 µL i.th.) at 60 minutes and lasted for 2 hours (Fig. 10A). The AUC was significantly increased by the intrathecal administration of t-CNRP1 (Fig. 10B). These results suggest that targeting CRMP2 interactions with neurofibromin, syntaxin 1A and Cav2.2 is beneficial for nociceptive pain behaviors in a rat model of HIV-SN.
Figure 10.
t-CNRP1 reduces gp120-induced nociceptive behaviors. Paw withdrawal thresholds were significantly reduced 15 days after 3 injections of glycoprotein 120 (gp120) in the intrathecal space. (A) Injection of TAT-CBD3-L5M (20 µg/5 µL) significantly reversed PWTs at the indicated times (n = 6; *P < 0.05; 2-way ANOVA with a Student–Newman–Keuls post hoc test). (B) Bar graph showing the area under the curve (AUC), using the trapezoid method for the data shown in (A). *P < 0.05, 1-way analysis of variance with Tukey post hoc analysis. TAT, transcription.
4. Discussion
An increased understanding of the PPI networks will likely lead to the identification of protein interaction hubs and nodes that are critical for acquisition and maintenance of characteristics of pain. Here, we identify a PPI framework that underpins NF1-related pain. The data reported here establish a protein interaction network with CRMP2 as a node and neurofibromin, syntaxin 1A, and Cav2.2 as interaction edges. Neurofibromin sequesters CRMP2 from syntaxin 1A. On loss of neurofibromin, as observed in patients with NF1, the CRMP2/neurofibromin interaction is uncoupled, which liberates CRMP2 to interact with both syntaxin 1A and Cav2.2, culminating in increased release of the pronociceptive neurotransmitter CGRP. Our work also identifies a novel tool–the CRMP2-derived aptamer CNRP, which is capable of uncoupling CRMP2’s interactions with neurofibromin, syntaxin 1A, and Cav2.2. Equally important, we demonstrate that t-CNRP1 inhibits CGRP release and attenuates pain in preclinical rodent models of inflammatory, postsurgical, and neuropathic pain.
More than 2 million people worldwide, including over a 100,000 Americans, suffer from neurofibromatosis type 1 (NF1),25 the most commonly inherited neurological disorder in humans. Prominent among the multisystemic manifestations related to the accumulation of neurofibromas in NF1 is painful peripheral SN; one in 4 persons with NF1 experiences chronic pain that can persist for months to years.46 Peripheral neuropathy constitutes a potentially severe complication in patients with NF1 associated with a frequent morbidity related to spinal complications.15 The pain from these neuropathies is often described as ongoing or paroxysmal stabbing, shooting, or electrical pain, and can be elicited by merely touching the skin overlying the tumor. Large nodular plexiform neurofibromas developed in deep peripheral nerves or roots are usually associated with permanent disabling pain, and subcutaneous neurofibromas are typically associated with episodes of paroxysmal pain. In NF1, pain is often overlooked,13,84 despite many studies reporting pain as a key symptom of patients with NF1 and a key component affecting their quality of life.13,47,84 Corollary to this, a survey from the National Cancer Institute reported 63% of the patient’s families hope for more clinical trials aiming at management of the pain related to NF1.84 Studies from a genetic model of NF1, a heterozygous mouse where only 1 allele of the Nf1 gene is deleted (Nf1+/−), revealed an upregulation of pathways associated with pain signal transmission.16,17,31,32,79,81 A potential mechanism postulated to underlie pain in people with NF1 is the increased sensitization of sensory neurons due to increased calcium currents in Nf1+/− mice.77 Involvement of ion channels, particularly N-type voltage-gated calcium channels has been suggested to underlie some of this neuronal sensitization and these channels are genetically39 validated. Of importance, these channels are also clinically8,49 validated targets for pain management. Cav2.2 participates in pain signal transmission by triggering the release of the neuropeptide (CGRP).40,44 This event correlates with findings from the Nf1+/− mouse, as evoked CGRP release was increased in sensory neuron cultures from these heterozygous mice.31 Taken together, these results highlight a dysregulation of Cav2.2 and, consequently, CGRP release, downstream of the loss of neurofibromin. Because CGRP from the central terminal of primary sensory neurons is important in pain signal propagation, modulation of release at this site may be a critically important component of pain processing/ hyperalgesia/nociception.
Two recent proteomics approaches identified collapsin response mediator protein 2 (CRMP2) as a novel target of neurofibromin.41,65 As CRMP2 is a major regulator of Cav2.25,6,11 and interacts with neurofibromin,41,65 we targeted the CRMP2 neurofibromin interaction as a novel approach to control Cav2.2 function. Because CRMP2 is a novel regulator of synaptic transmission and neurofibromin can regulate the activity of CRMP2,65 this implies that the tripartite neurofibromin–CRMP2– calcium channel complex might be an important determinant of neuronal excitability and thus, a new target for drug discovery. We identified the binding domain between CRMP2 and neurofibromin on the CNRP1 peptide sequence and then used this peptide to curb CaV2.2 activity. CNRP1 inhibited total calcium influx with an IC50 of 5.3 µM; at least 40% of this was due to inhibition of Cav2.2. This is within the same range as other strategies we used previously to target the CRMP2/Cav2.2 interplay: (S)-lacosamide,55,63,71 a molecule inhibiting CRMP2 phosphorylation,54–56 had an IC50 of 1.25 µM54; tat-CBD3, a peptide directly inhibiting CRMP2/CaV2.2 interaction, had an IC50 of 12.1 µM5; and a membrane-tethered version of the same peptide presented an IC50 of 2.8 µM.23 These IC50 values suggest that CNRP1 functions through the same signaling pathway as our previous CRMP2-targeting strategies. Indeed, CNRP1 did not change the biophysical properties of the N-type Ca2+ channel, which suggest that a trafficking mechanism is most likely responsible for the decreased peak current density observed in cells exposed to this peptide. Because t-CNRP1 acutely inhibits calcium influx, this sets it apart from gabapentin, a Cav2.2-targeted drug requiring >40 hours incubation to achieve channel inhibition.29,30
Here, we explored the mechanism of action of CNRP1 and found it to inhibit the CRMP2/Cav2.2 interaction and facilitate relocalization of the channel away from the membrane. The peptide also inhibited the CRMP2/neurofibromin interaction. In NF1, neurofibromin loss results in increased Cav2.2 currents16 and increased neurotransmitter release.31 Thus, the loss of the neurofibromin/CRMP2 interaction might contribute to this increase of Cav2.2 function, and the inhibition of this interaction by CNRP1 is therefore unlikely to explain the decreased Ca2+ currents. To explore further the mechanism of action of CNRP1, we screened a library of synaptic membrane-bound proteins embedded into nanodisc scaffolds.82 This strategy allowed us to explore PPIs in the absence of detergents that are classically used to solubilize the membrane proteins but may interfere with PPIs. Mass spectrometry analysis of the eluates from the CRMP2-interacting nanodiscs identified several proteins that bound to CRMP2. Not surprisingly, in these eluates we were unable to detect Cav2.2 or NaV1.7, 2 known CRMP2-binding proteins,6,18,19,38 likely because of their low expression level or because the large size of these channels (predicted to be 14–20 nm in diameter68,72,85) prevents their integration into the nanodiscs (13-nm maximum diameter48,82). Nevertheless, we identified syntaxin 1A as a novel CRMP2 protein partner. This protein is also known to bind to an intracellular region of the Cav2.2 channel, the so-called SYNaptic Protein INTeraction (“synprint”) region.2,73 The function of this interaction is to traffic the channel to the synaptic membrane53 and to promote synaptic vesicle docking,70 an event required before neurotransmitter release.86 We found CRMP2/syntaxin 1A interaction domain to be localized within the CNRP1 peptide. This suggested that the same region of CRMP2 can interact with either neurofibromin or syntaxin 1A. In NF1, the CRMP2/neurofibromin interaction is decreased (because of decreased expression of neurofibromin), and we found that deletion of neurofibromin increases CRMP2’s interaction with syntaxin 1A. Thus, we conclude that the augmented interaction between CRMP2 and syntaxin 1A is responsible for increased Cav2.2 currents and increased CGRP release in NF1. We propose that CNRP1 uncouples CRMP2’s interactions with Cav2.2 and syntaxin 1A, resulting in Cav2.2 relocalization away from the plasma membrane; decreased synaptic vesicle docking to the Ca2+ channel; and consequently, decreased CGRP release. This mechanism suggests that CRMP2’s interaction with syntaxin 1A might be a therapeutic target for pain management because this interaction directly controls neurotransmitter release. Our findings are in agreement with earlier studies which reported that interruption of the CaV2.2–syntaxin interaction with synprint peptides inhibits fast, synchronous transmitter release.53
Constellation pharmacology revealed an increased functional competence of the sensory neurons treated with CNRP1. A possible explanation for this could be that the loss of interaction between CRMP2 and syntaxin 1A may result in relocalization of syntaxin 1A to other protein partners, for instance TRPV1/TRPA150 or ATP receptors (eg, P2X, P2Y).1,34 Facilitating these interactions would promote the trafficking of these channels/receptors to the plasma membrane where they are active, thus explaining the increased number of neurons responding to the constellation pharmacology triggers. Of importance, CNRP1 inhibited depolarization-evoked Ca2+ influx in all cell populations except for menthol-sensitive neurons. Menthol has been reported to promote analgesia.42,64 Thus, CNRP1 curbs Ca2+ influx and neurotransmitter release from nociceptive neuron populations but not from neurons involved in antinociceptive signaling, explaining its efficacy in the pain models tested here.
The data reported here further solidify the importance of CRMP2 and its protein network in the etiology of inflammatory, postsurgical, and neuropathic pain. In addition to a direct coupling between Cav2.2 and CRMP2, the discovery of syntaxin 1A, a key protein in synaptic vesicle exocytosis, to CRMP2 suggests a more direct role for this PPI in nociceptive transmitter release. Uncoupling these interactions using the bifunctional peptide, CNRP1, may prove to be a useful antinociceptive strategy. Excitingly, as there is precedence for development of drugs against synaptic vesicle proteins with the development of levetiracetam targeting the synaptic vesicle protein SV2A,43 our findings support future efforts aimed at targeting the CRMP2– syntaxin 1A interaction for pain curbing drugs.
Supplementary Material
Acknowledgements
The authors thank Dr Stephen Sligar (University of Illinois at Urbana-Champaign, IL) for the MSP1E3D1 plasmid and Dr Samantha Perez-Miller for rendering the CRMP2 peptide structure in Pymol. This work was supported by a Neurofibromatosis New Investigator Award from the Department of Defense Congressionally Directed Military Medical Research and Development Program (NF1000099), National Institutes of Health awards (1R01NS098772 and 1R01DA042852), and a Children’s Tumor Foundation NF1 Synodos award (2015–04-009A) to R. Khanna. A. Moutal was supported by a Young Investigator’s Award from the Children’s Tumor Foundation. A. Dorame was supported by funds from the University of Arizona’s Border Latino and American Indian Summer Exposure to Research (BLAISER) program. S. S. Bellampalli and L. A. Chew were supported by funds to the University of Arizona’s Undergraduate Biology Research Program from the University of Arizona’s Senior Vice President for Research’s office and the Howard Hughes Medical Institute (L. A. Chew).
Footnotes
Conflict of interest statement
The authors have no conflict of interest to declare.
Appendix A. Supplemental digital content
Supplemental Digital Content associated with this article can be found online at http://links.lww.com/PAIN/A465, http://links.lww.com/PAIN/A466, http://links.lww.com/PAIN/A467, http://links.lww.com/PAIN/A469, http://links.lww.com/PAIN/A470.
References
- [1].Barden JA, Cottee LJ, Bennett MR. Vesicle-associated proteins and P2X receptor clusters at single sympathetic varicosities in mouse vas deferens. J Neurocytol 1999;28:469–80. [DOI] [PubMed] [Google Scholar]
- [2].Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 1992;257:255–9. [DOI] [PubMed] [Google Scholar]
- [3].Brennan TJ, Vandermeulen EP, Gebhart GF. Characterization of a rat model of incisional pain. PAIN 1996;64:493–501. [DOI] [PubMed] [Google Scholar]
- [4].Brittain JM, Chen L, Wilson SM, Brustovetsky T, Gao X, Ashpole NM, Molosh AI, You H, Hudmon A, Shekhar A, White FA, Zamponi GW, Brustovetsky N, Chen J, Khanna R. Neuroprotection against traumatic brain injury by a peptide derived from the collapsin response mediator protein 2 (CRMP2). J Biol Chem 2011;286:37778–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brittain JM, Duarte DB, Wilson SM, Zhu W, Ballard C, Johnson PL, Liu N, Xiong W, Ripsch MS, Wang Y, Fehrenbacher JC, Fitz SD, Khanna M, Park CK, Schmutzler BS, Cheon BM, Due MR, Brustovetsky T, Ashpole NM, Hudmon A, Meroueh SO, Hingtgen CM, Brustovetsky N, Ji RR, Hurley JH, Jin X, Shekhar A, Xu XM, Oxford GS, Vasko MR, White FA, Khanna R. Suppression of inflammatory and neuropathic pain by uncoupling CRMP-2 from the presynaptic Ca(2)(+) channel complex. Nat Med 2011;17:822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brittain JM, Piekarz AD, Wang Y, Kondo T, Cummins TR, Khanna R. An atypical role for collapsin response mediator protein 2 (CRMP-2) in neurotransmitter release via interaction with presynaptic voltage-gated calcium channels. J Biol Chem 2009;284:31375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Brittain JM, Wang Y, Eruvwetere O, Khanna R. Cdk5-mediated phosphorylation of CRMP-2 enhances its interaction with CaV2.2. FEBS Lett 2012;586:3813–18. [DOI] [PubMed] [Google Scholar]
- [8].Cao YQ. Voltage-gated calcium channels and pain. PAIN 2006;126:5–9. [DOI] [PubMed] [Google Scholar]
- [9].Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, Robertson M, Dunn D, Gesteland R, O’Connell P, White R. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell 1990;62:193–201. [DOI] [PubMed] [Google Scholar]
- [10].Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods 1994; 53:55–63. [DOI] [PubMed] [Google Scholar]
- [11].Chi XX, Schmutzler BS, Brittain JM, Hingtgen CM, Nicol GD, Khanna R. Regulation of N-type voltage-gated calcium (CaV2.2) channels and transmitter release by collapsin response mediator protein-2 (CRMP-2) in sensory neurons. J Cell Sci 2009;23:4351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Choe W, Messinger RB, Leach E, Eckle VS, Obradovic A, Salajegheh R, Jevtovic-Todorovic V, Todorovic SM. TTA-P2 is a potent and selective blocker of T-type calcium channels in rat sensory neurons and a novel antinociceptive agent. Mol Pharmacol 2011;80:900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Creange A, Zeller J, Rostaing-Rigattieri S, Brugieres P, Degos JD, Revuz J, Wolkenstein P. Neurological complications of neurofibromatosis type 1 in adulthood. Brain 1999;122:473–81. [DOI] [PubMed] [Google Scholar]
- [14].Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc 2004;126:3477–87. [DOI] [PubMed] [Google Scholar]
- [15].Drouet A, Wolkenstein P, Lefaucheur JP, Pinson S, Combemale P, Gherardi RK, Brugieres P, Salama J, Ehre P, Decq P, Creange A. Neurofibromatosis 1-associated neuropathies: a reappraisal. Brain 2004; 127:1993–2009. [DOI] [PubMed] [Google Scholar]
- [16].Duan JH, Hodgdon KE, Hingtgen CM, Nicol GD. N-type calcium current, Cav2.2, is enhanced in small-diameter sensory neurons isolated from Nf1+/− mice. Neuroscience 2014;270:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Duan JH, Wang Y, Duarte D, Vasko MR, Nicol GD, Hingtgen CM. Ras signaling pathways mediate NGF-induced enhancement of excitability of small-diameter capsaicin-sensitive sensory neurons from wildtype but not Nf1+/− mice. Neurosci Lett 2011;496:70–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dustrude ET, Moutal A, Yang X, Wang Y, Khanna M, Khanna R. Hierarchical CRMP2 posttranslational modifications control NaV1.7 function. Proc Natl Acad Sci U S A 2016;113:E8443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dustrude ET, Perez-Miller S, Francois-Moutal L, Moutal A, Khanna M, Khanna R. A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function. Channels (Austin) 2017. July 4;11:316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Feldman P, Khanna R. Challenging the catechism of therapeutics for chronic neuropathic pain: targeting CaV2.2 interactions with CRMP2 peptides. Neurosci Lett 2013;557:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Feng ZP, Hamid J, Doering C, Bosey GM, Snutch TP, Zamponi GW. Residue Gly1326 of the N-type calcium channel alpha 1B subunit controls reversibility of omega-conotoxin GVIA and MVIIA block. J Biol Chem 2001;276:15728–35. [DOI] [PubMed] [Google Scholar]
- [22].Field MJ, Holloman EF, McCleary S, Hughes J, Singh L. Evaluation of gabapentin and S-(+)-3-isobutylgaba in a rat model of postoperative pain. J Pharmacol Exp Ther 1997;282:1242–6. [PubMed] [Google Scholar]
- [23].Francois-Moutal L, Wang Y, Moutal A, Cottier KE, Melemedjian OK, Yang X, Wang Y, Ju W, Largent-Milnes TM, Khanna M, Vanderah TW, Khanna R. A membrane-delimited N-myristoylated CRMP2 peptide aptamer inhibits CaV2.2 trafficking and reverses inflammatory and postoperative pain behaviors. PAIN 2015;156:1247–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gosline SJ, Weinberg H, Knight P, Yu T, Guo X, Prasad N, Jones A, Shrestha S, Boone B, Levy SE, La Rosa S, Guinney J, Bakker A. A high-throughput molecular data resource for cutaneous neurofibromas. Sci Data 2017;4:170045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gutmann DH. Neurofibromin in the brain. J Child Neurol 2002;17: 592–601. [DOI] [PubMed] [Google Scholar]
- [26].Hargreaves K, Dubner R, Brown F, Flores C, Joris J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. PAIN 1988;32:77–88. [DOI] [PubMed] [Google Scholar]
- [27].Harkins AB, Cahill AL, Powers JF, Tischler AS, Fox AP. Deletion of the synaptic protein interaction site of the N-type (CaV2.2) calcium channel inhibits secretion in mouse pheochromocytoma cells. Proc Natl Acad Sci U S A 2004;101:15219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Harper AA, Lawson SN. Conduction velocity is related to morphological cell type in rat dorsal root ganglion neurones. J Physiol 1985;359:31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Heblich F, Tran Van Minh A, Hendrich J, Watschinger K, Dolphin AC. Time course and specificity of the pharmacological disruption of the trafficking of voltage-gated calcium channels by gabapentin. Channels (Austin) 2008;2:4–9. [DOI] [PubMed] [Google Scholar]
- [30].Hendrich J, Van Minh AT, Heblich F, Nieto-Rostro M, Watschinger K, Striessnig J, Wratten J, Davies A, Dolphin AC. Pharmacological disruption of calcium channel trafficking by the alpha2delta ligand gabapentin. Proc Natl Acad Sci U S A 2008;105:3628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hingtgen CM, Roy SL, Clapp DW. Stimulus-evoked release of neuropeptides is enhanced in sensory neurons from mice with a heterozygous mutation of the Nf1 gene. Neuroscience 2006;137: 637–45. [DOI] [PubMed] [Google Scholar]
- [32].Hodgdon KE, Hingtgen CM, Nicol GD. Dorsal root ganglia isolated from Nf1+/− mice exhibit increased levels of mRNA expression of voltage-dependent sodium channels. Neuroscience 2012;206:237–44. [DOI] [PubMed] [Google Scholar]
- [33].Hsueh YP, Roberts AM, Volta M, Sheng M, Roberts RG. Bipartite interaction between neurofibromatosis type I protein (neurofibromin) and syndecan transmembrane heparan sulfate proteoglycans. J Neurosci 2001;21:3764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hussl S, Kubista H, Boehm S. Autoregulation in PC12 cells via P2Y receptors: evidence for non-exocytotic nucleotide release from neuroendocrine cells. Purinergic Signal 2007;3:367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ibrahim MM, Patwardhan A, Gilbraith KB, Moutal A, Yang X, Chew LA, Largent-Milnes T, Malan TP, Vanderah TW, Porreca F, Khanna R. Long-lasting antinociceptive effects of green light in acute and chronic pain in rats. PAIN 2017;158:347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ju W, Li Q, Allette YM, Ripsch MS, White FA, Khanna R. Suppression of pain-related behavior in two distinct rodent models of peripheral neuropathy by a homopolyarginine-conjugated CRMP2 peptide. J Neurochem 2012;124:869–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Khanna R, Li Q, Bewersdorf J, Stanley EF. The presynaptic CaV2.2 channel-transmitter release site core complex. Eur J Neurosci 2007;26: 547–59. [DOI] [PubMed] [Google Scholar]
- [38].Khanna R, Zougman A, Stanley EF. A proteomic screen for presynaptic terminal N-type calcium channel (CaV2.2) binding partners. J Biochem Mol Biol 2007;40:302–14. [DOI] [PubMed] [Google Scholar]
- [39].Kim C, Jun K, Lee T, Kim SS, McEnery MW, Chin H, Kim HL, Park JM, Kim DK, Jung SJ, Kim J, Shin HS. Altered nociceptive response in mice deficient in the alpha(1B) subunit of the voltage-dependent calcium channel. Mol Cell Neurosci 2001;18:235–45. [DOI] [PubMed] [Google Scholar]
- [40].Kress M, Izydorczyk I, Kuhn A. N- and L- but not P/Q-type calcium channels contribute to neuropeptide release from rat skin in vitro. Neuroreport 2001;12:867–70. [DOI] [PubMed] [Google Scholar]
- [41].Lin YL, Hsueh YP. Neurofibromin interacts with CRMP-2 and CRMP-4 in rat brain. Biochem biophysical Res Commun 2008;369:747–52. [DOI] [PubMed] [Google Scholar]
- [42].Liu B, Fan L, Balakrishna S, Sui A, Morris JB, Jordt SE. TRPM8 is the principal mediator of menthol-induced analgesia of acute and inflammatory pain. PAIN 2013;154:2169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lynch BA, Lambeng N, Nocka K, Kensel-Hammes P, Bajjalieh SM, Matagne A, Fuks B. The synaptic vesicle protein SV2A is the binding site for the antiepileptic drug levetiracetam. Proc Natl Acad Sci U S A 2004; 101:9861–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Maggi CA, Tramontana M, Cecconi R, Santicioli P. Neurochemical evidence for the involvement of N-type calcium channels in transmitter secretion from peripheral endings of sensory nerves in Guinea pigs. Neurosci Lett 1990;114:203–6. [DOI] [PubMed] [Google Scholar]
- [45].Manji H Neuropathy in HIV infection. Curr Opin Neurol 2000;13:589–92. [DOI] [PubMed] [Google Scholar]
- [46].Marquez de PB, Hammond DL. Sex dependent enhancement of pain responses in a mouse model of neurofibromatosis. Proceedings of the Society for Neuroscience, 2011.
- [47].Martin S, Gillespie A, Wolters PL, Widemann BC. Experiences of families with a child, adolescent, or young adult with neurofibromatosis type 1 and plexiform neurofibroma evaluated for clinical trials participation at the National Cancer Institute. Contemp Clin Trials 2011;32:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Marty MT, Wilcox KC, Klein WL, Sligar SG. Nanodisc-solubilized membrane protein library reflects the membrane proteome. Anal Bioanal Chem 2013;405:4009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].McGivern JG, McDonough SI. Voltage-gated calcium channels as targets for the treatment of chronic pain. Curr Drug Targets CNS Neurol Disord 2004;3:457–78. [DOI] [PubMed] [Google Scholar]
- [50].Meng J, Wang J, Steinhoff M, Dolly JO. TNFalpha induces co-trafficking of TRPV1/TRPA1 in VAMP1-containing vesicles to the plasmalemma via Munc18–1/syntaxin1/SNAP-25 mediated fusion. Sci Rep 2016;6: 21226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Milligan ED, O’Connor KA, Nguyen KT, Armstrong CB, Twining C, Gaykema RP, Holguin A, Martin D, Maier SF, Watkins LR. Intrathecal HIV-1 envelope glycoprotein gp120 induces enhanced pain states mediated by spinal cord proinflammatory cytokines. J Neurosci 2001;21:2808–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mintz IM, Venema VJ, Swiderek KM, Lee TD, Bean BP, Adams ME. P-type calcium channels blocked by the spider toxin omega-Aga-IVA. Nature 1992;355:827–9. [DOI] [PubMed] [Google Scholar]
- [53].Mochida S, Sheng ZH, Baker C, Kobayashi H, Catterall WA. Inhibition of neurotransmission by peptides containing the synaptic protein interaction site of N-type Ca2+ channels. Neuron 1996;17:781–8. [DOI] [PubMed] [Google Scholar]
- [54].Moutal A, Chew LA, Yang X, Wang Y, Yeon SK, Telemi E, Meroueh S, Park KD, Shrinivasan R, Gilbraith KB, Qu C, Xie JY, Patwardhan A, Vanderah TW, Khanna M, Porreca F, Khanna R. (S)-lacosamide inhibition of CRMP2 phosphorylation reduces postoperative and neuropathic pain behaviors through distinct classes of sensory neurons identified by constellation pharmacology. PAIN 2016;157:1448–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Moutal A, Eyde N, Telemi E, Park KD, Xie JY, Dodick DW, Porreca F, Khanna R. Efficacy of (S)-Lacosamide in preclinical models of cephalic pain. PAIN Rep 2016:1:e565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Moutal A, Francois-Moutal L, Perez-Miller S, Cottier K, Chew LA, Yeon SK, Dai J, Park KD, Khanna M, Khanna R. (S)-Lacosamide binding to collapsin response mediator protein 2 (CRMP2) regulates CaV2.2 activity by subverting its phosphorylation by Cdk5. Mol Neurobiol 2016;53:1959–76. [DOI] [PubMed] [Google Scholar]
- [57].Moutal A, Li W, Wang Y, Ju W, Luo S, Cai S, Francois-Moutal L, Perez-Miller S, Hu J, Dustrude ET, Vanderah TW, Gokhale V, Khanna M, Khanna R. Homology-guided mutational analysis reveals the functional requirements for antinociceptive specificity of collapsin response mediator protein 2-derived peptides. Br J Pharmacol 2017. doi: 10.1111/bph.13737. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- [58].Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003;75: 4646–58. [DOI] [PubMed] [Google Scholar]
- [59].Newcomb R, Szoke B, Palma A, Wang G, Chen X, Hopkins W, Cong R, Miller J, Urge L, Tarczy-Hornoch K, Loo JA, Dooley DJ, Nadasdi L, Tsien RW, Lemos J, Miljanich G. Selective peptide antagonist of the class E calcium channel from the venom of the tarantula Hysterocrates gigas. Biochemistry 1998;37:15353–62. [DOI] [PubMed] [Google Scholar]
- [60].Newshan G HIV neuropathy treated with gabapentin. AIDS 1998;12: 219–21. [PubMed] [Google Scholar]
- [61].O’Brien DE, Brenner DS, Gutmann DH. Gereau RWt. Assessment of pain and itch behavior in a mouse model of neurofibromatosis type 1. J Pain 2013;14:628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Pacchioni AM, Vallone J, Worley PF, Kalivas PW. Neuronal pentraxins modulate cocaine-induced neuroadaptations. J Pharmacol Exp Ther 2009;328:183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Park KD, Yang XF, Lee H, Dustrude ET, Wang Y, Khanna R, Kohn H. Discovery of lacosamide affinity bait agents that exhibit potent voltage-gated sodium channel blocking properties. ACS Chem Neurosci 2013;4: 463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Patel R, Goncalves L, Leveridge M, Mack SR, Hendrick A, Brice NL, Dickenson AH. Anti-hyperalgesic effects of a novel TRPM8 agonist in neuropathic rats: a comparison with topical menthol. PAIN 2014;155: 2097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Patrakitkomjorn S, Kobayashi D, Morikawa T, Wilson MM, Tsubota N, Irie A, Ozawa T, Aoki M, Arimura N, Kaibuchi K, Saya H, Araki N. Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. J Biol Chem 2008;283:9399–413. [DOI] [PubMed] [Google Scholar]
- [66].Piekarz AD, Due MR, Khanna M, Wang B, Ripsch MS, Wang R, Meroueh SO, Vasko MR, White FA, Khanna R. CRMP-2 peptide mediated decrease of high and low voltage-activated calcium channels, attenuation of nociceptor excitability, and anti-nociception in a model of AIDS therapy-induced painful peripheral neuropathy. Mol Pain 2012;8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 2009;464:211–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sato C, Sato M, Iwasaki A, Doi T, Engel A. The sodium channel has four domains surrounding a central pore. J Struct Biol 1998;121:314–25. [DOI] [PubMed] [Google Scholar]
- [69].Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science 1999;285:1569–72. [DOI] [PubMed] [Google Scholar]
- [70].Seagar M, Takahashi M. Interactions between presynaptic calcium channels and proteins implicated in synaptic vesicle trafficking and exocytosis. J Bioenerg Biomembr 1998;30:347–56. [DOI] [PubMed] [Google Scholar]
- [71].Sheets PL, Heers C, Stoehr T, Cummins TR. Differential block of sensory neuronal voltage-gated sodium channels by lacosamide [(2R)-2-(acetylamino)-N-benzyl-3-methoxypropanamide], lidocaine, and carbamazepine. J Pharmacol Exp Ther 2008;326:89–99. [DOI] [PubMed] [Google Scholar]
- [72].Shen H, Zhou Q, Pan X, Li Z, Wu J, Yan N. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 2017; 355. [DOI] [PubMed] [Google Scholar]
- [73].Sheng ZH, Rettig J, Takahashi M, Catterall WA. Identification of a syntaxin-binding site on N-type calcium channels. Neuron 1994;13: 1303–13. [DOI] [PubMed] [Google Scholar]
- [74].Teichert RW, Memon T, Aman JW, Olivera BM. Using constellation pharmacology to define comprehensively a somatosensory neuronal subclass. Proc Natl Acad Sci U S A 2014;111:2319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Teichert RW, Schmidt EW, Olivera BM. Constellation pharmacology: a new paradigm for drug discovery. Annu Rev Pharmacol Toxicol 2015; 55:573–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang Y, Brittain JM, Jarecki BW, Park KD, Wilson SM, Wang B, Hale R, Meroueh SO, Cummins TR, Khanna R. In silico docking and electrophysiological characterization of lacosamide binding sites on collapsin response mediator protein-2 identifies a pocket important in modulating sodium channel slow inactivation. J Biol Chem 2010;285: 25296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wang Y, Brittain JM, Wilson SM, Hingtgen CM, Khanna R. Altered calcium currents and axonal growth in Nf1 haploinsufficient mice. Transl Neurosci 2010;1:106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wang Y, Duan JH, Hingtgen CM, Nicol GD. Augmented sodium currents contribute to the enhanced excitability of small diameter capsaicin-sensitive sensory neurons isolated from Nf1+/(−) mice. J Neurophysiol 2010;103:2085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang Y, Nicol GD, Clapp DW, Hingtgen CM. Sensory neurons from Nf1 haploinsufficient mice exhibit increased excitability. J Neurophysiol 2005; 94:3670–6. [DOI] [PubMed] [Google Scholar]
- [80].Wheeler DG, Groth RD, Ma H, Barrett CF, Owen SF, Safa P, Tsien RW. Ca(V)1 and Ca(V)2 channels engage distinct modes of Ca(2+) signaling to control CREB-dependent gene expression. Cell 2012; 149:1112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].White S, Marquez de Prado B, Russo AF, Hammond DL. Heat hyperalgesia and mechanical hypersensitivity induced by calcitonin gene-related peptide in a mouse model of neurofibromatosis. PLoS One 2014;9:e106767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wilcox KC, Marunde MR, Das A, Velasco PT, Kuhns BD, Marty MT, Jiang H, Luan CH, Sligar SG, Klein WL. Nanoscale synaptic membrane mimetic allows unbiased high throughput screen that targets binding sites for Alzheimer’s-associated abeta oligomers. PloS One 2015;10: e0125263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wilson SM, Schmutzler BS, Brittain JM, Dustrude ET, Ripsch MS, Pellman JJ, Yeum TS, Hurley JH, Hingtgen CM, White FA, Khanna R. Inhibition of transmitter release and attenuation of AIDS therapy-induced and tibial nerve injury-related painful peripheral neuropathy by novel synthetic Ca2+ channel peptides. J Biol Chem 2012;287: 35065–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wolkenstein P, Zeller J, Revuz J, Ecosse E, Leplege A. Quality-of-life impairment in neurofibromatosis type 1: a cross-sectional study of 128 cases. Arch Dermatol 2001;137:1421–5. [DOI] [PubMed] [Google Scholar]
- [85].Wu J, Yan Z, Li Z, Qian X, Lu S, Dong M, Zhou Q, Yan N. Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 A resolution. Nature 2016; 537:191–6. [DOI] [PubMed] [Google Scholar]
- [86].Wu MN, Fergestad T, Lloyd TE, He Y, Broadie K, Bellen HJ. Syntaxin 1A interacts with multiple exocytic proteins to regulate neurotransmitter release in vivo. Neuron 1999;23:593–605. [DOI] [PubMed] [Google Scholar]
- [87].Yaksh TL, Rudy TA. Chronic catheterization of the spinal subarachnoid space. Physiol Behav 1976;17:1031–6. [DOI] [PubMed] [Google Scholar]
- [88].Yu LC, Hou JF, Fu FH, Zhang YX. Roles of calcitonin gene-related peptide and its receptors in pain-related behavioral responses in the central nervous system. Neurosci Biobehav Rev 2009;33:1185–91. [DOI] [PubMed] [Google Scholar]
- [89].Yuan SB, Shi Y, Chen J, Zhou X, Li G, Gelman BB, Lisinicchia JG, Carlton SM, Ferguson MR, Tan A, Sarna SK, Tang SJ. Gp120 in the pathogenesis of human immunodeficiency virus-associated pain. Ann Neurol 2014;75: 837–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.