Abstract
Chromosomal rearrangements involving receptor tyrosine kinases (RTK) are a clinically relevant oncogenic mechanism in human cancers. These chimeric oncoproteins often contain the C-terminal kinase domain of the RTK joined in cis to various N-terminal, non-kinase fusion partners. The functional role of the N-terminal fusion partner in RTK fusion oncoproteins is poorly understood. Here we show that distinct N-terminal fusion partners drive differential subcellular localization, which imparts distinct cell signaling and oncogenic properties of different, clinically-relevant ROS1 RTK fusion oncoproteins. SDC4-ROS1 and SLC34A2-ROS1 fusion oncoproteins resided on endosomes and activated the MAPK pathway. CD74-ROS1 variants that localized instead to the endoplasmic reticulum (ER) showed compromised activation of MAPK. Forced re-localization of CD74-ROS1 from the ER to endosomes restored MAPK signaling. ROS1 fusion oncoproteins that better activate MAPK formed more aggressive tumors. Thus, differential subcellular localization controlled by the N-terminal fusion partner regulates the oncogenic mechanisms and output of certain RTK fusion oncoproteins.
Keywords: RTK, oncogene, ROS1, subcellular localization, MAPK
Precis:
ROS1 fusion oncoproteins exhibit differential activation of MAPK signaling according to subcellular localization, with ROS1 fusions localized to endosomes the strongest activators of MAPK signaling.
Introduction:
The aberrant hyper-activation of RTKs drives the growth of many cancers. This malignant RTK activation arises through a variety of mechanisms, including somatic genetic alterations such as missense mutations, small insertions and deletions, copy number alterations, and gene rearrangements (1–9). The latter class of genetic alterations comprise a clinically important group of cancer driver genes, prominent examples of which are fusions of the anaplastic lymphoma kinase (ALK) and ROS proto-oncogene 1 (ROS1), among other kinases, with various fusion partners (10). These gene rearrangements often lead to the generation of a chimeric protein with the non-kinase partner fused N-terminal to the RTK kinase domain (e.g. EML4-ALK, CD74-ROS1, SDC4-ROS1). While targeted kinase inhibitors (TKIs) are effective in many patients harboring cancers driven by these kinase fusions (e.g. crizotinib targeting ALK and ROS1 fusion oncoproteins), drug resistance remains a challenge that limits long-term patient survival (1,11–18). A better understanding of the mechanisms controlling the oncogenic signaling properties of these kinase fusion proteins is essential to identify complementary or alternative molecular strategies to enhance clinical outcomes.
The mechanisms by which a non-native N-terminal protein fused to the kinase domain of an RTK such as ROS1 cause kinase hyper-activation and cancer growth are partially understood. These mechanisms include overexpression of the C-terminal kinase as a result of the activity of the promoter of the N-terminal gene partner, constitutive oligomerization of the fusion kinase proteins, and release of kinase auto-inhibitory mechanisms (19). A relatively poorly-understood aspect of the regulation of oncoprotein kinase fusions is the extent to which the subcellular membrane localization of a particular fusion oncoprotein may contribute to its oncogenic properties. This is a particularly relevant unanswered cell biological question, as many oncoprotein fusion kinases present in human cancers, such as ALK and ROS1 variants, often gain the subcellular localization signals of the N-terminal partner in the context of the native RTK kinase domain (e.g. EML4 in EML4-ALK, CD74 in CD74-ROS1); it is plausible that abnormal subcellular localization (versus the native full-length RTK) could be an important feature of their oncogenic properties. While evidence is emerging that in certain cases the N-terminal fusion partner can cause aberrant subcellular localization of the fusion RTK compared to the native RTK (e.g. EML4-ALK), whether differential subcellular localization is a more general feature regulating oncogenesis across different oncoprotein fusions remains unclear (20).
Fusions involving the RTK ROS1 are found in 1–2% of lung adenocarcinomas, as well as in other tumor types (8,10). ROS1 is one of the last remaining orphan receptor tyrosine kinases, and little is known about the wildtype function of the protein. Wildtype ROS1 contains a substantial N-terminal extracellular domain, whose structure suggests extracellular matrix proteins may serve as ligands (21). In cancer-driving ROS1 gene fusions this extracellular domain is not included, leaving the transmembrane and entire kinase domain of ROS1 fused to a variety of N-terminal fusion partners (10,22). To date, 10 distinct N-terminal fusion partners for ROS1 kinase fusions have been identified in cancers (Figure S1A) (23). The most common ROS1 fusion partner is CD74 (found in ~50% of ROS1 fusions) (6). Other commonly observed ROS1 fusion partners include SDC4, SLC34A2, LRIG3, EZR, and TPM3 (22,24,25). These N-terminal partners lack clearly unifying protein domains or functions, raising the possibility that these fusion oncoproteins promote oncogenic signaling and cancer growth through non-identical mechanisms. Whether the N-terminal partner in ROS1 oncoprotein fusions regulates the subcellular localization and oncogenic properties of each kinase fusion or response to TKI treatment is not well-understood.
We tested the hypothesis that different ROS1 oncoprotein fusions engage distinct downstream signaling pathways and exhibit different oncogenic properties, and investigated the mechanistic role of specific N-terminal fusion partners in regulating such differential phenotypes through differential subcellular localization.
Materials and Methods:
Cell Culture.
All cell lines were maintained in a humidified incubator at 37°C, 5% CO2. The patient-derived ROS1-positive lung adenocarcinoma lines HCC78, CUTO-2, CUTO-23, and CUTO-33, and the normal lung epithelial line BEAS2-B were all maintained in RPMI-1640 supplemented with 10% FBS and 100ug/mL of penicillin/streptomycin. HEK-293T cells and NIH-3T3 cells were maintained in DMEM-High Glucose supplemented with 10% FBS and 100ug/mL of penicillin/streptomycin. HCC78 cells were a kind gift from John D. Minna (University of Texas Southwestern, Dallas, TX, USA). BEAS2-B and NIH-3T3 cells were purchased from ATCC (Manassas, VA, USA). CUTO-2, CUTO-23, and CUTO-33 cells were a generous gift from Dr. Robert Doebele (University of Colorado, Denver, CO, USA). All cell lines were tested for mycoplasma every 3 months using MycoAlert Mycoplasma Detection Kit (Lonza). All cells used were < 20 passages from thaw. Cell line authentication was performed for HCC78 using standard STR testing.
Compounds.
Crizotinib (Selleck Chemicals, Houston, TX, USA) and the SHP2 inhibitor RMC-4550 (Revolution Medicines, Redwood City, CA, USA) were dissolved in DMSO.
Antibodies.
The following Cell Signaling Technology (Danvers, MA, USA) antibodies were used: phospho-ROS1 (Y2274, #3078), ROS1 (#3287), phospho-ALK (Y1604, #3341), ALK (#3633), phospho-STAT3 (Y705, #9145), STAT3 (#9139), phospho-AKT (S473, #5012), AKT (#2920), phospho-ERK (Y202/204, #4370), ERK (#4694), phospho-MEK1/2 (Ser 217/221, #9121), MEK1 (#2352), Anti-rabbit IgG, HRP-linked Antibody (#7074), Anti-mouse IgG, HRP-linked Antibody (#7076). The following Sigma-Aldrich (St Louis, MO, USA) antibodies were used: Beta-Actin (#A2228). The following Santa Cruz Biotechnology (Santa Cruz, CA, USA) antibodies were used: EEA1 (sc-6415). The following Abcam (Cambridge, UK) antibodies were used: Calnexin-Alexa Fluor 488 (ab202574), PTP1B (ab201974). The following Life Technologies Thermo Fisher Scientific (Waltham, MA, USA) antibodies were used: Alexa Fluor 488 Donkey Anti-Mouse (#21202), Alexa Fluor 499 Donkey Anti-Goat (#11055), Alexa Fluor 594 Donkey Anti-Rabbit (#21207).
DNA transfections.
293T cells were transiently transfected using TransIt-LT1 transfection reagent (Mirus Bio LLC, Madison, WI, USA).
Immunoblotting.
For immunoblotting, cells were washed with ice-cold PBS and scraped in ice-cold RIPA buffer [25 mM Tris⋅HCl (pH 7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, supplemented with 1× HALT protease inhibitor cocktail and 1× HALT phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA)]. Lysates were clarified with sonication and centrifiugation. Lysates were subject to SDS/PAGE followed by blotting with the indicated antibodies. Signal was detected using Amersham ECL Prime reagent (GE Healthcare Life Sciences, Chicago, IL, USA) and chemiluminescnce on an ImageQuant LAS 4000 (GE Healthcare Life Science, Chicago, IL, USA). 293T cells were serum starved (0%S) for 5 hours and ROS1 BEAS2-B cells were serum starved (0%S) for 24 hours prior to lysate collection.
siRNA knockdown.
Cells were seeded in 6-well plates. The following day, siRNAs were resuspended to a final concentration of 25nM in serum-free medium with DharmaFECT transfection reagent (Thermo Fisher Scientific), then pipetted onto cells. Lysates were harvested 55 hours later. The following ROS1 siRNAs from Sigma-Aldrich were used: Hs01_00183685 (siROS1 #1) and Hs01_00183690 (siROS1 #2). Non-targeting control siRNA was purchased from Dharmacon (GE Life Sciences).
Constructs.
Lentiviral expression constructs for SDC4-ROS1 and CD74-ROS1 were generous gifts from Dr. Christine Lovly (Vanderbilt University, Nashville, TN, USA). Retroviral expression construct for SLC34A2-ROS1 was a generous gift from Dr. Monika Davare (OHSU, Portland, OR, USA). The retroviral expression constructs for MEK-DD (#15268) and CA-STAT3 (#24983) were purchased on Addgene.
Viral transduction.
293T viral packaging cells were plated in 10cm dishes the day prior to transfection. They were transfected with lentiviral or retroviral expression constructs and the appropriate packaging plasmids using TransIt-LT1 transfection reagent (Mirus Bio LLC, Madison, WI, USA). Viral supernatants were collected 48–72 hours post-transfection and used to transduce cell lines in the presence of 1× Polybrene for 24 hours. 72 hours post-infection, media was changed to standard growth media plus the appropriate selectable marker (1ug/mL puromycin for all lines except NIH-3T3, which were selected with 2ug/mL puromycin). CA-STAT3-infected cells were sorted on a BD FACSAria II (BD Biosciences, San Jose, CA) for GFP-positivity.
Crystal Violet Assays.
Cells were seeded in 12-well plates at 10% confluency and treated with drug the following day. They were grown for 6–8 days, then fixed with 4% paraformaldehyde and stained with crystal violet. Pictures of stained cells were taking using transillumination on an ImageQuant LAS 4000 (GE Healthcare, Chicago, IL, USA). Crystal violet was dissolved in 500ul 1% SDS and quantified based on 470nM absorbance using a SpectraMax spectrophotomer (Molecular Devices, Sunnyvale, CA, USA). Relative cell viability was determined by normalizing to DMSO-treated control. All crystal violet images are representative and quantification values arise from ≥ n = 3 experiments. Statistical significance was determined by multiple t-test analysis using Prism 6 (Graphpad Software, La Jolla, CA, USA).
Immunofluorescence.
Cells were seeded in 4-well Lab Tek II Chamber Slides (Thermo Fisher Scientific). The following day, cells were fixed for 15 minutes with 4% paraformaldehyde, washed, and incubated in blocking buffer for 1 hour (1× PBS with 1% BSA and 0.3% Triton X-100). Blocking buffer was aspirated and cells were incubated with primary antibody overnight in the dark at 4°C. The following day, cells were washed, incubated with fluorophore-conjugated secondary antibody for 1 hour at room temperature in the dark, washed, then mounted using ProLong Gold Antifade reagent with DAPI (Cell Signaling Technology, Danvers, MA). Slides were analyzed using a Nikon Ti microscope with a CSU-W1 spinning disk confocal using a 100×/1.4 Plan Apo VC objective (Nikon Imaging Center, UCSF). Images were acquired on MicroManager software and analyzed using Fiji software(26,27).
Tumor Xenograft Studies.
Xenografts were generated by injecting NIH-3T3 cells in matrigel in flanks of 8-week old NOD/SCID mice. All animal studies were conducted under the UCSF-approved IACUC protocol, AN107889.
Supplemental material:
This manuscript contains supplemental figures and legends that support the primary figures.
Results:
ROS1 fusion oncoproteins differentially activate the RAS/MAPK pathway
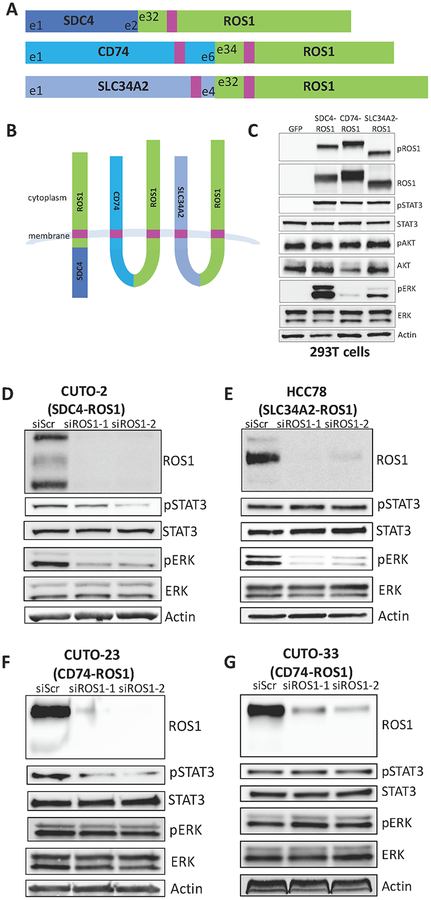
To investigate the potential differential functional properties of differential ROS1 oncoprotein fusions, first we engineered a genetically-controlled isogenic system to express some of the most common forms of ROS1 fusion oncoproteins that are present in patient tumors, including CD74-ROS1, SDC4-ROS1, and SLC34A2-ROS1 (Figure 1A) (23). An in silico topological analysis using an established computational method suggested that each of these fusions were predicted to result in a membrane-anchored cytoplasmic-facing kinase domain (Figure 1B) (28,29). All three ROS1 fusions showed constitutive activation of the kinase in cells, as measured by ROS1 phosphorylation (Figure 1C). While each ROS1 fusion that we tested activated JAK/STAT and IRS1 signaling (measured by STAT3 and IRS1 phosphorylation) to an equivalent degree, the ability of ROS1 fusions to activate the RAS/MAPK pathway (measured by ERK phosphorylation) varied significantly across the different ROS1 fusion proteins tested (Figure 1C). Both SDC4-ROS1 and SLC34A2-ROS1 fusions activated the MAPK pathway. In contrast, the CD74-ROS1 fusion variant studied failed to as substantially induce RAS/MAPK pathway signaling (Figure 1C, S2). To confirm whether this differential activation of the MAPK pathway by different ROS1 fusions was recapitulated in patient-derived NSCLC models, we conducted short-term siRNA-mediated knockdown of ROS1 in ROS1 fusion-positive patient-derived NSCLC cell lines that express the same fusions studied in our isogenic system. We observed that knockdown of SDC4-ROS1 and SLC34A2-ROS1 fusion proteins resulted in suppression of the MAPK pathway, whereas silencing CD74-ROS1 expression did not (Figure 1D–G). These findings corroborate our observations in the isogenic system.
Figure 1. ROS1 fusion partners dictate differential activation of downstream signaling pathways.
(A) Diagram of the commonly occurring ROS1 fusion oncoproteins, which were studied here. Pink denotes a transmembrane domain. (B) Topological configuration of ROS1 fusions based on CCTOP computational analysis (28). (C) Immunoblot analysis of 293T cells transiently transfected for 48h with GFP, SDC4-ROS1, CD74-ROS1, or SLC34A2-ROS1, with 5h serum starvation. The pROS1 antibody used recognizes Y2274 of the full-length ROS1 protein. (D-G) Immunoblot analysis of patient-derived cell lines expressing (D) SDC4-ROS1, (E) SLC34A2-ROS1, or (F-G) CD74-ROS1 with siRNA-mediated knockdown of ROS1 (55h after transfection). Data shown in (C-G) are representative of ≥ 3 independent experiments.
RAS/MAPK pathway signaling is necessary and sufficient for survival of cells expressing ROS1 fusion oncoproteins that specifically activate RAS/MAPK
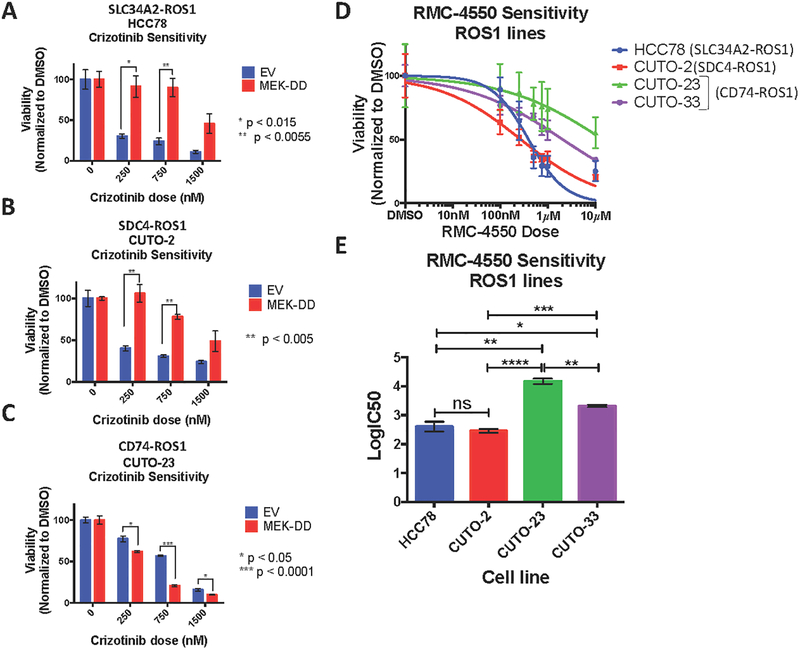
Based on these findings, we hypothesized that the MAPK pathway may play a more prominent role in controlling cell survival downstream of ROS1 fusion oncoproteins that we found can better engage this pathway, compared to those that are less capable. Indeed, we found that hyper-activation of the MAPK pathway by expression of a constitutively-active mutant form of MEK (MEK-DD) was sufficient to rescue cells expressing SDC4-ROS1 and SLC34A2-ROS1 fusions (which activate MAPK), but not the CD74-ROS1 fusion variant (that does not substantially activate MAPK), from ROS1 inhibitor (crizotinib) sensitivity (Figure 2A–C, S3). In contrast, hyper-activation of JAK/STAT signaling by expression of a constitutively-active mutant form of STAT3 (CA-STAT3) was unable to rescue cells from crizotinib sensitivity across all ROS1 fusion oncoproteins tested, suggesting a less prominent role for JAK/STAT signaling in regulating cell survival during ROS1 inhibitor treatment in these systems (Figure S4). The collective data suggest that IRS1 or STAT3 signaling could contribute to ROS1-fusion driven NSCLC oncogenesis, while STAT3 activation appears to be less potent at driving ROS1 inhibitor resistance in models of tumor maintenance (i.e. models established from NSCLC patients).
Figure 2. MAPK pathway signaling is necessary and sufficient for survival of SDC4-ROS1-positive and SLC34A2-ROS1-positive lines, but not a CD74-ROS1 positive line.
(A-C) Crystal violet quantification of ROS1 fusion-positive patient-derived cell lines (A) HCC78, (B) CUTO-2 and (C) CUTO-23, expressing empty vector or constitutively active MEK-DD, treated with DMSO or a dose-response of the ROS1 inhibitor crizotinib for 6 days. (D) Crystal violet quantification of HCC78 (SLC34A2-ROS1), CUTO-2 (SDC4-ROS1), CUTO-23 (CD74-ROS1), and CUTO-33 (CD74-ROS1) cell lines treated with DMSO or a dose-response of the SHP2 inhibitor RMC-4550 for 6 days. (E) Half-maximal inhibitory concentration (IC50) determination for the SHP2 inhibitor RMC-4550 in the indicated ROS1 patient-derived cell lines based on crystal violet quantification of the experiment in (D). Data represent three independent experiments. Data represented as mean +/− s.e.m.
An emerging mechanism linking oncogenic RTK activation to downstream RAS/MAPK signaling involves the protein phosphatase SHP2 (PTPN11), which is critical for augmenting RAS-GTP levels and RAF-MEK-ERK (MAPK pathway) activation (30). Interestingly, previous work identified SHP2 as a key factor in signaling mediated by the FIG-ROS1 fusion protein in glioblastoma, albeit not via the MAPK pathway in that context (31). Based on these collective findings, we hypothesized that SHP2 promotes MAPK pathway activation downstream of certain NSCLC ROS1 fusion oncoproteins. Indeed, SHP2 inhibition with an established allosteric SHP2 inhibitor, RMC-4550 (32) was highly effective in patient-derived NSCLC cell lines in which the MAPK pathway operated downstream of the ROS1 fusion (HCC78, CUTO-2), but was less effective in cells in which the MAPK pathway was disconnected from the ROS1 fusion oncoprotein (CUTO-23, CUTO-33) (Figure 2D–E, S5). Interestingly, the SHP2 inhibitor was still capable of suppressing some downstream MAPK signaling in cells driven by CD74-ROS1 (CUTO-23, CUTO-33), suggesting that MAPK signaling is still active in these cells, just not under the control of CD74-ROS1, and thus is not required for cell survival. Additionally, mutation of two of the major tyrosine phosphorylation sites within SDC4-ROS1 (Y591 and Y651), which are previously characterized SH2 domain binding sites, was sufficient to abrogate binding to SHP-2, suggesting a direct interaction between SDC4-ROS1 and SHP-2 (Figure S6). Our collective data show that in cells harboring SDC4-ROS1 and SLC34A2-ROS1 fusions, but not in those with the CD74-ROS1 fusion, MAPK pathway activation is necessary and sufficient for cell survival.
What mechanism could underlie this differential signaling pathway activation operating downstream of the different ROS1 fusion oncoproteins that we investigated? One possible mechanism is that the different exonic breakpoints that are present in the different fusion genes (e.g. ROS1 exon 32 versus exon 34 fused to an N-terminal partner) could contribute to differential pathway engagement. However, we found that the differential pathway activation observed downstream of a particular ROS1 fusion was similar whether the exonic breakpoint was in ROS1 exon 32 or 34 (Figure S7). Furthermore, we noted that the entire ROS1 kinase domain was retained and identical between the different fusion forms based on DNA sequence analysis. We further found no significant differences in protein expression levels across the different fusion oncoproteins that could readily explain differential pathway engagement (Figure 1C). Together, these data suggested a potential role for the N-terminal fusion partner in driving differential MAPK pathway activation.
ROS1 fusion oncoproteins display differential subcellular localizations
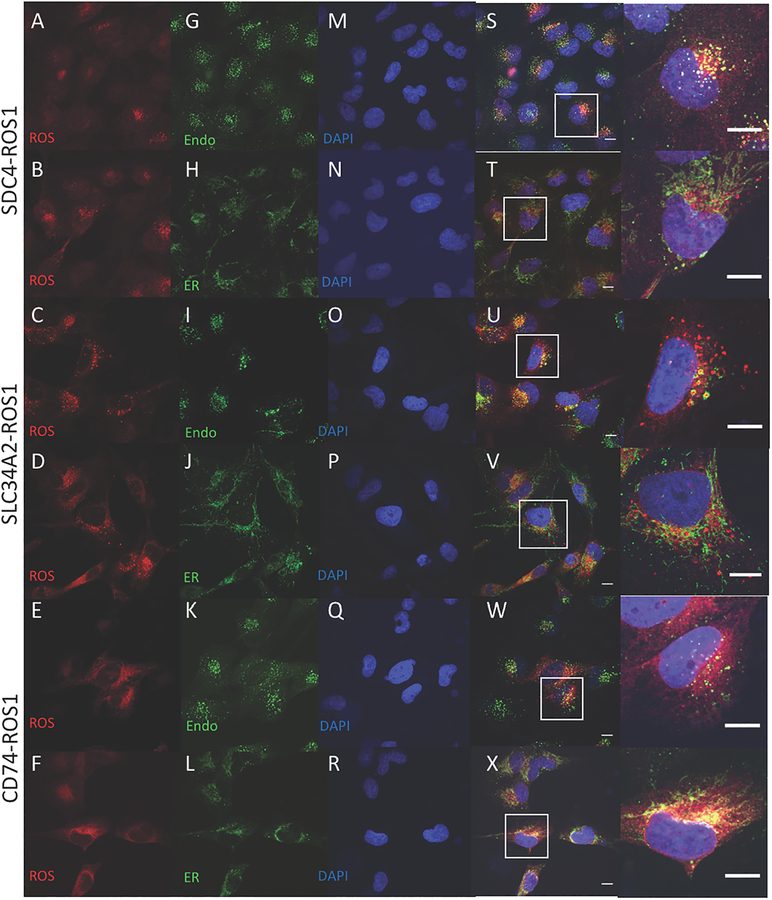
Previous studies of the FIG-ROS1 fusion, present primarily in glioblastoma, suggested that subcellular localization to the Golgi apparatus was important for the transforming ability of that particular fusion and that this localization was mediated by the coiled-coil domain contained within FIG (i.e. the N-terminal fusion partner) (33). Furthermore, our study examining EML4-ALK regulation showed that localization to cytoplasmic structures was dictated by the EML4 N-terminal partner in the EML4-ALK fusion and was essential for downstream RAS/MAPK pathway signaling (20). Based on these findings, we hypothesized that differential subcellular localization of distinct ROS1 oncoprotein fusion variants may regulate the differential signaling pathway activation that we observed. Using immunofluorescence and confocal microscopy analysis, we examined the subcellular localization of SDC4-ROS1, SLC34A2-ROS1, and CD74-ROS1 fusion proteins, both in isogenic BEAS2-B normal bronchial epithelial cell lines that we engineered to express these fusions (ROS1 B2Bs) and in the available ROS1 fusion patient-derived NSCLC cell lines (Figure 3, S8). While thought to be an integral membrane protein localized primarily to the plasma membrane, localization of the full-length, native ROS1 protein proved challenging due to the inability to ectopically express it given its large size (21). We found that there was distinct subcellular distribution of the different ROS1 fusions (localization of the full-length ROS1 protein was not possible due to difficulty with ectopic expression given its large size). SDC4-ROS1 and SLC34A2-ROS1, which activate the MAPK pathway, were found in punctate structures that co-localized with the established endosomal marker EEA-1 (34). The observation of these ROS1 fusion variants localizing on endosomes is consistent with the established principle that in mammalian cells RAS/MAPK signaling emanates exclusively from a lipid-membrane compartment, including endosomes in certain contexts (35,36). In contrast, the CD74-ROS1 variant studied, which does not substantially activate MAPK signaling, was localized in a different pattern that displayed perinuclear enhancement and co-localized with calnexin and PTP1B, established markers of the ER (37,38). Thus, the ability of the different ROS1 fusion protein variants to activate the MAPK pathway is associated with localization on endosomes (i.e. SDC4-ROS1 and SLC34A2-ROS1); localization on the ER (i.e. CD74-ROS1) is associated with compromised RAS/MAPK activation.
Figure 3. Localization of ROS1 protein in isogenic BEAS-2B system reveals different subcellular localization of the ROS1 fusion oncoproteins.
Immunofluorescence and confocal microscopy in BEAS-2B cells stably expressing SDC4-ROS1, SLC34A2-ROS1, and CD74-ROS1. Rows 1,2 = SDC4-ROS1; Rows 3,4 = SLC34A2-ROS1; Rows 5,6 = CD74-ROS1. Antibodies used were specific for: (A-F) = ROS1; (G,I,K) = EEA1; (H,J,L) = Calnexin; and (M-R) = DAPI; (S-X) = overlay image of the left 3 columns, with (right-most column) adjacent high magnification image of representative cells (outlined by white boxes). Scale bars shown indicate 10μM. Images are representative of ≥ 10 fields and at least 2 biological replicate experiments.
Re-localization of CD74-ROS1 to endosomes induces RAS/MAPK pathway activation
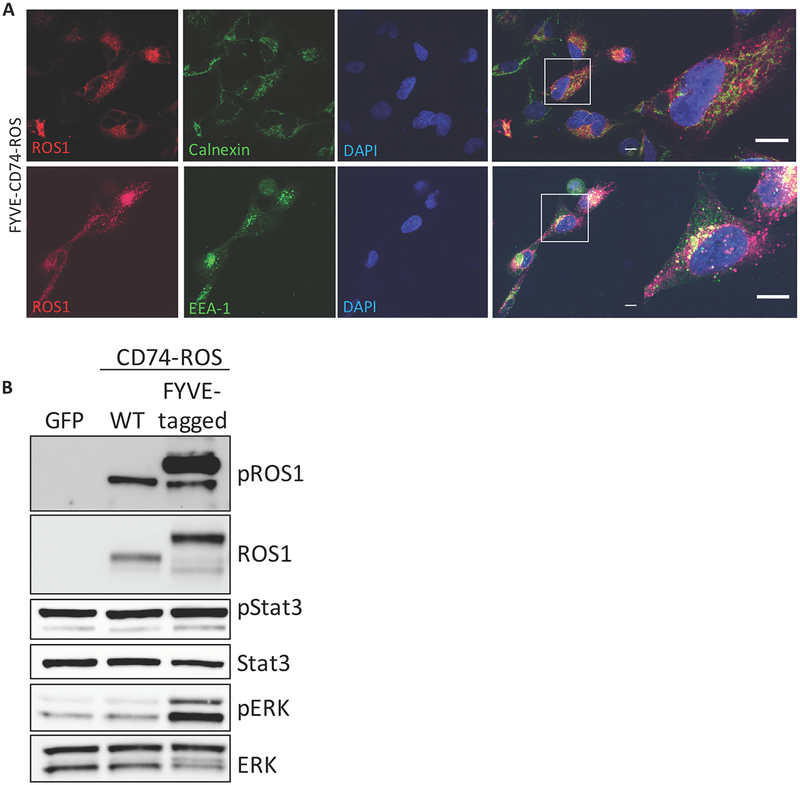
The data indicated that differential subcellular compartment localization correlated with differential MAPK pathway activation downstream of the different ROS1 oncoprotein fusions containing distinct N-terminal fusion partners. We next directly tested whether subcellular localization was required for pathway activation. Wildtype CD74 encodes the invariant chain, a type II transmembrane receptor which is involved in trafficking of MHC molecules through the ER to the endo-lysosome. CD74 contains a 15 amino acid N-terminal cytoplasmic extension, which anchors it into the ER and which is included in the CD74-ROS1 fusion gene used in our studies (39,40). We created a FYVE zinc finger domain-tagged CD74-ROS1 construct to re-localize the fusion protein to endosomes (41). Immunofluorescence analysis of ROS1 in BEAS2-B cells expressing this construct showed re-localization of the FYVE-CD74-ROS1 protein from the ER to punctate structures where it co-localized with the endosomal marker EEA-1, similar to SDC4-ROS1 and SLC34A2-ROS1 subcellular localization (Figure 4A). Furthermore, in contrast to CD74-ROS1, expression of the FYVE-CD74-ROS1 protein induced MAPK pathway activation, suggesting that the specific subcellular localization of ROS1 fusion oncoproteins is critical in mediating RAS/MAPK pathway signaling (Figure 4B). No difference in STAT3 phosphorylation was observed, suggesting pathway specificity in the signaling phenotype that is regulated via differential subcellular compartment localization (Figure 4B). Thus, the differential MAPK pathway activation that is observed between different ROS1 fusions is controlled by fusion-specific and distinct subcellular compartment localization, which is conferred by the N-terminal fusion partner.
Figure 4. Localization of ROS1 oncoproteins regulates engagement of downstream signaling pathways.
(A) Immunofluorescence and confocal microscopy of BEAS2-B cells stably expressing an endosome-targeted FYVE-tagged CD74-ROS1 construct and stained with the indicated antibodies. Far right panel = increased magnification of a representative individual cell. Confocal images are representative of ≥ 10 fields and at least 2 independent experiments. Scale bars shown indicate 10μM. (B) Immunoblot analysis of BEAS2-B cells transfected with GFP, WT CD74-ROS1, or FYVE-CD74-ROS1. Immunoblot is representative of 3 independent experiments.
ROS1 fusions that activate the RAS/MAPK pathway form more aggressive tumors in vivo
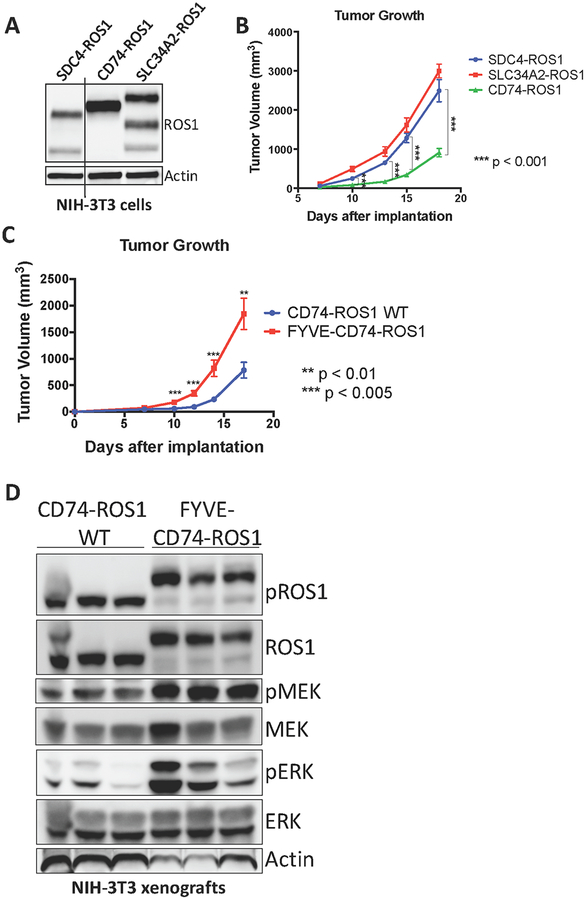
We next investigated the significance of the differential ability of these ROS1 fusions to activate the RAS/MAPK pathway. Despite multiple attempts, none of the limited number of ROS1 fusion-positive patient-derived cell lines that are currently available grew successfully as tumor xenografts in immunocompromised mice, and patient-derived xenograft (PDX) models are not currently available. Thus, to examine tumor growth in vivo, we generated a genetically-controlled isogenic system in which NIH-3T3 cells were engineered to express SDC4-ROS1 and SLC34A2-ROS1 (that activate MAPK signaling) and CD74-ROS1 (that does not activate MAPK signaling) (Figure 5A). While not of epithelial origin, NIH-3T3 cells are an established and controlled system in which tumor formation induced by an oncogenic protein can be assessed (42). Standard tumor xenograft studies in immunocompromised mice were conducted to assess for differential tumor growth in vivo. As expected, NIH-3T3 cells expressing all three ROS1 fusions formed tumors in mice, while control NIH-3T3 cells expressing an empty vector did not and those expressing an established lung cancer-derived oncogenic form of EGFR L858R did (Figure 5B, S9). Interestingly, the SDC4-ROS1 and SLC34A2-ROS1 fusion expressing cells formed more rapidly growing tumors in vivo compared to CD74-ROS1 fusion-driven tumors, and explants from SDC4-ROS1 and SLC34A2-ROS1 tumors demonstrated higher levels of MAPK activation (Figure 5B, S10). Additionally, we generated NIH-3T3 cells expressing the endosomal-targeted FYVE-CD74-ROS1 fusion protein, which is able to activate the MAPK pathway, and compared the in vivo growth rate of those cells to NIH-3T3 cells expressing wildtype CD74-ROS1, which do not demonstrate substantial MAPK pathway activation (Figure 5C). Intriguingly, we found that the FYVE-CD74-ROS1 tumors grow at a significantly faster rate than wildtype CD74-ROS1 (Figure 5D). These data suggest that expression of a ROS1 fusion oncoprotein that can activate the MAPK pathway as a consequence of localization to endosomes results in tumors with an increased growth capacity, compared to tumors expressing a ROS1 fusion oncoprotein that instead does not activate MAPK and localizes to the ER.
Figure 5. MAPK pathway activation in ROS1 fusion oncoprotein-driven cancer models is linked to increased tumorigenic properties in vivo.
(A) Immunoblot analysis of ROS1 fusion oncoprotein expression in isogenic NIH-3T3 cells. (B) Tumor growth rates of tumor xenografts of 1×106 NIH-3T3 ROS1 fusion oncoprotein-expressing cells described in (A) implanted into the flanks of immunocompromised mice. (C) Tumor growth rates of tumor xenografts of 5×105 cells NIH-3T3 cells expressing CD74-ROS1 WT or FYVE-tagged CD74-ROS1. (D) Immunoblot analysis of NIH-3T3 tumor xenograft explants expressing wild-type (WT) or FYVE-tagged CD74-ROS1. Each lane represents an individual tumor. Data in (B-C) are shown as the mean of 6 tumors +/− s.e.m.
Discussion:
Our findings provide evidence demonstrating that the specific N-terminal fusion partners present within gene rearrangements involving the same RTK partner can directly control differential MAPK signaling pathway engagement by causing alternative subcellular compartment localization. These findings have implications for the understanding of the molecular and cell biological basis of cancer growth and establish a link between differential subcellular localization of oncoprotein RTKs with their oncogenic mechanism and tumor growth phenotype. The data suggest that understanding how the subcellular localization of aberrant gene fusion oncoproteins is regulated and influences cell signaling warrants additional investigation more broadly.
Our study demonstrates that while all ROS1 fusions examined activate the JAK/STAT pathway to a similar degree, they vary in their ability to activate the MAPK pathway. SDC4-ROS1 and SLC34A2-ROS1 fusions activate the MAPK pathway while the CD74-ROS1 variant studied here does not. We found that this differential MAPK pathway activation is due to differential ROS1 fusion oncoprotein subcellular localization, which is dictated by the specific N-terminal fusion partner. Our data augment a growing body of evidence indicating that RAS/MAPK signaling can occur not only from the plasma membrane but also from endomembrane compartments such as endosomes in certain cellular contexts (35,43–47). Our observations uncover a link between endosome-based MAPK pathway activation and human disease.
What mediates the differential ability for pathway activation from endosomes (harboring SDC4-ROS1 and SLC34A2-ROS1) versus the ER (harboring CD74-ROS1) remains to be elucidated in future studies. One possibility is that there is differential, compartment-specific accessibility to a positive or a negative regulator of the MAPK pathway (e.g. a RAS guanine nucleotide exchange factor or GTPase activating protein or a MEK/ERK phosphatase). Another possibility is that the degree to which the ROS1 fusion oncoprotein is able to dimerize or associate with other MAPK pathway-activating factors (e.g. KSR or SHP2) may be influenced by subcellular localization.
The patient-derived CD74-ROS1 cDNA used in our studies contains the native ER-targeting motif of CD74 within the CD74-ROS1 fusion protein, thus providing the ER anchor and limiting MAPK pathway activation downstream of CD74-ROS1. Interestingly, a shorter isoform of CD74 can exist that lacks this innate N-terminal ER-targeting motif. Thus, it is possible that a shorter isoform of CD74-ROS1, if expressed in cells or tumors, may localize to endosomes or the plasma membrane instead of the ER and activate MAPK signaling (40,48).
Our findings prompt a series of future investigations that aim to better understand how subcellular localization regulates the ability of a fusion oncoprotein to activate specific signaling pathways. Such studies may help discern unrecognized and important co-factors driving oncogenic signaling, which could be used clinically as more precise diagnostic or therapeutic biomarkers. Genomic advances have led to the genetic classification of tumors including NSCLCs, with improvements in clinical outcomes through genotype-directed targeted therapy. One prominent example is the 19-month progression-free survival observed in patients harboring ROS1 fusion-positive NSCLC treated with ROS1 inhibitors such as crizotinib (11). In current typical clinical practice worldwide, the diagnosis of ROS1 fusions in cancers most commonly occurs via a break-apart FISH (Fluorescence In Situ Hybridization) assay. Consequently, the specific N-terminal fusion partner and its genetic sequence and domain structure are not identified typically in general clinic practice. Our findings suggest the potential importance of altering clinical practice to more routinely employ diagnostic assays with higher precision that detect the identity and domain features of the N-terminal fusion partner in RTK fusion oncoproteins. In our studies, the ability of individual fusion proteins to activate the MAPK pathway is linked to tumor growth capacity. This observation provides an additional rationale to suggest that current diagnostics identifying only the presence or absence of a fusion oncoprotein in a binary manner may be insufficient. More precise identification of the fusion partner may be critical to better stratify patients for prognosis or tailored treatment (either single-agent or rational combination therapy).
While most ROS1 fusion-positive tumors initially respond to crizotinib, virtually all of these tumors eventually develop resistance. We found that MAPK pathway activation is necessary and sufficient for survival of cells expressing SDC4-ROS1 and SLC34A2-ROS1, suggesting MAPK pathway reactivation may be a mechanism of resistance to crizotinib monotherapy. Consistent with this notion, there are limited reports of RAS activating mutations or upregulation driving resistance to crizotinib in the setting of ROS1 fusion driven cancers (49,50). Identification of the downstream pathways activated by an oncogene could be critical to predict what pathway a tumor may reactivate during the evolution of resistance, as we showed in EML4-ALK NSCLC (20). This knowledge could allow for prioritization of upfront combination therapy, such as a ROS1 inhibitor plus a MAPK pathway inhibitor (i.e. SHP2, MEK, or ERK inhibitor) to delay or prevent resistance from arising in tumors in which the RTK oncoprotein fusion is coupled to activation of the pathway.
Supplementary Material
Acknowledgements:
We thank C. Lovly and M. Davare for plasmids, and A. Tulpule, A. Sabnis, and F. Haderk for helpful discussions and suggestions. D.S. Neel was supported by the University of California, San Francisco Medical Scientist Training Program and the National Cancer Institute (F30 CA210444–03). T.G. Bivona acknowledges support from NIH / NCI U01CA217882, NIH / NCI U54CA224081, NIH / NCI R01CA204302, NIH / NCI R01CA211052, NIH / NCI R01CA169338, and the Pew-Stewart Foundations.
Footnotes
Potential competing interests: Trever G. Bivona has received a research grant and is a member of the SAB of Revolution Medicines. Robert Nichols is an employee of Revolution Medicines. The other authors declare no competing interests.
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. Massachusetts Medical Society; 2004;350:2129–39. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med. Nature Publishing Group; 2012;18:349–51. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Sun Y, Fang R, Han X, Luo X, Wang R, et al. Lung adenocarcinomas with HER2-activating mutations are associated with distinct clinical features and HER2/EGFR copy number gains. J Thorac Oncol. 2012;7:85–9. [DOI] [PubMed] [Google Scholar]
- 4.Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. Nature Publishing Group; 2004;431:525–6. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–9. [DOI] [PubMed] [Google Scholar]
- 6.Kohno T, Nakaoku T, Tsuta K, Tsuchihara K, Matsumoto S, Yoh K, et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res. 2015;4:156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. [DOI] [PubMed] [Google Scholar]
- 8.Bergethon K, Shaw AT, Ou S-HI, Katayama R, Lovly CM, McDonald NT, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. Nature Publishing Group; 2013;19:1469–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. Nature Publishing Group; 2012;18:378–81. [DOI] [PubMed] [Google Scholar]
- 11.Shaw AT, Solomon BJ. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. Massachusetts Medical Society; 2015;372:683–4. [DOI] [PubMed] [Google Scholar]
- 12.Shaw AT, Kim D-W, Mehra R, Tan DSW, Felip E, Chow LQM, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. Massachusetts Medical Society; 2014;370:1189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou S-HI, Ahn JS, De Petris L, Govindan R, Yang JC-H, Hughes B, et al. Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J Clin Oncol. 2016;34:661–8. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. Massachusetts Medical Society; 2005;352:786–92. [DOI] [PubMed] [Google Scholar]
- 15.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. Liu ET, editor. PLoS Med. Public Library of Science; 2005;2:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 18.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. Nature Publishing Group; 2017;17:637–58. [DOI] [PubMed] [Google Scholar]
- 19.Medves S, Demoulin J-B. Tyrosine kinase gene fusions in cancer: translating mechanisms into targeted therapies. J Cell Mol Med. Blackwell Publishing Ltd; 2012;16:237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. Nature Research; 2015;21:1038–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acquaviva J, Wong R, Charest A. The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. Biochim Biophys Acta. 2009;1795:37–52. [DOI] [PubMed] [Google Scholar]
- 22.Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19:4040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo J-S, Ju YS, Lee W-C, Shin J-Y, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. Nature Publishing Group; 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using μManager software. J Biol Methods. 2014;1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobson L, Reményi I, Tusnády GE. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43:W408–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobson L, Reményi I, Tusnády GE. The human transmembrane proteome. Biol Direct. BioMed Central; 2015;10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y-NP, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. Nature Publishing Group; 2016;535:148–52. [DOI] [PubMed] [Google Scholar]
- 31.Charest A, Wilker EW, McLaughlin ME, Lane K, Gowda R, Coven S, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 2006;66:7473–81. [DOI] [PubMed] [Google Scholar]
- 32.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol. Nature Publishing Group; 2018;20:1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charest A, Kheifets V, Park J, Lane K, McMahon K, Nutt CL, et al. Oncogenic targeting of an activated tyrosine kinase to the Golgi apparatus in a glioblastoma. Proc Natl Acad Sci USA. 2003;100:916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mu FT, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, et al. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–11. [DOI] [PubMed] [Google Scholar]
- 35.Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191:23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–9. [DOI] [PubMed] [Google Scholar]
- 37.Ahluwalia N, Bergeron JJ, Wada I, Degen E, Williams DB. The p88 molecular chaperone is identical to the endoplasmic reticulum membrane protein, calnexin. J Biol Chem. 1992;267:10914–8. [PubMed] [Google Scholar]
- 38.Frangioni JV, Beahm PH, Shifrin V, Jost CA, Neel BG. The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 1992;68:545–60. [DOI] [PubMed] [Google Scholar]
- 39.Khalil H, Brunet A, Thibodeau J. A three-amino-acid-long HLA-DRbeta cytoplasmic tail is sufficient to overcome ER retention of invariant-chain p35. J Cell Sci. The Company of Biologists Ltd; 2005;118:4679–87. [DOI] [PubMed] [Google Scholar]
- 40.Schröder B The multifaceted roles of the invariant chain CD74--More than just a chaperone. Biochim Biophys Acta. 2016;1863:1269–81. [DOI] [PubMed] [Google Scholar]
- 41.Hayakawa A, Hayes SJ, Lawe DC, Sudharshan E, Tuft R, Fogarty K, et al. Structural basis for endosomal targeting by FYVE domains. J Biol Chem. 2004;279:5958–66. [DOI] [PubMed] [Google Scholar]
- 42.Greulich H, Chen T-H, Feng W, Jänne PA, Alvarez JV, Zappaterra M, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. Rosen N, editor. PLoS Med. Public Library of Science; 2005;2:e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, et al. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. [DOI] [PubMed] [Google Scholar]
- 44.Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Molecular and Cellular Biology. American Society for Microbiology (ASM); 2000;20:2475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. Nature Publishing Group; 2002;4:343–50. [DOI] [PubMed] [Google Scholar]
- 46.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–93. [DOI] [PubMed] [Google Scholar]
- 47.Bivona TG, Pérez de Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, et al. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. Nature Publishing Group; 2003;424:694–8. [DOI] [PubMed] [Google Scholar]
- 48.McCoach CE, Le A, Gowan K, Jones KL, Schubert L, Doak A, et al. Resistance mechanisms to targeted therapies in ROS1+ and ALK+ non-small cell lung cancer. Clin Cancer Res. American Association for Cancer Research; 2018;:clincanres.2452.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Y-C, Lin X-P, Li X-F, Wu L-X, Chen H-F, Wang W-X, et al. Concurrent ROS1 gene rearrangement and KRAS mutation in lung adenocarcinoma: A case report and literature review. Thorac Cancer. 2017;30:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cargnelutti M, Corso S, Pergolizzi M, Mévellec L, Aisner DL, Dziadziuszko R, et al. Activation of RAS family members confers resistance to ROS1 targeting drugs. Oncotarget. 2015;6:5182–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.