Summary:
Patients with EGFR-mutant NSCLC achieve variable benefit from targeted therapy, and biomarkers to predict degree of benefit are not in clinical use. EGFR-mutant cancers with high tumor mutational burden demonstrate poorer outcomes on EGFR targeted therapy. Investigation into the mechanisms underlying this intriguing association is needed.
Body:
In this issue of Clinical Cancer Research, Offin and colleagues (1) report a negative association between tumor mutation burden (TMB) and clinical benefit from EGFR tyrosine kinase inhibitor (TKI) therapy in patients with EGFR-mutant NSCLC.
Increased nonsynonymous somatic mutation burden is positively associated with clinical benefit to anti-PD-1/PD-L1 immune checkpoint blockade in non-small cell lung cancer (NSCLC) and other solid tumors (2). Tumor mutation burden (TMB) can be readily calculated using both whole exome sequencing (WES) and targeted next-generation sequencing (NGS) (2), such that TMB is now commonly included in the clinical reports of targeted NGS assays available in both academic and commercial settings. As broad molecular testing is increasingly standard in the care of advanced NSCLC, TMB results will likely be increasingly available to physicians caring for NSCLC patients, including those with EGFR mutations or other targetable driver alterations. However, the significance of TMB as a biomarker has not previously been explored in EGFR-mutant NSCLC, and it is unknown how differences in TMB should inform clinical decision-making in this setting, if at all.
The authors creatively explore the impact of TMB in an institutional cohort of 153 patients with previously untreated metastatic NSCLC with the most common EGFR mutations (ex19del and L858R) treated with first or second-generation EGFR TKIs. Somatic TMB was calculated from targeted NGS results using pretreatment tumor tissue as well as matched normal DNA from peripheral blood sample collection, to control for white blood cell derived variants (e.g. germline and hematopoietic). In this retrospective analysis, an inverse relationship is identified between TMB and clinical outcomes, with patients in the highest TMB tertile demonstrating shorter time to treatment discontinuation (TTD) and overall survival (OS) compared to the low and intermediate tertiles. The survival difference was particularly striking, with a large difference in median OS between the highest TMB tertile (21 months) and the intermediate and low tertiles (37 and 41 months, respectively, p=0.02). The TTD and OS improvement for intermediate and low TMB tertiles persisted within clinical subgroups and in multivariable analysis including TP53 status and EGFR allele type (ex19del vs L858R). In a subset of 30 patients with paired tumor NGS performed prior to therapy as well as after TKI resistance, TMB was significantly higher at resistance (6.56 vs 3.42 mutations/Mb, p=0.008).
It should be noted that the criterion for high TMB in this paper differs from prior publications. In the above analysis, the highest tertile of TMB reflects a TMB above 4.85 mutations/Mb. The authors also repeated the analysis dichotomizing at median TMB (3.77 mutations/Mb) and found the OS difference was no longer statistically significant in multivariable analysis (HR 0.59, p=0.10). However, in a prior report using this same targeted NGS panel to study 84 patients with advanced NSCLC treated with immunotherapy, high TMB was defined as >7.4 mutations/Mb (2). In another study using the FoundationOne CDx targeted NGS panel (which does not sequence paired normal DNA to control for non-tumor variants), high TMB was defined as ≥10 mutations/Mb, though this was a subset analysis of a clinical trial which excluded EGFR-mutant and ALK-positive NSCLC (3). In this report by Offin et al, only 3% (5/153) of patients (and 10% [5/51] of patients in the highest TMB tertile) had ≥10 mutations/Mb. In addition, the use of targeted sequencing in this cohort of EGFR-mutant NSCLC with low absolute TMB values results in limited granularity in TMB values for analysis. Among 153 patients, there are only 36 unique TMB values (and likely fewer if controlling for differences in genomic coverage between the three versions of assay). For example, 24 patients have a TMB result of exactly 2.83.
The mechanistic explanation for why higher TMB is associated with worse outcomes is not clearly established in this article. Patients with EGFR-mutant NSCLC infrequently achieve durable clinical benefit to anti-PD-1/PD-L1 immune checkpoint blockade (2), and the low absolute TMB values observed in this cohort (even in the highest tertile) suggest that the differences in outcome are secondary to factors unconnected to antitumor immunity. One hypothesis might be that higher TMB represents a greater diversity of pre-existing subclones, some of which emerge under the selective pressure of TKI therapy. However, recent preclinical studies have found that pre-existing resistant subclones rarely lead to EGFR TKI resistance (4). One resistance mechanism that is understood to rarely pre-exist, T790M, was in fact seen less commonly in patients with higher TMB in this analysis. Another hypothesis might be an innate propensity towards mutagenesis in cancers with higher TMB, akin to the poorer prognosis seen in EGFR-mutant lung cancers with TP53 mutations (5). This could result in more heterogeneity at time of resistance, which we have found leads to early treatment failure in patients receiving osimertinib for acquired T790M (6). Unfortunately, the limited number of patients with sequencing of tissue at resistance does not allow pre-treatment TMB to be robustly connected to specific resistance mechanisms. TMB itself is likely an indirect marker of separate underlying biological mechanisms, which could be therapeutically leveraged if better understood, and perhaps studied beyond EGFR-mutant NSCLC.
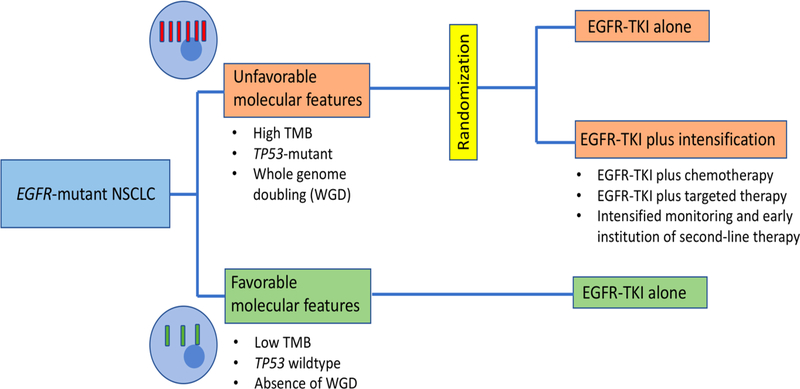
There is a clear clinical role for genomic biomarkers like TMB which could identify subgroups of patients with oncogene-driven NSCLC who have less favorable outcomes on TKI therapy. Such patients might benefit from more intensive monitoring with earlier institution of second-line therapy. Furthermore, such patients would be ideal for clinical trials of intensified first-line therapy (Figure 1) as there are now a number of TKI combination strategies under clinical investigation. However, intensified treatment for EGFR-mutant NSCLC with high TMB might be hard to support; in this paper, even the poorest prognosis tertile had a median TTD of 10 months, which is relatively favorable in the context of other lung cancer therapies. It must also be noted that other NGS assays, which cover different sets of genes and do not include sequencing of matched normal, would result in different TMB distributions such that establishing widely applicable thresholds for high TMB could prove difficult. For now, further investigation and validation is needed to understand whether measurement of TMB might be a scalable predictive or prognostic biomarker in EGFR-mutant lung cancer and other genotype-driven NSCLC subtypes.
Figure 1: Schema for the investigation of intensified therapy in EGFR-mutant lung cancer harboring poor prognosis genomic biomarkers.
Genomic biomarkers are not routinely employed to guide therapy in EGFR-mutant NSCLC. Yet there could be a clinical role for such biomarkers in differentiating patients who do well with standard single-agent TKI from those who might benefit from studies of intensified therapy Prospective clinical trials which investigate intensified therapy (such as TKI-based combinations) in cases with unfavorable molecular features could help establish the clinical utility of such biomarkers.
Acknowledgments:
G. R. Oxnard is supported by National Cancer Institute grant R01 CA114465 and by the Pamela Elizabeth Cooper Research Fund.
Footnotes
Conflict of Interest Disclosures:
Dr Oxnard receives consulting fees from AstraZeneca and Takeda. No other disclosures are reported.
References
- 1.Offin M, Rizvi H, Tenet M, Ni A, Sanchez-Vega F, Li BT, et al. Tumor Mutation Burden and Efficacy of EGFR-Tyrosine Kinase Inhibitors in Patients with EGFR-Mutant Lung Cancers. Clinical Cancer Research 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36(7):633–41 doi 10.1200/jco.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. The New England journal of medicine 2018;378(22):2093–104 doi 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oxnard GR. The cellular origins of drug resistance in cancer. Nature medicine 2016;22:232 doi 10.1038/nm.4058. [DOI] [PubMed] [Google Scholar]
- 5.Canale M, Petracci E, Delmonte A, Chiadini E, Dazzi C, Papi M, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clinical Cancer Research 2016. doi 10.1158/1078-0432.Ccr-16-0966. [DOI] [PubMed] [Google Scholar]
- 6.Oxnard GR, Hu Y, Mileham KF, et al. Assessment of resistance mechanisms and clinical implications in patients with egfr t790m–positive lung cancer and acquired resistance to osimertinib. JAMA Oncology 2018. doi 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]