Abstract
DNA methylation is instrumental for gene regulation. Global changes in the epigenetic landscape have been recognized as a hallmark of cancer. However, the role of DNA methylation in epithelial ovarian cancer (EOC) remains unclear. In this study, high density genetic and DNA methylation data in white blood cells from the Framingham Heart Study (N=1,595) were used to build genetic models to predict DNA methylation levels. These prediction models were then applied to the summary statistics of a genome-wide association study (GWAS) of ovarian cancer including 22,406 EOC cases and 40,941 controls to investigate genetically predicted DNA methylation levels in association with EOC risk. Among 62,938 CpG sites investigated, genetically predicted methylation levels at 89 CpG were significantly associated with EOC risk at a Bonferroni-corrected threshold of P<7.94×10−7. Of them, 87 were located at GWAS-identified EOC susceptibility regions and two resided in a genomic region not previously reported to be associated with EOC risk. Integrative analyses of genetic, methylation, and gene expression data identified consistent directions of associations across 12 CpG, five genes, and EOC risk, suggesting that methylation at these 12 CpG may influence EOC risk by regulating expression of these five genes, namely MAPT, HOXB3, ABHD8, ARHGAP27 and SKAP1. We identified novel DNA methylation markers associated with EOC risk and propose that methylation at multiple CpG may affect EOC risk via regulation of gene expression.
Keywords: DNA methylation, genetic models, gene expression, epithelial ovarian cancer risk
Introduction
Ovarian cancer is one of the most deadly cancers among women in the United States (1) and around the world (2). Approximately 90% of ovarian neoplasms are epithelial ovarian cancer (EOC) (1), a heterogeneous disease that can be categorized into five major histotypes (1). Genetic factors have an important impact on EOC etiology. Large-scale genome-wide association studies (GWAS) have identified 34 common risk loci for EOC to date (3). Of these, 27 are specific to the most common histotype, serous EOC (3). However, known loci are estimated to account for only a small proportion (~6.4%) of overall EOC risk (3). In addition, causal genes at most loci and the underlying pathogenic mechanisms are yet to be identified.
In addition to genetic susceptibility, cancer initiation and progression are also influenced by epigenetics (4). The most extensively studied epigenetic marker is DNA methylation, which regulates chromatin structure (5) and gene expression (6). DNA methylation patterns are generally programmed during normal development (7). Abnormal methylation has been observed in multiple malignancies, including EOC (8,9). Studies have identified multiple DNA methylation markers in tumor tissue samples as prognostic biomarkers for EOC (10,11). Several studies have also investigated the potential of DNA methylation from white blood cells to be early detection biomarkers for EOC and identified nearly 100 candidate CpGs for EOC risk (12-15). To date, only two CpGs, cg10061138 and cg10636246, were consistently observed across different studies (12-15). The lack of consistent findings may reflect the small sample sizes of prior studies (200-400 cases), an inadequate consideration of potential confounders and reverse causation.
DNA methylation is impacted by both environmental factors and genetic factors (6). High-throughput methylome profiling in both twin and familial studies has shown that methylation levels for a large number of CpGs are heritable (16,17). Furthermore, several studies (18,19) have revealed a large number of methylation quantitative trait loci (meQTL) in white blood cells. These results suggest that DNA methylation levels could be partially predicted by genetic variants. Indeed, meQTL single nucleotide polymorphisms (SNPs) appear to predict DNA methylation levels in white blood cells and the predicted methylation levels associated with disease risk (20,21). However, these studies only used single meQTL SNPs to predict methylation levels for each CpG site. The prediction accuracy is low because meQTL SNPs explain only a small proportion of variance. In the present study, we used a novel approach to overcome this limitation by building and validating statistical models to predict methylation levels based on multiple genetic variants in reference datasets. The prediction models were then applied to genetic data from 22,406 cases and 40,941 controls to test the hypothesis that genetically predicted DNA methylation is associated with EOC risk. This approach could overcome the selection bias and reverse causation in conventional epidemiological studies of DNA methylation and disease because alleles are randomly assigned during gamete formation.
Methods
Building DNA methylation prediction models using data from the Framingham Heart Study (FHS)
Genome-wide DNA methylation and genotype data from white blood cell samples from individuals in the FHS Offspring Cohort were obtained from dbGaP (accession numbers phs000724 and phs000342, respectively). Detailed descriptions of the FHS Offspring Cohort have been previously reported (22). Genotyping was conducted using the Affymetrix 500K mapping array and imputation was performed with the1000 Genome Phase I (version 3) data as reference. Only SNPs with a minor allele frequency (MAF) of ≥0.05 and an imputation quality (R2) of ≥0.80 were used to build prediction models. Genome-wide DNA methylation profiling was generated using the Illumina HumanMethylation450 BeadChip. We used the R package “minfi” (23) to filter low quality methylation probes, evaluate cell type composition for each sample and estimate methylation beta-values. Methylation data were then quantile-normalized across samples, rank-normalized to remove potential outliers, and then regressed on covariates including age, sex, cell type composition and top ten principal components (PCs) to eliminate potential experimental confounders and population structure. Finally, 1,595 unrelated individuals of European descent (883 females and 712 males, mean ± SD of age: 66.3 ± 9.0) with both genetic and DNA methylation data were included in prediction model building.
Using the elastic net method (α=0.50) implemented in the R package “glmnet” (24), we built a statistical model to predict methylation levels for each CpG site using the SNPs within its 2 megabase (Mb) flanking region. For each model, we performed tenfold cross-validation as internal validation and calculated the squared value of the correlation coefficient between measured and predicted methylation levels, i.e. RFHS2, to estimate prediction performance.
Evaluation of model performance using data from the Women’s Health Initiative (WHI)
Using data from white blood cell samples from 883 independent healthy women of European descent from the WHI, we evaluated the performance of the established genetic prediction models. Data from the WHI samples were obtained from dbGaP (accession numbers phs001335, phs000675 and phs000315). Genotyping was conducted using the HumanOmniExpress and HumanOmni1-Quad array. The data were quality control (QC)-ed and imputed using similar criteria and procedures as those described for the FHS data. The Illumina HumanMethylation450 BeadChip was used to profile DNA methylation and the data were then processed using the same pipeline as that for the FHS data. The prediction models established in FHS were applied to the genetic data in WHI to predict methylation levels at each CpG site for each sample. Then, the predicted and measured methylation levels for each CpG site were compared by estimating the squared value of the Spearman correlation coefficient, i.e. RWHI2.
We used the following criteria to select prediction models for association analyses: 1) a prediction RFHS2 of ≥0.01 (correlation between measured and predicted methylation levels of ≥0.10) in the FHS; 2) a RWHI2 of ≥0.01 in the WHI; and 3) methylation probes on the HumanMethylation450K BeadChip not overlapping with any SNP included in the dbSNP database (25) (Build 151), considering that SNPs on the probes may have a potential impact on the methylation level estimation (19). In total, models for 63,000 CpGs met these requirements and were included in the downstream association analyses for EOC risk.
Association between genetically predicted DNA methylation and EOC risk
MetaXcan (26) was used to estimate the associations between genetically predicted methylation levels and EOC risk. The methodology of MetaXcan has been described elsewhere (26,27). Briefly, the following formula was used to evaluate the association Z-score:
In the formula, Wsm represents the weight of SNP s on the methylation level of the CpG site m, estimated by the prediction model. and are the evaluated variances of SNP s and the predicted methylation level at CpG site m, respectively. and represent the beta coefficient and standard error of SNP s on EOC risk, respectively. For this study, the correlations between predicting SNPs for all CpGs were evaluated using the data from European participants in the 1000 Genomes Project Phase 3.
Beta coefficient and standard error for the association between SNP s and EOC risk were obtained from the Ovarian Cancer Association Consortium (OCAC), which includes 22,406 EOC cases and 40,941 controls of European ancestry (3). Details of this consortium have been described elsewhere (3). For EOC patients, some may have had neo-adj chemotherapy before surgery. They were not included in sub-type analyses but included in the analyses for overall EOC risk (3). Cases were classified as one of five histotypes: high-grade serous (N=13,037), endometrioid (N=2,810), mucinous invasive (N=1,417), clear cell (N=1,366) or low-grade serous (N=1,012). In addition, there were 2,764 EOC cases that could not be categorized into any histotypes. Genotyping was conducted using OncoArray and other GWAS arrays, followed by imputation, with the 1000 Genomes Project Phase 3 as reference. Association analyses were conducted within each dataset (different GWAS arrays) and the results were combined by a fixed-effect inverse-variance meta-analysis. Among the 751,157 SNPs included in the prediction models for 63,000 CpGs, summary statistics for associations between 751,031 (99.98%) of these SNPs and EOC risk were available from the OCAC. A total of 62,938 CpGs, corresponding to these 751,031 SNPs, were included in the final analyses. This study was approved by the OCAC Data Access Coordination Committee.
For risk analyses in OCAC, we used a Bonferroni-corrected threshold of P<7.94×10−7 (0.05/62,938) for statistical significance in assessing the association between each of the 62,938 CpGs and EOC risk. Associations of predicted methylation and EOC risk identified in the OCAC data were further evaluated using the summary statistics of two GWAS studies of ovarian cancer in the UK Biobank (28). However, the sample size of the EOC cases is very small, with only 440 histologically diagnosed and 579 self-reported ovarian cancer cases among nearly 337,000 unrelated individuals of European descent. GWAS analyses were conducted using a linear regression model. The summary statistics data are available at https://sites.google.com/broadinstitute.org/ukbbgwasresults/home.
We estimated whether the identified associations of predicted methylation with EOC risk were independent of GWAS-identified EOC susceptibility variants. For each SNP included in the prediction model, we used GCTA-COJO (29) to evaluate the and formula with EOC risk after adjusting for the GWAS-identified variants for EOC. Then we re-conducted the MetaXcan analyses to investigate the associations of the predicted methylation levels with EOC risk conditioning on the GWAS-identified EOC risk variants. We also performed stratification analyses by six EOC histotypes and estimated the heterogeneity across histotype groups by using Cochran’s Q test.
Functional annotation of methylation markers
Using ANNOVAR (30), all 62,938 investigated CpGs were classified into 11 functional categories: upstream, transcription start site upstream 1500bp (TSS1500), TSS200, 5’-untranslated region (UTR), exonic, intronic, 3’-UTR, downstream, intergenic, non-coding RNA (ncRNA) exonic and ncRNA intronic.
Correlation analyses of DNA methylation with gene expression in white blood cells
For those 89 CpGs with predicted methylation levels associated with EOC risk, we investigated those methylation levels in relation to the expression levels of genes flanking these CpGs. Individual-level DNA methylation and gene expression data of white blood cell samples from the FHS Offspring Cohort were accessed from dbGaP (accession numbers phs000724 and phs000363). The details of the Offspring Cohort of the FHS, the DNA methylation data and gene expression data have been described previously (22,31). In total, 1,367 unrelated participants with both methylation and gene expression data were included in correlation analyses. A threshold of P<0.05 was used to determine a nominally-significant correlation between methylation level and gene expression level. In addition, using data from the FHS, we investigated whether the methylation of those 89 EOC-associated-CpGs could regulate the expression of 19 homologous recombination (HR) genes (32,33).
Association analyses of genetically predicted gene expression with EOC risk
For genes with expression levels nominally correlated with methylation levels of CpGs that were associated with EOC, we investigated whether genetically predicted gene expression levels were associated with EOC risk following methods described elsewhere (27). Briefly, genome-wide genetic and gene expression data from 6,124 different tissue samples (donated by 369 participants of European ancestry) included in the Genotype-Tissue Expression (GTEx) release 6 (34) were used to build genetic models for gene expression prediction by following the elastic net method (27). The models were then applied to the OCAC data to estimate the associations between genetically predicted gene expression levels and EOC risk by using MetaXcan (26). We used Bonferroni correction to declare statistically significant associations.
Consistent directions of associations across methylation, gene expression and EOC risk
To infer potential mechanisms underlying the identified associations between DNA methylation and EOC risk, we conducted integrative analyses of the association results between predicted CpG methylation and EOC risk, correlations between CpG methylation and gene expression, and associations between gene expression and EOC risk. First, we examined whether the association directions among DNA methylation, gene expression and EOC risk were consistent. Then, we evaluated whether genetically predicted methylation might mediate associations between gene expression and EOC risk. Briefly, for each gene we used GCTA-COJO (35) to generate modified summary statistics of associations between SNPs in its expression prediction models and EOC risk after adjusting for SNPs included in the methylation prediction model of its corresponding CpG site. Finally, the prediction models of each gene were applied to the updated summary statistics using MetaXcan (26) to estimate the association between genetically predicted gene expression and EOC risk conditioning on the effects of the genetically predicted methylation level at each corresponding CpG site.
Results
DNA methylation prediction models
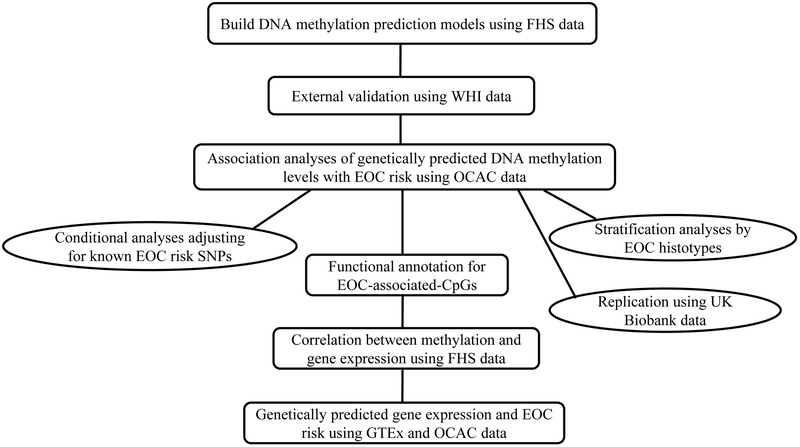
Figure 1 presents the overall workflow of this study. Data from the FHS Offspring Cohort were used to create methylation prediction models for 223,959 CpGs. Of these, 81,361 showed a prediction performance (RFHS2) of ≥0.01, representing at least a 10% correlation between predicted and measured methylation levels. For these 81,361 CpGs, the number of SNPs in prediction models ranged from 1 to 276, with a median of 25. Applying these 81,361 models to genetic data from the WHI, 70,269 (86.4%) models showed a correlation coefficient between predicted and measured methylation levels (RWHI) of >10%. Among these 70,269 CpGs, methylation probes of 7,269 on the HumanMethylation450 BeadChip overlapped with SNPs, which may have affected the estimation of the methylation levels (19). Hence, these CpGs were excluded. The remaining 63,000 CpGs were included in the downstream analyses.
Figure 1.
Study design flow chart
Associations of genetically predicted DNA methylation with EOC risk
The prediction models were applied to the data from a GWAS of 22,406 EOC cases and 40,941 controls included in OCAC. Summary statistics of associations between 751,031 of the 751,157 SNPs, corresponding to 62,938 of the 63,000 CpGs, and EOC risk were available in OCAC. For these 62,938 CpGs, a high correlation of prediction performance between models based on FHS (RFHS2) and WHI (RWHI2) data was observed, with a Pearson correlation coefficient of 0.95. This indicates that for each of these CpGs, a same set of predicting SNPs could predict a very similar methylation level, using either FHS or WHI data.
For most of these 62,938 CpGs, a large majority of predicting SNPs were available in OCAC (e.g., for 94% of the investigated CpGs, ≥95% of the SNPs in prediction models were available in OCAC). Supplementary Figure 1 is the Manhattan plot presenting the associations between genetically predicted methylation levels and EOC risk. Among the 62,938 CpGs, 89 were significantly associated with EOC risk at a Bonferroni-corrected threshold of P<7.94×10−7 (Table 1 and 2, Supplementary Table 1). Among these 89 CpGs, a higher predicted methylation level was associated with an increased risk of EOC at 48 CpGs, and with a decreased EOC risk in the other 41 CpGs. These indicates that the methylation levels were predicted to be higher for 48 CpGs and lower for 41 CpGs among EOC cases than among controls. For these 89 CpGs, we also re-built the prediction models only using data from females (N=833) in FHS. A very high correlation was observed, with a Pearson correlation coefficient of 0.99, between the prediction performance R2 values, based on data of all FHS participants (N=1,595) and those based on females only data (N=833). In the UK Biobank data, consistent associations were observed for 23 CpGs, including 12 at P<0.05, and 11 additional CpGs at P<0.10 (Supplementary Table 2). This relatively low replication rate is not unexpected, considering the very limited statistical power of the UK Biobank data because of a very small number of cases (N=400~600).
Table 1.
Two novel methylation-EOC associations for two CpGs located at a genomic region not yet reported for EOC risk
| CpG | Chr | Position | Closest gene | Classification | RFHS2 a | Histotype | Z score | OR (95% CI) b | P value |
|---|---|---|---|---|---|---|---|---|---|
| cg18139273 | 7 | 962,582 | ADAP1 | Intronic | 0.01 | Overall | −4.95 | 0.51 (0.39-0.66) | 7.25×10−7 |
| Serous c | −4.87 | 0.46 (0.34-0.63) | 1.13×10−6 | ||||||
| High-grade serous | −4.83 | 0.46 (0.33-0.63) | 1.39×10−6 | ||||||
| Endometrioid | −1.78 | 0.59 (0.33-1.06) | 0.08 | ||||||
| Mucinous | −0.99 | 0.67 (0.30-1.49) | 0.32 | ||||||
| Clear cell | −1.87 | 0.46 (0.21-1.04) | 0.06 | ||||||
| Low-grade serous | −0.97 | 0.62 (0.24-1.63) | 0.33 | ||||||
| cg03634833 | 7 | 965,534 | ADAP1 | Intronic | 0.09 | Overall | −5.00 | 0.84 (0.79-0.90) | 5.81×10−7 |
| Serous c | −4.85 | 0.83 (0.77-0.89) | 1.21×10−6 | ||||||
| High-grade serous | −4.85 | 0.82 (0.76-0.89) | 1.25×10−6 | ||||||
| Endometrioid | −2.21 | 0.83 (0.71-0.98) | 0.03 | ||||||
| Mucinous | −1.40 | 0.87 (0.71-1.06) | 0.16 | ||||||
| Clear cell | −1.76 | 0.84 (0.69-1.02) | 0.08 | ||||||
| Low-grade serous | −0.87 | 0.91 (0.73-1.13) | 0.39 |
Correlation between predicted and measured methylation levels.
OR, odds ratio per standard deviation increase in genetically predicted methylation level; CI, confidence interval.
Including high-grade serous and low-grade serous ovarian cancers.
Table 2.
Selected a seven methylation-EOC associations driven by previously identified EOC risk SNPs
| CpG | Chr | Position | Closest gene | Classification | Z score | OR (95% CI) b | P value | RFHS2 c | EOC risk SNPs | Distance to the risk SNPs (kb) |
P value adjusted for the risk SNPs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cg25137403 | 2 | 177,022,172 | HOXD4; HOXD3 | Intergenic | 7.51 | 1.24 (1.18-1.32) | 5.96×10−14 | 0.15 | rs6755777; rs711830 | 21;15 | 0.09 |
| cg26405475 | 3 | 156,324,038 | SSR3; TIPARP-AS1 | Intergenic | −9.45 | 0.69 (0.64-0.74) | 3.42×10−21 | 0.07 | rs62274041 | 111 | 0.34 |
| cg08478672 | 8 | 129,374,295 | MIR1208; LINC00824 | Intergenic | 5.08 | 1.29 (1.17-1.42) | 3.81×10−7 | 0.06 | rs1400482 | 167 | 0.05 |
| cg14653977 | 9 | 136,038,692 | GBGT1 | Intronic | 5.99 | 1.75 (1.46-2.09) | 2.04×10−9 | 0.03 | 9:136138765 d | 100 | 0.09 |
| cg04231319 | 10 | 21,824,447 | MLLT10 | Intronic | −5.72 | 0.88 (0.84-0.92) | 1.05×10−8 | 0.19 | rs144962376 | 54 | 0.94 |
| cg07067577 | 17 | 43,506,829 | ARHGAP27 | 3'UTR | −7.49 | 0.73 (0.67-0.79) | 6.86×10−14 | 0.07 | rs1879586 | 60 | 0.01 |
| cg21956434 | 19 | 17,377,697 | BABAM1 | TSS1500 | 7.07 | 1.13 (1.09-1.17) | 1.53×10−12 | 0.34 | rs4808075 | 12 | 0.39 |
Selected from 87 CpG-EOC associations. For each locus, only the most significantly associated CpG was presented. Complete list of results for all CpG-EOC associations is available in Supplementary Table 1.
OR, odds ratio per standard deviation increase in genetically predicted methylation level; CI, confidence interval.
Correlation between predicted and measured methylation levels.
GRCh37 position.
Among the 89 CpGs that were associated with EOC, two reside in a genomic region on chromosome 7 that has not yet been reported for EOC risk (500Kb away from any GWAS-identified EOC susceptibility variants) (Table 1). Given that there are no risk variants identified by previous GWAS on this chromosome, associations with EOC risk conditioning on proximally-located risk variants could not be conducted. Among the remaining 87 CpGs located in nine previously-identified EOC risk loci, no associations remained significant after an adjustment for all risk SNPs in the corresponding loci. This suggests that the associations of these 87 CpGs with EOC risk were all driven by known EOC risk SNPs in these loci (Table 2 and Supplementary Table 1).
Stratification analyses by EOC histotypes revealed that all 89 CpGs were associated with both serous ovarian cancer and high-grade serous ovarian cancer. Fewer CpGs were associated with the other histotypes, including endometrioid ovarian cancer (cg25137403, cg14454907 and cg25708328), mucinous ovarian cancer (cg25137403, cg14454907, cg10086659 and cg25708328) and low-grade serous ovarian cancer (cg01572694) (Supplementary Tables 3-4). Fourteen of these 89 CpGs showed more significant associations with the serous and the high-grade serous ovarian cancers than with other histotypes, with a heterogeneity test P<5.62×10−4, a Bonferroni-corrected threshold (0.05/89) (Supplementary Table 3). Among these 89 CpGs, a significant correlation of methylation and gene expression was identified for 91 CpG-HR gene pairs, including 22 CpGs and 11 HR genes, at a Bonferroni-corrected threshold of P<2.96×10−5 (0.05/1,691) (Supplementary Table 5). Interestingly, methylation levels of three CpGs, i.e. cg13568213 (9q34.2), cg10900703 (10p12.31) and cg23659289 (17q21.31) showed a strong correlation with the expression level of the ATM gene.
DNA methylation affecting EOC risk through regulating expression of a neighbor gene
For those 89 CpGs with predicted methylation levels associated with EOC risk, we conducted correlation analyses with gene expressions for 63 pairs of CpG-gene, including 58 CpGs with 21 flanking genes that were annotated by ANNOVAR (30). Nominally significant correlations were observed for 26 CpG-gene pairs, including 26 CpGs and 12 genes, at P<0.05 (Table 3, Supplementary Table 6). Among them, the most significant correlation was observed between the increased methylation at the CpG cg19139618, located in the promoter region of the SKAP1 gene, and the expression level of SKAP1, with a P value of 2.98×10−15 (Table 3). In addition, increased methylation levels at two CpGs, cg10900703 and cg04231319, located in the introns of the MLLT10 gene, were significantly correlated with an increased expression of MLLT10, with P values of 2.79×10−11 and 1.36×10−5, respectively. For the two CpGs located in a putative novel locus, a higher methylation level for one of them, cg03634833, was correlated with a lower expression of the ADAP1 gene in this locus, with a P value of 2.99×10−3 (Supplementary Table 6). As expected, methylation levels at CpGs located in promoter regions (TSS1500 and TSS200) were more likely to be negatively correlated with expressions of proximal genes. Nearly all CpGs located in downstream or in 3’UTR showed a negative regulatory effect on expression of neighbor genes. For CpGs residing in intronic regions, both positive and negative correlations were observed.
Table 3.
Selected a correlations between methylation levels at 26 CpGs and expression levels of 12 genes, data from the Framingham Heart Study
| CpG | Chr | Position | Classification | Closest gene | Rho | P value |
|---|---|---|---|---|---|---|
| cg25137403 | 2 | 177,022,172 | Downstream | HOXD4 | −0.06 | 0.02 |
| cg22211092 | 3 | 156,361,584 | Downstream | SSR3 | 0.09 | 9.43×10−4 |
| cg03634833 | 7 | 965,534 | Intronic | ADAP1 | −0.08 | 2.99×10−3 |
| cg14653977 | 9 | 136,038,692 | Intronic | GBGT1 | −0.06 | 0.02 |
| cg24267699 | 9 | 136,151,359 | TSS1500 | ABO | −0.09 | 8.07×10−4 |
| cg10900703 | 10 | 21,824,407 | Intronic | MLLT10 | 0.18 | 2.79×10−11 |
| cg23659289 | 17 | 43,472,725 | 3'UTR | ARHGAP27 | −0.19 | 9.89×10−13 |
| cg07368061 | 17 | 44,090,862 | Intronic | MAPT | 0.08 | 2.02×10−3 |
| cg19139618 | 17 | 46,504,791 | Intronic | SKAP1 | −0.21 | 2.98×10−15 |
| cg14285150 | 17 | 46,659,019 | Intronic | HOXB3 | 0.11 | 8.44×10−5 |
| cg22311200 | 17 | 46,695,514 | Downstream | HOXB8 | 0.08 | 2.59×10−3 |
| cg17941109 | 19 | 17,407,198 | Intronic | ABHD8 | −0.06 | 0.03 |
Selected from correlations between 26 CpGs and 12 genes. For each gene, only the most significantly correlated CpG was presented. Complete list of results for all CpG-EOC associations is available in Supplementary Table 6.
For the 12 genes with expression levels correlated with DNA methylation, expression prediction models were built for seven, with a prediction performance (R2) of ≥0.01, using GTEx data. Applying these seven models to the OCAC data, genetically predicted expression levels of three genes, namely MAPT, HOXB3 and ABHD8, were significantly associated with EOC risk after Bonferroni correction (Table 4). At 17q21.31 and 17q21.32, higher predicted expression levels of MAPT and HOXB3 were associated with a decreased EOC risk, with P values of 3.74×10−4 and 2.00×10−7, respectively. After adjusting for established EOC risk SNPs, the associations between these two genes and EOC risk disappeared. At 19p13.11, an increased predicted expression level for ABHD8 was associated with an increased EOC risk, with a P value of 9.93×10−6. Conditioning on the EOC risk SNP in this locus, the association disappeared as well (Table 4). Of the five genes without prediction models, two were previously reported to be associated with EOC susceptibility, including SKAP1 (36) and ARHGAP27 (37).
Table 4.
Three genes with genetically predicted expression levels associated with EOC risk
| Region | Gene | Type | Z score | P value | Adjusted a P value | R2 b |
|---|---|---|---|---|---|---|
| 17q21.31 | MAPT | Protein | −3.56 | 3.74×10−4 | 0.40 | 0.08 |
| 17q21.32 | HOXB3 | Protein | −5.20 | 2.00×10−7 | 0.71 | 0.12 |
| 19p13.11 | ABHD8 | Protein | 4.42 | 9.93×10−6 | 0.59 | 0.23 |
Adjusting for the EOC risk SNPs in the corresponding locus.
Correlation between predicted and measured gene expression levels.
We integrated the results for the associations between DNA methylation and EOC risk, the correlation between DNA methylation and gene expression, and the association between gene expression and EOC risk. We identified consistent directions of associations across seven CpGs, including cg18878992, cg00480298, cg07368061, cg01572694, cg14285150, cg24672833 and cg17941109, three genes, including MAPT, HOXB3 and ABHD8, and EOC risk (Table 5). The mechanism potentially underlying the associations of methylation at these seven CpGs and EOC risk may be their regulatory function on expression of these three genes. Among them, increased methylation at the CpG site cg14285150 was associated with an increased HOXB3 expression (P=8.44×10−5) and decreased EOC risk (P=5.53×10−8). As expected, an increased expression of HOXB3 was associated with a decreased EOC risk (P=2.00×10−7). Conditioning on SNPs included in the methylation prediction model for cg14285150, the association of HOXB3 expression and EOC risk disappeared (P=0.51) (Table 5).
Table 5.
Consistent directions of associations across CpG methylation, gene expression and EOC risk for 12 CpGs and five genes
| CpG | Chr | Position | Gene | Classification | CpG Vs. EOC risk | CpG Vs. Gex b | Gex bVs. EOC risk | Adjusted a Gex Vs. EOC risk | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dir b | P value | Dir b | P value | Dir b | P value | Dir | P value | |||||
| cg18878992 | 17 | 43,974,344 | MAPT | 5'UTR | + | 8.85×10−13 | − | 2.64×10−3 | − | 3.74×10−4 | − | 0.48 |
| cg00480298 | 17 | 44,068,857 | MAPT | Exonic | + | 6.39×10−9 | − | 3.98×10−3 | − | 3.74×10−4 | − | 0.65 |
| cg07368061 | 17 | 44,090,862 | MAPT | Intronic | − | 4.26×10−13 | + | 2.02×10−3 | − | 3.74×10−4 | − | 1.00 |
| cg01572694 | 17 | 46,657,555 | HOXB3 | Intronic | − | 5.52×10−9 | + | 7.49×10−3 | − | 2.00×10−7 | − | 0.82 |
| cg14285150 | 17 | 46,659,019 | HOXB3 | Intronic | − | 5.53×10−8 | + | 8.44×10−5 | − | 2.00×10−7 | − | 0.51 |
| cg24672833 | 17 | 46,659,318 | HOXB3 | Intronic | − | 9.00×10−8 | + | 5.51×10−3 | − | 2.00×10−7 | − | 0.41 |
| cg17941109 | 19 | 17,407,198 | ABHD8 | Intronic | − | 2.88×10−9 | − | 0.03 | + | 9.93×10−6 | − | 0.57 |
| cg19139618 | 17 | 46,504,791 | SKAP1 | Intronic | − | 7.08×10−7 | − | 2.98×10−15 | + | NA c | ||
| cg02957270 | 17 | 46,508,097 | SKAP1 | TSS1500 | + | 4.40×10−12 | + | 0.01 | + | |||
| cg07067577 | 17 | 43,506,829 | ARHGAP27 | 3'UTR | − | 6.86×10−14 | − | 1.20×10−3 | + | NA c | ||
| cg16281322 | 17 | 43,510,478 | ARHGAP27 | TSS200 | − | 6.82×10−13 | − | 1.14×10−9 | + | |||
| cg25708777 | 17 | 43,510,841 | ARHGAP27 | TSS1500 | − | 4.61×10−13 | − | 4.11×10−8 | + | |||
Adjusting for all the predicting SNPs included in prediction models of corresponding CpGs
Dir, direction of association/correlation; Gex, gene expression
SKAP1 and ARHGAP27 are previously identified EOC-susceptibility genes.
Expression prediction models could not be built for SKAP1 at 17q21.32 and ARHGAP27 at 17q21.31 in the present study. Hence, these two genes could not be investigated in association with EOC risk. However, higher expression levels of these two genes have been previously reported to be associated with an increased risk of EOC (36,37). This is expected, based on the association results of DNA methylation with EOC risk and DNA methylation with gene expression (Table 5). For example, a higher methylation at cg19139618 was associated with a lower expression of SKAP1 (P=2.98×10−15) and lower EOC risk (P=7.08×10−7). Hence, the potential mechanism underlying the association between cg19139618 and EOC risk may be the down-regulation effects on SKAP1 expression (Table 5).
Discussion
In this large study, we identified 89 CpGs that were significantly associated with EOC risk, including two CpGs located in a novel genomic region that have not yet been reported as a susceptibility locus for EOC. Integrating genetic, methylation and gene expression data suggested that methylation at 12 of 89 CpGs may exert their impacts on EOC risk through regulating the expression of five genes. These results provide new insights into the regulatory pathways that connect genetics, epigenetics, gene expression and EOC risk.
We identified two methylation markers, cg18139273 and cg03634833, located at 7p22.3, a novel genomic region that had not been reported as a risk locus for EOC. Both CpGs reside in the 3rd intron of the 1st transcript of the ADAP1 gene, which encodes an ADP-ribosylation factor GTPase-activating protein (ArfGAP) with dual PH domains 1. ADAP1 functions as a scaffolding protein in several signal transduction pathways. It is highly expressed in neurons, where it has roles in neuronal differentiation and neurodegeneration (38). This gene has also been reported to be involved in mitochondrial function (39), and is a target of the ErbB4 transcription factor in mammary epithelial cells (40). In the present study, we found that a higher methylation level at cg03634833 was significantly correlated with a lower ADAP1 expression, which was associated with a non-significantly decreased EOC risk. Thus, methylation at cg03634833 might be associated with EOC risk through a regulatory function on ADAP1 expression, or through other unidentified mechanisms.
Integrating the results of the associations between DNA methylation and EOC risk, the correlation between DNA methylation and gene expression, and the association between gene expression and EOC risk, we observed consistent directions of associations across 12 CpGs, five genes and EOC risk. For the MAPT gene (17q21.31), an increased methylation at two CpGs located in its exons, cg18878992 and cg00480298, were associated with a decreased MAPT expression and increased EOC risk. For the other CpG site, cg07368061, located at the 1st intron of MAPT, its increased methylation was associated with a higher MAPT expression and lower EOC risk. As expected, an increased MAPT expression was associated with decreased EOC risk. The MAPT gene has been linked to multiple neurodegenerative disorders, including progressive supranuclear palsy (41), Parkinson’s disease (42,43) and Alzheimer’s disease (42). In addition, a higher expression of a MAPT protein isoform (<70 kDa) was correlated with a lower sensitivity to taxanes in breast cancer cells (44). Methylation of the microRNA (miRNA) miR-34c-5p was shown to regulate the MAPT expression, which was related to paclitaxel-resistance in gastric cancer cells (45).
Increased methylation of three CpGs in the 1st intron of the HOXB3 gene (17q21.32), cg01572694, cg14285150 and cg24672833, were associated with an increased expression of HOXB3 and decreased EOC risk. As expected, an increased HOXB3 expression was associated with decreased EOC risk. A previous study reported that the expression of HOXB3 was up-regulated in EOC cell lines compared with normal samples (46). However, this study only included five patients and the results have not been replicated by an independent study. On the other side, we investigated the genetically predicted methylation levels in DNA from white blood cells, but not in ovary or fallopian tube epithelial cells. It is possible that the correlation between methylation levels of these CpGs and HOXB3 expression are different in ovary epithelial cells and white blood cells. For example, in the 5’UTR of HOXB3, a higher methylation at the CpG cg12910797 was significantly associated with an increased EOC risk. The increased methylation of this CpG was not correlated with the expression of HOXB3 in white blood cells samples from the FHS (Spearman correlation coefficient r=−0.02; P=0.43). Higher methylation of this CpG was significantly correlated with a decreased HOXB3 expression in ovarian serous cystadenocarcinoma samples from the Cancer Genome Atlas (TCGA) (Spearman correlation coefficient r=−0.27; P=2.01×10−6) (http://gdac.broadinstitute.org/runs/analyses__2016_01_28/reports/cancer/OV-TP/Correlate_Methylation_vs_mRNA/nozzle.html).
The higher methylation of the CpG site cg17941109, located at the 2nd intron of the ABHD8 gene, was associated with a lower ABHD8 expression and a lower EOC risk. This is consistent with the results of two recent studies that showed that a higher expression level of this gene was associated with an increased risk of EOC (47,48). This gene is located at 19p13.11, a susceptibility locus for both ovarian and breast cancers. Interestingly, in our unpublished data, the increased genetically predicted methylation level at cg17941109 was associated with decreased breast cancer risk, and the genetically predicted expression of ABHD8 was associated with an increased breast cancer risk. Increasing evidence also suggests that this protein family (ABHD) has a physiological significance in metabolism and disease (49).
For the ARHGAP27 gene, increased methylation of two CpGs in the promoter region, cg16281322 and cg25708777, and one CpG in the 3’-UTR, cg07067577, were associated with lower expression level of ARHGAP27 and lower EOC risk. For the SKAP1 gene, a higher methylation at the CpG cg02957270, located at the promoter region, was associated with a higher expression level and increased EOC risk. Increased methylation of the other intronic CpG, cg19139618, was associated with a lower SKAP1 expression and a decreased EOC risk. In the present study, the associations of expression levels of these two genes and EOC risk could not be investigated because the prediction models for them could not be built. However, two large GWAS studies have identified these two genes as EOC susceptibility genes with solid experimental evidence (36,37). Differential expression analyses showed a significantly higher expression of ARHGAP27 in ovarian cancer than in normal cells (37). It is suggested that the ARHGAP27 gene may play a role in carcinogenesis through the dysregulation of Rho/Rac/Cdc42-like GTPases (50). The expression of SKAP1 was significantly greater in ovarian cancer cells when compared to primary human ovarian surface epithelial cells (36). Our study is the first to suggest that these two genes may be associated with EOC risk through methylation regulation.
Several epidemiological studies have investigated the associations of CpG methylation and EOC risk in white blood cells and tumor tissue samples (12-15). Approximately 100 CpGs have been identified to be associated with EOC risk. However, only two CpGs, cg10061138 and cg10636246, showed consistent association directions in two or more studies. In the present study, prediction models could not be built for these two CpGs; hence, neither could be investigated in association with EOC risk. Among the remaining 98 reported CpGs, reliable prediction models were built for only 20 of them and only two, cg19399532 and cg21870884, could be replicated at P<0.10, with the same association directions as previously reported. Such a low replication rate is not unexpected because of several potential limitations in traditional epidemiological studies, which include possible false associations because of small sample size, lack of validation in other studies, potential confounders and reverse causation.
The methodology of this study is similar to that of transcriptome-wide association studies (TWAS), in which gene expression prediction models are established and applied to GWAS data to investigate genetically predicted gene expression in association with various diseases and traits. Of the five genes identified in the present study, the expression levels of two, HOXB3 and ABHD8, were significantly associated with EOC risk at the Bonferroni-corrected threshold (P<2.2×10−6) in our previous TWAS study for EOC (51). The MAPT gene showed an association with EOC at P=3.74×10−4 in the TWAS, however the association didn’t reach the Bonferroni-corrected threshold. For ARHGAP27 and SKAP1, gene expression prediction models could not be built, and they were not investigated in the TWAS. Expression levels for these two genes were reported to be associated with EOC (36,37). Some genes identified in TWAS were not tested in the present study because the methylation prediction models could not be built for CpGs flanking them. In addition, except DNA methylation, there are other biological processes that regulate gene expression. The regulation of DNA methylation on gene expression differs according to the locations of the CpGs. Therefore, integrating the results of methylation and gene expression analyses may help to understand the biological basis for EOC.
It would be ideal to build methylation prediction models using data from normal ovary or fallopian tube epithelial cells, but it is almost impossible to collect tissue samples from a large population of healthy women. However, as demonstrated by multiple studies, the large majority of the meQTLs identified in white blood cells were consistently detected across different tissue types (26,52,53). These results indicate that the genetically determined methylation at many CpGs are predictable and consistent among different tissues. Hence, it is reasonable to build methylation prediction models using data from white blood cell samples and then investigate predicted DNA methylation in association with EOC. It would be ideal to validate the findings in the present study by directly measuring methylation levels in pre-diagnosis blood samples in prospective studies to overcome reverse causation; however, the majority of the samples included in the present study were collected after cancer diagnosis. It is possible that DNA methylation regulation on gene expression differs across tissues. In the present study, data in white blood cell samples were used, which is another limitation. In the association analysis of predicted gene expression with EOC risk, the models were built using data from a limited sample size of GTEx. Thus, the number of genes evaluated in our study was small. More consistent associations across methylation, gene expression and EOC risk could be identified with a larger sample size to build gene expression prediction models.
Strengths of this study include the large number of samples in the reference dataset used in model building and that the model performance was evaluated in an independent dataset. Using genetic variants as study instruments, we can effectively overcome many limitations commonly encountered in conventional epidemiologic studies. In addition, this is the largest study of DNA methylation with EOC risk and a very stringent criterion was used, providing high statistical power to identify reliable associations between genetically predicted methylation and EOC risk. Finally, the integrative analyses of genetic, DNA methylation and gene expression data led to the identification of consistent evidence to support the hypothesis that DNA methylation could impact EOC risk through regulating gene expression.
In summary, in the largest study conducted to date that investigates DNA methylation in association with EOC risk, we identified multiple CpGs that were significantly associated with EOC risk and proposed that several CpGs may affect EOC risk through regulating expression of five genes. Our study demonstrates the feasibility of integrating multi-omics data to identify novel biomarkers for EOC risk and brings new insight into the etiology of this malignancy.
Supplementary Material
Significance: Identification of novel DNA methylation markers associated with EOC risk suggests that methylation at multiple CpG may affect EOC risk through regulation of gene expression.
Acknowledgments
We thank Jing He and Marshal Younger of Vanderbilt Epidemiology Center for their help with this study. The authors wish to thank all of the individuals who took part in the study, and all of the researchers, clinicians, technicians, and administrative staff who have enabled this work to be carried out. Data for the FHS Offspring Cohort were obtained from dbGaP (accession numbers phs000724, phs000342, and phs000363). Data for the WHI were obtained from dbGaP (accession numbers phs001335, phs000675 and phs000315). The data analyses were conducted using the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University.
Acknowledgements for individual studies: AOV: We thank Jennifer Koziak, Mie Konno, Michelle Darago, Faye Chambers and the Tom Baker Cancer Centre Translational Laboratories; AUS: The AOCS also acknowledges the cooperation of the participating institutions in Australia and acknowledges the contribution of the study nurses, research assistants and all clinical and scientific collaborators to the study. The complete AOCS Study Group can be found at www.aocstudy.org. We would like to thank all of the women who participated in these research programs; BEL: We would like to thank Gilian Peuteman, Thomas Van Brussel, Annick Van den Broeck and Joke De Roover for technical assistance; BGS: The BGS is funded by Breast Cancer Now and the Institute of Cancer Research (ICR). ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. We thank the study staff, study participants, doctors, nurses, health care providers and health information sources who have contributed to the study; BVU: The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU, which is supported by institutional funding, the 1S10RR025141-01 instrumentation award, and by the Vanderbilt CTSA grant UL1TR000445 from NCATS/NIH; CAM: This work was supported by Cancer Research UK; the University of Cambridge; National Institute for Health Research Cambridge Biomedical Research Centre; CHA: Innovative Research Team in University (PCSIRT) in China (IRT1076); CHN: To thank all members of Department of Obstetrics and Gynaecology, Hebei Medical University, Fourth Hospital and Department of Molecular Biology, Hebei Medical University, Fourth Hospital; COE: Gynecologic Cancer Center of Excellence (W81XWH-11-2-0131); CON: The cooperation of the 32 Connecticut hospitals, including Stamford Hospital, in allowing patient access, is gratefully acknowledged. This study was approved by the State of Connecticut Department of Public Health Human Investigation Committee. Certain data used in this study were obtained from the Connecticut Tumor Registry in the Connecticut Department of Public Health. The authors assume full responsibility for analyses and interpretation of these data; DKE: OCRF; EPC: To thank all members and investigators of the Rotterdam Ovarian Cancer Study. Dutch Cancer Society (EMC 2014-6699); GER: The German Ovarian Cancer Study (GER) thank Ursula Eilber for competent technical assistance; HOC: The study was supported by the Helsinki University Research Fund; JGO: JSPS KAKENHI grant; KRA: This study (Ko-EVE) was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), and the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (HI16C1127; 0920010); LUN: ERC −2011-AdG, Swedish Cancer Society, Swedish Research Council; MAS: We would like to thank Famida Zulkifli and Ms Moey for assistance in patient recruitment, data collection and sample preparation. The Malaysian Ovarian Cancer Genetic Study is funded by research grants from the Malaysian Ministry of Higher Education (UM.C/HIR/MOHE/06) and charitable funding from Cancer Research Initiatives Foundation; MCC: MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database; MOF: the Total Cancer Care™ Protocol and the Collaborative Data Services and Tissue Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292), Merck Pharmaceuticals and the state of Florida; NHS: The NHS/NHSII studies thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY; OPL: Members of the OPAL Study Group (http://opalstudy.qimrberghofer.edu.au/); RPC: National Institutes of Health (P50 CA159981, R01CA126841); SEA: SEARCH team, Craig Luccarini, Caroline Baynes, Don Conroy; SIS: The Sister Study (SISTER) is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES044005 and Z01-ES049033); SON: National Health Research and Development Program, Health Canada, grant 6613-1415-53; SRO: To thank all members of Scottish Gynaecological Clinical Trails group and SCOTROC1 investigators; SWE: Swedish Cancer foundation, WeCanCureCancer and årKampMotCancer foundation; SWH: The SWHS is supported primarily by NIH grant R37-CA070867. We thank the participants and the research staff of the Shanghai Women’s Health Study for making this study possible; UCI: The UCI Ovarian cancer study is supported by the National Institutes of Health, National Cancer Institute grants CA58860, and the Lon V Smith Foundation grant LVS-39420; UHN: Princess Margaret Cancer Centre Foundation-Bridge for the Cure; UKO: We particularly thank I. Jacobs, M.Widschwendter, E. Wozniak, A. Ryan, J. Ford and N. Balogun for their contribution to the study; UKR: Carole Pye; VAN: BC Cancer Foundation, VGH & UBC Hospital Foundation; WMH: We thank the Gynaecological Oncology Biobank at Westmead, a member of the Australasian Biospecimen Network-Oncology group, which is funded by the National Health and Medical Research Council Enabling Grants ID 310670 & ID 628903 and the Cancer Institute NSW Grants 12/RIG/1-17 & 15/RIG/1-16.
Financial support
This project was partially supported by the development fund from the Department of Medicine at Vanderbilt University Medical Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). The scientific development and funding for this project were, in part, supported by the US National Cancer Institute GAME-ON Post-GWAS Initiative (U19-CA148112). This study made use of data generated by the Wellcome Trust Case Control consortium, funded by the Wellcome Trust under award 076113. The results published here are, in part, based upon data generated by The Cancer Genome Atlas Pilot Project, established by the National Cancer Institute and National Human Genome Research Institute (dbGap accession number phs000178.v8.p7).
The OCAC OncoArray genotyping project was funded through grants from the U.S. National Institutes of Health (U19-CA148112 (T.A. Sellers), R01-CA149429 (C.M. Phelan) and R01-CA058598 (M.T. Goodman); Canadian Institutes of Health Research (MOP-86727 (L.E. Kelemen) and the Ovarian Cancer Research Fund (A. Berchuck). The COGS project was funded through a European Commission’s Seventh Framework Programme grant (agreement number 223175 - HEALTH-F2-2009-223175).
Funding for individual studies: AAS: National Institutes of Health (RO1-CA142081); AOV: The Canadian Institutes for Health Research (MOP-86727); AUS: The Australian Ovarian Cancer Study Group was supported by the U.S. Army Medical Research and Materiel Command (DAMD17-01-1-0729), National Health & Medical Research Council of Australia (199600, 400413 and 400281), Cancer Councils of New South Wales, Victoria, Queensland, South Australia and Tasmania and Cancer Foundation of Western Australia (Multi-State Applications 191, 211 and 182). The Australian Ovarian Cancer Study gratefully acknowledges additional support from Ovarian Cancer Australia and the Peter MacCallum Foundation; BAV: ELAN Funds of the University of Erlangen-Nuremberg; BEL: National Kankerplan; BGS: Breast Cancer Now, Institute of Cancer Research; the ICR acknowledges NHS funding to the NIHR Biomedical Research Centre; BVU: Vanderbilt CTSA grant from the National Institutes of Health (NIH)/National Center for Advancing Translational Sciences (NCATS) (ULTR000445); CAM: National Institutes of Health Research Cambridge Biomedical Research Centre and Cancer Research UK Cambridge Cancer Centre; CHA: Innovative Research Team at the University (PCSIRT) in China (IRT1076); CNI: Instituto de Salud Carlos III (PI 12/01319); Ministerio de Economía y Competitividad (SAF2012); COE: Department of Defense (W81XWH-11-2-0131); CON: National Institutes of Health (R01-CA063678, R01-CA074850; R01-CA080742); DKE: Ovarian Cancer Research Fund; DOV: National Institutes of Health R01-CA112523 and R01-CA87538; EMC: Dutch Cancer Society (EMC 2014-6699); EPC: The coordination of EPIC is financially supported by the European Commission (DG-SANCO) and the International Agency for Research on Cancer. The national cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF) (Germany); the Hellenic Health Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPIC-Oxford) (United Kingdom); GER: German Federal Ministry of Education and Research, Programme of Clinical Biomedical Research (01 GB 9401) and the German Cancer Research Center (DKFZ); GRC: This research has been co-financed by the European Union (European Social Fund - ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) - Research Funding Program of the General Secretariat for Research & Technology: SYN11_10_19 NBCA. Investing in knowledge society through the European Social Fund; GRR: Roswell Park Cancer Institute Alliance Foundation, P30 CA016056; HAW: U.S. National Institutes of Health (R01-CA58598, N01-CN-55424 and N01-PC-67001); HJO: Intramural funding; Rudolf-Bartling Foundation; HMO: Intramural funding; Rudolf-Bartling Foundation; HOC: Helsinki University Research Fund; HOP: Department of Defense (DAMD17-02-1-0669) and NCI (K07-CA080668, R01-CA95023, P50-CA159981 MO1-RR000056 R01-CA126841); HUO: Intramural funding; Rudolf-Bartling Foundation; JGO: JSPS KAKENHI grant; JPN: Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labour and Welfare; KRA: This study (Ko-EVE) was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), and the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (HI16C1127; 0920010); LAX: American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124; LUN: ERC-2011-AdG 294576-risk factors cancer, Swedish Cancer Society, Swedish Research Council, Beta Kamprad Foundation; MAC: National Institutes of Health (R01-CA122443, P30-CA15083, P50-CA136393); Mayo Foundation; Minnesota Ovarian Cancer Alliance; Fred C. and Katherine B. Andersen Foundation; Fraternal Order of Eagles; MAL: Funding for this study was provided by research grant R01- CA61107 from the National Cancer Institute, Bethesda, MD, research grant 94 222 52 from the Danish Cancer Society, Copenhagen, Denmark; and the Mermaid I project; MAS: Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation; MAY: National Institutes of Health (R01-CA122443, P30-CA15083, P50-CA136393); Mayo Foundation; Minnesota Ovarian Cancer Alliance; Fred C. and Katherine B. Andersen Foundation; MCC: Cancer Council Victoria, National Health and Medical Research Council of Australia (NHMRC) grants number 209057, 251533, 396414, and 504715; MDA: DOD Ovarian Cancer Research Program (W81XWH-07-0449); MEC: NIH (CA54281, CA164973, CA63464); MOF: Moffitt Cancer Center, Merck Pharmaceuticals, the state of Florida, Hillsborough County, and the city of Tampa; NCO: National Institutes of Health (R01-CA76016) and the Department of Defense (DAMD17-02-1-0666); NEC: National Institutes of Health R01-CA54419 and P50-CA105009 and Department of Defense W81XWH-10-1-02802; NHS: UM1 CA186107, P01 CA87969, R01 CA49449, R01-CA67262, UM1 CA176726; NJO: National Cancer Institute (NIH-K07 CA095666, R01-CA83918, NIH-K22-CA138563, and P30-CA072720) and the Cancer Institute of New Jersey; NOR: Helse Vest, The Norwegian Cancer Society, The Research Council of Norway; NTH: Radboud University Medical Centre; OPL: National Health and Medical Research Council (NHMRC) of Australia (APP1025142) and Brisbane Women’s Club; ORE: OHSU Foundation; OVA: This work was supported by Canadian Institutes of Health Research grant (MOP-86727) and by NIH/NCI 1 R01CA160669-01A1; PLC: Intramural Research Program of the National Cancer Institute; POC: Pomeranian Medical University; POL: Intramural Research Program of the National Cancer Institute; PVD: Canadian Cancer Society and Cancer Research Society GRePEC Program; RBH: National Health and Medical Research Council of Australia; RMH: Cancer Research UK, Royal Marsden Hospital; RPC: National Institute of Health (P50 CA159981, R01CA126841); SEA: Cancer Research UK (C490/A10119 C490/A10124); UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge; SIS: NIH, National Institute of Environmental Health Sciences, Z01 ES044005 and Z01-ES049033; SMC: The Swedish Research Council; SON: National Health Research and Development Program, Health Canada, grant 6613-1415-53; SRO: Cancer Research UK (C536/A13086, C536/A6689) and Imperial Experimental Cancer Research Centre (C1312/A15589); STA: NIH grants U01 CA71966 and U01 CA69417; SWE: Swedish Cancer foundation, WeCanCureCancer and årKampMotCancer foundation; SWH: NIH (NCI) grant R37-CA070867; TBO: National Institutes of Health (R01-CA106414-A2), American Cancer Society (CRTG-00-196-01-CCE), Department of Defense (DAMD17-98-1-8659), Celma Mastery Ovarian Cancer Foundation; TOR: NIH grants R01 CA063678 and R01 CA063682; UCI: NIH R01-CA058860 and the Lon V Smith Foundation grant LVS-39420; UHN: Princess Margaret Cancer Centre Foundation-Bridge for the Cure; UKO: The UKOPS study was funded by The Eve Appeal (The Oak Foundation) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre; UKR: Cancer Research UK (C490/A6187), UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge; USC: P01CA17054, P30CA14089, R01CA61132, N01PC67010, R03CA113148, R03CA115195, N01CN025403, and California Cancer Research Program (00-01389V-20170, 2II0200); VAN: BC Cancer Foundation, VGH & UBC Hospital Foundation; VTL: NIH K05-CA154337; WMH: National Health and Medical Research Council of Australia, Enabling Grants ID 310670 & ID 628903. Cancer Institute NSW Grants 12/RIG/1-17 & 15/RIG/1-16; WOC: National Science Centren (N N301 5645 40) The Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology, Warsaw, Poland.
We are grateful to the family and friends of Kathryn Sladek Smith for their generous support of the Ovarian Cancer Association Consortium through their donations to the Ovarian Cancer Research Fund. The OncoArray and COGS genotyping projects would not have been possible without the contributions of the following: Per Hall (COGS); Douglas F. Easton, Kyriaki Michailidou, Manjeet K. Bolla, Qin Wang (BCAC), Marjorie J. Riggan (OCAC), Rosalind A. Eeles, Douglas F. Easton, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Antonis Antoniou, Lesley McGuffog, Fergus Couch and Ken Offit (CIMBA), Alison M. Dunning, Andrew Lee, and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Anna Gonzalez-Neira and the staff of the CNIO genotyping unit, Jacques Simard and Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, Stig E. Bojesen, Sune F. Nielsen, Borge G. Nordestgaard, and the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility. We pay special tribute to the contribution of Professor Brian Henderson to the GAME-ON consortium and to Olga M. Sinilnikova for her contribution to CIMBA and for her part in the initiation and coordination of GEMO until she sadly passed away on the 30th June 2014. We thank the study participants, doctors, nurses, clinical and scientific collaborators, health care providers and health information sources who have contributed to the many studies contributing to this manuscript.
Footnotes
Conflict of interest
The authors declare no potential conflicts of interest.
References
- 1.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA: a cancer journal for clinicians 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. The Lancet 2014;384:1376–88 [DOI] [PubMed] [Google Scholar]
- 3.Phelan CM, Kuchenbaecker KB, Tyrer JP, Kar SP, Lawrenson K, Winham SJ, et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nature genetics 2017;49:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarkar S, Horn G, Moulton K, Oza A, Byler S, Kokolus S, et al. Cancer development, progression, and therapy: an epigenetic overview. International journal of molecular sciences 2013;14:21087–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome biology 2011;12:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer research 2016;76:3446–50 [DOI] [PubMed] [Google Scholar]
- 8.Earp MA, Cunningham JM. DNA methylation changes in epithelial ovarian cancer histotypes. Genomics 2015;106:311–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koukoura O, Spandidos DA, Daponte A, Sifakis S. DNA methylation profiles in ovarian cancer: implication in diagnosis and therapy. Molecular medicine reports 2014;10:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan KY, Ozçelik H, Cheung AN, Ngan HY, Khoo U-S. Epigenetic factors controlling the BRCA1 and BRCA2 genes in sporadic ovarian cancer. Cancer research 2002;62:4151–6 [PubMed] [Google Scholar]
- 11.Widschwendter M, Jiang G, Woods C, Müller HM, Fiegl H, Goebel G, et al. DNA hypomethylation and ovarian cancer biology. Cancer research 2004;64:4472–80 [DOI] [PubMed] [Google Scholar]
- 12.Koestler DC, Chalise P, Cicek MS, Cunningham JM, Armasu S, Larson MC, et al. Integrative genomic analysis identifies epigenetic marks that mediate genetic risk for epithelial ovarian cancer. BMC medical genomics 2014;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridley BL, Armasu SM, Cicek MS, Larson MC, Wang C, Winham SJ, et al. Methylation of leukocyte DNA and ovarian cancer: relationships with disease status and outcome. BMC medical genomics 2014;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winham SJ, Armasu SM, Cicek MS, Larson MC, Cunningham JM, Kalli KR, et al. Genome‐Wide Investigation of Regional Blood‐Based DNA Methylation Adjusted for Complete Blood Counts Implicates BNC2 in Ovarian Cancer. Genetic epidemiology 2014;38:457–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D, Yang H, Winham SJ, Natanzon Y, Koestler DC, Luo T, et al. Mediation analysis of alcohol consumption, DNA methylation, and epithelial ovarian cancer. Journal of human genetics 2018:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundberg E, Meduri E, Sandling JK, Hedman ÅK, Keildson S, Buil A, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. The American Journal of Human Genetics 2013;93:876–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McRae AF, Powell JE, Henders AK, Bowdler L, Hemani G, Shah S, et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome biology 2014;15:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, et al. Systematic identification of genetic influences on methylation across the human life course. Genome biology 2016;17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McRae A, Marioni RE, Shah S, Yang J, Powell JE, Harris SE, et al. Identification of 55,000 replicated DNA methylation QTL. bioRxiv 2017:166710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson TG, Zheng J, Smith GD, Timpson NJ, Gaunt TR, Relton CL, et al. Mendelian randomization analysis identifies CpG sites as putative mediators for genetic influences on cardiovascular disease risk. The American Journal of Human Genetics 2017;101:590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson TG, Haycock PC, Zheng J, Timpson NJ, Gaunt TR, Davey Smith G, et al. Systematic Mendelian randomization framework elucidates hundreds of CpG sites which may mediate the influence of genetic variants on disease. Human molecular genetics 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families: the Framingham Offspring Study. American journal of epidemiology 1979;110:281–90 [DOI] [PubMed] [Google Scholar]
- 23.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman J, Hastie T, Tibshirani R. glmnet: Lasso and elastic-net regularized generalized linear models. R package version 2009;1 [Google Scholar]
- 25.Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research 2001;29:308–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nature communications 2018;9:1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Shi W, Long J, Guo X, Michailidou K, Beesley J, et al. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nature genetics 2018:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. Genome-wide genetic data on~ 500,000 UK Biobank participants. bioRxiv 2017:166298 [Google Scholar]
- 29.Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature genetics 2012;44:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research 2010;38:e164–e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin H, Yin X, Xie Z, Lunetta KL, Lubitz SA, Larson MG, et al. Methylome-wide association study of atrial fibrillation in Framingham Heart Study. Scientific reports 2017;7:40377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.da Cunha Colombo Bonadio RR, Fogace RN, Miranda VC, Diz MdPE. Homologous recombination deficiency in ovarian cancer: a review of its epidemiology and management. Clinics 2018;73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frey MK, Pothuri B. Homologous recombination deficiency (HRD) testing in ovarian cancer clinical practice: a review of the literature. Gynecologic oncology research and practice 2017;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium G. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015;348:648–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature genetics 2012;44:369–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nature genetics 2010;42:874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Permuth-Wey J, Lawrenson K, Shen HC, Velkova A, Tyrer JP, Chen Z, et al. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21. 31. Nature communications 2013;4:1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stricker R, Reiser G. Functions of the neuron-specific protein ADAP1 (centaurin-α1) in neuronal differentiation and neurodegenerative diseases, with an overview of structural and biochemical properties of ADAP1. Biological chemistry 2014;395:1321–40 [DOI] [PubMed] [Google Scholar]
- 39.Galvita A, Grachev D, Azarashvili T, Baburina Y, Krestinina O, Stricker R, et al. The brain‐specific protein, p42IP4 (ADAP 1) is localized in mitochondria and involved in regulation of mitochondrial Ca2+. Journal of neurochemistry 2009;109:1701–13 [DOI] [PubMed] [Google Scholar]
- 40.Wali VB, Haskins JW, Gilmore-Hebert M, Platt JT, Liu Z, Stern DF. Convergent and divergent cellular responses by ErbB4 isoforms in mammary epithelial cells. Mol Cancer Res 2014:molcanres. 06372013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borroni B, Agosti C, Magnani E, Di Luca M, Padovani A. Genetic bases of Progressive Supranuclear Palsy: the MAPT tau disease. Current medicinal chemistry 2011;18:2655–60 [DOI] [PubMed] [Google Scholar]
- 42.Desikan RS, Schork AJ, Wang Y, Witoelar A, Sharma M, McEvoy LK, et al. Genetic overlap between Alzheimer’s disease and Parkinson’s disease at the MAPT locus. Molecular psychiatry 2015;20:1588–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Mullersman J, Liu X. Family-based association analysis of theMAPT gene in Parkinson. Journal of applied genetics 2010;51:509–14 [DOI] [PubMed] [Google Scholar]
- 44.Ikeda H, Taira N, Hara F, Fujita T, Yamamoto H, Soh J, et al. The estrogen receptor influences microtubule-associated protein tau (MAPT) expression and the selective estrogen receptor inhibitor fulvestrant downregulates MAPT and increases the sensitivity to taxane in breast cancer cells. Breast Cancer Research 2010;12:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu H, Huang M, Lu M, Zhu W, Shu Y, Cao P, et al. Regulation of microtubule-associated protein tau (MAPT) by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel. Cancer chemotherapy and pharmacology 2013;71:1159–71 [DOI] [PubMed] [Google Scholar]
- 46.Yamashita T, Tazawa S, Yawei Z, Katayama H, Kato Y, Nishiwaki K, et al. Suppression of invasive characteristics by antisense introduction of overexpressed HOX genes in ovarian cancer cells. International journal of oncology 2006;28:931–8 [PubMed] [Google Scholar]
- 47.Lawrenson K, Kar S, McCue K, Kuchenbaeker K, Michailidou K, Tyrer J, et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13. 1 breast–ovarian cancer susceptibility locus. Nature communications 2016;7:12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kar SP, Tyrer JP, Li Q, Lawrenson K, Aben KK, Anton-Culver H, et al. Network-based integration of GWAS and gene expression identifies a HOX-centric network associated with serous ovarian cancer risk. Cancer Epidemiology and Prevention Biomarkers 2015;24:1574–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lord CC, Thomas G, Brown JM. Mammalian alpha beta hydrolase domain (ABHD) proteins: lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids 2013;1831:792–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh Y, Katoh M. Identification and characterization of ARHGAP27 gene in silico. International journal of molecular medicine 2004;14:943–7 [PubMed] [Google Scholar]
- 51.Lu Y, Beeghly-Fadiel A, Wu L, Guo X, Li B, Schildkraut JM, et al. A transcriptome-wide association study among 97,898 women to identify candidate susceptibility genes for epithelial ovarian cancer risk. Cancer research 2018;78:5419–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stueve TR, Li W-Q, Shi J, Marconett CN, Zhang T, Yang C, et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Human molecular genetics 2017;26:3014–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hannon E, Weedon M, Bray N, O’Donovan M, Mill J. Pleiotropic effects of trait-associated genetic variation on DNA methylation: utility for refining GWAS loci. The American Journal of Human Genetics 2017;100:954–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.