Abstract
The faithful inheritance of the epigenome is critical for cells to maintain gene expression programs and cellular identity across cell divisions. We mapped strand-specific DNA methylation after replication forks, and show maintenance of the vast majority of the DNA methylome and inheritance of some hemi-methylated CpGs (hemiCpGs) within 20 minutes of replication. Mapping the nascent DNA methylome targeted by each of the three DNA methyltransferases (DNMTs) reveals interactions between DNMTs and substrate daughter Cs en route to maintenance methylation or hemi-methylation. Finally, we show the inheritance of hemiCpGs at short regions flanking CTCF/cohesin binding sites in pluripotent cells. Elimination ofhemi-methylation causes reduced frequency of chromatin interactions emanating from these sites, suggesting a role for hemi-methylation as a stable epigenetic mark regulating CTCF-mediated chromatin interactions.
One Sentence Summary:
Stably inherited hemi-methylation surrounding CTCF motifs is required for CTCF-mediated chromatin interactions.
Cytosine DNA methylation in mammals is maintained mainly by the canonical maintenance methyltransferase DNMT1 during each cell cycle (1, 2). By interacting with PCNA and UHRF1 during DNA replication, DNMT1 is recruited to replication foci and loaded onto hemiCpGs to methylate the nascent cytosines (Cs) (3–5). While the onset of this process is closely coupled with the entry into S phase, the kinetics of maintenance methylation and the content of the nascent DNA methylome have never been studied quantitatively in a genome-wide scale (6). Furthermore, although various biochemical (7, 8) and genetic perturbation experiments (6, 9–12) have strongly suggested the involvement of the de novo methyltransferase DNMT3A/3B in maintenance methylation, direct evidence of in vivo interaction between DNMTs and hemiCpGs is missing.
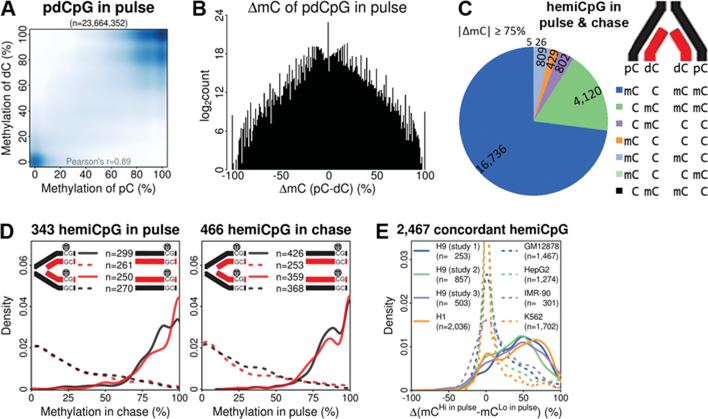
To gain insights into these key aspects of maintenance methylation, we used nascent DNA Bisulfite sequencing (nasBS-seq) to measure cytosine methylation frequency strand-specifically on nascent chromatin across the genome (fig. S1,A and B, and Methods). We first labeled H9 human embryonic stem cells (H9-hESCs) with the nucleotide analog EdU for 20 min as a pulse condition. Libraries for a chase condition were also made by labeling the cells with EdU for 20 min and growing them for another 8hrin the absence of EdU to monitor the maintenance methylation at a later time point within the cell cycle. We obtained 357–544 million uniquely mapped and deduplicated alignments from libraries for each strand, covering 5.6–9.8 billion Cs (fig. S2, A and B), converging to 24 million parent-daughter CpG dyads (pdCpGs) from either pulse or chase (fig. S2C). The methylation frequency was highly reproducible between replicates for each library (fig. S2D). Methylation frequency between parental Cs (pC) and daughter Cs (dC) in the same pdCpGs (Fig. 1A and fig. S3A), and between the same Cs in pulse and chase (fig. S3B), were highly correlated. Although Cs in the context of CH are not symmetrically methylated in mammals (13), their methylation was also maintained on the dCs in the other nascent DNA duplex (fig. S3C). The methylation frequency of dCs was globally maintained in both pulse and chase irrespective of the nature of the genomic features investigated (fig. S3D). These results suggest that the vast majority of the DNA methylome is maintained within 20 min after passage of replication forks and thereafter.
Fig. 1. The vast majority of the DNA methylome is maintained 20 min after replication.
(A) Correlation of methylation frequency between pCs and dCs within the same pdCpGs in pulse. (B) Count of pdCpGs in pulse with differential ΔmC values. (C) Different types of hemiCpGs with all 4 Cs mapped at least 4 times. (D) For concordant hemiCpGs in pulse, the distribution of methylation frequency of4Csin chase is shown (left), and vice versa (right). (E) All concordant hemiCpGs were intersected with WGBS datasets from other human cells. The distribution of ΔmC values is shown for each dataset.
Despite the high correlation of methylation frequency between pC and dC, the two Cs in many CpGs showed differential methylation frequency (Fig. 1B and fig. S4A), suggesting the existence of hemiCpGs with a spectrum of frequencies (fig. S4B). The strand-specific nascent DNA methylome enables the resolution of different types of hemiCpGs with respect to the parent-daughter axis. Using a highly stringent cutoff (ΔmC≤−75%, or ≥75%, ΔmC=m(pC-dC)), we obtained a list of 23,305 CpGs with at least one dyad showing hemi-methylation either in pulse or chase (Fig. 1C). The vast majority (96%) of them were hemi-methylated in only one dyad, and failed to reproduce the methylation pattern in the other condition (fig. S4, C to F), suggesting they may represent the rapid DNA methylation turnover events abundant in pluripotent cells (14). In contrast, the methylation pattern of CpGs hemi-methylated in both dyads in a concordant way (Cs on either two Watson or two Crick strands are methylated.) was highly consistent between pulse and chase (Fig. 1D), suggesting that concordant hemiCpGs were stably inherited through S phase. By concatenating the data from pulse and chase (Methods), we expanded the category of concordant hemiCpGs to include 2,467 CpGs, and confirmed their stable inheritance across up to 6 passages (>12 cell divisions) (fig. S4, G and H). The ΔmC values of these CpGs were compared with several whole-genome BS-seq (WGBS) datasets in various human cells. Surprisingly, the majority of them are conserved in other pluripotent cells, but are absent in non-pluripotent cells (Fig. 1E), suggesting hemi-methylation could be cell type-specific and well conserved across related cell lineages.
WGBS only reports the independent methylation frequency of two Cs in the same CpGs. To obtain methylation status of CpGs per se, we developed a computational method called in silico Strand Annealing (iSA) to resolve the nasBS-seq data and identify pairs of alignments sharing exactly the same two ends between strands of parentWatson and daughterCnck, and between daughterWatson and parentCnck (Fig. 2A and Methods). We employed a “moving-ends” statistical test to justify that most oi these pairs (26–111 -fold enrichment over random pairing) represent distinct nascent dsDNA fragments (fig. S5A). iSA enabled us to call intra-molecule CpGs (intraCpGs) from single dsDNA fragments and to determine their methylation state to be one of four types: methylation (intraCpGme), unmethylation (intraCpGunme), or pC- or dC- hemi-methylation (intraCpGhemi-pC or intraCpGhemi-dc). Approximately 4.5 and 2.1 million intraCpGs were called from all replicates in pulse and chase, respectively (Fig. 2B). The two conditions showed nearly identical fractions for all four types, including a surprisingly high and consistent 14% combined fraction of intraCpGhemi. Next, we used iSA to resolve published WGBS datasets in mouse early embryonic stages (15), and showed that hemiCpGs account for 4–18% of the DNA methylome (Fig. 2C), and is relatively depleted at TSSs (Fig. 2D). Notably, inner cell mass cells have the highest frequency of hemiCpGs on gene bodies, where it correlates slightly with transcription level, although it anti-correlates with transcription at promoters (fig. S5B), suggesting a pleiotropic role of hemiCpGs on gene expression. iSA was also used to resolve WGBS and TAB-seq datasets in Hl-hESCs (fig. S5C) (16), and showed that although 5hmC preferentially exists in hemi-methylated form (fig. S5D), the vast majority of hemiCpGs discovered by nasBS-seq/WGBS is contributed by 5mC (fig. S5E). The ΔmC values from WGBS highly correlate with the frequency of hemiCpGs resolved by iSA (fig. S5F), suggesting ΔmC values from WGBS can serve as proxy for frequency of hemi-methylation.
Fig. 2. HemiCpG is a significant component of the DNA methylome.
(A) Schematic representation of the principles underlying the iSA method. (B) The fraction of all 4 types of intraCpGs in pulse and chase. (C) The frequency of 3 types of intraCpGs (with 2 types of intraCpGshemi combined) at different mouse embryonic stages. ICM: inner cell mass. MEF: mouse embryonic fibroblast. mPGC/fPGC: male/female primordial germ cell. (D) The frequency of 3 types of intraCpGs at genic regions at different mouse embryonic stages.
The use of 20 min EdU labeling achieved a synchronization of genomic fragments by their replicative “age” of l0 min on average (0–20 min after passage of the local replication fork). The same frequency of hemiCpGs in pulse and chase suggests that the maintenance methylation reaction happens in a sub-minute scale (a one-minute-long methylation reaction would result in the pulse sample to have 10% more hemiCpGs than chase), preventing nasBS-seq from revealing the rich plethora of pC-methylated hemiCpGs en route to maintenance methylation. We thus postulated that an enrichment of binding events between DNMTs and nascent chromatin would achieve both spatial and temporal enrichment of such transient interactions, and would help identify cognate substrate CpGs maintained by a certain DNMT. Hence, we developed nasChIP-BS-seq to specifically map the nascent DNA methylome targeted by DNMT1, DNMT3A, or DNMT3B in both pulse and chase (fig. S6, A and B). Surprisingly, in pulse but not in chase, all three DNMT-targeted methylomes of the two daughter strands showed incomplete methylation, which was most significant at centers of alignments, indicative of the precise location of DNMTs (fig. S6C). Analysis of the data using iSA revealed that approximately 42%, 46% and 44% of all DNMT1-, DNMT3A- and DNMT3B-targeted CpGs in pulse were intraCpGhemi-pC, respectively, while the same category only contributed 7%, 6% and 5% in chase, respectively (Fig. 3A and fig. S6D). Furthermore, binding sites for all three DNMTs showed an enrichment of CpGs over flanking sequences in pulse (fig. S6E). In chase, the enrichment diminished for DNMT1 and DNMT3B, while DNMT3A showed an enrichment of methylated CpGs (fig. S6E), suggesting DNMTs may have differential occupancy preferences on nascent and mature chromatin (17, 18). When viewed through the pulse nasBS-seq, the vast majority of DNMT-targeted intraCpGhemi”pC were fully methylated (Fig. 3B), suggesting they were methylated shortly after the binding events. In DNMT1 knockout (KO) cells (12), both DNMTl-targeted intraCpGhemi”pC and intraCpGme showed higher than average reduction of methylation, suggesting their methylation state is maintained by DNMT1 (Fig. 3C). Under DNMT3A/3B double KO (12), both DNMT3A- and 3B-targeted intraCpGhemi”pC showed significantly higher reduction of methylation than the targeted intraCpGme (Fig. 3C), suggesting these intraCpGhemi”pC are better candidate for DNMT3-maintained CpG than the targeted intraCpG™6. Indeed, these two types of CpGs showed mutually exclusive distribution, suggesting they are subject to different regulation (fig. S6F). We next asked if nasChIP-BS-seq can also capture the substrate state of dCs en route to de novo methylation by examining the methylation state of targeted dCs in either inherited hemiCpGs or maintained CH methylation. In both cases, the yet-to-be methylated dCs showed extensive hypomethylation in DNMT3A/3B nasChIP-BS-seq but not in nasBS-seq (Fig. 3D and fig. S6G). These results suggest that nasChIP-BS-seq can visualize the transient interactions between a certain DNMT and substrate dCs in both maintenance and de novo methylation (fig. S6H).
Fig. 3. Transient interactions between DNMTs and substrate dCs in both maintenance and de novo methylation.
(A) The fraction of all 4 types ofDNMT- targeted intraCpGs in pulse and chase. (B) Counts of all DNMT-targeted intraCpGshemi-pC in pulse allocated to the appropriate cells according to their methylation frequency in pulse nasBS-seq. (C) Reduction of methylation under DNMT1 KO (24 hr) or DNMT3A/3B double KO (late) is shown for all CpGs, DNMT-targeted intraCpGshemi-pC and intraCpGsme in pulse and chase, and unmapped CpGs. ***, p<0.001. NS, not significant. (D) Distribution of methylation frequency of the 4 Cs in concordant hemiCpGs viewed through nasBS-seq and DNMT3A/3B nasChIP-BS-seq. The asterisks mark the 2 Cs inspected in each panel.
To identify chromatin features associated with hemiCpGs, we examined the frequency of different types of intraCpG at different genomic features inHl-hESC. Surprisingly, CTCF binding sites showed a very high ratio of intraCpGhemi over intraCpGme (fig. S7A). CTCF/cohesin binding sites orchestrate 3-D chromatin interactions across the mammalian genome (19). We then developed nasChIP-seq to map the binding landscape of CTCF and SMC1A (a cohesin subunit) on nascent chromatin in H9-hESC (fig. S7,BtoE). By examining the average methylation profiles of the strands harboring the CTCF motif and the opposite strands around oriented CTCF motifs, we found two short regions flanking occupied CTCF motifs exhibiting an apparent spectrum of ΔmC with opposing orientation on the two sides (Fig. S8A). The same pattern exists in Hl- hESC, naive H9-hESC, mouse ESC (mESC) (fig. S8A), and mouse embryos as early as the 8-cell stage (fig. S8B). Two independent methods, iSA and ChlP-hairpinBS-seq, confirmed at the single-molecule level that this spectrum of ΔmC indeed reflects an enrichment of hemiCpGs (Fig. 4A, fig. S8, C and D). The flanking hemiCpGs adopt a conformation of rotational symmetry with respect to CTCF motifs (fig. S8E), enabling us to search for the same pattern by screening the published ChIP-seq datasets of 60 chromatin-binding proteins in Hl-hESC (20). This pattern is only exhibited by sites co-occupied by CTCF and RAD21 (a cohesin subunit) (fig. S8F). We also determined that this pattern is contributed by 5mC more than by 5hmC (fig. S8G) (16). Next, we built a Hemi Index (HI) to quantitatively rank all CTCF motifs by the degree they associate with this pattem (Methods). Surprisingly, the CTCF motifs from the two nascent DNA duplexes are highly concordant in HI (Fig. 4B), suggesting these hemiCpGs were inherited during DNA replication. To confirm this, the methylation frequency was compared between the two dyads in pulse, and between the same dyads in pulse and chase. The inheritance of ΔmC was observed in both cases, only from the enriched type of hemiCpGs (Fig. 4C). We also confirmed the inheritance at the level of CTCF motifs by comparing their HI between 1) two dyads, 2) pulse and chase, 3) two cell populations with >10 cell divisions apart (Fig. 4D).
Fig. 4. Inherited hemiCpGs flanking CTCF/cohesin sites may regulate chromatin interactions.
(A) Frequency of motif or opposite strand-methylated (same me or oppo me) intraCpGshemi around oriented CTCF motifs co-occupied by CTCF/SMC1A from the two nascent DNA duplexes. Frequency of hairpinCpGhemi from CTCF ChIP-hairpinBS-seq is also shown. (B) All CTCF motifs co-occupied by CTCF/SMC1A in pulse were ranked by their Hemi Index. ΔmC of CpGs from the two nascent DNA duplexes, and reads per million (RPM) for CTCF and SMC1A nasChIP-seq within al kb window surrounding the motifs are shown. Black in the ΔmC heatmaps represents missing data points. (C) All hemiCpGs (ΔmC≥67% or ≤−67%) from two flanking regions in (B) were retrieved. Methylation frequency of the 2 Cs in the other dyad in pulse (left) or the same dyad in chase (right) are shown. ***,_p<0.001. NS, not significant. (D) The Hemi Index of CTCF motifs showing HI>50 in the pooled data were compared between two dyads, from pulse to chase, and across 5 passages. (E) Occupancy of WT and R133C mutant MeCP2 in WT mESC, and MeCP2 in DNMT1/3A/3B triple KO (TKO) mESCs was profiled around CTCF motifs showing upstream- or downstream-only hemi-methylation in mESCs. (F) The ratio between interaction contacts from Hi-ChIP in WT and DNMT3B-KO HUES64 hESCs emanating from occupied CTCF motifs and extending up to ±l Mb window is shown.
Methyl-CpG-binding domain (MBD) proteins can bind to both mCpG and mCA (21, 22), suggesting their binding to mC is not selective for the methylation state of the other strand. To test their putative association with hemiCpGs, we analyzed published WGBS and MBD ChIP-seq in mESC (23). To overcome the insufficient resolution of ChIP-seq, we profiled the occupancy of MBD proteins at CTCF motifs showing inherited hemi-methylation either upstream-only or downstream-only. Indeed, MeCP2, Mbdla, Mbdlb, Mbd2a and Mbd2t all showed orientation-specific co-localization with hemi-methylation (Fig. 4E and fig. S9). An MeCP2 mutant in the MBD domain (R133C) prominent in Rett syndrome showed significantly reduced co-localization with hemi-methylation (Fig. 4E). Interestingly, in the absence of DNA methylation, MeCP2 loses the orientation- specific occupancy with no changes in occupancy level (Fig. 4E), while all other MBD proteins show reduced occupancy (fig. S9), suggesting binding ofMeCP2 shifts from a hemiCpG-dependent mode to a methylation-independent mode in the absence of DNA methylation. MeCP2 physically interacts with cohesin and regulates chromatin looping (24–26), compelling us to investigate the relationship between hemi-methylation and CTCF-mediated chromatin interactions. We first determined that hemi-methylation is not significantly altered under an acute and near complete loss of CTCF protein (fig. S10, A and B) (27), suggesting the inheritance of hemi-methylation is CTCF-independent. In hESC, DNMT3B-KO alone is sufficient to eliminate most of the inherited hemi-methylation at CTCF motifs with minimal impact on surrounding DNA methylation (fig. S10C) (12). ChIP-seq revealed that DNMT3B-KO led to no changes in RAD21 occupancy, and a mild increase (~1.3-fold) in CTCF occupancy at CTCF motifs (fig. SIO, D and E). We then performed RAD21 Hi-ChIP in both WT and DNMT3B-KO hESC, and found that loss ofDNMT3B causes reduced interactions emanating from these CTCF motifs, extending up to 1 Mb apart (Fig. 4F), with no changes in interaction directionality bias (fig. S10D). This suggests that loss of hemi-methylation renders the CTCF/cohesin complex to a less productive state, possibly through altered mechanism of physical interaction with MeCP2.
Our results provide unprecedented temporal and strand resolution of the nascent DNA methylome, identifying hemiCpGs with distinct methylation kinetics during DNA replication. Several studies have observed hemiCpGs in cells under heterogeneous cell cycle conditions using a hairpin adaptor-based strategy (11, 28, 29). Our study adds the resolution of the parent-daughter axis, the dimension of replication timing, and integrates a single-molecule perspective to the understanding of hemi-methylation. The efficient re-occupancy by CTCF/cohesin and inheritance of flanking hemi-methylation during DNA replication, and the co-localization with MBD proteins, support a model suggesting that CTCF sites actively engaged in chromatin interactions are marked by hemiCpGs shortly after passage of the local replication forks, which may facilitate timely assembly of the interaction complex, possibly with the involvement of MBD proteins, to ensure the proper inheritance of chromatin interactome and gene expression programs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Corees lab for critical feedback and discussion, A. Meissner for providing the WT and DNMT3B-KO hESC, and E. P. Nora for providing the CTCF-AID mESC. All sequence data have been deposited in GEO under accession number GSE97394. This work was supported by U.S. Public Health Service Award 5P01 GM085354. The content is solely the responsibility o the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES AND NOTES
- 1.Holliday R, Pugh JE, DNA modification mechanisms and gene activity during development. Science 187, 226–232 (1975). [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE, Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuang LS et al. , Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAFl. Science 277, 1996–2000 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Bostick M et al. , UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Sharif J et al. , The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmtl to methylated DNA. Nature 450, 908–912 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Liang G et al. , Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements.Mol Cell Biol 22, 480–491 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowher H, Jeltsch A, Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J Mol Biol 309, 1201–1208 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Aoki A et al. , Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res 29, 3506–3512 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen T, Ueda Y, Dodge JE, Wang Z, Li E, Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 23, 5594–5605 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsumura A et al. , Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmtl, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Arand J et al. , In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet 8, el002750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao J et al. , Targeted disruption of DNMTl, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet 47, 469–478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Ecker JR, Non-CG Methylation in the Human Genome. Annu Rev Genomics Hum Genet 16, 55–77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shipony Z et al. , Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature 513, 115–119 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Wang L et al. , Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu M et al. , Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong S et al. , Selective anchoring ofDNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol 29, 5366–5376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G, Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet 7, el001286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong CT, Corees VG, CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15, 234–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium EP, An integrated encyclopedia ofDNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP, Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58, 499–507 (1989). [DOI] [PubMed] [Google Scholar]
- 22.Gabel HW et al. , Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baubec T, Ivanek R, Lienert F, Schubeler D, Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell 153, 480–492 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Kemohan KD et al. , ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell 18, 191–202 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Kemohan KD, Vemimmen D, Gloor GB, Berube NG, Analysis of neonatal brain lacking ATRX or MeCP2 reveals changes in nucleosome density, CTCF binding and chromatin looping. Nucleic Acids Res 42, 83568368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T, Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37, 31–40 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Nora EP et al. , Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944 e922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laird CD et al. , Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc Natl Acad Sci U S A 101, 204–209 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L et al. , The dynamics of DNA methylation fidelity during mouse embryonic stem cell self-renewal and differentiation. Genome Res 24, 1296–1307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.