Abstract
Prostate cancer bone metastasis remains lethal and incurable, and often arises years after elimination of the primary tumor. It is unclear what underlies the decades-long clinical latency before recurrence, but evidence points to the existence of dormant residual tumor cells that disseminated before the primary tumor was eliminated. To design therapies to prevent progression of disseminated tumor cells (DTCs) into lethal metastases, it is crucial to understand the mechanism(s) underlying this dormancy. The current study functionally validated our previous observation that implicated the GAS6/AXL axis in mediating DTC dormancy in the bone marrow. AXL-null and AXL-overexpressing prostate cancer cell lines were generated to determine if AXL was necessary and/or sufficient for dormancy. Characterization of these cells in vitro and using in vivo mouse models of DTC growth demonstrated that AXL was indeed sufficient to induce dormancy, but was unable to maintain it long-term and was not absolutely required for a dormancy period. Clinically, AXL expression correlated with longer survival in prostate cancer patients, and AXL was not expressed by cancer cells in primary or metastatic tissue. These data point to a tumor suppressive role for AXL in prostate cancer, and future work is required to determine if AXL is expressed on human bone marrow DTCs.
Keywords: AXL, dormancy, prostate, cancer, bone
Introduction
Almost 165,000 men are diagnosed with prostate cancer every year in the U.S., making it the second-most common cancer among men [1]. While approximately 70% of patients are cured with primary local treatment, the remaining 30% experience biochemical recurrence (BCR; detection of circulating prostate-specific antigen (PSA)), and approximately 29,000 men will die each year from progression to fully metastatic disease [1,2]. The most common site for prostate cancer metastasis is the bone, and virtually all men who die from metastatic disease have some level of bone involvement [3]. There is currently no cure for metastatic prostate cancer, and the five year survival rate for advanced cases with bone involvement is 1% compared to 56% for those without [4].
The majority of metastatic prostate cancer cases occur years after elimination of the primary tumor followed by a long clinical ‘disease-free’ period. It is widely accepted that cancer cells disseminate early during primary tumor development, and it is thought that upon entering the secondary site (e.g. bone marrow) disseminated tumor cells (DTCs) can undergo a period of dormancy, allowing them to evade therapy and clinical detection for years to decades [5,6]. The concept of tumor dormancy has expanded to include multiple definitions but is not well studied due to the limitations of robust in vitro models. Tumor mass dormancy, referring to both angiogenic and immunologic control of growth by a balance of cellular proliferation and death, is likely not able to be sustained long term [6,7]. Cellular dormancy, on the other hand, the reversible quiescence of individual cells, likely underlies the observed clinical latency observed over a decade or more [8–10]. Since dormant bone marrow DTCs are thought to be the seeds for lethal prostate cancer metastasis, understanding the mechanism(s) that govern cellular dormancy would provide an opportunity for therapeutic intervention to prevent recurrent disease altogether [11].
It has been demonstrated that prostate cancer DTCs preferentially home to the hematopoietic stem cell (HSC) endosteal niche within the bone marrow and bind to osteoblasts, displacing resident HSCs and co-opting their microenvironment [12]. Since the role of the HSC niche is to support the quiescence and self-renewal properties of HSCs, niche factors may similarly support the quiescence of DTCs [13,14]. We previously showed that one of these niche factors, growth arrest-specific 6 (GAS6), is secreted by osteoblasts and restricts the growth of prostate cancer cells [13]. GAS6 is a ligand for the TAM (TYRO3, AXL, MER) family of receptor tyrosine kinases and plays various roles to support normal physiologic functions such as platelet aggregation, apoptotic cell clearance, and, importantly, the self-renewal potential of HSCs [15]. In many types of cancer, the TAM receptors become upregulated and contribute to various hallmarks of cancer [15]. The functional consequences of GAS6 binding to each of its receptors is context-dependent and wide-ranging. We previously showed that two of the receptors, AXL and TYRO3, play opposing roles in prostate cancer. In a metastatic prostate cancer mouse model, AXL expression is associated with dormant DTCs while TYRO3 expression is associated with proliferating tumors [16].
In this study, we functionally validated the role of GAS6/AXL in mediating dormancy of DTCs with the overall goal of determining its clinical utility in preventing lethal metastases. We generated AXL-overexpression and AXL-null prostate cancer cell lines and characterized their growth in vitro and in vivo. We found that AXL alone was not necessary for a dormant phase and is therefore likely not a suitable stand-alone target to prevent metastatic outgrowth. Importantly, however, AXL was sufficient to restrict proliferation of prostate cancer cells. Further, we found that cancer cells in outgrown human primary prostate tumors and metastatic lesions are AXL-negative. Our findings highlight a potential tumor suppressor role for AXL as an inducer of dormancy in prostate cancer.
Materials and Methods
Generation of AXL knockout cells using CRISPR/Cas9
A gRNA targeting the second exon of human AXL (5’-AACCTGGAGCTGACACCGAA-3’), designed using CHOPCHOP (https://chopchop.rc.fas.harvard.edu/), was generated by annealing and amplifying 2 overlapping oligos (designed using http://crispr.technology/) [17]. The gRNA was cloned into the pH1v1 by Gibson assembly40 (NEB) with AvrII digestion [18]. PC3luc cells were transfected with the gRNA plasmid and Cas9-P2A-GFP plasmid. GFP+ cells were sorted and seeded at a low density to isolate and pick colonies. Clones were screened by PCR (primers forward/reverse: 5’-CTGTTTCTCTCTCTTTCACAGTCTC-3’/5’-TAGAGGTTCCATCACATGCTCAAAG-3’) followed by Sanger sequencing (primers forward/reverse: 5’-TCTCTCTCTCTCTTCTCAGCCTC-3’/ 5’-ACAAGTGGTCAAACTGGGGT-3’) of the gRNA target site (Johns Hopkins Synthesis and Sequencing Facility). Sequencing reads were aligned to PC3luc (SnapGene). Clonality of the alterations was confirmed using TIDE analysis [19]. The Cntl KO clone was isolated from clonal expansion and was confirmed to have no genetic alterations around the gRNA target region by sequencing and TIDE analysis. The pH1v1 and Cas9-P2A-GFP plasmids were a gift from Donald Zack.
Generation of AXL overexpression cells
The Gateway® pENTR221 vector containing the AXL CDS from the Ultimate™ORF Clones collection (Invitrogen, clone ID IOH22600) was obtained through the Johns Hopkins High Throughput Biology Center. Sanger sequencing confirmed a known SNP at position 2456 of the corresponding mRNA sequence (BC032229). Site-directed mutagenesis was used to correct the SNP to match the gDNA sequence of human AXL (primers forward/reverse: 5’-CTATCTGCGCCAGGGAAATCGCCTGAAG-3’/5’-CTTCAGGCGATTTCCCTGGCGCAGATAG-3’) as described previously, and validated by Sanger sequencing (primers: 5’-CAGGAAACAGCTATGACC-3’, 5’-TACTACCGCCAGGGACGTAT-3’) [20]. The corrected AXL CDS was subcloned into a pLenti CMV Neo DEST (705–1) vector gifted from Eric Campeau & Paul Kaufman (Addgene plasmid #17392) and confirmed by Sanger Sequencing (primers: 5’-ACGGGTCTGTGTCCAATCTG-3’, 5’-CTTATCCCCACTTGCAGCCC-3’, 5’-CGCAAATGGGCGGTAGGCGTG-3’) [21].
For the Tet-On vector, PCR-based cloning of the corrected pENTR221-AXL CDS vector via restriction sites EcoRI and AgeI was performed (primers forward/reverse: 5’-TAAGCAGAATTCATGGCGTGGCGGTGCCCCAGGAT-3’/5’-TGCTTAACCGGTTCAGGCACCATCCTCCTGCCCT-3’) according to https://www.addgene.org/protocols/pcr-cloning/ (Addgene) and validated by Sanger sequencing (primers: 5’-TACTACCGCCAGGGACGTAT-3’, 5’-CAGGAAACAGCTATGACC-3’). The amplified EcoRI-AXL CDS-AgeI amplicon was cloned into the pLVX-Tet-One-Puro vector (Clontech, #631847) and confirmed by Sanger sequencing (primers: 5’-GCTTGGCAGCTCAGGTTGAA-3’, 5’-TACTACCGCCAGGGACGTAT-3’, 5’-AACGGACGTGAAGAATGTG-3’).
Lentivirus was made by co-transfection of HEK293T cells with the appropriate expression plasmid, an envelope plasmid (pMD2.G, Addgene plasmid #12259), and a packaging plasmid (psPAX2, Addgene plasmid #12260). pMD2.G and psPAX2 plasmids were gifts from Didier Trono. For viral transduction, virus-containing media was removed from the HEK293T cells and applied to the C42Bluc target cells with polybrene (Sigma-Aldrich, #H9268). Target cells were selected: pDEST plasmids with Neomycin (0.5 mg/mL, ThermoFisher, #10131035), and Tet-One plasmids with Puromycin (1 μg/mL, Sigma-Aldrich, #P8833). Cells remained under selection for 5 passages.
Cell culture
Cells were cultured at 37ºC and 5% CO2. HEK293T cells (ATCC) were maintained in DMEM with 10% FBS and 1% penicillin/streptomycin. PC3luc cells, generated as described previously, and C42Bluc cells (gift from Evan Keller, University of Michigan) were maintained in RPMI with 10% FBS and 1% penicillin/streptomycin [22]. All cell lines were authenticated and tested for mycoplasma (Genetica) by passage 20 and were not used past passage 40. Tet-On cell lines were maintained in media containing Tet-free FBS (Clontech, #631107) and in the absence of doxycycline. When necessary, doxycycline (Sigma-Aldrich, #D9891) was administered at the time of seeding at 100 ng/mL unless otherwise indicated. For GAS6 treatment, rhGAS6 (R&D Systems, #885-GS-050) was administered at 100 ng/mL (unless otherwise indicated) for 30 min. Each rhGAS6 lot was tested for activity using pAKT as a readout in H1299 cells (gift from Kolltan Pharmaceuticals Inc.; not authenticated or tested for mycoplasma since receival) which have high AXL expression. Prior to all experiments, cells were synchronized by serum deprivation (0.1% FBS) overnight prior to seeding.
Western blotting
Lysates were collected in Frackelton lysis buffer with Halt Protease and Phosphatase Inhibitor Cocktail (ThermoFisher, #78442) [23]. Protein was fractionated on a 4–20% SDS-PAGE gel (BioRad, #456–1093) and transferred to a nitrocellulose membrane (BioRad, #1704158; Trans-Blot® Turbo™ Transfer System). After blocking, membranes were incubated with AXL (Cell Signaling, #4939) and ACTIN (Sigma-Aldrich, #A5441) antibodies or TYRO3 (Cell Signaling, #5585) and ACTIN antibodies (diluted 1:1000). Anti-rabbit and anti-mouse secondary antibodies (Licor, #926–32211 and #926–68070) were combined for secondary detection. Blots were imaged using the LI-COR Odyssey® scanner (LI-COR Biosciences). PC3 samples were prepared for 2 simultaneous immunoblots for AXL/ACTIN and TYRO3/ACTIN.
Reverse transcription and quantitative PCR
RNA was extracted using RNeasy Mini Kit (Qiagen) and RNA was reverse transcribed with the iScript™ cDNA Synthesis Kit (BioRad, #170–8891). qRT-PCR was performed in triplicate with SsoFast EvaGreen Supermix (BioRad, #1725201). qRT-PCR analysis was done using the delta-delta Ct method normalizing to ACTIN. Primer sequences are listed in Supplementary File 1.
Flow cytometric analysis
General processing for flow cytometry analysis was as follows: cells were dissociated using enzyme-free Cell Dissociation Buffer (ThermoFisher, #13151014). Cells were stained with AXL-488 antibody (R&D Systems, #FAB154G). Data was collected using a BioRad S3™ Cell Sorter and analysis was performed using FlowJo®. When indicated, 7-AAD (BioLegend, #420404) was added to identify dead cells. For EdU incorporation analysis, seeded cells were cultured +/− doxycycline, +/− R428 for 4 days and EdU was added to cultures 2 hrs prior to sample collection and Click-iT EdU detection (ThermoFisher, #C10425) was followed according to the manufacturer’s protocol except for a further 1:10 dilution of the azide. EdU was detected with an AF488 azide and AXL was detected with AXL-PE (R&D Systems, #FAB154P).
Immunofluorescence EdU proliferation assay
Cells were seeded on chamber slides (Corning, #354114) +/− GAS6. On Day 3, GAS6 was respiked and EdU added (final concentration 10 μM). 24 hrs later, cells were stained by IF using the Click-iT™ EdU Alexa Fluor™ 555 Imaging Kit (ThermoFisher, #C10338) and stained for DAPI. Stained slides were imaged manually as described previously [24]. Slides from the same experiment were imaged using the same settings. 3 fields of view (FOV) were captured per sample and the percent of EdU+ cells was manually counted for each FOV and averaged.
Plate-based BrdU proliferation assay
BrdU incorporation over 24 hrs was detected using a colorimetric ELISA (Sigma-Aldrich, #11647229001). GAS6 was added at time of seeding and respiked on Day 4. 5 technical replicates were averaged and fold changes were calculated based on Day 2 signal.
Soft agar assay
The soft agar assay was performed according to [25]. Cells were seeded (10,000 cells per well PC3 and constitutive AXL overexpression; 5,000 cells per well for Tet-On cells) and doxycycline or GAS6 was included in the top layer of media and was added twice a week. After incubation of cultures in nitroblue tetrazolium chloride (Sigma-Aldrich, #N5514) overnight at 37ºC, colonies were imaged. FIJI software (http://fiji.sc, particle analysis function using a pixel size range of 10-infinity and a circularity range of 0.8–1.0) was used to quantify the number and size of colonies.
Human platelet isolation
Blood from a healthy volunteer was drawn into a Vacutainer® containing ACD buffer (BD Biosciences, #364606) and processed according to http://www.abcam.com/protocols/isolation-of-human-platelets-from-whole-blood. All recommended steps included Prostaglandin E1 (Sigma-Aldrich, #P7527) to prevent premature platelet aggregation. Platelets were labeled with DiI (ThermoFisher, #V22885) and filtered prior to co-culture. Cells were exposed +/− doxycycline for 3 days, dissociated using enzyme-free buffer, and labeled with NucBlue™ Live ReadyProbes™ Reagent (ThermoFisher, #R37605) prior to plating. Co-culture (1,000 C42B cells:20×106 platelets) was plated in technical duplicates +/− doxycycline, +/− R428 (50 nM or DMSO vehicle control; Selleckchem.com, #S2841) as indicated. Images (phase, DAPI, DsRed) were taken daily (EVOS™ FL Auto Imaging System). Colocalization was quantified using the FIJI Colocalization Threshold feature (percent of DAPI volume covered by DsRed signal). Results were averaged between technical replicates, and treated samples were normalized to untreated.
Characterization of aggregates with R428
Cells were seeded in duplicate +/− doxycycline, +/− R428 and images taken daily. On Day 3 NucLight Rapid Red Reagent (1:500; IncuCyte, #4717) was added. CellTiter96 (Promega, #G3580) was used to assess viability on Day 5. Aggregates were quantified using FIJI 3D Object Counter (threshold: 3140; min/max: 10/122880. Numbers of aggregate size were combined for technical duplicates, and skewness was measured (GraphPad Prism 6).
Mouse model of intracardiac injection and tissue harvesting
The Johns Hopkins Institutional Animal Care and Use Committee approved all experiments involving mice (protocol #M016M41). 4–8 week old male NOD/SCID/gamma null (NSG) mice were obtained from an immunodeficient mouse colony housed at Johns Hopkins. 200,000 PC3, 1×106 C42B, or DPBS (negative control) cells were injected into the left ventricle of the heart. For C42B AXL overexpression experiments, half of the mice received 0.1 mg doxycycline intraperitoneal (IP) 1–2 hrs prior to intracardiac injection and doxycycline was administered in the drinking water (2 mg/mL + 1% sucrose) for the duration of the experiment. Mice were imaged weekly to assess tumor burden (IVIS Spectrum In Vivo Imaging System; PerkinElmer). Whole body total flux (photons/sec) was quantified (Living Image® 4.4). At 109 flux, mice were injected IP with EdU (1 mg; ThermoFisher, #A10044) and 24 hrs later euthanized. Soft tissues were fixed in 10% NBF and stored in 70% ethanol. Hindlimb bone marrow (femur and tibia) was collected/processed/stored as described previously [26,27].
Immunofluorescent DTC detection in mice
Bone marrow slides were stained as previously detailed for human bone marrow [24]. Instead of human, mouse Fc block (BioLegend, #101320) was used. A cocktail of human/epithelial-specific antibodies was combined to detect DTCs (HLA-A: Abcam, #ab52922; pan-cytokeratin: Abcam, #ab9377; human nucleolin-488: Abcam, #ab154028) and detected by Alexa Fluor (AF) 488 F(ab) fragment goat anti-rabbit secondary antibody (Jackson Immuno Labs, #111–547-003). A cocktail of WBC markers (CD45: BioLegend #103102; CD11b: BioLegend #101201) was detected by an AF555 goat anti-rat secondary antibody (ThermoFisher, #A-21434). EdU was detected using the Click-iT™ EdU Alexa Fluor™ 647 Imaging Kit (ThermoFisher, #C10340). DTCs were counted using the Metafer5 (MetaSystems, V3.11.8) automated scanning system as described previously [24]. A 4-color classifier was used (minimum/maximum exposure times, seconds: DAPI 0.0012/0.0092, AF488 0.0092/0.16, AF555 0.0092/0.08, AF647 0.0092/0.16).
Immunohistochemical (IHC) staining of mouse samples
Tissue was paraffin-embedded and sectioned by the Johns Hopkins Reference Histology Laboratory. Sections were stained according to https://www.cellsignal.com/contents/resources-protocols/immunohistochemistry-protocol-(paraffin)/ihc-paraffin for chromogenic staining with slight modification: deparaffinization with Citrisolv (Fisher Scientific, #22–143-975); antigen unmasking with steaming in Target Retrieval Solution (Dako, #S1699); AXL antibody (1:300; Cell Signaling, #8661); goat anti-rabbit biotinylated secondary antibody (1:200; Vector Labs, #BA-1000) detected by ABC HRP reagent (Vector Labs, #NC9313719).
Sections stained by multiplex IF were blocked with TrueBlack (Chemometech, #23007) and stained with the Click-iT™ EdU Alexa Fluor™ 647 Imaging Kit. Reagents: Human nucleolin AF488 (1:200), AXL antibody (1:300; Cell Signaling, #8661), Cy3 F(ab) fragment goat anti-rabbit secondary antibody (Jackson Immuno Labs, #111–167-003).
Metastatic patient tissue IHC
The rapid autopsy program was approved by the Institutional Review Board (IRB number NA_00036610) at The Johns Hopkins University School of Medicine. Metastatic castrate-resistant prostate cancer samples were obtained from different patients. Tissues were isolated 0–24 hrs post mortem and fixed in 10% neutral-buffered formalin for 48 hrs and paraffin embedded and sectioned by the Johns Hopkins Reference Histology Laboratory. Sections were stained as above for mouse tissues with AXL antibody.
Human prostate tissue microarray (TMA) staining
This study was approved by the Johns Hopkins institutional internal review board and followed the U.S. Common Rule. Prostate TMAs were prepared as in [28]. AXL staining (1:400, Cell Signaling, #8661) was performed on 4 TMAs using the Discovery anti-Rabbit HQ kit (Roche). One TMA included 52 cases of matched normal/benign, tumor, and lymph node metastases from prostatectomies of untreated patients. The 3 other TMAs each contained 40 cases of normal/benign and tumor tissue from prostatectomies.
Microarray sample collection and analysis
RNA was extracted from Tet-ø, Tet-Axl 1, and Tet-Axl 2 cells seeded +/− doxycycline for 4 days. Quality assessment, cDNA conversion, and the Agilent Human Gene Expression 4×44K Microarray were conducted at The Sidney Kimmel Cancer Center Microarray Core Facility at Johns Hopkins University. Samples were background-corrected and normalized between arrays. Probe sets without gene names were removed, and batch effects were corrected before transforming the data to log fold change. While accounting for batch, limma package in R tested for differential expression. Differentially-expressed genes had logFC ≥ 1.5 and BH adjusted P value ≤ 0.05. Raw data can be found on the NCBI Gene Expression Omnibus database with the accession number GSE119003. Relevant code is available at Code Ocean: https://codeocean.com/2018/07/02/axl-putative-tumor-supressor-and-dormancy-regulator-in-prostate-cancer/.
AXL expression analysis in patient tumor datasets
The expression profiles (n = 1,405) of retrospective patients were extracted from the Decipher GRID registry (NCT02609269). The retrospective GRID cohort was pooled from five published microarray studies: Cleveland Clinic (PMID: 25466945), Johns Hopkins (PMID: 26058959), Mayo Clinic (PMID: 23826159, 23770138) and Thomas Jefferson University (PMID: 25667284). Institutional Review Board approval was obtained from the participating institutions prior to initiating the current study.
Survival curves:
recurrence-free survival was determined for patients with high (above median) and low (below median) AXL expression. P-value was calculated using the Log-rank test (reanalysis of GSE21032 Taylor et al. 2010; visualized using Project Betastasis http://www.betastasis.com).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6. For comparisons of fold change to control samples analysis for test samples was performed using a one-sample t-test against the null hypothesis, log fold change = 0. For mouse experiments, mice were randomized into groups. 15 mice from each group were injected intracardiac to obtain 10 successfully injected mice. Successful injections were classified as having no BLI signal in the heart. BLI imaging and analysis and bone marrow DTC counting were all performed blinded to sample type. All comparisons are not considered significant (ns, P > 0.05) unless otherwise indicated. Significance: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001, ****, p ≤ 0.0001.
Results
AXL is not necessary for prostate cancer cell dormancy
We initially observed an increase in AXL expression in PC3 prostate cancer cells when grown at low density as individual cells (‘early DTCs’) relative to full confluence (‘proliferating tumor mass’) (Fig. 1A). We tested if loss of AXL in these cells would prevent dormancy and/or increase proliferation by generating AXL-null PC3 cells using CRISPR. We isolated two AXL knockout clones (Axl KO 1, Axl KO 2) and a control clone from the same population (Cntl KO) and confirmed loss of expression by western blot, qPCR, and Sanger sequencing (Fig. 1B, Supplementary Fig. S1A-B). Interestingly, AXL knockout increased expression of TYRO3, but only at the protein level (Supplementary Fig. S1A, C). Loss of AXL did not affect the genes involved in the epithelial to mesenchymal transition (EMT) or stemness, two common phenotypes known to be influenced by AXL (Supplementary Fig. S1D-E).
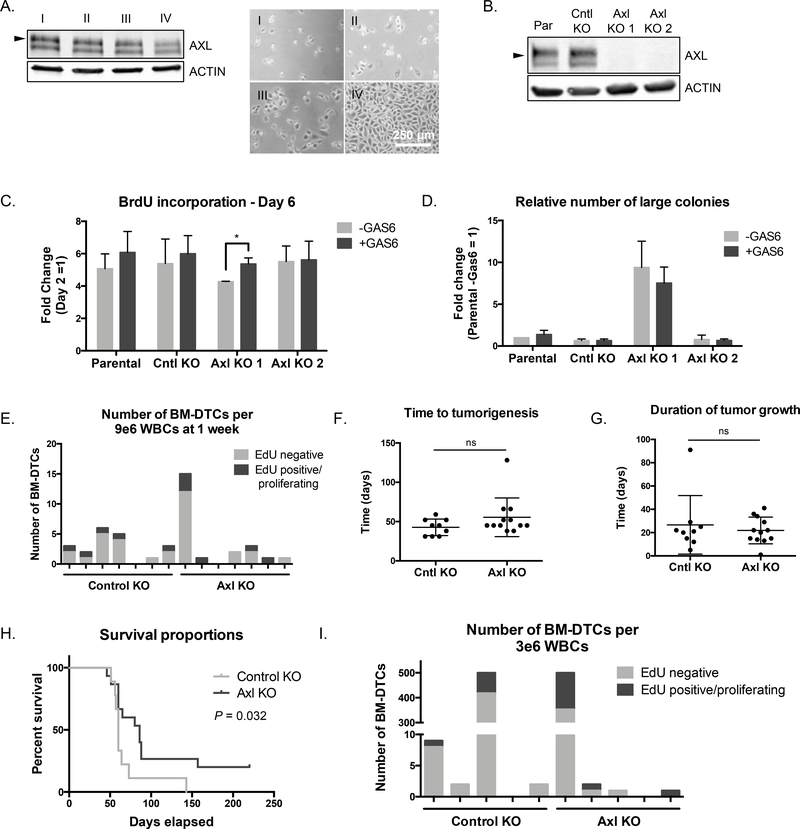
Figure 1.
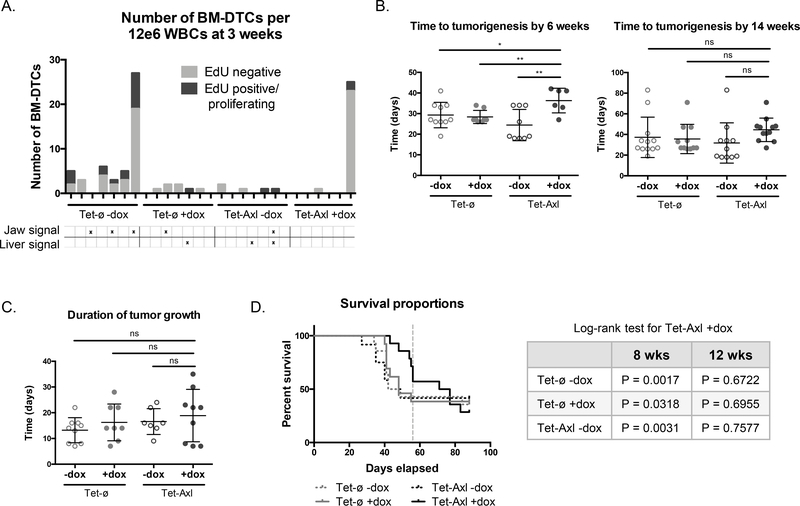
AXL knockout does not prevent dormancy. A, Western blot showing AXL expression (left) in PC3 cells seeded at various densities (right, phase images) (n = 1). Arrow represents 125 kDa. Scale bar, 500 μm. B, AXL expression by western blot in PC3 cells (Par), a control knockout clone (Cntl KO), and two AXL knockout clones (Axl KO1, Axl KO 2) (n = 1). Arrow represents 125 kDa. C, BrdU incorporation over 24 hrs in control and knockout cell lines +/− GAS6. Results represent fold change between the incorporation readout on day 6 to day 2 (n = 3). Error bars, mean ± SEM; P values calculated using t-test between Axl KO and Cntl KO, and within each cell line between treatments. D, Quantification of the number of large colonies (number of colonies in the top 65% of the range of colony size) in soft agar +/− GAS6, relative to reference sample Par -GAS6 (n = 3). Error bars, mean ± SEM; P values calculated using multiple t-tests to assess differences due to treatment. E, Number and proliferation status of bone marrow DTCs detected in mice at 1 week after intracardiac injection. Each bar represents 1 mouse (n = 7 per group). F, Time in days to tumorigenesis after intracardiac injection. Tumorigenesis was defined as BLI signal over control PBS-injected mice (Cntl KO, n = 9; Axl KO, n = 12; each dot represents one mouse; mean ± SD). G, Duration of tumor growth of each group after intracardiac injection. Duration of growth was defined as the time between the onset of tumorigenesis and time to lethal tumor burden (Cntl KO, n = 9; Axl KO, n = 12; each dot represents one mouse; mean ± SD). H, Survival proportions for each group of mice over time (Cntl KO, n = 9 mice; Axl KO, n = 12 mice). P value calculated by Log-rank test. For relevant experiments GAS6 was added at 100 ng/mL at the time of seeding. For mouse tumor growth P values were calculated by t-test. I, Number and proliferation status of bone marrow DTCs detected in mice at the time of death. Counting stopped after the first 500 BM-DTCs (n = 5 for each group).
There was no change in the proliferation rate of Axl KO compared to controls as assessed by BrdU incorporation, and upon rhGAS6 treatment only one Axl KO clone actually increased proliferation (Fig. 1C). When grown in 3D culture in soft agar, Axl KO clones did not display any difference in their ability to outgrow into large colonies compared to PC3 parental or Cntl KO cells, independent of GAS6 treatment (Fig. 1D, Supplementary Fig. S1F).
Because in vitro studies are a limited model to study dormancy and are only an approximation of the dormancy phase in vivo, we next directly tested whether loss of AXL would prevent the dormant phase of bone marrow DTCs in a mouse model of DTC growth. Cntl KO or Axl KO 2 cells were inoculated intracardiac in NSG mice. Mice were injected with EdU one day prior to sacrifice to detect proliferating cells. At the one-week timepoint there was no difference in the number or proliferation status of DTCs that reached the bone marrow. The presence of a similar number of EdU-DTCs in each group indicated the dormancy period was not bypassed by the loss of AXL (Fig. 1E). A separate cohort of mice from each group was monitored over time by weekly bioluminescent imaging (BLI) until whole body tumor signal reached a lethal threshold. There was no difference between the two groups in terms of either the time it took to reach first detectable tumor burden or the duration of the tumor growth from first detectable signal to lethal threshold (Fig. 1F,G). However, there was a statistically significant difference indicating better survival overall for mice with AXL KO tumors; this is likely due to the greater number of mice in the KO group with a successful intracardiac injection despite randomizing the groups, indicating this may not be biologically significant (Fig. 1H). Focusing on bone-specific signal there was not a statistically significant increase in the number of mice with hindleg and/or jaw signal in the Axl KO group, and there was no difference in the number or proliferation status of bone marrow DTCs at the time of death (Supplementary Fig. S1G, Fig. 1I).
AXL overexpression decreases proliferation in vitro
To determine if AXL is sufficient to induce dormancy we generated C42B prostate cancer cells with constitutive overexpression of AXL. C42B cells expressing a control Neomycin resistance vector (Neo) did not express endogenous AXL, and AXL expression was only observed in two overexpression clones (Axl 1, Axl 2) (Fig. 2A, Supplementary Fig. S2A-B). EMT and stemness genes were largely unaffected by AXL overexpression (Supplementary Fig. S2C-D).
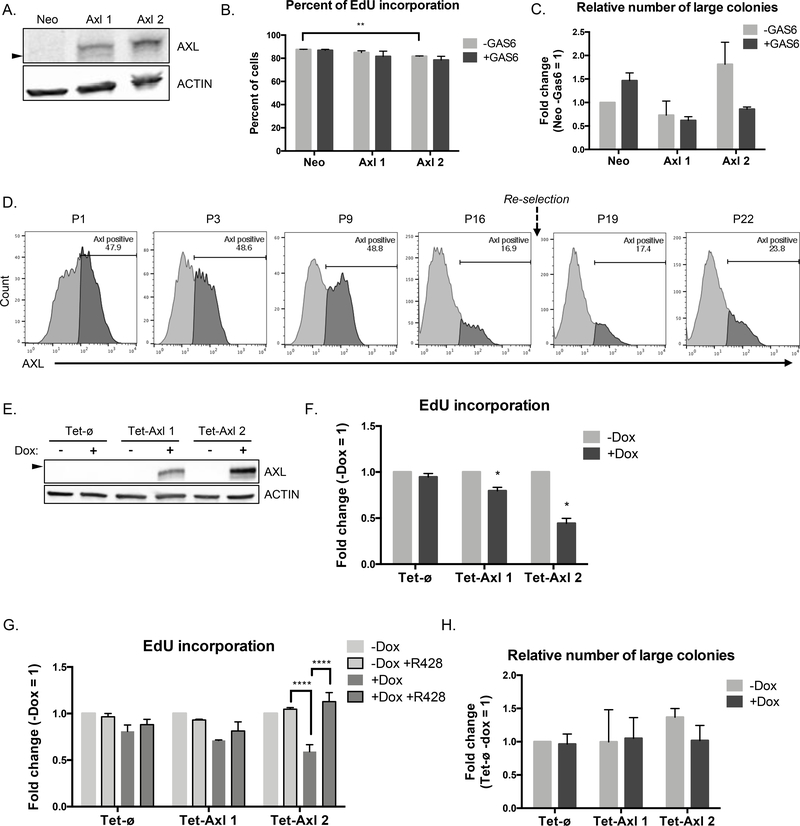
Figure 2.
AXL overexpression decreases proliferation in vitro. A, AXL expression by western blot in C42B control Neomycin resistant cells (Neo), and two AXL overexpression clones (Axl 1, Axl 2) (n = 1). Arrow represents 125 kDa. B, EdU incorporation over 24 hrs in C42B Axl 1 and 2 cells compared to Neo control +/− GAS6 by IF. Individual cells were counted manually in 3 fields of view per sample (n = 2). P values calculated using t-test between cell lines and their control, and within each cell line between treatments. C, Quantification of the number of large colonies (number of colonies in the top 16% of the range of colony size) in soft agar +/− GAS6, relative to reference sample Neo -GAS6 (n = 2). D, AXL expression after dead cell exclusion by flow cytometry in an AXL overexpression clone over increasing passage number. Cells were re-selected with Neomycin over 2 passages between passage 16 and 19 (n = 1). E, AXL expression by western blot in conditionally overexpressing AXL clones (Tet-Axl 1, Tet-Axl 2) compared to empty vector control (Tet-ø) +/− doxycycline (n = 1). Arrow represents 125 kDa. F, EdU incorporation over 2 hrs in Tet-Axl 1 and 2 cells compared to Tet-ø control +/− doxycycline by flow cytometry (n = 3). P values calculated using one-sample t-test against the null hypothesis (log fold change = 0) was performed between treated samples. G, EdU incorporation over 2 hrs in all cell lines +/− doxycycline and AXL inhibitor R428 by flow cytometry; fold change is relative to untreated (Tet-ø and Tet-Axl 2, n = 3; Tet-Axl 1, n = 2). P values calculated using Dunnett’s multiple comparison test against +Dox samples for each cell line, excluding -Dox samples. H, Quantification of the number of large colonies (number of colonies in the top 13% of the range of colony size) in soft agar +/− doxycycline, relative to reference sample Tet-ø -Dox (n = 2). For each experiment treatments were initiated at the time of cell seeding; GAS6: 100 ng/mL, doxycycline: 100 ng/mL, R428: 50 nM. Error bars, mean ± SEM. For soft agar assays, P values were calculated using multiple t-tests to assess differences due to treatment.
To determine if AXL overexpression decreased the rate of proliferation we performed EdU labeling. We observed a slight decrease in the number of EdU+ cells by immunofluorescent (IF) detection in AXL overexpressing cell lines relative to Neo control (Fig. 2B). This was also confirmed by detection of BrdU incorporation by a plate-based ELISA assay, however this population-based method was not as sensitive as individual cell counting (Supplementary Fig. S2E). Surprisingly, when grown in soft agar AXL overexpressing cells did not show significantly reduced outgrowth compared to Neo control cells, regardless of GAS6 (Fig. 2C, Supplementary Fig. S2F).
We hypothesized that the modest proliferation changes observed may have been due to the loss of AXL expression over time in the population, wherein AXLhi cells do not proliferate, leading to the overrepresentation of AXLneg or AXLlow cells. We did in fact observe a decrease in the percent of AXL-positive cells with increasing passage of an AXL overexpression clone that was unaffected by re-selection of the cells with Neomycin (Fig 2D). Therefore, to improve our model of AXL overexpression, we cloned the AXL ORF into a tetracycline-On system (Tet-Axl 1, Tet-Axl 2) in which AXL was only expressed in the presence of doxycycline (‘AXL-on’) and cells were maintained without doxycycline (‘AXL-off’) (Fig 2E, Supplementary Fig. S2G-H). Again, AXL overexpression did not alter EMT or stemness gene expression (Supplementary Fig. S2I-J).
When we assessed the percent of cells that incorporated EdU in these cell lines, Tet-Axl clones proliferated at a slower rate when treated with doxycycline, compared to Tet-ø control cells that were unaffected by doxycycline treatment (Fig. 2F). The decrease in proliferation was more evident in the Tet-Axl 2 clone that had higher AXL expression. Inhibition of AXL signaling with R428, an AXL-specific kinase inhibitor, increased proliferation rates of Tet-Axl cells with doxycycline back to baseline levels (Fig. 2G). Surprisingly, these cells did not exhibit any differences in outgrowth ability when grown in soft agar (Fig. 2H, Supplementary Fig. S2K).
High AXL expression leads to cell aggregation
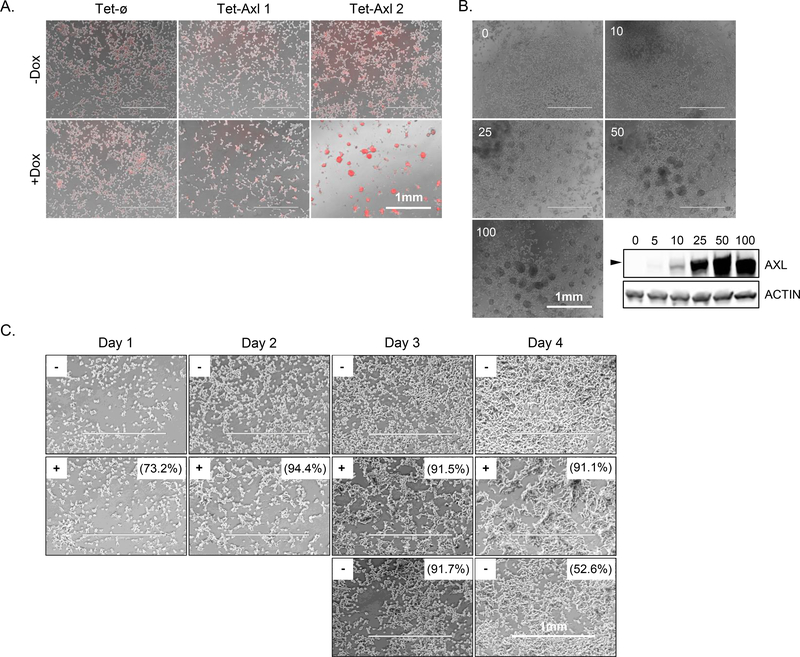
Upon doxycycline treatment of Tet-Axl cells we consistently observed a clear aggregation phenotype. This was more evident in the Tet-Axl 2 clone that had higher AXL expression (Fig. 3A). The aggregation only occurred in Tet-Axl cells and in the presence of doxycycline. Weak aggregation became apparent with 25 ng/mL of doxycycline and reached full ability by 100 ng/mL (the concentration used for all other experiments), correlating with AXL expression (Fig. 3B). Removal of doxycycline after two days of exposure and aggregation formation led to the rapid dissociation of aggregates by the following day but did not correlate with a significant decrease in AXL expression (Fig. 3C). The formation of aggregates by addition of doxycycline in Tet-Axl cells was blocked with simultaneous AXL kinase inhibition, indicating the requirement of AXL signaling (Fig 3D). Treatment did not affect viability (Supplementary Fig. S3A).
Figure 3.
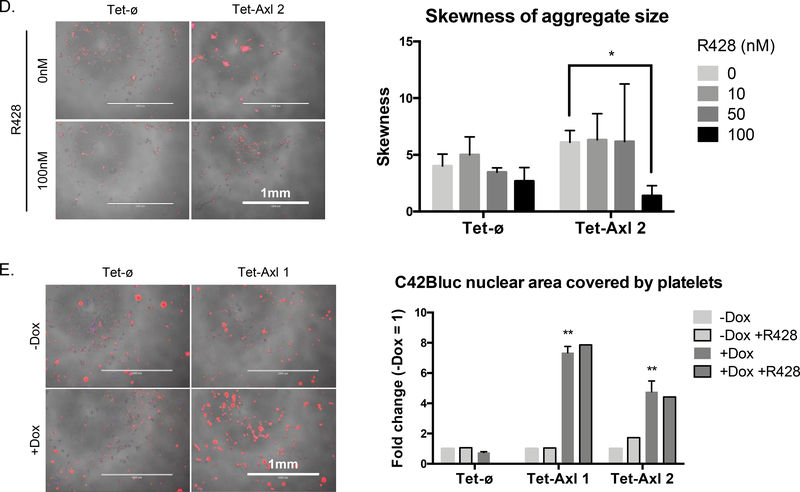
High AXL expression leads to cell aggregation. A, Overlay of phase and nuclear labeling (NucLight Rapid Red Reagent) images of conditional AXL overexpression cell lines seeded +/− doxycycline for 4 days (n = 1). B, Tet-Axl cells (pooled population prior to Tet-Axl 1 and 2 cloning) seeded with increasing concentrations of doxycycline as indicated (ng/mL) were grown for 3 days and were imaged or collected for protein expression analysis by western blot (n = 1). Arrow represents 125 kDa. C, Phase images of Tet-Axl 1 cells seeded without doxycycline (−) over 4 days (top row), with 100 ng/mL doxycycline (+) over 4 days (middle row), and after doxycycline withdrawal on day 2 (bottom row). Numbers indicate the percent of AXL-positive cells in each condition by flow cytometry (n = 1). Images were taken on indicated days and contrast was enhanced to the same degree for each image to more easily visualize cells. D, Representative phase and nuclear labeling (NucLight Rapid Red Reagent) overlay images of Tet-ø and Tet-Axl 2 cells seeded with doxycycline (100 ng/mL) for 5 days +/− AXL inhibitor R428 (left) and quantification of skewness of aggregate size with increasing concentrations of R428 (right) (n = 3). P values calculated using t-test between R428 treated and untreated. E, Left: Representative images of Tet-ø and Tet-Axl 1 cells (phase, NucBlue Live Cell Stain) +/− doxycycline (100 ng/mL) and cocultured with human platelets (red). Right: Fold change of quantified colocalization of nuclear area (blue) and platelet signal (red) relative to -Dox (n = 2 for samples without R428 treatment, n = 1 for samples with R428 treatment (50 nM)). P values calculated using t-test to compare Tet-Axl to Tet-ø for +Dox samples only. Error bars, mean ± SEM. Scale bars, 1 mm.
As AXL signaling plays a critical role in platelet aggregation, and cancer cell clustering with platelets has been demonstrated to promote their survival in the blood of prostate cancer patients, we next investigated the ability of AXL overexpressing cells to aggregate with platelets [29–31]. Upon co-culture, human platelets preferentially coated the cell surface of doxycycline pretreated AXL-on cells, compared to co-culture with Axl-off cells (Fig. 3E). Interestingly, microarray analysis comparing genes upregulated in doxycycline-treated Tet-Axl 2 cells (robust aggregate formation) relative to Tet-Axl 1 cells (weak aggregate formation) revealed an upregulation of less than 30 genes, two of which (AGR1, ELF3) were also found to be upregulated in breast cancer patient circulating tumor cell (CTC) clusters compared to single CTCs (Supplementary Fig. S3B, Supplementary Table 1) [32].
AXL expression delays but is not sufficient to prevent tumorigenesis in vivo
To determine if AXL overexpression could induce dormancy in vivo, we injected either Tet-ø or Tet-Axl 2 cells intracardiac into NSG mice that were administered doxycycline in their drinking water (‘AXL-on’, refers only to Tet-Axl 2 cells exposed to doxycycline) or received normal water (‘AXL-off’). After three weeks, a cohort with representative mice from each group was injected with EdU to label proliferating cells and sacrificed a day later to capture differences in bone marrow-specific dormancy periods that occur prior to tumorigenesis. There was a greater number of total and proliferating bone marrow DTCs detected in AXL-off mice, compared to almost no DTCs and no proliferating DTCs found in AXL-on mice (Fig. 4A). One AXL-on mouse was not representative of the AXL-on group, harboring a high number of total and proliferating DTCs. Notably, only AXL-off mice developed tumor signal either in the jaw or liver at the three-week timepoint. We monitored a larger cohort of mice over time by BLI and observed that at six weeks there was a significant delay in tumorigenesis in the AXL-on mice. By fourteen weeks, however, this data was no longer significant, indicating that almost all mice eventually developed tumors (Fig. 4B). AXL had no effect on the duration of tumor growth once tumors were initiated (Fig. 4C). This translated to the overall survival differences mimicking those of the onset of tumorigenesis, with a statistically significant increased survival rate for AXL-on mice compared AXL-off groups at eight weeks post injection, but by 12 weeks they had reached a lethal tumor burden (Fig. 4D). At the time of death, there was no difference in bone marrow DTC number between AXL-on or AXL-off groups. The increased bone marrow DTC number in the Tet-ø groups compared to Tet-Axl groups is likely due to unintentional selection for intrinsic phenotypes during the clonal expansion of these cell lines and is unrelated to AXL, especially as this data was not recapitulated in the AXL-on group compared to the AXL-off controls (Supplementary Fig. S4A). Overt bone-specific metastasis was observed at low frequency across all groups, and therefore no conclusions about AXL preventing bone metastasis could be made (Supplementary Fig. S4B).
Figure 4.
AXL overexpression delays but does not prevent tumorigenesis in a mouse model of disseminated tumor cell growth. A, Number and proliferation status of bone marrow DTCs detected in mice at 3 weeks after intracardiac injection. Each bar represents 1 mouse; positive BLI signal is indicated below (AXL-off groups, n = 7; AXL-on group, n = 6). B, Time in days to tumorigenesis of each group after intracardiac injection at the 6 week (Tet-ø -Dox, n = 10; Tet-ø +Dox, n = 8; Tet-Axl -Dox, n = 9; Tet-Axl +Dox, n = 6) and 14 week (Tet-ø -Dox, n = 12; Tet-ø +Dox, n = 11; Tet-Axl -Dox, n = 11; Tet-Axl +Dox, n = 12) timepoints. Tumorigenesis was defined as BLI signal over control PBS-injected mice; each dot represents one mouse. C, Duration of tumor growth after intracardiac injection. Duration of growth was defined as the time between the onset of tumorigenesis and time to lethal tumor burden; each dot represents one mouse (Tet-ø -Dox, n = 9; Tet-ø +Dox, n = 8; Tet-Axl -Dox, n = 7; Tet-Axl +Dox, n = 9). D, Survival proportions for each group of mice over time (left) with tabulated Log-rank test results for each group compared to Tet-Axl +Dox group at the 8 or 12 week timepoint (right). Vertical dashed line indicates 8 weeks (Tet-ø -Dox, n = 14; Tet-ø +Dox, n = 13; Tet-Axl -Dox, n = 12; Tet-Axl +Dox, n = 14). Error bars, mean ± SD. For mouse tumor growth P values were calculated by t-test.
Tumors from Tet-Axl mice on doxycycline are heterogeneous for AXL expression and EdU incorporation
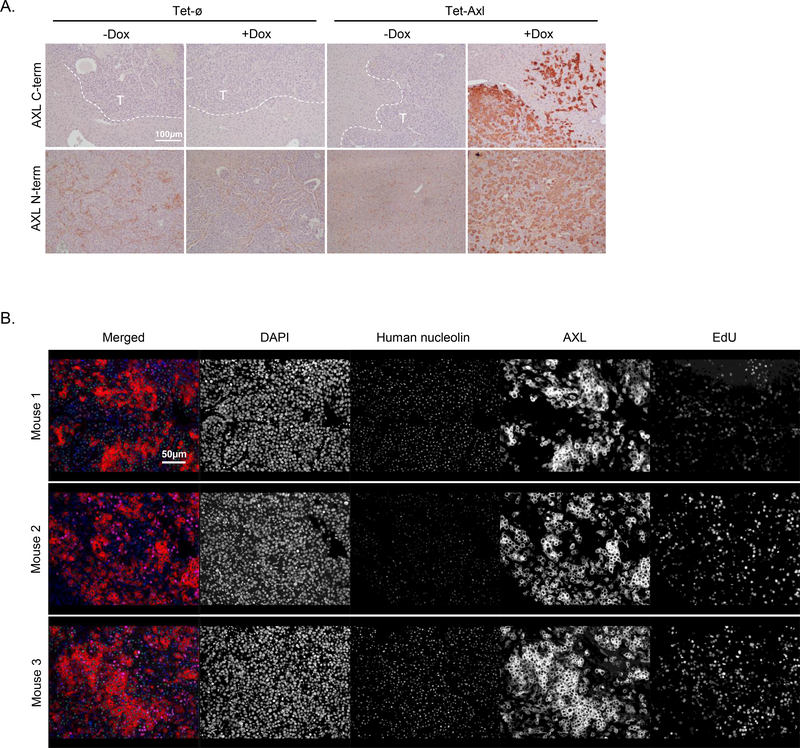
One explanation for the development of tumors in AXL-on mice could simply be that some of the DTCs lost AXL expression and that the delay in tumorigenesis represented the time it took for those cells to outgrow, similar to the loss of AXL-positive cells in the constitutively driven AXL model (Fig. 2D). To determine if this was true, we sectioned and stained liver tumors (the most common tumor site and the site with the greatest tumor burden, regardless of group) from each group for AXL expression. Unexpectedly, there was robust AXL expression in tumors from AXL-on mice (Fig. 5A, upper panel). To rule out that AXL was being inactivated by extracellular cleavage, we stained the same tumors with an antibody recognizing the N-terminal extracellular region and still observed strong AXL staining (Fig. 5A, lower panel). To determine if AXL-positive cells were perhaps not proliferating, or were only a fraction of the entire tumor, we used multiplex IF to stain for AXL (C-term Ab), human nucleolin as a cancer cell marker, and EdU to indicate proliferating cells. AXL was not expressed on every cancer cell yet did not show any correlation with EdU status (Fig. 5B).
Figure 5.
AXL-on tumors are heterogeneous for AXL expression and EdU incorporation. A, Representative IHC staining to detect either the C-terminus or N-terminus of AXL in liver tumors from each group. Dashed white lines depict the border between tumor and liver tissue; ‘T’ indicates the tumor region (n = 3 per group; scale bar, 100 μm). B, Multiplex IF staining for DAPI, human nucleolin, AXL (C-terminus), and EdU incorporation in liver tumors from 3 different AXL-on mice; scale bar, 50 μm.
AXL is not expressed in human prostate tumors
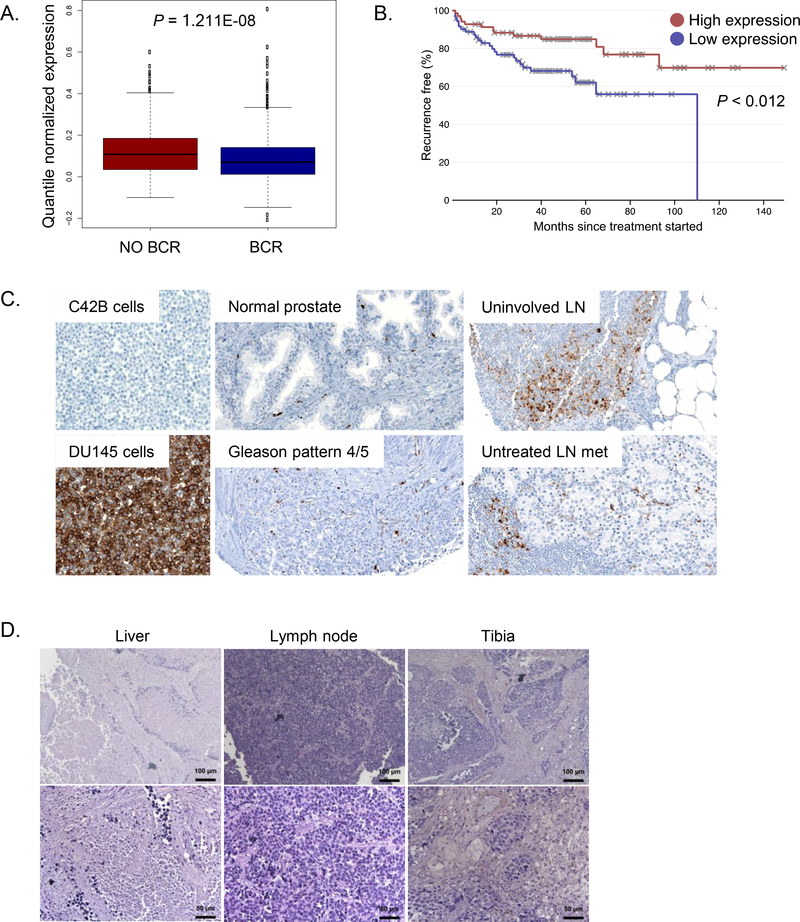
If AXL is sufficient to induce dormancy in human prostate cancer, we would hypothesize that it would have low or no expression in primary and metastatic tumors and would correlate with better disease outcome. Indeed, prostate cancer patients who did not have a BCR had higher AXL expression in their primary tumors compared to those who did recur (GenomeDx Decipher GRID database; Fig. 6A). Furthermore, higher AXL expression also correlated with better progression-free survival (Taylor et al. 2010 dataset; Fig. 6B) [33]. We stained a prostate cancer tissue microarray comprised of healthy prostate tissue, primary cancer tissue of varying Gleason scores, and metastatic tissue for AXL expression. We found no AXL staining in any cancer cells of any corresponding tissue; all positive AXL expression is localized to macrophages and/or endothelial cells (Fig. 6C). To query additional metastatic tumors, we stained tissue sections from liver, lymph node, and tibia metastases and again found no AXL expression (Fig. 6D).
Figure 6.
AXL is not expressed in human prostate tumors. A, AXL expression prostate cancer patients who did and did not experience a biochemical recurrence (n = 1405 patients, GenomeDx GRID dataset). P value was calculated using the Wilcoxon rank test. B, Recurrence-free survival of patients with high (above the median) and low (below the median) AXL expression (n = 571 patients, Taylor et al, 2010 visualized and analyzed with Project Betastasis). C, Representative images of AXL IHC staining of a prostate tissue microarray; LN, lymph node. C42B cells served as a negative control, DU145 as a positive control. All images at original magnification x 200. D, AXL IHC staining in metastatic patient tissue; upper panel scale bar, 100 μm; lower panel scale bar, 50 μm.
Discussion
Dormancy of bone marrow DTCs is thought to underlie the emergence of lethal prostate cancer metastases years after elimination of the primary tumor [34–36]. Treatment strategies aimed at eliminating the primary tumor are not able to prevent metastasis because cancer cells escape the primary tumor early during its development and dormant DTCs are not susceptible to chemotherapy. Therefore, in order to prevent the outgrowth of lethal metastases we must understand the mechanism(s) by which their DTC ‘seeds’ persist in a dormant state. Previously we have shown that the bone marrow HSC niche factor GAS6 restricts prostate cancer cell growth, and that dormant bone marrow DTCs express AXL, a receptor for GAS6 [13,16]. In most cancer types, AXL and the other GAS6 receptors are known to play pro-tumorigenic roles by promoting EMT phenotype, migration, stemness, angiogenesis, therapeutic resistance, and proliferation [15]. Interestingly, the TAM receptors have been widely characterized to promote cancer progression, but there have been no activating mutations identified, unlike those in cancer-driving oncogenes [37]. Early investigation of the role of AXL in cancer described the transforming potential of AXL as being cell line-specific [38]. There is increasing evidence that the functional output of GAS6/TAM receptor signaling is heavily context-dependent [15]. In the work presented here, we provide evidence that AXL signaling mediates prostate DTC dormancy in the bone marrow.
Our initial evidence demonstrated that AXL was not necessary for prostate cancer cell dormancy. AXL loss did not have a significant effect on tumor initiation and growth, or on overall survival in our in vivo model. Importantly, loss of AXL did not affect the number or proliferation status of bone marrow DTCs after intracardiac injection in mice, indicating that the dormancy phase in this model was not bypassed. In other processes, including platelet aggregation, EMT, and therapeutic resistance, AXL and the other TAM receptors are redundant with, and often potentiate, other signaling pathways [15,39,40]. Indeed, mice null for all three TAM receptors or for GAS6 alone are viable, in contrast with the lethal phenotype of mice null for other essential RTKs [41–43]. Similarly, it is likely that other dormancy mechanisms exist to compensate for the loss of AXL in this setting. Our data supports the paradigm that the TAM receptors are dispensable in many processes and participate as actively signaling passengers and collaborators rather than as drivers.
While AXL is non-necessary, we found that overexpression of AXL was sufficient to induce, though not maintain, dormancy in our models. AXL overexpression decreased proliferation in vitro, but only at high levels, and delayed tumorigenesis in vivo. High AXL overexpression alone was sufficient to drive the proliferation phenotype independent of GAS6. The reversal of this phenotype by AXL kinase inhibition suggests it was autoactivated by overexpression, as has previously been shown with high expression levels of AXL and other RTKs [44,45]. Since activation of AXL was thus cell-intrinsic, assessment of AXL-specific effects in vivo is independent of GAS6 availability at a particular organ site. AXL was able to prolong the dormancy phase of cells across the entire body, including in the bone. The more evident in vivo phenotype compared to effects of AXL modulation in vitro is not surprising, as this has been reported for another dormancy regulator [46]. Also consistent with these results, AXL was shown to be significantly upregulated in dormant myeloma cells in mouse bone marrow compared to proliferating cells [10].
That AXL-on cells eventually outgrew into overt tumors in the liver while retaining full length AXL expression indicated that AXL is not able to maintain dormancy long-term, and that other factors are required to maintain dormancy. Perhaps these additional dormancy factors are present in the bone marrow microenvironment. In our experience, intracardiac injection of C42B cells does not frequently yield bone metastases before mice die from tumor burden at other sites, yet we were able to detect individual viable bone marrow DTCs. This suggests there are indeed other signaling axes that can regulate dormancy. The growth behavior of individual AXL-on cells with and without additional factors can be assessed by comparing the in vivo bone marrow DTC data with the 3D growth of the cells in soft agar. While we observed a decrease in cell number in the bone marrow, this was not replicated in vitro, supporting the idea that AXL requires additional bone marrow factors for dormancy maintenance.
The striking aggregation phenotype we observed upon high AXL overexpression is not unprecedented: AXL overexpression has been shown to cause aggregation via homophilic binding both in the presence and the absence of GAS6 [44,47]. GAS6/AXL promotes platelet aggregation and activation and AXL−/− mice are protected against thrombosis [39,48]. The extracellular domain of AXL contains two immunoglobulin and two fibronectin III domains, features shared by cell adhesion molecules such as NCAM. It is possible, therefore, that artificially high AXL overexpression may simply cause cells to ‘stick’ to each other by virtue of their extracellular adhesion features, rather than due to any AXL-specific signaling effects. However, when cells were simultaneously treated an AXL kinase inhibitor and with doxycycline to induce AXL expression, aggregates failed to form, indicating a requirement for active AXL signaling. CTC clustering with platelets has been demonstrated to promote cancer cell survival in the circulation, allowing for successful homing to secondary sites [30,49,50]. While it remains unknown whether AXL is expressed on prostate CTCs, we demonstrated here that human platelets preferentially coated AXL expressing cells. Initial results suggested the coating was independent of AXL kinase inhibition, implying this interaction was structurally-mediated.
In many types of cancer AXL expression becomes upregulated in the primary tumor and/or metastases and correlates with advanced disease and poor survival [15]. If AXL plays a pro-tumor role regardless of cancer type, we would expect to see similar associations in prostate cancer. We found, however, that AXL expression correlates with decreased BCR and better recurrence-free survival. Moreover, gain or loss of AXL in our prostate cancer cell lines did not consistently alter EMT or stemness gene expression, unlike what has been reported for other cancer cell types. Furthermore, we show that AXL is not expressed by cancer cells in primary tumors of any grade or in metastases. Interestingly, AXL expression seems to localize to macrophages and/or endothelial cells. These cells may be the source of AXL signal in the correlation data that predicts better prognosis, indicating a potential anti-tumor role for AXL-positive cells in the tumor microenvironment. It is worth considering that AXL may be regulated carefully at the protein level while its RNA expression does not change (as was observed for TYRO3 upon AXL KO; Supplementary Fig. S1A,C), in which case its transient protein expression would be difficult to capture. The ability of the cell to quickly adapt through regulation of protein stability would allow it to easily respond to microenvironmental signals that would induce the aggregation or dormant phenotypes we observed. Overall this data supports a tumor-suppressive role for AXL in human prostate cancer but does not determine if AXL induces dormancy of human bone marrow DTCs. It is currently not feasible to accurately detect and characterize bone marrow DTCs, as more sensitive methods and disease-specific markers are required.
Our study functionally validates a role for AXL in promoting prostate tumor cell dormancy. While AXL is not necessary for dormancy, it is sufficient to induce short-term dormancy in prostate cancer models. The inability of AXL to maintain dormancy coupled with the idea that there are likely other dormancy-inducing factors, indicates that AXL alone is not a suitable target for preventing lethal metastases. More importantly, this study demonstrates the pleotropic nature of AXL and further highlights the complexity of GAS6/TAM receptor signaling and downstream functions. Careful observation should be made when studying the roles of the TAM receptors in various contexts, and caution should be taken when considering targeting AXL for therapeutic purposes.
Supplementary Material
Implications:
The ability of AXL to initiate but not maintain dormancy, coupled with its dispensability, suggests that targeting AXL alone will not prevent lethal metastatic outgrowth and likely a cooperative network of factors exist to mediate long-term cellular dormancy.
ACKNOWLEDGEMENTS
The authors thank Dr. Russell Taichman for his guidance and thoughtful discussion during this work, and members of the Pienta Lab for helping to process mice. Thanks to Wayne Yu at the SKCCC Microarray Core, and members of the Johns Hopkins Synthesis and Sequencing Facility and the Johns Hopkins Reference Histology Laboratory.
Financial support
This work was supported by the Prostate Cancer Foundation (Young Investigator Award to SRA), the American Cancer Society (Postdoctoral Fellowship PF-16–025-01-CSM to SRA), the NIH (Grants F32-CA206394 to KC Valkenburg, P01-CA093900 to KJ Pienta, P30 CA006973 to The Sidney Kimmel Cancer Center Microarray Core Facility at Johns Hopkins University), Department of Defense (Prostate Cancer Research Program Award W81XWH-14–2-0182 to AMD), and National Cancer Institute/National Institutes of Health (NCI/NIH) Prostate SPORE Grant P50CA58236 and Cancer Center Support Grant 5P30CA006973 to AMD.
ABBREVIATIONS
- BCR
biochemical recurrence
- BLI
bioluminescent imaging
- CTC
circulating tumor cell
- DTC
disseminated tumor cell
- EMT
epithelial-to-mesenchymal transition
- HSC
hematopoietic stem cell
- IF
immunofluorescence
- PSA
prostate-specific antigen
- TAM
TYRO3, AXL, MER
Footnotes
Disclosure of potential conflicts of interest
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Prostate Cancer: Statistics. Vol. 2018. (ASCO, Cancer.Net).
- 2.Pound CR, Partin AW, Epstein JI & Walsh PC. Prostate-specific antigen after anatomic radical retropubic prostatectomy. Patterns of recurrence and cancer control. Urol Clin North Am. 1997;24:395–406. [DOI] [PubMed] [Google Scholar]
- 3.Mehra R, Kumar-Sinha C, Shankar S, Lonigro RJ, Jing X, Philips NE, et al. Characterization of bone metastases from rapid autopsies of prostate cancer patients. Clin Cancer Res. 2011;17:3924–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norgaard M, Jensen AO, Jacobsen JB, Cetin K, Fryzek JP & Sorensen HT. Skeletal related events, bone metastasis and survival of prostate cancer: a population based cohort study in Denmark (1999 to 2007). J Urol. 2010;184:162–167. [DOI] [PubMed] [Google Scholar]
- 5.Willis RA. The Spread of Tumours in the Human Body. London: J&A: Churchill; 1934; [Google Scholar]
- 6.Sosa MS, Bragado P & Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nature reviews. Cancer. 2014;14:611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez AG, Seoane JM & Sanjuan MAF. Dynamics of the cell-mediated immune response to tumour growth. Philos Trans A Math Phys Eng Sci. 2017;375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller V, Stahmann N, Riethdorf S, Rau T, Zabel T, Goetz A, et al. Circulating tumor cells in breast cancer: correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin Cancer Res. 2005;11:3678–3685. [DOI] [PubMed] [Google Scholar]
- 9.Naumov GN, MacDonald IC, Weinmeister PM, Kerkvliet N, Nadkarni KV, Wilson SM, et al. Persistence of solitary mammary carcinoma cells in a secondary site: a possible contributor to dormancy. Cancer Res. 2002;62:2162–2168. [PubMed] [Google Scholar]
- 10.Lawson MA, McDonald MM, Kovacic N, Hua Khoo W, Terry RL, Down J, et al. Osteoclasts control reactivation of dormant myeloma cells by remodelling the endosteal niche. Nat Commun. 2015;6:8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paget S THE DISTRIBUTION OF SECONDARY GROWTHS IN CANCER OF THE BREAST. The Lancet. 1889;133:571–573. [PubMed] [Google Scholar]
- 12.Shiozawa Y, Pedersen EA, Havens AM, Jung Y, Mishra A, Joseph J, et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest. 2011;121:1298–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung Y, Shiozawa Y, Wang J, McGregor N, Dai J, Park SI, et al. Prevalence of Prostate Cancer Metastases after Intravenous Inoculation Provides Clues into the Molecular Basis of Dormancy in the Bone Marrow Microenvironment. Neoplasia (New York, N.Y.). 2012;14:429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen EA, Shiozawa Y, Mishra A & Taichman RS. Structure and function of the solid tumor niche. Frontiers in bioscience (Scholar edition). 2012;4:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Axelrod H & Pienta KJ. Axl as a mediator of cellular growth and survival. Oncotarget. 2014;5:8818–8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taichman RS, Patel LR, Bedenis R, Wang J, Weidner S, Schumann T, et al. GAS6 receptor status is associated with dormancy and bone metastatic tumor formation. PLoS One. 2013;8:e61873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaskula-Ranga V & Zack DJ. grID: A CRISPR-Cas9 guide RNA Database and Resource for Genome-Editing. bioRxiv. 2016; [Google Scholar]
- 18.Ranganathan V, Wahlin K, Maruotti J & Zack DJ. Expansion of the CRISPR–Cas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nature Communications. 2014;5:4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brinkman EK, Chen T, Amendola M & van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laible M & Boonrod K. Homemade site directed mutagenesis of whole plasmids. J Vis Exp. 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4:e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roca H, Hernandez J, Weidner S, McEachin RC, Fuller D, Sud S, et al. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One. 2013;8:e76773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mooney SM, Grande JP, Salisbury JL & Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. [DOI] [PubMed] [Google Scholar]
- 24.Axelrod HD, Pienta KJ & Valkenburg KC. Optimization of Immunofluorescent Detection of Bone Marrow Disseminated Tumor Cells. Biological Procedures Online. 2018;20:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, et al. The soft agar colony formation assay. J Vis Exp. 2014:e51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amend SR, Valkenburg KC & Pienta KJ. Murine Hind Limb Long Bone Dissection and Bone Marrow Isolation. J Vis Exp. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valkenburg KC, Amend SR, Verdone JE, van der Toom EE, Hernandez JR, Gorin MA, et al. A simple selection-free method for detecting disseminated tumor cells (DTCs) in murine bone marrow. Oncotarget. 2016;7:69794–69803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baena-Del Valle JA, Zheng Q, Hicks JL, Fedor H, Trock BJ, Morrissey C, et al. Rapid Loss of RNA Detection by In Situ Hybridization in Stored Tissue Blocks and Preservation by Cold Storage of Unstained Slides. Am J Clin Pathol. 2017;148:398–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol. 2012;9:016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erpenbeck L & Schon MP. Deadly allies: the fatal interplay between platelets and metastasizing cancer cells. Blood. 2010;115:3427–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karpatkin S, Pearlstein E, Ambrogio C & Coller BS. Role of adhesive proteins in platelet tumor interaction in vitro and metastasis formation in vivo. The Journal of Clinical Investigation. 1988;81:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhr JW & Pantel K. Controversies in clinical cancer dormancy. Proceedings of the National Academy of Sciences. 2011;108:12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantel K, Brakenhoff RH & Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nature Reviews Cancer. 2008;8:329. [DOI] [PubMed] [Google Scholar]
- 36.Shiozawa Y, Pedersen EA, Patel LR, Ziegler AM, Havens AM, Jung Y, et al. GAS6/AXL axis regulates prostate cancer invasion, proliferation, and survival in the bone marrow niche. Neoplasia. 2010;12:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma A, Warner SL, Vankayalapati H, Bearss DJ & Sharma S. Targeting Axl and Mer Kinases in Cancer. Molecular Cancer Therapeutics. 2011;10:1763. [DOI] [PubMed] [Google Scholar]
- 38.Burchert A, Attar EC, McCloskey P, Fridell Y-WC & Liu ET. Determinants for transformation induced by the Axl receptor tyrosine kinase. Oncogene. 1998;16:3177. [DOI] [PubMed] [Google Scholar]
- 39.Angelillo-Scherrer A, Burnier L, Flores N, Savi P, DeMol M, Schaeffer P, et al. Role of Gas6 receptors in platelet signaling during thrombus stabilization and implications for antithrombotic therapy. J Clin Invest. 2005;115:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vouri M & Hafizi S. TAM Receptor Tyrosine Kinases in Cancer Drug Resistance. Cancer Res. 2017;77:2775–2778. [DOI] [PubMed] [Google Scholar]
- 41.Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM & Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes & Development. 1994;8:3045–3057. [DOI] [PubMed] [Google Scholar]
- 42.Jones N, Voskas D, Master Z, Sarao R, Jones J & Dumont DJ. Rescue of the early vascular defects in Tek/Tie2 null mice reveals an essential survival function. EMBO Reports. 2001;2:438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Q, Gore M, Zhang Q, Camenisch T, Boast S, Casagranda F, et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature. 1999;398:723. [DOI] [PubMed] [Google Scholar]
- 44.Bellosta P, Costa M, Lin DA & Basilico C. The receptor tyrosine kinase ARK mediates cell aggregation by homophilic binding. Mol Cell Biol. 1995;15:614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlessinger J Cell Signaling by Receptor Tyrosine Kinases. Cell. 2000;103:211–225. [DOI] [PubMed] [Google Scholar]
- 46.Gao H, Chakraborty G, Lee-Lim AP, Mo Q, Decker M, Vonica A, et al. The BMP Inhibitor Coco Reactivates Breast Cancer Cells at Lung Metastatic sites. Cell. 2012;150:764–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCloskey P, Fridell YW, Attar E, Villa J, Jin Y, Varnum B, et al. GAS6 mediates adhesion of cells expressing the receptor tyrosine kinase Axl. J Biol Chem. 1997;272:23285–23291. [DOI] [PubMed] [Google Scholar]
- 48.Angelillo-Scherrer A, de Frutos PG, Aparicio C, Melis E, Savi P, Lupu F, et al. Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nature Medicine. 2001;7:215. [DOI] [PubMed] [Google Scholar]
- 49.Palumbo JS, Talmage KE, Massari JV, La Jeunesse CM, Flick MJ, Kombrinck KW, et al. Platelets and fibrin(ogen) increase metastatic potential by impeding natural killer cell-mediated elimination of tumor cells. Blood. 2005;105:178–185. [DOI] [PubMed] [Google Scholar]
- 50.Gay LJ & Felding-Habermann B. Contribution of platelets to tumour metastasis. Nature Reviews Cancer. 2011;11:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.