Abstract
DNA sequencing has identified a limited number of driver mutations in metastatic breast cancer beyond single base-pair mutations in the estrogen receptor (ESR1). However, our previous studies and others have observed that structural variants, such as ESR1 fusions, may also play a role. Therefore, we expanded upon these observations by performing a comprehensive and highly sensitive characterization of copy number (CN) alterations in a large clinical cohort of metastatic specimens. NanoString DNA hybridization was utilized to measure CN gains, amplifications and deletions of 67 genes in 108 breast cancer metastases, and in 26 cases, the patient-matched primary tumor. For ESR1, a copyshift algorithm was applied to identify CN imbalances at exon-specific resolution and queried large datasets (>15,000 tumors) that had previously undergone next-generation sequencing (NGS). Interestingly, a subset of ER+ tumors showed increased ESR1 CN (11/82, 13%); three had CN amplifications (4%) and eight had gains (10%). Increased ESR1 CN was enriched in metastatic specimens versus primary tumors and this was orthogonally confirmed in a large NGS dataset. ESR1-amplified tumors showed a site-specific enrichment for bone metastases and worse outcomes than non-amplified tumors. No ESR1 CN amplifications and only one gain was identified in ER− tumors. ESR1 copyshift was present in five out of the 11 ESR1-amplified tumors. Other frequent amplifications included ERBB2, GRB7 and cell cycle pathway members CCND1 and CDK4/6, which showed mutually exclusivity with deletions of CDKN2A, CDKN2B, and CDKN1B.
Keywords: metastases, breast cancer, copy number, estrogen receptor, amplification, cyclin dependent kinase
Background
Breast cancer is a genetic disease driven by accumulations of single nucleotide mutations and structural alterations [1]. The latter include gene fusions, deletions, tandem duplications, and copy number (CN) amplifications as seen in ERBB2 (HER2), CCND1, and MYC. Numerous studies have shown varying levels of estrogen receptor (ERalpha) gene (ESR1) CN amplifications in primary breast cancer. In 2007, Holst et al used fluorescence in situ hybridization (FISH) and identified ESR1 CN amplifications (20.6%) and gains (15.3%) in primary breast tumors [2]. Subsequently, multiple groups reported ESR1 CN amplifications at much lower frequency (0–10%), using FISH and array comparative genomic hybridization (aCGH) [3–8]. Analysis of The Cancer Genome Atlas (TCGA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) datasets using cBioPortal [9–11] revealed that ESR1 CN amplifications are present in only 2.5% and 2.3% of breast cancer samples heavily enriched for primary disease, respectively. More recently, Desmedt et al showed ESR1 CN gains and corresponding increased ESR1 mRNA levels in 25% of primary invasive lobular breast cancers (ILC) [12]. Thus, the true frequency as well as the relevance of these ESR1 CNV events, particularly in recurrent and metastatic disease, has yet to be determined.
The landscape of structural variants in metastatic breast cancer (MBC) is largely unknown, particularly after therapy resistance. Gene rearrangements and CN amplifications of ESR1, FOXA1, CYP19A1, and ERBB2 have been reported in relatively small cohorts of endocrine resistant breast cancers [13–16]. Li et al [17] originally described an ESR1 fusion arising in a patient-derived xenograft (PDX) model and we have recently identified nine additional ESR1 fusions in advanced disease [18]. Importantly, the location between the N-terminal ESR1 gene and the C-terminal fusion partner is often conserved between ESR1 exons 6 and 7. Gene fusion events are often associated with CN alterations adjacent to the fusion junction that results in an imbalance of DNA CN between the breakpoint flanking exons [19,20]. We therefore designed and applied a computational method (copyshift) that identifies CN imbalances and found imbalances were significantly enriched in ER+ metastases compared to primary breast tumors [18].
Despite overwhelming evidence for a critical role of ESR1 mutations and increasing evidence for ESR1 fusions in endocrine resistant breast cancer, a comprehensive study of ESR1 CN variations in recurrent and metastatic disease is lacking. In this study, we set out to characterize ESR1 CN alterations with exon-level resolution in a large cohort of well curated metastatic lesions using a highly sensitive nanoString-based approach. Additionally, we assessed the CN alteration landscape of 66 genes with known roles in breast cancer progression and therapy resistance.
Materials and methods
Sample collection
We obtained 141 formalin-fixed paraffin-embedded (FFPE) and frozen sections from eligible metastatic cases from the University of Pittsburgh Health Sciences Tissue Bank (HSTB) and Charite Universitätsmedizin, Berlin. Collection and analysis of specimens was approved under the University of Pittsburgh IRB and Charite Universitätsmedizin Berlin guidelines. When available, patient-matched treatment naïve primary tumors were included in the study (n=26). Seven samples were excluded from the study due to unknown ER status. In total, we had 134 samples from 100 patients that met the study criteria (82 ER+ and 52 ER−), including 26 primary tumors (15 ER+ and 11 ER−) and 108 metastases (67 ER+ and 41 ER−). In some cases, multiple primary tumors or metastases were from the same patient. More than 85% of the samples had tumor cellularity > 40%, that was determined by surface area of tumor cells. If it was lower than 30–40%, we macrodissected the tumor area. Therefore, we circled an area of interest on the slide (i.e., a subregion of the slide that contains the highest concentration of invasive cancer), and from within this region, we estimated the percentage of nuclei that are tumor cell nuclei.
Clinicopathological characteristics are provided in Additional file 1: Supplementary Table S1. We collected information on ER status, source of tissue, site of metastasis, histologic subtype. A subset of the samples has recently been used in another, unrelated study [21] (indicated in Supplementary Figure S1).
Immunohistochemistry (IHC)
Hematoxylin and eosin (H&E) staining procedures were performed at the Histology and Micro-imaging Core (HMC) facility at Magee-Women’s Research Institute (MWRI). Analysis of histological classification was performed by two pathologists (PL and EE) (Additional file 1: Supplementary Table S1).
DNA isolation
DNA was isolated from 4–6 FFPE macrodisected sections (10 μm) per sample depending on tumor size and cellularity using the Qiagen AllPrep FFPE kit (cat# 80234) as per manufacturer’s instructions. DNA from frozen samples was extracted using the Qiagen DNeasy kit (cat# 69506) according to the manufacturer’s instructions. All DNA quantifications were done using Qubit dsDNA HS/BR assay kits (ThermoFisher).
Control samples for nanoString and Digital droplet PCR (ddPCR) comparison
We isolated DNA from MCF-7 long-term estrogen-deprived (LTED) cells, BT-474, and MCF10A cells for validation of CN calls. MCF-7 LTED cells have ESR1 amplifications [17] and BT-474 cells have been described to have a heterozygous ESR1 deletion (Cancer Cell Line Encyclopedia (CCLE)) [22]. MCF10A is a non-tumorigenic epithelial cell line with normal (2N) ESR1 CN [23].
Digital droplet PCR (ddPCR)
Primers and probes were designed and ordered through Integrated DNA Technologies (IDT) for ESR1 and Bio-Rad for two reference genes recommended by the manufacturer (EIF2C and AP3B1) (Additional file 1: Supplementary Table S2). 60 ng of control samples were processed for ddPCR analysis as previously described [24]. Briefly, DNA samples were combined with primers, probes, and supermix, and then added to cartridge. Droplets were generated using Biorad QX100 Droplet Generator and transferred into a 96-well plate for PCR amplification, and a droplet reader (Biorad QX100 Droplet Reader) was used to count PCR+ve and PCR-ve droplets. Data were analyzed using QuantaSoft™ software (Bio-Rad) where target concentration was normalized to reference concentration and multiplied by the number of reference loci in the genome (assumed to be 2) to generate copy number calls.
nanoString
We designed 100bp DNA hybridization probes for a total of 67 genes (Additional file 1: Supplementary Table S3), including ESR1 (n=10 probes) (Additional file 2: Supplementary Figure S1), and 66 genes with known roles in breast cancer development and progression (n=3 probes per gene). For the latter, we queried TCGA breast cancer dataset and previous breast cancer metastasis studies [25–28], with a focus on potentially druggable genes as identified in drug gene interaction database (DGIdb) [29]. Processing of CN data was performed as per manufacturer recommendations. Briefly, DNA samples were fragmented at 37 ⁰C using Alu1 restriction enzyme, denatured at 95 ⁰C, and hybridized overnight with target probes. Post-hybridization sample processing was done using the automated nCounter Prep Station. Raw counts were then collected from the nCounter Digital Analyzer and transferred to the nSolver™ software (v 2.5) for data analysis [30]. Raw counts were normalized to 10 invariant reference probes (intra-sample count normalization) and to reference samples pools of normal breast FFPE (n=13) and frozen (n=4) DNA to generate CN calls (inter-sample normalization; Additional file 1: Supplementary Tables S3-S5). Average CN estimate value was calculated per gene based on all probes for that gene relative to the CN estimate in the normal sample pool. We used the 67 genes of the nanoString code set for comprehensive quality control assessment of copy number calls between FFPE and cell pellet DNA.
Preparation and sequencing of RNA-seq libraries
RNA sequencing data was available for a subset of 66 samples (46 ER+), and was used to associate correlation between ESR1 CN and mRNA expression. TruSeq RNA Access library preparation (illumina) and sequencing was performed at the sequencing core facility at Children’s Hospital of University of Pittsburgh Medical Center (UPMC) using the NextSeq500 platform that produced paired-end reads (2X75bp). Gene expression values for ESR1 were calculated using counts per million normalized to trimmed-means of M-values (TMM-normalized CPM) using DESeq2 and edgeR packages in R [31,32].
Genomic copy number (CN) analysis of RNAseq data
Detection of chromosomal aberrations was based on measuring allelic bias in RNAseq data using the package eSNP-Karyotyping in R [33]. Briefly, we first called variants using GATK package Haplotype caller [34]. Variants were then ordered according to their genomic coordinates and the ratio between major and minor alleles was calculated. To increase confidence, variants with coverage below 20 reads and frequency below 0.2 were excluded from the analysis. Statistical significance was calculated with a one tailed t-test comparing the SNPs major/minor ratio values in each window with the total SNP pool and false discovery rate (FDR) correcting for multiple testing. Moving median plots were generated using windows of 151 SNPs as suggested by the package developers.
ESR1 CN validation datasets
Comprehensive genomic profiling was performed on breast tumors in a CLIA-certified, New York State- and CAP-accredited laboratory (Foundation Medicine [FM], Cambridge, MA) [35,36]. In addition, tumor data from the American Association for Cancer Research (AACR) GENIE and Memorial Sloan Kettering Cancer Center (MSKCC) was accessed and analyzed through cBioPortal [37,38]. Statistical analyses with p value significance of 0.05 were performed in R and GraphPad, and figures were plotted using ggplot2.
Statistical and bioinformatics analysis
R environment was used for statistical computing and graphics [39]. Oncoprints visualizing multiple genomic alterations were generated using ComplexHeatmap package [40]. Given the important role of ESR1 in breast cancer development and progression, and prior evidence for low level amplifications, copy number changes of ESR1 were a major focus of our study. Thus, we used 10 probes to cover the ESR1, and only 2–3 probes for the other 66 genes. ESR1 CN increase by 35% (CN ≥2.7) and decrease by 50% (CN ≤ 1) were considered gains/amplifications and deletions, respectively. CN calls above ≥10 were considered high amplifications. Giving the lower resolution for other genes in the panel, we used one CN cutoff of ≥5 as amplification. Plots and heatmap of actual CN imbalance and shifts were generated using ggplot2 and heatmap.3 packages, respectively [41,42]. For CN imbalance, we reported only shifts of ≥ 30% difference in CN (ratio of ≥ 1.3) between ESR1 5’ and 3’ exons, which is similar to the 35% CN increase used to define gains. Multiple correlations between the different ESR1 exons and mRNA expression were clustered by first principal component (FPC) scores. Spearman and Pearson correlations, Fisher’s exact, and Wilcoxon rank sum tests were performed in R and GraphPad Prism with significance cutoff of 0.05.
Results
Measurement of DNA CN changes using nanoString technology
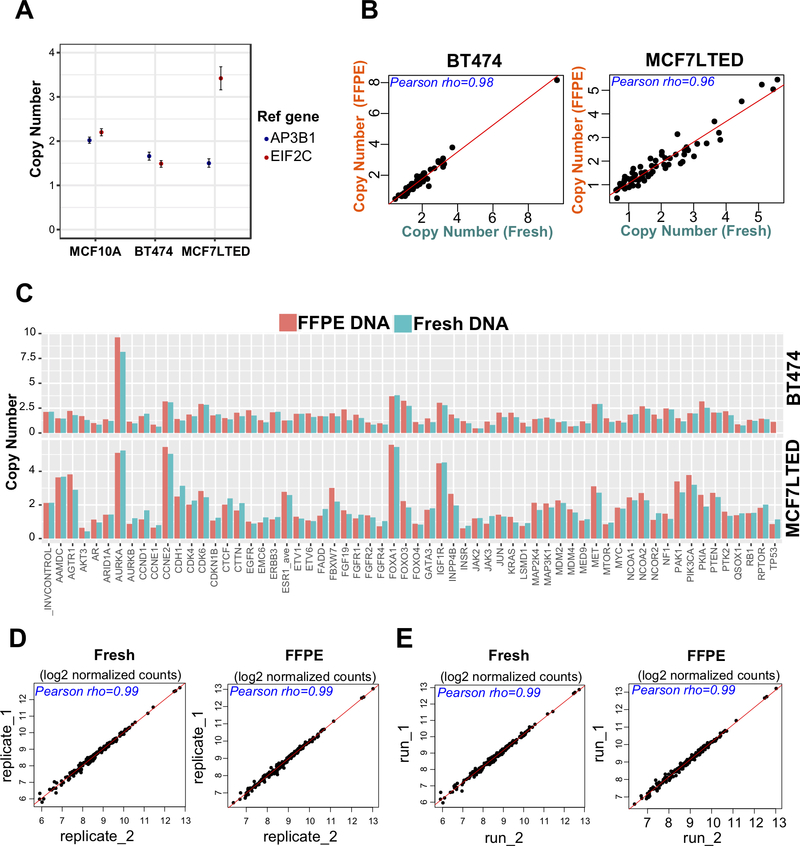
We set out to determine which platform (ddPCR or nanoString) is best suited for quantifying CN alterations. Using ddPCR, we determined ESR1 CN status in MCF7-LTED, BT-474, and MCF10A cells, with EIF2C and AP3B1 reference probes. Our CN calls were consistent between the two probes for the ESR1 CN normal (MCF10A) and deletion (BT-474) models, but not MCF7-LTED cells, that have a known ESR1 amplification (Figure 1A and Additional file 1: Supplementary Table S6). These data suggested that use of two reference probes was not sufficient for accurate measurement of CN. Next, we measured ESR1 CN using nanoString technology, where ten reference probes were included in the library. This analysis correctly identified CN in the three control cell lines (Figure 1C). We then performed quality control experiments to test sensitivity and reproducibility of the CN calls within and between the nanoString runs using DNA from fresh cell pellets and from processed FFPE sections. There was a high correlation between DNA isolated from fresh and fixed samples (Figure 1B-C, and Additional file 1: Supplementary Table S8; rho > 0.9, p < 2.2e-16) and excellent reproducibility between different nanoString runs using the same DNA (Figure 1D-E and Table S9; rho > 0.99, p < 2.2e-16). We therefore proceeded using nanoString technology for the characterization of CN alterations in a large set of well curated breast tumors.
Figure 1: nanoString and ddPCR CN analysis in control cell lines.
A. ddPCR ESR1 copy number analysis of MCF7-LTED, BT-474, and MCF10A cell lines that represent amplification, deletion, and normal copy number models, respectively. The y-axis represents CN status for the ESR1 gene that was normalized separately using two reference genes normalizers (AP3B1 and EIF2C). Error bars of the droplet Poisson distribution for the 95% Confidence Interval are indicated for each data point. B-C. Comparison of nanoString CN calls from FFPE vs immediately processed high quality (HQ) samples of the same cell lines (left; BT474, right; MCF7LTED) showed very high correlation (Pearson’s rho > 0.9, p < 2.2e-16). Data points at extreme ends (i.e. very low or very high expressed genes, n=6) are not included in the graph for better visualization of data. Plots of 61 genes and invariant controls shows highly correlated absolute CN calls for HQ and FFPE DNA of each cell line (top; BT474, bottom; MCF7LTED). Y-axis represent CN calls for each gene. D-E. MCF7LTED HQ and FFPE DNA was analyzed twice within the same run (D) and in two separate runs (E) to assess reproducibility. Reproducibility was very high within and between the runs (Pearson’s rho > 0.99, p < 2.2e-16). CN data points shown as log2 normalized counts.
Frequent ESR1 amplifications in metastatic breast cancer
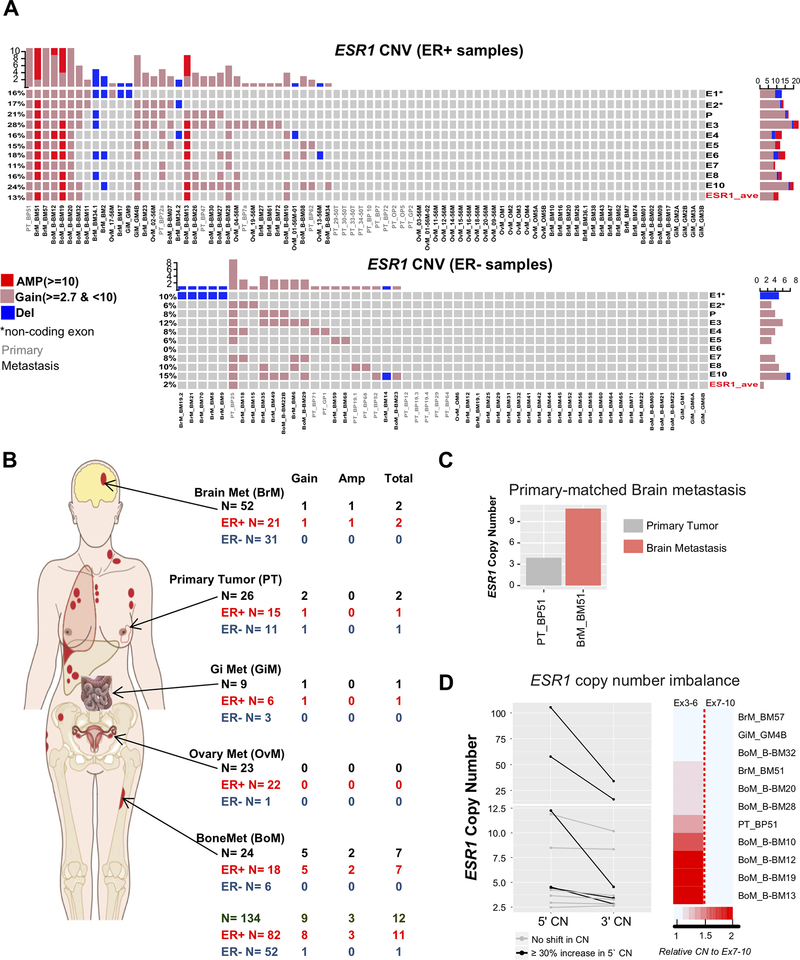
Our probe design targeted the ESR1 promoter region, untranslated exons (E1 and E2), and coding exons (E3-E10) (Additional file 2: Supplementary Figure S1). Given the high resolution of our coverage, we were able to call low level amplifications/gain (CN ≥ 2.7), in addition to amplification (CN ≥ 10). We measured ESR1 CN in a total of 134 tumor samples (82 ER+ and 52 ER−) (Figure 2). When averaging CN calls from all ESR1 probes, we detected ESR1 CN alterations in eleven (13.4%) out of 82 ER+ tumor samples (67 metastases and 15 primary) (Figure 2A and Table 1). Specifically, eight (9.8%) samples had CN gains and three (3.7%) samples had amplifications. There was a trend for enrichment of ESR1 CN alterations in metastatic samples (14.9%) compared to primary samples (6.6%). Bone metastases showed significant enrichment for ESR1 CN alterations vs primary breast cancer and ovarian metastases (Figure 2B and Table 1; p <0.05).
Figure 2: Frequent ESR1 amplifications in metastatic breast cancer.
A. Oncoprint visualization of ESR1 copy number alterations in ER+ (top) and ER− (bottom) samples. Levels of amplification and deletion are color coded. Each column represents a single sample (sample IDs labelled as: grey; primary tumor, black; metastasis). Each row indicates copy number call of the correspondent single exon probe. Untranslated exons (E1 and E2) are annotated with ‘*’ symbol. P; Promoter probe. ESR1_ave: average copy number call of all probes. B. Distribution of ESR1 copy number amplifications by site (primary, brain, bone, GI, and ovaries as indicated by arrows) and ER status (color coded). Bone metastases showed significant enrichment for amplifications vs primary and ovaries (Fisher’s exact test, p <0.05). C. Single pair where the brain metastasis had higher level ESR1 amplifications compared to its primary tumor. D. Graphical representation (left; actual copy number) and heatmap (right; ratio) of 5’−3’ copy number imbalance among the ESR1 amplified samples. The dark red color indicates higher copy number amplification toward the 5’ side of ESR1 (Wilcoxon matched pairs signed-rank test p = 0.0024).
Table 1:
ESR1 copy number alterations by site in ER+ samples
| Site | N | n (%) |
||
|---|---|---|---|---|
| AMP | Gain | Total | ||
| Primary | 15 | 0.0 | 1 (6.7) | 1 (6.7) |
| Brain | 21 | 1 (4.8) | 1 (4.8) | 2 (9.5) |
| Bone | 18 | 2 (11.1) | 5 (27.8) | 7 (38.9)* |
| Ovaries | 22 | 0.0 | 0.0 | 0.0 |
| GI | 6 | 0.0 | 1 (16.7) | 1 (16.7) |
| Total | 82 | 3 (3.7) | 8 (9.8) | 11 (13.4) |
significant enrichment for amplifications vs Primaries and ovaries (Fisher’s exact test, p <0.05)
N= number of samples included in the cohort; n and (%)= count and percentage of samples with alterations for the indicated site, respectively.
In ER− tumors (41 metastases and 11 primary), we did not detect any ESR1 amplifications, and found only one primary tumor with a CN gain (Figure 2A and Additional file 1: Supplementary Table S10). ESR1 CN alterations were significantly enriched in ER+ vs ER− tumors (Table 2; p = 0.0192).
Table 2:
ESR1 copy number alterations by ER status
| ER status | N | n (%) |
||
|---|---|---|---|---|
| AMP | Gain | Total | ||
| ER+ | 82 | 3 (3.7) | 8 (9.8) | 11 (13.4)* |
| ER− | 52 | 0.00 | 1 (1.9) | 1 (1.9) |
| Total | 134 | 3 (2.2) | 9 (6.7) | 12 (9) |
Fisher’s exact test, p =0.0192
N= number of samples included in the cohort; n and (%)= count and percentage of samples with alterations for the indicated site, respectively.
Since our cohort included 13 ER+ patient-matched primary-metastatic tumor pairs, we were able to explore if any metastatic lesions demonstrated an increase in CN as compared to their matched primary lesions. Indeed, we observed this for one pair, where the ER+ primary tumor BP51 had a ESR1 CN gain (CN 3.9), and the patient-matched brain metastases BM51 showed amplifications (CN 10.9) (Figure 2C and Additional file 2: Supplementary Figure S3).
We have recently described a novel algorithm that helps identify potential gene fusions from hybrid capture DNA sequencing based upon CN exon imbalances at fusion breakpoints [18]. The high resolution of nanoString exon-level CN detection within ESR1 allowed us to adapt this approach to determine ESR1 CN imbalance by calculating the ratio of CN signal of 5’ exon probes (3–6) to 3’ exon probes (7–10). Our analysis revealed that five out of 11 amplified samples (45.5%) exhibited ≥ 30% increase in CN (ratio of ≥ 1.3) in the 5’ exons (Figure 2D and Additional file 1: Supplementary Table s12, p= 0.0024), while no samples harbored an increase CN of 3’ exons relative to 5’ exons.
In summary, we observed enrichment of ESR1 amplifications in ER+, recurrent samples and bone metastases versus other metastatic sites. We also identified frequent ‘−3’ exonic imbalances indicating potential rearrangements in ESR1.
Correlation of ESR1 CN with ER mRNA expression
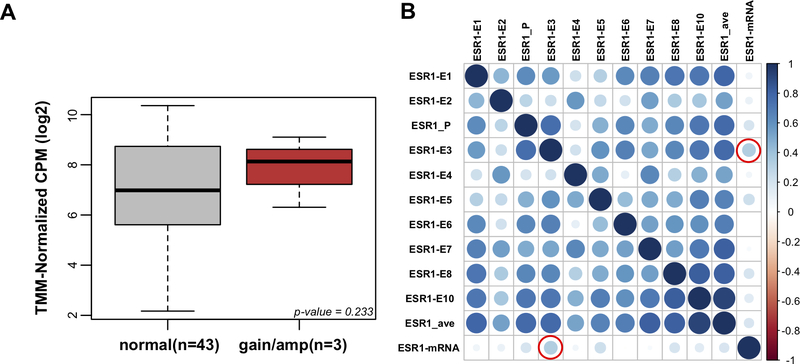
To determine whether ESR1 CN gains and amplifications were correlated with ER mRNA expression, we utilized expression data from RNAseq. As expected, we observed higher ESR1 mRNA expression in samples with ESR1 gains/amplifications (Figure 3A), although this association did not reach statistical significance, likely due to the limited number of samples in the CN gains/amplification group for which RNAseq data was available (n=3). An exploratory analysis within this group of ESR1 amplified samples showed a correlation between CN and expression, but this was also not significant (Additional file 2: Supplementary Figure S2). The first translated ESR1 exon E3 was most predictive for mRNA expression (Figure 3B and Additional file 1: Supplementary Table S13; rho=0.34, p= 0.0219) which supports the higher CN on the 5’ side.
Figure 3: Correlation of ESR1 copy number with mRNA expression.
A. Box plot comparison of ESR1 mRNA expression between normal CN vs gain/amplification ER+ groups (Wilcoxon rank-sum test, p=0.233). Y-axis: TMM-normalized log2 of count per million (CPM). B. Correlation matrix plot of ESR1 copy number calls by different exons and mRNA expression. Multiple correlations were clustered by first principal component (FPC) scores. Bigger and darker blue circles represent higher correlation. Red outline represents significant p-value for correlation with mRNA expression (p < 0.05).
Validation of ESR1 CN amplifications enrichment in additional cohorts
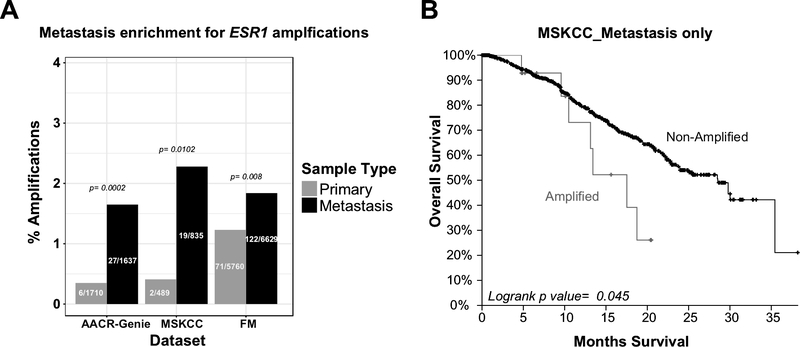
To validate our findings of ESR1 CN amplifications, we queried three additional datasets (AACR-GENIE, MSKCC, and FM) that used sequencing to characterize breast cancer metastases [35,36,43,44]. This analysis revealed significant enrichment for ESR1 amplifications in metastatic versus primary tumors (Figure 4A). The frequency of ESR1 amplifications was 1.65% (n=27/1637), 2.28% (n=19/835), and 1.84% (n=122/6629) in the AACR-GENIE, MSKCC, and FM metastatic cohorts, respectively. Lower frequency of amplifications in these cohorts are likely due to the use of techniques less sensitive for measurement of amplifications.
Figure 4: Metastasis enrichment for ESR1 amplifications in validation datasets.
A. Barplot for the percentage of ESR1 amplifications in three different genomic datasets (grey, primary; black, metastasis). Actual counts for amplified samples are indicated within each bar, with p values for metastatic enrichment on top. AACR, American Association for Cancer Research; MSKCC, Memorial Sloan Kettering Cancer Center; FM, Foundation Medicine. MSKCC is also a major contributor to the AACR_GENIE dataset. (For the FM dataset, samples denoted as “Primary” tumors can contain local recurrences since disease is defined by the site of biopsy). B. Kaplan-Meier curves for MSKCC survival data grouped by ESR1 CN status for patients with metastastic breast cancer.
In the FM cohort, where we have had access to ER status for a subset of the tumors, we observed a significant enrichment of ESR1 amplifications in ER+ (36/1272; 2.8%) compared to ER− samples (3/963; 0.3%) (p=1.9e-06). Analysis of survival data from MSKCC cohort showed significantly worse overall survival (OS) for patients with ESR1 amplified vs non-amplified tumors (p value <0.05, Figure 4B), pointing towards potential clinical relevance of the amplification.
CN alterations of known and potential breast cancer driver genes other than ESR1
In addition to ESR1, we extended our CN analysis to 66 genes with described roles in breast cancer progression and metastasis. For these genes, we had slightly lower resolution due to lower coverage (2–3 probes per gene) compared to the ESR1 study (10 probes), which did not allow for high confidence detection of low level amplifcations, and thus we limited the analysis to binary calls (yes or no CNV, defined as CN ≥ 5). Across all ER+ and ER− tumors, the genes with most frequent CN amplifications are shown in Table 3 and Additional file 2: Supplementary Figure S4. The most amplified genes were ERBB2 and GRB7, harboring amplifications of 44% and 21%, respectively, in brain and bone metastases, respectively. Previous studies have reported co-amplifications of ERBB2 and GRB7 on amplicon 17q12 [45–47]. This was confirmed by our eSNP-karyotyping analysis using RNAseq data from a subset of our samples that predicts the amplification to cover a broader region rather than being limited to a focal event (Additional file 2: Supplementary Figure S5). Metastasic site-unique or enriched CN alterations were seen for a number of genes, including higher rates of FADD amplifications (17%) in bone metastasis compared to other metastatic sites, and higher rates of PTK2 and PKIA amplifications (~10–20%) in brain and GI metastases. Additionally, comparison of ER+ vs ER− brain metastases showed significant enrichment of FGFR1 amplifications in the ER+ group (Additional file 2: Supplementary Figure S6; p= 0.0221). The comparative analysis between ER+ and ER− tumors was only possible in the brain metastases cohort where we had balanced distribution of ER+ vs ER− tumors. The most frequently deleted gene was TP53 (10%) and this loss was mainly observed in brain metastases (15.4%) (Table 3 and Additional file 2: Supplementary Figure S4).
Table 3:
Most frequent copy number alterations by site
| Alteration | Gene | n (%) | |||||
|---|---|---|---|---|---|---|---|
| Primary | All Mets | Brain | Bone | Ovary | GI | ||
| Amplifications | ERBB2 | 9 (34.6) | 31 (28.7) | 23 (44.2) | 5 (20.8) | 2 (8.6) | 1 (11.1) |
| GRB7 | 9 (34.6) | 29 (26.8) | 24 (46.1) | 2 (8.3) | 2 (8.6) | 1 (11.1) | |
| CCND1 | 1 (3.8) | 13 (12) | 6 (11.5) | 5 (20.8) | 2 (8.6) | 0 (0) | |
| MYC | 4 (15.3) | 13 (12) | 8 (15.3) | 4 (16.6) | 0 (0) | 1 (11.1) | |
| FGF19 | 3 (11.5) | 10 (9.2) | 6 (11.5) | 2 (8.3) | 2 (8.6) | 0 (0) | |
| CTTN | 0 (0) | 9 (8.3) | 5 (9.6) | 2 (8.3) | 2 (8.6) | 0 (0) | |
| FADD | 1 (3.8) | 8 (7.4) | 3 (5.7) | 4 (16.6) | 1 (4.3) | 0 (0) | |
| PTK2 | 0 (0) | 8 (7.4) | 6 (11.5) | 0 (0) | 0 (0) | 2 (22.2) | |
| PKIA | 0 (0) | 7 (6.4) | 5 (9.6) | 1 (4.1) | 0 (0) | 1 (11.1) | |
| FGFR1 | 4 (15.3) | 6 (5.5) | 4 (7.6) | 0 (0) | 2 (8.6) | 0 (0) | |
| Deletions | TP53 | 0 (0) | 11 (10.1) | 8 (15.3) | 1 (4.1) | 1 (4.3) | 1 (11.1) |
| AURKB | 0 (0) | 9 (8.3) | 7 (13.4) | 2 (8.3) | 0 (0) | 0 (0) | |
| CDKN1B | 3 (11.5) | 9 (8.3) | 9 (17.3) | 0 (0) | 0 (0) | 0 (0) | |
| CDKN2B | 3 (11.5) | 9 (8.3) | 6 (11.5) | 1 (4.1) | 2 (8.6) | 0 (0) | |
| CDKN2A | 4 (15.3) | 7 (6.4) | 5 (9.6) | 1 (4.1) | 1 (4.3) | 0 (0) | |
| MAP2K4 | 0 (0) | 7 (6.4) | 7 (13.4) | 0 (0) | 0 (0) | 0 (0) | |
| JUN | 0 (0) | 7 (6.4) | 6 (11.5) | 0 (0) | 1 (4.3) | 0 (0) | |
| LSMD1 | 1 (3.8) | 7 (6.4) | 4 (7.6) | 0 (0) | 3 (13) | 0 (0) | |
| PTEN | 2 (7.6) | 6 (5.5) | 6 (11.5) | 0 (0) | 0 (0) | 0 (0) | |
n and (%)= count and percentage of samples with alterations for the indicated site, respectively.
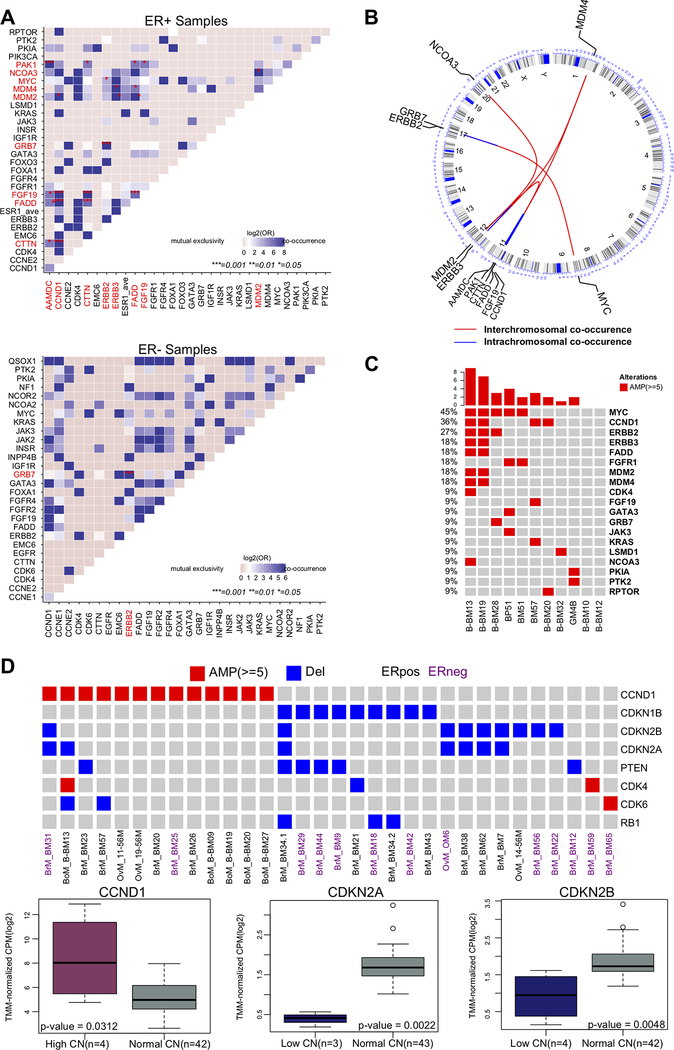
Co-occurrence analysis identified gene combinations that are enriched in ER+ tumors (Figure 5A, top panel; p < 0.05). For example, we confirmed previously reported co-amplifications of genes (CCND1, CTTN, FADD, PAK1, AAMDC, and FGF19) at the 11q13 amplicon [48–53], of MYC (8q24) and ERBB2 (17q12) [54–56], and of NCOA3 (20q13) and MDM2 (12q15) [57] (Figure 5B). We also observed co-amplification of MDM2 and ERBB3 (12q13), which had not been previously described. Finally, there was an enrichment of MYC (45%) and CCND1 (36%) in tumors with ESR1 amplifications (Figure 5C; p < 0.05). We identified only one event of co-occurrence in ER− tumors, this involved CN amplifications of ERBB2 and GRB7 at the 17q12 amplicon (Figure 5A; bottom panel).
Figure 5: Co-occurrence and mutual exclusivity of CN alterations in metastatic breast cancer.
A. Tile plots of ER+ (top) and ER− (bottom) samples for copy number amplifications co-occurrences. Gene pairs with significant co-occurrence are colored in red and marked with ‘*’ in the plot (Multiple Fisher’s exact test with FDR adjusted p <0.05). B. Circos plot for genomic co-occurrence events. Circular tracks from outside to inside: genome positions by chromosomes (black lines are cytobands); Inside arcs connects genes with co-occurrence (red; inter-chromosomal, blue; intra-chromosomal). C. Oncoprint shows enrichment of MYC (45%) and CCND1 (36%) amplifications in tumors with ESR1 amplifications (Fisher’s exact test; MYC p= 0.0083, CCND1, p= 0.0365). D. Top: Oncoprint of samples with CN alterations in the CDKs pathway and its inhibitors. Amplifications of the activators CCND1 and CDK4/6 of the pathway were mutually exclusive to the deletions of the inhibitory members CDKN1B, CDKN2A, and CDKN2B (Fisher’s exact test p < 0.0001). Bottom: RNAseq data for a subset of samples (n=46) shows significant correlation of mRNA expression with copy number status for genes of interest (p indicated for Wilcoxon rank-sum test).
Among the most frequent recurrent deletions, we identified mutually exclusive deletions of CDKN2A, CDKN2B, and CDKN1B with amplifications of CCND1 and CDK4/6 (Figure 5D; p < 0.0001). Expression analysis from RNAseq data showed significant correlation with these CN alterations (Figure 5D).
Discussion
Breast cancer is increasingly being recognized as a disease that can be driven by DNA structural variation, in addition to well described single base pair mutations. The majority of CN studies have been performed in primary tumors, at least in part due to difficulties in accessing metastatic and recurrent tissue. However, it is now well recognized that expression of key biomarkers such as ER and HER2 can change between a primary and metastatic breast cancer [58], and that this has clinical significance for therapeutic decision making.
Recent studies have documented ESR1 base pair mutations and fusions in breast cancer. ESR1 amplification has also been identified, although there have been conflicting reports on the frequency, ranging from 0 to 30% [3–8]. CN has been explored using FISH, aCGH, and qPCR, and sequencing is currently evolving as promising approach. Due to the potential clinical relevance, we performed a comprehensive measurement of ESR1 CN (by examining all exons) in a unique cohort of metastatic, and when available patient-matched primary tissues.
In total, we detected ESR1 gene gains or amplifications in 13% of ER+ metastatic breast tumors. Given the sensitivity and specificity of our method, and careful sampling of tumors with cellularity of at least 40%, we are confident that this is a representative number for ER+ metastatic tumors. Previously reported wide ranges in detected CN alterations are due, at least in part, to the use of technologies with limited sensitivities and specificity such as aCGH, FISH, and qPCR. Our study clearly shows PCR approaches using a limited number of reference genes need to be interpreted with caution. Additional reasons for variability include tumor cellularity and threshold for calling gain and amplification, as previously discussed [59].
We observed significant enrichment of ESR1 gains and amplification in ER+ metastatic lesions compared to the primary tumors, and we were able to validate this finding in three large datasets. In one out of 15 ER+ paired samples, ESR1 CN increased from 3.9 in the primary tumor to 10.9 in the patient-matched brain metastasis. This was not due to cellularity, since both the primary tumor and the metastatic lesion for sample #51 had similar tumor cellularities (~70%). In support of our findings, a recent study by Ullah et al showed an ESR1 amplifcation in an ER+ metastatic lesion, but not in the paired primary tumor [60]. This enrichment suggests selection of ESR1-amplified clones under endocrine treatment, similar to that described for ESR1 mutations [25,26] and HER2 amplifications in brain metastases [16]. Although somewhat limited by small numbers, we did observe a correlation between ESR1 amplifications and ESR1 mRNA expression, a finding that has previously been reported by others [2,61]. However, there are reports in which ESR1 amplification does not correlate with expression level [5,59,62], and ESR1 amplifications have been previously detected in ER− tumors with poor prognosis [63]. Of note, in our study, we observed an ESR1 gain (CN= 2.7) in only one ER− sample. Additional studies are necessary to decipher the functional consequences of low level ESR1 amplifications.
We [18] and others [17,64] have recently identified ESR1 gene fusions in metastatic breast cancer, with fusions generally maintaining the DNA binding and N-terminal transcriptional activation domains, but deleting the ligand-binding domain. We recently described a copyshift algorithm that determines imbalance in DNA CN of exons that are 5’ or 3’ to the break [18]. The high resolution of our nanoString assay allowed us to detect CN imbalances at the exon level, and we found that five out of 11 amplified samples (45.5%) showed at least 30% increase in CN at the 5’ side of ESR1. We did not have RNAseq data for four out of these five cases, and we were unable to detect an ESR1 fusion in the sample with available RNA seq data, likely due to inadequate sequencing coverage. Other mechanisms for generation of structural variants could be involved, such as fusions resulting from tandem duplications. An example is the ESR1-CCDC170 fusion [65] in which CCDC170 is truncated but the coding region of ESR1 is intact. Overall, these imbalanceles are indicative of genetic rearrangements, therefore future studies should determine whether tumors with significant exon imbalances harbor ESR1 fusion genes, and if so, which ones might be non-functional vs drivers of endocrine resistance.
The clinical relevance of ESR1 CN is unclear at this point in time. ESR1 amplifications have been associated with improved [2,59,61] as well as worse outcome [66] in patients treated with endocrine therapy. In endocrine resistant cell line models [67] and in a patient-derived xenografts (PDX) model [17] with ESR1 amplifications, estradiol treatment resulted in tumor regression. We recently described a metastatic breast cancer case with ESR1 amplifications that showed sustained partial response to high dose estradiol treatment as measured by CA 27–29 level and by decrease in liver metastasis burden [68]. These findings might deserve further exploration in a clinical trial setting including prospective measurement of ESR1 CN.
Analysis of 66 other genes revealed frequent ERBB2 and GRB7 amplifications in different metastatic sites, with an enrichment in brain metastases. These results support the increasingly growing need for testing HER2 in the metastatic settings [69,70]. Amplification of genes at the 11q13 locus has been reported in about 15% of primary breast cancer cases and is associated with poor prognosis [48–53]. In our analysis, amplifications of multiple genes at this locus (CCND1, CTTN, FADD, PAK1, AAMDC, and FGF19) were also frequent in multiple metastatic sites. Intriguingly, we observed co-amplification of ERBB3 and MDM2, which has not been previously described. Given prior evidence for functional interaction between ERBB3 signaling and MDM2 complex formation, co-amplification may be positively selected in some tumors. Similar to ESR1, some amplifications also showed organ-specific enrichments suggesting CN alterations may be driving metastatic tropisms. For example, brain and GI metastases showed higher PTK2 and PKIA amplifications (~10–20%), while FADD amplifications were more frequent in bone metastases (17%). In our patient-matched samples, most of the CN alterations were maintained in the pairs except for two pairs with a slight increase in CN for PKIA, PTK2, and FGFR4 in the metastases (Additional file 2: Supplementary Figure S7). Moreover, FGFR1 amplifications were enriched in ER+ vs ER− brain metastases.
Cyclin-dependent kinases (CDKs), which control transitions through the different stages of cell cycle, have been considered as promising targets for cancer therapy. Amplifications/overexpression of CCND1 (CDK4/6 activator) and loss of CDKN2A (p16, CDK4/6 inhibitor) have been described in primary breast cancer patients [71–77]. Preclinical and in vitro data across multiple cancers support that loss of CDK4/6 inhibitory members may serve as biomarkers for CDK4/6 inhibition sensitivity [78–83]. On the other hand, clinical evidence in breast cancer trials performed thus far showed that CCND1 amplifications or p16 loss are unlikely to predict treatment benefit [84]. However, these trials assessing p16 as a biomarker were earlier phase with small sample size, and didn’t show if concurrent alterations in other genes existed, which can alter response to therapy. In our analysis, 32 out of 108 metastases (29%) showed aberrations in the CDKs pathway and 24 samples (22%) had alterations in the CCND1-CDK4/6 axis specifically. Among those, deletions of the CDK4/6 inhibitors CDKN2A/B were significantly mutually exclusive with CCND1 amplifications, and correlated with mRNA expression, suggesting functional consequences for the alterations. Those deletions were maintained in the paired metastases, which supports the concept of using CDK4/6 inhibitors in metastatic settings as previously shown [85–88]. Moreover, it is important to note that these deletions also occur in ER− tumors and tumors with HER2 amplifications. Most clinical trials have been conducted in patients with ER+/HER2- breast cancer. Our data suggests that testing of CDK4/6 inhibitors in other subgroups (e.g. ER−) may deserve further investigation.
Conclusion
Our findings suggest that, in addition to ESR1 mutations, ESR1 amplifications and exon imbalances might play a role in endocrine resistance, but further studies are necessary to test this hypothesis. Our analysis also defines CDK pathway alterations as common in metastatic disease—which could be especially relevant to recently adopted CDK4/6 inhibitor therapy. Taken together, copy number alterations in druggable genes are common in metastatic breast cancer and could serve as biomarkers for precision therapies in advanced disease.
Supplementary Material
Implications: Copy number alterations of ESR1 and key CDK pathway genes are frequent in metastatic breast cancers, and their clinical relevance should be tested further.
Acknowledgements
This project used the UPMC HCC Tissue and Research Pathology/Health Sciences Tissue Bank shared resource, which is supported in part by award P30CA047904.
Funding
We kindly acknowledge the funding support from Breast Cancer Research Foundation (AVL, SO), National Cancer Institute of the National Institutes of Health (P30CA047904), Fashion Footwear Association of New York (FFANY), the Nicole Meloche Memorial Breast Cancer Research Foundations, the Penguins Alumni Foundation, the Shear Family Foundation, and Magee-Womens Research Institute and Foundation. AB is supported by full fellowship from KSU. AVL and SO are recipients of Scientific Advisory Council awards from Susan G. Komen for the Cure, and Hillman Foundation Fellows. NP was supported by a training grant from the NIH/NIGMS (2T32GM008424–21) and an individual fellowship from the NIH/NCI (5F30CA203095). Kurt Weiss is supported by K08 CA177927.
List of abbreviations
- aCGH
array comparative genomic hybridization
- CCLE
the cell cancer cell line encyclopedia
- CN
copy number
- ddPCR
digital droplet polymerase chain reaction
- DGIdb
drug gene interaction database
- ER
estrogen receptor
- FDR
false discovery rate
- FFPE
formalin-fixed paraffin-embedded
- FISH
fluorescence in situ hybridization
- IDC
invasive ductal breast cancer
- IHC
immunohistochemistry
- ILC
invasive lobular breast cancer
- LTED
long-term estrogen-deprived
- MBC
metastatic breast cancer
- METABRIC
Molecular Taxonomy of Breast Cancer International Consortium
- qPCR
quantitative polymerase chain reaction
- TCGA
The Cancer Genome Atlas
Footnotes
Declarations
Ethics approval and consent to participate
Collection and analysis of specimens were approved under the University of Pittsburgh IRB # PRO 11100645, 14040193, 1500502, 16020301, and Charite Universitätsmedizin Berlin guidelines.
Consent for publication
Not applicable
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. [Internet]. 2013. [cited 2018 Feb 19];45:1127–33. Available from: http://www.nature.com/doifinder/10.1038/ng.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat. Genet. [Internet] 2007. [cited 2016 May 9];39:655–60. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17417639 [DOI] [PubMed] [Google Scholar]
- 3.Brown LA, Hoog J, Chin S-F, Tao Y, Zayed AA, Chin K, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. [Internet] Nature Publishing Group; 2008. [cited 2017 Feb 28];40:806–7. Available from: http://www.nature.com/doifinder/10.1038/ng0708-806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horlings HM, Bergamaschi A, Nordgard SH, Kim YH, Han W, Noh D-Y, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. [Internet] 2008. [cited 2016 May 9];40:807–8–2. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18583965 [DOI] [PubMed] [Google Scholar]
- 5.Reis-Filho JS, Drury S, Lambros MB, Marchio C, Johnson N, Natrajan R, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. [Internet] Nature Publishing Group; 2008. [cited 2017 Feb 28];40:809–10. Available from: http://www.nature.com/doifinder/10.1038/ng0708-809b [DOI] [PubMed] [Google Scholar]
- 6.Vincent-Salomon A, Raynal V, Lucchesi C, Gruel N, Delattre O. ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. [Internet] Nature Publishing Group; 2008. [cited 2017 Feb 28];40:809–809. Available from: http://www.nature.com/doifinder/10.1038/ng0708-809a [DOI] [PubMed] [Google Scholar]
- 7.Albertson DG. Conflicting evidence on the frequency of ESR1 amplification in breast cancer Nat. Genet. [Internet] Nature Publishing Group; 2008. [cited 2017 Feb 28];40:821–2. Available from: http://www.nature.com/doifinder/10.1038/ng0708-821 [DOI] [PubMed] [Google Scholar]
- 8.Holst F, Singer CF. ESR1-Amplification-Associated Estrogen Receptor α Activity in Breast Cancer. Trends Endocrinol. Metab. [Internet] 2016. [cited 2018 Feb 19];27:751–2. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1043276016300911 [DOI] [PubMed] [Google Scholar]
- 9.cBioPoratal for cancer genomics [Internet]. [cited 2017 Jan 3]. Available from: http://www.cbioportal.org
- 10.Cancer T, Atlas G. Comprehensive molecular portraits of human breast tumours. Nature [Internet]. 2012. [cited 2014 Jul 9];490:61–70. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3465532&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature [Internet] 2012. [cited 2018 Feb 18];486:346–52. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22522925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desmedt C, Zoppoli G, Gundem G, Pruneri G, Larsimont D, Fornili M, et al. Genomic Characterization of Primary Invasive Lobular Breast Cancer. J. Clin. Oncol. [Internet] 2016. [cited 2016 May 9]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/26926684 [DOI] [PubMed] [Google Scholar]
- 13.Fumagalli D, Wilson TR, Salgado R, Lu X, Yu J, O’Brien C, et al. Somatic mutation, copy number and transcriptomic profiles of primary and matched metastatic estrogen receptor-positive breast cancers. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. [Internet] 2016. [cited 2017 Dec 29];27:1860–6. Available from: https://academic.oup.com/annonc/article-lookup/doi/10.1093/annonc/mdw286 [DOI] [PubMed] [Google Scholar]
- 14.Fu X, Jeselsohn R, Pereira R, Hollingsworth EF, Creighton CJ, Li F, et al. FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc. Natl. Acad. Sci. U. S. A. [Internet] 2016. [cited 2017 Dec 29];113:E6600–9. Available from: http://www.pnas.org/lookup/doi/10.1073/pnas.1612835113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matissek KJ, Onozato ML, Sun S, Zheng Z, Schultz A, Lee J, et al. Expressed Gene Fusions as Frequent Drivers of Poor Outcomes in Hormone Receptor Positive Breast Cancer. Cancer Discov. [Internet] 2017. [cited 2017 Dec 29];CD-17–0535 Available from: http://cancerdiscovery.aacrjournals.org/lookup/doi/10.1158/2159-8290.CD-17-0535 [DOI] [PubMed] [Google Scholar]
- 16.Priedigkeit N, Hartmaier RJ, Chen Y, Vareslija D, Basudan A, Watters RJ, et al. Intrinsic Subtype Switching and Acquired ERBB2 / HER2 Amplifications and Mutations in Breast Cancer Brain Metastases. JAMA Oncol. [Internet] 2016. [cited 2017 Mar 2]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/27926948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. [Internet] 2013. [cited 2016 Dec 14];4:1116–30. Available from: http://linkinghub.elsevier.com/retrieve/pii/S2211124713004634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmaier RJ, Trabucco SE, Priedigkeit N, Chung JH, Parachoniak CA, Vanden Borre P, et al. Recurrent hyperactive ESR1 fusion proteins in endocrine therapy resistant breast cancer. Ann. Oncol. [Internet] 2018. [cited 2018 Feb 19]; Available from: http://academic.oup.com/annonc/advance-article/doi/10.1093/annonc/mdy025/4817340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. [Internet] 2011. [cited 2018 Jan 17];12:R6 Available from: http://genomebiology.biomedcentral.com/articles/10.1186/gb-2011-12-1-r6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwakawa R, Takenaka M, Kohno T, Shimada Y, Totoki Y, Shibata T, et al. Genome-wide identification of genes with amplification and/or fusion in small cell lung cancer. Genes. Chromosomes Cancer [Internet] 2013. [cited 2018 Jan 17];52:802–16. Available from: http://doi.wiley.com/10.1002/gcc.22076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varešlija D, Priedigkeit N, Fagan A, Purcell S, Cosgrove N, O’Halloran PJ, et al. Transcriptome Characterization of Matched Primary Breast and Brain Metastatic Tumors to Detect Novel Actionable Targets. J. Natl. Cancer Inst. [Internet] 2018. [cited 2018 Sep 17]; Available from: https://academic.oup.com/jnci/advance-article/doi/10.1093/jnci/djy110/5046632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature [Internet] 2012. [cited 2016 May 9];483:603–307. Available from: http://www.nature.com/doifinder/10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soule H, McGrath CM. Immortal human mammary epithelial cell lines. US Pat. 5,026,637. 1991;
- 24.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. [Internet] 2011. [cited 2016 May 9];83:8604–10. Available from: http://pubs.acs.org/doi/abs/10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson DR, Wu Y-M, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. [Internet] 2013. [cited 2016 Aug 9];45:1446–51. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24185510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. [Internet] 2013. [cited 2016 Aug 9];45:1439–45. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24185512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig DW, O’Shaughnessy JA, Kiefer JA, Aldrich J, Sinari S, Moses TM, et al. Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol. Cancer Ther. [Internet] 2013. [cited 2016 Aug 9];12:104–16. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23171949 [DOI] [PubMed] [Google Scholar]
- 28.Chang K, Creighton CJ, Davis C, Donehower L, Drummond J, Wheeler D, et al. The Cancer Genome Atlas Pan-Cancer analysis project Nat. Genet. [Internet] Nature Research; 2013. [cited 2016 Aug 9];45:1113–20. Available from: http://www.nature.com/doifinder/10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffith M, Griffith OL, Coffman AC, Weible JV, McMichael JF, Spies NC, et al. DGIdb: mining the druggable genome. Nat. Methods [Internet] 2013. [cited 2016 May 9];10:1209–10. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24122041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.nSolverTM Analysis Software | NanoString Technologies [Internet] Available from: http://www.nanostring.com/products/nSolver/
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. [Internet] 2014. [cited 2016 Aug 9];15:550 Available from: http://www.ncbi.nlm.nih.gov/pubmed/25516281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data Bioinformatics [Internet]. Oxford University Press; 2010. [cited 2016 Dec 15];26:139–40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19910308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weissbein U, Schachter M, Egli D, Benvenisty N. Analysis of chromosomal aberrations and recombination by allelic bias in RNA-Seq. Nat. Commun. [Internet]. 2016. [cited 2017 Jun 15];7:12144 Available from: http://www.nature.com/doifinder/10.1038/ncomms12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DePristo MA, Banks E, Poplin R, Garimella K V, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. [Internet] 2011. [cited 2017 Jun 15];43:491–8. Available from: http://www.nature.com/doifinder/10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. [Internet] 2013. [cited 2018 Mar 26];31:1023–31. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24142049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung JH, Pavlick D, Hartmaier R, Schrock AB, Young L, Forcier B, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann. Oncol. [Internet] 2017. [cited 2018 Mar 26];28:2866–73. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28945887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. [Internet] American Association for Cancer Research; 2012. [cited 2018 Mar 13];2:401–4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22588877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. [Internet] 2013. [cited 2018 Mar 13];6:pl1–pl1. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23550210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Core Team (2013). R: The R Project for Statistical Computing [Internet]. Available from: https://www.r-project.org/
- 40.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data Bioinformatics [Internet]. Oxford University Press; 2016. [cited 2016 Aug 9];btw313 Available from: http://bioinformatics.oxfordjournals.org/lookup/doi/10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 41.Wickham H ggplot2: Elegant Graphics for Data Analysis [Internet]. Springer-Verlag; New York; 2009. Available from: http://ggplot2.org [Google Scholar]
- 42. https://raw.githubusercontent.com/obigriffith/biostar-tutorials/master/Heatmaps/heatmap.3.R.
- 43.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. [Internet] 2017. [cited 2018 Mar 13];7:818–31. Available from: http://cancerdiscovery.aacrjournals.org/lookup/doi/10.1158/2159-8290.CD-17-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients Nat. Med. [Internet] NIH Public Access; 2017. [cited 2018 Mar 13];23:703–13. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28481359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito M, Kato Y, Ito E, Fujimoto J, Ishikawa K, Doi A, et al. Expression screening of 17q12–21 amplicon reveals GRB7 as an ERBB2-dependent oncogene. FEBS Lett. [Internet] 2012. [cited 2018 Jan 18];586:1708–14. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22584052 [DOI] [PubMed] [Google Scholar]
- 46.Sahlberg KK, Hongisto V, Edgren H, Mäkelä R, Hellström K, Due EU, et al. The HER2 amplicon includes several genes required for the growth and survival of HER2 positive breast cancer cells. Mol. Oncol. [Internet] 2013. [cited 2018 Jan 18];7:392–401. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23253899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacot W, Fiche M, Zaman K, Wolfer A, Lamy P-J. The HER2 amplicon in breast cancer: Topoisomerase IIA and beyond. Biochim. Biophys. Acta - Rev. Cancer [Internet] 2013. [cited 2018 Jan 18];1836:146–57. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23628726 [DOI] [PubMed] [Google Scholar]
- 48.Hui R, Campbell DH, Lee CS, McCaul K, Horsfall DJ, Musgrove EA, et al. EMS1 amplification can occur independently of CCND1 or INT-2 amplification at 11q13 and may identify different phenotypes in primary breast cancer. Oncogene [Internet] 1997. [cited 2017 Jun 20];15:1617–23. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9380415 [DOI] [PubMed] [Google Scholar]
- 49.Shibata T, Uryu S, Kokubu A, Hosoda F, Ohki M, Sakiyama T, et al. Genetic Classification of Lung Adenocarcinoma Based on Array-Based Comparative Genomic Hybridization Analysis: Its Association with Clinicopathologic Features. Clin. Cancer Res. [Internet] 2005. [cited 2017 Jun 20];11:6177–85. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16144918 [DOI] [PubMed] [Google Scholar]
- 50.Zaharieva BM, Simon R, Diener P-A, Ackermann D, Maurer R, Alund G, et al. High-throughput tissue microarray analysis of 11q13 gene amplification (CCND1, FGF3, FGF4, EMS1) in urinary bladder cancer. J. Pathol. [Internet] 2003. [cited 2017 Jun 20];201:603–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14648664 [DOI] [PubMed] [Google Scholar]
- 51.Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res. Treat. [Internet] 2003. [cited 2017 Jun 20];78:323–35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12755491 [DOI] [PubMed] [Google Scholar]
- 52.Hui R, Ball JR, Macmillan RD, Kenny FS, Prall OW, Campbell DH, et al. EMS1 gene expression in primary breast cancer: relationship to cyclin D1 and oestrogen receptor expression and patient survival. Oncogene [Internet] 1998. [cited 2017 Jun 20];17:1053–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9747885 [DOI] [PubMed] [Google Scholar]
- 53.Borg A, Sigurdsson H, Clark GM, Fernö M, Fuqua SA, Olsson H, et al. Association of INT2/HST1 coamplification in primary breast cancer with hormone-dependent phenotype and poor prognosis. Br. J. Cancer [Internet] 1991. [cited 2017 Jun 20];63:136–42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/1989653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borg A, Baldetorp B, Fernö M, Olsson H, Sigurdsson H. c-myc amplification is an independent prognostic factor in postmenopausal breast cancer. Int. J. cancer [Internet] 1992. [cited 2018 Feb 19];51:687–91. Available from: http://www.ncbi.nlm.nih.gov/pubmed/1612775 [DOI] [PubMed] [Google Scholar]
- 55.Mitsui F, Dobashi Y, Imoto I, Inazawa J, Kono K, Fujii H, et al. Non-incidental coamplification of Myc and ERBB2 and Myc and EGFR, in gastric adenocarcinomas. Mod. Pathol. [Internet] 2007. [cited 2018 Feb 19];20:622–31. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17431415 [DOI] [PubMed] [Google Scholar]
- 56.Ooi A, Suzuki S, Nakazawa K, Itakura J, Imoto I, Nakamura H, et al. Gene amplification of Myc and its coamplification with ERBB2 and EGFR in gallbladder adenocarcinoma. Anticancer Res. [Internet] 2009. [cited 2018 Feb 19];29:19–26. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19331129 [PubMed] [Google Scholar]
- 57.Courjal F, Cuny M, Rodriguez C, Louason G, Speiser P, Katsaros D, et al. DNA amplifications at 20q13 and MDM2 define distinct subsets of evolved breast and ovarian tumours Br. J. Cancer [Internet] Nature Publishing Group; 1996. [cited 2018 Feb 19];74:1984–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8980401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aurilio G, Disalvatore D, Pruneri G, Bagnardi V, Viale G, Curigliano G, et al. A meta-analysis of oestrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 discordance between primary breast cancer and metastases. Eur. J. Cancer [Internet] 2014. [cited 2018 Apr 20];50:277–89. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24269135 [DOI] [PubMed] [Google Scholar]
- 59.Holst F Estrogen receptor alpha gene amplification in breast cancer: 25 years of debate World Journal of Clinical Oncology. World J Clin Oncol World J Clin Oncol April [Internet] 2016. [cited 2017 Mar 1];7:160–73. Available from: http://www.wjgnet.com/2218-4333/full/v7/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ullah I, Karthik G-M, Alkodsi A, Kjällquist U, Stålhammar G, Lövrot J, et al. Evolutionary history of metastatic breast cancer reveals minimal seeding from axillary lymph nodes. J. Clin. Invest. [Internet] 2018. [cited 2018 Apr 23];128:1355–70. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29480816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomita S, Zhang Z, Nakano M, Ibusuki M, Kawazoe T, Yamamoto Y, et al. Estrogen receptor alpha gene ESR1 amplification may predict endocrine therapy responsiveness in breast cancer patients. Cancer Sci. [Internet] 2009. [cited 2017 Mar 2];100:1012–7. Available from: http://doi.wiley.com/10.1111/j.1349-7006.2009.01145.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ooi A, Inokuchi M, Harada S, Inazawa J, Tajiri R, Kitamura SS, et al. Gene amplification of ESR1 in breast cancers-fact or fiction? A fluorescence in situ hybridization and multiplex ligation-dependent probe amplification study. J. Pathol. [Internet] 2012. [cited 2017 Mar 1];227:8–16. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22170254 [DOI] [PubMed] [Google Scholar]
- 63.Markiewicz A, Wełnicka-Jaśkiewicz M, Skokowski J, Jaśkiewicz J, Szade J, Jassem J, et al. Prognostic significance of ESR1 amplification and ESR1 PvuII, CYP2C19*2, UGT2B15*2 polymorphisms in breast cancer patients. Medeiros R, editor. PLoS One [Internet]. 2013. [cited 2017 Mar 1];8:e72219 Available from: http://dx.plos.org/10.1371/journal.pone.0072219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lei JT, Shao J, Zhang J, Iglesia M, Chan DW, Cao J, et al. Functional Annotation of ESR1 Gene Fusions in Estrogen Receptor-Positive Breast Cancer. Cell Rep. [Internet] 2018. [cited 2018 Aug 30];24:1434–1444.e7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30089255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veeraraghavan J, Tan Y, Cao X-X, Kim JA, Wang X, Chamness GC, et al. Recurrent ESR1–CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat. Commun. [Internet] 2014. [cited 2018 Aug 8];5:4577 Available from: http://www.ncbi.nlm.nih.gov/pubmed/25099679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen KV, Ejlertsen B, Müller S, Møller S, Rasmussen BB, Balslev E, et al. Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res. Treat. [Internet] 2011. [cited 2017 Mar 2];127:345–55. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20556506 [DOI] [PubMed] [Google Scholar]
- 67.Song RX, Mor G, Naftolin F, McPherson RA, Song J, Zhang Z, et al. Effect of long-term estrogen deprivation on apoptotic responses of breast cancer cells to 17beta-estradiol. J. Natl. Cancer Inst. [Internet]. 2001. [cited 2018 Feb 19];93:1714–23. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11717332 [DOI] [PubMed] [Google Scholar]
- 68.Kota K, Brufsky A, Oesterreich S, Lee A. Estradiol as a Targeted, Late-Line Therapy in Metastatic Breast Cancer with Estrogen Receptor Amplification. 2017. [cited 2017 Aug 21]; Available from: http://assets.cureus.com/uploads/case_report/pdf/7550/1503088326-20170818-4-vzwbsi.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palmieri D, Bronder JL, Herring JM, Yoneda T, Weil RJ, Stark AM, et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. [Internet]. 2007. [cited 2017 Jun 20];67:4190–8. Available from: http://cancerres.aacrjournals.org/cgi/doi/10.1158/0008-5472.CAN-06-3316 [DOI] [PubMed] [Google Scholar]
- 70.Wolff AC, Hammond MEH, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. J. Clin. Oncol. [Internet] 2013. [cited 2017 Jun 20];31:3997–4013. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24101045 [DOI] [PubMed] [Google Scholar]
- 71.Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, et al. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. [Internet] 1994. [cited 2017 Jun 20];54:1812–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8137296 [PubMed] [Google Scholar]
- 72.Weinstat-Saslow D, Merino MJ, Manrow RE, Lawrence JA, Bluth RF, Wittenbel KD, et al. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat. Med. [Internet] 1995. [cited 2017 Jun 20];1:1257–60. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7489405 [DOI] [PubMed] [Google Scholar]
- 73.Kenny FS, Hui R, Musgrove EA, Gee JM, Blamey RW, Nicholson RI, et al. Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin. Cancer Res. [Internet] 1999. [cited 2017 Jun 20];5:2069–76. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10473088 [PubMed] [Google Scholar]
- 74.McIntosh GG, Anderson JJ, Milton I, Steward M, Parr AH, Thomas MD, et al. Determination of the prognostic value of cyclin D1 overexpression in breast cancer. Oncogene [Internet]. 1995. [cited 2017 Jun 20];11:885–91. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7675447 [PubMed] [Google Scholar]
- 75.Yu Q, Sicinska E, Geng Y, Ahnström M, Zagozdzon A, Kong Y, et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell [Internet] 2006. [cited 2017 Jun 20];9:23–32. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16413469 [DOI] [PubMed] [Google Scholar]
- 76.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. [Internet] 2016. [cited 2017 Jun 20];13:417–30. Available from: http://www.nature.com/doifinder/10.1038/nrclinonc.2016.26 [DOI] [PubMed] [Google Scholar]
- 77.Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo A, et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat. Genet. [Internet] 1995. [cited 2017 Jun 20];11:210–2. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7550353 [DOI] [PubMed] [Google Scholar]
- 78.Cen L, Carlson BL, Schroeder MA, Ostrem JL, Kitange GJ, Mladek AC, et al. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro. Oncol. [Internet] 2012. [cited 2017 Jun 20];14:870–81. Available from: https://academic.oup.com/neuro-oncology/article-lookup/doi/10.1093/neuonc/nos114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Konecny GE, Winterhoff B, Kolarova T, Qi J, Manivong K, Dering J, et al. Expression of p16 and Retinoblastoma Determines Response to CDK4/6 Inhibition in Ovarian Cancer. Clin. Cancer Res. [Internet] 2011. [cited 2017 Aug 29];17:1591–602. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21278246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Logan JE, Mostofizadeh N, Desai AJ, VON Euw E, Conklin D, Konkankit V, et al. PD-0332991, a potent and selective inhibitor of cyclin-dependent kinase 4/6, demonstrates inhibition of proliferation in renal cell carcinoma at nanomolar concentrations and molecular markers predict for sensitivity. Anticancer Res. [Internet] 2013. [cited 2017 Aug 29];33:2997–3004. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23898052 [PubMed] [Google Scholar]
- 81.Pal SK, Ali SM, Ross J, Choueiri TK, Chung JH. Exceptional Response to Palbociclib in Metastatic Collecting Duct Carcinoma Bearing a CDKN2A Homozygous Deletion JCO Precis. Oncol. [Internet]. American Society of Clinical Oncology; 2017. [cited 2017 Aug 29];1–5. Available from: http://ascopubs.org/doi/10.1200/PO.17.00017 [DOI] [PubMed] [Google Scholar]
- 82.Gao J, Adams RP, Swain SM. Does CDKN2A loss predict palbociclib benefit? Curr. Oncol. [Internet] Multimed Inc.; 2015. [cited 2017 Aug 29];22:e498–501. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26715889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. [Internet] 2009. [cited 2017 Aug 29];11:R77 Available from: http://breast-cancer-research.biomedcentral.com/articles/10.1186/bcr2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. [Internet] 2015. [cited 2017 Aug 29];16:25–35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25524798 [DOI] [PubMed] [Google Scholar]
- 85.Turner NC, Ro J, André F, Loi S, Verma S, Iwata H, et al. Palbociclib in Hormone-Receptor–Positive Advanced Breast Cancer. N. Engl. J. Med. [Internet] 2015. [cited 2018 Apr 23];373:209–19. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26030518 [DOI] [PubMed] [Google Scholar]
- 86.Hortobagyi GN, Stemmer SM, Burris HA, Yap Y-S, Sonke GS, Paluch-Shimon S, et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N. Engl. J. Med. [Internet] 2016. [cited 2018 Apr 23];375:1738–48. Available from: http://www.nejm.org/doi/10.1056/NEJMoa1609709 [DOI] [PubMed] [Google Scholar]
- 87.Finn RS, Martin M, Rugo HS, Jones S, Im S-A, Gelmon K, et al. Palbociclib and Letrozole in Advanced Breast Cancer N. Engl. J. Med. [Internet] Massachusetts Medical Society; 2016. [cited 2018 Jan 24];375:1925–36. Available from: http://www.nejm.org/doi/10.1056/NEJMoa1607303 [DOI] [PubMed] [Google Scholar]
- 88.Wolff AC. CDK4 and CDK6 Inhibition in Breast Cancer - A New Standard. N. Engl. J. Med. [Internet] 2016. [cited 2018 Apr 23];375:1993–4. Available from: http://www.nejm.org/doi/10.1056/NEJMe1611926 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.