Abstract
Tumor metastasis is a major contributor to cancer patient mortality, but the process remains poorly understood. Molecular comparisons between primary tumors and metastases can provide insights into the pathways and processes involved. Here, we systematically analyzed and cataloged molecular correlates of metastasis using The Cancer Genome Atlas (TCGA) datasets across 11 different cancer types, these data involving 4,473 primary tumor samples and 395 tumor metastasis samples (including 369 from melanoma). For each cancer type, widespread differences in gene transcription between primary and metastasis samples were observed. For several cancer types, metastasis-associated genes from TCGA comparisons were found to overlap extensively with external results from independent profiling datasets of metastatic tumors. While some differential expression patterns associated with metastasis were found to be shared across multiple cancer types, by and large each cancer type showed a metastasis signature that was distinctive from those of the other cancer types. Functional categories of genes enriched in multiple cancer type-specific metastasis over-expression signatures included cellular response to stress, DNA repair, oxidation-reduction process, protein deubiquitination, and receptor activity. The TCGA-derived prostate cancer metastasis signature in particular could define a subset of aggressive primary prostate cancer. Transglutaminase 2 protein and mRNA were both elevated in metastases from breast and melanoma cancers. Alterations in microRNAs and in DNA methylation were also identified.
Introduction
Metastases are formed by cancer cells that have left the primary tumor mass to form new colonies at sites throughout the human body(1). Tumor metastasis remains a major contributor to cancer patient deaths(2). Metastasis is a multi-step process, which includes localized invasion, intravasation into lymphatic or blood vessels, traversal of the bloodstream, extravasation from the bloodstream, formation of micrometastasis, and colonization(1, 2). The process of metastasis and the factors governing cancer spread and establishment at secondary locations remains poorly understood(3). Only a small fraction of cancer cells from the primary tumor may go on to successfully establish distant, macroscopic metastasis, and while the tumor microenvironment is understood to play an important role(3), the molecular state of the cancer cells in a macroscopic metastasis may widely differ from that of the cancer cells in the associated primary tumor.
Molecular comparisons between primary tumors and metastases can potentially provide insights into the pathways and processes involved with cancer disease progression(4, 5). Numerous independent studies have carried out gene expression profiling of metastasis versus primary cancer for individual cancer types(4–18). In addition to individual studies by cancer type, “pan-cancer” molecular analyses would allow for examining similarities and differences among the molecular alterations that may be associated with metastasis across diverse cancer types. The recently published “MET500” dataset includes transcriptome profiling data for metastasis samples from ~500 patients, involving over 30 primary sites and biopsied from over 22 organs(19); however, the MET500 dataset does not include any data on primary cancers. The Cancer Genome Atlas (TCGA), a large-scale initiative to comprehensively profile over 10,000 cancer cases at the molecular level, includes data on some metastasis samples as well as on primary samples. Other than the TCGA-sponsored melanoma marker study(20), the metastasis samples were not featured in the respective marker analyses by cancer type that were led by TCGA network, as the project as a whole was focused on primary disease. The advantages of analyzing TCGA data for metastasis-associated molecular correlations include the multiple cancer types having been profiled on a common platform that involves multiple levels of molecular data in addition to mRNA expression.
In this present study, we systematically analyzed and cataloged molecular correlates of metastasis using TCGA datasets, across 11 different cancer types for which metastasis versus primary data were available. Molecular profiling data platforms analyzed included mRNA expression, protein expression, microRNA expression, and DNA methylation. Significantly altered genes, as identified in a given cancer type, were compared across the other cancer types, as well as across results from other profiling datasets from studies outside of TCGA.
Materials and methods
TCGA patient cohort
Results are based upon data generated by TCGA Research Network (https://gdc.cancer.gov/). Molecular data were aggregated from public repositories. Tumors analyzed in this study spanned 11 different TCGA projects, each project representing a specific cancer type, listed as follows: BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CRC, Colorectal adenocarcinoma (combining COAD and READ projects); ESCA, Esophageal carcinoma; HNSC, Head and Neck squamous cell carcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; THCA, Thyroid carcinoma. Cancer molecular profiling data were generated through informed consent as part of previously published studies and analyzed in accordance with each original study’s data use guidelines and restrictions. Metastasis versus primary samples were inferred using the TCGA sample code (“06” versus “01,” respectively), which is the two digit code following the TCGA legacy sample name (e.g. metastasis sample “TCGA-V1-A9O5–06” and primary sample “TCGA-ZG-A9L9–01”).
Datasets
RNA-seq data were obtained from The Broad Institute Firehose pipeline (http://gdac.broadinstitute.org/). All RNA-seq samples were aligned using the by UNC RNA-seq V2 pipeline(21). Expression of coding genes was quantified for 20531 features based on the gene models defined in the TCGA Gene Annotation File (GAF). Gene expression was quantified by counting the number of reads overlapping each gene model’s exons and converted to Reads per Kilobase Mapped (RPKM) values by dividing by the transcribed gene length, defined in the GAF and by the total number of reads aligned to genes. Proteomic data generated by RPPA across 7663 patient tumors (“Level 4” data) were obtained from The Cancer Proteome Atlas (http://tcpaportal.org/tcpa/)(22). The miRNA-seq dataset was obtained from TCGA PanCanAtlas project (https://gdc.cancer.gov/about-data/publications/pancanatlas)(23), which dataset involved batch correction according to Illumina GAIIx or HiSeq 2000 platforms. DNA methylation profiles for 450K Illumina array platform were obtained from The Broad Institute Firehose pipeline (http://gdac.broadinstitute.org/).
Differential analyses by molecular feature
For mRNA, miRNA, and RPPA data platforms, differential expression between comparison groups was assessed using Pearson’s correlation on log-transformed values (base 2). For cancer types with more than one metastasis profile, the Pearson’s correlation p-value is equivalent to a t-test; for cancer types with just one metastasis profile, significant genes in effect represented outliers with large differences at the edge or outside of the distribution as defined by the primary samples. Differential analyses between metastasis and primary by alternate methods for RNA-seq data were found to be largely concordant with results by the Pearson’s method (Supplementary Figure S1). For DNA methylation platform, differential expression between comparison groups was assessed using Pearson’s correlation on logit-transformed values (natural log). For SKCM datasets, a linear regression model was also carried out for each gene, with dependent variable (continuous variable) of expression and with independent variables: metastasis/primary (categorical variable) + estimated tumor purity(24) (continuous variable). False Discovery Rates (FDRs) were estimated using the method of Storey and Tibshirini(25). For selecting top features for a given data platform and cancer type, FDR<10% was used as a cutoff; for SKCM datasets, top features were also significant with p<0.05 for linear model incorporating tumor purity as a covariate. Visualization using heat maps was performed using both JavaTreeview (version 1.1.6r4)(26) and matrix2png (version 1.2.1)(27). R software (version 3.1.0) was used for generation of box plots.
Pathway and network analyses
Enrichment of GO annotation terms within sets of differentially expressed genes was evaluated using SigTerms software(28) and one-sided Fisher’s exact tests, with FDRs estimated using the method of Storey and Tibshirini(25). Protein interaction network analysis used the entire set of human protein–protein interactions cataloged in Entrez Gene (downloaded June 2017). Entrez gene interactions with yeast two-hybrid experiments providing the only support for the interaction were not included in the analysis. Graphical visualization of networks was generated using Cytoscape(29).
Analysis of external expression profiling datasets
We examined the following external gene expression profiling datasets of metastasis versus primary samples (listed by Gene Expression Omnibus or ArrayExpress accession number): BRCA studies E-MTAB-4003(8), GSE100534(9), and GSE110590(5); CRC studies GSE50760(10), GSE22834(11), and GSE41258(12); PAAD studies GSE42952(13) and GSE19281(14); PRAD studies GSE21034(7), GSE3933(6), and GSE6099(4); SKCM studies GSE65904(15), GSE17275(16), and GSE46517(17); and THCA study GSE60542(18). Differential expression between comparison groups was assessed using t-test on log-transformed values (base 2). For the purposes of comparing results of external datasets with TCGA metastasis signatures, where multiple expression array features referred to the same gene, the feature with the smallest p-value for differences between metastasis and primary tumors (either direction) was used to represent the gene. For patient survival associations involving the TCGA PRAD metastasis signature, we examined external gene expression profiling datasets of primary prostate cancer from Taylor et al. (GSE21034)(7), Sboner et al. (GSE16560)(30), and Nakagawa et al. (GSE10645)(31), assigning a metastasis signature score to each external tumor profile using our previously described “t-score” metric(21); log2-transformed values within each dataset were for normalized to standard devations from the median across the primary sample profiles. In the same way, the t-score metric was also used in applying the TCGA metastasis gene signature for a given cancer type to the primary sample mRNA profiles in TCGA for that cancer type.
We also examined tissue specific mRNA signatures, in order to determine whether these might overlap with the cancer metastasis-specific mRNA signatures that were identified. Gene expression data (TPM values) from GTEx Analysis version 7 release were obtained from the GTEx Portal (https://www.gtexportal.org). Genes with average TPM values greater than five units across the normal tissue samples were used in this analysis, which involved 12769 unique genes in total. Using log-transformed values, for each tissue in GTEx dataset that would be associated with one of the cancer types analyzed in the present study (Breast:BRCA, Skin:SKCM, Cervix Uteri:CESC, Colon:CRC, Esophagus:ESCA, Muscle:SARC, Nerve:PCPG, Pancreas:PAAD, Prostate:PRAD, Thyroid:THCA), the top 500 genes positively correlated with that tissue as compared to all other tissues were determined (t-test using log-transformed data). For a given cancer type, both the genes over-expressed in metastasis and the genes under-expressed in metastasis were each compared with the set of tissue-specific mRNA markers from GTEx corresponding to that cancer type, with the significant of overlap determined using one-sided Fisher’s exact tests. In the same way, we examined GTEx-derived markers of tissues representing common sites of metastasis (adrenal gland, brain, liver, lung) for significant overlap with TCGA-derived metastasis over-expressed genes.
Statistical analysis
All p-values were two-sided unless otherwise specified.
Results
TCGA cohort of primary and metastasis samples
Our study utilized 4,473 primary tumor samples and 395 tumor metastasis samples, involving 4,839 human cancer cases representing 11 different major types, for which TCGA generated data on one or more of the following molecular characterization platforms (Supplementary Data S1): RNA sequencing (4446 primaries and 393 metastases), reverse-phase protein array (RPPA, 3194 and 267), microRNA sequencing (4350 and 378), and DNA methylation arrays (3913 and 391). Of the cancer types studied, TCGA SKCM (melanoma) data involved the most metastasis samples (n=369), followed by THCA (thyroid, n=8), and BRCA (breast, n=7); CESC (cervical), HNSC (head and neck), and PCPG (pheochromocytoma and paraganglioma) cancer types each involved two metastasis samples; CRC (colorectal), ESCA (esophageal), PAAD (pancreas), PRAD (prostate), and SARC (sarcoma) each involved one metastatic sample. Just 29 of the 395 metastasis samples had a primary pair from the same patient, and so unpaired analyses between primary and metastasis were made the focus of this study. In terms of somatic DNA copy by SNP array platform, only SKCM metastasis samples had available data, with no data generated on primaries. Somatic mutation calls by whole-exome sequencing were considered too sparse for carrying out comparisons within each cancer type, with the exception of SKCM, which data have been studied previously(20).
Differential mRNA patterns associated with metastasis by TCGA cancer type
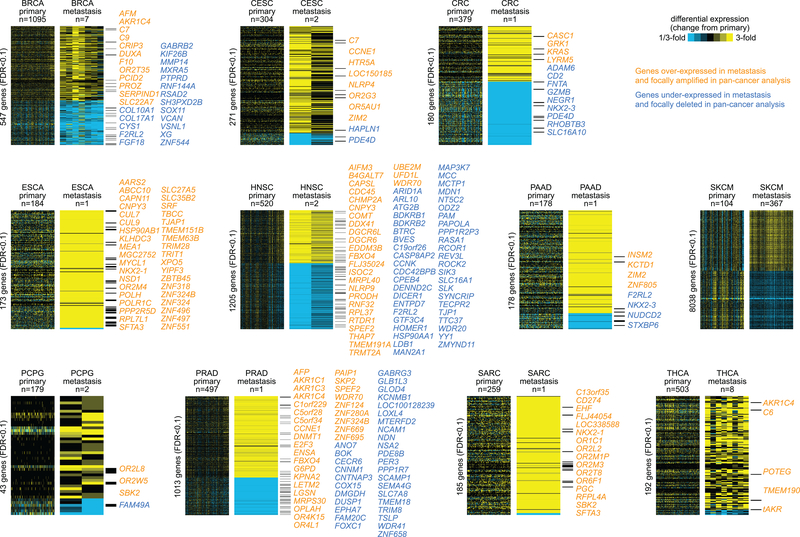
We first set out to define differentially expressed mRNAs (based on RNA sequencing platform) between primary and metastasis samples for each cancer type. For each cancer type, the top differentially expressed mRNAs (genes) in metastasis greatly exceeded chance expected (Figure 1, Supplementary Data S2, and Supplementary Data S3). Using a False Discovery Rate (FDR) cutoff of 10%, the numbers of top significant genes ranged from 43 for PCPG to 10,084 for SKCM, with the other cancer types having between 178 and 1205 top genes. For cancer types with only one metastasis sample, significant genes in effect represented outliers with large differences at the edge or outside of the distribution as defined by the primary samples (Supplementary Figure S1). The limitations with metastasis signatures as defined by a single sample would include false negatives (e.g. in cases where the distributions between primary and metastasis would overlap) and questions as to the generalizability of the signature to other metastasis cases, where the latter may be partially addressable by comparisons with results from external datasets (see below). We examined differences involving estimated tumor purities(24), as gene expression patterns in cancer can reflect non-cancer as well as cancer cells(32). Of all the cancer types examined, only SKCM showed significantly lower tumor purity in metastasis versus primary (p=7.6E-7, t-test, Supplementary Figure S2). Using linear models incorporating purity as a covariate, on the order of 8,038 genes remained significantly differentially expressed in SKCM, out of the above 10,084 genes (Figure 1).
Figure 1. Top differentially expressed mRNAs in metastasis versus primary samples for each of 11 cancer types in The Cancer Genome Atlas (TCGA).
Top genes for each cancer type were selected using Pearson’s correlation (on log-transformed values) with Storey and Tibshirini estimate of False Discovery Rate (FDR) of <10% (for SKCM, FDR<10% and significant with p<0.05 for linear model incorporating tumor purity as a covariate). Yellow denotes high expression relative to the average of primary samples; blue denotes low expression. Genes listed individually are either over-expressed in metastasis and focally amplified in a previous pan-cancer analysis(44) or under-expressed in metastasis and focally deleted in pan-cancer analysis. For SKCM, hundreds of genes involved regions of focal amplification or deletion, and so these are not listed here. BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CRC, Colorectal adenocarcinoma (combining COAD and READ projects); ESCA, Esophageal carcinoma; HNSC, Head and Neck squamous cell carcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; THCA, Thyroid carcinoma.
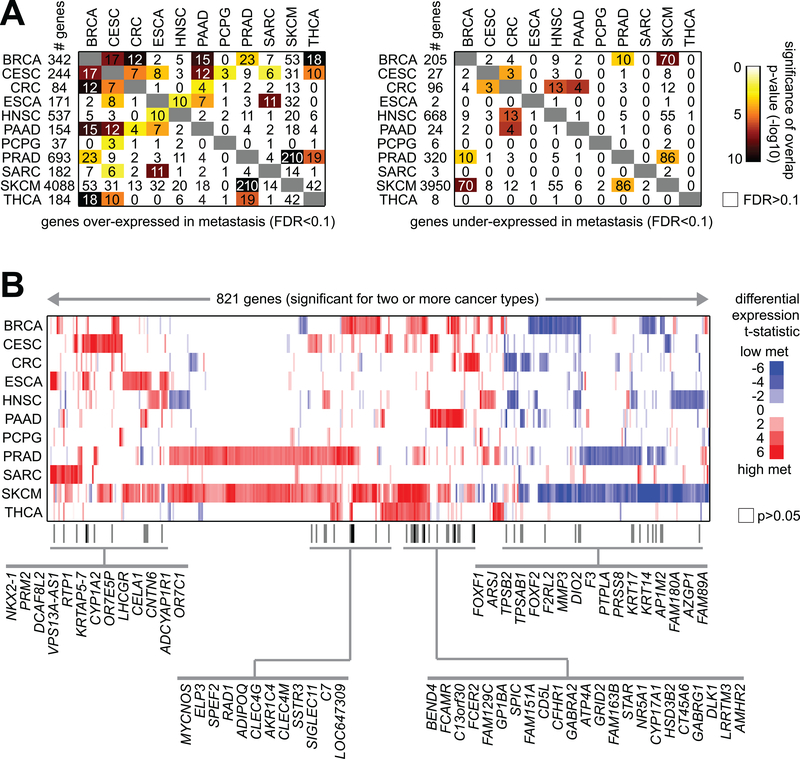
While some differential expression patterns associated with metastasis were found to be shared across multiple cancer types, by and large each cancer type showed a metastasis signature that was distinctive from those of the other cancer types. In comparing the respective expression signatures of metastasis from each cancer type to each other, some amount of gene set overlap was observed (Figure 2A). In a number of cases, the overlap in signatures between any two cancer types was statistically significant, even if the overlap itself involved a fraction of genes (e.g. on the order of 10%). A set of 821 genes were found significant (FDR<10%) with same direction of change for two or more cancer types (Figure 2B). Of these genes, 65 were significant for three or more cancer types, including genes with previously demonstrated functional roles in metastasis such as EPL3(33), MYCNOS(34), and FOXF2(35). Just eight genes (BEND4, CD5L, CELA1, CLEC4M, CYP17A1, DCAF8L2, FAM151A, SPIC) were over-expressed in metastasis (FDR<10%) for four or more cancer types. We furthermore examined whether any of the metastasis signature genes (considering over-expressed and under-expressed gene sets separately) would be enriched for normal tissue-specific mRNA markers associated with the given cancer type (as obtained using GTEx data). Of 10 different tissue specific marker gene sets, only a nominally significant association (p<0.001, one-sided Fisher’s exact test) was observed between SKCM metastasis under-expressed genes and gene markers associated with GTEx mRNA markers of normal skin tissues.
Figure 2. Genes shared among the cancer type-specific metastasis mRNA signatures.
(A) For both the genes over-expressed in metastasis for at least one cancer type (left, genes from Figure 1) and the genes under-expressed in metastasis for at least one cancer type (right, genes from Figure 1), the numbers of overlapping genes between any two cancer types are indicated, along with the significance of overlap (using colorgram, by one-sided Fisher’s exact test). (B) Heat map of differential t-statistics (Pearson’s correlation on log-transformed data), by cancer type, comparing metastasis versus primary (red, higher in metastasis; white, not significant with p>0.05), for 821 genes significant for two or more cancer types (FDR<10%; for SKCM, FDR<10% and significant with p<0.05 for linear model incorporating tumor purity as a covariate). Genes significant for three or more cancer types are indicated by name.
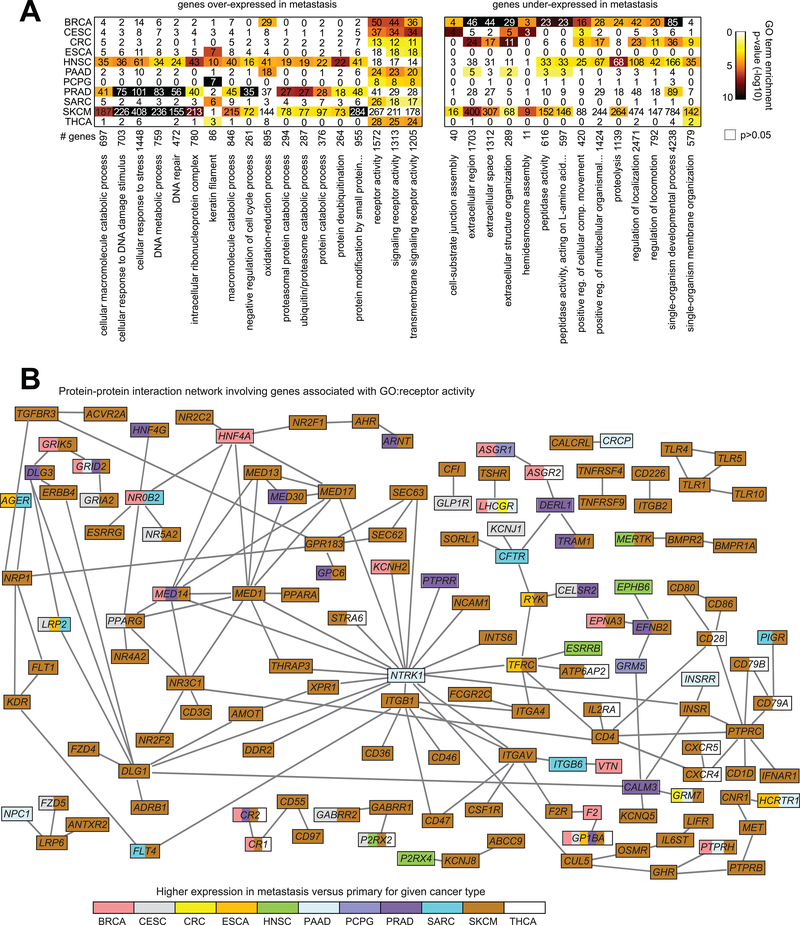
Functional categories of genes represented by the cancer type-specific metastasis expression signatures were examined, using the Gene Ontology (GO) annotation terms (Supplementary Data S4). Specific GO term categories were found enriched within the corresponding metastasis signatures of multiple cancer types (Figure 3A). Significantly enriched GO terms (FDR<10% using one-sided Fisher’s exact tests) found with the metastasis over-expressed genes for at least three cancer types included “cellular response to stress,” “DNA repair,” “oxidation-reduction process,” “protein deubiquitination,” and “receptor activity,” and significant GO terms within the under-expressed genes for at least three cancer types included “extracellular region,” “proteolysis,” and “regulation of locomotion.” We took the genes related to receptor activity and genes high in metastasis (FDR<10%) for at least one cancer type, and we integrated these with public databases of protein–protein interactions to generate a protein interaction network (Figure 3), which allowed us to visualize the potential relationships involving these genes. While most of the genes in this network involved SKCM, a number of other genes involved a trend (p<0.05, Pearson’s on log-transformed data) of higher expression in metastasis in two or more cancer types, and ten genes in the network were high (p<0.05) in three or more cancer types: CR1, CR2, GP1BA, GRID2, GRM7, LHCGR, LRP2, MED14, P2RX2, and PTPRH. Similar types of interaction networks were also generated involving genes related to oxidation-reduction process or protein deubiquitination (Supplementary Figure S3). Genes involved in the immune checkpoint pathway were also examined in TCGA metastasis profiles (Supplementary Figure S4), with these being elevated across SKCM metastasis samples as expected(20), as well as elevated in a portion of metastasis samples from other cancer types.
Figure 3. Functional gene classes shared among the cancer type-specific metastasis mRNA signatures.
(A) Left, Gene Ontology (GO) terms significantly enriched for at least three cancer types (enrichment for cancer type defined as FDR<10% using one-sided Fisher’s exact test) within the respective sets of genes over-expressed in metastasis (based on the gene sets represented in Figure 1); right, GO terms significantly enriched for at least three cancer types within the respective sets of genes under-expressed in metastasis. For both sets of enriched GO terms, the numbers of genes involved for each cancer type and overall significance of enrichment (by colorgram; black, highly significant) are indicated. (B) Protein-protein interaction network involving genes over-expressed in metastasis, with focus on genes involved in receptor activity. Nodes represent genes with GO annotation “receptor activity” and which were found over-expressed in metastasis for at least one cancer type (FDR<10%; for SKCM, FDR<10% and significant with p<0.05 for linear model incorporating tumor purity as a covariate). Nodes are colored according to the individual cancer types in which a trend (p<0.05, Pearson’s correlation on log-transformed data) of higher expression in metastasis versus primary samples was observed. A line between two nodes signifies that the corresponding protein products of the genes can physically interact (according to the literature, from Entrez gene interactions database).
Metastasis-associated mRNA patterns as observed in datasets external to TCGA
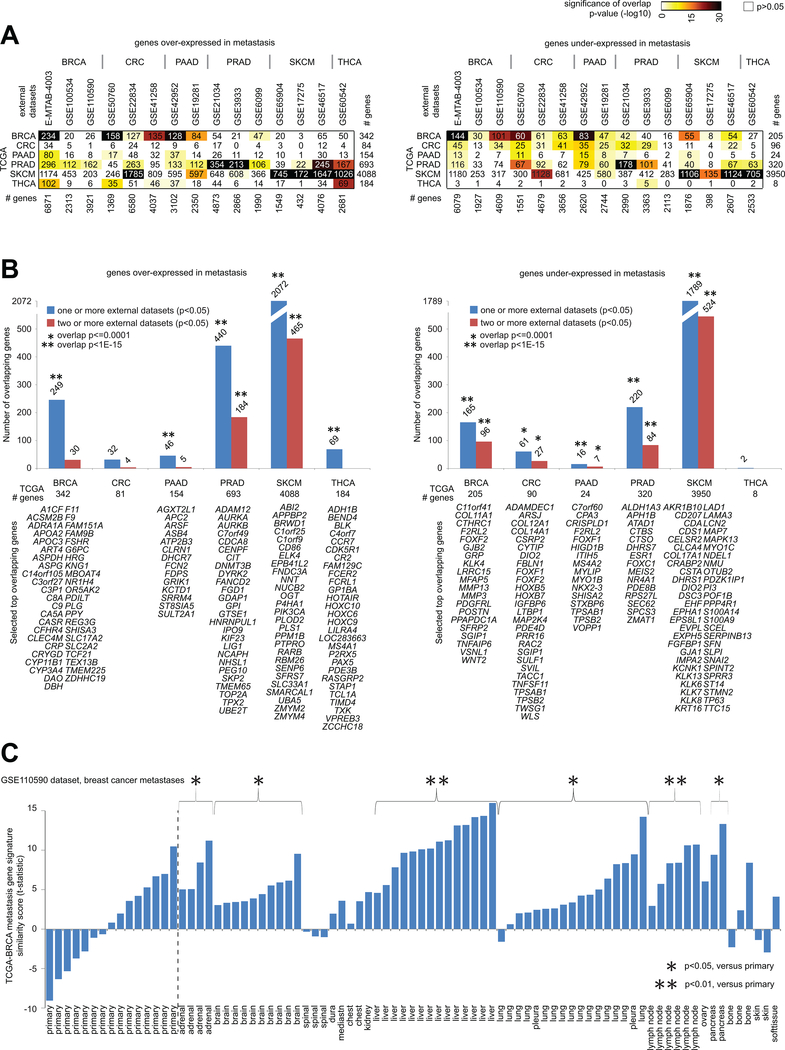
To help assess their generalizability, we compared the gene expression signatures of metastasis, as defined for each cancer type using TCGA data, with metastasis expression signatures obtained from external datasets made available by previously published studies. We examined 15 external gene expression profiling datasets of metastasis versus primary samples, involving six cancer types (BRCA, CRC, PAAD, PRAD, SKCM, and THCA). For each of the cancer types surveyed, a significant number of genes where found to overlap with the results of at least one external dataset of the given cancer type, for either the metastasis over-expressed or under-expressed genes (Figure 4A). Perhaps in part because the CRC and THCA metastasis signatures each involved fewer genes, the CRC over-expressed genes showed some overlap but not significant overlap with CRC over-expressed genes from external datasets, and THCA under-expressed genes by TCGA did not show significant overlap with external dataset results. For each cancer type, on the order of 35–70% of genes comprising the corresponding TCGA metastasis signature showed a similar significant trend (p<0.05) in at least one external dataset of that cancer type (Figure 4B).
Figure 4. Significance of overlap between TCGA metastasis mRNA signatures and metastasis mRNA signatures from datasets external to TCGA.
(A) For both the genes over-expressed in metastasis for a given cancer type (left) and the genes under-expressed in metastasis for a given cancer type (right), the numbers of overlapping genes between the TCGA mRNA signatures (rows, signatures from Figure 1) and the genes over- or under-expressed in metastasis (p<0.05, t-test) in the indicated external datasets from previously published gene expression profiling studies (columns), along with the corresponding significances of overlap (using colorgram, by one-sided Fisher’s exact test, chi-squared test for TCGA SKCM gene sets). (B) For each indicated cancer type, numbers of genes overlapping between the TCGA metastasis signature genes (left, genes over-expressed in metastasis; right, genes under-expressed in metastasis) and the genes significantly high or low in metastasis (p<0.05, t-test) in the published external datasets corresponding to the given cancer type. Significance of overlap (by one-sided Fisher’s exact test; chi-squared test for SKCM genes) is indicated for TCGA genes found in one or more external datasets (blue bars) and in two or more external datasets (red bars). Selected top genes overlapping between TCGA and results from other datasets are listed (BRCA over-expressed: TCGA p<1E-6 and p<1E-6 for E-MTAB-4003 dataset; BRCA under-expressed: TCGA p<1E-6 and p<1E-6 for E-MTAB-4003 dataset; CRC under-expressed: TCGA FDR<10% and p<0.05 for two or more external datasets; PAAD over-expressed: TCGA FDR<10% and p<0.01 for GSE42952 dataset; PAAD under-expressed: TCGA FDR<10% and p<0.05 for one or more external datasets; PRAD over-expressed: TCGA FDR<10% and p<0.01 for all three external datasets; PRAD under-expressed: TCGA FDR<10% and p<0.05 for all three external datasets; SKCM over-expressed: TCGA FDR<10% and p<0.05 for all three external datasets; SKCM under-expressed: TCGA FDR<10% and p<0.05 for all three external datasets; THCA over-expressed: TCGA FDR<10% and p<0.001 for GSE60542 dataset; P-values by Pearson’s correlation or t-test on log-transformed data). (C) TCGA-BRCA metastasis gene expression signature similarity score (t-statistic as derived from the “t-score” metric(21, 45)), as applied to the sample profiles in the GSE110590 breast cancer metastases RNA-seq dataset(5). For selected groups of metastasis according to site, comparisons with the primary group are indicated (t-test as applied to the signature t-scores).
Notably, the external datasets often involved different sites of metastasis for a given cancer type; for example, the external PRAD datasets involved samples taken from various sites including lymph node, bone, lung, testes, and brain(4, 6, 7), implying that the TCGA PRAD signature, while derived from a single metastasis sample, would not be specific to a single site. Similarly, breast metastasis in the GSE110590 dataset(5) involved a number of different sites, with the TCGA BRCA metastasis signature being manifested in samples from most of these sites (Figure 4C). Furthermore, we examined GTEx-derived markers of tissues representing common sites of metastasis (adrenal gland, brain, liver, lung) for significant overlap with TCGA-derived metastasis over-expressed genes; after multiple testing correction(25), only the GTEx liver signature was found to significantly overlap with metastasis genes associated with BRCA (p<1E-7, one-sided Fisher’s exact test, with 20 of the 342 BRCA metastasis over-expressed genes also included in the top 500 genes highly expressed in normal liver), but not with genes from the other cancer types.
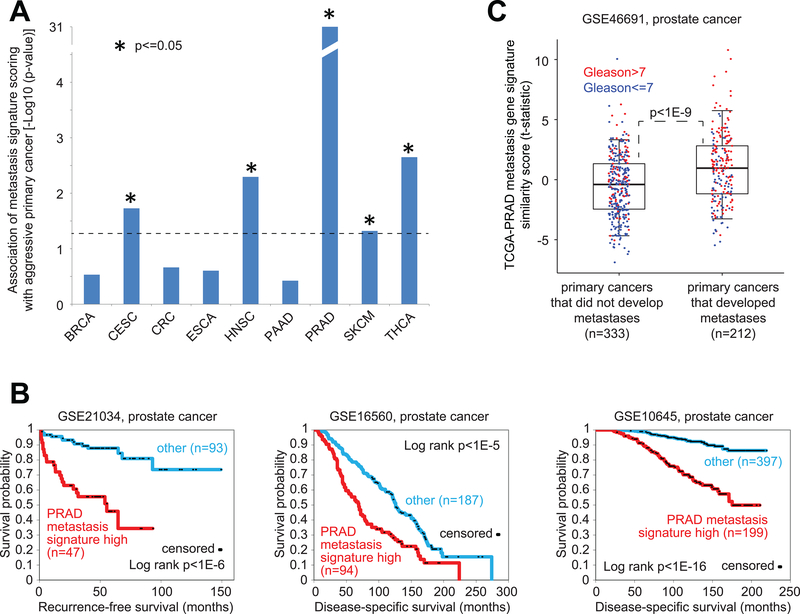
Previous studies have suggested that a subset of primary tumors resemble metastatic tumors with respect to gene expression patterns(36). For each cancer type in our TCGA cohort, we investigated the corresponding metastasis expression signatures in primary tumors. TCGA expression profiles of primary tumors were each scored for manifestation of the metastasis signature. Out of nine cancer types for which pathological stage or grade information were provided, five (CSEC, HNSC, PRAD, SKCM, and THCA) showed some statistical trend for positive correlation between the signature score and stage or grade across primary cancers (one-sided p<=0.05, Pearson’s, Figure 5A). This association was notably strongest for PRAD (prostate) cancer type (p<1E-30), to the extent that clear differences in time to adverse events between patients with primary prostate tumors manifesting the PRAD metastasis signature as compared to the rest of the were observable, when applying the signature to profiles from multiple external cohorts (Figure 5B). In addition, in another prostate cancer dataset, consisting of primary prostate cancer samples from patients for which the early onset of metastasis following radical prostatectomy was recorded(37), PRAD metastasis signature scores were significantly elevated (p<1E-9) in the early onset group (Figure 5C).
Figure 5. For specific cancer types, gene expression signatures of metastasis found present within a subset of primary samples and associated with more aggressive disease.
(A) For each of the indicated cancer types, the corresponding TCGA metastasis gene signature was applied to the primary sample mRNA profiles for that cancer type; across the primary samples, the metastasis signature similarity scores (t-statistic as derived from the “t-score” metric(21, 45)) were correlated with the cancer stage or grade (Gleason grade for PRAD, pathologic stage for the other cancer types). One-sided p-values indicate the Pearson’s correlation between the signature score and stage or grade (numerical 1–4 for stage, 6–10 for Gleason grade). (B) For each of three independent mRNA expression profiling datasets of primary prostate cancer(7, 30, 31), differences in survival between patients with tumors manifesting the TCGA-PRAD metastasis signature (top third of signature similarity scores across the samples) and the other patients. P-values by log-rank test. (C) The TCGA PRAD metastasis gene signature was applied to the primary sample mRNA profiles for the GSE46691 prostate cancer dataset (37), consisting of primary prostate cancer samples from patients for which the early onset metastasis following radical prostatectomy was recorded. Box plot represents 5%, 25%, 50%, 75%, and 95%. P-value for differences in signature scores between groups with or without metastasis by t-test.
Molecular patterns associated with metastasis involving protein, microRNAs, and methylation
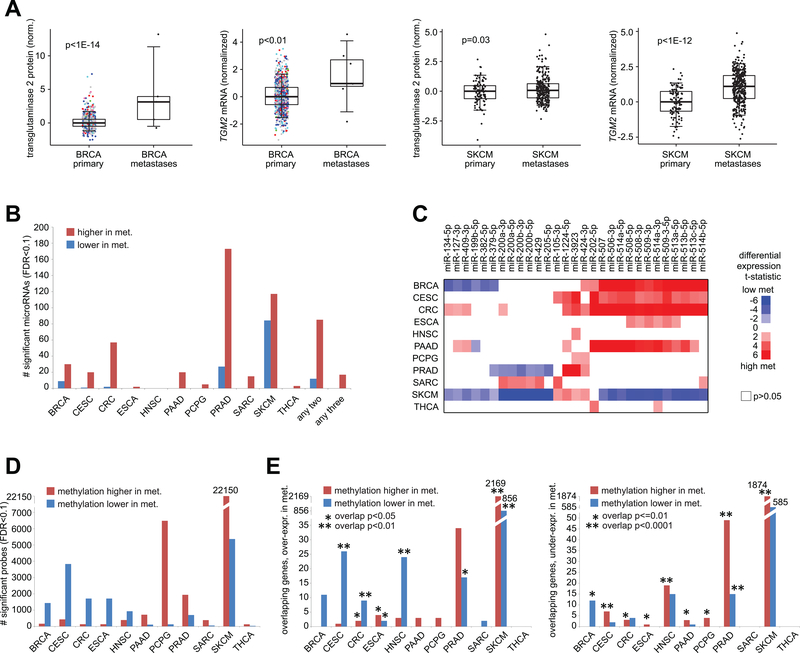
We went on to examine protein, microRNA, and DNA methylation datasets in TCGA, in order to define differentially expressed features between primary and metastasis samples for each cancer type. RPPA proteomic data involved 218 features and four cancer types (BRCA, PCPG, SKCM, THCA) with metastasis profiles. For SKCM, a large portion of RPPA features examined were differentially expressed in metastasis (94 features at FDR<10%, Pearson’s correlation on log-transformed data, 83 features significant after corrections for tumor purity, Supplementary Data S5), analogous to results from mRNA expression. No RPPA features with globally significant (FDR<10%) were found for PCPG or THCA, likely in part due to limited sample power. For BRCA, one protein feature, transglutaminase 2, was elevated in metastasis and globally significant at FDR<10% (FDR<1E-12), corresponding to mRNA-level differences (Figure 6A). Transglutaminase 2 protein and mRNA were also elevated in SKCM (Figure 6A), and the protein is known to promote metastasis(38). For most cancer types, widespread differences in microRNA (miRNA) expression between metastasis and primary were observed (Figure 6B and Supplementary Data S5). Most of the significant miRNAs detected were over-expressed versus under-expressed in metastasis, with 85 over-expressed miRNAs and 12 under-expressed miRNAs significant (FDR<10%) in two or more cancer types, and with 17 over-expressed miRNAs significant in three or more cancer type (Figure 6C). For a number of cancer types, mRNA:microRNA pairings, as defined by both a previously identified miRNA-target interaction (as cataloged by miRTarBase(39)) and significant differential expression in metastasis for both mRNA and microRNA (in opposite directions), could also be identified (Supplementary Data S5).
Figure 6. Molecular correlates of metastasis by protein, microRNA, or DNA methylation profiling.
(A) Transglutaminase 2 protein and mRNA (TGM2 gene) were significantly elevated in TCGA BRCA metastases as well as in TCGA SKCM metastases. Box plots represent 5%, 25%, 50%, 75%, and 95%. P-values by t-test on log-transformed data. (B) Numbers of significant microRNAs between metastasis (“met.”) and primary samples for each cancer type (FDR<10%, based on Pearson’s correlation using log-transformed values; for SKCM, FDR<10% and significant with p<0.05 for linear model incorporating tumor purity as a covariate), along with the numbers or microRNAs significant (FDR<10%) for two or three cancer types. (C) Heat map of differential t-statistics (Pearson’s correlation on log-transformed data), by cancer type, comparing metastasis versus primary (red, higher in metastasis; white, not significant with p>0.05), for 29 microRNAs that were either significantly over-expressed for three or more cancer types (FDR<10%) or significantly under-expressed for two or more cancer types. (D) Numbers of significant DNA methylation array probes located within CpG Islands (by Illumina 450K array, ~150K CpG Island probes) between metastasis (“met.”) and primary samples for each cancer type (FDR<10%, based on Pearson’s correlation using logit-transformed values; for SKCM, FDR<10% and significant with p<0.05 for linear model incorporating tumor purity as a covariate). (E) For each cancer type, numbers of genes overlapping between the RNA-seq and DNA methylation results (FDR<10% for each platform, with top features in SKCM corrected for tumor purity as described above). Left plot represents genes over-expressed in metastases (“met.”), and right plot represents genes under-expressed in metastases. Significance of overlap by one-sided Fisher’s exact test (chi-squared test for SKCM results).
Using TCGA data from DNA methylation arrays, we examined 150,253 CpG Island probes, finding widespread differences in methylation between metastasis and primary samples for each cancer type studied (Figure 6D and Supplementary Data S6). The numbers of top significant methylation features (FDR<10%, Pearson’s on logit-transformed data) ranged from 163 for THCA to 27,530 for SKCM (after corrections for tumor purity), with the other cancer types having between 441 and 6,611 top features. As increased methylation of regulatory regions in proximity to genes can lead to epigenetic silencing, we integrated DNA methylation results with mRNA expression results, defining sets of gene associated with both altered methylation and expression (Figure 6E and Supplementary Data S6). For all cancer types except SARC and THCA, significant inverse correspondences between methylation and expression results were observed (p<0.05, one-sided Fisher’s exact test or chi-squared test), either involving genes over-expressed and with lower associated methylation in metastasis or involving genes under-expressed and with higher associated methylation in metastasis. The significantly overlapping results involved, for example, 2730 genes for SKCM (both over-expressed and under-expressed genes, with inverse patterns of DNA methylation), 66 genes for PRAD, 43 genes for HNSC, and 33 genes for CESC.
Discussion
Our study of TCGA data on cancer metastasis samples had three overall objectives: 1) to obtain a preliminary global view of metastasis versus primary molecular differences across several cancer types; 2) to provide a resource for future studies investigating the role of specific genes in metastasis; and 3) to help provide direction for future genomics studies of metastasis, e.g. by showcasing the utility of examining molecular differences across cancer types and across other molecular profiling platforms in addition to RNA-sequencing. A clear limitation of the present study involves the limited number of metastasis samples profiled as part of TCGA consortium, as the main focus of TCGA was to examine genomic and molecular patterns of primary rather than of metastatic cases. For several of the cancer types examined, this limitation is mitigated somewhat by comparing the results from TCGA transcriptomic data to existing data from other studies, thereby demonstrating the relevance of differential gene patterns as observed across sample cohorts. Our results would support the need for future multiplatform-based and pan-cancer genomics studies profiling larger numbers of metastasis with primary samples, which would allow us to further define and refine the molecular signatures of metastasis as put forth in this present study. Nevertheless, our study demonstrates that, even on the basis of a single metastasis sample, there would be molecular information contained here representing real biological differences that may involve at least some metastasis cases for a given cancer type.
Our study has identified widespread molecular differences in metastasis versus primary tumors for 11 different cancer types, with each cancer type having a signature of metastasis that is distinct from that of the other cancer types. This would suggest that there are different molecular pathways to metastasis involved in different cancers. Our findings would seemingly differ with those of two early studies of gene expression patterns of metastasis, one from Ramaswamy et al. (36), which defined a single 128-gene signature of metastasis across multiple cancer types (lung, breast, prostate, colorectal, uterus, ovary, etc.), and one from Weigelt et al.(40), which could not find any global significant differences over chance expected between breast cancer primary and metastasis samples. Studies subsequent to the Weigelt study have been able to define widespread differences associated with breast cancer metastasis versus primary tumors(5, 8, 9). Interestingly, when surveying TCGA data, none of the Ramaswamy signature genes showed consistent high or low expression patterns in metastasis across the different cancer types (Supplementary Data S2). While the Ramaswamy study found that a subset of primary tumors from various cancer types expressing the 128-gene metastasis signature were associated with worse outcome, we find in our present study that aggressive prostate cancers in particular appear to express a metastasis signature pattern, but other cancer types such as breast cancer do not show a similar phenomenon. One salient feature of this present study was to survey available data from multiple external sources in addition to TCGA data. Where genes are found to show consistent patterns across multiple datasets and studies, we may place the most confidence in these gene patterns, at least given the currently available data.
The results of this present study (e.g. as provided in the supplementary materials) may serve as a resource for future studies investigating the role of specific genes in metastasis. The various gene signatures of metastasis, as identified in each cancer type by TCGA data, may be mined to help identify candidates for functional studies. For cancer types with only a single metastasis sample with TCGA, there would be potential limitations with the associated metastasis signature, including questions as to the generalizability of the signature to other metastasis cases. Integration of TCGA results with results of external public datasets can considerably strengthen the metastasis associations as identified for specific genes. For example, the TCGA PRAD metastasis signature was based on a single metastasis profile, but this signature showed highly significant overlap with results with each of three independent profiling datasets of prostate cancer metastasis versus primary disease(4, 6, 7), and the TCGA PRAD signature could also define a subset of aggressive primary prostate cancer. Integration between mRNA data and data from other platforms in TCGA may also be used to select genes of particular interest, such as genes showing concordant alterations involving both expression and DNA methylation. Genes that appear significant in multiple cancer types, including genes encoding cell receptors, may also be of interest for further investigation.
While successfully defining molecular signatures of metastasis across several different cancer types, our present study points to the need for more molecular data on metastasis in human tumors. Much could be gained by generating molecular data on larger numbers of metastasis and primary cancers, using multiple “omics” data platforms in addition to mRNA expression profiling. The global molecular patterns involved in metastasis would entail proteomic and DNA methylation levels in addition to transcriptomic levels. For many cancer types with metastasis data in TCGA, few or no relevant external molecular profiling datasets were found to be available. For cancer types where a large number of expression outliers could be associated with a single metastasis sample profile, profiling more metastasis cases would enable us to define more robust molecular signatures that would presumably be generalizable to the disease as a whole. Profiling larger numbers of cases would also allow for paired analyses by patient between primary and metastasis samples, as well as offering the possibility of subtype discovery within metastatic tumors according to differential patterns being found within some but not all metastasis cases. Molecular data from human tumors may be combined with molecular data from experimental models of metastasis(41), in order to identify genes common to both, which may help pinpoint critical targets relevant in both the laboratory and human disease settings. The top gene correlates of metastasis by and large do not appear to represent canonical oncogenes(32, 42) or frequent targets of point mutation(43), but rather appear indicative of complex processes at work involving multiple internal and external factors. The molecular signatures of metastasis for each cancer type have the potential to lead to new discoveries into the disease process.
Supplementary Material
Implications:
Our findings suggest that there are different molecular pathways to metastasis involved in different cancers. Our catalog of alterations provides a resource for future studies investigating the role of specific genes in metastasis.
Acknowledgements
This work was supported in part by National Institutes of Health (NIH) grant P30CA125123 (C. Creighton).
Abbreviations:
- TCGA
The Cancer Genome Atlas
- RNA-seq
RNA sequencing
- RPPA
reverse-phase protein arrays
Footnotes
Disclosure of potential conflicts of interest: The authors have no conflicts of interest.
References
- 1.Weinberg RA. The Biology of Cancer. New York: Garland Science; 2006. [Google Scholar]
- 2.Steeg P Targeting metastasis. Nat Rev Cancer. 2016;16(4):201–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang W, Sanders A, Katoh M, Ungefroren H, Gieseler F, Prince M, Thompson S, Zollo M, Spano D, Dhawan P, et al. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin Cancer Biol. 2015;35 Suppl(S244-S75). [DOI] [PubMed] [Google Scholar]
- 4.Tomlins S, Mehra R, Rhodes D, Cao X, Wang L, Dhanasekaran S, Kalyana-Sundaram S, Wei J, Rubin M, Pienta K, et al. Integrative molecular concept modeling of prostate cancer progression. Nature genetics. 2007;39(1):41–51. [DOI] [PubMed] [Google Scholar]
- 5.Siegel M, He X, Hoadley K, Hoyle A, Pearce J, Garrett A, Kumar S, Moylan V, Brady C, Van Swearingen A, et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest. 2018;E-pub Feb 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lapointe J, Li C, Giacomini C, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks J, and Pollack J. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67(18):8504–10. [DOI] [PubMed] [Google Scholar]
- 7.Taylor B, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver B, Arora V, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawler K, Papouli E, Naceur-Lombardelli C, Mera A, Ougham K, Tutt A, Kimbung S, Hedenfalk I, Zhan J, Zhang H, et al. Gene expression modules in primary breast cancers as risk factors for organotropic patterns of first metastatic spread: a case control study. Breast Cancer Res. 2017;19(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulten H, Bangash M, Karim S, Dallol A, Hussein D, Merdad A, Al-Thoubaity F, Al-Maghrabi J, Jamal A, Al-Ghamdi F, et al. Comprehensive molecular biomarker identification in breast cancer brain metastases. J Transl Med. 2017;15(1):269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Kim S, Kim J, Roh S, Cho D, Kim Y, and Kim J. A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients. Mol Oncol. 2014;8(8):1653–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin A, Chua M, Choi Y, Yeh W, Kim Y, Azzi R, Adams G, Sainani K, van de Rijn M, So S, et al. Comparative profiling of primary colorectal carcinomas and liver metastases identifies LEF1 as a prognostic biomarker. PloS one. 2011;6(2):e16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheffer M, Bacolod M, Zuk O, Giardina S, Pincas H, Barany F, Paty P, Gerald W, Notterman D, and Domany E. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A. 2009;106(17):7131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van den Broeck A, Vankelecom H, Van Eijsden R, Govaere O, and Topal B. Molecular markers associated with outcome and metastasis in human pancreatic cancer. J Exp Clin Cancer Res. 2012;31(68). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barry S, Chelala C, Lines K, Sunamura M, Wang A, Marelli-Berg F, Brennan C, Lemoine N, and Crnogorac-Jurcevic T. S100P is a metastasis-associated gene that facilitates transendothelial migration of pancreatic cancer cells. Clin Exp Metastasis. 2013;30(3):251–64. [DOI] [PubMed] [Google Scholar]
- 15.Cirenajwis H, Ekedahl H, Lauss M, Harbst K, Carneiro A, Enoksson J, Rosengren F, Werner-Hartman L, Törngren T, Kvist A, et al. Molecular stratification of metastatic melanoma using gene expression profiling: Prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget. 2015;6(14):12297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins W, Esteves G, Almeida O, Rezze G, Landman G, Marques S, Carvalho A, L Reis L, Duprat J, and Stolf B. Gene network analyses point to the importance of human tissue kallikreins in melanoma progression. BMC Med Genomics. 2011;4(76). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabbarah O, Nogueira C, Feng B, Nazarian R, Bosenberg M, Wu M, Scott K, Kwong L, Xiao Y, Cordon-Cardo C, et al. Integrative genome comparison of primary and metastatic melanomas. PloS one. 2010;5(5):e10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarabichi M, Saiselet M, Trésallet C, Hoang C, Larsimont D, Andry G, Maenhaut C, and Detours V. Revisiting the transcriptional analysis of primary tumours and associated nodal metastases with enhanced biological and statistical controls: application to thyroid cancer. Br J Cancer. 2015;112(10):1665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson D, Wu Y, Lonigro R, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer_Genome_Atlas_Network. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The_Cancer_Genome_Atlas_Research_Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Kwok-Shing Ng P, Kucherlapati M, Chen F, Liu Y, Tsang Y, de Velasco G, Jeong K, Akbani R, Hadjipanayis A, et al. A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell. 2017;E-pub May 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoadley K, Yau C, Hinoue T, Wolf D, Lazar A, Drill E, Shen R, Taylor A, Cherniack A, Thorsson V, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173(2):291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aran D, Sirota M, and Butte A. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6(8971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD, and Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(3246–8. [DOI] [PubMed] [Google Scholar]
- 27.Pavlidis P, and Noble W. Matrix2png: A Utility for Visualizing Matrix Data. Bioinformatics. 2003;19(2):295–6. [DOI] [PubMed] [Google Scholar]
- 28.Creighton C, Nagaraja A, Hanash S, Matzuk M, and Gunaratne P. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14(11):2290–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga N, Wang J, Ramage D, Amin N, Schwikowski B, and Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sboner A, Demichelis F, Calza S, Pawitan Y, Setlur S, Hoshida Y, Perner S, Adami H, Fall K, Mucci L, et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics. 2010;3(8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa T, Kollmeyer T, Morlan B, Anderson S, Bergstralh E, Davis B, Asmann Y, Klee G, Ballman K, and Jenkins R. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PloS one. 2008;3(5):e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen F, Zhang Y, Gibbons D, Deneen B, Kwiatkowski D, Ittmann M, and Creighton C. Pan-cancer molecular classes transcending tumor lineage across 32 cancer types, multiple data platforms, and over 10,000 cases. Clin Cancer Res. 2018;24(9):2182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaunay S, Rapino F, Tharun L, Zhou Z, Heukamp L, Termathe M, Shostak K, Klevernic I, Florin A, Desmecht H, et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med. 2016;213(11):2503–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao X, Li D, Pu J, Mei H, Yang D, Xiang X, Qu H, Huang K, Zheng L, and Tong Q. CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene. 2016;35(27):3565–76. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, Kong P, Li X, Yang F, and Feng Y. FOXF2 deficiency promotes epithelial-mesenchymal transition and metastasis of basal-like breast cancer. Breast Cancer Res. 2015;17(30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramaswamy S, Ross K, Lander E, and Golub T. A molecular signature of metastasis in primary solid tumors. Nature genetics. 2003;33(1):49–54. [DOI] [PubMed] [Google Scholar]
- 37.Erho N, Crisan A, Vergara I, Mitra A, Ghadessi M, Buerki C, Bergstralh E, Kollmeyer T, Fink S, Haddad Z, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PloS one. 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang L, Xu A, and Liu W. Transglutaminase 2 in cancer. Am J Cancer Res. 2015;5(9):2756–76. [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu S, FM L, Wu W, Liang C, Huang W, Chan W, Tsai W, Chen G, Lee C, Chiu C, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39(Database issue):D163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weigelt B, Glas A, Wessels L, Witteveen A, Peterse J, and van’t Veer L. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci U S A. 2003;100(26):15901–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibbons D, Lin W, Creighton C, Zheng S, Berel D, Yang Y, Raso M, Liu D, Wistuba I, Lozano G, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PloS one. 2009;4(4):e5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanahan D, and Weinberg R. The hallmarks of cancer. Cell. 2000;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- 43.Lawrence M, Stojanov P, Mermel C, Robinson J, Garraway L, Golub T, Meyerson M, Gabriel S, Lander E, and Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zack T, Schumacher S, Carter S, Cherniack A, Saksena G, Tabak B, Lawrence M, Zhsng C, Wala J, Mermel C, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45(10):1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Creighton C, Hernandez-Herrera A, Jacobsen A, Levine D, Mankoo P, Schultz N, Du Y, Zhang Y, Larsson E, Sheridan R, et al. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PloS one. 2012;7(3):e34546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.