Abstract
Keratin 8 (KRT8), a type II basic intermediate filament (IF) protein, is essential for the development and metastasis of various cancers. In this study, by analyzing RNA-seq data from the Cancer Genome Atlas (TCGA)-lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), we have determined the expression profile of KRT8, and assessed its prognostic significance and the possible mechanism underlying the dysregulation. Our results showed that KRT8 mRNA expression was significantly up-regulated in both LUAD and LUSC tissues compared with normal lung tissues. The high KRT8 expression group for LUAD patients significantly reduced overall survival (OS) and recurrence-free survival (RFS). Univariate and multivariate analysis revealed that KRT8 expression was an independent prognostic indicator for poor OS and RFS in LUAD patients. However, KRT8 expression had no prognostic value in terms of OS and RFS for LUSC. By exploring DNA copy number alterations (CNAs) of the KRT8 gene in LUAD, we found that DNA low copy gain (+1 and +2) was associated with elevated KRT8 mRNA expression. From the above findings, we have deduced that KRT8 is aberrantly expressed in LUAD tissues and that its expression might independently predict poor OS and RFS for LUAD patients, but not for LUSC patients.
Keywords: KRT8, LUAD, LUSC, prognostic marker
1. Introduction
Keratins (KRTs) predominantly expressed in epithelial tissues can be divided into two types—I (acidic forms) and II (basic forms) of intermediate filament (IF) proteins—based on their biochemical characteristics [1,2]. KRTs contain 54 identified members, that is, 28 of type I (K9–K28) and 26 of type II (K1–K8 and K71–K74) [3]. KRTs comprising of a majority of intermediate filament (IF) are the key components of cytoskeleton [4] and are involved in many cellular processes, including mitosis, differentiation, and apoptosis [3]. For instance, Li et al. have demonstrated that overexpression of KRT13 directly promotes primary prostate cancer to metastasize to bone and brain tissue [5]. Escobar-Hoyos et al. have found that overexpression of KRT17 is identified as a novel unfavorable diagnostic and prognostic biomarker with regard to cervical cancer [6]. Recently, Liu et al. have demonstrated that a significant upregulation of KRT17 in lung adenocarcinoma (LUAD) tissues can promote cell proliferation and invasion, and its high expression is associated with poor overall survival in LUAD patients [7]. In addition, van Sprundel et al. have shown that KRT19 expression is correlated with poor differentiated histology and more aggressive behavior in hepatocellular carcinoma (HCC) [8].
Previous studies have shown that KRT8 expression is significantly elevated in various human cancers, including bladder cancer [9], breast cancer [10], kidney cancer [11], pancreatic cancer [12], and lung cancer [13]. KRT8 mRNA and protein expression has been found to be upregulated in gastric cancer (GC) tissues, and its high expression has been observed to promote the cell progression and metastasis of GC cells and produce unfavorable outcomes for patients with GC [14]. KRT8 upregulated expression can promote the metastasis of clear cell renal cell carcinoma (ccRCC) cells by up-regulating IL-11 expression, inducing IL-11 autocrine, and initiating the STAT3 signaling pathway [15]. Loss of keratin 8/18 can regulate oncogenic potential by controlling various signaling pathways, including TMS1-NF-κB signaling and MARCKSL1-Paxillin1-Rac axis, in skin squamous cell carcinomas (SCC) [16]. The KRT8 protein can bind to annexin A2 and mediate both the apoptosis and the redox pathway in anaplastic thyroid carcinoma (ATC) [17]. KRT8 overexpression mediates resistance to cadmium-induced adaptation and carcinogenesis [18]. In some cancers, upregulation of KRT8 has been considered a valuable prognostic marker, including for gastric cancer [14], oral squamous cell carcinomas [19], and ccRCC [15]. It has also been considered a promising indicator for differentiating between the diagnoses of leukoplakia [20] and head-and-neck carcinomas (HNC) [21]. A recent study has shown that a high KRT8/18 ratio correlates with an aggressive HCC phenotype and can be treated as a novel biomarker for HCC patients [22]. However, little is known regarding the effects of KRT8 on the pathological processes of lung adenocarcinoma (LUAD) and the prognostic values associated with its expression.
In this study, by using large sequencing data with patient information from TCGA, we aimed to examine the expression profile of KRT8 in LUAD and LUSC, and to investigate its prognostic significance in these subtypes.
2. Materials and Methods
2.1. Retrospective Analysis Using TCGA Data
This study is a retrospective study which used TCGA level 3 data, with access provided by the University of California, Santa Cruz (UCSC) Xena browser (https://xenabrowser.net/) [23]. Molecular, clinicopathological, and more than 10 years’ survival data of over 1000 TCGA_LUAD and LUSC patients were recorded. Primary tumor tissues from 514 LUAD patients and 501 LUSC patients were collected for RNA-seq. 502 of the 514 LUAD patients and 494 of the 501 LUSC patients had complete survival data. Clinicopathological parameters of LUAD and LUSC patients with primary tumors, including age at diagnosis, gender, smoking history, pathologic stage, living status, recurrence-free survival (RFS), and OS were downloaded for survival-curve analysis. Kaplan-Meier curves of OS and RFS were mapped to assess the survival difference between patients grouped with high or low KRT8 expression. The gene level thresholded genomic identification of significant targets in cancer 2 (GISTIC2)-processed copy number alterations (CNAs) data of LUAD were also downloaded and examined using the UCSC Xena browser.
2.2. Immunohistochemistry (IHC) Staining
Immunohistochemistry images of KRT8 protein expression in normal lung tissues and lung cancer tissues including LUAD and LUSC were downloaded and examined from the Human Protein Atlas (HPA) (http://www.proteinatlas.org/) [24].
2.3. Kaplan-Meier Plotter Data Mining Analysis
The association between the prognostic value of KRT8 and OS in 720 LUAD patients and in 524 LUSC patients was examined by data-mining an online survival analysis software Kaplan-Meier plotter using transcriptomic data [25]. The patients were divided into two groups by using the 50% cut-off value for KRT8 expression; the hazard ratio (HR) with 95% CI and log-rank p-value were directly calculated and generated using the website.
2.4. Oncomine Data-Mining Analysis
Oncomine (https://www.oncomine.org/resource/main.html) is a cancer microarray database and data-mining platform which can help researchers easily acquire gene expression information for human cancer tissues and cells [26]. The selected parameters for inclusion of studies, including p-value < 0.05, fold change ≥ 2, and gene ranking in the top 10%, were set up in this database.
2.5. Data-Mining Analysis of Gene Expression Omnibus (GEO) Database
Treatment-related transcriptome microarray datasets with accession numbers GSE21656 [27], GSE6400 [28], and GSE6914 [29] were downloaded from the GEO database, and normalized raw transcriptome data were reanalyzed to assess the effect of KRT8 mRNA expression on responses to chemotherapy.
2.6. Statistical Analysis
SPSS 19.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Inc., La Jolla, CA, USA) software were used for statistical analysis. χ2 tests were conducted to explore the correlation between KRT8 expression and clinicopathological factors. The mRNA expression of KRT8 in LUAD and LUSC cancer tissues was compared with that in normal tissues, using a Students’ t-test to calculate a p value. Kaplan-Meier curves of OS and RFS (using TCGA-LUAD data) were mapped by GraphPad Prism 5.0 by setting the median KRT8 expression as the cut-off. A log-rank test was performed to examine the significant differences between the survival curves of patients. Univariate and multivariate Cox regression models were performed to assess the prognostic role of KRT8 in terms of OS and RFS for LUAD and LUSC patients using SPSS. Risk factors (p < 0.2) analyzed by univariate analysis were selected for multivariate Cox regression analysis using a stepwise regression method. A value of p < 0.05 was considered statistically significant.
3. Results
3.1. Both LUAD & LUSC Tissues Had Significantly Increased KRT8 Expression Compared with Normal Tissues
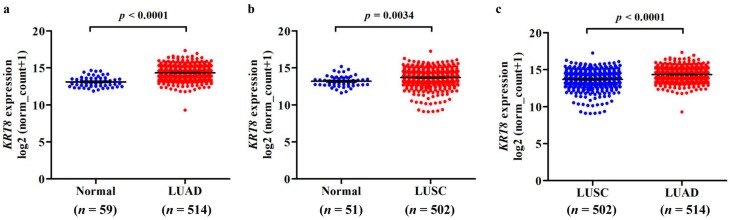
Using TCGA RNA-seq data, we first analyzed KRT8 mRNA expression in both lung cancer tissues and normal tissues. Results showed significantly elevated KRT8 expression in both LUAD (n = 514) and LUSC (n = 502) tissues compared with the normal controls (Figure S1 and Figure 1a–b). In addition, KRT8 expression in LUAD was higher than that in LUSC (Figure 1c).
Figure 1.
KRT8 mRNA expression was up-regulated in both lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) in comparison with normal lung tissues. (a,b) Plots charts of KRT8 expression in LUAD, LUSC, and their corresponding normal lung tissues. (c) Comparison of KRT8 expression in LUAD and LUSC tissues.
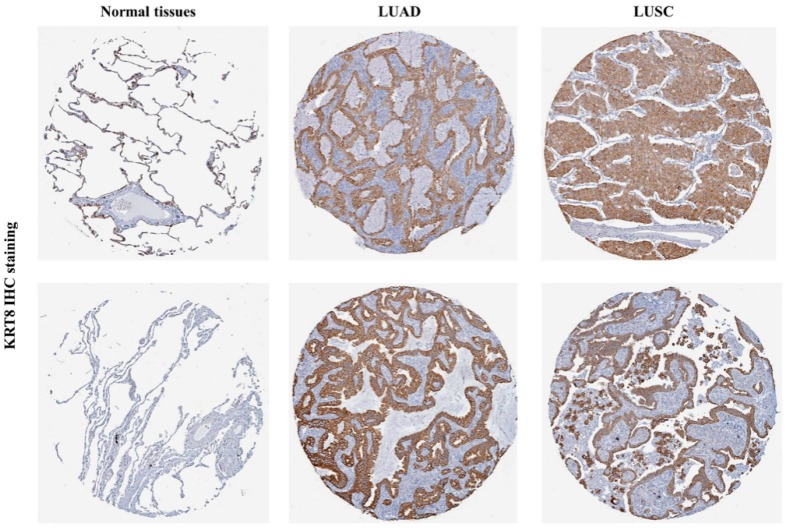
Following this, we tried to characterize KRT8 protein expression in normal tissues and in lung cancer tissues by analyzing IHC staining images from the HPA database [24] and found that normal tissues had no KRT8 staining (Figure 2, left). Conversely, LUAD and LUSC had moderate-to-strong KRT8 staining (Figure 2, middle and right).
Figure 2.
KRT8 protein expression was significantly higher in both LUAD and LUSC tissues in comparison with normal respiratory epithelial tissues. KRT8 immunohistochemistry (IHC) staining images: in normal respiratory epithelial tissues (left), in LUAD tissues (middle), and in LUSC tissues (right). Images were downloaded from the Human Protein Atlas (HPA) (http://www.proteinatlas.org/).
3.2. KRT8 Increased Expression Was Correlated with Poor OS & RFS in LUAD Patients, but not in LUSC
To further assess the KRT8 expression profile in different pathologic stages, we downloaded data of this kind for both LUAD and LUSC. Results showed that there was no significant difference between stages I and II, between stages II and III, and between stages III and IV, for both LUAD and LUSC (Figure S2). Following this, we compared KRT8 mRNA expression in patients with different survival outcomes. For LUAD, we discovered that the deceased cases (n = 183) had significantly higher KRT8 expression in comparison with the living cases (n = 319; p < 0.0001, Figure 3a). In addition, patients with recurrence (n = 151) also had increased KRT8 expression compared to patients without recurrence (n = 275; p = 0.0361, Figure 3b). However, these associations were not found in LUSC patients (Figure 3c,d).
Figure 3.
Comparison of KRT8 mRNA expression in LUAD and LUSC patients with different survival outcomes. Comparison of KRT8 expression in LUAD (a,b) and LUSC (c,d) patients, according to living status (a,c) and recurrence status (b,d).
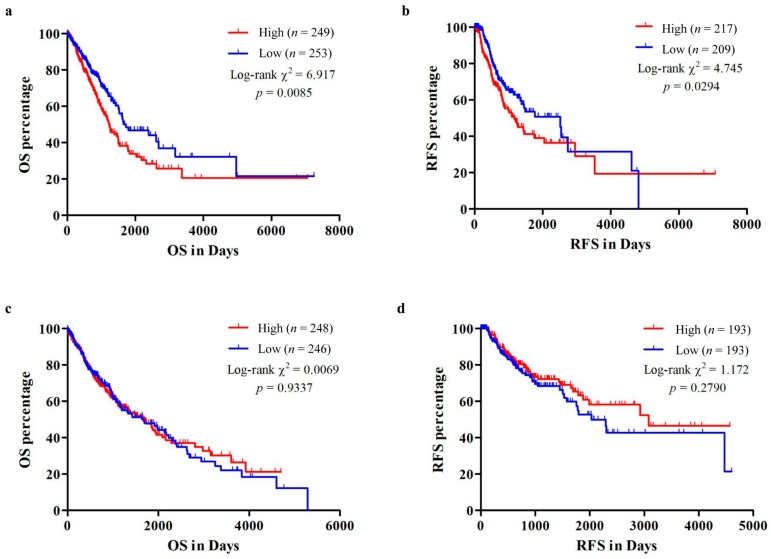
By mapping Kaplan-Meier curves of OS and RFS, we found that LUAD patients with high KRT8 expression had inferior OS (p = 0.0085) and RFS (p = 0.0294) in comparison to patients who had low KRT8 expression (Figure 4a,b). In comparison, these associations were not present in LUSC patients (p = 0.9337 and 0.2790, respectively, Figure 4c,d). To validate these associations, we also performed data mining in the Kaplan-Meier plotter. Results showed that the high KRT8 expression group had inferior OS (HR: 1.29; 95% CI: 1.02–1.63; p = 0.031) for LUAD (Figure S3a), but not for LUSC (Figure S3b). Many new treatment strategies have been developed in the past decade and these treatment strategies have had a strong effect on the RFS and OS of LUAD patients. For instance, LUAD patients with or without epidermal growth factor receptor (EGFR) mutation or anaplastic lymphoma kinase (ALK) or c-ros oncogene 1 (ROS-1) rearrangement had different outcomes whether they received tyrosine kinase inhibitors (TKIs) or not. Therefore, to minimize bias for patients harboring EGFR mutation or ALK or ROS-1, we excluded these types of patients (31, four, and three patients with EGFR mutation, ALK rearrangement, and ROS-1 rearrangement, respectively) [30] and mapped Kaplan-Meier curves of OS and RFS for the remaining LUAD patients by setting the median KRT8 mRNA expression. These results showed that LUAD patients with high KRT8 expression also had worse OS and RFS (Figure S4).
Figure 4.
Kaplan-Meier curves of overall survival (OS) and recurrence-free survival (RFS) in LUAD and LUSC. Kaplan-Meier curves of OS (a,c) and RFS (b,d) in LUAD (a,b) and LUSC (c,d) patients.
3.3. KRT8 Expression Was an Independent Prognostic Biomarker for Poor OS and RFS in LUAD
Consequently, we determined the independent prognostic value of KRT8 in LUAD. The association between KRT8 expression and the clinicopathological parameters of LUAD patients is shown in Table 1. Results show that the high KRT8 expression group had a significantly higher ratio of patients in advanced stages (III/IV) (63/187 vs. 43/200; p = 0.048), male gender (128/125 vs. 103/146; p = 0.040), and death (108/145 vs. 75/174; p = 0.004) compared to the low KRT8 expression group. By performing univariate analysis, we found that advanced stages and increased KRT8 expression correlated with inferior OS and RFS for LUAD (Table 2). The following multivariate analysis confirmed that increased KRT8 expression was an independent prognostic indicator in terms of OS (HR: 1.416; 95% CI: 1.050–1.909; p = 0.022; Table 2) and RFS (HR: 1.512; 95% CI: 1.077–2.122; p = 0.017; Table 2). However, there was no significant association between KRT8 expression and OS/RFS in LUSC patients (p = 0.934 and 0.280 respectively, Table S1).
Table 1.
The association between KRT8 expression and the demographic and clinicopathological parameters of patients with primary LUAD in the Cancer Genome Atlas (TCGA).
| Parameters | KRT8 Expression | KRT8 Expression | χ2 | p-Value | |
|---|---|---|---|---|---|
| High (n = 253) | Low (n = 249) | ||||
| Age (Mean ± SD) | 64.56 ± 10.11 | 66.10 ± 9.74 | 0.08 | ||
| Gender | Female | 125 | 146 | 4.301 | 0.04 |
| Male | 128 | 103 | |||
| Smoking history | 1 | 31 | 41 | 1.629 | 0.251 |
| 2/3/4/5 | 213 | 203 | |||
| Null | 9 | 5 | |||
| Clinical stage | I/II | 187 | 200 | 4.112 | 0.048 |
| III/IV | 63 | 43 | |||
| Discrepancy + null | 3 | 5 | |||
| Recurrence status | No | 125 | 150 | 3.056 | 0.086 |
| Yes | 82 | 69 | |||
| Null | 46 | 24 | |||
| Living status | Living | 145 | 174 | 8.566 | 0.004 |
| Dead | 108 | 75 |
Smoking history: 1. lifelong non-smoker; 2. current smoker; 3. current reformed smoker (for >15 years); 4. current reformed smoker (for ≤ 15 years); 5. current reformed smoker (duration not specified); Null. no data.
Table 2.
Univariate and multivariate analysis of OS and RFS in patients with primary LUAD.
| Parameters | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| p | HR | 95% Cl (Lower/Upper) |
p | HR | 95% Cl (Lower/Upper) |
|||
| OS | ||||||||
| Age >65 vs. ≤65 |
0.210 | 1.207 | 0.899 | 1.620 | ||||
| Female vs. Male | 0.670 | 0.939 | 0.702 | 1.256 | ||||
| Smoking history 2/3/4/5 vs. 1 |
0.684 | 0.918 | 0.608 | 1.386 | ||||
| Clinical stage III/IV vs. I/II |
<0.001 | 2.651 | 1.945 | 3.612 | <0.001 | 2.535 | 1.856 | 3.461 |
| KRT8 expression High vs. Low |
0.004 | 1.540 | 1.147 | 2.068 | 0.022 | 1.416 | 1.050 | 1.909 |
| RFS | ||||||||
| Age >65 vs. ≤65 |
0.089 | 1.330 | 0.957 | 1.849 | 0.023 | 1.482 | 1.056 | 2.079 |
| Female vs. Male | 0.574 | 1.097 | 0.794 | 1.516 | ||||
| Smoking history 2/3/4/5 vs. 1 |
0.435 | 1.207 | 0.752 | 1.939 | ||||
| Clinical stage III/IV vs. I/II |
0.006 | 1.699 | 1.160 | 2.489 | 0.021 | 1.580 | 1.070 | 2.333 |
|
KRT8 expression High vs. Low |
0.03 | 1.428 | 1.035 | 1.972 | 0.017 | 1.512 | 1.077 | 2.122 |
3.4. KRT8 mRNA Expression Was Regulated by Its DNA CNAs
By analyzing KRT8 DNA CNAs in 511 cases of LUAD, we found that although low copy gain (+1 and +2, n = 148, 28.96%) was not frequent, it was still associated with significantly increased KRT8 expression compared with the copy-neutral (0) cases (Figure 5). In comparison, 93 cases (18.2%) had low-level copy loss (−1), which resulted in significantly decreased KRT8 expression (Figure 5).
Figure 5.
KRT8 expression was modulated by its DNA copy number alterations (CNAs).
3.5. The Role of KRT8 in LUAD Therapies
In addition, we used three treatment-related GEO microarray datasets to explore the possible role of KRT8 in the therapeutic response of LUAD patients. In the GSE21656 dataset [27], we found that when compared with the parental H460 cells, KRT8 was significantly up-regulated in the cisplatin-resistant cell line (CDDP-R) (p = 0.0331) (Figure S5a). In the GSE6400 dataset [28], we found that KRT8 was significantly up-regulated in the A549 cell line treated with 1.25 μM and 2.5 μM of the anti-cancer agent sapphyrin PCI-2050 (p = 0.0006 and p = 0.0005, respectively) (Figure S5b). Additionally, from the GSE6914 dataset [29], KRT8 mRNA expression was significantly increased in the gemcitabine resistant Calu3 cell line treated with bexarotene, gemcitabine, or a two-drug combination (p = 0.0046, p = 0.0031, and p = 0.0328, respectively) (Figure S5c). These results show that aberrant KRT8 expression levels might be involved in LUAD cancer treatment.
4. Discussion
Recently, several promising and potential prognostic biomarkers have been identified in LUAD and LUSC by secondary analysis of the TCGA data. For instance, Zhou et al. have found that epithelial cell transforming 2 (ECT2) expression is increased in LUAD and may predict poor OS and RFS for LUAD patients, but not for LUSC patients [31]. Using the UCSC Xena browser to analyze the same TCGA-LUAD dataset [32], Chen et al. and Yu et al. have demonstrated that high expression of lactate dehydrogenase-A (LDHA) [33] and the S100 protein family member S100A16 [34] might be independent prognostic markers of inferior OS for LUAD patients.
KRT8, predominantly expressed in epithelial cells, and its aberrant expression in multiple types of tumors is associated with cell migration [35], cell adhesion [36], and drug resistance [37]. Nevertheless, little has been reported about the expression and potential prognostic functions of KRT8 in LUAD and LUSC. In this study, using the RNA-seq data from TCGA and protein data from the HPA, we found that when in comparison to normal lung tissues, KRT8 expression was significantly up-regulated in both LUAD and LUSC tissues. Oncomine analysis of lung cancer versus normal tissue also confirmed that KRT8 was significantly overexpressed in LUAD (Table 3).
Table 3.
Datasets of KRT8 gene expression in LUAD.
| Gene | Dataset | Normal (Cases) | Tumor (Cases) | Fold Change | t-Test | p-Value |
|---|---|---|---|---|---|---|
| Selamat [13] | Lung (58) | LUAD (58) | 3.846 | 14.527 | 7.59 × 10−26 | |
| Landi [38] | Lung (49) | LUAD (58) | 2.538 | 9.991 | 3.29 × 10−17 | |
| KRT8 | Beer [39] | Lung (10) | LUAD (86) | 2.183 | 5.838 | 8.30 × 10−6 |
| Su [40] | Lung (30) | LUAD (27) | 2.100 | 4.126 | 8.56 × 10−5 | |
| Hou [41] | Lung (65) | LUAD (45) | 2.144 | 6.904 | 8.77 × 10−10 | |
| Okayama [42] | Lung (20) | LUAD (226) | 2.676 | 9.060 | 3.65 × 10−10 |
In addition, we observed that KRT8 upregulation was associated with late clinical stages and higher ratios of recurrence and death (Figure 3). More importantly, we found that patients with high KRT8 expression had worse OS and RFS by setting a median KRT8 mRNA expression. Additionally, by performing univariate and multivariate analysis, we discovered that high KRT8 expression was an independent prognostic indicator of poor OS (HR: 1.416, 95% CI: 1.050–1.909, p < 0.022) and RFS (HR: 1.512, 95% CI: 1.077–2.122, p < 0.017) for LUAD. On the contrary, although we found that KRT8 was up-regulated in LUSC, its expression had no prognostic value in terms of OS and RFS. These results indicate that KRT8 might be a specific and promising prognostic biomarker for LUAD patients. A recent study showed that KRT8 over-expression led to resistance to cadmium-induced adaptation and carcinogenesis [18]. KRT8 can mediate resistance to apoptosis in granulosa cell tumors by intervening in cell surface death receptor Fas (FAS) expression [43]. These studies may help to clarify the correlation between KRT8 expression and inferior overall survival for LUAD patients.
In non-small cell lung cancer (NSCLC), the potential mechanisms of abnormal gene expression are quite complex. Genetic alterations regulating gene expression are frequently common in LUAD and have significant impacts on tumor phenotypes and patients’ survival. For instance, de novo ERBB2 amplification confers intrinsic resistance to erlotinib in EGFR-L858R mutated TKI-naive LUAD [44]. We also explored possible clues to KRT8 dysregulation and found that DNA low copy gain might contribute to increased KRT8 expression in LUAD (Figure 5). Therefore, it would be worthwhile to exploring the mechanism for genetic alterations of KRT8 influencing LUAD cell behaviors in the future. In 2017, Ricciardelli et al. found that KRT5 mRNA expression was consistently higher in chemotherapy-resistant cells compared to chemotherapy-sensitive primary serous ovarian cancer cells and that the number of serous ovarian carcinomas with high KRT5/KRT6 or high KRT5 protein expression significantly increased following carboplatin chemotherapy [45]. Recent research has shown that KRT8/KRT18 protein levels are markedly increased in TNF-related apoptosis inducing ligand (TRAIL)-resistant cells compared to TRAIL-sensitive breast cancer cells, and they may limit TRAIL-induced apoptosis signaling via negatively regulating death receptors’ (DR5) protein stability and surface expression [46]. Through analysis of datasets obtained from the GEO database, significantly high mRNA levels of KRT were identified in cisplatin-resistant ADC cells. Moreover, KRT8 was significantly increased in the ADC cell line A549 treated with sapphyrin PCI-2050, and in the gemcitabine resistant Calu3 cell line treated with bexarotene, gemcitabine, or a two-drug combination. This implies that KRT8 may be a potential therapeutic target for LUAD. However, it is worth noting that we have only demonstrated the relationship between the prognosis of LUAD and expression of KRT8 mRNA, but not KRT8 protein expression. In addition, knowledge of the detailed biological function of the KRT8 gene in LUAD is lacking. Hence, the prognosis values of the KRT8 protein need further exploration, and further intensive in vitro and in vivo investigations will help clarify the underlying mechanism of KRT8 within the pathogenesis and development of LUAD.
5. Conclusions
Although KRT8 is significantly increased with regards to both RNA and protein levels in LUAD and LUSC compared to normal tissues, its expression might only serve as an independent prognostic indicator of inferior OS and RFS in LUAD and not in LUSC.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/10/1/36/s1: Figure S1: Heatmap of KRT8 expression in LUAD, LUSC, and their corresponding normal lung tissues; Figure S2: Comparison of KRT8 expression in LUAD (a) and LUSC (b) patients, according to pathologic stages; and Figure S3: The association between KRT8 expression and overall survival for LUAD and LUSC. Kaplan-Meier curves of OS in LUAD (a) and LUSC (b) patients. Results were generated using a Kaplan-Meier plotter; Figure S4: Kaplan-Meier curves of overall survival and recurrence-free survival in LUAD patients without EGFR mutation or ALK or ROS-1 rearrangement; Figure S5: The influence of KRT8 on the therapeutic responses of LUAD patients. Three treatment-related microarray datasets (GSE21656, GSE6400, and GSE6914) from the GEO database were used to assess the potential roles of KRT8 expression on therapeutic effects for LUAD (a, b, and c); Table S1: Univariate and multivariate analysis of OS and RFS in patients with primary LUSC.
Author Contributions
Designed the experiments: L.X. and X.G.; Acquisition of data: L.X., Y.D., J.G., and X.G.; Analysis and interpretation of data: L.X., Y.D., J.G., X.S., T.X., L.Z., Z.Y., and X.G.; Draft of the manuscript: L.X., Y.D., and X.G.; Critical revision of the manuscript for intellectual content: L.X., Y.D., J.G., and X.G.
Funding
This study was supported by the Kaifeng Science and Technology Major Project 18ZD008, Innovative National Natural Science Foundation of China (No. 81602362), China Postdoctoral Science Foundation (No. 2017M62237), Post-doctoral Research Grant in Henan Province (No. 001702052), Projects for College Students in Henan University(No. 201819002), Program for Science and Technology Development in Henan Province (No. 162102310391), the Program for Young Key Teacher of Henan Province (2016GGJS-214), supporting grants of Henan University (No. 2015YBZR048; No. B2015151), the Yellow River Scholar Program (No. H2016012), and the Program for Innovative Talents of Science and Technology in Henan Province (No. 18HASTIT048).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hesse M., Zimek A., Weber K., Magin T.M. Comprehensive analysis of keratin gene clusters in humans and rodents. Eur. J. Cell Biol. 2004;83:19–26. doi: 10.1078/0171-9335-00354. [DOI] [PubMed] [Google Scholar]
- 2.Schweizer J., Coulombe P.A., Langbein L., Lane E.B., Magin T.M., Omary M.B. New Consensus Nomenclature for Mammalian Keratins. J. Cell Biol. 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karantza V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulombe P.A., Omary M.B. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 2002;14:110–122. doi: 10.1016/S0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- 5.Li Q., Yin L., Jones L.W., Chu G.C., Wu J.B., Huang J.M., Li Q., You S., Kim J., Lu Y.-T., et al. Keratin 13 expression reprograms bone and brain metastases of human prostate cancer cells. Oncotarget. 2016;7:84645–84657. doi: 10.18632/oncotarget.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escobar-Hoyos L.F., Yang J., Zhu J., Cavallo J.A., Zhai H., Burke S., Koller A., Chen E.I., Shroyer K.R. Keratin 17 in premalignant and malignant squamous lesions of the cervix: Proteomic discovery and immunohistochemical validation as a diagnostic and prognostic biomarker. Mod. Pathol. 2014;27:621–630. doi: 10.1038/modpathol.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Liu L., Cao L., Wen Q. Keratin 17 promotes lung adenocarcinoma progression by enhancing cell proliferation and invasion. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018;24:4782–4790. doi: 10.12659/MSM.909350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprundel R.G.V., Ingh T.S.V.D., Desmet V.J., Katoonizadeh A., Penning L.C., Rothuizen J., Roskams T., Spee B. Keratin 19 marks poor differentiation and a more aggressive behaviour in canine and human hepatocellular tumours. Comp. Hepatol. 2010;9:4. doi: 10.1186/1476-5926-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchezcarbayo M., Socci N.D., Lozano J., Saint F., Cordoncardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. Off. J. Am. Soc.Clin. Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 10.Christina C., Shah S.P., Suet-Feung C., Gulisa T., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yusenko M.V., Kuiper R.P., Boethe T., Ljungberg B., Kessel A.G.V., Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huadong P., Liang L., Fridley B.L., Jenkins G.D., Kalari K.R., Wilma L., Gloria P., Zhen K., Wang L., et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selamat S.A., Chung B.S., Girard L., Zhang W., Zhang Y., Campan M., Siegmund K.D., Koss M.N., Hagen J.A., Lam W.L., et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jian F., Hao W., Yun L., Fangfang D., Ying N., Shihe S. High KRT8 expression promotes tumor progression and metastasis of Gastric Cancer. Cancer Sci. 2017;108:178–186. doi: 10.1111/cas.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan H.S., Jiang W.H., He Y., Wang D.S., Wu Z.J., Wu D.S., Gao L., Bao Y., Shi J.-Z., Liu B., et al. KRT8 upregulation promotes tumor metastasis and is predictive of a poor prognosis in clear cell renal cell carcinoma. Oncotarget. 2017;8:76189–76203. doi: 10.18632/oncotarget.19198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiwari R., Sahu I., Soni B.L., Sathe G.J., Thapa P., Patel P., Sinha S., Vadivel C.K., Patel S., Jamghare S.N., et al. Depletion of keratin 8/18 modulates oncogenic potential by governing multiple signaling pathways. Febs J. 2018;285:1251–1276. doi: 10.1111/febs.14401. [DOI] [PubMed] [Google Scholar]
- 17.Guo D., Xu Q., Pabla S., Koomen J., Biddinger P., Sharma A., Pabla S., Pacholczyk R., Chang C.H., Friedrich K., et al. Cytokeratin-8 in anaplastic thyroid carcinoma: More than a simple structural cytoskeletal protein. Int. J. Mol. Sci. 2018;19:577. doi: 10.3390/ijms19020577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau A.T., Chiu J.F. The possible role of cytokeratin 8 in cadmium-induced adaptation and carcinogenesis. Cancer Res. 2007;67:2107–2113. doi: 10.1158/0008-5472.CAN-06-3771. [DOI] [PubMed] [Google Scholar]
- 19.Fillies T., Werkmeister R., Packeisen J., Brandt B., Morin P., Weingart D., Joos U., Buerger H. Cytokeratin 8/18 expression indicates a poor prognosis in squamous cell carcinomas of the oral cavity. BMC Cancer. 2006;6:10. doi: 10.1186/1471-2407-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gires O., Mack B., Rauch J., Matthias C. CK8 correlates with malignancy in leukoplakia and carcinomas of the head and neck. Biochem. Biophys. Res. Commun. 2006;343:252–259. doi: 10.1016/j.bbrc.2006.02.139. [DOI] [PubMed] [Google Scholar]
- 21.Matthias C., Mack B., Berghaus A., Gires O. Keratin 8 expression in head and neck epithelia. BMC Cancer. 2008;8:267. doi: 10.1186/1471-2407-8-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golob-Schwarzl N., Bettermann K., Mehta A.K., Kessler S.M., Unterluggauer J., Krassnig S., Kojima K., Chen X., Hoshida Y., Bardeesy N.M., et al. High keratin 8/18 ratio predicts aggressive hepatocellular cancer phenotype. Transl. Oncol. 2018;12:256–268. doi: 10.1016/j.tranon.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman M., Craft B., Zhu J., Swatloski T., Cline M., Haussler D. Abstract 5270: The UCSC Xena system for integrating and visualizing functional genomics. Cancer Res. 2016;76:5270. doi: 10.1158/1538-7445.AM2016-5270. [DOI] [Google Scholar]
- 24.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 25.Győrffy B., Surowiak P., Budczies J., Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pander A., Chinnaiyan A.M., et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y., Zheng S., Torossian A., Speirs C.K., Schleicher S., Giacalone N.J., Carbone D.P., Zhao Z., Lu B. Role of insulin-like growth factor-1 signaling pathway in cisplatin-resistant lung cancer cells. Int. J. Radiat. Oncol. Biol. Phys. 2012;82:e563–e572. doi: 10.1016/j.ijrobp.2011.06.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Lecane P.S., Thiemann P., Fan Q., Cortez C., Ma X., Tonev D., Miles D., Naumovski L., Miller R.A., et al. Synthesis and biologic properties of hydrophilic sapphyrins, a new class of tumor-selective inhibitors of gene expression. Mol. Cancer. 2007;6:9. doi: 10.1186/1476-4598-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tooker P., Yen W.C., Ng S.C., Negro-Vilar A., Hermann T.W. Bexarotene (LGD1069, Targretin), a selective retinoid X receptor agonist, prevents and reverses gemcitabine resistance in NSCLC cells by modulating gene amplification. Cancer Res. 2007;67:4425–4433. doi: 10.1158/0008-5472.CAN-06-4495. [DOI] [PubMed] [Google Scholar]
- 30.Lander E.S. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou S., Wang P., Su X., Chen J., Chen H., Yang H., Fang A., Xie L., Yao Y., Yang J., et al. Correction: High ECT2 expression is an independent prognostic factor for poor overall survival and recurrence-free survival in non-small cell lung adenocarcinoma. PLoS ONE. 2018;12:e0187356. doi: 10.1371/journal.pone.0196354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.John N.W., Eric A.C., Gordon B.M., Mills Shaw K.R., Brad A.O., Kyle E., Shmulevich I., Sander C. The Cancer Genome Atlas Pan-Cancer analysis project. Chin. J. Lung Cancer. 2015;45:1113–1120. [Google Scholar]
- 33.Yu C., Hou L., Cui H., Zhang L., Tan X., Leng X., Li Y. LDHA upregulation independently predicts poor survival in lung adenocarcinoma, but not in lung squamous cell carcinoma. Future Oncol. 2018;14:2483–2492. doi: 10.2217/fon-2018-0177. [DOI] [PubMed] [Google Scholar]
- 34.Chen D., Luo L., Liang C. Aberrant S100A16 expression might be an independent prognostic indicator of unfavorable survival in non-small cell lung adenocarcinoma. PLoS ONE. 2018;13:e0197402. doi: 10.1371/journal.pone.0197402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortier A.M., Asselin E., Cadrin M. Keratin 8 and 18 loss in epithelial cancer cells increases collective cell migration and cisplatin sensitivity through claudin1 up-regulation. J. Biol. Chem. 2013;288:11555. doi: 10.1074/jbc.M112.428920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galarneau L., Loranger A., Gilbert S., Marceau N. Keratins modulate hepatic cell adhesion, size and G1/S transition. Exp. Cell Res. 2007;313:179–194. doi: 10.1016/j.yexcr.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., He Q.Y., Tsao S.W., Cheung Y.H., Wong A., Chiu J.F. Cytokeratin 8 silencing in human nasopharyngeal carcinoma cells leads to cisplatin sensitization. Cancer Lett. 2008;265:188–196. doi: 10.1016/j.canlet.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Maria Teresa L., Tatiana D., Melissa R., Figueroa J.D., Huaitian L., Abhijit D., Mann F.E., Fukuoka J., Hames M., Bergen A.W., et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beer D.G., Kardia S.L., Huang C.C., Giordano T.J., Levin A.M., Misek D.E., Lin L., Chen G., Gharib T.G., Thomas D.G., et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 40.Su L.J., Chang C.W., Wu Y.C., Chen K.C., Lin C.J., Liang S.C., Lin C.-H., Whang-Peng J., Hsu S.-L., Chen C.-H., et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jun H., Joachim A., Bianca D.H., Wilfred V.I., Michael D.B., Peter R., van der Leest C., van der Spek P., Foekens J.A., Hoogsteden H.C., et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirokazu O., Takashi K., Yuko I., Yoko S., Kouya S., Reika I., Furuta K., Tsuta K., Shibata T., Yamamoto S., et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 43.Trisdale S.K., Schwab N.M., Hou X., Davis J.S., Townson D.H. Molecular manipulation of keratin 8/18 intermediate filaments: Modulators of FAS-mediated death signaling in human ovarian granulosa tumor cells. J. Ovar. Res. 2016;9:8. doi: 10.1186/s13048-016-0217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carney B.J., Rangachari D., Vanderlaan P.A., Gowen K., Schrock A.B., Ali S.M., Costa D.B. De novo ERBB2 amplification causing intrinsic resistance to erlotinib in EGFR-L858R mutated TKI-naïve lung adenocarcinoma. Lung Cancer. 2017;114:108. doi: 10.1016/j.lungcan.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 45.Ricciardelli C., Lokman N.A., Pyragius C.E., Ween M.P., Macpherson A.M., Ruszkiewicz A., Hoffmann P., Oehler M.K. Keratin 5 overexpression is associated with serous ovarian cancer recurrence and chemotherapy resistance. Oncotarget. 2017;8:17819–17832. doi: 10.18632/oncotarget.14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bozza W.P., Zhang Y., Zhang B. Cytokeratin 8/18 protects breast cancer cell lines from TRAIL-induced apoptosis. Oncotarget. 2018;9:23264–23273. doi: 10.18632/oncotarget.25297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.