Abstract
3D genome structures play a key role in gene regulation and cell functions. Characterization of genome structures necessitates single-cell measurements. This has been achieved for haploid cells but remained a challenge for diploid cells. Here we report a single-cell chromatin conformation capture method, termed Dip-C, which combines a transposon-based whole-genome amplification method, called Multiplex End-tagging Amplification (META), to detect many chromatin contacts, and an algorithm to impute the two chromosome haplotypes linked by each contact. We reconstructed the genome structures of single diploid human cells from a lymphoblastoid cell line and from primary blood cells with high spatial resolution, locating specific single-nucleotide and copy-number variations in the nucleus. The two alleles of imprinted loci and the two X chromosomes were structurally different. Cells of different types displayed statistically distinct genome structures. Such structural cell typing is crucial for understanding cell functions.
The nucleus of a human diploid cell contains 46 chromosomes — 23 maternal and 23 paternal, together carrying 6 Gb of genomic DNA. The 3D genome structure is thought to be crucial for the regulation of gene expression and other cellular functions (1). For example, the nuclei of sensory neurons assume unusual architectures in the mouse visual (2) and olfactory systems (3). Chromatin conformation capture assays, such as 3C (4) and Hi-C (5), allow for studies of 3D genome structures in bulk samples through proximity ligation of DNA (6). However, the difference between cells can only be observed by single-cell measurements. Single-cell chromatin conformation capture methods avoid ensemble averaging (7–12) and have yielded 3D genome structures of haploid mouse cells (10, 11). However, characterizing the 3D genome structures of diploid mammalian cells remains challenging (13). Here we use an improved chromatin conformation capture method and phased (haplotype-resolved) single-nucleotide polymorphisms (SNPs) to distinguish between the two haplotypes of each chromosome. This allows us to examine the cell-type dependence of 3D genome structures of diploid cells.
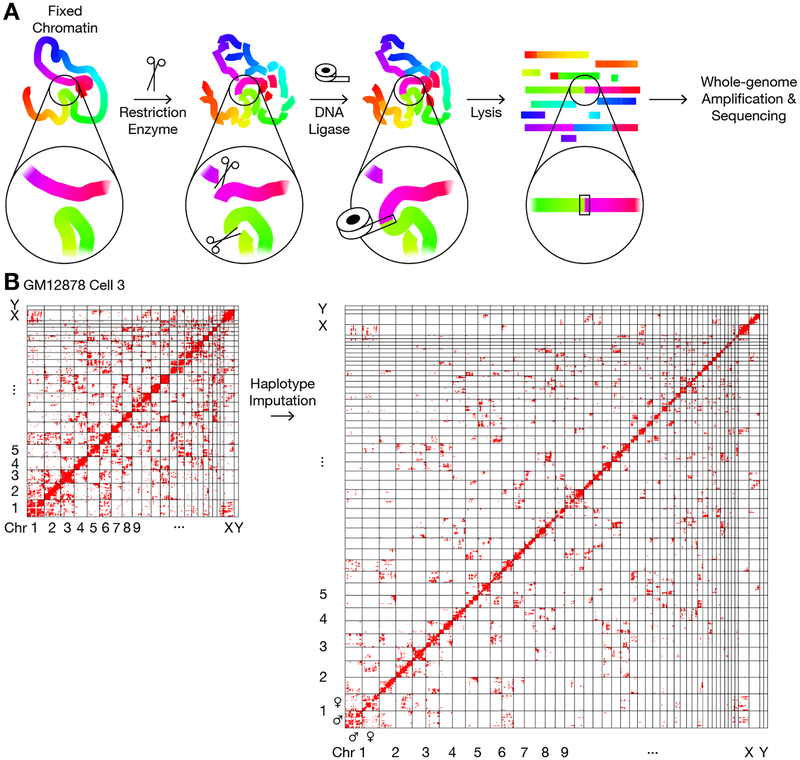
Obtaining high-resolution 3D genome structures of single diploid cells requires resolving a large number of chromatin “contacts” — pairs of genomic loci that are joined by proximity ligation. We developed a chromatin conformation capture method, termed Dip-C (Fig. 1A), that can detect more contacts than existing methods with minimal false positives. Compared to previous methods, special care was taken to avoid losing contacts by omitting biotin pulldown (8, 9) and conducting high-coverage whole-genome amplification with Multiplex End-tagging Amplification (META) (14, 15), which introduced few artefactual chimera (Materials and Methods). We detected a median of 1.04 million contacts per single cell (n = 17, min = 0.71 million, max = 1.48 million) from GM12878, a female human lymphoblastoid cell line, and a median of 0.84 million contacts (n = 18, min = 0.67 million, max = 1.08 million) from peripheral blood mononuclear cells (PBMCs) of a male human donor (16). This is four times greater than the medians of existing methods (Fig. S4, Table S1). Most cells were in the G1 or G0 phase of the cell cycle. In addition, we simultaneously detected copy-number variations (CNVs), loss of heterozygosity (LOH), DNA replication, and V(D)J recombination with a 10-kb bin size (Fig. S2, Fig. S3).
Fig. 1. Single-cell chromatin conformation capture and haplotype imputation by Dip-C.
(A) Schematics of the chromatin conformation capture protocol. The 3D information of chromatin structure was encoded in the linear genome through proximity ligation of chromatin fragments, as in 3C (4) and Hi-C (5, 19). Ligation product was then amplified by META (15) and sequenced. Colors represented genomic coordinates. Note that ligation products may be linear (illustrated here) or circular (not shown). (B) Imputation of the two chromosome haplotypes linked by each chromatin “contact” (red dot) in a representative single cell.
Another challenge in reconstructing diploid genomes is knowing which haplotypes are involved in each chromatin contact (17–20) (Table S1). To assign haplotypes, we developed an imputation algorithm (Fig. 1B). We reasoned that unknown haplotypes can be imputed from “neighboring” (in terms of genomic distances) contacts based on the assumption that the two homologs would typically contact different partners. Using a statistical property of interchromosomal and long-range intrachromosomal contacts that contacts tended to cluster along the linear genome (Materials and Methods), we defined a contact neighborhood as a superellipse with an exponent of 0.5 and a radius of 10 Mb, where haplotypes of nearby contacts were weighted in imputing the haplotypes of each contact (Fig. S7). In the Dip-C algorithm, after removing 3C/Hi-C artefacts (contacts with few neighbors (11)) and initial imputation, haplotypes may be optionally refined through a series of draft 3D models (Materials and Methods, Fig. S5). Imputation accuracy was estimated to be ~ 96% for each haplotype by cross-validation (Materials and Methods, Table S1). Regions harboring CNVs or LOHs, as well as an apparently damaged GM12878 cell, were excluded from reconstruction (Table S1).
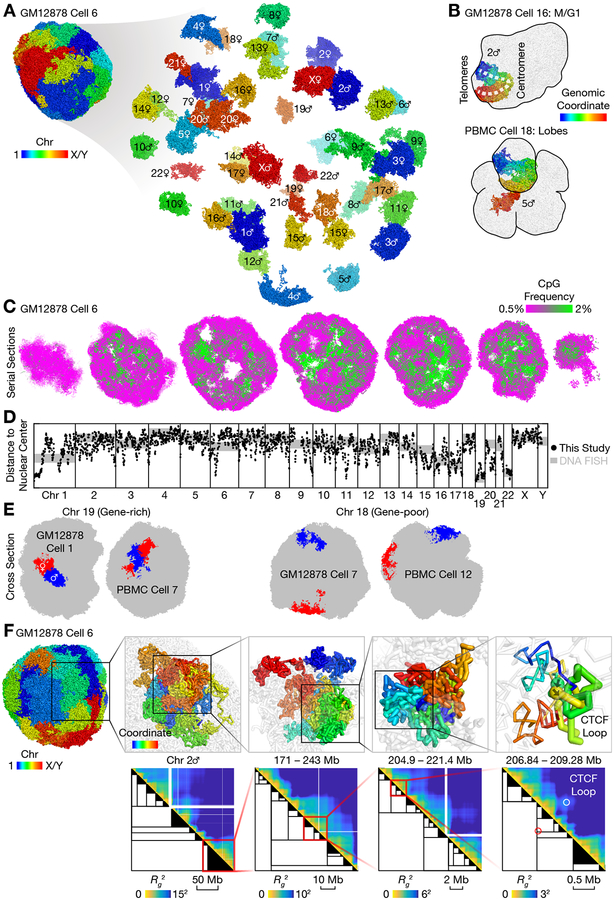
We reconstructed the 3D diploid human at a 20-kb resolution. Reconstruction was successful without supervision for 94% (15 out of 16) of the GM12878 cells and 67% (12 out of 18) of the PBMCs, and after removal of small problematic regions for 6% (one out of 16) of the GM12878 cells and 22% (4 out of 18) of the PBMCs (Table S1, Fig. S8, Materials and Methods). Note that because chromatin conformation capture — the process of converting 3D coordinates to chromatin contacts — is intrinsically lossy and noisy, our 3D structures harbored additional uncertainties including perturbations of chromatin structures during the experiments, inaccuracies in the energy function used by 3D modeling, and nuclear volumes inaccessible to DNA sequencing (for example centromeres, nucleoli, and nuclear speckles). These uncertainties were general to all 3C/Hi-C studies and difficult to estimate; and imputation was less successful when two homologs were nearby or adopted similar shapes. Therefore, other problematic regions might persist even after manual removal. Fig. 2A shows a representative cell. Each particle, displayed as a colored point, represents 20 kb of chromatin, or a radius of ~ 100 nm. A lower bound for reconstruction uncertainty was estimated from the median deviation of ~ 0.4 particle radii (~ 40 nm) across all 20-kb particles between three replicates (Fig. S9, Table S1). Well known nuclear morphologies were observed in an M/G1-phase GM12878 cell, where chromosomes retained their characteristic V shapes after recent mitosis, and in several PBMCs, where multiple nuclear lobes were reminiscent of the partially segmented nuclei of low-density neutrophils and other blood cell types (Fig. 2B).
Fig. 2. 3D genome structures of single diploid human cells.
(A) 3D genome structure of a representative GM12878 cell. Each particle represents 20 kb of chromatin, or a radius of ~ 100 nm. (B) Peculiar nuclear morphology in a cell that recently exited mitosis (upper panel) and in a cell with multiple nuclear lobes (lower panel). (C) Serial cross sections of a single cell showed compartmentalization of euchromatin (green) and heterochromatin (magenta), visualized by CpG frequency as a proxy (21). (D) Radial preferences across the human genome, as measured by average distances to the nuclear center of mass. Our results (black dots, smoothed by 1-Mb windows) agreed well with published DNA FISH data (gray lines) on whole chromosomes (22) (shifted and rescaled), and provided novel fine-scale information. Axis limits were 20 and 50 particle radii for the black dots. GM12878 Cell 4 (extensive chromosomal aberrations) and Cell 16 (M/G1 phase) was excluded. (E) Example radial preferences of two chromosomes. The gene-rich chromosome 19 preferred the nuclear interior (left), while the gene-poor chromosome 18 almost always resided on the nuclear surface (right). (F) Stochastic fractal organization of chromatin was quantified by a matrix of radii of gyration of all possible subchains of each chromosome (heatmaps). We identified a hierarchy of single-cell domains across genomic scales (black trees). A subtree was simplified as a black triangle if either of its two subtrees was below a certain size (from left to right: 10 Mb, 2 Mb, 500 kb, 100 kb). In each panel, the region from the previous panel was shown in transparent gray. In the rightmost panel, thick sticks (top) and circles (bottom) highlighted the formation of a known CTCF loop (19). Spheres with arrows (top) heighted the positions and orientations of the two converging CTCF sites. Genomic coordinates were in hg19.
We also used published data on mouse embryonic stem cells (mESCs) (10) to reconstruct 3D diploid mouse genomes despite fewer contacts (~ 0.3 million per cell, or ~ 0.2 million under our definition) (Table S1), because the mouse line harbored more SNPs than humans (Materials and Methods).
Similar to previously described haploid mouse genomes (10, 11), the diploid human genomes exhibited chromosome territories (Fig. 2A) and chromatin compartments (visualized by CpG frequency as a proxy (21)), with the heterochromatic compartment B (5) concentrated at the nuclear periphery and around foci in the nuclear center (Fig. 2C). Spatial clustering of DNA sequences with similar CpG frequencies suggests a correlation between primary sequence features and 3D genome folding (1).
Our 3D structures revealed different radial preferences across the human genome (black dots in Fig. 2D). Our results agree well with whole-chromosome painting data by DNA fluorescent in situ hybridization (FISH) (22) (gray lines in Fig. 2D). Both methods show that the gene-rich chromosome 19 prefers the nuclear interior, while the gene-poor chromosome 18 prefers the nuclear periphery (Fig. 2E). Within each chromosome, different segments could have distinctly different radial preferences, which were correlated with chromatin compartments (Fig. S11A). For example, the CpG-rich, euchromatic end (left) of chromosome 1 was heavily biased towards the nuclear center, while some other regions on the same chromosome were biased towards the nuclear periphery (Fig. 2D). Such fine-scale information cannot be obtained from whole-chromosome painting (22, 23) experiments.
Our Dip-C results provide a holistic view of the stochastic, fractal organization of chromatin across different genomic scales. Bulk Hi-C suggests that chromatin forms a “fractal globule” with compartments (5, 19) and domains such as topologically associating domains (TADs) (24) and CTCF loop-domains (19). However, such fractal organization has not been visualized in single human cells in a genome-wide manner. We observed spatial clustering (globules) and segregation (insulation) of consecutive chromatin particles along each chromosome (upper panels in Fig. 2F). Such organization could be quantified by a matrix of radii of gyration of all possible subchains in each chromosome (lower panels in Fig. 2F). Single-cell domains could then be identified as squares that had relatively small radii (partly similar to (8)) (Materials and Methods). We found single-cell domains across all genomic scales and therefore identified them through hierarchical merging, yielding a tree of domains (partly similar to (25, 26) in bulk Hi-C) (Fig. 2F). On the smallest scale, some domains coincided with CTCF loop-domains from bulk Hi-C (19) (rightmost panels in Fig. 2F). Single-cell domains were highly heterogeneous between cells, frequently breaking and merging bulk domains (Fig. S19), consistent with a recent study on tetraploid mouse cells (8).
Traditional methods such as bulk Hi-C and two-color DNA FISH are pairwise measurements and thus cannot study multi-chromosome intermingling. In our 3D models, we quantified multi-chromosome intermingling by the diversity of chromosomes (Shannon’s index) near each 20-kb particle (Fig. S20A), revealing genomic regions that frequently contacted multiple chromosomes (Fig. S20B). These regions were similar between the human cell types despite their different average extents of intermingling (Fig. S10), and were mostly euchromatic (CpG-rich) (Fig. S11B) for two reasons: (1) euchromatin more frequently resided on the surface of chromosomes than heterochromatin (consistent with (7)) (Fig. S11D), and (2) even when heterochromatin resided on the surface, it tended to face the nuclear periphery (11) (Fig. S11A) and thus had no partners to intermingle with. The intermingling regions partially overlapped with “hubs” identified by a recent report (27).
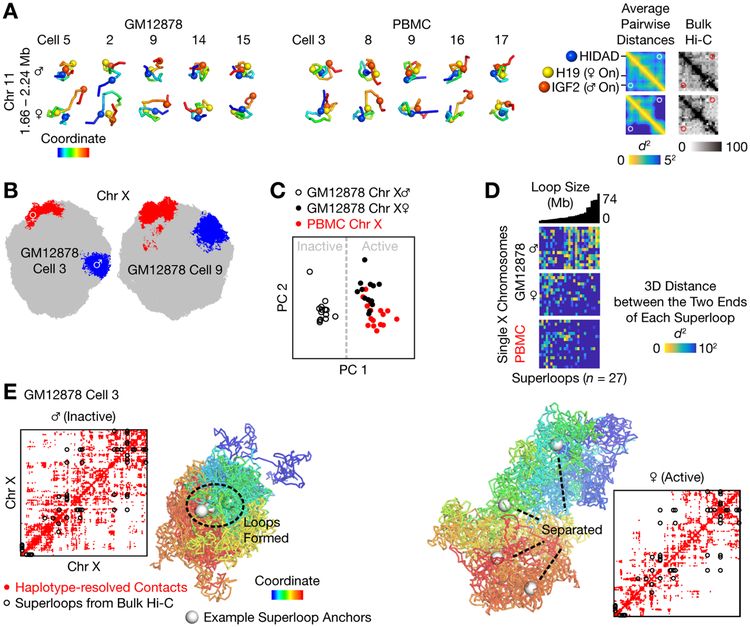
We examined the structural relationship between the maternal and paternal alleles, which can only be studied in diploid cells. Our data captured the structural difference between the two alleles caused by genomic imprinting. At imprinted loci, the two alleles can differ drastically in transcriptional activity (28). Near the maternally transcribed H19 gene and the paternally transcribed IGF2 gene, bulk Hi-C identified different contact profiles and different use of CTCF loops between the two homologs (19). We directly visualized this ~ 0.6-Mb region in single cells (Fig. 3A). Despite cell-to-cell heterogeneity, the maternal allele more frequently separated IGF2 from both H19 and the nearby HIDAD site and disrupted the IGF2-HIDAD CTCF loop, while the paternal allele more frequently stayed fully intermingled.
Fig. 3. Distinct 3D structures of the maternal and the paternal alleles.
(A) Structural difference between the two alleles of the imprinted H19/IGF2 locus. Despite cell-to-cell heterogeneity, the maternal allele more frequently separated IGF2 from both H19 and the nearby HIDAD site and disrupted the IGF2-HIDAD CTCF loop (white and red circles). Spheres highlighted three CTCF sites from bulk Hi-C. Heatmaps showed the r.m.s. average pairwise distances between all 20-kb particles. Haplotype-resolved bulk Hi-C (black heatmap, with 25-kb bins) was from Fig. 7C of (19). (B) The active (red) and inactive (blue) X chromosomes preferred extended and compact morphologies, respectively, as shown by cross sections of two representative cells. (C) Individual active and inactive X chromosomes could be distinguished by principal component analysis (PCA) of single-cell chromatin compartments, defined for each 20-kb particle as the average CpG frequency of nearby particles. (D) The inactive X chromosome tended to form the previously reported “superloops” — 27 very-long-range (5–74 Mb) chromatin loops identified by bulk Hi-C (19, 20, 29). Superloops were sorted by sizes (Mb). (E) Haplotype-resolved contact maps (red dots) and 3D structures of the two X chromosomes in an example cell. Black circles denoted all superloops (19). White spheres denoted 4 example superloop anchors (DXZ4, x75, ICCE, and FIRRE). GM12878 Cells 4 and 16 were excluded from (C) and (D).
X chromosome inactivation (XCI) presents a striking example of the difference between two homologs (28). As expected, we found in the female GM12878 cell line that the active X chromosome (the maternal allele based on RNA expression, Materials and Methods) tended to exhibit an extended morphology, and the inactive X a compact one (Fig. 3B), although in some cells this morphological difference was not obvious. More consistently, the two X chromosomes in each cell were characterized by their distinct patterns of chromatin compartments. The active X featured clear compartmentalization of euchromatin and heterochromatin, resembling that of the male X (in PBMCs); in contrast, compartments along the inactive X were more uniform (Fig. S12E). Individual X chromosomes could be clearly separated into two clusters — active and inactive — by principal component analysis (PCA) of single-cell compartments (Fig. 3C). Our conclusion held if single-cell compartments were defined based on contacts (partly similar to (10)) rather than 3D structures (Fig. S15A and Fig. S15B). We also visualized the simultaneous formation of multiple “superloops” (19, 20, 29) in the inactive X chromosome (Fig. 3D and Fig. 3E). Averaged contact matrices of the inactive and active X chromosomes agreed well with bulk Hi-C (19) (Fig. S15C and Fig. S15D).
In contrast to XCI, it is unknown whether single-cell compartments of two autosomal alleles may vary in a coordinated manner. By decomposing the variability of single-cell compartments into between-cell and within-cell differences (Fig. S12A), we found that autosomal alleles fluctuate (with respect to their median compartments) almost independently from each other, exhibiting on average near-zero Spearman’s correlation (Fig. S12D). Our conclusion held if compartments were defined based on contacts (Fig. S16).
We can pinpoint genomic changes, such as SNPs and CNVs, to their precise spatial locations in the cell nucleus. The donor of the GM12878 cell line carried a heterozygous G-to-A mutation (rs4244285) in a cytochrome P450 gene CYP2C19, leading to a truncated, non-functional protein variant CYP2C19*2 and affecting metabolism of hormones and drugs (30). Fig. S18A shows the 3D localization of this drug-response SNP on the paternally inherited chromosome 10 of a GM12878 cell. In addition to inherited mutations, single cells also harbor somatic changes. In lymphocytes, somatic V(D)J recombination generates diversity of immunoglobulins and T-cell receptors by DNA deletions and inversions. Fig. S18B shows the 3D localization of two V(D)J recombinations at a T-cell receptor locus, leading to two different DNA deletions on the two alleles of chromosome 14 of a T lymphocyte. The capability to spatially localize genomic changes is important for studying cancers and inherited diseases, where mutations can have dramatic consequences and may disrupt the chromatin structure of nearby regions.
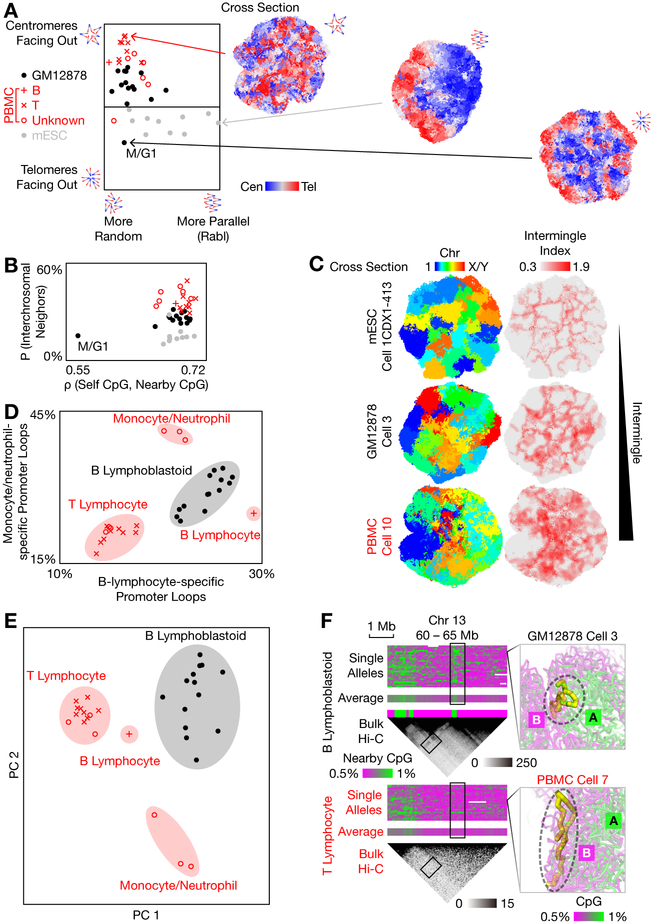
We also examined the cell-type dependency of 3D genome structures. Similar to haploid mESCs (11), chromosomes in diploid mESCs preferred the Rabl configuration (centromeres pointing towards one side of the nucleus, and telomeres towards the other), albeit to a different extent in each cell (Fig. 4A). In contrast, Rabl configuration is weak in most GM12878 cells and PBMCs. Most PBMCs pointed their centromeres towards the nuclear periphery and telomeres towards the nuclear center, consistent with previously reported arrangements in human lymphocytes (31). On the contrary, the M/G1-phase GM12878 cell pointed centromeres towards the outer rim of a characteristic mitotic rosette.
Fig. 4. Cell-type-specific chromatin structures.
(A) Quantification of the organization of centromeres and telomeres. The mESCs exhibited stronger Rabl configuration (horizontal axis; the length of summed centromere-to-telomere vectors normalized by the total particle number, which was different between human and mouse; axis limit = 0.005 particle radii), while the PBMCs tended to point centromeres outwards relative to telomeres (vertical axis; the summed centromere-to-telomere difference in distances from the nuclear center of mass normalized by the total particle number; axis limit = 0.007 particle radii). Each marker represented a single cell and was inferred by V(D)J recombination in PBMCs (Table S1, Fig. S3B). (B) Quantification of chromosome intermingling (vertical axis; the average fraction of nearby particles that were not from the same chromosome) and chromatin compartmentalization (horizontal axis; Spearman’s correlation between each particle’s own CpG frequency and the average of nearby particles). (C) Example cross sections of 3 cell types, colored by chromosomes (left) or by the multi-chromosome intermingle index (right). (D) Among the human cells, 4 cell-type clusters (shaded) — B lymphoblastoid cells, presumable T lymphocytes, B lymphocytes, and presumable monocytes/neutrophils (PBMC Cells 9, 14, and 18) — could be distinguished from the differential formation (defined as end-to-end distance ≤ 3 particle radii) of known cell-type-specific promoter-enhancer loops from published bulk promoter capture Hi-C (35). (E) The same 4 clusters could also be distinguished by unsupervised clustering via PCA of single-cell chromatin compartments, without the need for bulk data. The two alleles of each locus were treated as two different loci. (F) An example region that was differentially compartmentalized between two cell types (black: B lymphoblastoid cells; red: presumable T lymphocytes). Right panels visualized the configuration of the ~ 0.5-Mb region (chr 13: 62.5 – 63 Mb, thick yellow sticks) with respect to the rest of the genome (transparent, colored by CpG frequencies) in two representative cells. Only the paternal alleles were shown. Bulk Hi-C (black heatmap, with 50-kb bins) was from (19, 40). GM12878 Cell 4 was excluded. GM12878 Cell 16 was excluded from (D) and (E).
The overall extent of chromosome intermingling also differed among the cell types. Chromosomes tended to intermingle less in mESCs and more in PBMCs, with GM12878 intermediate (Fig. 4B, Fig. 4C), consistent with previous reports that chromosomes intermingled less in the pluripotent mESCs than in terminally differentiated fibroblasts (32), and that chromosomes intermingled more in resting human lymphocytes than in activated ones (which resembled GM12878) (33). As expected (10, 34), the M/G1-phase cell exhibited a low level of chromosome intermingling and the lowest level of chromatin compartmentalization.
Cell-type dependent promoter-enhancer looping is suggested to underlie differential gene expression (35). Among the human cells, differential formation of known cell-type-specific promoter-enhancer loops (based on cell-type purified bulk Hi-C (35), Materials and Methods) clearly separated the single cells into four cell-type clusters — B lymphoblastoid cells (GM12878), presumable T lymphocytes, B lymphocytes, and presumable monocytes/neutrophils (Fig. 4D). Defining loop formation based on contacts rather than 3D structures yielded similar results (Fig. S17A).
Cell-type clusters could be equally well separated in an unsupervised manner, without prior knowledge of the cell types. Unlike ensemble-averaged structures such as protein crystal structures, single-cell 3D genomes are intrinsically stochastic and dynamic, blended with measurement uncertainties. Statistical characterization such as PCA is necessary to distinguish different cell types, in which clusters of single cells correspond to valleys in a Waddington landscape (36) of certain cellular phenotypes. This kind of cell typing has been carried out based on phenotype variables such as single-cell transcriptomes (37) and open chromatin regions (38, 39), each of which must have underlying structural differences in the 3D genome.
With Dip-C, we are in a position to carry out cell typing with genome structure as the sole variable. Given the high information content of 3D structures, many possible features might be used in cluster analysis. Here we chose single-cell chromatin compartments as the input variable of PCA. The four cell-type clusters were clearly separated (Fig. 4E), with one of the most differentially compartmentalized regions shown in Fig. 4F. Our conclusion held if compartments were defined based on contacts (Fig. S17B and Fig. S17C). Previous reports (7, 8, 10–12) had focused on defining the width, or spread, of a single Waddington valley, studying for example cell-cycle dynamics within a cell type and domain stochasticity within a cell-cycle phase. Our PCA result, in contrast, highlighted the consistent difference among cell types, signifying the separation between Waddington valleys.
Our initial examination of only a handful of cell types has clearly shown the tissue-dependence of 3D genome structures. A systematic survey of more cell types under various conditions will likely lead to new discoveries in cell differentiation, genesis of cancer, learning and memory, and aging.
Supplementary Material
Acknowledgments
The authors thank Chenghang Zong (Harvard University, currently Baylor College of Medicine) for his involvement at the early stage, Erez Lieberman Aiden (Baylor College of Medicine) and Elena Stamenova (Broad Institute) for advice about their in situ Hi-C protocol, Tim Stevens (University of Cambridge) for help on their simulated annealing software (“nuc_dynamics”), Mingyu Yang (Peking University) for published phased genotypes of the blood donor, Takashi Nagano and Steven Wingett (Babraham Institute) for answering questions about published raw data, and Yiqin Gao, Luming Meng, and Sirui Liu (Peking University) for helpful discussion.
Funding: This work was supported by Beijing Advanced Innovation Center for Genomics at Peking University, an NIH Director’s Pioneer Award (DP1 CA186693), a Harvard Brain Initiative (HBI) Collaborative Seed Grant, and two grants from the National Science Foundation of China (21390412 and 21327808) to X.S.X.. L.T. was supported by an HHMI International Student Research Fellowship. H.L. was supported by an NHGRI grant (R01 HG010040).
Footnotes
Competing interests: L.T., D.X., C.H.C., and X.S.X. are inventors on a provisional patent application US 62/509,981 filed by Harvard University that covers META and Dip-C.
Data and materials availability: Raw sequencing data were deposited at the National Center for Biotechnology Information with accession number SRP149125 at https://www.ncbi.nlm.nih.gov/sra/SRP149125. Processed data were deposited with GEO Series accession number GSE117876 at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117876. Code is available at GitHub (https://github.com/tanlongzhi/dip-c and https://github.com/lh3/hickit). Chromatin contacts can be viewed interactively in the Juicebox web browser (41); please see the manual page of our codes.
References and Notes
- 1.Cremer T, Cremer C, Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2, 292–301 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B, Nuclear Architecture of Rod Photoreceptor Cells Adapts to Vision in Mammalian Evolution. Cell 137, 356–368 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S, Nuclear Aggregation of Olfactory Receptor Genes Governs Their Monogenic Expression. Cell 151, 724–737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker J, Rippe K, Dekker M, Kleckner N, Capturing chromosome conformation. Science 295, 1306–1311 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J, Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen KE, Kladde MP, Seyfred MA, Interaction between transcription regulatory regions of prolactin chromatin. Science 261, 203–206 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P, Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flyamer IM, Gassler J, Imakaev M, Brandao HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K, Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, ZHANG J, Zhao H, PEI Z, Xuan Z. (Google Patents, 2017).
- 10.Nagano T, Lubling Y, Varnai C, Dudley C, Leung W, Baran Y, Mendelson Cohen N, Wingett S, Fraser P, Tanay A, Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O’Shaughnessy-Kirwan A, Cramard J, Faure AJ, Ralser M, Blanco E, Morey L, Sanso M, Palayret MGS, Lehner B, Di Croce L, Wutz A, Hendrich B, Klenerman D, Laue ED, 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, Noble WS, Duan Z, Shendure J, Massively multiplex single-cell Hi-C. Nat Methods 14, 263–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carstens S, Nilges M, Habeck M, Inferential Structure Determination of Chromosomes from Single-Cell Hi-C Data. PLoS Comput Biol 12, e1005292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, Xie XS, Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI). Science 356, 189–194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie XS, Xing D, Chang C-H, Tan L. Multiplex end-taggin amplification. Provisional Patent Application US 62/509,981 (2017).
- 16.Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, Hu X, Xu L, Yan L, Bai F, Qiao J, Tang F, Li R, Xie XS, Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 338, 1627–1630 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, Diao Y, Liang J, Zhao H, Lobanenkov VV, Ecker JR, Thomson JA, Ren B, Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E, HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL, A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, Dekker J, Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie WJ, Meng L, Liu S, Zhang L, Cai X, Gao YQ, Structural Modeling of Chromatin Integrates Genome Features and Reveals Chromosome Folding Principle. Sci Rep 7, 2818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA, The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10, 211–219 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, Wang P, Tian F, Gao G, Huang L, Wei W, Xie XS, Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging. Cell Res 27, 298–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B, Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraser J, Ferrai C, Chiariello AM, Schueler M, Rito T, Laudanno G, Barbieri M, Moore BL, Kraemer DC, Aitken S, Xie SQ, Morris KJ, Itoh M, Kawaji H, Jaeger I, Hayashizaki Y, Carninci P, Forrest AR, Consortium F, Semple CA, Dostie J, Pombo A, Nicodemi M, Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol 11, 852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinreb C, Raphael BJ, Identification of hierarchical chromatin domains. Bioinformatics 32, 1601–1609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, Trinh V, Aznauryan E, Russell P, Cheng C, Jovanovic M, Chow A, Cai L, McDonel P, Garber M, Guttman M, Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 174, 744–757 e724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JT, Bartolomei MS, X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308–1323 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC, Sun Z, Huang SC, Sanborn AL, Machol I, Shamim M, Seberg AP, Lander ES, Chadwick BP, Aiden EL, Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A 113, E4504–4512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coriell Insitute. vol. 2017. [Google Scholar]
- 31.Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, Solovei I, Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res 11, 485–502 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Maharana S, Iyer KV, Jain N, Nagarajan M, Wang Y, Shivashankar GV, Chromosome intermingling-the physical basis of chromosome organization in differentiated cells. Nucleic Acids Res 44, 5148–5160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branco MR, Branco T, Ramirez F, Pombo A, Changes in chromosome organization during PHA-activation of resting human lymphocytes measured by cryo-FISH. Chromosome Res 16, 413–426 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J, Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Varnai C, Thiecke MJ, Burden F, Farrow S, Cutler AJ, Rehnstrom K, Downes K, Grassi L, Kostadima M, Freire-Pritchett P, Wang F, Consortium B, Stunnenberg HG, Todd JA, Zerbino DR, Stegle O, Ouwehand WH, Frontini M, Wallace C, Spivakov M, Fraser P, Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384 e1319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waddington CH, The strategy of the genes; a discussion of some aspects of theoretical biology. (Allen & Unwin, London,, 1957), pp. ix, 262 p. [Google Scholar]
- 37.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA, mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ, Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J, Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong M, Huang X, Zhang X, Su J, Shamim M, Bochkov I, Reyes J, Jung H, Heikamp E, Presser Aiden A, Li W, Aiden E, Goodell MA, A Cell Type-Specific Class of Chromatin Loops Anchored at Large DNA Methylation Nadirs. bioRxiv, (2017). [Google Scholar]
- 41.Robinson JT, Turner D, Durand NC, Thorvaldsdottir H, Mesirov JP, Aiden EL, Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Syst 6, 256–258 e251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aronesty E, Comparison of sequencing utility programs. The Open Bioinformatics Journal 7, (2013). [Google Scholar]
- 43.Li H, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv e-prints. 2013. [Google Scholar]
- 44.LaSalle JM, Lalande M, Homologous association of oppositely imprinted chromosomal domains. Science 272, 725–728 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.