Abstract
Background
Newborns affected with congenital pulmonary airway malformations (CPAMs) may present with severe respiratory distress or remain asymptomatic. While surgical resection is the definitive treatment for symptomatic CPAMs, prophylactic elective surgery may be recommended for asymptomatic CPAMs owing to the risk of tumour development. However, the implementation of prophylactic surgery is quite controversial on the grounds that more evidence linking CPAMs and cancer is needed. The large gap in knowledge of CPAM pathogenesis results in uncertainties and controversies in disease management. As developmental genes control postnatal cell growth and contribute to cancer development, we hypothesised that CPAMs may be underlain by germline mutations in genes governing airways development.
Methods
Sequencing of the exome of 19 patients and their unaffected parents.
Results
A more than expected number of mutations in cancer genes (false discovery rate q-value <5.01×10−5) was observed. The co-occurrence, in the same patient, of damaging variants in genes encoding interacting proteins is intriguing, the most striking being thyroglobulin (TG) and its receptor, megalin (LRP2). Both genes are highly relevant in lung development and cancer.
Conclusions
The overall excess of mutations in cancer genes may account for the reported association of CPAMs with carcinomas and provide some evidence to argue for prophylactic surgery by some surgeons.
Short abstract
Congenital pulmonary airway malformation (CPAM) patients have more than expected numbers of damaging variants in genes involved in lung carcinoma; this may provide evidence for clinicians choosing to adopt prophylactic excision in CPAM http://ow.ly/h1AE30n4DIe
Introduction
Congenital pulmonary airway malformation (CPAM) defines those conditions involving fetal pulmonary lesions that result from abnormal overgrowth of tracheal, bronchial, bronchiolar or alveolar tissues. CPAMs have not been linked to race, maternal age or exposures and it appears that males are slightly more affected than females [1]. The majority of CPAMs present antenatally (detected by routine ultrasound examination) and persist postnatally. The clinical presentation is variable, ranging from the most serious phenotype that may result in hydrops fetalis and termination of pregnancy to asymptomatic. Affected newborns present with severe respiratory distress or remain asymptomatic until later in life. CPAMs are classified in five different types: type 0 (3% of all CPAMs) denotes global arrest of lung development and is therefore lethal; type 1 (60–70% of all CPAMs) presents with large (>2 cm) intercommunicating cysts surrounded by multiple smaller cysts; type 2 (15–20% of all CPAMs) consists of a mixture of solid tissue and small cysts (<2 cm), and is the sole CPAM known to be associated with other congenital anomalies such as tracheo-oesophageal fistula, renal agenesis, intestinal atresia and diaphragmatic hernia (overall poor prognosis); type 3 (5–10% of all CPAMs) consists of a solid large adenomatoid malformation with minor cystic components; and type 4 (10–15% of all CPAMs) also presents with large (>10 cm) cysts with unique associations with pneumothorax or pleuropulmonary blastoma, a malignant condition often part of an inherited cancer syndrome for which germline mutations in the coding sequence of the DICER1 gene have been identified [2].
While surgical resection is the definitive treatment for symptomatic CPAMs [1, 3], prophylactic elective surgery is recommended for asymptomatic CPAMs by some surgeons owing to risk of tumour development within the malformation, i.e. rhabdomyosarcoma, pleuropulmonary blastoma, adenocarcinoma, squamous cell carcinoma and mesenchymoma [4]. However, this approach remains controversial and not universally accepted on the grounds that more evidence on the link between CPAMs and cancer is needed. Notwithstanding, those cancers can be underlain by germline or hereditary mutations, as recently reported [5, 6].
CPAMs result from a defective branching morphogenesis of the lung at different developmental stages and at different levels of the tracheobronchial tree, thus explaining the different types. What triggers this developmental defect is unknown, but an imbalance between cell proliferation and apoptosis during organogenesis has been suggested [7]. Data on the molecular basis underlying CPAMs are scant and consist of gene expression analyses in fetal or postnatal resected human CPAM tissues or in animal models. However, these studies have identified deregulation of genes/proteins crucial for lung morphogenesis and patterning, including Fabp-7 [8], members of the fibroblast growth factor family [9], cell adhesion molecules [10] and developmental genes such as Hoxb5, which regulates lung airway development through interactions among the aforementioned molecules. Thus, alteration of the signalling pathways controlling lung development is likely the mechanism underlying CPAMs [11–13]. Plausibly, DNA alterations in gene members of the involved pathways could lead to CPAMs.
No genetic study has ever been conducted on CPAM, probably because there is no evidence for a classic genetic inheritance of the disorder (as the disease presents mainly sporadically), which may be due to the fact that some CPAMs may go undetected or manifest later in life when a malignancy has already developed. Notwithstanding, a few cases of familial aggregation/segregation have been reported [14], advocating genetic transmission of the disorder. Indeed, CPAM appears mainly sporadically with a very low incidence (ranging from 1 per 8300 to 1 per 35 000 live births); locally, the incidence is 1 per 7200 live births (EUROCAT prevalence tables 2017: www.eurocat-network.eu) [15].
Thus, we set out to investigate the genetics underlying CPAMs under the hypothesis that rare de novo or recessive inherited damaging genetic variants in genes governing the development of airways could trigger the disorder, and account for the sporadic presentation and scarcity of CPAM and its controversial link with malignancies.
Methods
To test our hypothesis, we adopted the “trio-based” based whole-exome sequencing (WES) and copy number variants (CNVs) approach whereby the exomes and CNVs of the patient and his/her unaffected parents are analysed and compared. Generation and analysis of WES and CNV data is described in the supplementary material [16, 17].
Common DNA variants were filtered out, leaving only variants whose minor allele frequency (MAF) in the East and South Asian population is ≤1% according to the Exome Aggregation Consortium (exac.broadinstitute.org), and variant-based prediction methods were adapted (see supplementary material). This is consistent with recessive transmission of the disorder by homozygous or compound heterozygous inheritance. For digenic damaging variants analyses, variants with MAF ≥5% were filtered out. Comparison of parental and offspring sequences was used to identify de novo variants and recessively inherited mutations.
The study was approved by the Institutional Review Board of The University of Hong Kong (Hong Kong SAR, China) together with the Hospital Authority (UW 12-469). Blood samples were drawn from all participants after obtaining informed parental consent.
Patients
A total of 19 CPAM ethnic Chinese patients (nine male and 10 female; 19×3=57 samples) and their parents were included in the study. Patients were recruited at the Dept of Surgery of Queen Mary Hospital (Hong Kong SAR, China), to which patients from all the territory are referred, and at the Dept of Pediatric Surgery, Guangzhou Women and Children's Medical Center (Guangzhou, China). Clinical characteristics of the patients, including associated anomalies, are listed in supplementary table S1. One patient, CPAM9, was diagnosed with type 4 CPAM with pleuropulmonary blastoma, but bore no DICER1 mutations. All patients had a normal karyotype. Patient management was done as previously described [3]. Pathological assessment of the resected specimens was done in the Dept of Pathology (Queen Mary Hospital or Guangzhou). No family history of CPAM was reported for any of the patients. All babies had been full term.
DNA was extracted from blood using a DNA extraction kit (Qiagen, Hilden, Germany) and 3 μg submitted to the Center for Genomic Sciences of The University of Hong Kong where genotypes were obtained.
Controls
For quality assessment and potential population stratification, principal component analysis was conducted. The common variants (MAF >5%) from 699 local Chinese subjects participating in a degenerative disc disease cohort as part of an in-house exome sequencing project were used as controls.
Results
After strict WES quality control (supplementary table S2), one trio (CPAM21) had to be excluded due to contamination. Downstream analyses were conducted on 18 trios (supplementary table S1). In brief, 85.6% of the capture regions reached at least 10 times coverage with average target coverage of 45.4 times. Similarly, after quality control of the single nucleotide polymorphism array for CNV analysis, three trios had to be excluded (supplementary table S1). Principal component analysis indicated no major biases due to population stratification or technical issues (supplementary figure S1).
As pathological mechanisms, we considered the effect of damaging de novo germline variants (either exonic and/or CNVs) and rare recessive inherited variants (either exonic and/or CNVs) in 1) homozygosis, 2) compound heterozygosis (different variants in the same gene: one paternally and one maternally inherited), and 3) a “di/oligogenic model” assuming that variants in different genes of related pathways coexist in the patient through maternal and paternal inheritance. If the same digenic gene pair occurred in any of the parents, the pair was excluded. Genes with “qualifying” variants (according to the criteria described) were queried for gene and/or protein expression in the developing lung, involvement in lung morphogenesis, and, given the alleged correlation between CPAMs and lung cancer, lung cancer development. In addition, the germline/constitutive variants identified were checked against the somatic variants of the COSMIC (Catalogue of Somatic Mutations in Cancer) database (https://cancer.sanger.ac.uk/cosmic).
De novo single nucleotide variant/indel and CNV analysis
WES analysis identified 14 de novo damaging variants (12 missense and two stop-gain) in 14 genes distributed among 12 patients (supplementary table S3). The de novo damaging coding sequence variant rate was 0.78 (14 out of 18) per patient, which is in line with reported rates [18]. Only the de novo mutations in genes known to be expressed in fetal lung are considered. Importantly, the de novo variants identified in LRRC16A and in SPG11 (loss of function) are reported in COSMIC as somatic cancer mutations, with that in SPG11 being reported in a lung adenocarcinoma patient. It is noteworthy that the variants in these two genes are not present in the South or East Asian population. Following the cancer angle, the inhibitor of growth gene ING, involved in head and neck squamous cell carcinoma, is also de novo mutated. Although not reported in the current databases, an exhaustive literature search described TRIP12 as a potential lung adenocarcinoma gene [19]. CEP295, a key gene in centriole formation, has a loss-of-function variant. Defects in centriole number and structure are associated with congenital disorders and cancers [20].
In addition, a total of five genes with de novo variants linked to autosomal recessive developmental phenotypes were identified. Although the variants identified in this study are heterozygous de novo, CPAM patients carrying variants in these genes were clinically re-assessed. No other conjunctional congenital disorder could be observed in the patients examined (supplementary table S1).
We only considered de novo genic CNVs, i.e. CNVs that overlap with gene coding sequences. None were identified, suggesting that CNVs do not play a major role in CPAMs.
Inherited exonic variants: homozygosis, hemizygosis, compound heterozygosis and gene interacting models
We then assessed the patients for inherited damaging variants acting in a recessive manner including homozygous, X-hemizygous (unpaired X chromosome in males), compound heterozygous as well as for di/oligogenic inheritance whereby two or more interacting genes are mutated in the same patient
Three genes (LCN1, SMAD7 and AKAP13) carried rare predicted damaging variants in homozygosis and six in hemizygosis, of which three (DMD, KDM6A and HEPH) were identified in patient CPAM6 (supplementary table S4). DMD and KDM6A are known to underlie X-linked disorders, which were not manifest in the patient. It is, however, important to emphasise that KDM6A has causally been implicated in cancer and is one of the genes underlying Kabuki syndrome. Many Kabuki syndrome patients are also affected with anatomical abnormalities of the lungs and/or diaphragm hernia that are also present in CPAM patients [21]. The Kdm6a mouse model exhibits respiratory distress and atelectasis. SMAD7, also homozygously mutated in patient CPAM6, is involved in cancer susceptibility.
For compound heterozygotes, events with at least one deleterious allele were identified in 21 genes distributed among 14 CPAM families (supplementary table S5). One of the constitutional/germline variants detected in LZTS1, which underlies hereditary oesophageal squamous cell carcinoma, has been reported as a somatic change in one lung adenocarcinoma patient. Likewise for one of the ZNF804B variants. Germline variants identified in PLEKHA6, UBR1 and PCDH15 have been reported as somatic variants in cancers other than lung. MUC17 and SYNE1 are somatically mutated in >10% of lung adenocarcinomas.
Similarly, we identified 10 recurrent, physical protein–protein interaction pairs (supplementary table S6 and table 1) implicating a total of 20 genes and distributed among 14 families. The selection criterion was the presence in the patient of at least one damaging rare nonsynonymous variant in one interacting protein inherited from one parent and another inherited rare nonsynonymous variant in the other interacting protein inherited from the other parent. Lung abnormalities are detected in the mouse models of four of these genes; one of the variants in PRKG1 is also reported as a somatic mutation in carcinoma.
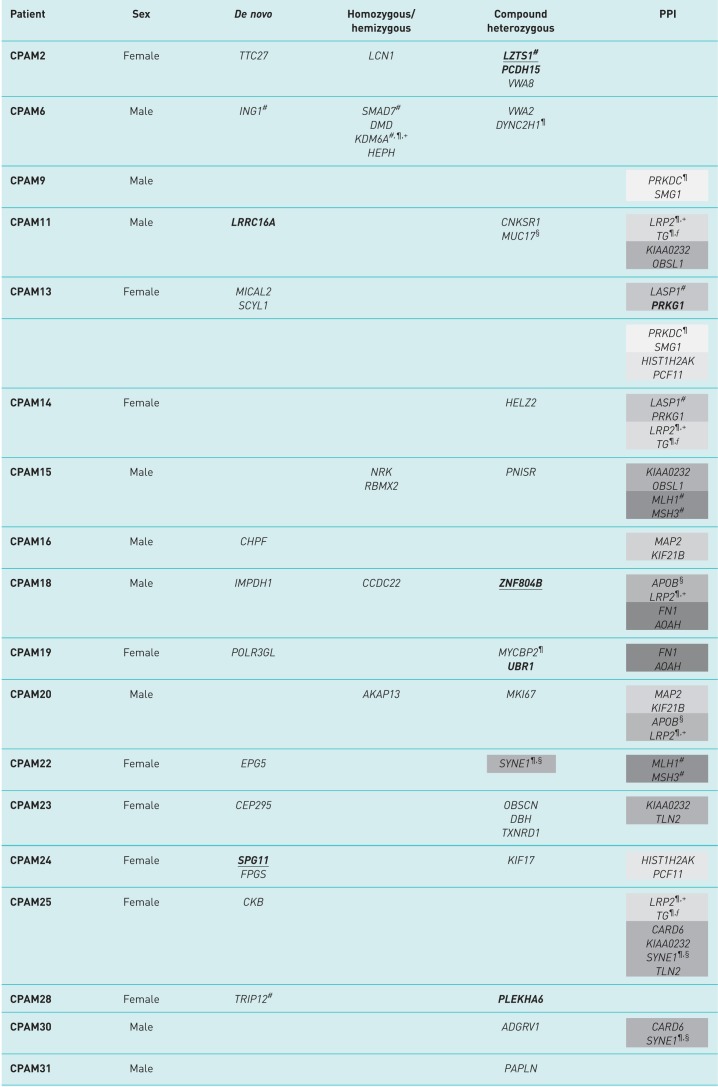
TABLE 1.
Genetic profile of the congenital pulmonary airway malformation (CPAM) patients included in this study
PPI: protein–protein interaction (or genes encoding interacting proteins). Bold: same variant identified in cancer (somatic); underlined: same variant identified in lung cancer (somatic). Recurrently mutated gene pairs (encoding interacting proteins) are grouped under the same grey shade. Each shade represents a different group of genes encoding interacting proteins. #: gene causally implicated in any human cancer; ¶: mouse model with lung abnormalities; +: gene implicated in congenital human disorders with features shared with those commonly found in CPAM patients; §: gene mutated in >10% lung cancer samples; ƒ: gene implicated in lung development.
A very interesting and suggestive mutated gene pair is that formed by LRP2 and TG, which was identified in three out of 18 CPAM patients (CPAM11: type 2; CPAM14: left upper lobe extralobar sequestration; CPAM25: type 2; supplementary table S1), each with different alleles for both genes. LRP2 encodes megalin, a cell receptor mainly active in epithelial cells and involved in several roles due to its variety of ligands. It is noteworthy that data from megalin-deficient mice imply developmental roles in the kidney, central nervous system and lung [22]. Lungs of newborn Lrp2 knockout mice are characterised by emphysematous and atelectic areas, features seen in other human congenital lung malformations.
Homozygous mutations in the human LRP2 gene cause Donnai–Barrow syndrome (supplementary table S6), a multiple congenital malformation syndrome that, as with many CPAMs, includes diaphragm hernia. TG encodes thyroglobulin, a precursor of the thyroid hormones that when homozygously mutated cause thyroid dyshormonogenesis. Thyroid hormones are involved in the regulation of fetal lung development [23], as shown in animal models where lung maturation is accelerated by antepartum exposure to such hormones [24, 25]. Intriguingly, megalin binds thyroglobulin to mediate its transport and thus regulate the extent of thyroid hormone release [26, 27]. It is therefore tempting to speculate that heterozygous mutations in the LPR2–TG pair play a role in CPAMs by either affecting the thyroid hormone levels necessary for proper lung development or by directly interfering with airway structure formation, as seen in megalin mouse models. Importantly, the thyroid transcription factor 1 (TTF1/NKX2.1), which controls the production of thyroid hormones, plays a key role in the development of lung structures and it is heavily involved in lung carcinomas [28].
The MLH1/MSH3 pair was found mutated in two patients. Both genes are associated with hereditary cancers as well as with young-onset lung cancer risk [29]. Both inherited as well as acquired deficiencies in DNA mismatch repair components are associated with the development of a wide range of benign and malignant neoplasms of the lung [30]. The other recurrently mutated genes encoding interacting proteins appear to play roles in DNA damage signalling and cell cycle regulation, which are extremely relevant in cancer development and well as in fetal growth and development. Of particular interest is SYNE1 with a high prevalence of nonsilent somatic mutations in multiple cancers and a role in fetal development (together with CARD6), with mutant mice displaying lung anomalies.
The search for functional overlap among genes and patients implicates cancer-related pathways
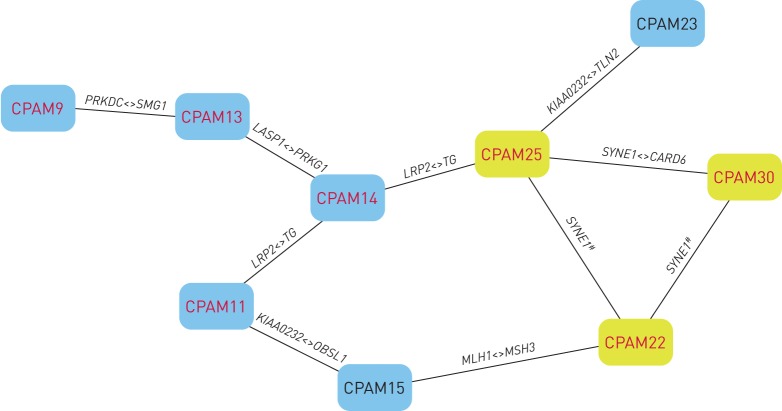
In order to fully understand how and how many of this myriad of mutated genes identified could contribute to the CPAM phenotype and to consider whether our findings fit into any pathological process, we performed a careful examination of the genetic profile or mutational load of each patient in search of shared genetic components masked by the apparent genetic heterogeneity observed. Table 1 presents the genetic profile of each of the 18 CPAM patients with the genes containing damaging alleles and figure 1 represents the genetic components shared among patients. The data advocates for the idea of CPAM being an oligogenic and genetically heterogeneous disorder; as such, the phenotype will be variable and result from gene interactions.
FIGURE 1.
Genetic components shared by congenital pulmonary airway malformation (CPAM) patients. The nodes represent CPAM patients and the edges represent selected “shared mutated genes”. The yellow nodes indicate CPAM patients with mutations in SYNE1. The red font denotes patients with mutations in genes that cause lung abnormalities in mouse models. Nine out of 18 (50%) patients were covered in this genetic component network. #: compound heterozygous and/or digenic inheritance; <>: shared interacting genes.
We attempted to account for biological or functional relatedness among genes by grouping rare variants for association tests. Gene enrichment tests suggest that the genes with damaging rare variants were over-represented in cancer-related pathways (e.g. deficiency of nucleotide excision repair pathway (http://software.broadinstitute.org/gsea/msigdb: MSigDB#M4500); false discovery rate q-value <5.01×10−5) (supplementary figure S2) as well as in proven paediatric disease gene sets as described in the Clinical Genomic Database (https://research.nhgri.nih.gov/CGD; false discovery rate q-value 1.4×10−3).
Discussion
We set out to investigate the genomic lesions that might underlie CPAM, a congenital disorder that has hardly been investigated at genomic level. Bearing in mind that this is a developmental disorder that may occur in conjunction with other congenital anomalies and/or paediatric cancers, and which may represent a risk factor for cancer development later in life [4, 31], we hypothesised that CPAMs might be triggered by germline defects in genes governing the formation of the airways during gestation. Notably, many of those genes also control postnatal cell growth and differentiation, thus contributing to cancer development when mutated [32]. An individual with germline defects involved in both cancer and embryonic lung development may be predisposed to CPAMs. Importantly, the reported association between CPAMs and lung tumours is unlikely to be random, and it seems to apply to all histological types [4].
Based on the presentation and incidence of the disease in the paediatric population, we used a straightforward design whereby we studied coding sequence mutations and CNVs in patients and their unaffected parents. Variants were filtered by frequency and deleteriousness, and the plausibility of the gene was inquired in multiple databases and through an exhaustive literature search.
Overall, the data generated in this study suggest that CPAM is genetically heterogeneous and oligogenic, i.e. mutations in more than one gene are required for the manifestation of the disorder, thus hampering drawing genotype–phenotype correlations. However, in spite of the genetic diversity there are some genetic features shared by or common to many patients. 1) Seven out of the 18 patients (38.8%) studied have damaging mutations in genes causally implicated in adenocarcinomas (table 1: CPAM2, CPAM6, CPAM13, CPAM14, CPAM15, CPAM22 and CPAM28). 2) Also, seven out of the 18 patients (38.8%) have identical mutations in genes that appear somatic mutated in other carcinomas, even though causality is not yet established (CPAM2, CPAM11, CPAM13, CPAM18, CPAM19, CPAM24 and CPAM28), or even have variants also reported as somatic in lung cancer (CPAM2, CPAM11, CPAM18 and CPAM24).
Importantly, several of these genes are recurrently mutated at different sites in independent patients (table 1 and figure 1) as gene pairs and/or in compound heterozygotes, as is the case of SYNE1. The identification of a gene with different damaging variants in different individuals (i.e. a recurrently mutated gene) also evidences pathogenicity [33].
Taken together, the presence of damaging variants in genes involved in cancer processes seems to be higher in CPAM patients than what would be expected by chance, which is of particular interest given the reported association of CPAMs with primary adenocarcinoma and other carcinomas of the lung in paediatric patients [34]. The recurrence of mutations in the LRP2/TG pair, where both genes encode interacting proteins well known for their role in lung development, is also striking.
An inherent limitation to this study (and to any involving a rare genetic heterogenic and oligo/polygenic disorder) is the small sample size and/or the lack of large pedigrees where the disease segregates, thus establishing causality by statistically association is virtually unfeasible due to the rarity of the disorder. However, in spite of the limitations and in spite of the apparent heterogeneity of our findings, we believe that the data presented here are encouraging as they provide a “common ground” in the form of a pathway involving cancer genes, vindicating the idea that CPAMs could eventually develop into tumours, and therefore these patients should be carefully followed up and never underestimated [4]. We are aware that, ideally, the affected lung tissue should also be sequenced in case the CPAMs arose from somatic lesions. However, given that many of the germline variants are also identified as somatic in tumours, and the presence of a few familiar cases, we believe that CPAMs are indeed the result of constitutional genetic variants.
We are aware that ours is a small contribution to the field as our data need to be replicated and functional assays are required (although it is difficult to link so many lesions); however, it is a start, a lead that hopefully the rest of the scientific community will follow as this is the first ever genetic study of CPAMs.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00196-2018_supp (336.2KB, pdf)
Supplementary table 00196-2018_supp_table (141.8KB, pdf)
Acknowledgements
We are grateful to the numerous patients, their families and referring physicians who have participated in these studies in our laboratories, and the numerous members of our laboratories for their valuable contributions over many years.
Footnotes
This article has supplementary material available from openres.ersjournals.com
Conflict of interest: J.S. Hsu has nothing to disclose.
Conflict of interest: R. Zhang has nothing to disclose.
Conflict of interest: F. Yeung has nothing to disclose.
Conflict of interest: C.S.M. Tang has nothing to disclose.
Conflict of interest: K.K.Y. Wong has nothing to disclose.
Conflict of interest: M-T. So has nothing to disclose.
Conflict of interest: H. Xia has nothing to disclose.
Conflict of interest: P. Sham has nothing to disclose.
Conflict of interest: P.K. Tam has nothing to disclose.
Conflict of interest: M. Li has nothing to disclose.
Conflict of interest: J.K.L. Wong has nothing to disclose.
Conflict of interest: M-M. Garcia-Barcelo has nothing to disclose.
Support statement: This work was supported by the Human Medical Research Fund 01121576 to P.K. Tam and M-M. Garcia-Barcelo. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Sfakianaki AK, Copel JA. Congenital cystic lesions of the lung: congenital cystic adenomatoid malformation and bronchopulmonary sequestration. Rev Obstet Gynecol 2012; 5: 85–93. [PMC free article] [PubMed] [Google Scholar]
- 2.Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009; 325: 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong KKY, Flake AW, Tibboel D, et al. Congenital pulmonary airway malformation: advances and controversies. Lancet Child Adolesc Health 2018; 2: 290–297. [DOI] [PubMed] [Google Scholar]
- 4.Casagrande A, Pederiva F. Association between congenital lung malformations and lung tumors in children and adults: a systematic review. J Thorac Oncol 2016; 11: 1837–1845. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 2015; 373: 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan SH, Lim WK, Ishak NDB, et al. Germline mutations in cancer predisposition genes are frequent in sporadic sarcomas. Sci Rep 2017; 7: 10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson RD, Hedrick HL, Liechty KW, et al. Cystic adenomatoid malformation of the lung: review of genetics, prenatal diagnosis, and in utero treatment. Am J Med Genet A 2006; 140: 151–155. [DOI] [PubMed] [Google Scholar]
- 8.Wagner AJ, Stumbaugh A, Tigue Z, et al. Genetic analysis of congenital cystic adenomatoid malformation reveals a novel pulmonary gene: fatty acid binding protein-7 (brain type). Pediatr Res 2008; 64: 11–16. [DOI] [PubMed] [Google Scholar]
- 9.Jancelewicz T, Grethel EJ, Chapin CJ, et al. Vascular endothelial growth factor isoform and receptor expression during compensatory lung growth. J Surg Res 2010; 160: 107–113. [DOI] [PubMed] [Google Scholar]
- 10.Volpe MV, Chung E, Ulm JP, et al. Aberrant cell adhesion molecule expression in human bronchopulmonary sequestration and congenital cystic adenomatoid malformation. Am J Physiol Lung Cell Mol Physiol 2009; 297: L143–L152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volpe MV, Wang KT, Nielsen HC, et al. Unique spatial and cellular expression patterns of Hoxa5, Hoxb4, and Hoxb6 proteins in normal developing murine lung are modified in pulmonary hypoplasia. Birth Defects Res Part A Clin Mol Teratol 2008; 82: 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochieng JK, Schilders K, Kool H, et al. Sox2 regulates the emergence of lung basal cells by directly activating the transcription of Trp63. Am J Respir Cell Mol Biol 2014; 51: 311–322. [DOI] [PubMed] [Google Scholar]
- 13.Gonzaga S, Henriques-Coelho T, Davey M, et al. Cystic adenomatoid malformations are induced by localized FGF10 overexpression in fetal rat lung. Am J Respir Cell Mol Biol 2008; 39: 346–355. [DOI] [PubMed] [Google Scholar]
- 14.DeBoer EM, Keene S, Winkler AM, et al. Identical twins with lethal congenital pulmonary airway malformation type 0 (acinar dysplasia): further evidence of familial tendency. Fetal Pediatr Pathol 2012; 31: 217–224. [DOI] [PubMed] [Google Scholar]
- 15.Lau CT, Kan A, Shek N, et al. Is congenital pulmonary airway malformation really a rare disease? Result of a prospective registry with universal antenatal screening program. Pediatr Surg Int 2017; 33: 105–108. [DOI] [PubMed] [Google Scholar]
- 16.Hsu JS, Kwan JS, Pan Z, et al. Inheritance-mode specific pathogenicity prioritization (ISPP) for human protein coding genes. Bioinformatics 2016; 32: 3065–3071. [DOI] [PubMed] [Google Scholar]
- 17.Du Y, Kitzmiller JA, Sridharan A, et al. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 2017; 72: 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347. [DOI] [PubMed] [Google Scholar]
- 19.Li G, Yi S, Yang F, et al. Identification of mutant genes with high-frequency, high-risk, and high-expression in lung adenocarcinoma. Thorac Cancer 2014; 5: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonczy P. Centrosomes and cancer: revisiting a long-standing relationship. Nat Rev Cancer 2015; 15: 639–652. [DOI] [PubMed] [Google Scholar]
- 21.van Haelst MM, Brooks AS, Hoogeboom J, et al. Unexpected life-threatening complications in Kabuki syndrome. Am J Med Genet 2000; 94: 170–173. [DOI] [PubMed] [Google Scholar]
- 22.Fisher CE, Howie SE. The role of megalin (LRP-2/Gp330) during development. Dev Biol 2006; 296: 279–297. [DOI] [PubMed] [Google Scholar]
- 23.Hume R, Richard K, Kaptein E, et al. Thyroid hormone metabolism and the developing human lung. Biol Neonate 2001; 80: Suppl. 1, 18–21. [DOI] [PubMed] [Google Scholar]
- 24.van Tuyl M, Blommaart PE, de Boer PA, et al. Prenatal exposure to thyroid hormone is necessary for normal postnatal development of murine heart and lungs. Dev Biol 2004; 272: 104–117. [DOI] [PubMed] [Google Scholar]
- 25.Ballard PL. Hormonal influences during fetal lung development. Ciba Found Symp 1980; 78: 251–274. [DOI] [PubMed] [Google Scholar]
- 26.Marino M, Zheng G, Chiovato L, et al. Role of megalin (gp330) in transcytosis of thyroglobulin by thyroid cells. A novel function in the control of thyroid hormone release. J Biol Chem 2000; 275: 7125–7137. [DOI] [PubMed] [Google Scholar]
- 27.Zheng G, Marino M, Zhao J, et al. Megalin (gp330): a putative endocytic receptor for thyroglobulin (Tg). Endocrinology 1998; 139: 1462–1465. [DOI] [PubMed] [Google Scholar]
- 28.Morotti RA, Gutierrez MC, Askin F, et al. Expression of thyroid transcription factor-1 in congenital cystic adenomatoid malformation of the lung. Pediatr Dev Pathol 2000; 3: 455–461. [DOI] [PubMed] [Google Scholar]
- 29.Landi S, Gemignani F, Canzian F, et al. DNA repair and cell cycle control genes and the risk of young-onset lung cancer. Cancer Res 2006; 66: 11062–11069. [DOI] [PubMed] [Google Scholar]
- 30.Downey CM, Jirik FR. DNA mismatch repair deficiency accelerates lung neoplasm development in K-rasLA1/+ mice: a brief report. Cancer Med 2015; 4: 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David M, Lamas-Pinheiro R, Henriques-Coelho T. Prenatal and postnatal management of congenital pulmonary airway malformation. Neonatology 2016; 110: 101–115. [DOI] [PubMed] [Google Scholar]
- 32.Moore SW. Developmental genes and cancer in children. Pediatr Blood Cancer 2009; 52: 755–760. [DOI] [PubMed] [Google Scholar]
- 33.O'Roak BJ, Stessman HA, Boyle EA, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun 2014; 5: 5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim MY, Kang CH, Park SH. Multifocal synchronous mucinous adenocarcinomas arising in congenital pulmonary airway malformation: a case report with molecular study. Histopathology 2014; 65: 926–932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00196-2018_supp (336.2KB, pdf)
Supplementary table 00196-2018_supp_table (141.8KB, pdf)