Abstract
An oncolytic herpes simplex virus (oHSV) has proven amenable in oncolytic virotherapy and was approved to treat melanoma. The immediate-early (IE) protein ICP27 encoded by gene UL54 is essential for HSV infection. Post-transcriptional modification of UL54 would increase tumor targeting of oHSVs. However, UL54 gene transcription regulatory sequences and factors were not reported yet. Here we isolated a new strain LXMW of type 1 HSV (HSV-1-LXMW) in China and found it's closely related to HSV-1 strains Patton and H129 in the US by the first and next generation DNA sequencing viral DNA phylogenetic analysis. Using a weight matrix-based program Match, we found the UL54 transcription regulatory sequences binding to the transcription factors Oct-1, v-Myb and Pax-6 in HSV-1-LXMW, while the sequences binding to Oct-1 and Hairy in a HSV-2 strain. Further validation showed that HSV-1 and HSV-2 shared the common sequence binding to Oct-1, but had unique sequences to bind v-Myb and Pax-6, or Hairy, respectively, by DNA sequence alignment of total 11 HSV strains. The published results howed that the expression of transcription factors is consistent with the tissue tropism of HSV-1 and HSV-2. In the current article a new HSV-1 strain LXMW was isolated and its putative HSV UL54 transcription regulatory sequences and factors were identified for the first time. Our findings highlight the new understanding of the principles of transcriptional regulation in HSV biology and oncolytic virotherapy.
Keywords: Oncolytic Herpes Simplex Virus (oHSV), Long Unique Region 54 (UL54), Infected Cell Polypeptide 27 (ICP27), Transcription Regulatory Sequence, Transcription Factor (TF), RNA polymerase II (RNAP II)
Introduction
Tumors are originated from transformed cells in tissues or organs, which contain heterogeneous cancer cells, such as tumor stem cells etc 1-5, and tumor stromal cells, such as immune cells etc., in tumor microenvironment 6. Chromosome rearrangement and deletion in cancer cells create cancer-specific fusion genes. Such fusion proteins and mutated proteins can be targeted therapeutically 7. Oncolytic viruses have been used for tumor virotherapy 8-10. An oncolytic herpes simplex virus (oHSV) expressing GM-CSF was approved to treat melanoma in 2016. However, novel oHSVs are necessary to increase their anti-tumor efficiency and safety.
Human herpesviruses have three subfamilies α, β and γ 11. The γ herpesviruses include HSV type 1 (HSV-1, causes cold sores) and type 2 (HSV-2, causes genital herpes) and varicella-zoster virus (causes chickenpox and shingles), and can establish lifelong latent infections within the peripheral nervous systems of their hosts 11. HSVs express immediate- early (IE or α) genes, early (E or β) genes and late (L or γ) genes, which are important during virus replication 12. There are five immediate-early proteins of HSV, such as infected cell polypeptide 0, 4 and 27 (ICP0, ICP4 and ICP27) 13. ICP0 is an ubiquitin E3 ligase, which has been reported to be central for viral evasion of type I IFN responses in human fibroblasts 14. ICP4 is responsible for regulatory functions by initiating expression of a set of viral genes, encoding enzymes necessary for DNA replication, such as the E gene UL23. γ34.5 is a L gene encoding ICP34.5, important for viral entry, antagonizing antiviral immune responses and neural virulence 12.
The multifunctional ICP27 is the only protein that has counterparts in all the herpes virus families 13. ICP27 is a 512-amino acid IE protein and is post-translationally modified by phosphorylation and arginine methylation 15, 16. ICP27 is phosphorylated at serine 63 in its kinase-inducible domain by serine/threonine kinases, cAMP-dependent protein kinase A, calmodulin-dependent protein kinase I/II, mitogen-and stress-activated protein kinase and cyclin-dependent kinase 3 (cdk-3). Its phosphorylation enhances its transactivation and transcriptional activities.
ICP27 can re-localize the cellular protein Upstream Binding Factor (UBF) from the nucleolus to viral replication compartments (VRCs), which to promote viral DNA replication 17, 18. HSV ICP27 functions in all stages of viral mRNA biogenesis from transcription, RNA processing and export to translation machinery 15, 16. ICP27 interacts with the C-terminal domain of RNA polymerase II (RNAP II) and recruits RNAP II to viral replication transcription sites, stimulates viral gene transcription 19. ICP27 can both enhance and repress expression of HSV early and late genes driven by ICP0 and ICP4. ICP27 affects post-transcriptional processing by redistributing splicing components, inhibiting host pre-mRNA splicing and facilitating viral mRNA processing and trafficking 13. In the early infection time ICP27 inhibits cellular splicing resulting in accumulation of un-spliced and unprocessed 3' end pre-mRNAs transcripts in the nucleus and suppression of host mRNA export 18, 20. Later in the infection, ICP27 can bind in vivo to seven HSV transcripts through its RGG box-like arginine-glycine region and export the RNAs by the interaction of ICP27 leucine-rich nuclear export signal with the cellular mRNA export receptor TAP/NXF and the mRNA export adaptor protein Aly/ REF20, 21. ICP27 could be crosslinked to poly(A)+ RNA for RNA export 20. Nuclear pore glycoprotein p62 (Nup62) provides additional binding sites of ICP27, promoting viral mRNA export and inhibiting cellular mRNA export 18. ICP27 can also regulate the metabolic process of mRNAs. Absence of ICP27 resulted in decreased accumulation of viral replication proteins and reduction in viral DNA synthesis 13. ICP27 may also stimulate translation of viral transcripts, repress host gene expression, block the cell cycle at the G1 phase and prevents apoptosis 18.
Specific and dynamic gene expression strongly depends on transcription factors (TFs) 10, 19. TFs bind to core promoters, short sequences of ~100 base pair (bp) surrounding the transcription start-sites at the 5' ends of genes 22. Synergistic interaction of several TFs bound to the transcription regulatory sequences or enhancers and arranged as separate modules, also known as enhanceosomes, is a form of cooperative regulation of transcription 19. This dependency leads to an on/off mode of transcriptional regulation 19.
The UL54 gene encodes the essential protein ICP27 of HSV. Until now only one study showed that replacing the whole sequence upstream of the UL54 start codon but downstream of the UL53 stop codon with the telemerase promoter increased the mutant HSV targeting selectively to tumor cells 23. There have been no other reports on UL54 gene transcription regulatory sequences and factors in HSV. Here we isolated a new HSV-1 strain LXMW, and identified novel putative HSV UL54 transcription regulatory sequences binding to Oct-1, v-Myb, Pax-6 or Hairy for the first time. These findings may have significant impact in HSV biology and oncolytic virotherapies.
Materials and Methods
Cells
Experiments were performed with Vero cells which were cultured in Dulbecco's modified Eagle's medium (DMEM) with antibiotics and 5% serum at 37 °C. For viral infection, the vero cells were grown in DMEM with 2% serum. The HSV-1 clinical strain was grown on Vero cell monolayers.
HSV-1 isolation and identification
Sample collection
A 45 years old male patient with acute oral herpes was admitted for HSV-1 isolation. The herpes lesion was punctured with sterile syringe, the liquid was dipped onto sterile cotton swab, the swab was placed in the virus collection tube (Youkang Keye Biotechnology (Beijing) Co., Ltd., Cat. No. MT0301) and shipped at 4°C to the laboratory.
Virus culture
Vero cells were incubated in a 25 ml cell culture bottle (Corning-Costar company) to 70%-80% confluence. Then the culture medium was removed and the cells were washed twice with PBS. A 1.0 ml sample was inoculated in the culture and incubated at 37°C for 2 hours. The cells were washed with PBS twice and cultured with 6 ml maintenance fluid (2% FBS). The cells were observed daily. After 3 days of culture, most of the cells became round and floated. The culture medium was collected and cryo-preserved at -80°C.
HSV-1 identification
A 200 uL of the culture medium supernatant was used to isolate DNA with DNeasy Blood & Tissue Kit (QIAGEN, Cat No69506) according to the instruction. PCR primers (forward primer was 5'-GCCAGCGAGACGCTGAT-3', the reverse primer was 5'-ACGCAGGTACTCGTGGTGA-3') were synthesized by Bioengineering (Shanghai) Company according to the reference 24. The PCR reaction was carried out in 25 uL reaction system using Platinum PCR SuperMix premix (Invitrogen Company, USA, No. 11306-016). Reaction conditions are as following: pre-denaturation at 95°C for 4 minutes followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds and extension 72°C for 30 seconds and followed with a final extention step at 72°C for 7 Min. PCR product 173 bps was delivered to Bioengineering (Shanghai) Limited for DNA sequencing.
HSV genomic DNA sequencing analysis
High-quality genomic DNA (500 ng) was submitted to Beijing Genomics Institute (BGI, http:// www.genomics.cn) for sequence analysis. The qualified genomic DNA sample was randomly fragmented by Covaris technology and the fragments of 350 bp were obtained after fragment selection. The end repair of DNA fragments was performed and an “A” base was added at the 3'-end of each strand. Adapters were then ligated to both ends of the end repaired/dA tailed DNA fragments. The ligated fragments were amplified by ligation-mediated PCR (LM-PCR). The rolling circle amplification (RCA) was performed to produce DNA Nanoballs (DNBs). The qualified DNBs were loaded into the patterned nanoarrays and pair-end read were read through on the BGISEQ-500 platform and high-throughput sequencing are performed for each library to ensure that each sample meet the average sequencing coverage requirement. Sequencing-derived raw image files were processed by BGISEQ-500 basecalling Software for basecalling with default parameters and the sequence data of each individual is generated as paired-end reads, which is defined as “raw data” and stored in FASTQ format. Then Bioinformatics Analysis Overview was used to analyze the sequencing data (raw data from the BGISEQ machine). Burrows-Wheeler Aligner (BWA) software was used to do the alignment. The sequencing depth and coverage for each individual were calculated based on the alignments. In addition, the strict data analysis quality control system (QC) in the whole pipeline was built to guarantee qualified sequencing data.
PCR verification of the DNA sequence upstream of the UL54 gene
To verify the DNA sequence upstream of the UL54 gene of HSV-1 by PCR, three pairs of primers were designed using Snapgene offline software. Forward-1 primer GCGATTGTGTTATATCGCCGT, and reverse primer-1 AGGCCGAGGTCAATTAGCAT, Forward primer-2 TGCGATTGTGTTATATCGCCG, and reverse primer -2 TCCAGGCCGAGGTCAATTAG, Forward primer-3 AGGCGCCTGTTTGATGTATGA and reverse primer-3 TGTCCGATTCCAGGTCGTC. The primers were synthesized at Bioengineering (Wuhan) Company. Reaction conditions are as following: pre-denaturation at 95°C for 4 minutes followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds and extension 72°C for 30 seconds and followed with a final extention step at 72°C for 7 Min. The PCR products 667 bps, 671bps and 674 bps for the above primer pairs 1, 2 and 3 were subjected to gel electrophoresis on 1% agarose gel with and visualized by staining with ethidium bromide. The agarose gel which contained the target DNA was cut down under ultraviolet lamp and the PCR product was purified with DNA Gel Extraction Kit from Axygen® AxyPrep™ according to the instruction. The extracted PCR product was sent to GENECREATE at Wuhan for DNA sequencing (http://www.genecreate.cn).
Phylogenetic analysis of the sequences upstream of the UL54 gene
For phylogenetic analysis of the sequences upstream of the UL54 gene of 11 HSV strains, the online program MEGA7 was used. The evolutionary history was inferred by using the Maximum Likelihood method based on the General Time Reversible model. The bootstrap consensus tree is taken to represent the evolutionary history of the taxa analyzed.
Prediction of UL54 transcription regulatory sequences and factors
The online program Match was used to predict UL54 transcription regulatory sequences and factors. Match is a weight matrix-based program for predicting transcription factor binding sites (TFBS) in DNA sequences. It uses a library of positional weight matrices from TRANSFAC® Public 6.0. The stringent conditions were set as default by the online program.
Alignment of the sequences upstream of UL54
The online program ApE (A plasmid Editor) was used to do alignment of the sequences upstream of UL54 gene. The conserved sequences (>5 base pairs) were marked.
Results
Isolation and DNA sequencing of a New HSV-1 strain LXMW
A new HSV strain from an oral herpes lesion of a patient in Beijing, named as HSV-1 strain LXMW (HSV-1-LXMW), was isolated. LXMW stands for the last name initials of the authors YNL, HWX, ZWM and YYW representing all the authors. The initial identification was carried out by PCR amplification and DNA sequencing. The sequence was determined as follows: GCGTAAAGCCTTTGTGGTGCTGGTTCCCAGGGAACCCCCGACGTTCAGTTGCGCCTGACGAGGCCGCAGCTCACCAAGGTCCTTAACGCGACCGGGGCCGATAGTGCCACGCCCACCACGTTCGAGCTCGGGGTTAACGGCAAATTTTCCGTGTTCACCACGAGTACCTGCGT. The sequence has only 1 nucleotide different from HSV-1-17 93603 CTTTGTGGTGCTGGTTCCCCAGGGAACCCCCGACGTTCAGTTGCGCCTGACGAGGCCGCAGCTCACCAAGGTCCTTAACGCGACCGGGGCCGATAGTGCCACGCCCACCACGTTCGAGCTCGGGGTTAACGGCAAATTTTCCGTGTTCACCACGAGTACCTGCGT 93767. However, there is no significant similarity between the sequences of our HSV-1-LXMW and other HSV-2 strains. Our data support that HSV-1-LXMW is a HSV-1 strain.
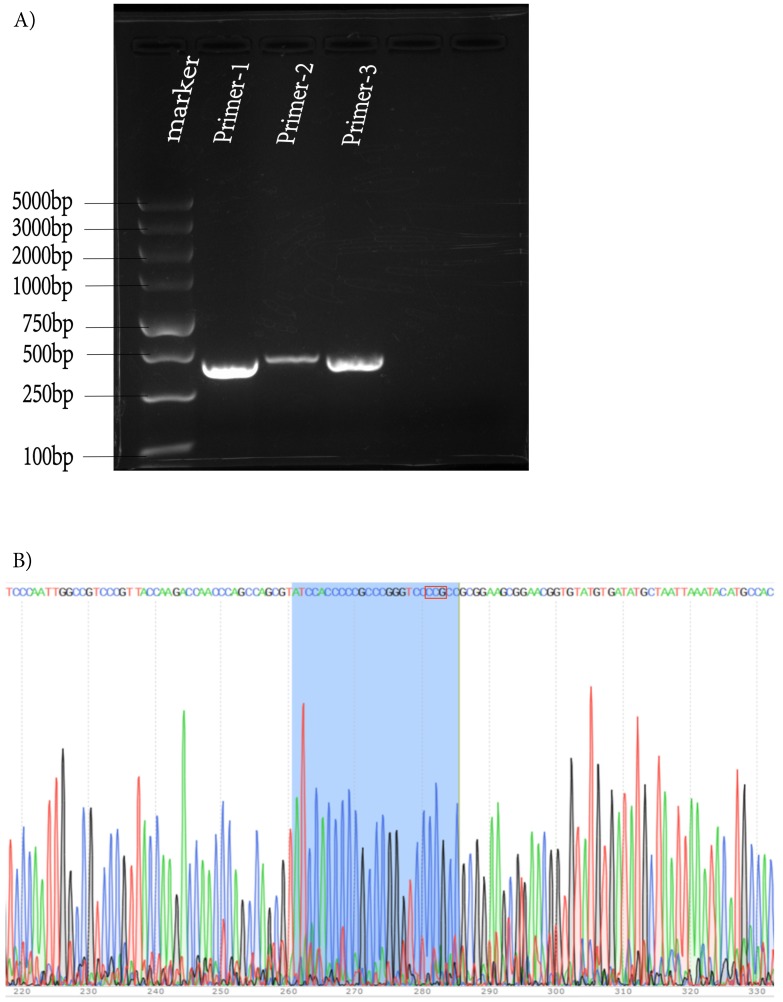
To further identify the new strain, HSV-1-LXMW DNA was then subjected to next generation genomic DNA sequencing. The DNA sequence (from 113211 to 113752, or from the stop codon of UL53 to the start codon of UL54) upstream of the UL54 gene was determined as follows: ACGTCACATCCAGGCCGGCGGAAACCGGAACG-GCATATGCAAATTGGAAACTGTCCTGTCTTGGGGCCCACCCACCCGACGCGTCATATGCAAATGAAAATCGGTCCCCCGAGGCCACGTGTAGCCTGGATCCCAACGACCCCGCCCATGGGTCCCAATTGGCCGTCCCGTTACCAAGACCAACCCAGCCAGCGTATCCACCCCCGCCCGGGTCCCCGCCGCGGAAGCGGAACGGTGTATGTGATATGCTAATTAAATACATGCCACGTACTTATGGTGTCTGATTGGTCCTTGTCTGTGCCGGAGGTGGGGCGGGGGCCCCGCCCGGGGGGCGGAACGAGGAGGGGTTTGGGAGAGCCGGCCCCGGCACCACGGGTATAAGGACATCCACCACCCGGCCGGTGGTGGTGTGCAGCCGTGTTCCAACCACGGTCACGCTTCGGTGCCTCTCCCCGATTCGGGCCCGGTCGCTCGCTACCGGTGCGCCACCACCAGAGGCCATATCCGACACCCCAGCCCCGACGGCAGCCGACCGCCCGGTC. Then the sequence was verified by PCR and first-generation sequencing again (Figure 1A, B).
Figure 1.
Verification of the DNA sequence upstream of UL54 gene of HSV-1-LXMW by PCR and sequencing. (A) The three bands in lanes 2-4 were the 483, 447, 490 base pairs of PCR products. (B) The Sanger sequencing result for UL54 transcription regulatory sequences. The three additional nucleotides were in the red box.
Phylogenetic analysis of HSV-1-LXMW and 10 other HSV strains
To understand the evolutionary relationship of the new HSV strain with other HSV-1 and HSV-2 strains, a phylogenetic analysis based on the DNA sequences upstream of the UL54 gene of HSV-1-LXMW together with 7 HSV-1 strains (17, F, H129, RH2, SC16, Patton and KOS) and 3 HSV-2 strains (G, HG52 and SD90e) were performed (Table 1). The DNA sequences upstream of UL54 gene, from the stop codon of UL53 gene to the start codon of UL54 gene, were chosen for the analysis. In Table 1, it showed that the DNA sequence upstream of UL54 gene of HSV-1-LXMW strain reads 542 base pairs, which is 3 base pairs longer than those (539 bp) of all the other HSV1 strains. The DNA sequences upstream of UL54 gene of HSV-2 strains G, HG 52, SD 90 are all 546 base pairs, 4 base pairs longer than that of HSV-1-LXMW and 7 base pairs longer than those of all other seven HSV-1 strains. These results support that our HSV-1-LXMW is a new HSV-1 strain.
Table 1.
HSV strains studied in this article
| HSV Strain | Gene Bank ID | Tax-ID | Sub-Date | Sequence upstream UL54 | University, Country |
|---|---|---|---|---|---|
| HSV-1, strain LXMW | 113211/113752 | Yangtze University, Jingzhou, China | |||
| HSV-1 strain 17 | JN555585 | 10299 | 2011-08-02 | 113,196/113,735 | RC University, Glasgow, UK |
| HSV-1, strain H129 | GU734772 | 744249 | 2010-01-18 | 113,135/113,674 | Princeton University, USA |
| HSV-1, strain RH2 | AB618031 | 946522 | 2014-03-12 | 111,534/112,073 | Osaka University, Japan |
| HSV-1, isolate SC16 | KX946970 | 10309 | 2016-10-30 | 113,629/114,168 | Severo Ochoa, Spain |
| HSV-1, strain Patton isolate | MF959544 | 10308 | 2017-10-11 | 113,085/113,624 | NYU, New York, USA |
| HSV-1, strain F | GU734771 | 10304 | 2010-01-18 | 113,086/113,625 | Princeton University, USA |
| HSV-1, strain KOS | JQ673480 | 10306 | 2012-03-10 | 113,117/113,656 | University of Kansas, USA |
| HSV-2, strain SD90e | KF781518 | 1177628 | 2013-10-25 | 113,998/114,544 | Harvard Medical School, Boston, US |
| HSV-2, strain HG52 | JN561323 | 10310 | 2011-08-05 | 114,078/114624 | University of Glasgow, UK |
| HSV-2, strain G | KU310668.1 | 10314 | 2016-02-06 | 118,041/118587 | Einstein College, USA |
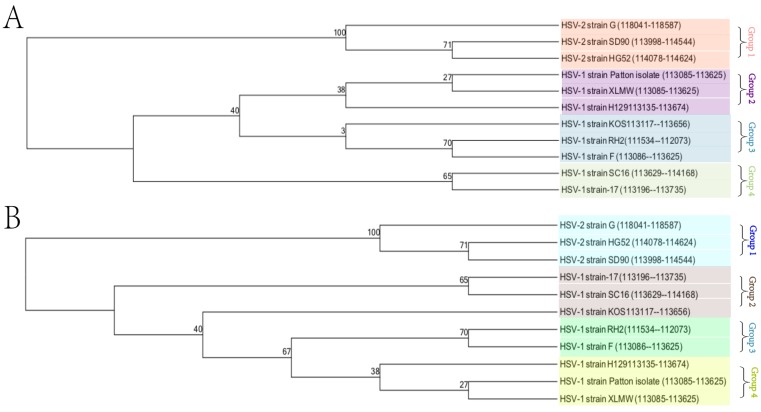
Both the phylogenetic tree data (Figure 2A) and neighbor network data (Figure 2B) showed the presence of four groups of clustering structures. The analysis involved 11 nucleotide sequences of HSV1 and HSV2. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The bootstrap consensus tree is taken to represent the evolutionary history of the taxa analyzed. Evolutionary analyses were conducted in MEGA7.
Figure 2.
Phylogenetic analysis of HSV-1-LXMW together with 10 other HSV strains. A.The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. B. The bootstrap consensus tree is taken to represent the evolutionary history of the taxa analyzed. Evolutionary analyses were conducted in MEGA7.
HSV-1 strains were clustered into 3 groups and HSV-2 strains were clustered into 1 group. Our new strain HSV-1-LXMW in Beijing, China is close to strains HSV-1-Patton in New York, US and HSV-1-H129 in Princeton, US, and far from the strain HSV-1-17 in Glasgow, UK. The data showed a mean distance of approximately 8% among the strains tested collectively.
Identification of the UL54 transcription regulatory sequences and factors
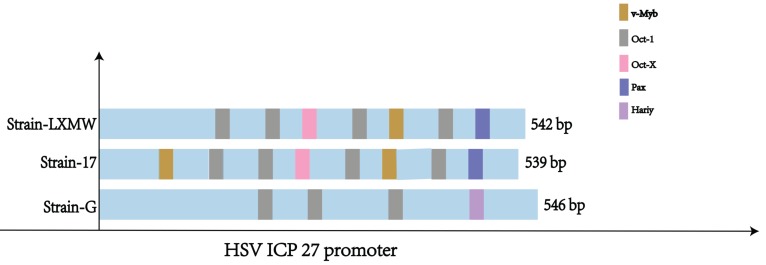
Understanding the UL54 transcription regulatory sequences and factors is important for HSV biology and transcriptional tumor targeting of oncolytic HSVs. Using the weight matrix-based program Match, we found seven major UL54 transcription regulatory sequences of HSV-1-LXMW strain, which bind to transcription factors Oct-1, v-Myb and Pax-6, and eight major UL54 transcription regulatory sequences of HSV-1-17, which bind to transcription factors Oct-1, v-Myb and Pax-6 (Table 2, Figure 3). There were five binding sites for Oct-1/Oct-x, one for v-Myb and one for Pax-6 in HSV-1-LXMW, and there was an additional binding site for v-Myb in HSV-1-17. The difference of binding sites for v-Myb would be related to strain-specific features.
Table 2.
The UL54 transcription regulatory sequences and factors in HSVs
| HSV strain | Matrix identifier | Position strand | Core match | Matrix match | Sequence | Factor name |
|---|---|---|---|---|---|---|
| HSV-1 strain LXMW |
V$OCT1_Q6 | 33(+) | 1.000 | 0.956 | gcatatGCAAAttgg | Oct-1 |
| V$OCT1_Q6 | 84(+) | 1.000 | 0.978 | tcatatGCAAAtgaa | Oct-1 | |
| V$OCT_C | 86(-) | 1.000 | 0.991 | atatGCAAAtgaa | OCT-X | |
| V$OCT1_Q6 | 90(+) | 0.893 | 0.891 | gcaaatGAAAAtcgg | Oct-1 | |
| V$VMYB_01 | 166(-) | 1.000 | 0.962 | tcCCGTTacc | v-Myb | |
| V$OCT1_Q6 | 242(+) | 0.883 | 0.909 | tgatatGCTAAttaa | Oct-1 | |
| V$PAX6_01 | 428(+) | 1.000 | 0.822 | cacggTCACGcttcggtgcct | Pax-6 | |
| HSV-1 strain 17 |
V$VMYB_01 | 26(+) | 1.000 | 0.962 | cgtAACGGca | v-Myb |
| V$OCT1_Q6 | 33(+) | 1.000 | 0.956 | gcatatGCAAAttgg | Oct-1 | |
| V$OCT1_Q6 | 84(+) | 1.000 | 0.978 | tcatatGCAAAtgaa | Oct-1 | |
| V$OCT_C | 86(+) | 1.000 | 0.991 | atatGCAAAtgaa | OCT-X | |
| V$OCT1_Q6 | 90(+) | 0.893 | 0.891 | gcaaatGAAAAtcgg | Oct-1 | |
| V$VMYB_01 | 166(+) | 1.000 | 0.962 | tcCCGTTacc | v-Myb | |
| V$OCT1_Q6 | 239(+) | 0.883 | 0.909 | tgatatGCTAAttaa | Oct-1 | |
| V$PAX6_01 | 425(+) | 1.000 | 0.822 | cacggTCACGcttcggtgcct | Pax-6 | |
| HSV-2 | V$OCT1_Q6 | 39(+) | 1.000 | 0.981 | gcccatGCAAAttaa | Oct-1 |
| V$OCT1_Q6 | 91(+) | 1.000 | 0.976 | tcgcatGCAAAttaa | Oct-1 | |
| V$OCT1_Q6 | 257(+) | 0.883 | 0.908 | ccgtatGCTAAttaa | Oct-1 | |
| I$HAIRY_01 | 492(+) | 1.000 | 0.973 | ccgaCACGCgcccc | Hairy |
Figure 3.
The UL54 transcription regulatory sequences and factors in HSVs
The study found only four major UL54 transcription regulatory sequences binding to transcription factors Oct-1 and Hairy in HSV-2 strain G. There were three sites for Oct-1 and one for Hairy in HSV-2-G (Table 2, Figure 3). These data suggest that HSV-1 and HSV-2 would share the common UL54 transcription regulatory sequence binding to Oct-1, but would have unique sequences binding to v-Myb and Pax-6, or Hairy, respectively.
The UL54 transcription regulatory sequences are conserved
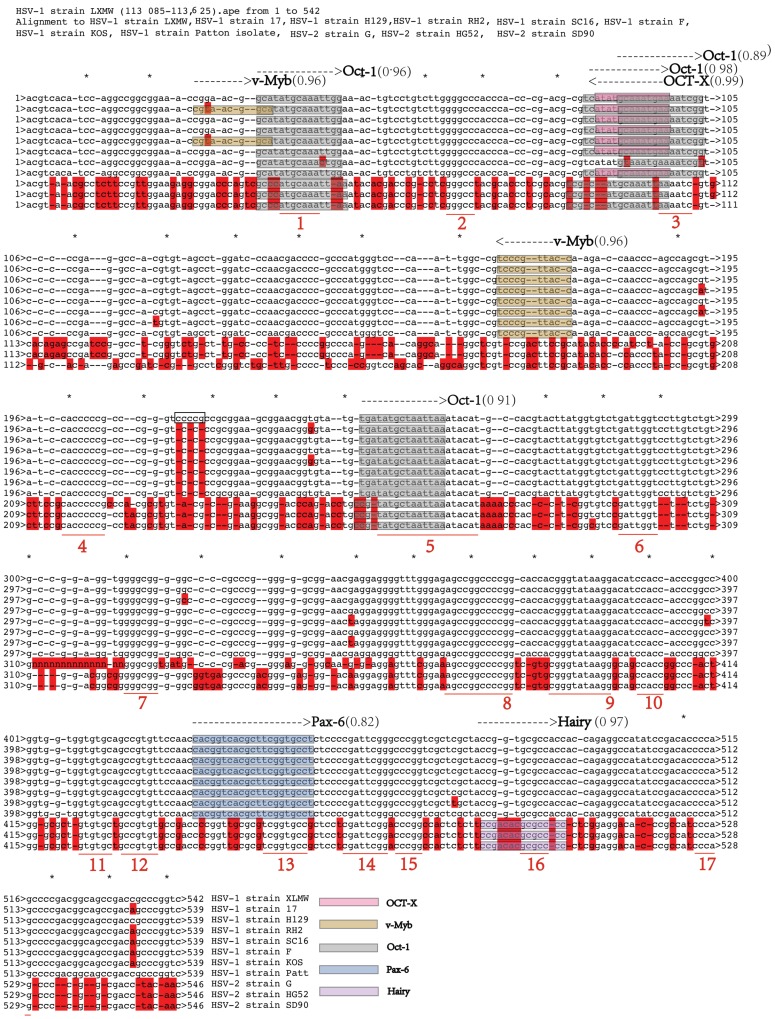
To understand if the UL54 transcription regulatory sequences are conserved among HSVs and thus likely to be biologically functional, the sequence alignment of the 11 above-described sequences of HSVs were performed (Figure 3).
Our results showed that there were less mutations among HSV-1 strains in the transcription regulatory sequences, but more mutations between HSV-1 and HSV-2. In our HSV-1-LXMW strain there exist three more base pairs in the non-conserved region of the transcription regulatory sequences ( HSV-1 strain LXMW: 200 CCCCCGCCCGGGTCCCCGCCGCGGAAGCGGAACGGTGTAT 240) than those of the other seven 7 HSV-1 strains (HSV-1 strain 17: 200 CCCCCGC-CCGGGTCC***CCGCGGAAGCGGAACGGGGTAT 237). Specific function of the additional base pairs needs further exploration. Between HSV-1-LXMW and HSV-1-17 the 538 base pairs of the transcription regulatory sequences are matched and the 3 base pairs are mismatched in addition to the 3 gaps above. Between the HSV-1-LXMW and HSV-2-G there are 391-401 matched base pairs and 80-88 mismatched base pairs. These results support that our HSV-1-LXMW is a new HSV-1 strain.
We defined the sequences with 5 conserved base pairs or more as conserved regions. From the alignment analysis of the 11 HSV strains 17 conserved regions were found. The conserved regions of the transcription regulatory sequences in HSV-1-LXMW started respectively at 38 bp, 65 bp, 95 bp, 199 bp, 245 bp, 285 bp, 310 bp, 355 bp, 375 bp, 388 bp, 408 bp, 413 bp, 441 bp, 455 bp, 485 bp and 511 bp. We found that the UL54 transcription regulatory sequences identified above were mostly overlapped with the conserved regions.
Importantly, the study found that Oct-1, v-Myb, Pax-6 or Hairy binding sites of UL54 transcription regulatory sequences are conserved among HSV-1 or HSV-2 strains, thus supporting that the UL54 transcription regulatory sequences are likely to be biologically functional. We further validated that HSV-1 and HSV-2 shared the common sequence binding to Oct-1, but had unique sequences binding to v-Myb and Pax-6, or Hairy, respectively. These results support that the type-specific UL54 transcription regulatory sequences are likely to be related to their different biological functions between HSV-1 and HSV-2. HSV-1-LXMW contains one v-Myb less than HSV-1-17. Further studies may be needed to see if this difference represents a type-specific or strain-specific function.
The UL54 transcription regulatory sequences may determine type-specific tissue tropism
Our data showed that v-Myb and Pax-6 are conserved in HSV-1, while Hairy is conserved in HSV-2, suggesting that they may determine type-specific tissue tropism. In all published articles the transcription factors Oct-1 and v-Myb have been reported to be expressed in HSV-1 infected cells, which is consistent with HSV-1 tissue tropism (Table 3). In the HSV-1 infected tissues, Oct-1 was reported to activate VP16 gene transcription, v-Myb was reported to activate γ34.5 and suppress ICP6 gene transcription. There is no report of Oct-1 or v-Myb regulating transcription of HSV UL54. There were no reports on expression of the two transcription factors in HSV-2 infected tissues, and no report on expression of the other two transcription factors, Pax-6 and Hairy, in HSV infected tissue. The above unresolved issues will need to be further studied in the future.
Table 3.
The expression of the identified transcription factors in HSV-1 or HSV-2 infected tissues
| Tissue type | HSV1 | HSV2 | Oct-1 | Myb | Pax-6 | Hairy | TF Function | Ref. |
|---|---|---|---|---|---|---|---|---|
| Brain: Human glioblastoma cells, brain | HSV1-F | — | — | B-Myb | — | — | activate/γ34.5 | 32 |
| Urinary: Human PDAC-derived Capan-2 cells, BxPC-3, pancreatic, | HSV1-F | — | — | Myb | — | — | suppress/ICP6 | 33 |
| Colon: Human HT29 carcinoma cells, | HSV1-F | — | — | Myb | — | — | activate /γ34.5 | 34 |
| Embryonic: WT and dOct MEF cells | HSV1-F | — | Oct-1 | — | — | — | activate/VP16 | 35 |
| Genital: HeLa cells | HSV1-F | — | Oct-1 | — | — | — | not described | 27 |
| Digestive: Hep2 cells | HSV-1 KOS | — | Oct-1 | — | — | — | activate /VP16 | 36 |
| Genital: HeLa cells | HSV-1 KOS | — | Oct-1 | — | — | — | activate /VP16 | 37 |
| Urinary: COS-7 cells | HSV-1 KOS | — | Oct-1 | — | — | — | activate /VP16 | 38 |
| Genital: HFF | HSV-1 KOS | — | Oct-1 | — | — | — | activate /VP16 | 39 |
PDAC: pancreatic ductal adenocarcinoma; MEF: mouse embryonic fibroblas; HFF: human foreskin fibroblast. COS-7: are fibroblast-like cell lines derived from monkey kidney tissue.
Discussion
Transcriptional modification of UL54 would increase tumor targeting of oHSVs. However, UL54 gene transcription regulatory sequences and factors were not reported yet. Here a new strain HSV-1-LXMW was isolated in China and found to be closely related to HSV-1 strains Patton and H129 in the US. We identified the HSV UL54 transcription regulatory sequences and factors for the first time. Further analysis found that both HSV-1 and HSV-2 shared the common sequence binding to Oct-1, but had unique sequences binding to v-Myb and Pax-6, or Hairy, respectively. The study predicted that the HSV-1 or HSV-2 specific transcription regulatory sequences would be associated with HSV-1 and HSV-2 tissue tropism. The findings may have significant impact in HSV biology and oncolytic virotherapy.
Our novel HSV-1 strain and it genetic relationship with other HSV strains may reflect the real HSV evolution. Clinical studies showed that the mutations in either the thymidine kinase gene or DNA polymerase gene of HSVs have caused resistance to the anti-HSV drug aciclovir. Most of the mutations occurred in the thymidine kinase gene rather than the DNA polymerase gene 25. The mutation rate in HSV-1 genome is 1.82×10-8 nucleotide substitution per site per year 26. With such a mutation rate the most recent HSV-1 strains could be originated from a common ancestor in ~710,000 years ago.
The functions and their relationship with HSV-1/2 tissue tropism of the identified transcription factors need to be further studied. We summarized the HSV-1/2 tissue tropism and the transcription factor expression in different tissues in Table 4. Oct binding DNA sequence motif ATTTGCAT (octamer) or its reverse complement has been identified as an evolutionarily conserved element in the promoter region of immunoglobulin genes, and is an important transcriptional control element 27. The motif is located at 36 base pairs upstream of the functional TATA box. Two major DNA-binding proteins that bind in a sequence-specific manner to the octamer DNA sequence have been identified in mammalian species-a ubiquitously expressed protein (Oct-1) and a lymphoid-specific protein (Oct-2) 27. The ubiquitous expression of Oct-1 is consistent with our finding that both HSV-1 and HSV-2 contain Oct-1 binding sites.
Table 4.
The HSV- 1/2 tissue tropism and the transcription factor expression in different tissues
| System | cell | HSV1 | HSV2 | Oct-1 | Myb | Pax-6 | Hairy |
|---|---|---|---|---|---|---|---|
| Blood system | CD34+ stem cell | + | — | H | M | L | N |
| 721 B lymphoblasts | + | — | H | M | L | N | |
| CD19+ B cell | + | — | H | M | L | H | |
| Leukemia lymphoblastic | + | — | L | H | M | M | |
| Bonemarrow | + | — | H | M | L | M | |
| Pituitariy | + | — | M | H | M | H | |
| Head | Prefrontal Cortex | + | — | H | M | M | H |
| Pineal | + | — | M | H | H | H | |
| Tongue | + | — | M | H | L | M | |
| Tonsil | + | — | L | M | L | M | |
| Retina | + | — | M | H | H | H | |
| Cerebellum | + | — | L | M | H | M | |
| viscera | Heart | + | — | M | M | L | H |
| Lung | + | — | M | M | L | H | |
| Liver | + | — | M | M | L | H | |
| Kidney | + | — | L | M | L | H | |
| Smooth Muscles | + | — | M | M | L | H | |
| Adipocyte | + | — | L | M | L | H | |
| Secretory system | Adrenalgland, | + | — | L | M | L | H |
| Pancreaticlstet | + | — | M | M | H | H | |
| Genital system |
Placenta | + | + | M | H | L | H |
| Fetalthyroid | + | + | M | M | L | H | |
| Uterus | + | + | L | M | L | M | |
| Testis | + | + | L | M | L | M |
H: high-expression M: middle expression L: little expression N:no-expression.
The result from: http://biogps.org. Grading was based on fold increases compared to median fluorescence intensity on Affymetrix microarray chips at 0-2.5 (L), >2.5-<5 (M), >5 (H).
The viral Myb (v-Myb) regulates proliferation and differentiation of hematopoietic cells 28. It contains three domains, a DNA-binding domain (DBD), a transcription activation domain and a transcriptional repression domain 28. v-Myb can cooperate with the CAAT-enhancer binding protein (C/EBP) family, the Ets family, and core binding factors (CBFs)28. v-Myb may block or reverse the differentitation of nearly mature macrophages into myeloblast-like cells, suggesting v-Myb is an oncogene 28. Pax proteins are important in organ development and stem cell biology. Several Pax proteins, including Pax6, possess an additional DNA-binding domain, which recognizes the sequences containing a TAAT core motif. Hairy is a developmental repressor pair-rule gene, required for proper body plan and peripheral nervous system development 29. Hairy binds to the sequences (ggCACGCGA/ CC) with the core Hairy site (CACGCG) 29. Hairy has transcriptional cofactors Groucho, Drosophila C-terminal binding protein (dCtBP), and Drosophila silent information regulator 2 (dSir2) 29.
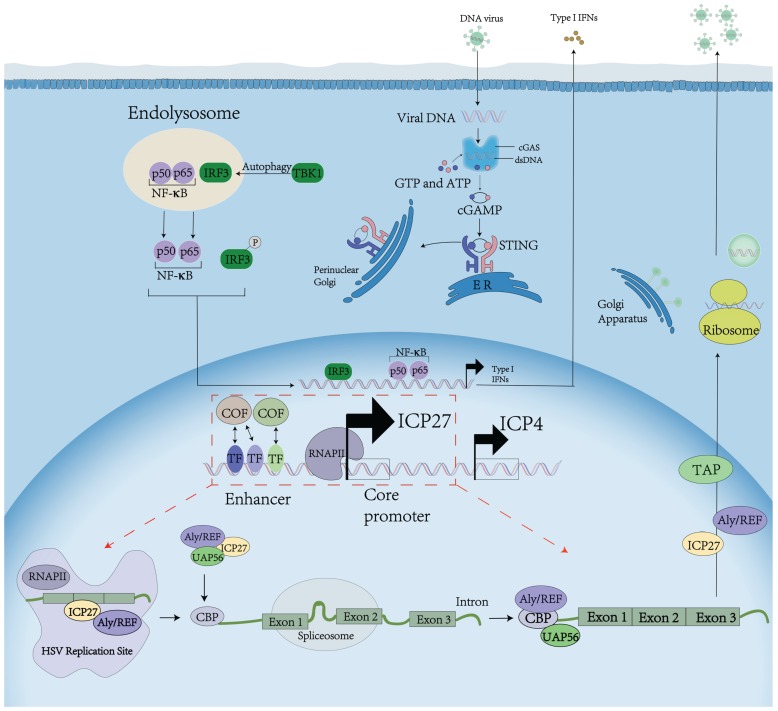
To better understand the significance of our identification of the novel UL54 transcription regulatory sequences and factors, we summarize the UL54 encoded ICP27 function during HSV infection (Figure 5). When a HSV fuses into a cell, viral cytosolic DNA is sensed mainly through cGAMP synthase cGAS. cGAS catalyzes the production of a secondary messenger 2′,3′ cGAMP, which binds to and activates the signaling adaptor STING. Activated STING in turn recruits and activates the transcription factor NF-κB inhibitor IκB kinase IKK and the kinase TBK1, which leads to activation of NF-κB and the transcription factor IRF3, respectively. Then type I IFN is expressed and secreted to inhibit host protein translation. ICP27 interacts with the C‐terminal domain of RNAP II and recruits RNAP II to viral transcription sites, which stimulates viral gene transcription.
Figure 5.
ICP27 function during HSV infection. ICP27 affects post-transcriptional processing by redistributing splicing components, inhibiting host pre-mRNA splicing, and facilitating viral mRNA processing. ICP27 orchestrates viral mRNA export by interacting with cellular mRNA export adaptors Aly/REF, SRp20 and 9G8.
oHSV-1 is one of the most promising oncolytic viruses 8, 30. The human telomerase reverse transcriptase promoter (hTERT) has been used to replace the promoters of ICP4 and HSV thymidine kinase (HSV-TK) 10. The resulting oHSVs elicited significant antitumor effects 30, 31. CEA-ICP4 was generated by placing the ICP4 gene under the CEA promoter12, and HIF-E6L-HSV and HIF-V6R-HSV were generated by expressing the ICP4 gene under the hypoxia-inducible factor (HIF)-responsive promoter E6L or V6R respectively12. The whole sequence upstream of the UL54 start codon but downstream of the UL53 stop codon was replaced with the telemerase promoter, which increased the oHSV targeting selectively to tumor cells 23. These recombinant viruses may be safe and effective therapeutic agents for cancer treatment, warranting clinical trials in humans 9, 30.
Future studies should further validate the UL54 transcription regulatory sequences and factors identified here, and their relationship with HSV tissue tropism. These may lead to our better understanding of HSV biology and better transcriptional targeting of tumors for oncolytic virotherapy.
Figure 4.
The UL54 transcription regulatory sequences are conserved. The conserved regions are marked and numbered as 1-17. The three nucleotides addition CCG in HSV-1-LXMW was marked In the black box. The conserved transcription regulatory sequences and factors are shown in different shade colors.
Acknowledgments
This work was partly supported by grants from the National Natural Science Foundation of China (81872412 to XHW, 81772223 to RBX, 81602303 to XY, 31700736 to WXW). We thank Hubei Province Natural Science Foundation of China (2016CFB180 to WXW), Foundation of Health and Family Planning Commission of Hubei Province (WJ2016-Y-02 To MZ, WJ2016Y07 To WXW), Hubei Province Scientific and Technological Research Project (Q20171306 to XWW), Jingzhou Science and Technology Development Planning Project (JZKJ15063 to WXW) and Yangtze University Fellowship to graduate student WYY and Beijing Jing-Meng Stem Cell Technology, Co. Ltd. (not for intellectual properties or materials). Our new HSV-1 strain HSV-1-LXMW is our protected intellectual property and material.
Abbreviations
- oHSV
oncolytic herpes simplex virus
- IE
immediate-early
- HSV-1-LXMW
strain LXMW of type 1 HSV
- ICP
Infected Cell Polypeptide
- UL54
Long Unique Region 54
- TF
Transcription Factor
- RNAP II
RNA polymerase II
- cdk-3
cyclin-dependent kinase 3
- UBF
Upstream Binding Factor
- VRCs
viral replication compartments
- Nup62
Nuclear pore glycoprotein p62
- BGI
Beijing Genomics Institute
- LM-PCR
ligation-mediated PCR
- RCA
rolling circle amplification
- TFBS
transcription factor binding sites
- ApE
A plasmid Editor
- dCtBP
Drosophila C-terminal binding protein
- dSir2
Drosophila silent information regulator 2
- hTERT
human telomerase reverse transcriptase promoter
- HSV-TK
HSV thymidine kinase
- HIF
hypoxia-inducible factor.
References
- 1.Xin HW, Ambe CM, Miller TC, Chen JQ, Wiegand GW, Anderson AJ. et al. Liver Label Retaining Cancer Cells Are Relatively Resistant to the Reported Anti-Cancer Stem Cell Drug Metformin. Journal of Cancer. 2016;7:1142–51. doi: 10.7150/jca.10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xin HW, Ambe CM, Ray S, Kim BK, Koizumi T, Wiegand GW. et al. Wnt and the cancer niche: paracrine interactions with gastrointestinal cancer cells undergoing asymmetric cell division. Journal of Cancer. 2013;4:447–57. doi: 10.7150/jca.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xin HW, Ambe CM, Hari DM, Wiegand GW, Miller TC, Chen JQ. et al. Label-retaining liver cancer cells are relatively resistant to sorafenib. Gut. 2013;62:1777–86. doi: 10.1136/gutjnl-2012-303261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xin HW, Hari DM, Mullinax JE, Ambe CM, Koizumi T, Ray S. et al. Tumor-initiating label-retaining cancer cells in human gastrointestinal cancers undergo asymmetric cell division. Stem Cells. 2012;30:591–8. doi: 10.1002/stem.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hari D, Xin HW, Jaiswal K, Wiegand G, Kim BK, Ambe C. et al. Isolation of live label-retaining cells and cells undergoing asymmetric cell division via nonrandom chromosomal cosegregation from human cancers. Stem cells and development. 2011;20:1649–58. doi: 10.1089/scd.2010.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Yu C, Wu Y, Sun X, Su Q, You C. et al. CD44(+) fibroblasts increases breast cancer cell survival and drug resistance via IGF2BP3-CD44-IGF2 signalling. Journal of cellular and molecular medicine. 2017;21:1979–88. doi: 10.1111/jcmm.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ZH, Yu YP, Zuo ZH, Nelson JB, Michalopoulos GK, Monga S. et al. Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene. Nature biotechnology. 2017;35:543–50. doi: 10.1038/nbt.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Wang XW, Peng XC, Xiang Y, Song SB, Wang YY. et al. CRISPR/Cas9 genome editing technology significantly accelerated herpes simplex virus research. Cancer gene therapy. 2018;25:93–105. doi: 10.1038/s41417-018-0016-3. [DOI] [PubMed] [Google Scholar]
- 9.Wu ZJ, Tang FR, Ma ZW, Peng XC, Xiang Y, Zhang Y. et al. Oncolytic Viruses for Tumor Precision Imaging and Radiotherapy. Human gene therapy. 2018;29:204–22. doi: 10.1089/hum.2017.189. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Bao L, Yang S, Qian Z, Dong M, Yin L. et al. Tumor-selective replication herpes simplex virus-based technology significantly improves clinical detection and prognostication of viable circulating tumor cells. Oncotarget. 2016;7:39768–83. doi: 10.18632/oncotarget.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai X, Zhou ZH. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science; 2018. p. 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lou W, Ji F, Fu J, Han Z, Di W, Zhang N. Transcriptional retargeting of herpes simplex virus for cell-specific replication to control cancer. Journal of cancer research and clinical oncology; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qing G, Weili W, Fanqin Z, Rongchang Z, Yijin L, Jianqun D. Research of UL54-specific siRNA on herpes simplex virus type II replication. Int J Dermatol. 2011;50:362–6. doi: 10.1111/j.1365-4632.2010.04732.x. [DOI] [PubMed] [Google Scholar]
- 14.Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS. et al. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J. 2016;35:1385–99. doi: 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iversen MB, Reinert LS, Thomsen MK, Bagdonaite I, Nandakumar R, Cheshenko N. et al. An innate antiviral pathway acting before interferons at epithelial surfaces. Nature immunology. 2016;17:150–8. doi: 10.1038/ni.3319. [DOI] [PubMed] [Google Scholar]
- 16.Park D, Lalli J, Sedlackova-Slavikova L, Rice SA. Functional comparison of herpes simplex virus 1 (HSV-1) and HSV-2 ICP27 homologs reveals a role for ICP27 in virion release. Journal of virology. 2015;89:2892–905. doi: 10.1128/JVI.02994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouellet Lavallee G, Pearson A. Upstream binding factor inhibits herpes simplex virus replication. Virology. 2015;483:108–16. doi: 10.1016/j.virol.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Malik P, Tabarraei A, Kehlenbach RH, Korfali N, Iwasawa R, Graham SV. et al. Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. The Journal of biological chemistry. 2012;287:12277–92. doi: 10.1074/jbc.M111.331777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bemer M, van Dijk ADJ, Immink RGH, Angenent GC. Cross-Family Transcription Factor Interactions: An Additional Layer of Gene Regulation. Trends Plant Sci. 2017;22:66–80. doi: 10.1016/j.tplants.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Sandri-Goldin RM. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes & development. 1998;12:868–79. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian X, Devi-Rao G, Golovanov AP, Sandri-Goldin RM. The interaction of the cellular export adaptor protein Aly/REF with ICP27 contributes to the efficiency of herpes simplex virus 1 mRNA export. Journal of virology. 2013;87:7210–7. doi: 10.1128/JVI.00738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zabidi MA, Stark A. Regulatory Enhancer-Core-Promoter Communication via Transcription Factors and Cofactors. Trends Genet. 2016;32:801–14. doi: 10.1016/j.tig.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CY, Bu LX, DeBenedetti A, Williams BJ, Rennie PS, Jia WW. Transcriptional and translational dual-regulated oncolytic herpes simplex virus type 1 for targeting prostate tumors. Mol Ther. 2010;18:929–35. doi: 10.1038/mt.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Yi Y, Chen W, Si S, Yin M, Jin H. et al. Development and evaluation of the quantitative real-time PCR assay in detection and typing of herpes simplex virus in swab specimens from patients with genital herpes. International journal of clinical and experimental medicine. 2015;8:18758–64. [PMC free article] [PubMed] [Google Scholar]
- 25.Hussin A, Md Nor NS, Ibrahim N. Phenotypic and genotypic characterization of induced acyclovir-resistant clinical isolates of herpes simplex virus type 1. Antiviral research. 2013;100:306–13. doi: 10.1016/j.antiviral.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Norberg P, Tyler S, Severini A, Whitley R, Liljeqvist JA, Bergstrom T. A genome-wide comparative evolutionary analysis of herpes simplex virus type 1 and varicella zoster virus. PloS one. 2011;6:e22527. doi: 10.1371/journal.pone.0022527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Advani SJ, Durand LO, Weichselbaum RR, Roizman B. Oct-1 is posttranslationally modified and exhibits reduced capacity to bind cognate sites at late times after infection with herpes simplex virus 1. Journal of virology. 2003;77:11927–32. doi: 10.1128/JVI.77.22.11927-11932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahirov TH, Sato K, Ichikawa-Iwata E, Sasaki M, Inoue-Bungo T, Shiina M. et al. Mechanism of c-Myb-C/EBP beta cooperation from separated sites on a promoter. Cell. 2002;108:57–70. doi: 10.1016/s0092-8674(01)00636-5. [DOI] [PubMed] [Google Scholar]
- 29.Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS biology. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Ge K, Zhao Q, Zhuang X, Deng Z, Liu L. et al. A novel oHSV-1 targeting telomerase reverse transcriptase-positive cancer cells via tumor-specific promoters regulating the expression of ICP4. Oncotarget. 2015;6:20345–55. doi: 10.18632/oncotarget.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higashi K, Hazama S, Araki A, Yoshimura K, Iizuka N, Yoshino S. et al. A novel cancer vaccine strategy with combined IL-18 and HSV-TK gene therapy driven by the hTERT promoter in a murine colorectal cancer model. Int J Oncol. 2014;45:1412–20. doi: 10.3892/ijo.2014.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung RY, Saeki Y, Chiocca EA. B-myb promoter retargeting of herpes simplex virus gamma34.5 gene-mediated virulence toward tumor and cycling cells. Journal of virology. 1999;73:7556–64. doi: 10.1128/jvi.73.9.7556-7564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gayral M, Lulka H, Hanoun N, Biollay C, Selves J, Vignolle-Vidoni A. et al. Targeted oncolytic herpes simplex virus type 1 eradicates experimental pancreatic tumors. Human gene therapy. 2015;26:104–13. doi: 10.1089/hum.2014.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura H, Kasuya H, Mullen JT, Yoon SS, Pawlik TM, Chandrasekhar S. et al. Regulation of herpes simplex virus gamma(1)34.5 expression and oncolysis of diffuse liver metastases by Myb34.5. The Journal of clinical investigation. 2002;109:871–82. doi: 10.1172/JCI10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nogueira ML, Wang VE, Tantin D, Sharp PA, Kristie TM. Herpes simplex virus infections are arrested in Oct-1-deficient cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1473–8. doi: 10.1073/pnas.0307300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akhova O, Bainbridge M, Misra V. The neuronal host cell factor-binding protein Zhangfei inhibits herpes simplex virus replication. Journal of virology. 2005;79:14708–18. doi: 10.1128/JVI.79.23.14708-14718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ottosen S, Herrera FJ, Doroghazi JR, Hull A, Mittal S, Lane WS. et al. Phosphorylation of the VP16 transcriptional activator protein during herpes simplex virus infection and mutational analysis of putative phosphorylation sites. Virology. 2006;345:468–81. doi: 10.1016/j.virol.2005.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misra V, Walter S, Yang P, Hayes S, O'Hare P. Conformational alteration of Oct-1 upon DNA binding dictates selectivity in differential interactions with related transcriptional coactivators. Molecular and cellular biology. 1996;16:4404–13. doi: 10.1128/mcb.16.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suk H, Knipe DM. Proteomic analysis of the herpes simplex virus 1 virion protein 16 transactivator protein in infected cells. Proteomics. 2015;15:1957–67. doi: 10.1002/pmic.201500020. [DOI] [PMC free article] [PubMed] [Google Scholar]