Abstract
Recent studies show that dysregulated miRNAs play an important role in breast cancer initiation and progression. Here, we identified upregulated expression of miR-1307-3p in breast cancer tissues and that increased level of miR-1307-3p was closely correlated with lower survival rate in breast cancer patients. Consistent with clinical data, our in vitro data show that expression level of miR-1307-3p was significantly increased in breast cancer cell lines compared to human mammary epithelial cell line MCF10A. Overexpression of miR-1307-3p in MCF10A stimulated cell proliferation and caused their growth in soft agar and tumor formation in nude mice. In contrast, inhibition of miR-1307-3p suppressed breast cancer cell proliferation and their growth in soft agar and inhibited tumor formation in nude mice. Further, we identified that miR-1307-3p plays its oncogenic role through targeting SET and MYND domain-containing 4 (SMYD4) expression in breast cancer. Taken together, our findings suggest that miR-1307-3p is a oncogenic miRNA that significantly contributes to breast cancer development and progression, and inhibition of miR-1307-3p may be a novel strategy for inhibits breast cancer initiation and progression.
Keywords: breast cancer, tumorigenesis, miR-1307-3p, SMYD4
Introduction
Improved surveillance and early detection have improved breast cancer mortality statistics over the past 30 years 1. However, breast cancer still is the most frequently diagnosed cancer and the leading cause of cancer-related death in women worldwide 2. In 2018, estimated 260,000 new cases diagnosed with breast cancer and that breast cancer caused over 40,000 deaths in the United States 3. However, the molecular mechanism of breast cancer development is not fully understood.
miRNAs are small non-coding RNAs, regulates gene transcription by base pairing with the 3'-untranslated regions (3'-UTR) of target genes' mRNA, leading to the inhibition of gene expression. Accumulating evidences show that miRNAs expression are dysregulated in most cancers, including breast cancer and these aberrantly expressed miRNAs are closely involved in the breast cancer development and progression 4-6. Notably, even one single miRNA dysregulation can cause cancer development and progression 7. Because, one miRNA can target hundreds of target genes, thereby affecting a large cellular signaling networks 8. Therefore, investigating the role and mechanism function of each individual miRNA that is dysregulated in cancer is important for cancer prevention and therapy.
In breast cancer, a number of dysregulated miRNAs have been detected by many research groups and miR-1307-3p is one of the deregulated miRNA in breast cancer 9. Shimomura et al. reported that miR-1307-3p was increased in breast cancer patients' serum 9. In addition, although not in breast cancer, reports show that increased expression of miR-1307- 3p significantly promotes prostate cancer progression 10 and ovarian cancer cell chemoresistance 11. These findings suggesting that miR-1307-3p may be an oncogenic miRNA. But, the detailed role and function mechanism of miR-1307-3p in breast cancer is not clear.
In the present study, we identified significantly increased expression of miR-1307-3p in breast cancer specimens compared to matched normal breast tissues and that increased expression level of miR-1307-3p was significantly associated with lower overall survival rate in breast cancer patients. In addition, we demonstrated that miR-1307-3p causes tumor formation of nontumorigenic human mammary epithelial cells in nude mice and promotes cell proliferation by targeting cancer suppressor gene SET and MYND domain containing 4 (SMYD4).
Materials and methods
Cell lines and clinical samples
Cell lines MCF10A, MCF-7, MDA-MB-468 and MDA-MB-231 were obtained from the American Type Culture Collection (Manassas, VA, USA), and these cells were cultured in Dulbecco's Modified Eagle's Medium, supplemented with 10% FBS, at 37°C in an atmosphere of 95% air and 5% CO2.
Tumor and matched adjacent normal tissues were collected from 10 patients with newly diagnosed with early stage breast cancer at the Chuncheon Sacred Heart Hospital, College of Medicine, Hallym University. This research was approved by the Research Ethics Board of the Chuncheon Sacred Heart Hospital.
Reverse transcriptase real-time quantitative polymerase chain reaction analysis (RT-qPCR)
Total RNA was isolated from cells and tissues using TRIzol reagent according to the manufacturer's protocol Life Technologies (Carlsbad, CA, USA). Mature miR-1307-3p and the RNU6 endogenous control were analyzed using the TaqMan microRNA Assay Kit. For analysis of SMYD4 expression, RT and PCR were performed with a High-Capacity cDNA Reverse Transcription Kit and QuantiTect SYBR Green PCR kit, respectively. All agents used for RT-qPCR were purchased from Life Technologies. The expression of SMDY4 was quantified in relation to the expression of β-actin. Primers sequences are as follows. For SMYD4 forward, 5'-GGTGGGAAAGGACTCGGAC-3' and reverse, 5'-GGTTAGCATGACACAGTGACAT-3'; for Actin, forward 5′-AGAGCTACGAGCTGCCTGAC-3′ and reverse, 5′-AGCACTGTGTTGGCGTACAG-3′.
Luciferase reporter assay
3'-UTR segments of SMYD4 that was predicted to interact with miR-1307-3p were amplified by PCR from human genomic DNA and inserted into the Mlu I and Hind III sites of the miRNA Expression Reporter Vector (Life Technologies). Luciferase reporter assay was performed as described previously 12. Briefly, cells were seeded into 24-well cell culture plates at a concentration of 1×104 per well. Next day, cells were transfected with the indicated reporter plasmids containing firefly luciferase and either miR-1307-3p mimics or control nucleotides. The Renilla luciferase plasmid was co-transfected as a transfection control. Cells were lysed 48 hours after transfection, and luciferase activity was measured by a Dual-Luciferase Assay System according to the manufacturer's protocol (Promega, Madison, WI, USA). The firefly luciferase activity was normalized by the activity of Renilla luciferase. All transfection were conducted using lipofectamin 3000 according to manufacturer's instruction (Invitrogen).
Western blot and immunohistochemistry (IHC) assays
Western blotting and immunohistochemical assays were performed as described by Xu et al. 13. Briefly, for Western blot, 30 ug of proteins were separated on sodium dodecyl sulfate polyacrylamide gel and transferred to nitrocellulose (NC) membranes. The NC membranes were blocked in tris-buffered saline with Tween 20 (TTBS), then incubated with primary antibody for overnight at 4°C. Membranes were washed with TTBS and incubated with secondary antibody for 3 hours at room temperature. After washing the bands were analyzed by the luminescent image analyzer. For the IHC, tissue sections were deparaffinized and quenched endogenous peroxidase activity using 3% hydrogen peroxide. After washing, tissue sections were incubated with 5% bovine serum albumin in PBS, then incubated with primary antidody for overnight at 4°C. The following day, tissue sections were washed and incubated with secondary antibodies for 1 hour at room temperature. After washing, the substrate color was developed using the liquid 3,30-kiaminobenzidine substate chromogen system (DAKO) and the slides were reviewed by an expert panel of pathologists. All primary and secondary antibodies that used in present study were purchased from Abcam (Cambridge, MA, USA).
Soft agar and proliferation assay
Cells were trypsinized and resuspended in 0.5 ml 0.35% agar in growth medium at a density of 2,500 cells/well (6-well plate). Then, the agar-cell mixture was plated on the top of a solid layer of 0.8% agar in growth medium. Colonies were counted 2 weeks later. For proliferation assay, indicated cells were transfected with indicated nucleotides or plasmid. After 24 hours of transfection, cells were plated into 6-well plate at 5×105 cells per well density and cell counts were estimated by trypsinizing the cells and performing analysis in triplicate with a coulter counter.
Stable cell line selection
The lentivirus expressing miR-1307-3p (pHBLV-h-miR1307-3p-puro) or miR-1307-3p inhibitor (pHBLV-h-shmiR1307-3p-puro) were purchased from Hanbio (Shanghai, China). Cells were infected with lentivirus that expressing miR-1307-3p or miR-1307- 3p antisense according to manufacturer's instruction. After 72 hours of infection, cells were incubated with 2 mg/mL of puromycin for 1 week. Then, the expression of miR-1307-3p was measured and frozen down in aliquots for later use.
The SMYD4 expression plasmid (Pcmv6-Entry-Neo) was obtained from Origene Technologies, Inc (Rockville, MD, USA). Indicated cells were transfected with SMYD4 expressing vector or empty vector. After 48 hours of transfection, cells were incubated with 500ug/mL neomycin for 3 weeks. Then, the expression of SMYD4 was measured and frozen down in aliquots for later use.
Animal experiments
Stably expressing miR-1307-3p or miR-1307-3p-antisense cells and their vector control cells were used to generate the animal model. Indicated cells in 0.1 mL of phosphate-buffered saline (PBS) were subcutaneously injected into 6-week-old female nude mice. The tumor volume was measured every week.
Statistical analysis
All data are presented as the mean ± standard deviation (SD), and significant differences between treatment groups were analyzed by Student's t-test or one-way analysis of variance (ANOVA) and Duncan's multiple range test using SAS statistical software version 6.12 (SAS Institute). Differences were considered statistically significant at a p value of less than 0.05.
Results
Increased expression of miR-1307-3p was correlated with breast cancer progression
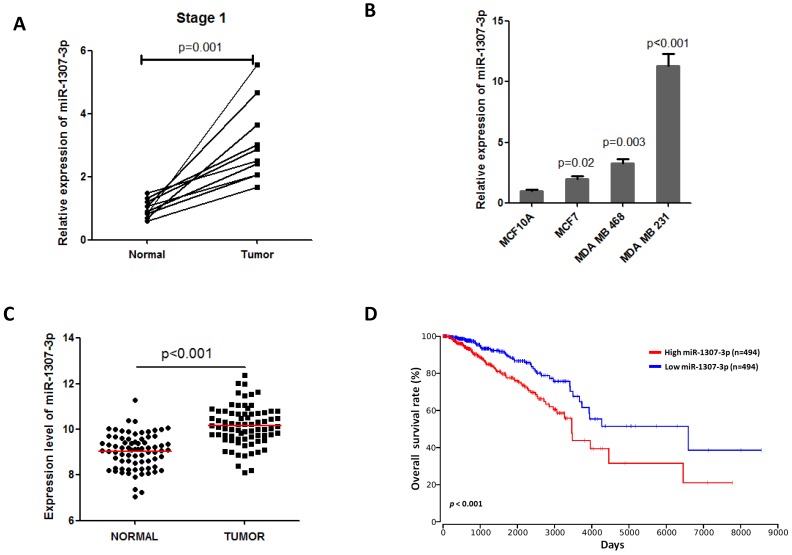
Previous a report show that miR-1307-3p was increased in serum of breast cancer patients and it is a candidate biomarker for detecting early stage breast cancer 9. Thus, we first investigated the expression of miR-1307-3p in early stage breast tumor tissues. Our data show that miR-1307-3p expression was significantly increased in early stage breast cancer specimens compared to that in their matched adjacent normal breast tissues (Fig. 1A). These findings suggest that miR-1307-3p may play a role in breast cancer development. Consistent with clinical data, we identified that dramatically increased expression of miR-1307-3p in human breast cancer cell lines compared to human mammal epithelial cell MCF10A (Fig. 1B). In addition, TCGA data set also show that miR-1307-3p was significantly increased in breast cancer tissues compared to their matched adjacent normal tissues (Fig. 1C). More importantly, TCGA data set analysis results show that increased expression level of miR-1307-3p closely associated with lower overall survival rate in breast cancer patients (Fig. 1D). Taken together, these findings suggest that increased expression of miR-1307-3p was significantly correlated with breast cancer development and progression.
Figure 1.
Increased expression of miR-1307-3p was closely associated with breast cancer progression. (A) miR-1307-3p expression was significantly increased in human early stage breast tumor tissues compare to that in matched adjacent normal tissues (n=10). (B) miR-1307-3p expression level was significantly increased in breast cancer cell lines compared to mammary epithelial cell MCF10A. (C) TCGA data set analysis show that miR-1307-3p expression was significantly increased in human breast tumor tissues compare to that in matched adjacent normal tissues (n=70). (D) TCGA data set analysis show that increased expression of miR-1307-3p level was significantly correlated with lower overall survival of breast cancer patients.
Overexpression of miR-1307-3p stimulates breast tumorigenesis
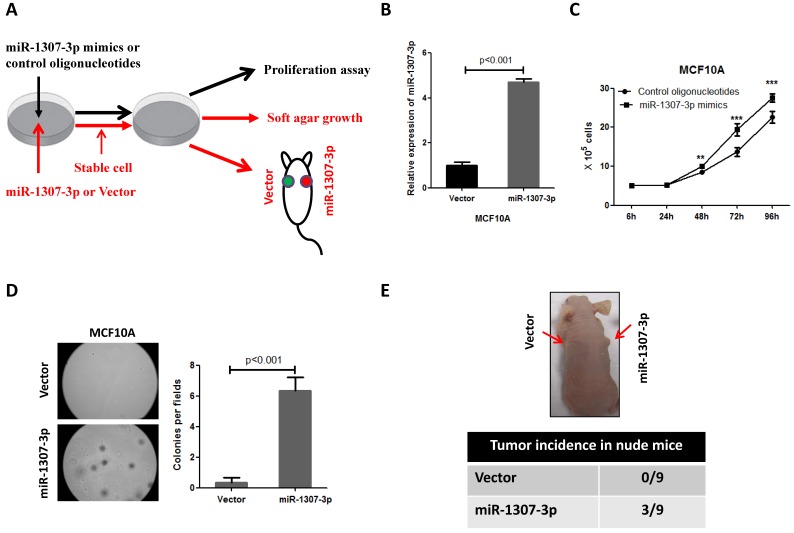
To investigate whether the increased expression of miR-1307-3p was involved in breast cancer development, we overexpressed miR-1307-3p in a nontumorigenic human mammary epithelial cell MCF10A, then subjected to cell proliferation, anchorage-independent growth, and tumor formation assays (Fig. 2A). Our results show that overexpression of miR-1307-3p (Fig. 2B) significantly stimulated MCF10A cell proliferation compared to vector control (Fig. 2C). In addition, overexpression of miR-1307-3p caused MCF10A cells readily forming foci in soft agar, indicating that miR-1307-3p promotes the transformation of mammary epithelial cells (Fig. 2D). More importantly, overexpression of miR-1307-3p lead to nontumorigenic mammary epithelial cell MCF10A formed tumor in 33% of nude mice after 6 months of cell injection (Fig. 2E). Taken together, these findings suggest that increased expression of miR-1307-3p significantly contributes to breast cancer development.
Figure 2.
Overexpression of miR-1307-3p stimulates transformation of MCF10A mammary epithelial cells. (A) Schematic diagram of the experiments. (B) miR-1307-3p expression was significantly increased in miR-1307-3p stably expressing MCF10A cells compared to vector control cells. (C) Overexpression of miR-1307-3p accelerated MCF10A cell proliferation. Cells were transfected with control oligonucleotides or miR-1307-3p mimics. After 12 hours of transfection, cells were plated in 6-well plate, then counted cell numbers at indicated times. **, p<0.01 compared to control oligonucleotides group; ***, p<0.001 compared to control oligonucleotides group. (D) Overexpression of miR-1307-3p caused anchorage-independent growth of MCF10 mammary epithelial cells in soft agar. Soft agar assay was performed using MCF10A cells that stably expressing miR-1307-3p (miR-1307-3p) or their vector control cells (vector). (E) Overexpression of miR-1307-3p caused tumor formation of MCF10A cells in nude mice. 5×106 MCF10A cells, which stably express miR-1307-3p (right back, miR-1307-3p) and their vector control cells (left back, vector) were injected subcutaneously into nude mice (n=9). This experiment was conducted 6 months.
Inhibition of miR-1307-3p suppressed breast cancer cell tumor formation and inhibited proliferation
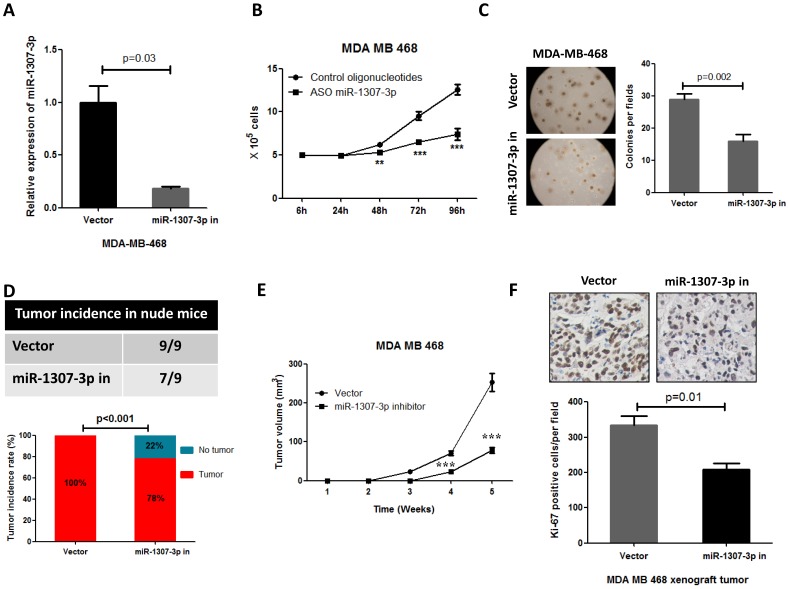
Our observations that the overexpression of miR-1307-3p stimulates breast tumorigenesis and proliferation in turn prompted us to investigate whether miR-1307-3p inhibition could suppress breast cancer. As expected, our data show that inhibition of miR-1307-3p (Fig. 3A) dramatically suppressed breast cancer cell MDA-MB-468 proliferation (Fig. 3B) and anchorage-independent growth (Fig. 3C). Consistent with in vitro experiment results, inhibition of miR-1307-3p blocked MDA-MB-468 breast cancer cell tumor formation in 22% of nude mice when subcutaneously injected with 5×104 cells per mouse (Fig. 3D). When mice were injected with 5×106 cells per mouse, miR-1307-3p inhibition did not affected tumor formation of MDA-MB-468 cells in nude mice. However, inhibition of miR-1307-3p dramatically delayed MDA-MB-468 cell tumor formation time in nude mice (Fig. 3E) and significantly inhibited cancer cell proliferation compared to vector control (Fig. 3F). These data suggest that inhibition of miR-1307-3p may be a useful strategy for the inhibition of breast tumorigenesis and breast cancer progression.
Figure 3.
Inhibition of miR-1307-3p suppressed breast cancer cell proliferation, tumor growth and tumor formation in nude mice. (A) miR-1307-3p expression was significantly decreased by transfection of miR-1307-3p-antisense expression plasmid (miR-1307-3p in). (B) Inhibition of miR-1307-3p significantly suppressed breast cancer cell MDA-MB-468 proliferation. Cells were transfected with control oligonucleotides or antisense oligonucleotides of miR-1307-3p (ASO miR-1307-3p). After 12 hours of transfection, cells were plated in 6-well plate, then counted cell numbers at indicated times. **, p<0.01 compared to control oligonucleotides group; ***, p<0.001 compared to control oligonucleotides group. (C) Inhibition of miR-1307-3p inhibited anchorage-independent growth of MDA-MB-468 cells in soft agar. Soft agar assay was performed using MDA-MB-468 cells that stably expressing antisense of miR-1307-3p (miR-1307-3p in) or their vector control cells (vector). (D) Inhibition of miR-1307-3p affected tumor formation of MDA-MB-468 cells in nude mice. 5×104 cells, which stably express miR-1307-3p antisense (right back, miR-1307-3p in) and their vector control cells (left back, vector) were injected subcutaneously into nude mice (n=9). This experiment was conducted 2 months. (E) Inhibition of miR-1307-3p delayed tumor formation of MDA-MB-468 cells in nude mice. 5×106 cells, which stably express miR-1307-3p antisense (miR-1307-3p in) and their vector control cells (Vector) were injected subcutaneously into nude mice (n=9 mice/group). ***, p<0.001 compared to vector control group. (F) Inhibition of miR-1307-3p significantly suppressed cancer cell proliferation in MDA-MB-468 breast cancer cell xenograft tumors. Tumor tissues that from MDA-MB-468 xenograft model experiments were stained using cell proliferation marker protein Ki-67 antibody.
miR-1307-3p targeting SMYD4 in breast cancer
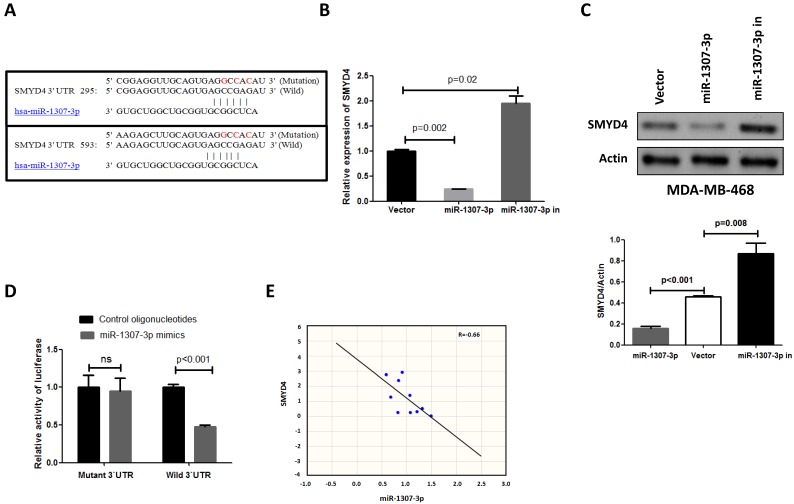
To investigate the underlying mechanism of the effect of miR-1307-3p on breast tumorigenesis, we used miRNA target prediction algorithms (targetscan.org) to screen for miR-1307-3p target genes, and we identified SMYD4 as a tentative target of miR-1307-3p. Because previous a report show that SMYD4 was involved in breast cancer development as a potential tumor suppressor 14, and our target prediction result show that 3'UTR of SMYD4 contain two binding sites of miR-1307-3p (Fig. 4A). To investigate the association between miR-1307-3p and target gene expression, we examined the expression levels of SMYD4 in miR-1307-3p-overexpressing or inhibited MDA-MB-468 breast cancer cells. As shown in Figs. 4B and 4C the expression of SMYD4 was significantly decreased in miR-1307-3p-overexpressing cells at both the mRNA and protein levels compared to that in vector control cells. In contrast, inhibition of miR-1307-3p led to an increase in the expression of SMYD4 at both the mRNA and protein levels (Figs. 4B and C). Further, to determine whether the regulation of the expression of target luciferase reporter genes depended on the binding of their complementary 3'-UTR sequences to the miR-1307-3p seed sequence, a three-nucleotide mutation was inserted into the target genes' 3'-UTR, as indicated in Fig. 4A. Our data show that overexpression of miR-1307-3p significantly repressed the luciferase activity associated with the wild-type 3'-UTR, respectively. However, the 3'-UTR mutation completely abrogated the effect of miR-1307-3p overexpression on luciferase activity in MDA-MB-468 cells (Fig. 4D). Finally, we investigated the correlation between miR-1307-3p and SMYD4 expression in breast cancer patients' specimens. Data show that expression of miR-1307-3p and SMYD4 was inversely correlated in breast cancer specimens (Fig. 4E). Cumulatively, these data suggest that miR-1307-3p suppress the expression of SMYD4 in breast cancer cells by directly targeting their 3'-UTR sequence.
Figure 4.
SMYD4 is a target of miR-1307-3p in breast cancer. (A) Sequence alignment of miR-1307-3p with the 3'-UTR of SMYD4. (B-C) miR-1307-3p negatively regulates the expression of SMYD4 in MDA-MB-468 cells at both mRNA and protein levels. MDA-MB-468 cells (vector) and stably expressing miR-1307-3p (miR-1307-3p) or miR-1307-3p-antisense (miR-1307-3p in) MDA-MB-468 cells were subjected to RT-qPCR and Western blot analysis. (D) MDA-MB-468 cells were co-transfected with the 3'-UTR luciferase reporter construct (wild-type or mutant type) of SMYD4 and the indicated nucleotides. After 48 hours of transfection, the cells were subjected to luciferase activity assay. Ns, no significance. (E) The expression of miR-1307-3p and SMYD4 present inversed correlation in breast cancer cell. MiR-1307-3p and SMYD4 mRNA expression levels are measured in 10 breast cancer patients specimens and analyzed their correlation.
miR-1307-3p stimulated breast tumorigenesis through SMYD4
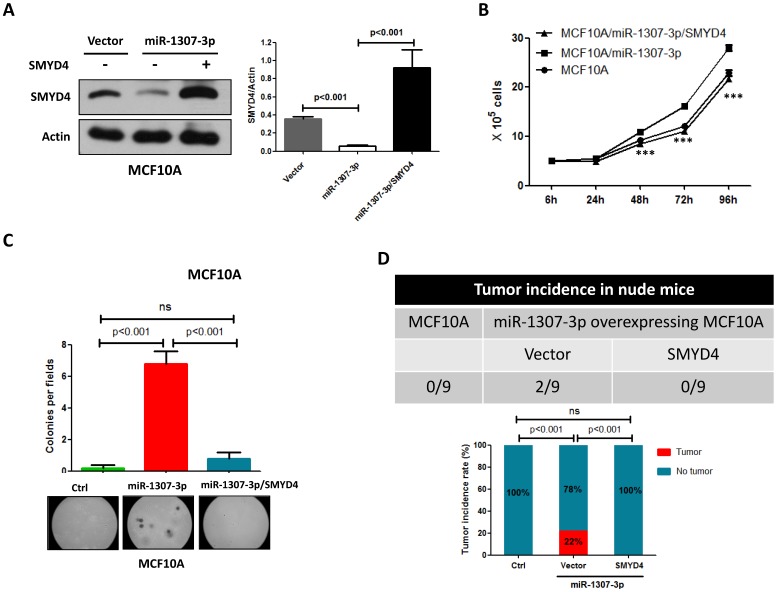
To investigate whether SMYD4 was involved in miR-1307-3p induced breast tumorigenesis, we overexpressed SMYD4 in stably overexpressing-miR-1307-3p MCF10A cells and then performed cell proliferation, soft agar and in vivo tumor formation assay. Our data show that overexpression of SMYD4 (Fig. 5A) significantly weakened miR-1307-3p-induced stimulation of MCF10A cell proliferation (Fig. 5B), foci formation in soft agar (Fig. 5C) and tumor formation in nude mice (Fig. 5D). These findings suggesting that miR-1307-3p plays its oncogenic roles through SMYD4 in breast cancer.
Figure 5.
miR-1307-3p plays its oncogenic role through SMYD4. (A) SMYD4 protein level was dramatically increased by transfection of SMYD4 expression plasmid. Stable expressing miR-1307-3p MCF10A cells were transfected with MYD4 expression plasmid or vector. After 72 hours of transfection subjected to Western blot. (B) miR-1307-3p induced stimulation of cell proliferation was abrogated by SMYD4 overexpression in MCF10A cells. Stably expressing miR-1307-3p MCF10A cells were transfected with SMYD4 expression plasmid (MCF10A/miR-1307-3p/SMYD4) or empty vector (MCF10A/miR-1307-3p), then subjected to cell proliferation assay. ***, p<0.001 compared to MCF10A/miR-1307-3p group. (C) miR-1307-3p induced foci forming in soft agar was blocked by SMYD4 overexpression in MCF10A cells. Stably expressing miR-1307-3p MCF10A cells were transfected with SMYD4 expression plasmid (miR-1307-3p/SMYD4) or empty vector (miR-1307-3p), then subjected to cell proliferation assay. Ctrl, MCF10A cells that infected with lentivirus empty vector. (D) miR-1307-3p stimulated tumor formation of MCF10A cell in nude mice was blocked by SMYD4 overexpression in MCF10A cells. 5×105 indicated cells were subcutaneously injected to per mouse and this experiment continued 6 months.
Discussion
In present study, we demonstrated that aberrantly upregulated expression of miR-1307-3p is closely associated with breast cancer development and progression. TCGA data set analysis results show that miR-1304-3p was increased in breast cancer tissues and closely associated with poor clinical outcomes. In addition, our clinical data show that miR-1307-3p was significantly increased in early stage of breast tumor tissues, and overexpression of miR-1307-3p significantly stimulated nontumorigenic mammary epithelial cell MCF10A proliferation, transformation and tumor formation in nude mice. These findings indicate that miR-1307-3p plays important role in breast cancer development and progression as an oncogenic miRNA. Of course, our findings are supported by other research groups. Shimomura et al. reported that miR-1307-3p was significantly increased in serum of breast cancer patients compared to healthy control and miR-1307-3p combine with other four miRNAs that increased in breast cancer patients' serum can detect early stage breast cancer 9.
Furthermore, present study suggesting that inhibition of miR-1307-3p may be a useful novel strategy for breast cancer treatment. In vitro experiments show that inhibition of miR-1307-3p significantly inhibits breast cancer cell proliferation and anchorage-independent growth. Notably, animal experiments show that inhibition of miR-1307-3p not only inhibited cancer cell proliferation, but also delayed tumor formation time of breast cancer cells in nude mice. Moreover, inhibition of miR-1307-3p even blocked breast cancer cells tumor formation in part of nude mice.
In this study, we also clarified the oncogenic mechanism of miR-1307-3p in breast cancer. SMYD4 is a member of SYMD family. Hu et al. reported that SYMD4 is a tumor suppressor gene involved in breast cancer development 14. According to Hu et al. report that silencing of SMYD4 in nontumorigenic mammary cells stimulated the proliferation, the anchorage-independent growth, and tumor formation in nude mice. In this study, for the first time, we found that SMYD4 expression was negatively regulated by miR-1307-3p in breast cancer cells. Our data show that miR-1307-3p and SMYD4 expression was inversely correlated in breast cancer patients and the overexpression of miR-1307-3p in breast cancer cells leads to the suppression of SMYD4 expression; conversely, inhibition of miR-1307-3p further upregulates SMYD4 expression. Also, luciferase reporter gene experiments show that miR-1307-3p directly targets the 3'-UTR of SMYD4. In addition, our data show that overexpression of SMYD4 in MCF10A cell dramatically abrogated miR-1307-3p-overexpression induced stimulation of cell proliferation, anchorage-independent growth in soft agar, and tumor formation in nude mice. These findings clearly suggest that miR-1307-3p mediates tumorigenesis and cell proliferation via inhibition of SMYD4 in breast.
In summary, present study clarified a new mechanism of breast cancer development and progression. Our findings suggest that miR-1307-3p is an oncogenic miRNA significantly contribute to breast cancer development and progression by targeting SMYD4. Also, miR-1307-3p may be a useful target for breast cancer therapy.
Acknowledgments
Haesung Kim and Hua Zou designed this study. Sanghak Han and Hua Zou performed in vitro and animal experiments. Jin-Won Lee, Heung Cheol Kim, and Jeong Jin Cheol analyzed data. Jeong Jin Cheol and Lee-Su Kim collected patients' samples. Haesung Kim wrote the paper.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 3.Crabtree JS, Miele L. Breast Cancer Stem Cells. Biomedicines; 2018. p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 5.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T. et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–8. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isobe T, Hisamori S, Hogan DJ, Zabala M, Hendrickson DG, Dalerba P, miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. Elife; 2014. p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C. et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya A, Ziebarth JD, Cui Y. SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013;41:D977–82. doi: 10.1093/nar/gks1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J. et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326–34. doi: 10.1111/cas.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu X, Dou Y. miR-1307 promotes the proliferation of prostate cancer by targeting FOXO3A. Biomed Pharmacother. 2017;88:430–435. doi: 10.1016/j.biopha.2016.11.120. [DOI] [PubMed] [Google Scholar]
- 11.Chen WT, Yang YJ, Zhang ZD, An Q, Li N, Liu W. et al. MiR-1307 promotes ovarian cancer cell chemoresistance by targeting the ING5 expression. J Ovarian Res. 2017;10:1. doi: 10.1186/s13048-016-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JW, Guan W, Han S, Hong DK, Kim LS, Kim H. MicroRNA-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Sci. 2018;109:1404–1413. doi: 10.1111/cas.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu CX, Jere D, Jin H, Chang SH, Chung YS, Shin JY. et al. Poly(ester amine)-mediated, aerosol-delivered Akt1 small interfering RNA suppresses lung tumorigenesis. American Journal Of Respiratory And Critical Care Medicine. 2008;178:60–73. doi: 10.1164/rccm.200707-1022OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu L, Zhu YT, Qi C, Zhu YJ. Identification of Smyd4 as a potential tumor suppressor gene involved in breast cancer development. Cancer Res. 2009;69:4067–72. doi: 10.1158/0008-5472.CAN-08-4097. [DOI] [PubMed] [Google Scholar]