Abstract
Background: Cervical cancer (CCa) is a multifactorial gynecologic disease worldwide. Effects of HER2 polymorphisms, especially those in exonic region, have been investigated in many gynecologic diseases. In this study, we evaluated the influence of functional HER2 polymorphisms on susceptibility and survival of CCa in a Chinese population.
Methods: We genotyped the HER2 exonic polymorphisms by TaqMan in both case-control study (413 CCa patients vs. 396 controls) and survival study (413 CCa patients). Logistic regression and Cox regression were adopted to evaluate the genetic association with the risk and outcomes of CCa, respectively.
Results: In the case-control study, there was no significant difference between patients and controls in either HER2 rs1136201 or rs1058808. However, when combined, these two polymorphisms demonstrated a significant hazardous effect for CCa (P = 0.012). Besides, number of variants was also influential (P trend =0.002). In survival analysis, dominant model of rs1136201 and co-dominant modelof rs1058808 were significantly associated with the survival (P = 0.037 and P =0.028). The combination of rs1136201 and rs1058808 also negatively impacted CCa survival (P = 0.009). Cox regression further revealed the significance of the polymorphism combination (β = 0.38, P = 0.025, HR= 1.47, 95%CI = 1.05-2.05). Functional assay of these polymorphisms demonstrated that rs1058808 G allele was associated with stronger expression of HER2 gene.
Conclusions: Our results suggested that the combination of HER2 rs1136201and rs1058808 was significantly associated with the susceptibility of CCa. Besides, this combination of polymorphism s also substantially impacted the survival of CCa patients.
Keywords: HER2, cervical cancer, tag SNP, polymorphism, susceptibility, survival
Introduction
Cervical cancer (CCa) is the third most common and third lethal gynecologic cancer globally. In 2015, approximate 12,900 CCa incident cases and 4,100 deaths were reported in the U.S 1. In China, CCa is the most severe genital malignancy cancer among women, with 98,900 incident cases and 30,500 deaths in the same year 2. The squamous cervical carcinoma (SCCa), derived from squamous intraepithelial lesions (SILs), is the most frequent histological CCa type 3. The human papillomavirus (HPV) has been the known factor for CCa incidence and mortality. However, HPV is only a necessary but insufficient factor for the onset and development of CCa. As reported previously, genetic variation, including single nucleotide polymorphism (SNP), also play an important role in the progression of CCa 4-6.
With the introduction of the cytological screening, CCa mortality rate has been substantially reduced in clinics. Recently, the investigations of CCa incident and treatment have been improved, and some ideal strategies have been figured out soundly. As described by Dueñas-Gonzalez et.al., pre-invasion lesions such as high-grade squamous intraepithelial lesions (including CIN2 and CIN3 could be the best targets for healing 7. However, because early diagnosis and outcome of standard therapy of CCa is still unsatisfactory, the precious right time for CCa treatment is always wasted. Hence, there is an urgent need for more thorough and comprehensive understanding of the molecular mechanisms and corresponding biomarkers and targets for prediction, diagnose, treatment, and survival of the CCa susceptible population.
erb-b2 receptor tyrosine kinase 2 (ERBB2, more widely known as HER2), is one of the epidermal growth factor receptors. As a sensitive target for cancer therapy, its antibody, Herceptin, is widely recommended for breast cancer and CCa treatment with an eutherapeutic effect 8-11. Therefore, HER2 is an ideal candidate to screen for genetic variability on CCa susceptibility and survival. In this study, we tested the effects of SNPs in HER2 exonic region in case-control study for susceptibility of CCa, and further longitudinal study for survival of the patients. The results of this study provide more evident molecular basis for both diagnosis and treatment of CCa in the Chinese populations.
Method and Materials
Ethic principle
The study was approved by the ethics committee of Jiangsu Provincial Center for Disease Control and Prevention (no.2012025), in accordance with the principles of the Helsinki Declaration. Each participant signed a written informed consent before donating 5 ml venous blood for further analyses.
Study population
413 CCa patients (confirmed with histopathological evidence) and 396 controls without history of gynecologic diseases were enrolled in this study. The patients (in the case group) were followed for the further survival study. All patients' survival information was collected using the medical insurance system during follow-ups. All patients were recruited from the first affiliated hospital of Soochow University and the Suzhou Center for Disease Prevention and Control between Match 2004 and Jan 2010, along with their corresponding demographic and/or clinical characteristics (including age, menarche age, menopause age, first birth age, smoking status, HPV status, menopausal status, family cancer history, histological types and cancer stage for patients). The control participants were free of cancer history, who participated in a community-based chronic disease program of Suzhou Center for Disease Prevention and Control during the same period as the cases were collected with the help of Liuhe Hospital Affiliated to Medical College of Yangzhou University. People who smoked daily for 1 year were defined as smokers 12. All participants were genetically unrelated Han Chinese.
SNP selection criteria
We established the following three criteria to identify the target SNPs: a) located in exonic region of the HER2 gene; b) MAF (minor allele frequency) of Han Chinese in Beijing (HCB) > 0.05; c) a linkage disequilibrium value of r2 < 0.8 for each target SNPs.
DNA isolation and Genotyping
Genomic DNA was extracted from participants' peripheral venous blood by the QIAcube HT Plasticware with QIAamp 96 DNA QIAcube HT Kit (Qiagen, Dusseldorf, Germany) following the manufacturer's protocol and then stored at-80℃ before genotyping. The A260/A280 of the purified DNA, tested by the NanodropOneC Ultramicro ultraviolet spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA), was between 1.8 and 2.0, indicating no external DNA contamination. The TaqMan SNP Genotyping Assay with ABI7900HT real-time PCR System (Applied Biosystems, Foster City, CA, USA) was used for genotyping HER2 polymorphisms. These samples were added in each plate for quality control of genotyping. Two staff operated the genotyping assay independently. More than 10% of the samples were randomly selected for validation, and the results were exactly same between the two sets of assays.
Construction of HER2 expression plasmid and transient transfection
The total cDNA sequence of HER2 was synthesized and constructed into the pIRES2-EGFP by Generay Company (Shanghai, China). And the single-point mutations were performed on the original plasmid to evaluate the impact of variants of both rs1136201 and rs1058808 by the same company. All plasmids were confirmed by DNA sequencing. After transformation and purification, these plasmids were transiently transfected into the HeLa cell line. Each kind of cells was seeded on 24-well plates overnight to ensure the sufficient amount of cells for further transfection. With the help of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), 0.8 μg of each plasmid were then transfected into HeLa cells, respectively. All the experiments were conducted by the same technician following standardized protocol, to eliminate the impact of inequable transfection efficiencies and the total amount of each plasmid. The expression of EGFP was tested as the internal control for transfections in each plate.
RNA expression and Western blot of HER2 expression plasmids
After transformation and purification, these plasmids were transiently transfected into the HeLa cell line. Each kind of cells was seeded on 24-well plates overnight to ensure the sufficient amount of cells for further transfection. With the help of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), 0.8 μg of each plasmid were then transfected into HeLa cells, respectively. All the experiments were conducted by the same technician following standardized protocol, to eliminate the impact of inequable transfection efficiencies and the total amount of each plasmid. The expression of EGFP was tested as the internal control for transfections in each plate.
RNA expression and Western blot of HER2 expression plasmids
After a 48h transfection, RNA and protein of HER2 were collected from each plate. The total RNA of cells was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Quantitative real-time PCR were performed by ABI7900 HTreal-time PCR System (Applied Biosystems) after reverse transcription, and the glyceraldehydes 3-phosphate dehydrogenase (GAPDH) was adopted as the internal control. The primer for HER2 and GAPDH were listed as follows: HER2 sense primer: 5′-CTGATGGGTTAATGAGCAAACTGA-3′, and HER2 antisense primer: 5′-CCAAATTCTGTGCTGGAGGTAGAG-3′; GAPDH sense primer: 5′-CCACCCATGG CAAATTCCATGGCA-3′, and GAPDH sense primer: 5′-TCTAGACGGCAGGTCAGGTCCACC-3′. All experiments were performed in triplicate, and the relative quantification of HER2 mRNA was calculated using the 2-ΔΔCt method.
Cells were harvested and washed with PBS and lysed in RIPA with protease and phosphatase inhibitors on ice and centrifugated at 4°C. The protein contents were determined using Bradford protein assay (Bio-Rad Hercules, CA, USA). Total protein (25 μg) of each sample transfected with different plasmids was separated by SDS-PAGE (12.5%), and then transferring onto PVDF membrane. After blocking by 5% low-fat powder for eliminating nonspecific binding, the membrane was incubated with primary antibody (dilution =1:1000, HER2 antibody, ab134182, Abcam, Cambridge, UK; dilution = 1:5000, GAPDH antibody, ab9485, Abcam) at 4°C overnight. In the second day, the transferred membrane was incubated with secondary antibody (dilution = 1:5000) for 2 h at 4℃. The Bio-rad ChemiDoc XRS+ system (Bio-Rad Hercules) was used to visualize the protein signals on the membrane.
Immunohistochemistry (IHC)
The IHC for HER2 was carried out on representative paraffin sections of CCa tissues. Immunohistochemical staining was performed by using the Boster SABC (rabbit IgG)-POD kit (Wuhan, China) according to the recommended protocol. The HER2 antibody (ab134182, Abcam) was used to incubate the preparing sections overnight at 4°C, and the 3, 3'-diaminobenzidine was used to get a brown precipitate for scoring. The evaluation of PSCA expression was performed by the Department of Pathology, the first affiliated hospital of Nanjing Medical University with a blind fashion by two independent experienced pathologists. The final HER2 score was calculated by multiplying the intensity and the percentage of positive cells. The categorization criteria were as follows: ≤ 3, negative or weak; > 3 and ≤6, moderate; > 6, strong.
Statistical Analysis
Goodness-of-fit χ2 test was adopted to test for the Hardy-Weinberg equilibrium (HWE). In the case-control study, the Student-t test and/or χ2 test were used to demonstrate the how distributions of demographic, clinical characteristics, and frequency of genotypes differ between case and control groups. Unconditional univariate and multivariate logistic regressions were applied to estimate the adjusted odds ratios (ORs), with 95% confidence intervals (CIs), which quantify the effects of the SNPs.
Furthermore, Kaplan-Meier estimator was computed to evaluate the effect of HER2 polymorphisms on CCa patients' survival, demographic, and clinical characteristics. Mean survival time (MSTb) was provided when MST could not be calculated. Final predictive factors of CCa patients' survival were revealed with the univariate or multivariate Cox regression by estimating the hazard ratios (HRs) and corresponding 95%CIs. The Cox step wise regression was performed for CCa prognosis, with a significant level of 0.05 for entering and 0.10 for removing the respective explanatory variables.
Results
Characteristics of study participants
The characteristics of all participants, including 413 CCa patients (cases) and 396 controls, were provided in Table 1. There was no significant difference between case and control group in age (56.12 ±12.48 vs. 56.28 ± 11.91, case and control, respectively, P = 0.844), as well as in smoking status. However, the ages at menarche, menopause, first live birth showed significant differences between these two groups (P < 0.05). In clinical indices, the characters of HPV infection and family cancer history in patients dominated high proportions (87.65% vs. 80.30%, P < 0.001 and 67.31% vs. 32.69, P < 0.001, respectively). Also, in menopausal status (P = 0.007) and parity (P = 0.010), patients presented enormous discrepancies.
Table 1.
Demographic and Selected Variables in Cervical Cancer Cases and Controls
| Variables | Cases (n=413) | Controls (n=396) | Pa | |||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Age, year (mean±SD) | 56.12±12.48 | 56.28±11.91 | 0.844 | |||
| Age at menarche, year (mean±SD) | 14.55±2.36 | 14.84±1.78 | 0.006 | |||
| Age at menopause, year (mean±SD)b | 49.75±2.83 | 49.26±3.52 | 0.010 | |||
| Age at first live birth, year (mean±SD)c | 25.36±1.79 | 24.82±2.34 | <0.001 | |||
| Smoking status | ||||||
| Smoker | 27 | 6.54 | 14 | 3.54 | 0.052 | |
| Nonsmoker | 386 | 93.46 | 382 | 96.46 | ||
| HPV infection | ||||||
| Yes | 362 | 87.65 | 78 | 19.70 | <0.001 | |
| No | 51 | 12.35 | 318 | 80.30 | ||
| Menopausal status | ||||||
| Premenopausal | 180 | 43.58 | 136 | 34.34 | 0.007 | |
| Postmenopausal | 233 | 56.42 | 260 | 65.66 | ||
| Parity | ||||||
| 0-1 | 141 | 34.14 | 170 | 42.93 | 0.010 | |
| ≥2 | 272 | 65.86 | 226 | 57.07 | ||
| Family history of cancer | ||||||
| Yes | 278 | 67.31 | 87 | 21.97 | <0.001 | |
| No | 135 | 32.69 | 309 | 78.03 | ||
| Histological types | ||||||
| CIN3d | 12 | 2.91 | ||||
| Squamous cell carcinoma | 344 | 83.29 | ||||
| Adenocarcinomas | 50 | 12.11 | ||||
| Adenosquamous carcinoma | 7 | 1.69 | ||||
| Stage | ||||||
| CIN3d | 4 | 0.97 | ||||
| I | 101 | 24.46 | ||||
| II | 238 | 57.63 | ||||
| III | 65 | 15.74 | ||||
| IV | 5 | 1.21 | ||||
a two-side test. b Information was available in postmenopausal women (233 cases and 260 controls). c Information was available in 407 cases and 389 controls with parity. d CIN, cervical intraepithelial neoplasia.
The survival information of the 413 patients was provided in Table 2. Among all the 413 CCa patients, 156 had passed away. There were no statistically significant association between MST (median survival time) and patients' age (log-rank P = 0.262), age at menarche (log-rank P = 0.884), age at menopause (log-rank P = 0.354), age at first live birth (log-rank P = 0.840), or smoking status (log-rank P = 0.053). On the other hand, there was significant association between menopausal status and CCa patients' survival (log-rank P = 0.023): the postmenopausal patients had a higher mortality rate than premenopausal ones (MST = 52.03, HR = 1.47, 95%CI = 1.06-2.04). Besides, there was also significant difference among histological types (log-rank P = 0.007).
Table 2.
Cervical cancer patients' characteristics and clinical features
| Variables | Patients n=413 (%) | Deaths n=156 (%) | MST (months)a | Log- rank P | HR (95%CI) |
|---|---|---|---|---|---|
| Age (years) | |||||
| ≤55 | 198 (47.94) | 77 (38.89) | 45.03 | 0.262 | 1.00 (Ref.) |
| >55 | 215 (52.06) | 79 (36.74) | 52.36 | 0.83 (0.60-1.15) | |
| Age at menarche (years) | |||||
| <15 | 227 (54.96) | 85 (37.44) | 51.20 | 0.884 | 1.00 (Ref.) |
| ≥15 | 186 (45.04) | 71 (38.17) | 52.35 | 1.02 (0.74-1.41) | |
| Age at menopause (years)b | |||||
| <50 | 119 (51.07) | 42 (35.29) | 51.76 | 0.354 | 1.00 (Ref.) |
| ≥50 | 114 (48.93) | 49 (42.86) | 52.63 | 1.17 (0.84-1.62) | |
| Age at first live birth (years)c | |||||
| <25 | 190 (46.68) | 73 (38.02) | 52.53 | 0.840 | 1.00 (Ref.) |
| ≥25 | 217 (53.32) | 81 (37.33) | 45.92 | 1.03 (0.75-1.42) | |
| Smoking status | |||||
| Smoker | 27 (6.54) | 7 (25.93) | 46.17d | 0.053 | 1.00 (Ref.) |
| Nonsmoker | 386 (93.48) | 149 (38.60) | 51.75 | 2.14 (0.99-4.61) | |
| HPV infection | |||||
| Yes | 362 (87.65) | 142 (39.23) | 52.31 | 0.819 | 1.00 (Ref.) |
| No | 51 (12.35) | 14 (27.45) | 43.77 | 1.07 (0.61-1.87) | |
| Menopausal status | |||||
| Premenopausal | 180 (43.58) | 60 (33.33) | 52.03 | 0.023 | 1.00 (Ref.) |
| Postmenopausal | 233 (56.42) | 96 (41.20) | 45.14 | 1.47 (1.06-2.04) | |
| Parity | |||||
| 0-1 | 141 (34.14) | 60 (42.55) | 42.94 | 0.116 | 1.00 (Ref.) |
| ≥2 | 272 (65.86) | 96 (35.29) | 52.38 | 0.77 (0.55-1.07) | |
| Family history of cancer | |||||
| Yes | 278 (67.31) | 107 (38.49) | 52.91 | 0.492 | 1.00 (Ref.) |
| No | 135 (32.69) | 49 (36.30) | 51.90 | 0.89 (0.63-1.25) | |
| Histological types | |||||
| CIN3e | 12 (2.91) | 4 (33.33) | 32.69d | 0.007 | 1.00 (Ref.) |
| Squamous cell carcinoma | 344 (83.29) | 141 (40.99) | 45.93 | 1.29 (0.47-3.53) | |
| Adenocarcinomas | 50 (12.11) | 10 (20.00) | 51.05d | 0.52 (0.16-1.72) | |
| Adenosquamous carcinoma | 7 (1.69) | 1 (14.29) | 41.00d | 0.44 (0.05-3.98) | |
| Stage | |||||
| CIN3e | 4 (0.97) | 3 (75.00) | 56.62 | 0.174 | 1.00 (Ref.) |
| I | 101 (24.46) | 44 (43.56) | 44.31 | 1.32 (0.38-4.55) | |
| II | 238 (57.63) | 87 (36.55) | 52.12 | 1.10 (0.33-3.71) | |
| III | 65 (15.74) | 21 (32.31) | 51.01 | 0.81 (0.22-2.89) | |
| IV | 5 (1.21) | 1 (20.00) | 52.00 | ||
| Chemotherapy | |||||
| Yes | 273 (66.10) | 99 (36.26) | 52.69 | 0.456 | 1.00 (Ref.) |
| No | 140 (33.90) | 57 (40.71) | 44.35 | 1.13 (0.81-1.58) | |
a MST: median survival time; b Information was available in 91/233 (deaths/patients) postmenopausal women; c Information was available in 153/407 (deaths/patients) cases with parity; d Mean survival time was provided when MST could not be calculated; e CIN, cervical intraepithelial neoplasia.
Effects of HER2 polymorphisms on the risk of CCa in the case-control study
According to the SNP selection criteria we had mentioned above, there were only two candidate SNPs finally identified as our objects: rs1136201 and rs1058808 in the HER2 gene.
The frequencies of participants' genotypes confirmed the HWE (χ2 = 0.716, P= 0.437). There was no significant effect for either SNP (rs1136201 or rs1058088) in any model (Table 3). However, with the increase of alleles, these SNPs displayed their harmful (P trend = 0.036 for rs1136201) and protective (P trend = 0.012 for rs1058808) trends. The combined effects of these two SNPs were shown in Table 4. Participants with 3 variants in combination of rs1136201 and rs1058088 were associated with significantly higher susceptibility to CCa (OR = 1.98, 95%CI= 1.18-3.35). Although individuals having 4 variants showed no significant association with CCa occurrence, there was still a risky trend among individuals having the combination of these two SNPs (P trend = 0.002). Using 0-2 variants as the reference group, individuals with 3-4 variants of these two SNPs showed significantly higher susceptibility to CCa (P = 0.028, OR = 1.70, 95%CI = 1.06-2.74; Table 4).
Table 3.
Genotype and allele frequencies of rs1136201 and rs1058808 polymorphisms among cervical cancer cases and controls
| Genotype | Cases (n=413) | Controls (n=396) | Pa | Adjusted OR (95%CI)b | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| rs1136201 | |||||||
| AA | 218 | 52.78 | 237 | 59.85 | 1.00 (Ref.) | ||
| AG | 161 | 38.98 | 135 | 34.09 | 0.126 | 1.39 (0.91-2.12) | |
| GG | 34 | 8.23 | 24 | 6.06 | 0.169 | 1.66 (0.80-3.44) | |
| AG+GG | 195 | 47.22 | 159 | 40.15 | 0.074 | 1.44 (0.96-2.14) | |
| G allele | 229 | 27.72 | 183 | 23.11 | 0.069 | 1.33 (0.98-1.81) | |
| P trend | 0.036 | ||||||
| rs1058808 | |||||||
| GG | 172 | 41.65 | 131 | 33.08 | 1.00 (Ref.) | ||
| CG | 181 | 43.83 | 192 | 48.48 | 0.104 | 0.70 (0.45-1.08) | |
| CC | 60 | 14.53 | 73 | 18.43 | 0.112 | 0.62 (0.35-1.12) | |
| CG+CC | 241 | 58.35 | 265 | 66.92 | 0.059 | 0.68 (0.45-1.02) | |
| C allele | 301 | 36.44 | 338 | 42.68 | 0.067 | 0.77 (0.58-1.02) | |
| P trend | 0.012 | ||||||
a two-side chi-square test. b Adjusted for age, age at menarche, age at menopause, age at first live birth, smoking status, menopausal status, parity and family history of cancer.
Table 4.
Frequency distributions of the combined genotypes of rs1136201 and rs1058808 polymorphisms among the cases and controls, and their correlation to risk of cervical cancer
| Genotype | Cases (n=413) | Controls (n=396) | Pa | Adjusted OR (95%CI)b | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| Number of risk allelesc | |||||||
| 0 | 42 | 10.17 | 52 | 13.13 | 0.012 | 1.00 (Ref.) | |
| 1 | 105 | 25.42 | 121 | 30.56 | 1.22 (0.60-2.46) | ||
| 2 | 161 | 38.98 | 159 | 40.15 | 1.52 (0.78-2.97) | ||
| 3 | 93 | 22.52 | 58 | 14.65 | 2.23 (1.02-4.91) | ||
| 4 | 12 | 2.91 | 6 | 1.52 | 4.48 (0.94-21.29) | ||
| P trend | 0.002 | ||||||
| Recombined groupsc | |||||||
| 0-2 | 308 | 74.58 | 332 | 83.84 | 1.00 (Ref.) | ||
| 3-4 | 105 | 25.42 | 64 | 16.16 | 0.028 | 1.70 (1.06-2.74) | |
a two-side chi-square test. b Adjusted for age, age at menarche, age at menopause, age at first live birth, smoking status, menopausal status, parity and family history of cancer. c The 0-4 represents the numbers of risk alleles within the combined genotypes; the risk alleles used for the calculation were the rs1136201 G and rs1058808 G alleles.
The association between HER2 polymorphisms and CCa survival
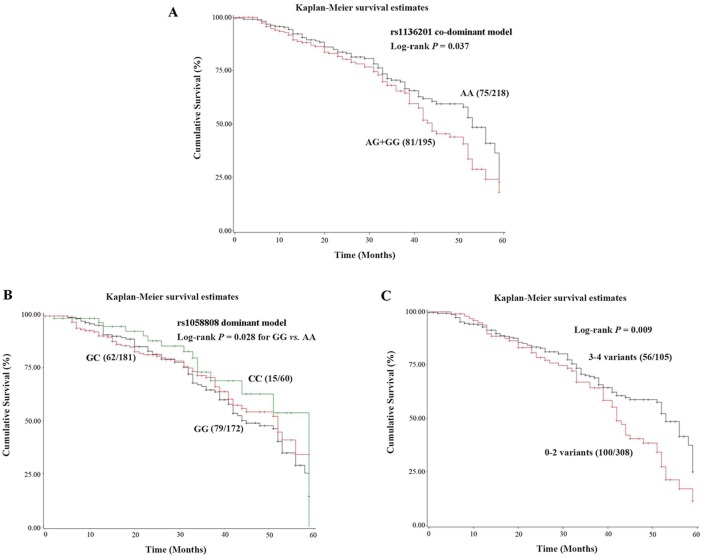
Kaplan-Meier survival curve was constructed, and log-rank test were performed for the 413 patients (Table 5). In SNP rs1136201, the co-dominant model revealed significantly higher risk of death (HR = 1.53, 95%CI = 1.04-2.28) in AG/GG genotypes, which coincided with the hazardous effect of the G allele (log-rank P trend = 0.040). The Kaplan-Meier survival curves for rs1136201 and rs1058088 were showed in Figure 1A and 1B, respectively.
Table 5.
Genotypes of rs1136201 and rs1058808 polymorphisms and cervical cancer survival
| Variable | Patients [n=413 (%)] |
Deaths [n=156 (%)] |
MST (months)a | Log- rank P | HR (95%CI)b |
|---|---|---|---|---|---|
| rs1136201 | |||||
| AA | 218 (52.78) | 75 (34.40) | 53.96 | 1.00 (Ref.) | |
| AG | 161 (38.98) | 64 (39.75) | 44.45 | 0.052 | 1.51 (1.00-2.30) |
| GG | 34 (8.23) | 17 (50.00) | 43.24 | 0.275 | 1.52 (0.72-3.21) |
| AG+GG | 195 (47.22) | 81 (41.54) | 44.28 | 0.037 | 1.53 (1.03-2.28) |
| P trend | 0.040 | ||||
| rs1058808 | |||||
| GG | 172 (41.65) | 79 (45.93) | 45.34 | 1.00 (Ref.) | |
| CG | 181 (43.83) | 62 (34.25) | 52.14 | 0.558 | 0.88 (0.56-1.36) |
| CC | 60 (14.53) | 15 (25.00) | 59.16 | 0.028 | 0.40 (0.18-0.90) |
| CG+CC | 241 (58.35) | 77 (31.95) | 52.09 | 0.189 | 0.75 (0.49-1.15) |
| P trend | 0.140 | ||||
a MST: median survival time. b Adjusted for age, age at menarche, age at menopause, age at first live birth, smoking status, menopausal status, parity and family history of cancer.
Figure 1.
The Kaplan-Meier survival curves of HER2 polymorphisms. (A). Co-dominant model of rs1136201; (B). Dominant model of rs1058808; (C).Combined analysis of rs1136201 and 1058808.
The combined analysis results for rs1136201 and rs1058808 were shown in Table 6 and Figure 1C. The patients with 3 variants in these two polymorphisms had the shortest survival time (MST = 42.85) and the highest mortality rate (HR =2.42, 95%CI = 1.02-5.79). Group with 3-4 variants had much shorter MST and higher mortality rate (MST = 42.24, log-rank P = 0.009, HR = 1.74, 95%CI = 1.15-2.65), comparing to group with 0-2 variants (MST = 53.67, as the reference group).
Table 6.
Frequency distributions of the combined genotypes of rs1136201 and rs1058808 polymorphisms in cervical cancer patients, and their correlation to survival of cervical cancer patients
| Genotype | Patients [n=413 (%)] |
Deaths [n=156 (%)] |
MST (months)a | Log- rank P | HR (95%CI)b |
|---|---|---|---|---|---|
| Number of risk allelesc | |||||
| 0 | 42 (10.17) | 13 (30.95) | 51.24 | 0.014 | 1.00 (Ref.) |
| 1 | 105 (25.42) | 30 (28.57) | 56.79 | 1.12 (0.44-2.82) | |
| 2 | 161 (38.98) | 57 (35.40) | 52.57 | 1.25 (0.50-3.08) | |
| 3 | 93 (22.52) | 50 (53.76) | 42.85 | 2.42 (1.02-5.79) | |
| 4 | 12 (2.91) | 6 (50.00) | 45 | 4.54 (0.42-48.88) | |
| P trend | 0.018 | ||||
| Recombined groupsc | |||||
| 0-2 | 308 (74.58) | 100 (32.47) | 53.67 | 0.009 | 1.00 (Ref.) |
| 3-4 | 105 (25.42) | 56 (53.33) | 42.24 | 1.74 (1.15-2.65) | |
a MST: median survival time. b Adjusted for age, age at menarche, age at menopause, age at first live birth, smoking status, menopausal status, parity and family history of cancer. c The 0-4 represents the numbers of risk alleles within the combined genotypes; the risk alleles used for the calculation were the rs1136201 G and rs1058808 G alleles.
The stratified analysis and Cox regression model for survival in CCa patients
Based on the significant difference between 0-2 variants group and 3-4 variants group in the CCa survival study (see above paragraph), we validated the impacts of HER2 polymorphisms on CCa survival. The results were shown in Table 7. 3-4 SNP variants more substantially impacted the subgroups of age above 55 (log-rank P= 0.004, HR=2.06, 95%CI = 1.26-3.37), menarche age below 15 (log-rank P = 0.020, HR = 1.99, 95%CI = 1.11-3.56), menopause age below 50 (log-rank P = 0.016, HR= 1.71, 95%CI = 1.10-2.64), first live birth age below 25 (log-rank P =0.017, HR =2.14, 95%CI = 1.15-4.01), and non-smokers (log-rank P = 0.045, HR = 1.56, 95%CI = 1.01-2.42). In clinical, the combined perniciousness of HER2 SNP variants existed in HPV infection group (log-rank P =0.018, HR = 1.69, 95%CI = 1.09-2.61), ≥2 parity (log-rank P = 0.003, HR = 1.76, 95%CI = 1.08-2.86), adenocarcinomas (log-rank P =0.004, HR = 21.12, 95%CI = 2.72-163.66), and clinical stage II of CCa (log-rank P = 0.008, HR = 2.24, 95%CI = 1.23-4.07; Table 7). Considering all potential demographic characters, clinical feature, and combination of HER2 polymorphisms (rs1136201 and rs1058808), four variables: smoking status (P = 0.024), menopausal status (P = 0.020), histological types (P = 0.006), and number of variants (P =0.025) were selected based on the Cox regression model (Table 8).
Table 7.
Stratified analysis of different number of risk alleles associated with cervical cancer patients' survival
| Variables | Number of risk allelesa
(deaths/patients) |
Log- rank P | HR (95%CI)b | |
|---|---|---|---|---|
| 0-2 | 3-4 | |||
| Age (years) | ||||
| ≤55 | 50/149 | 25/49 | 0.842 | 0.90 (0.31-2.58) |
| >55 | 48/159 | 31/56 | 0.004 | 2.06 (1.26-3.37) |
| Age at menarche (years) | ||||
| <15 | 54/169 | 21/58 | 0.020 | 1.99 (1.11-3.56) |
| ≥15 | 46/139 | 25/47 | 0.179 | 1.52 (0.83-2.79) |
| Age at menopause (years) c | ||||
| <50 | 27/91 | 15/28 | 0.016 | 1.71 (1.10-2.64) |
| ≥50 | 31/81 | 18/33 | 0.272 | 1.38 (0.78-2.44) |
| Age at first live birth (years) d | ||||
| <25 | 48/145 | 24/45 | 0.017 | 2.14 (1.15-4.01) |
| ≥25 | 50/158 | 31/59 | 0.470 | 1.24 (0.70-2.20) |
| Smoking status | ||||
| Smoker | 2/17 | 5/10 | 0.225 | 5003.51 (0.00-4.70E9) |
| Nonsmoker | 98/291 | 51/95 | 0.045 | 1.56 (1.01-2.42) |
| HPV infection | ||||
| Yes | 90/265 | 52/97 | 0.018 | 1.69 (1.09-2.61) |
| No | 10/43 | 4/8 | 0.436 | 2.43 (0.26-22.61) |
| Menopausal status | ||||
| Premenopausal | 41/136 | 19/44 | 0.125 | 1.82 (0.85-3.91) |
| Postmenopausal | 59/172 | 37/61 | 0.052 | 1.70 (1.00-2.91) |
| Parity | ||||
| 0-1 | 43/107 | 17/34 | 0.993 | 1.00 (0.45-2.19) |
| ≥2 | 57/201 | 39/71 | 0.003 | 2.21 (1.31-3.74) |
| Family history of cancer | ||||
| Yes | 68/205 | 39/73 | 0.024 | 1.76 (1.08-2.86) |
| No | 32/103 | 17/32 | 0.200 | 1.74 (0.75-4.04) |
| Histological types | ||||
| CIN3e | 2/10 | 2/2 | - | - |
| Squamous cell carcinoma | 92/251 | 49/93 | 0.261 | 1.30 (0.82-2.04) |
| Adenocarcinomas | 5/40 | 5/10 | 0.004 | 21.12 (2.72-163.66) |
| Adenosquamous carcinoma | 1/7 | 0/0 | - | - |
| Stage | ||||
| CIN3e | 2/3 | 1/1 | - | - |
| I | 30/75 | 14/26 | 0.501 | 1.34 (0.57-3.11) |
| II | 56/183 | 31/55 | 0.008 | 2.24 (1.23-4.07) |
| III | 12/44 | 9/21 | 0.408 | 0.53 (0.12-2.41) |
| IV | 0/3 | 1/2 | - | - |
| Chemotherapy | ||||
| Yes | 65/210 | 34/63 | 0.167 | 1.51 (0.84-2.73) |
| No | 35/98 | 22/42 | 0.118 | 1.76 (0.87-3.59) |
a The 0-4 represents the numbers of risk alleles within the combined genotypes; the risk alleles used for the calculation were the rs1136201 G and rs1058808 G alleles.
b Adjusted for age, age at menarche, age at menopause, age at first live birth, smoking status, menopausal status, parity and family history of cancer.
c Information was available in 91/233 (deaths/patients) postmenopausal women.
d Information was available in 153/407 (deaths/patients) cases with parity.
e CIN, cervical intraepithelial neoplasia.
Table 8.
Results of stepwise Cox regression analysis of cervical cancer survival
| Variables | β | SE | HR | 95%CI | P |
|---|---|---|---|---|---|
| Smoking status (Nonsmoker vs. Smoker) | 0.89 | 0.39 | 2.44 | 1.13-5.29 | 0.024 |
| Menopausal status (Postmenopausal vs. Premenopausal) | 0.39 | 0.17 | 1.48 | 1.06-2.06 | 0.020 |
| Histological types (CIN3a vs. Squamous cell carcinoma vs. Adenocarcinomas vs. Adenosquamous carcinoma) | 0.56 | 0.20 | 1.75 | 1.71-2.61 | 0.006 |
| Number of risky allelesb (3-4 vs. 0-2) | 0.38 | 0.17 | 1.47 | 1.05-2.05 | 0.025 |
a CIN, cervical intraepithelial neoplasia. b The 0-4 represents the numbers of risk alleles within the combined genotypes; the risk alleles used for the calculation were the rs1136201 G and rs1058808 G alleles.
The functional analysis of HER2 polymorphisms and HER2 expression
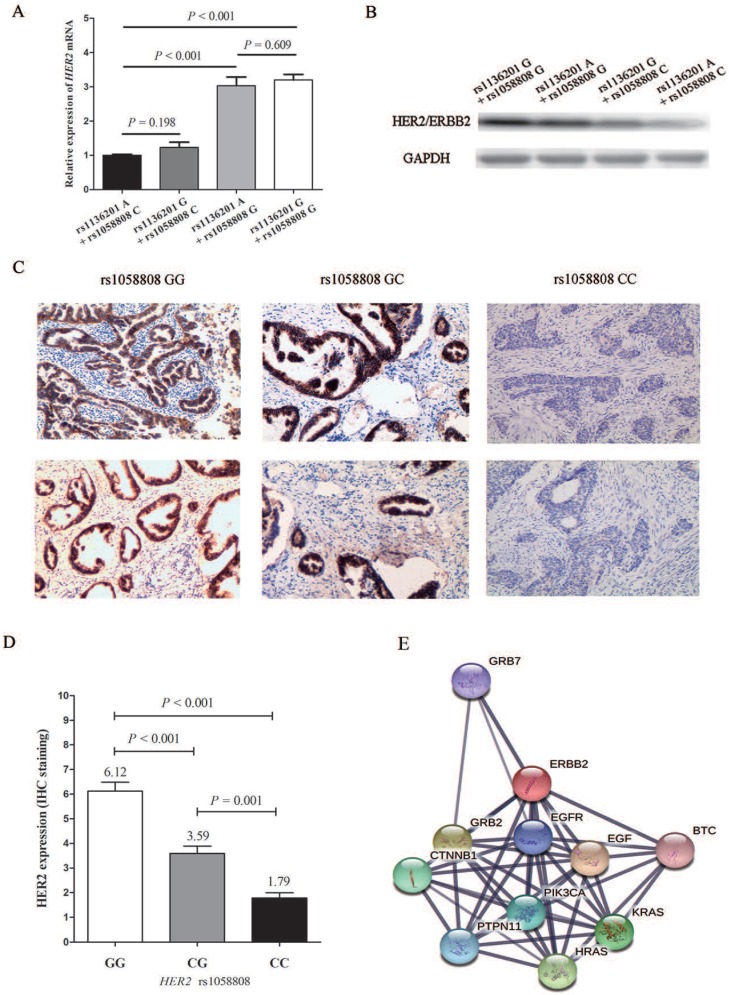
After the population study of HER2 polymorphisms, we further investigated the biological function of these two SNPs. As shown in Figure 2A and 2B, both mRNA level and protein level of HER2 expression plasmid and its corresponding mutations of rs1136201 and rs1058808 displayed that plasmids with rs1058808 G allele was associated with higher products of HER2 in Hela cell lines (P < 0.001), but those with rs1136201 genotype did not influence the expression of HER2 (P = 0.198) (Figure 2A and B). Based on these consequences, we performed the further IHC assay of HER2 in CCa tissues. We firstly genotyped the SNP rs1058808 in 157paraffin sections of CCa tissues, and the frequency distribution of GG, CG, and CC genotype was 58,66 and 33, respectively.
Figure 2.
The functional analysis of HER2 polymorphisms. (A).Expression of HER2 mRNA levels in Hela cell lines transfected with different HER2 expression plasmids. (B). Expression of HER2 protein levels in Hela cells transfected with different HER2 expression plasmids. (C). Immunohistochemical staining of HER2in human cervical cancer tissues with different rs1058802 genotype. (D). Prediction of HER2 possible pathway and corresponding associated genes.
The results of HER2 IHC staining in CCa tissues with different genotypes of HER2 rs1058808 was shown in Figure 2C. And the corresponding competetion of scores among GG, CG and CC were listed in Figure 2D. The dramatic significance between GG, CG and CC (average staining score: 6.12 vs. 3.59 vs. 1.79, P < 0.001 for One-way ANOVA) also suggested the transcriptional function of HER2 rs1058808 in CCa.
Discussion
In this study, we have demonstrated that the combination of the HER2 exonic SNPs (rs1136201 and rs1058808) was significantly associated with cervical carcinogenesis in the Chinese Han population. Besides, AG/GG genotype of rs1136201 conspicuously impacted the survival time of CCa patients, and combined model of above two SNPs also emerged its hazard and independence in CCa patients' survival. Therefore, these results have revealed the potential predictive and diagnostic values for the HER2 SNPs in CCa.
The HER2 gene, located in the 17th chromosome, usually acts as an oncogene and is closely related to the dysregulation of cell growth, differentiation, migration, and even apoptosis 13, 14. In the gynecologic oncology, the amplification or over-expression of HER2 has been detected in approximately 20% to 30% of breast cancer patients 15, 16. In ovarian cancer patients, HER2 is also reported to induce poorer clinical outcomes and survival 17, 18. Therefore, HER2 testing have become a standard procedure in feminine tumors; the corresponding HER2-targeted medicines (such as Herceptin or Trastuzumab) is provided as specific remedies, and some studies demonstrates its significant effects on tumor suppress 19-21. Although evidences haven shown that overexpression of HER2 is also present in CCa patients 22, the anti-HER2 therapy does not draw enough attention. According to a previous report from Doo-Yi Ohet al., the treatment of HER2 could be considered as a novel and efficient target for CCa, especially the patient-derived xenograft model 11.
Both rs1136201 and rs1058808 SNPs have been deeply investigated for the occurrence, development and survival of several carcinomas 23-25. rs1136201, also named HER2 Ile655Val, locates within a consensus sequence that encodes the TM helix segment's N-terminus. Variant of rs1136201 might lead to destabilization of the active HER2 heterodimers, consequently increases signal transduction for HER2 expression, and finally stimulates kinase activity and malignant transformation 26, 27. Although the function of HER2 exonic polymorphism is clear, the association between HER2 SNP rs1136201 and CCa is still controversial 28,29. Our results show that rs1136201 alone does not significantly influence the risk of CCa, which coincide to the previous report in a Japanese population 28; however our results do not align with the population in Poland 29. Such difference might be due to the distinction of Asian and Caucasian populations. As for the combined effect of rs1136201 and rs1058808, the patients with three or even more variants showed significantly higher risk of CCa (P = 0.028, OR = 1.70, 95%CI = 1.06-2.74 in Table 4). This finding suggests that there might be synergistic interaction between these two SNPs.
For the survival of CCa, we have demonstrated that patients with rs1136201 AG/GG genotypes (1.53-fold, MST = 44.28) had significantly higher mortality rate and shorter survival time. And patients with such genotype with 3-4 variants of rs1136201 and rs1058808 face further risk of CCa, especially in adenocarcinomas (HR = 21.12). The Cox regression analysis of CCa has also identified the role of joint genotypes of HER2 polymorphisms, which could be a potential indicator for poor CCa survival. However, the HPV infection is not a significant factor according to the results of Cox regression, which suggests that HPV infection is an incentive for the onset of CCa but not an important (oreven determinant) factor for the CCa survival.
In functional analysis, the corresponding assays displayed that the variant of G allele of rs1058808 significantly decreased the expression of HER2 in both mRNA level and protein level, but variant of rs1136201 did not emerge any strong transcriptional activity (Figure 2A and B). These results partly coincided with the data reported by Suet al. 30. Beyond our expectations, the combine effect of rs1058808and rs1136201 did not reflect on expressions of HER2 (both mRNA and protein levels, Figure 2A and B), when we transfected cells with plasmids including different alleles of these two SNPs. As some mutations could influence the protein dynamics or mis-folding instead of the its expression 31-33, it was rational to hypothesis that the variant of rs1136201 could also perform its function in a special method. Therefore, the further investigation of rs1136201 was expected. Finally, we detected the possible signal pathway involving in HER2 (Figure 2E), the string software (https://string-db.org/) reminded us that the HER2 was closely associated with GRB7, EGFR, EGF, PIK3CA, HRAS, KRAS, PTPN11, CTNNB1, GRB2 and BTC genes; a deep research of SNPs on the interactivity of these genes will be helpful to re-discover the importance of these SNPs, including rs1058808 and rs1136201.
There are, however, some limitations in this study. First, all the patients have been recruited in the hospital. Thus, the results and conclusions from this study should be further validated with larger sample sizes from other study areas and populations. Second, environmental factors, such as diet and physical activity, have not been considered in our study because of the lack of detailed background information of the participants. These factors may also influence CCa occurrence and/or survival. Finally, as a retrospective hospital-based study, the inherent selection bias should not be neglected.
Conclusion
In conclusion, this study demonstrated that CCa risk was associated with either HER2 polymorphism (rs1136201 and rs1058808) alone, but the combination of these two SNPs significantly aggravated the onset of CCa. Besides, combination of rs1136201 and rs1058808 significantly jeopardized the survival of CCa patients, and would further reduce CCa survival rate and shorten MST.
Acknowledgments
This study was supported by the National Science Foundation for Young Scientists of China (81602919, 81502796, and 81703201), Natural Science Foundation for Young Scientists of Jiangsu Province (BK20171076), Suzhou Key Technology Support Program (SS201111), Jiangsu Provincial Medical Innovation Team (CXTDA2017029), the Jiangsu Provincial Medical Youth Talent (QNRC2016548, QNRC2016536 and QNRC2016231), the 9th Suzhou Special projects for industrial and technological innovation (SYS201581), the project of Jiangsu province preventive medicine (Y2015019), the Key technology in prevention and control of serious diseases and infectious diseases (Gwzx201601), the Program of Jiangsu Preventive Medicine Association (Y2018086), the Lifting Program of Jiangsu Provincial Scientific and Technological Association, and the Jiangsu Government Scholarship for Overseas Studies.
References
- 1.Siegel RL MK, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Wells M, Östor AG, Crum CP, Tumors of the uterine cervix: Epithelial tumors. In World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Breast and Female Genital Organs. WHO, IARC Press, Lyon; 2003. pp. 262–279. [Google Scholar]
- 4.Hasan S, Hameed A, Saleem S. et al. The association of GSTM1 and GSTT1 polymorphisms with squamous cell carcinoma of cervix in Pakistan. Tumour biology: the journal of the International Society for Onco developmental Biology and Medicine. 2015;36(7):5195–5199. doi: 10.1007/s13277-015-3175-y. [DOI] [PubMed] [Google Scholar]
- 5.Jeon YT, Kim JW, Song JH. et al. Cyclin D1 G870 A polymorphism and squamous cell carcinoma of the uterine cervix in Korean women. Cancer letters. 2005;223(2):259–263. doi: 10.1016/j.canlet.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 6.Sobti RC, Shekari M, Kordi Tamandani DM. et al. Effect of NBS1gene polymorphism on the risk of cervix carcinoma in a northern Indian population. The International journal of biological markers. 2008;23(3):133–139. doi: 10.1177/172460080802300301. [DOI] [PubMed] [Google Scholar]
- 7.Duenas-Gonzalez A, Cetina L, Mariscal I. et al. Modern management of locally advanced cervical carcinoma. Cancer treatment reviews. 2003;29(5):389–399. doi: 10.1016/s0305-7372(03)00068-9. [DOI] [PubMed] [Google Scholar]
- 8.Pohlmann PR, Mayer IA, Mernaugh R. Resistance to Trastuzumab in Breast Cancer. Clinicalcancer research: an official journal of the American Association for Cancer Research. 2009;15(24):7479–7491. doi: 10.1158/1078-0432.CCR-09-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slamon DJ, Leyland-Jones B, Shak S. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England journal of medicine. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 10.Bellone S, Palmieri M, Gokden M. et al. Selection of HER-2/neu-positive tumor cells in early stage cervical cancer: implications for Herceptin-mediated therapy. Gynecologic oncology. 2003;91(1):231–240. doi: 10.1016/s0090-8258(03)00460-8. [DOI] [PubMed] [Google Scholar]
- 11.Oh DY, Kim S, Choi YL. et al. HER2 as a novel therapeutic target for cervical cancer. Oncotarget. 2015;6(34):36219–36230. doi: 10.18632/oncotarget.5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Tang J, Wang M. et al. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis. 2010;31(4):621–624. doi: 10.1093/carcin/bgp323. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Castillo J, Pedersen K, Angelini PD. et al. HER2 carboxyl-terminal fragments regulate cell migration and cortactin phosphorylation. The Journal of biological chemistry. 2009;284(37):25302–25313. doi: 10.1074/jbc.M109.001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FaltusT Yuan J, Zimmer B et al. Silencing of the HER2/neu gene by siRNA inhibits proliferation and induces apoptosis in HER2/neu-overexpressing breast cancer cells. Neoplasia. 2004;6(6):786–795. doi: 10.1593/neo.04313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MM, Allen P, Gonzalez-Angulo AM. et al. Pathologic complete response to neoadjuvant chemotherapy with trastuzumab predicts for improved survival in women with HER2-overexpressing breast cancer. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2013;24(8):1999–2004. doi: 10.1093/annonc/mdt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slamon DJ, Clark GM, Wong SG. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 17.Cai Y, Wang J, Zhang L. et al. Expressions of fatty acid synthase and HER2 are correlated with poor prognosis of ovarian cancer. Medical oncology. 2015;32(1):391. doi: 10.1007/s12032-014-0391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao WR, Lee MY, Lin WL. et al. HER2 amplification and overexpression are significantly correlated in mucinous epithelial ovarian cancer. Human pathology. 2014;45(4):810–816. doi: 10.1016/j.humpath.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Vogel CL, Cobleigh MA, Tripathy D. et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20(3):719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 20.Wilken JA, Webster KT, Maihle NJ. Trastuzumab Sensitizes Ovarian Cancer CellstoEGFR-targeted Therapeutics. Journal of ovarian research. 2010;3:7. doi: 10.1186/1757-2215-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heyerdahl H, Krogh C, Borrebaek J. et al. Treatment of HER2-expressingbreastcancer and ovarian cancer cells with alpha particle-emitting 227Th-trastuzumab. International journalof radiation oncology, biology, physics. 2011;79(2):563–570. doi: 10.1016/j.ijrobp.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 22.Monk BJ, Mas Lopez L, Zarba JJ. et al. Phase II, open-label study of pazopanib or lapatinib monotherapy compared with pazopanib plus lapatinib combination therapy in patients with advanced and recurrent cervical cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(22):3562–3569. doi: 10.1200/JCO.2009.26.9571. [DOI] [PubMed] [Google Scholar]
- 23.Ananiev J, Aleksandrova E, Skerleva D. et al. Impact of HER2 codon 655 polymorphism and expression of HER2 and HER3 in non small cell lung cancer patients. Wiener medizinische Wochenschrift. 2015;165(15-16):315–321. doi: 10.1007/s10354-015-0366-5. [DOI] [PubMed] [Google Scholar]
- 24.Watrowski R, Castillo-Tong DC, Schuster E. et al. Association of HER2 codon 655 polymorphism with ovarian cancer. Tumour biology: the journal of the International Society for Onco developmental Biology and Medicine. 2016;37(6):7239–7244. doi: 10.1007/s13277-015-4609-2. [DOI] [PubMed] [Google Scholar]
- 25.Satiroglu-Tufan NL, Bir F, Calli-Demirkan N. Investigation of HER-2 codon 655 single nucleotide polymorphism frequency and c-ErbB-2 protein expression alterations in gastric cancer patients. World journal of gastroenterology. 2006;12(20):3283–3287. doi: 10.3748/wjg.v12.i20.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(25):15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sternberg MJ, Gullick WJ. A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. Protein engineering. 1990;3(4):245–248. doi: 10.1093/protein/3.4.245. [DOI] [PubMed] [Google Scholar]
- 28.Ueda M, Hung YC, Terai Y. et al. HER-2 codon 655 polymorphism in cervical carcinogenesis. International journal of gynecological cancer: officialjournal of the International Gynecological Cancer Society. 2006;16(1):325–328. doi: 10.1111/j.1525-1438.2006.00349.x. [DOI] [PubMed] [Google Scholar]
- 29.Kruszyna L, Lianeri M, Roszak A. et al. HER2 codon 655 polymorphism is associated with advanced uterine cervical carcinoma. Clinical biochemistry. 2010;43(6):545–548. doi: 10.1016/j.clinbiochem.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Su Y, Jiang Y, Sun S. et al. Effects of HER2 genetic polymorphisms on its protein expression in breast cancer. Cancer epidemiology. 2015;39(6):1123–1127. doi: 10.1016/j.canep.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Larif S, BenSalem C, Soua Z. et al. Insight into TPMT (*)23 mutation mis-folding using molecular dynamics simulation and protein structure analysis. Journalofbiomolecular structure & dynamics. 2013;31(10):1066–1076. doi: 10.1080/07391102.2012.721495. [DOI] [PubMed] [Google Scholar]
- 32.Lam AR, Teplow DB, Stanley HE. et al. Effects of the Arctic (E22->G) mutation on amyloid beta-protein folding: discrete molecular dynamics study. Journal of the American Chemical Society. 2008;130(51):17413–17422. doi: 10.1021/ja804984h. [DOI] [PubMed] [Google Scholar]
- 33.Peng XN, Wang J, Zhang W. Molecular dynamics simulation analysis of the effect of T790M mutation on epidermal growth factor receptor protein architecture in non-small cell lung carcinoma. Oncology letters. 2017;14(2):2249–2253. doi: 10.3892/ol.2017.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]